Abstract
Ferroptosis, a form of regulated cell death caused by lipid peroxidation, was recently identified as a natural tumor suppression mechanism. Here, we show that ionizing radiation (IR) induces ferroptosis in cancer cells. Mechanistically, IR induces not only reactive oxygen species (ROS) but also the expression of ACSL4, a lipid metabolism enzyme required for ferroptosis, resulting in elevated lipid peroxidation and ferroptosis. ACSL4 ablation largely abolishes IR-induced ferroptosis and promotes radioresistance. IR also induces the expression of ferroptosis inhibitors, including SLC7A11 and GPX4, as an adaptive response. IR- or KEAP1 deficiency-induced SLC7A11 expression promotes radioresistance through inhibiting ferroptosis. Inactivating SLC7A11 or GPX4 with ferroptosis inducers (FINs) sensitizes radioresistant cancer cells and xenograft tumors to IR. Furthermore, radiotherapy induces ferroptosis in cancer patients, and increased ferroptosis correlates with better response and longer survival to radiotherapy in cancer patients. Our study reveals a previously unrecognized link between IR and ferroptosis and indicates that further exploration of the combination of radiotherapy and FINs in cancer treatment is warranted.
Subject terms: Radiotherapy, Cell death
Introduction
Regulated cell death, most notably apoptosis, serves as a natural barrier to tumor development; resistance to cell death is a hallmark of cancer.1–3 Apoptosis can be induced by stress stimuli during tumor development or in the presence of cancer therapies.1 While apoptosis is clearly induced by and involved in many cancer therapies, whether and how cancer therapies induce other forms of regulated cell death remains less clear.
Ferroptosis, an iron-dependent form of regulated cell death that is induced by excessive lipid peroxidation, is morphologically and mechanistically distinct from apoptosis.4–6 Current studies indicate that ferroptosis is mainly induced by peroxidation of phospholipids (PLs) that contain polyunsaturated fatty acids (PUFAs), most notably arachidonic acid; correspondingly, ferroptosis can be significantly attenuated by inactivation of ACSL4, a long-chain fatty-acid-coenzyme A ligase that is required for PUFA-PL biosynthesis and preferentially utilizes arachidonic acid as its substrate.7–10 GPX4, a glutathione peroxidase, utilizes reduced glutathione to convert lipid hydroperoxides to lipid alcohols, thereby mitigating lipid peroxidation and inhibiting ferroptosis.11–13 Glutathione is synthesized from glycine, glutamate, and cysteine, among which cysteine is the rate-limiting precursor. Most cells obtain cysteine through the import of extracellular cystine — an oxidized dimeric form of cysteine — via the amino acid transporter SLC7A11 (also known as xCT).5,14,15, Correspondingly, inactivation of GPX4 or SLC7A11 by genetic or pharmacologic means induces ferroptosis.4,12,13 Previous studies have identified several classes of ferroptosis inducers (FINs) that potently inhibit either SLC7A11 or GPX4.16 For example, class 1 FINs (such as erastin and sulfasalazine) inhibit SLC7A11-mediated cystine transport, class 2 FINs (such as RSL3 and ML162) act by inhibiting GPX4 activity, and class 3 FINs (such as FIN56) induce ferroptosis by depleting both GPX4 protein and coenzyme Q10.4,13,16 These FINs not only provide important tools for ferroptosis studies, but also are potential therapeutic agents for treating diseases caused by insufficient ferroptosis.5
Like other forms of regulated cell death, ferroptosis is tightly regulated in normal cells, and its dysregulation has been associated with multiple pathologic conditions and diseases. For example, excessive ferroptosis has been linked to ischemia-reperfusion injury, kidney failure, and neurodegeneration, whereas insufficient ferroptosis can lead to cancer.5 We and others recently identified ferroptosis as a natural tumor suppression mechanism and showed that inactivation of ferroptosis, like inactivation of apoptosis, contributes to tumor development.17–22 These studies also raise the intriguing questions of whether ferroptosis can be induced by any cancer therapy currently in clinical use and whether ferroptosis induction might enhance the therapeutic response to cancer treatments in patients.
Radiotherapy is a cornerstone of the treatment of many cancers,23,24 but radioresistance remains a major factor leading to the failure of radiotherapy. Radiotherapy uses high-energy ionizing radiation (IR) to produce DNA double-strand breaks that induce cell cycle arrest, senescence, and various modes of cell death, including apoptosis, necrosis, autophagy, and mitotic catastrophe.25 In addition to directly damaging DNA, IR can induce indirect cellular effects. For instance, IR causes cellular damage through the radiolysis of cellular water and the stimulation of oxidases to generate reactive oxygen species (ROS), such as the hydroxyl radical and hydrogen peroxide, which may damage nucleic acids, proteins, and lipids.26 Whether IR induces forms of cell death that are independent of DNA damage still remains largely unknown. In this study, we characterized the role of ferroptosis in IR-induced cell death and tumor suppression and evaluated the therapeutic strategy of combining FINs and radiotherapy for cancer treatment.
Results
IR induces ferroptosis in cancer cells
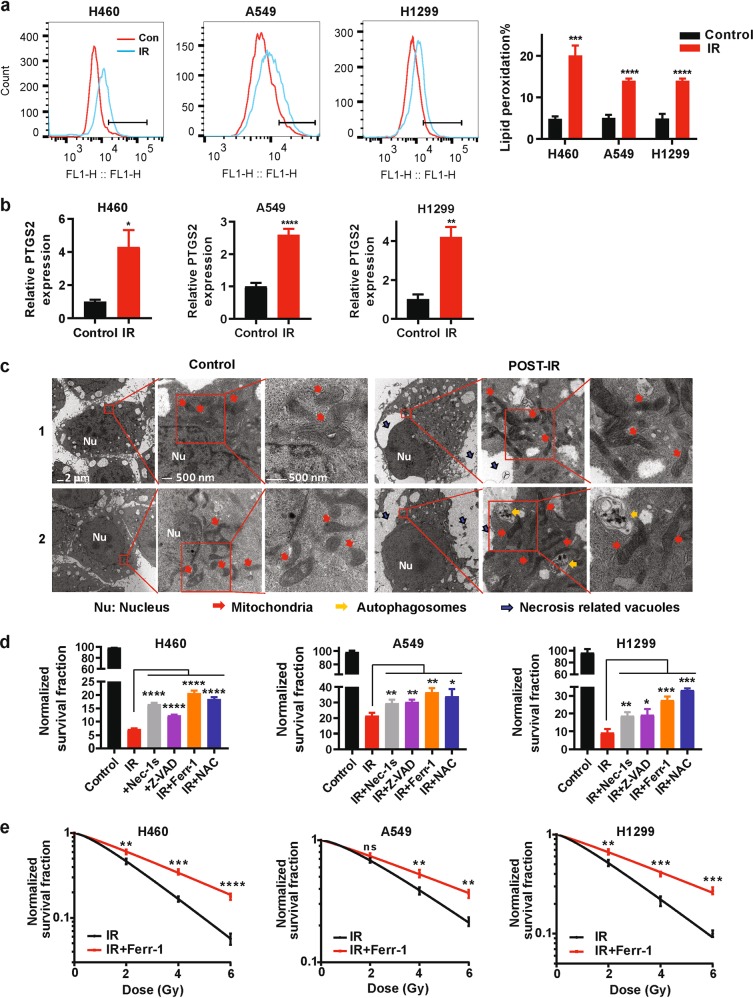
IR is widely used to treat cancer. Given that both IR and ferroptosis are linked to ROS, we determined whether IR could induce ferroptosis in cancer cells. As expected, we confirmed (by DCFDA staining, details in Methods and Materials) that IR increased total ROS in cancer cells (Supplementary information, Fig. S1a). A hallmark of ferroptosis is the accumulation of lipid peroxidation.5 Indeed, we observed (by C11-BODIPY staining, details in Methods and Materials) that IR induced lipid peroxidation in the tested cancer cells (Fig. 1a). We then extended these observations to additional cell lines representing a variety of cancer types in which radiotherapy is commonly used, including lung cancer, breast cancer, fibrosarcoma, and esophageal cancer (Supplementary information, Fig. S1b). Ferroptosis induction is associated with the increased expression of ferroptosis marker genes such as PTGS2.5 Consistent with this, IR induced the expression of PTGS2 in all tested cancer cell lines except MCF-7 and UMRC6, in which PTGS2 expression was undetectable (Fig. 1b; Supplementary information, Fig. S1c). Finally, transmission electron microscopy revealed that cancer cells after IR treatment exhibited shrunken mitochondria with enhanced membrane density, a morphologic feature of ferroptosis5 (Fig. 1c); as expected, IR-treated cells also showed morphologic features of autophagy and necrosis (such as autophagosomes and necrosis-related vacuoles; Fig. 1c).
Fig. 1. Ionizing radiation induces ferroptosis in cancer cells.
a Lipid peroxidation assessment in H460, A549, and H1299 cell lines at 24 h after exposure to 6 Gy of IR. Bar graph showing relative levels of lipid peroxidation by C11-BODIPY staining in the indicated cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. b qRT-PCR analysis of PTGS2 expression in H460, A549, and H1299 cell lines at 24 h after exposure to 6 Gy of IR. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. c Transmission electron microscopy images of H460 cells without radiation (control) or at 24 h after exposure to 6 Gy of IR (post-IR). Nu, nucleus; red arrows, mitochondria; yellow arrows, autophagosomes; black arrows, necrosis-related vacuoles. Scale bars: left, 2 µm; right, 500 nm. d Clonogenic survival assay in H460, A549, and H1299 cell lines that were pretreated with 2 μM necrostatin-1s, 5 μM Z-VAD-fmk, 5 μM ferrostatin-1, 5 mM N-acetyl-L-cysteine, or DMSO for 24 h followed by exposure to 6 Gy of IR. The survival data were normalized to those of unirradiated control cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. e Clonogenic survival curves for H460, A549, and H1299 cell lines that were pretreated with 5 μM ferrostatin-1 or DMSO for 24 h followed by exposure to IR at doses from 0 to 6 Gy. The survival data were normalized to those of unirradiated control cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test.
