Dear Editor,
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) systems confer RNA-guided adaptive immunity against invading genetic elements in prokaryotic cells.1 These systems employ CRISPR RNA (crRNA) containing surveillance complexes for sequence-specific recognition and degradation of foreign DNA or RNA. Several nuclease-deficient type I-F, I-B, and V-K systems have been reported to be evolutionarily and functionally associated with Tn7-like transposons lacking a key gene for DNA targeting, implying a new mode of RNA-guided DNA insertion.2,3 Recent studies characterized CRISPR-associated transposase in type I-F and V-K systems and established genome-wide programmable site-specific DNA transposition in E. coli cells, providing the prospect of genome editing strategy without requirement of double-strand breaks and endogenous DNA repair pathways.4,5 All these CRISPR-associated transposons contain transposase subunits, CRISPR effector, and a CRISPR array. Furthermore, subunit TniQ, a homolog of E. coli TnsD, forms a stable complex with Vibrio cholerae Tn6677 type I-F effector Cascade (also called Csy complex) and plays an essential role during DNA insertion.5
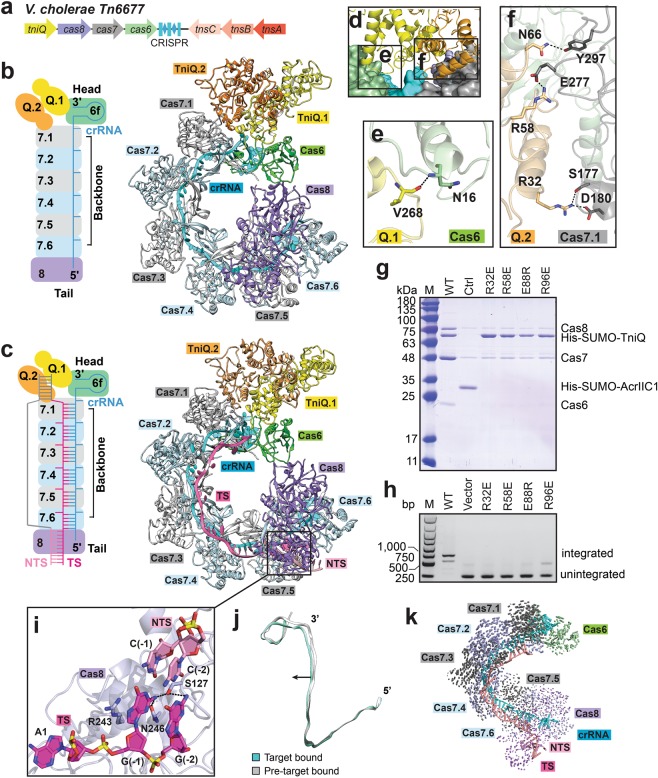
To better understand the assembly and mechanisms of CRISPR-associated transposase machineries, we determined the crystal structure of apo-TniQ and cryo-EM structures of type I-F Cascade-TniQ complex in pre-target-bound and DNA target-bound states at an average resolution of 3.29 Å and 3.18 Å, respectively (Fig. 1a–c; Supplementary information, Figs. S1-2 and Tables S1-2). Unlike canonical type I-F system, V. cholerae Cascade consists of Cas6, Cas7, naturally fused Cas5-Cas8 (hereafter referred to as Cas8), and a 60-nucleotide (nt) crRNA including 32-nt spacer region and 28-nt repeat region.3,5 Before DNA loading, Cascade adopts a similar architecture to the reported type I-F Cascade6–8 and TniQ homodimer binds to Cas6 and Cas7.1 (Fig. 1b). The region in TniQ that is distal to the TniQ-Cascade interface has poor density especially in TniQ-Cascade-dsDNA complex. For the sake of completeness and consistency, we modeled the missing region according to our crystal structure of apo-TniQ. All Cas proteins are arranged around the crRNA in a helically twisted “G” shape, with Cas6 binding 3’ stem-loop of crRNA in head region and Cas8 binding 5’ handle of crRNA in tail region. Ribonuclease Cas6 is responsible for precursor crRNA processing during expression stage and necessary for RNA-guided DNA transposition.9,10 Mutation of the highly conserved key residue His29 in Cas6 to alanine abolishes crRNA maturation,9 Cascade assembly,10 and DNA integration.5 In our structure, the scissile phosphate group at 3’-end of crRNA is located at catalytic pocket of Cas6 and makes hydrogen bonds with H29 (Supplementary information, Fig. S3a, b), which is consistent with previously reported data.5,9,10 Cas8 contains two subdomains and shares relatively similar architecture to the reported P. aeruginosa Cas5-Cas8 heterodimer (Supplementary information, Fig. S3c, d). As observed in other type I-F Cascade, six interlocking subunits of Cas7 are located along the spacer region of crRNA and interact with Cas6 and Cas8 at two ends (Fig. 1b; Supplementary information, Fig. S3e, f), and the Cas7 thumb domains kink crRNA following a periodic “5 + 1” pattern (Supplementary information, Fig. S4a).
