Abstract
The kidney is subject to a wide range of abnormalities, many of which have a significant hereditable component. Next generation sequencing is increasingly bringing the genetic drivers of Mendelian disease into focus at the base pair level, whereas inexpensive genotyping arrays have surveyed hundreds of thousands of individuals to identify common variants that predispose to kidney dysfunction. In this first article in a CJASN series on kidney genomics, we review how both rare and common variants contribute to kidney disease, explore how evolution may influence the genetic variants that affect kidney function, consider how genetic information is and will be used in the clinic, and identify some of the most important future directions for kidney disease research. Forthcoming articles in the series will elaborate on many of these themes.
Keywords: humans, high-throughput nucleotide sequencing, genotype, base pairing, genomics, kidney diseases, kidney, urinary tract physiological phenomena, architecture, Kidney Genomics Series, renal insufficiency
Introduction
The kidney is an intricate organ, an extraordinary machine consisting of a million filters and miles of vasculature that perform a myriad of functions. It is thus susceptible to a wide range of abnormalities from altered tubule transport of sodium and other ions to abnormal growth patterns leading to structural defects. We typically divide the kidney into functional elements, such as the nephron, the vasculature, and the interstitium, each of which can be altered by genetic variation. At even smaller scale, we subdivide the nephron into glomerulus and tubule, and we further divide these structures by cell type, with each highly specialized cell population susceptible to structural and functional dysfunction caused by genetic variation.
The Variety of Inherited Kidney Disorders
The number of genes that have been implicated in human kidney diseases or variation in kidney function is very large (several hundred). Even those of us interested in inherited disorders of the podocyte cannot remember the names of all the 50+ genes that, when mutated, can cause the nephrotic syndrome. Nevertheless, it is important for all of those involved in either the care of patients with kidney disease and/or the study of the kidney to have a basic understanding of the spectrum of variation and the breadth of phenotypes relevant to kidney health and disease. It is also important to develop familiarity with the technologies that are enabling our expanding knowledge and increasingly, informing the care of patients with kidney disease.
Kinds of Genetic Variation
The human nuclear genome contains about 3 billion bp of DNA per haploid genome. Except for the sex chromosomes, humans normally inherit two copies of the genome, one from each parent. Meiosis shuffles the arrangement of genes in such a way that, with the exception of genes that are physically very close, the copies of any two genes inherited from each parent are independent. The deviation from independence (known as linkage disequilibrium [LD]) at short distances forms the basis of genetic mapping methods both in the case of linkage studies in families and in the case of association studies in populations. The genomes of all humans are almost identical. Older texts often cite differences of about 1 in 1000 bp on average between two random people. When all types of variation are included in estimating these differences, including changes other than single-base pair differences, the number is probably closer to 16 in 1000 (1). Some base pairs in the genome are more variable than others. The more common variants in the genome are referred to as single-nucleotide polymorphisms (SNPs). Although it is generally thought that most SNPs do not have direct effects on human phenotypes, the presence of this common variation is what has enabled a large body of research. At the level of either families or populations, we can identify regions of the genome that influence specific phenotypes. After such regions are identified, additional studies can be performed to define the precise variation that is causally related to disease.