Next, we studied the potential role of ferroptosis in the IR-induced cell death response. To this end, we examined the impact of the ferroptosis inhibitor ferrostatin-1, the apoptosis inhibitor Z-VAD-fmk, the necroptosis inhibitor necrostatin-1s, and the ROS scavenger N-acetyl-L-cysteine (NAC), on the clonogenic survival of a variety of IR-treated cancer cells. We found that treatment with ferrostatin-1 or NAC partially restored clonogenic survival that had been reduced by exposure to IR in the tested cancer cells (Fig. 1d, e; Supplementary information, Fig. S1d, e); importantly, the restoration of cell survival induced by ferrostatin-1 or NAC was comparable to or even more pronounced than that induced by Z-VAD-fmk or necrostatin-1s (Fig. 1d, e; Supplementary information, Fig. S1e). Further analysis revealed that combining ferrostatin-1 with Z-VAD-fmk (or necrostatin-1s) resulted in more restoration of clonogenic survival under IR (Supplementary information, Fig. S1f). Taken together, our data strongly suggest that IR induces ferroptosis in cancer cells and that ferroptosis represents a part of the IR-induced cell death response.
IR induces ferroptosis partly through upregulating ACSL4
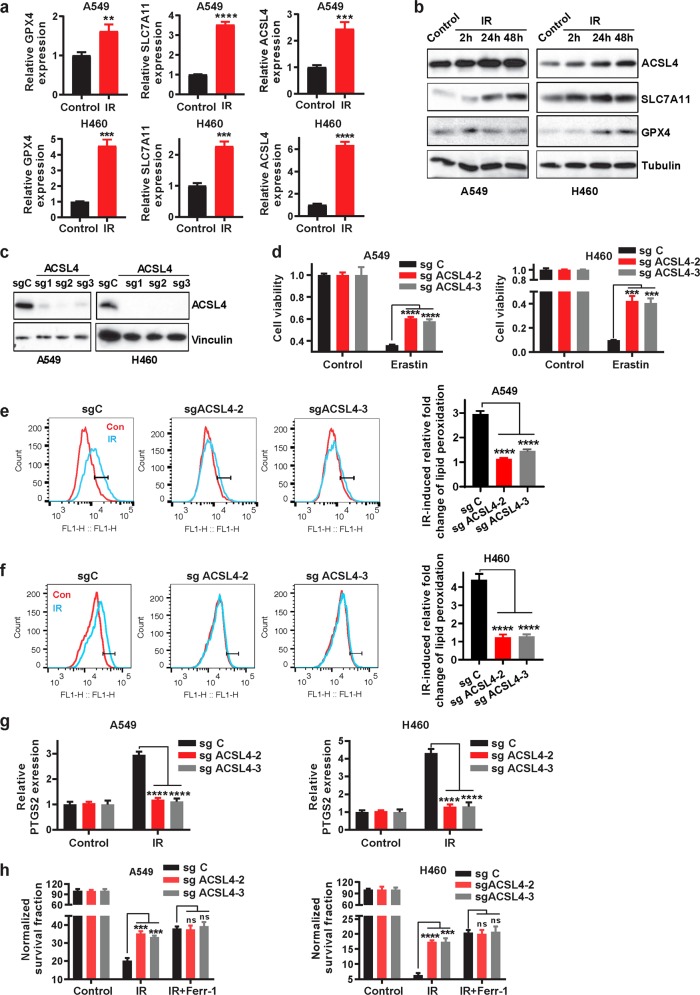
Next, we sought to study how IR induces ferroptosis. One such mechanism could involve IR-induced ROS production, which presumably further increases lipid peroxidation to promote ferroptosis. In support of this hypothesis, our data showed that IR induced total ROS (Supplementary information, Fig. S1a) and that the ROS scavenger NAC partially restored clonogenic survival that had been reduced upon IR exposure (Fig. 1d). We reasoned that IR-induced ferroptosis might also involve other mechanisms that modulate the expression or activity of proteins involved in ferroptosis pathways. To test this hypothesis, we first examined the expression levels of several key components involved in ferroptosis pathways in response to IR. As shown in Fig. 2a, b, IR induced expression of both SLC7A11 and GPX4. Since both SLC7A11 and GPX4 function to inhibit ferroptosis,4,12,13 it seems unlikely that IR-induced SLC7A11 or GPX4 expression would play a role in IR-induced ferroptosis. Interestingly, we found that IR also significantly induced the expression of ACSL4 (Fig. 2a, b), which promotes ferroptosis by regulating PUFA-PL biosynthesis.7,8,10 Of note, IR-induced ACSL4 expression preceded the induction of GPX4 and SLC7A11 (Supplementary information, Fig. S2a), suggesting that IR-induced expression of SLC7A11 and GPX4 may function as a negative feedback loop to restore cell survival upon IR.
Fig. 2. Ionizing radiation induces ferroptosis partly through upregulating ACSL4.
a qRT-PCR analysis of GPX4, SLC7A11, and ACSL4 expression in A549 and H460 cell lines at 24 h after exposure to 6 Gy of IR. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. b Western blotting analysis of ACSL4, SLC7A11, and GPX4 expression in A549 and H460 cell lines at 2 h, 24 h, and 48 h after exposure to 6 Gy of IR or no radiation. c Western blotting analysis of ACSL4 expression in sg Control (sg C), sg ACSL4-1, sg ACSL4-2, and sg ACSL4-3 A549 and H460 cell lines. d Cell viability measurements in sg C, sg ACSL4-2, and sg ACSL4-3 A549 and H460 cell lines treated with or without 10 μM erastin for 24 h. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. e, f Lipid peroxidation assessment in sg C, sg ACSL4-2, and sg ACSL4-3 A549 (e) and H460 (f) cell lines at 24 h after exposure to 6 Gy of IR. Bar graph showing IR-induced relative fold changes of lipid peroxidation by C11-BODIPY staining in the indicated cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. g qRT-PCR analysis of PTGS2 expression in sg C, sg ACSL4-2, and sg ACSL4-3 A549 and H460 cell lines at 24 h after exposure to 6 Gy of IR. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. h Clonogenic survival assay in sg C, sg ACSL4-2, and sg ACSL4-3 A549 and H460 cell lines that were pretreated with 5 μM ferrostatin-1 or DMSO for 24 h followed by exposure to 6 Gy of IR. The survival data were normalized to those of unirradiated control cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test.
To study the potential role of ACSL4 in IR-induced ferroptosis, we used CRISPR-Cas9 technology with at least 2 independent single guide RNAs (sgRNAs) to generate ACSL4 knockout (KO) H460 and A549 cells (Fig. 2c). Consistent with previous reports,7,10 ACSL4 deletion significantly attenuated erastin-induced ferroptosis in these cells (Fig. 2d). Notably, IR-induced lipid peroxidation and PTGS2 expression were almost completely abolished in ACSL4 KO cells (Fig. 2e–g). It should be noted that ACSL4 deletion did not significantly affect IR-induced total ROS in these cells (Supplementary information, Fig. S2b-c), which is consistent with the current model that ACSL4 promotes lipid peroxidation and ferroptosis through regulating the biosynthesis of PUFA-PLs but not ROS per se.7,8
We further studied whether ACSL4 modulates radiosensitization in cancer cells through regulating ferroptosis. We showed that ACSL4 ablation significantly restored clonogenic survival in IR-treated cells to a level similar to that caused by ferrostatin-1 treatment; importantly, ACSL4 deletion did not further promote radioresistance in ferrostatin-1–treated cells (Fig. 2h), suggesting that ACSL4 promotes radiosensitization largely through promoting ferroptosis. Finally, to study whether IR-induced ACSL4 expression plays a role in ferroptosis and radiosensitization, we used shRNA to moderately knock down ACSL4 under IR conditions to the level similar to that in control cells under basal conditions (Supplementary information, Fig. S2d). Further analysis revealed that ACSL4 knockdown attenuated erastin-induced ferroptosis (Supplementary information, Fig. S2e); importantly, normalization of ACSL4 level to that under basal conditions promoted radioresistance, and the effect of ACSL4 knockdown on radioresistance was abolished under ferrostatin-1 treatment (Supplementary information, Fig. S2f). Together, our data suggest that IR likely induces ferroptosis through increasing both ROS production and ACSL4 levels (see Discussion).
SLC7A11 and GPX4 promote radioresistance largely through inhibiting ferroptosis
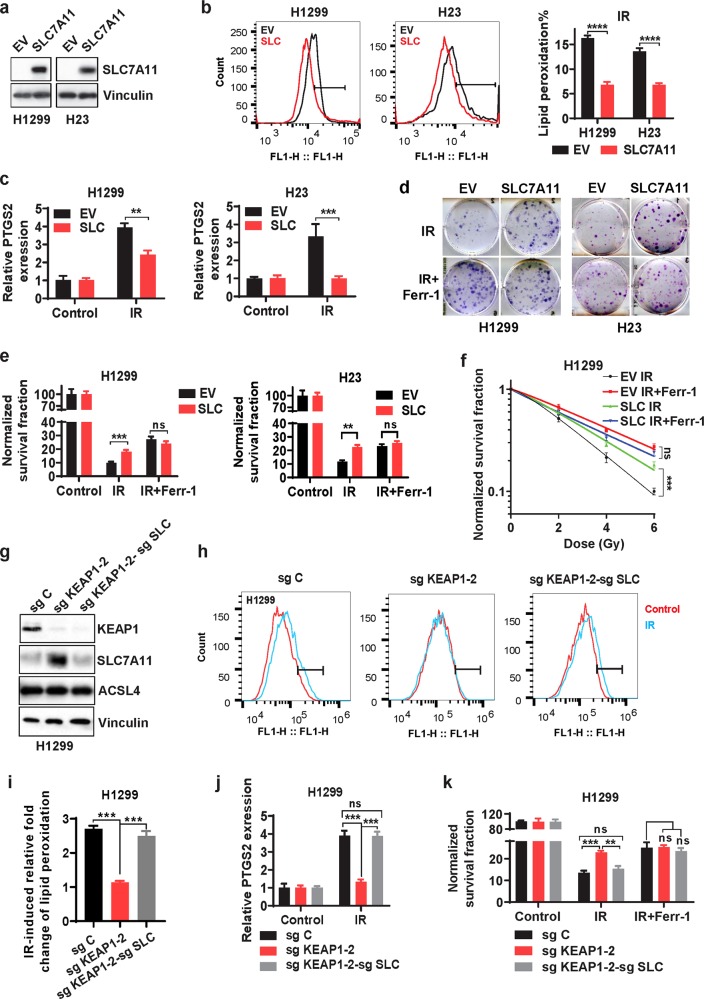
In response to cancer therapy, various cell survival pathways are often activated as an adaptive response, resulting in therapy resistance.27 As noted above (Fig. 2a, b), we observed that IR induced the expression of SLC7A11 and GPX4, both inhibiting ferroptosis.4,5,12,13 We reasoned that IR-induced SLC7A11 or GPX4 expression likely represents an adaptive response wherein cancer cells attempt to restore cell survival in response to IR, which might contribute to radioresistance. Similarly, we previously showed that erastin treatment or cystine deprivation, while inducing ferroptosis, also potently induces SLC7A11 expression as an adaptive response.18 We therefore examined whether inhibiting ferroptosis by enforced expression of SLC7A11 or GPX4 would promote radioresistance in cancer cells. We found that overexpression of SLC7A11 in SLC7A11-low cancer cell lines H1299 and H23 cells significantly mitigated IR-induced lipid peroxidation and PTGS2 expression (Fig. 3a–c) and promoted radioresistance (Fig. 3d–f). Consistent with our data shown in Fig. 1, ferrostatin-1 treatment also promoted radioresistance; importantly, the effect of SLC7A11 overexpression on radioresistance was abrogated under ferrostatin-1 treatment (Fig. 3d–f), suggesting that SLC7A11 promotes radioresistance largely through inhibiting ferroptosis. Similarly, we found that GPX4 overexpression also mitigated IR-induced lipid peroxidation and PTGS2 expression and promoted radioresistance in a ferroptosis-dependent manner (Supplementary information, Fig. S3a–c).