Fig. 1. Assembly and target DNA recognition of type I-F Cascade-TniQ complex.
a Schematic diagram of genes involved in transposon-encoded type I-F CRISPR-Cas systems in V. cholerae Tn6677. b, c Schematic rendering of arrangement of subunits and ribbon representation of overall structures of Cascade-TniQ complex in pre-target-bound (b) and target dsDNA-bound (c) states. d–f Recognition of TniQ homodimer by subunits Cas6 and Cas7.1 of Cascade. Detailed hydrophilic interactions between TniQ and Cas6 (e) and TniQ and Cas7.1 (f) are shown in the expanded panels. g Mutation analysis of the TniQ residues involved in binding to Cascade and dimerization by His pull-down assay. Unrelated anti-CRISPR protein AcrIIC1 is used as a negative control. h PCR analysis of in vivo transposition with TniQ mutants. The plasmid without Cascade genes is used as a negative control. i PAM recognition by Cas8 subunit. j Elongation of helical pitch of crRNA after DNA loading. k Domain movements occur upon target dsDNA loading. Vector lengths are correlated with the domain motion scale.
TniQ exists as a homodimer in solution and forms a head-to-tail homodimer in our crystal structure (Supplementary information, Fig. S5a–c and Table S1). The symmetric dimer interfaces are mainly formed by the linkers between α3 helix and β1 strand at the N-terminus in one monomer and helix α13 and the following region at the C-terminus in the other monomer, and stabilized by numerous hydrophilic interactions (Supplementary information, Fig. S5b). Specifically, mutations of E88R, R96E, E379R, and R387E impair the dimer formation (Supplementary information, Fig. S5d). There are two tandem zinc finger motifs at the C-terminal region (Supplementary information, Fig. S5b), of which the first zinc finger motif formed by C128, C131, C150, and H153 are highly conserved in TnsD family proteins.5 When bound to Cascade, the head-to-tail TniQ homodimer is located at the surface formed by Cas6 and Cas7.1 in the head region of Cascade (Fig. 1d; Supplementary information, Fig. S5c). Monomer TniQ.1 binds to Cas6 mainly through hydrogen bonds contributed by main chain of the loop between helices α8 and α9 (Fig. 1e). Monomer TniQ.2 binds to Cas7.1 mainly through side-chain hydrophilic interactions contributed by helix α5 and the following loop (Fig. 1f). Mutations of the TniQ residues involved in ionic interactions reduce its binding to Cascade in vitro (Fig. 1g). We further transformed E. coli cells with genes encoding the components of transposon including TnsA, TnsB, TnsC, TniQ together with Cascade as described previously,5 and found that these mutations in TniQ abolish RNA-guided DNA integration in vivo (Fig. 1h). We further investigated the effect of TniQ mutations causing reduced dimerization and found that these mutations lower TniQ binding affinity to Cascade, resulting in deficiency in RNA-guided DNA integration (Fig. 1g, h).