Fig. 3. SLC7A11 overexpression or KEAP1 deficiency promotes radioresistance largely through inhibiting ferroptosis.
a Western blotting analysis showing SLC7A11 levels in H1299 and H23 cell lines with stable expression of empty vector (EV) and SLC7A11. b Lipid peroxidation assessment in EV- and SLC7A11-expressing H1299 or H23 cells at 24 h after exposure to 6 Gy or 2 Gy of IR, respectively. Bar graph showing the relative levels of lipid peroxidation by C11-BODIPY staining in the indicated cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. c qRT-PCR analysis of PTGS2 expression in EV- and SLC7A11-expressing H1299 or H23 cells at 24 h after exposure to 6 Gy or 2 Gy of IR, respectively. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. d Representative images of clonogenic survival assays in EV- and SLC7A11-expressing H1299 or H23 cells that were pretreated with 5 μM ferrostatin-1 or DMSO for 24 h followed by exposure to 6 Gy or 2 Gy of IR, respectively. e The quantified clonogenic survival assay in EV- and SLC7A11-expressing H1299 or H23 cells that were pretreated with 5 μM ferrostatin-1 or DMSO for 24 h followed by exposure to IR at a dose of 6 Gy or 2 Gy, respectively. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. f The quantified clonogenic survival curve in EV- and SLC7A11-expressing H1299 cells that were pretreated with 5 μM ferrostatin-1 or DMSO for 24 h followed by exposure to IR at the indicated doses. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using multiple t-test. g Western blotting analysis of KEAP1, SLC7A11 and ACSL4 expression in sg Control (sg C), sg KEAP1-2, and sg KEAP1-2-sg SLC7A11 (sg SLC) H1299 cell lines. h, i Lipid peroxidation assessment in sg C, sg KEAP1-2, and sg KEAP1-2-sg SLC H1299 cells at 24 h after exposure to 6 Gy of IR h. Bar graph showing IR-induced relative fold changes of lipid peroxidation by C11-BODIPY staining in the indicated cells (i). Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. j qRT-PCR analysis of PTGS2 expression in sg C, sg KEAP1-2, and sg KEAP1-2-sg SLC H1299 cells at 24 h after exposure to 6 Gy of IR. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. k Clonogenic survival assays in sg C, sg KEAP1-2, and sg KEAP1-2-sg SLC H1299 cells that were pretreated with 5 μM ferrostatin-1 or DMSO for 24 h followed by exposure to 6 Gy of IR. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test.
Many cancer cells also exhibit de novo resistance to cancer therapies due to mutations or genetic alterations in oncogenic pathways.27 Thus, we further studied whether SLC7A11 promotes radioresistance through ferroptosis in a specific cancer context with aberrant SLC7A11 expression: KEAP1-mutant lung cancer. The tumor suppressor KEAP1 is frequently mutated in lung cancers,28,29 and it is known that KEAP1-mutant lung cancers are resistant to radiotherapy.30 KEAP1 normally targets NRF2, a master transcription factor of antioxidant response, for ubiquitination and proteasomal degradation. KEAP1 mutation or deficiency in lung cancer cells leads to constitutive activation of NRF2 and aberrant expression of NRF2 transcriptional targets, including SLC7A11.31,32 Consistent with this, analysis of data from The Cancer Genome Atlas revealed that KEAP1-mutant lung tumors exhibit significantly higher expression of SLC7A11 than do KEAP1-wild-type (WT) lung tumors (Supplementary information, Fig. S3d). We showed that KEAP1 deletion by CRISPR/Cas9 technology in H1299 cells (a KEAP1-WT lung cancer cell line) substantially increased SLC7A11 expression (Fig. 3g; Supplementary information, Fig. S3e), dampened IR-induced lipid peroxidation and PTGS2 expression (Fig. 3h–j; Supplementary information, Fig. S3f-g), and promoted radioresistance (Fig. 3k; Supplementary information, Fig. S3h). We made similar observations in H23 cells with KEAP1 deletion (Supplementary information, Fig. S3i-l). Importantly, SLC7A11 deletion in KEAP1-KO H1299 cells restored IR-induced lipid peroxidation and PTGS2 expression and resensitized cells to IR (Fig. 3g–k), and the effect of KEAP1 or SLC7A11 deficiency on clonogenic survival upon IR was abolished with ferrostatin-1 treatment (Fig. 3k; Supplementary information, Fig. S3h, l). These data together suggest that KEAP1 deficiency in lung cancer cells promotes radioresistance at least partly through SLC7A11-mediated ferroptosis inhibition. Collectively, our data suggest that aberrant expression of SLC7A11 or GPX4, either induced by IR as an adaptive response or caused by KEAP1 deficiency, promotes radioresistance largely through inhibiting ferroptosis.
Ferroptosis perturbation does not affect IR-induced DNA damage and repair
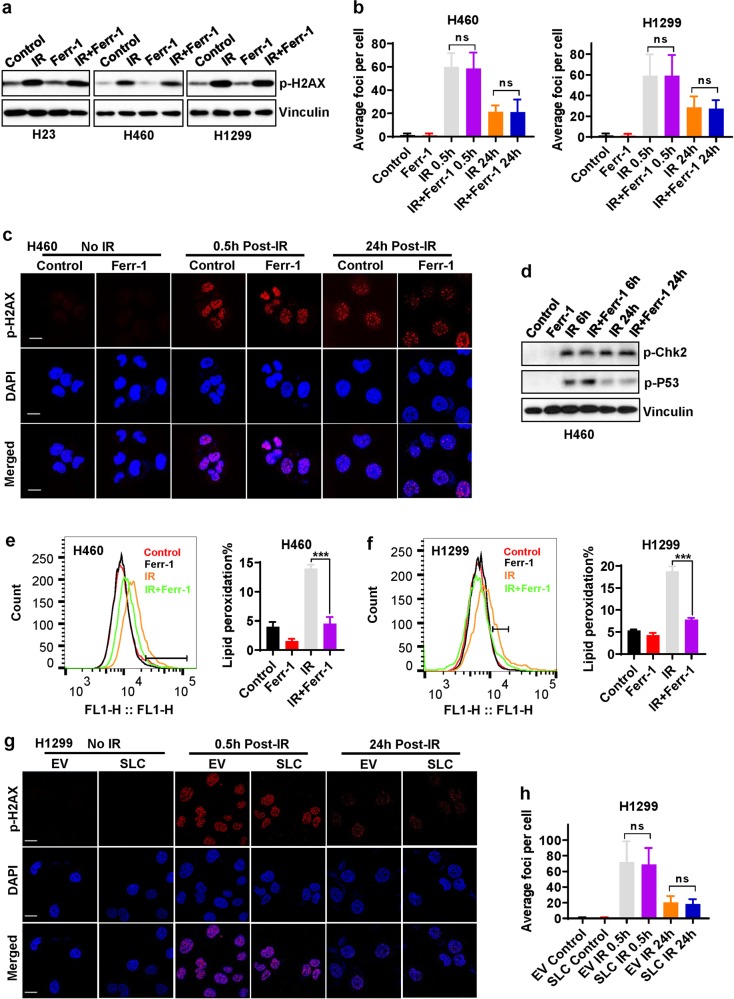
Our aforementioned data from both pharmacologic (ferrostatin-1 treatment) and genetic (SLC7A11 or GPX4 overexpression) approaches revealed that ferroptosis inhibition promotes radioresistance in cancer cells. Since DNA damage and repair represent a major IR-induced cellular effect, we next examined whether the radioresistance mediated by ferroptosis inhibition involves the DNA damage response and subsequent DNA repair. Figure 4a shows that, as expected, IR potently induced H2AX phosphorylation, a marker of DNA damage; however, ferrostatin-1 treatment did not affect either basal or IR-induced H2AX phosphorylation in these cancer cells. Time-course analyses revealed that ferrostatin-1 did not affect phosphorylated H2AX foci formation at 30 min or 24 h after IR (Fig. 4b, c), which correspond to IR-induced DNA double-strand breaks and subsequent DNA repair, respectively. In addition, ferrostatin-1 treatment did not obviously affect IR-induced DNA damage response signaling, such as Chk2 or p53 phosphorylation (Fig. 4d). As a control, we confirmed that ferrostatin-1 treatment mitigated IR-induced lipid peroxidation in these cancer cell lines (Fig. 4e, f). Consistent with this, we showed that SLC7A11 or GPX4 overexpression or KEAP1 deletion, while inhibiting IR-induced ferroptosis and promoting radioresistance (Fig. 3; Supplementary information, Fig. S3), did not obviously affect the IR-induced phosphorylated H2AX foci formation or their subsequent disapperance (Fig. 4g, h; Supplementary information, Fig. S4). Collectively, our data revealed that ferroptosis inhibition does not affect IR-induced DNA damage or repair, indicating that neither the DNA damage response nor DNA repair is involved in ferroptosis inhibition-mediated radioresistance in cancer cells.
Fig. 4. Ferroptosis is not involved in IR-induced DNA damage and repair.
a Western blotting analysis of phospho-H2AX levels in H460, H1299, and H23 cell lines pretreated with DMSO (control) or 5 μM ferrostatin-1 for 24 h followed by exposure to IR at a dose of 6 Gy (for H460 and H1299 cells) or 2 Gy (for H23 cells). b The numbers of phospho-H2AX foci per nucleus were counted on the basis of phospho-H2AX immunofluorescence at 30 min or 24 h after exposure to 6 Gy of IR in cells that had been pretreated with DMSO or 5 μM ferrostatin-1 for 24 h. Error bars are means ± SD, n = 20 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. c Representative immunofluorescence images showing staining of phospho-H2AX foci (red) and nuclei counterstained with DAPI (blue) in the indicated cells. d Western blotting analysis of phospho-Chk2 and phospho-p53 levels in H460 cells pretreated with DMSO or 5 μM ferrostatin-1 for 24 h followed by exposure to 6 Gy of IR. e, f Lipid peroxidation assessment in H460 (e) and H1299 cells (f) pretreated with DMSO or 5 μM ferrostatin-1 for 24 h followed by exposure to 6 Gy of IR. Bar graph showing relative levels of lipid peroxidation by C11-BODIPY staining in the indicated cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. g Representative immunofluorescence images showing staining of phospho-H2AX foci (red) and nuclei counterstained with DAPI (blue) in the indicated cells with or without exposure to IR. h The numbers of phospho-H2AX foci per nucleus were counted on the basis of immunofluorescence at 30 min or 24 h after exposure to 6 Gy of IR in H1299 cells with stable expression of empty vector (EV) or SLC7A11 (SLC). Error bars are means ± SD, n = 50 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test.
FINs sensitize cancer cells to IR
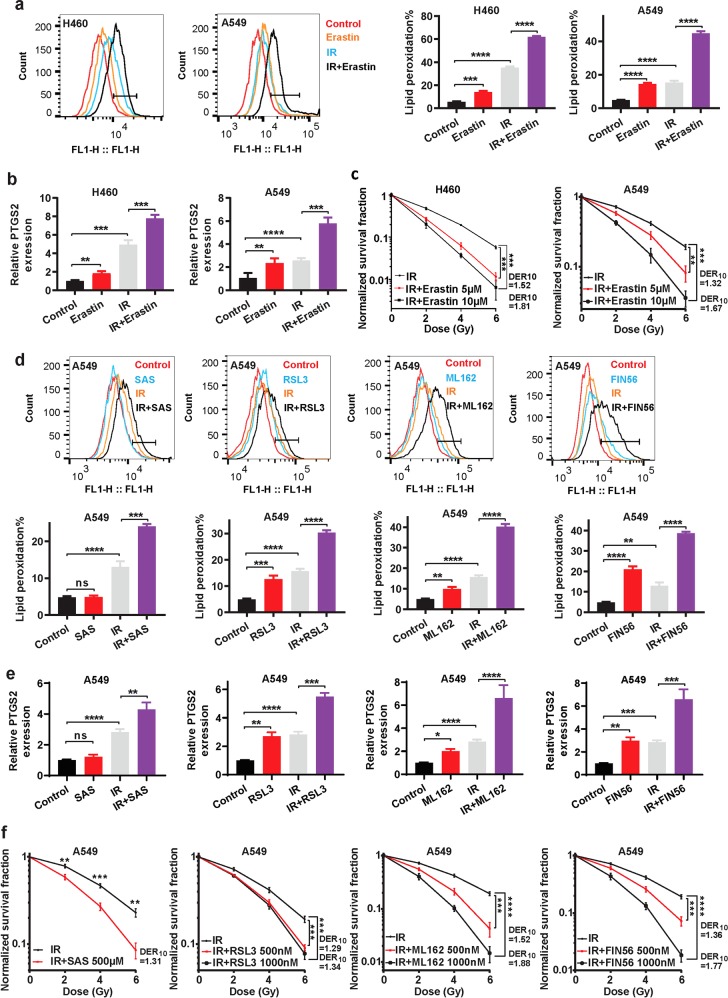
Ferroptosis can be induced by either class 1 FINs that inhibit SLC7A11 or class 2 or 3 FINs that inhibit or deplete GPX4.16 Our aforementioned data prompted us to examine whether inactivating SLC7A11 or GPX4 with the corresponding FINs would potentiate IR-induced lipid peroxidation and ferroptosis, thus sensitizing cancer cells to IR. In these experiments, we pretreated cancer cells with FINs, exposed them to IR, and then cultured them without FINs. We found that IR or treatment with erastin, a class 1 FIN, induced lipid peroxidation (Fig. 5a) and PTGS2 expression (Fig. 5b) in H460 and A549 cells. Importantly, combined treatment with IR and erastin resulted in synergistic increases of lipid peroxidation and PTGS2 expression in the tested cancer cells (Fig. 5a, b). Further analyses revealed that erastin significantly sensitized these cancer cells to IR (with DER10 > 1.2 or combination index < 1; Fig. 5c and Supplementary information, Fig. S5a).
Fig. 5. FINs sensitize cancer cells to IR.
a Lipid peroxidation assessment in H460 and A549 cells pretreated with DMSO or 10 μM erastin for 24 h followed by exposure to 4 Gy of IR. Bar graph showing relative levels of lipid peroxidation by C11-BODIPY staining in the indicated cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. b qRT-PCR analysis of PTGS2 expression in H460 and A549 cells pretreated with DMSO or 10 μM erastin for 24 h followed by exposure to 4 Gy of IR. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. c Clonogenic survival curves in H460 and A549 cells pretreated with DMSO or 10 μM erastin for 24 h followed by exposure to 4 Gy of IR. The survival data were normalized to those of unirradiated control cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. d Lipid peroxidation assessment in A549 cells pretreated with DMSO, 500 mM sulfasalazine, 10 μM RSL3, 10 μM ML162, or 10 μM FIN56 for 24 h followed by exposure to 4 Gy of IR. Bar graphs showing relative levels of lipid peroxidation by C11-BODIPY staining in the indicated cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. e qRT-PCR analysis of PTGS2 expression in A549 cells pretreated with DMSO, 500 mM sulfasalazine, 10 μM RSL3, 10 μM ML162, or 10 μM FIN56 for 24 h followed by exposure to 4 Gy of IR. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. f Clonogenic survival curves for A549 cells pretreated with DMSO, 500 mM sulfasalazine, 10 μM RSL3, 10 μM ML162, or 10 μM FIN56 for 24 h followed by exposure to 4 Gy of IR. The survival data were normalized to those of unirradiated control cells. Error bars are means ± SD, n = 3 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test.
Similarly, treatment with sulfasalazine (another class 1 FIN), RSL3 or ML162 (class 2 FINs), or FIN56 (a class 3 FIN), when combined with IR, had synergistic effects on inducing lipid peroxidation and PTGS2 expression (Fig. 5d, e) and significantly radiosensitized A549 cells (Fig. 5f; Supplementary information, Fig. S5a). It should be noted that the effects of IR on lipid peroxidation and PTGS2 induction were even more dramatic than those of most FINs in the cell lines we studied (Fig. 5a, b, d, e), further strengthening the evidence that IR is a potent ferroptosis inducer. We made similar observations in H460 cells (Supplementary information, Fig. S5b-e). Together, our data strongly suggest that FINs sensitize cancer cells to IR and that combining FINs and IR synergistically induces lipid peroxidation and ferroptosis.
Ferroptosis is involved in IR-induced tumor suppression in vivo
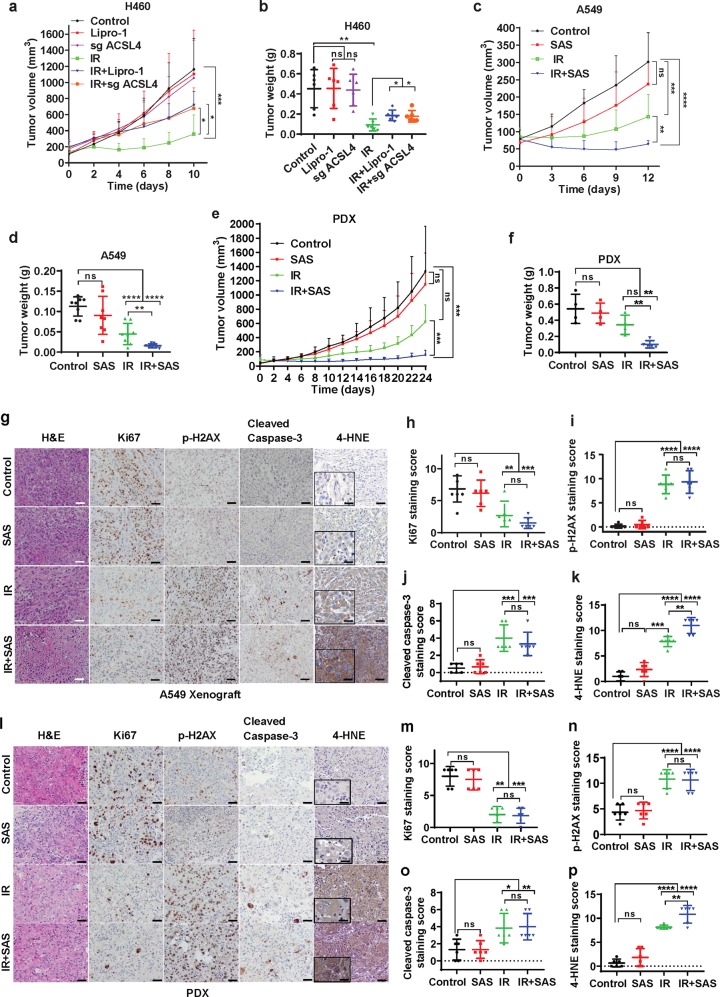
Next, we sought to determine the relevance of ferroptosis in IR-induced tumor suppression in vivo. To this end, we first studied the impact of ferroptosis inhibition, by either ACSL4 deletion or treatment with the ferroptosis inhibitor liproxstatin-1, on the growth of xenograft tumors derived from the radiosensitive cell line H460 with or without IR (Supplementary information, Fig. S6a). Although ferrostatin-1 is commonly used as the ferroptosis inhibitor in cell line studies, it is not suitable for long-term treatment in animal studies. The ferroptosis inhibitor liproxstatin-1, unlike ferrostatin-1, is stable in plasma and exhibits excellent in vivo efficacy in animal studies, including tumor treatment studies.12,18 As expected, IR potently suppressed H460 xenograft tumor growth; while neither ACSL4 deletion nor liproxstatin-1 treatment alone affected tumor growth, both ACSL4 deletion and liproxstatin-1 treatment partially restored tumor growth after IR (Fig. 6a, b). As expected, IR decreased Ki67 staining (a marker of cell proliferation) and increased phospho-H2AX or cleaved caspase-3 (a apoptosis marker) staining in tumor tissues (Supplementary information, Fig. S6c-f). IR also significantly increased the staining of 4-hydroxy-2-noneal (4-HNE), a lipid peroxidation marker,20,33 in tumor samples (Supplementary information, Fig. S6c, g); Importantly, ACSL4 deletion or liproxstatin-1 treatment significantly decreased 4-HNE staining without affecting Ki67, phospho-H2AX, or cleaved caspase-3 staining under IR treatment (Supplementary information, Fig. S6c-g). Thus, mirroring our in vitro data (Figs. 1e, 2h), our in vivo data further suggest that ferroptosis is at least partly responsible for IR-induced tumor suppression in vivo.