The recognition of target double-strand DNA (dsDNA) was initialized from protospacer adjacent motif (PAM) recognition, which allows self/non-self discrimination. To clarify how the Cascade-TniQ sequence-specifically recognizes target dsDNA, we have determined the cryo-EM structure of Cascade-TniQ bound to 62-bp dsDNA containing 32-bp target sequence, 10-bp PAM-proximal duplex, and 20-bp PAM-distal duplex (Fig. 1c). However, only 7-bp PAM duplex and 32-nt target DNA strand could be observed, but there is no clear density for PAM-distal duplex and loop-out non-target DNA strand. The 5’-CC-3’ PAM is recognized by Cas8 from the minor groove in a similar manner as previously reported (Fig. 1i).7 The side chain of Arg243 stacks with the base of dG(–1) of PAM, facilitating the formation of the guide:target heteroduplex and looping-out of the non-target DNA strand (Fig. 1i). The RNA:DNA hybrid follows the periodic “5 + 1” pattern, as observed in I-F Cascade bound to dsDNA, with a β-hairpin of the adjacent Cas7 protruding through the flipped-out base pair (Supplementary information, Fig. S4b). Interestingly, minimal conformational changes are observed upon target dsDNA loading, except for slight increase of the helical pitch of crRNA, resulting in relative extension of the complex (Fig. 1j, k). Because the distal region of TniQ homodimer to the interface between Cascade and TniQ has poor density, at this stage we are not sure whether DNA loading induces any conformational changes in this region. Taken together, our data elucidate the assembly of the transposon subunit TniQ to type I-F Cascade and target DNA recognition by Cascade-TniQ complex, providing the hints of the cooperation between CRISPR-Cas system and Tn7-like transposon. During revision of our manuscript, a related paper was published online in Nature presenting cryo-EM structures of Cascade-TniQ in the presence or the absence of dsDNA;11 their structural studies and our data reach the common conclusions on recruitment of TniQ by Cascade. More research remains to be done to illustrate the recruitments and roles of all transposon subunits including TniQ during RNA-guided DNA integration when associated with different CRISPR-Cas systems.
The density maps have been deposited to the EM Data Bank with entry code: EMD-0929 and EMD-0930. The atomic coordinates have been deposited to Protein Data Bank with entry code: 6LNB, 6LNC, and 6LND.
Supplementary information
Supplementarty information, Materials and Figures
Acknowledgements
We thank the staff of the BL18U and BL19U1 beamlines of National Center for Protein Science in Shanghai (NCPSS) and BL17U of Shanghai Synchrotron Radiation Facility (SSRF). We thank the staff of Electron Microscopy facility of NCPSS for technical assistance in data collection. This study was supported by the National Natural Science Foundation of China (31971135) to H.Y.
Author contributions
B.W. performed all the experiments. W.X. helped with the data collection. H.Y. designed the study, wrote the manuscript, and supervised the research.
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0274-0.
References
- 1.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters JE, Makarova KS, Shmakov S, Koonin EV. Proc. Natl Acad. Sci. USA. 2017;114:E7358–E7366. doi: 10.1073/pnas.1709035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faure G, et al. Nat. Rev. Microbiol. 2019;17:513–525. doi: 10.1038/s41579-019-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strecker J, et al. Science. 2019;365:48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH. Nature. 2019;571:219–225. doi: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 6.Chowdhury S, et al. Cell. 2017;169:47–57. doi: 10.1016/j.cell.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo TW, et al. Cell. 2017;171:414–426. doi: 10.1016/j.cell.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rollins MF, et al. Mol. Cell. 2019;74:132–142. doi: 10.1016/j.molcel.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haurwitz RE, Sternberg SH, Doudna JA. EMBO J. 2012;31:2824–2832. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halpin-Healy, T.S., Klompe, S.E., Sternberg, S.H. & Fernández, I.S. Nature, 10.1038/s41586-019-1849-0 (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementarty information, Materials and Figures