Fig. 6. Ferroptosis is involved in IR-induced tumor suppression in vivo.
a Volumes of H460 xenograft tumors with the indicated genotypes and treatments at different time points (days) after exposure to 10 Gy of IR. Error bars are means ± SD, n = 8 independent repeats. P values were determined using 2-way ANOVA. b Individual value plot showing the weights of H460 tumor xenografts in the indicated genotype and treatment groups. Error bars are means ± SD, n = 8 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. c Volumes of A549 xenograft tumors with the indicated treatments at different time points (days) after exposure to 10 Gy of IR. Error bars are means ± SD, n = 8 independent repeats. P values determined using 2-way ANOVA. d Individual value plot showing the weights of A549 tumor xenografts from the indicted treatments. Error bars are means ± SD, n = 8 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. e Volumes of lung cancer patient-derived xenograft tumors with the indicated treatments at different time points (days) after exposure to 8 Gy of IR. Error bars are means ± SD, n = 4-5 independent repeats. P values were determined using 2-way ANOVA. f Individual value plot showing the weights of lung cancer patient-derived xenograft tumor xenografts from the indicted treatments. Error bars are means ± SD, n = 4-5 independent repeats. P values were calculated using two-tailed unpaired Student’s t-test. g Representative images of hematoxylin and eosin and immunohistochemical staining (Ki67, phospho-H2AX, cleaved caspase-3 and 4-HNE) of A549 xenograft tumors with the indicated treatments. Scale bars, 50 µm/20 µm (inset). h–k Immunochemistry scoring of Ki67, phospho-H2AX, cleaved caspase-3 and 4-HNE staining. Error bars are means ± SD, n = 6 randomly selected magnification fields. P values were calculated using two-tailed unpaired Student’s t-test. l Representative images of hematoxylin and eosin and immunohistochemical staining (Ki67, phospho-H2AX, cleaved caspase-3 and 4-HNE) of lung cancer patient-derived xenograft tumors with the indicated treatments. Scale bars, 50 µm/20 µm (inset). m–p Immunochemistry scoring of Ki67, phospho-H2AX, cleaved caspase-3 and 4-HNE staining. Error bars are means ± SD, n = 6 randomly selected magnification fields. P values were calculated using two-tailed unpaired Student’s t-test.
We then examined whether inducing ferroptosis with FINs can sensitize tumors to IR in vivo (Supplementary information, Fig. S6a). Figure 5 shows the results of experiments using a series of FINs that inhibit either SLC7A11 or GPX4. It should be noted that GPX4 is an essential gene; germline or postnatal Gpx4 deletion in mice leads to embryonic or adult lethality.11 In contrast, Slc7a11 KO mice are viable with no obvious phenotype.34 Therefore, SLC7A11 likely represents a better therapeutic target for cancer treatment than GPX4 because inhibiting SLC7A11 would presumably cause less toxicity in patients than inhibiting GPX4. Considering this, we have primarily focused on class 1 FINs (which inhibit SLC7A11) in our in vivo studies. Although erastin is the most widely used class 1 FIN in cell line studies, erastin is not suitable for in vivo treatment because of its low solubility and poor metabolic stability; on the other hand, sulfasalazine, another class 1 FIN, has been used for in vivo treatment.35–38 For these reasons, we used sulfasalazine in the following animal studies.
We found that, while sulfasalazine treatment alone did not significantly affect tumor growth of A549 xenografts, it did dramatically sensitize tumors to IR, and the combination of sulfasalazine and IR potently suppressed tumor growth (Fig. 6c, d; Supplementary information, Fig. S6b). Sulfasalazine treatment exhibited a similar radiosensitizing effect in lung cancer patient-derived xenografts (PDXs) with KEAP1 mutation (Fig. 6e, f). Further analyses of both A549 xenograft and PDX tumor samples revealed that sulfasalazine treatment did not obviously affect Ki67, phospho-H2AX, or cleaved caspase-3 staining under either control or IR treatment condition (Fig. 6g–p). Importantly, IR significantly increased 4-HNE staining and sulfasalazine treatment combined with IR further increased 4-HNE staining in tumor samples (Fig. 6g, k, l, p). Of note, sulfasalazine treatment alone also moderately increased 4-HNE staining, although the increase was not statistically significant (Fig. 6k, p). This is consistent with the notion that sulfasalazine is considered a weak FIN16 (e.g., compare the effects of sulfasalazine and other FINs on inducing PTGS2 expression and lipid peroxidation in cell line studies shown in Fig. 5). Together, our data suggest that while sulfasalazine is less effective in inducing lipid peroxidation and suppressing tumor growth when used alone, it significantly sensitizes tumors to IR likely by synergizing with IR-induced lipid peroxidation and ferroptosis.
Ferroptosis induction correlates with cancer patient response to radiotherapy
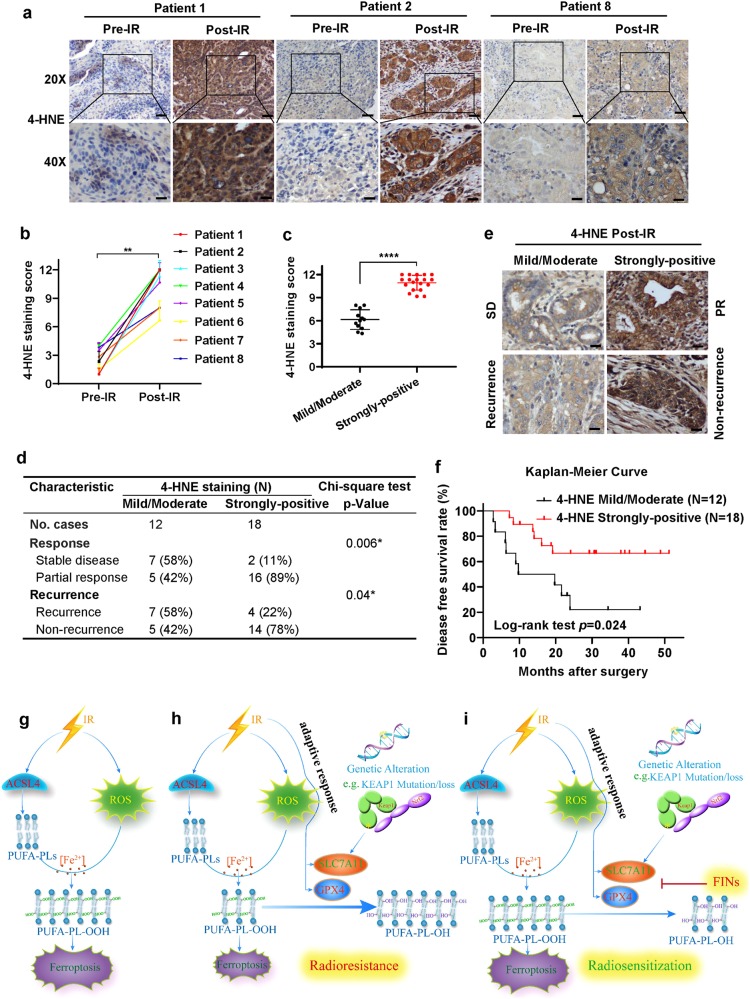
To further study the clinical relevance of ferroptosis to radiotherapy in cancer patients, we enrolled 30 esophageal cancer patients who had undergone surgical resection after radiotherapy into our study, and obtained 30 post-radiotherapy and 8 matched pre-radiotherapy esophageal tumor samples from these cancer patients (of note, we confirmed that IR induced ferroptosis in esophageal cancer cell line FLO-1 cells; Supplementary information, Fig. S1b-d). Analyses of 8 pairs of pre- and post-radiotherapy tumor samples revealed very weak 4-HNE staining in all pre-radiotherapy tumor samples; notably, radiotherapy moderately or strongly induced 4-HNE levels in all tumor samples (Fig. 7a–c), indicating that radiotherapy induces lipid peroxidation and therefore likely ferroptosis in cancer patients. We then correlated 4-HNE levels in post-radiotherapy tumor samples with patient outcomes. Our analyses showed that, while 4-HNE levels did not correlate with patient baseline characteristics, such as age, sex, or tumor stage (Supplementary information, Table S1), strongly positive 4-HNE staining significantly correlated with better clinical outcomes in cancer patients (with partial response or non-recurrence) (Fig. 7d–e). Disease free survival analysis also revealed that patients with strongly-positive 4-HNE staining survived significantly longer than those with mild/moderate 4-HNE staining (Fig. 7f). Together, our data suggest that ferroptosis likely contributes to radiotherapy response in cancer patients.
Fig. 7. Ferroptosis induction correlates with cancer patient response to radiotherapy.
a Representative images of 4-HNE immunohistochemical staining of matched esophageal tumor samples from the same patients before and after radiotherapy. Scale bars, 50 µm/20 µm (inset). b Immunochemistry scoring of 4-HNE staining of matched esophageal tumor samples before and after radiotherapy from 8 cancer patients. Error bars are means ± SD, n = 6 randomly selected magnification fields. P values calculated using two-tailed unpaired Student’s t-test. c Immunochemistry scoring of 4-HNE staining of post-radiotherapy esophageal tumor samples from 30 esophageal cancer patients. Error bars are means ± SD, n = 6 randomly selected magnification fields. P values were calculated using two-tailed unpaired Student’s t-test. d Correlations between radiotherapy response (or recurrence) and 4-HNE staining scores in post-radiotherapy esophageal tumor samples from 30 esophageal cancer patients. SD: stable disease; PR: partial response; P values were calculated using χ2 test. e Representative images of 4-HNE immunohistochemical staining of post-radiotherapy esophageal tumor samples in 4-HNE mild/moderate group or 4-HNE strongly positive group with the indicated status of patient response and recurrence. Scale bars, 20 µm (inset). f Disease-free survival Kaplan–Meier curves of esophageal cancer patients treated with radiotherapy from 4-HNE mild/moderate group and 4 -HNE strongly positive group. P values were calculated using log-rank test. g–i The working model depicting the roles and mechanisms of ferroptosis in IR response and radioresistance. See Discussion for detailed description.
Discussion
Previously, we and others have shown that like apoptosis, ferroptosis serves as a natural barrier to tumor development.17–22 In this study, we show that ferroptosis can also be induced by radiotherapy, one of the most widely used cancer therapies. Mechanistically, we propose that IR induces lipid peroxidation, a hallmark of ferroptosis, through at least 2 parallel pathways (Fig. 7g). First, IR-induced ROS promote lipid peroxidation. Presumably, IR-induced ROS can remove electrons from PUFAs to form fatty-acid radicals (PUFA•). These unstable carbon-centered radicals then react rapidly with molecular oxygen to generate lipid peroxyl radicals (PUFA-OO•), which can abstract H• by Fenton reactions from other molecules to ultimately form lipid hydroperoxides (PUFA-OOH).26,39 In addition, we showed that IR induces ACSL4 expression, which is critical for mediating the biosynthesis of PUFA-PLs, the type of lipids that are particularly susceptible to peroxidation.7–9 Consistent with this, ACSL4 deletion blocked IR-induced lipid peroxidation (but not total ROS) and inhibited ferroptosis (Fig. 2). Presumably, IR-induced ACSL4 expression increases the biosynthesis of PUFA-PL, which, together with IR-induced ROS, drives PUFA-PL peroxidation (PUFA-PL-OOH) and ferroptosis (Fig. 7g). How IR induces ACSL4 expression remains unclear and potentially involves transcriptional factors or chromatin modifying enzymes that regulate both radiation response and ferroptosis, such as p53 and BAP1.17,18 It will be interesting to study this in future explorations.
It is important to emphasize that our study identifies IR as a potent ferroptosis inducer, equivelant to or even more potent than most of the FINs we have tested (see Fig. 5). We speculate that the 2 parallel responses induced by IR (i.e., IR-induced ROS and ACSL4 expression) ensure robust ferroptosis induction by IR. It is possible that additional mechanisms are also involved in IR-induced ferroptosis; clearly, dissecting such mechanisms will remain an important area of future investigation.
We propose that IR also induces an adaptive response involving SLC7A11 or GPX4 induction that dampens IR-induced ferroptosis and promotes cancer cell survival during radiotherapy, leading to radioresistance (Fig. 7h). This radioresistance mechanism is analogous to the pathway reactivation mechanism that leads to cancer cell resistance to targeted therapies27 in that both resistance mechanisms restore the original downstream signaling output. Alternatively, certain genetic alterations in tumor cells (such as mutations in KEAP1-NRF2 signaling in lung or esophageal cancer) lead to aberrant expression of SLC7A11 (or presumably GPX4 in other contexts), resulting in de novo radioresistance (Fig. 7h), much like de novo drug resistance to targeted therapies. In support of this, our clinical analyses showed that the levels of ferroptosis marker 4-HNE correlate with patient outcomes to radiotherapy (Fig. 7c–f). We further propose that treatment with FINs that inhibit either SLC7A11 or GPX4 promotes lipid peroxidation and resensitizes radioresistant cancer cells to IR-induced ferroptosis, resulting in radiosensitization (Fig. 7i). We have provided several lines of evidence to support this model. First, IR induces not only ACSL4, but also SLC7A11 and GPX4, which function to protect cells from ferroptosis by converting PUFA-PL-OOH to PUFA-PL-OH. In addition, aberrant expression of SLC7A11 or GPX4 by either enforced overexpression or KEAP1 deficiency suppressed IR-induced lipid peroxidation and ferroptosis and promoted radioresistance in a ferroptosis-dependent manner. Finally, combined treatment of IR and FINs synergistically increased lipid peroxidation and PTGS2 expression, and FINs exerted significant radiosensitizing effects both in vitro and in vivo.
In this study, we tested various FINs for their radiosensitization effects, including the class 1 FINs erastin and sulfasalazine, the class 2 FINs RSL3 and ML162, and the class 3 FIN FIN56. Our data showed that all these FINs exhibited significant radiosensitizing effects in vitro. Our analysis further revealed that sulfasalazine sensitizes cell line xenograft and PDX tumors to IR in vivo. Sulfasalazine is a Food and Drug Administration-approved drug commonly used for the treatment of rheumatoid arthritis; it has been previously identified as an inhibitor of SLC7A11 transporter activity.40 Thus, our study may motivate further testing of the combination of sulfasalazine and radiotherapy in clinical trials. Given that sulfasalazine is considered a relatively weak FIN and that other FINs either are not suitable for in vivo treatment (such as erastin) or raise toxicity concerns (such as FINs that inactivate GPX4), our findings also support the development of more potent and specific FINs suitable for in vivo use and testing of their use as radiosensitizers in cancer treatment. Finally, we envision that a detailed understanding of the exact genetic or tumor contexts that maximize treatment efficacy will also likely govern the therapeutic utility of FINs in radiotherapy.
Together, our data reveal that IR can potently induce lipid peroxidation and ferroptosis in cell lines, xenograft tumors, and cancer patients, indicate that ferroptosis is at least as important as other forms of cell death in IR-induced cell death and tumor suppression, and suggest the use of FINs as radiosensitizers alongside radiotherapy. Our findings are consistent with another report which was published during our manuscript submission.41 These results together provide definitive evidence linking IR with lipid peroxidation and ferroptosis as well as a broad framework for further understanding and targeting ferroptosis in cancer therapy.
Materials and Methods
Cell line-derived xenograft model
Xenograft mouse model experiments were conducted in accordance with a protocol reviewed and approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center. The study adhered to all relevant ethical regulations regarding animal research. Female 4- to 6-week-old athymic nude mice (Foxn1nu/Foxn1nu) were purchased from the Experimental Radiation Oncology Breeding Core Facility at MD Anderson Cancer Center and housed in the Animal Care Facility at the Department of Veterinary Medicine and Surgery at MD Anderson. Cancer cell lines were suspended and counted in cold phosphate-buffered saline (PBS), and 3 × 106 H460 or 5 × 106 A549 cells were injected into mice subcutaneously. When the tumor reached 50–100 mm3, the mice were assigned randomly into different treatment groups. Tumors were irradiated with a JL Shepherd Mark I-68A irradiator with a 137Cs source at a dose of 10 Gy. Sulfasalazine was dissolved in dimethyl sulfoxide (DMSO) and diluted in PBS, then intraperitoneally injected into mice at a dose of 250 mg/kg once a day. Liproxstatin-1 diluted in PBS was intraperitoneally injected daily at a dose of 10 mg/kg. Sulfasalazine or liproxstatin-1 was injected three times before irridation followed by continued daily injection until the endpoint as indicated in the corresponding figures. The tumor volume was measured 2 or 3 times per week until the endpoint and calculated according to the equation volume = length × width2 × 1/2. Details for the reagents and resources in this experiment and others below are listed in Table 1.
Table 1.
Reagents and resources.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Vinculin | Sigma-Aldrich | Cat#V4505 |
| SLC7A11/xCT (D2M7A) | Cell Signaling Technology | Cat#12619 |
| phospho-p53 (Ser15) | Cell Signaling Technology | Cat#9284 |
| ACSL4 | Santa Cruz | Cat#sc-271800 |
| KEAP1 | Santa Cruz | Cat#sc-365626 |
| GPX4 | R&D Systems | Cat#MAB5457 |
| Tubulin | Cell Signaling Technology | Cat#2144 |
| phospho-Chk2 (Thr68) | Cell Signaling Technology | Cat#2197 |
| phospho-histone H2A.X (Ser139) | EMD Millipore | Cat#05-636 |
| Anti-4 Hydroxynonenal (4-HNE) | Abcam | Cat#ab46545 |
| Ki-67 (D2H10) | Cell Signaling Technology | 9027S |
| Cleaved Caspase-3 (Asp175) | Cell Signaling Technology | 9661s |
| Chemicals | ||
| Erastin | Sigma | Cat#E7781 |
| Ferrostatin-1 | Sigma | Cat#SML0583 |
| Liproxstatin-1 | Sigma | Cat#SML1414 |
| N-acetyl-L-cysteine | Sigma | Cat#A9165 |
| Sulfasalazine | Sigma | Cat#S0883 |
| Z-VAD-fmk | R&D Systems | Cat#FMK001 |
| Necrostatin-1s | BioVision | Cat#2263 |
| RSL3 | Selleckchem | Cat#S8155 |
| FIN56 | Cayman Chemical | Cat#25180 |
| ML162 | Cayman Chemical | Cat#20455 |
| CM-H2DCFDA | ThermoFisher | Cat#C6827 |
| BODIPY 581/591 C11 | Invitrogen | Cat#D3861 |
| Critical Commercial Assays | ||
| Cell Counting Kit-8 (CCK8) reagent | Dojindo Molecular Technologies | Cat#CK04 |
| Experimental Models: Cell Lines | ||
| A549 | ATCC | Cat#CCL-185 |
| H460 | ATCC | Cat#HTB-177 |
| H1299 | ATCC | Cat#CRL-5803 |
| H23 | ATCC | Cat#CRL-5800 |
| MCF7 | ATCC | Cat#HTB-22 |
| HT-1080 | ATCC | Cat#CCL-121 |
| FLO-1 | Dr. Steven H. Lin at MD Anderson Cancer Center | |
| UMRC6 | Dr. William G. Kaelin at Dana-Farber Cancer Institute | |
| HEK293T | ATCC | Cat#CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| Mouse: athymic nude: Foxn1nu/nu | The facility in the Department of Experimental Radiation Oncology at MD Anderson Cancer Center | |
| NOD scid gamma (NSG) mice | The facility in the Department of Experimental Radiation Oncology at MD Anderson Cancer Center | |
| Sequence-Based Reagents | ||
| For primer and guide RNA sequences, please see Supplementary information, Table S2. | ||
| Software and Algorithms | ||
| GraphPad Prism 6 | GraphPad Software, Inc. | |
| FlowJo_V10 | FlowJo, LLC | |
| CompuSyn | CompuSyn, Inc. | |
| ImageJ | NIH | |
| SPSS 25.0 | SPSS Inc. | |
Patient-derived xenograft (PDX) model
For PDX experiments, all the NOD scid gamma (NSG) mice were purchased from the Experimental Radiation Oncology Breeding Core Facility at MD Anderson Cancer Center and housed in the Animal Care Facility at the Department of Veterinary Medicine and Surgery at MD Anderson Cancer Center. PDX model used in this study was originally obtained from lung cancer PDX platform at MD Anderson Cancer Center. PDX experiments were performed, as previously described.42 Briefly, PDX tumors in cold DMEM media were minced into fragments 1–2 mm3 in volume. Then each PDX tumor fragment was subcutaneously inoculated into the dorsal flank of NSG mice. When the tumors reached 50–100 mm3 in volume, the mice were randomized into four groups and treated with PBS, sulfasalazine, ionizing radiation or sulfasalazine plus ionizing radiation, respectly. Ionizing radiation (X-ray) was applied locally to the tumor in the flank of mice at 8 Gy. Sulfasalazine was dissolved in dimethyl sulfoxide (DMSO) and diluted in PBS, then intraperitoneally injected into mice three times before irridation at a dose of 250 mg/kg followed by continued injection once every two days until the endpoint as indicated in the corresponding figures. The tumor volume was measured 3 times per week until the endpoint and calculated according to the equation volume = length × width2 × 1/2.
Cancer cell lines
All cells were cultured in a 37 °C incubator in an atmosphere of 5% CO2. The human non-small cell lung cancer cell line A549, human breast adenocarcinoma cell line MCF-7, human fibrosarcoma cell line HT-1080, human esophageal adenocarcinoma cell line FLO-1, human renal carcinoma cell line UMRC6, and human embryonic kidney 293 (HEK293T) cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 10,000 U/mL of penicillin-streptomycin. The human non-small cell lung cancer cell lines H460, H1299, and H23 were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 10,000 U/mL of penicillin-streptomycin.
Irradiation and clonogenic survival assay
For all clonogenic survival assays, irradiations were conducted with an X-RAD 320 cabinet irradiator (Precision X-Ray) at doses from 0 to 6 Gy and a dose rate of 250 MU/min. To determine the effects of cell death inhibitors on IR-induced cell death, 200–1000 cells per well were incubated in triplicate in 6-well plates overnight. The cells were then pretreated with different cell death inhibitors at appropriate doses or with DMSO for 24 h. The cells were then irradiated. Fresh medium containing different inhibitors was added to the plates every 2 days. To determine the synergistic effects of ferroptosis inducers with IR, cells were pretreated with erastin, sulfasalazine, RSL3, FIN56, or ML162 for 24 h, then irradiated and cultured in normal medium. After incubation for 1–2 weeks, cells were stained with 0.5% crystal violet (Sigma, #C0775) dissolved in 20% methanol. The colonies in each well were counted visually. The surviving fraction was calculated using GraphPad Prism 6 and normalized to that of unirradiated control cells. The dose survival curve was plotted using the linear quadratic model (Y = exp(−(a × x + b × (x2)))). The dose enhancement ratio (DER10) for 10% survival was calculated as follows: DER10 = (Radiation dose correlated with 10% survival fraction in DMSO-treated cells)/(Radiation dose correlated with 10% survival fraction in drug treated cells). DER10 > 1 indicates that the drug radiosensitizes cancer cells.
Cell viability assay and treatment combination analysis
For cell viability assays, 5000 cells per well were seeded in replicates of 4 in 96-well plates and incubated for 24 h. Cells were treated with or without 10 μM erastin for 24 h, and then the medium was replaced with 100 μL fresh medium containing 10 μL Cell Counting Kit-8 (CCK8) reagent (Dojindo Molecular Technologies, CK04). After incubation for 1 h in a humidified incubator (at 37 °C, 5% CO2), absorbance was measured at 450 nm using a FLUOstar Omega microplate reader (BMG Labtech). Cell viability of samples was calculated according to the manufacturer’s instructions.
For treatment combination analysis, a series of doses and effects (FA -fraction affected) were entered into the CompuSyn software tool. For each treatment alone and their combinations (nonconstant-ratio combinations), the software automatically calculated combination index (CI) values at different FA levels based on the CI algorithm. The resulting CI theorem of Chou–Talalay offers quantitative definition for additive effects (CI = 1), synergism (CI < 1), and antagonism (CI > 1) of drug combinations.
ROS and lipid peroxidation assay
ROS and lipid peroxidation levels were measured, as previously described.18,43 Cells were seeded in triplicate in 12-well plates 24 h prior to treatment, pretreated with or without drugs for 24 h, and then irradiated. After the cells had incubated for 24 or 48 h, fresh medium containing 4 μM CM-H2DCFDA (ThermoFisher, C6827) for ROS measurements or 5 μM BODIPY 581/591 C11 dye (Invitrogen, D3861) for lipid peroxidation measurements was added to each well. After incubation for 30 min in a humidified incubator (at 37 °C, 5% CO2), the cells were washed with PBS and trypsinized to obtain a cell suspension. ROS and lipid peroxidation levels were analyzed by flow cytometry using an Accuri 6 cytometer (BD Bioscience).
Transmission electron microscopy
Transmission electron microscopy analyses were conducted, as previously described.18 Briefly, cells cultured in a 6-well plate were fixed with a solution containing 3% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.3). After being washed in 0.1 M sodium cacodylate buffer, cells were treated with 0.1% Millipore-filtered cacodylate-buffered tannic acid, postfixed with 1% buffered osmium, and stained with 1% Millipore-filtered uranyl acetate. After dehydration and embedding, samples were incubated in a 60 °C oven for ~3 days. Ultrathin sections were prepared and examined with a JEM 1010 transmission electron microscope (JEOL). Digital images were obtained using an AMT Imaging System (Advanced Microscopy Techniques Corp.) at the High Resolution Electron Microscopy Facility at MD Anderson Cancer Center.
qRT-PCR
qRT-PCR was performed, as previously described.44,45 Total RNA was extracted using TRIzol reagent (Invitrogen), and cDNA was synthesized with SuperScript II Reverse Transcriptase (Invitrogen, #18064014). Quantitative real-time PCR was performed using SYBR GreenER qPCR SuperMix Universal (Invitrogen, # 11762500), and triplicate samples were run on a Stratagene MX3000P qPCR system according to the manufacturer’s protocol. The threshold cycle (Ct) values for each gene were normalized to those of β-actin, and the 2−ΔΔCt method was used for quantitative analysis. The primers used are listed in Supplementary information, Table S2.
Western blot analysis
Western blotting was performed according to the standard protocol, as previously described.46,47 Cell lysates were prepared in NP-40 buffer (150 mM sodium chloride, 1.0% NP-40, 50 mM Tris, pH 8.0) supplemented with complete mini protease inhibitors (Roche, 11836170001) and a phosphatase inhibitor cocktail (Calbiochem, 524625). Protein expression was analyzed by western blot using primary antibodies against the following target proteins: Vinculin (Sigma Aldrich, V4505, 1:10,000 dilution), SLC7A11/xCT (D2M7A) (Cell Signaling Technology, 12619, 1:5000 dilution), phospho-p53 (Ser15) (Cell Signaling Technology, 9284, 1:1,000 dilution), ACSL4 (Santa Cruz, sc-271800, 1:2000 dilution), KEAP1 (Santa Cruz, sc-365626, 1:1000 dilution), GPX4 (R&D Systems, MAB5457, 1:1000 dilution), tubulin (Cell Signaling Technology, 2144, 1:5000 dilution), phospho-Chk2 (Thr68) (Cell Signaling Technology, 2197, 1:1000 dilution), and phospho-histone H2A.X (Ser139) (EMD Millipore, 05–636, 1:1000 dilution). Following incubation with horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature, proteins were visualized with chemiluminescence using Pierce ECL 2 Western Blotting Substrate (ThermoFisher Scientific, 80196).
Immunofluorescence
Immunofluorescence analyses were performed as previously described.48,49 Cells seeded on coverslips were fixed in 3.7% formaldehyde in PBS for 15 min at different time points (30 min and 24 h after 6-Gy irradiation). Then cells were washed with PBS and permeabilized for 10 min in 0.1% TritonX-100/PBS. After permeabilization, blocking buffer (5% bovine serum albumin in 0.1% Triton/PBS) was added for 1 h, followed by incubation with a phospho-histone H2A.X (Ser139) mouse monoclonal antibody (EMD Millipore, 05–636, 1:500 dilution) overnight at 4 °C. After being washed with PBS, cells were incubated with Alexa 594 anti-mouse fluorescent secondary antibodies (Life Technologies, 1:200 dilution) in the dark for 2 h at room temperature. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, ThermoFisher Scientific). Fluorescence was monitored using a Leica confocal microscope, and foci of phospho-H2AX were quantified using ImageJ software (NIH).
Constructs and generation of overexpression, knockdown, or knockout cell lines
CRISPR-mediated knockout plasmids containing guide RNAs targeting ACSL4, KEAP1, and SLC7A11 were generated in LentiCRISPR-V2 (Addgene, #52961) according to the standard protocol. The sequences of ACSL4 sh RNA and guide RNAs are listed in Supplementary information, Table S2. The expression vectors containing SLC7A11 cDNA are described in our previous publication.50 The pcDNA3.1-flag-GPX4 construct was a kind gift from Dr. Aikseng Ooi from The University of Arizona Health Sciences. Stable cell lines were generated as described previously.18
Histology and immunohistochemistry
Xenograft tumor samples were collected and immediately fixed in 10% neutral-buffered formalin (ThermoFisher Scientific) overnight. After being washed once with PBS, samples were transferred into 70% ethanol and subjected to embedding, sectioning, and haematoxylin and eosin staining. For immunohistochemical staining, tissue sections were processed according to methods described in our previous publications.51–53 The primary antibody used for immunohistochemistry was anti-4-HNE (1:400, Abcam, ab46545), anti-phospho-histone H2A.X (1:500, EMD Millipore, Cat#05–636), anti-Ki-67 (D2H10) (1:500, Cell Signaling Technology, 9027 S), anti-cleaved caspase-3 (1:500, Cell Signaling Technology, 9661 s). Images were obtained at 400× magnification using an Olympus BX43 microscope. Immunohistochemical staining was semiquantitatively analyzed using the immunoreactive score (IRS) system. The percentage of positive cells was scored as follows: no stained cells: 0; 1–10% staining: 1; 10–50% staining: 2; 51–80% staining: 3; and 81–100% staining: 4. The staining intensity was scored as follows: no color reaction: 0; mild reaction: 1; moderate reaction: 2; and intense reaction: 3. Final IRS scores of immunohistochemistry = (scores of staining intensity) × (scores of percentage of positive cells).
Patient samples
Tumor samples of esophageal cancer patients were collected from the Department of Radiation Oncology, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School Of Medicine, Central South University at Changsha, China (25 of 30 patients) and the Department of Radiation Oncology, Devision of Radiation Oncology, The University of Texas MD Anderson Cencer Center at Houston, USA (5 of 30 patients). Patients who underwent surgical resection after 30–60 Gy radiotherapy from 2014 to 2018 were enrolled in the study. Post-radiotherapy tumor samples from all 30 patients and pre-radiotherapy samples from 8 of them were obtained. The procedures of human sample collection were approved by the Ethic Committee of Hunan Cancer Hospital & The Affiliated Cancer Hospital of Xiangya School Of Medicine, Central South University, and The University of Texas MD Anderson Cencer Center, respectively. Informed consent was obtained from all subjects. All available patient samples were evaluated by experienced pathologists for confirmation of histological type. Tumor response was assessed every 8 weeks according to the Response Evaluation Criteria in Solid Tumor (RECIST, version 1.1).
Qantification and statistical analyses
Results of cell culture experiments were collected from at least 2 or 3 independent replicates. Tumor volumes or weights from at least 6 individual mice in each group were plotted. Data are presented as means ± standard deviation (SD). Statistical significance (P values) was calculated using unpaired Student’s t-tests, χ2 test or log-rank test by GraphPad Prism 7.0 (GraphPad Software, Inc.) or SPSS 25.0 (SPSS Inc.). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., non-significant.
Supplementary information
Acknowledgements
We thank J. Chen for helpful suggestions and discussions and A. Ninetto from the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for manuscript editing. This research was supported by the Andrew Sabin Family Fellow Award, the Sister Institution Network Fund, and a Radiation Oncology Strategic Initiatives (ROSI) Platform Seed Grant from The University of Texas MD Anderson Cancer Center (to B.G.). B.G. is an Andrew Sabin Family Fellow. Y.Z. and P.K. were Scholars at the Center for Cancer Epigenetics at The University of Texas MD Anderson Cancer Center. P.K. is also supported by CPRIT Research Training Grant (RP170067) and Dr. John J. Kopchick Research Award from The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences.
Author contributions
G.L. and Y.Z. performed most of the experiments with assistance from P.K., J.Z., and X.L.; J.A.A., Q.X., Z.L., and H.W. provided patient samples. S.H.L. provided resources for local radiation in animals. B.G. designed the experiments. B.G. and H.W. supervised the study. B.G. wrote most of the manuscript with assistance from other co-authors. All authors commented on the manuscript.
Competing interests
S.H.L. receives grant funding from Hitachi Chemical Diagnostics, Genentech, Beyond Spring Pharmaceuticals, New River Labs; honorarium from Varian Medical Systems; and serves in Advisory Board for AstraZeneca and Beyond Spring Pharmaceuticals. Other authors declare no competing interests.
Footnotes
Further information and requests for reagents should be directed to the lead contact, Boyi Gan (bgan@mdanderson.org).
These authors contributed equally: Guang Lei, Yilei Zhang
Contributor Information
Hui Wang, Email: wanghui@hnszlyy.com.
Boyi Gan, Email: bgan@mdanderson.org.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0263-3.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat. Rev. Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 3.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/S1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 4.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stockwell BR, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao M, Jiang X. To eat or not to eat-the metabolic flavor of ferroptosis. Curr. Opin. Cell Biol. 2017;51:58–64. doi: 10.1016/j.ceb.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doll S, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagan VE, et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang WS, et al. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl Acad. Sci. USA. 2016;113:E4966–E4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan H, Li X, Zhang X, Kang R, Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem. Biophys. Res Commun. 2016;478:1338–1343. doi: 10.1016/j.bbrc.2016.08.124. [DOI] [PubMed] [Google Scholar]
- 11.Seibt TM, Proneth B, Conrad M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 2019;133:144–152. doi: 10.1016/j.freeradbiomed.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Friedmann Angeli JP, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koppula P, Zhang Y, Zhuang L, Gan B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. (Lond.) 2018;38:12. doi: 10.1186/s40880-018-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad M, Sato H. The oxidative stress-inducible cystine/glutamate antiporter, system x (c) (-): cystine supplier and beyond. Amino Acids. 2012;42:231–246. doi: 10.1007/s00726-011-0867-5. [DOI] [PubMed] [Google Scholar]
- 16.Feng H, Stockwell BR. Unsolved mysteries: How does lipid peroxidation cause ferroptosis? PLoS Biol. 2018;16:e2006203. doi: 10.1371/journal.pbio.2006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. cell Biol. 2018;20:1181–1192. doi: 10.1038/s41556-018-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T., Jiang L., Tavana O., Gu W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 79, 1749 (2019). [DOI] [PMC free article] [PubMed]
- 20.Zhang Y, Zhuang L, Gan B. BAP1 suppresses tumor development by inducing ferroptosis upon SLC7A11 repression. Mol. Cell. Oncol. 2019;6:1536845. doi: 10.1080/23723556.2018.1536845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao M, et al. Role of mitochondria in ferroptosis. Mol. Cell. 2019;73:354–363 e353. doi: 10.1016/j.molcel.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jennis M, et al. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30:918–930. doi: 10.1101/gad.275891.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer. 2005;104:1129–1137. doi: 10.1002/cncr.21324. [DOI] [PubMed] [Google Scholar]
- 24.Jaffray DA. Image-guided radiotherapy: from current concept to future perspectives. Nat. Rev. Clin. Oncol. 2012;9:688–699. doi: 10.1038/nrclinonc.2012.194. [DOI] [PubMed] [Google Scholar]
- 25.Baidoo KE, Yong K, Brechbiel MW. Molecular pathways: targeted alpha-particle radiation therapy. Clin. Cancer Res. 2013;19:530–537. doi: 10.1158/1078-0432.CCR-12-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azzam EI, Jay-Gerin JP, Pain D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012;327:48–60. doi: 10.1016/j.canlet.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konieczkowski DJ, Johannessen CM, Garraway LA. A convergence-based framework for cancer drug resistance. Cancer Cell. 2018;33:801–815. doi: 10.1016/j.ccell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeong Y, et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Disco. 2017;7:86–101. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rojo de la Vega M., Chapman E., Zhang D. D. NRF2 and the Hallmarks of Cancer. Cancer Cell34, 21–43 (2018). [DOI] [PMC free article] [PubMed]
- 33.Young O, Crotty T, O'Connell R, O'Sullivan J, Curran AJ. Levels of oxidative damage and lipid peroxidation in thyroid neoplasia. Head. Neck. 2010;32:750–756. doi: 10.1002/hed.21247. [DOI] [PubMed] [Google Scholar]
- 34.Sato H, et al. Redox imbalance in cystine/glutamate transporter-deficient mice. J. Biol. Chem. 2005;280:37423–37429. doi: 10.1074/jbc.M506439200. [DOI] [PubMed] [Google Scholar]
- 35.Lo M, Ling V, Low C, Wang YZ, Gout PW. Potential use of the anti-inflammatory drug, sulfasalazine, for targeted therapy of pancreatic cancer. Curr. Oncol. 2010;17:9–16. doi: 10.3747/co.v17i3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sehm T, et al. Sulfasalazine impacts on ferroptotic cell death and alleviates the tumor microenvironment and glioma-induced brain edema. Oncotarget. 2016;7:36021–36033. doi: 10.18632/oncotarget.8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robe PA, et al. In vitro and in vivo activity of the nuclear factor-kappaB inhibitor sulfasalazine in human glioblastomas. Clin. Cancer Res. 2004;10:5595–5603. doi: 10.1158/1078-0432.CCR-03-0392. [DOI] [PubMed] [Google Scholar]
- 38.Cobler L, Zhang H, Suri P, Park C, Timmerman LA. xCT inhibition sensitizes tumors to gamma-radiation via glutathione reduction. Oncotarget. 2018;9:32280–32297. doi: 10.18632/oncotarget.25794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shadyro OI, Yurkova IL, Kisel MA. Radiation-induced peroxidation and fragmentation of lipids in a model membrane. Int J. Radiat. Biol. 2002;78:211–217. doi: 10.1080/09553000110104065. [DOI] [PubMed] [Google Scholar]
- 40.Gout PW, Buckley AR, Simms CR, Bruchovsky N. Sulfasalazine, a potent suppressor of lymphoma growth by inhibition of the x(c)- cystine transporter: a new action for an old drug. Leukemia. 2001;15:1633–1640. doi: 10.1038/sj.leu.2402238. [DOI] [PubMed] [Google Scholar]
- 41.Lang X. et al. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 9, 2159–8290 (2019). [DOI] [PMC free article] [PubMed]
- 42.Yan X, et al. Inhibition of thioredoxin/thioredoxin reductase induces synthetic lethality in lung cancers with compromised glutathione homeostasis. Cancer Res. 2019;79:125–132. doi: 10.1158/0008-5472.CAN-18-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai F, et al. BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc. Natl Acad. Sci. USA. 2017;114:3192–3197. doi: 10.1073/pnas.1619588114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat. cell Biol. 2016;18:431–442. doi: 10.1038/ncb3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao ZD, et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat. Commun. 2017;8:783. doi: 10.1038/s41467-017-00902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin A. et al. FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacological inhibition of the PI3K-AKT pathway. Cancer Res. 74, 1682–1693 (2014). [DOI] [PMC free article] [PubMed]
- 47.Lin A, et al. The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene. 2014;33:3183–3194. doi: 10.1038/onc.2013.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee H, et al. BAF180 regulates cellular senescence and hematopoietic stem cell homeostasis through p21. Oncotarget. 2016;7:19134–19146. doi: 10.18632/oncotarget.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chauhan AS, et al. STIM2 interacts with AMPK and regulates calcium-induced AMPK activation. FASEB J. 2019;33:2957–2970. doi: 10.1096/fj.201801225R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koppula P, Zhang Y, Shi J, Li W, Gan B. The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J. Biol. Chem. 2017;292:14240–14249. doi: 10.1074/jbc.M117.798405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gan B, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gan B, et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell. 2010;18:472–484. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gan B, et al. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J. Cell Biol. 2006;175:121–133. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.