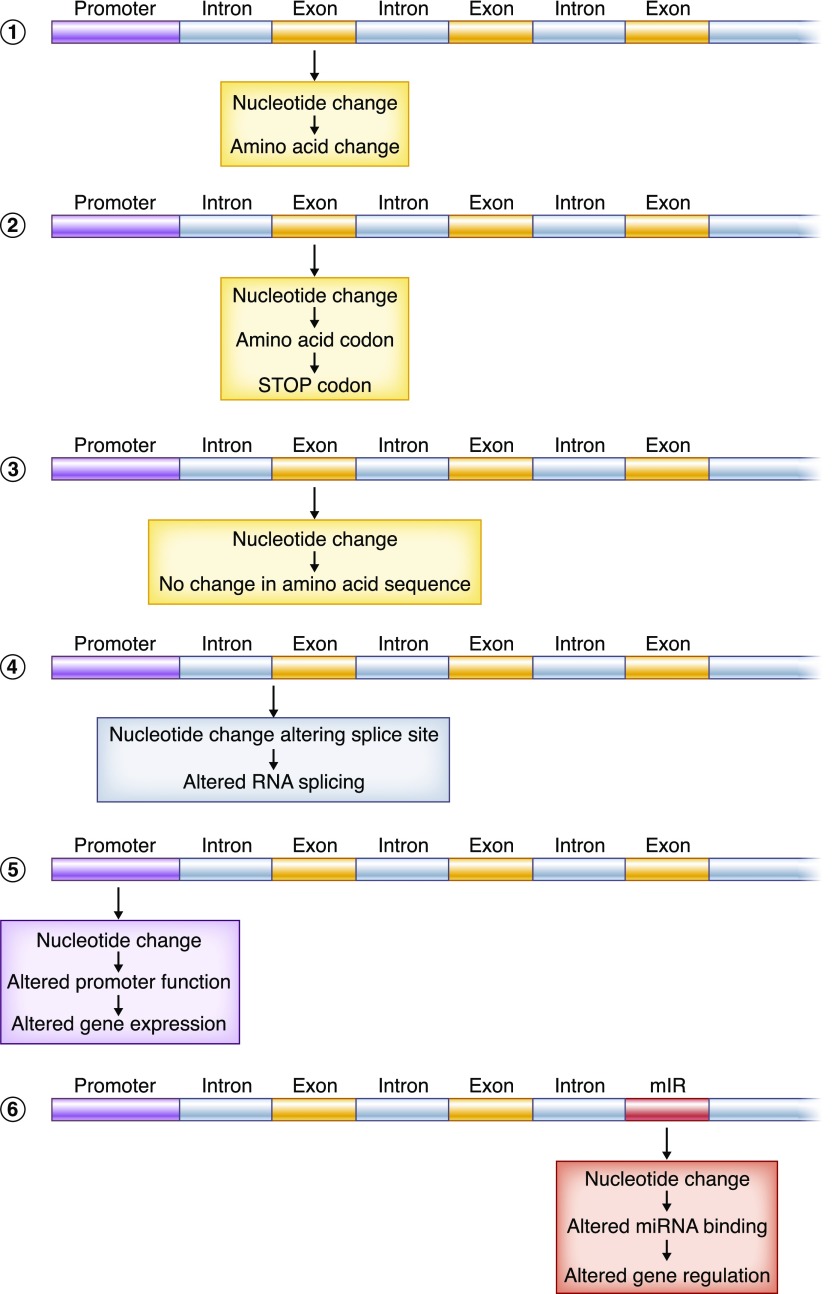
People typically use the word “mutation” for the variations in the genome that are very rare and have phenotypic consequences, whereas “polymorphism” is used to describe those differences that are more common. “Variant” is a more neutral term that simply recognizes genetic differences between individuals. There are multiple types of genetic variation. There can be cytogenetically detectable alterations in the structure of parts of or entire chromosomes (as in trisomies, translocations, and other large-scale alterations), smaller deletions or insertions of a segment of DNA (from just a few base pairs to millions), and variants that alter a single nucleotide. These single-nucleotide variants can change the sequence of an encoded protein, lead to an early truncation of a protein, alter the expression of a gene or genes, or have yet other effects (Figure 1, Table 1). It is remarkable that an alteration of a single nucleotide among the set of 3 billion can sometimes lead to severe lethal phenotypes and other times have no phenotypic consequence whatsoever.
Figure 1.
Single-nucleotide change have a range of possible effects. Single-nucleotide substitutions are perhaps the most studied class of genetic variant. The term single-nucleotide polymorphism (SNP) is usually used for those single-nucleotide changes at specific positions in the genome where the less common allele is present with a frequency of 1% or greater. The more general terms single-nucleotide substitution and single-nucleotide variants can be used to describe exceedingly rare changes (perhaps having occurred only once in human history) or variants where both the wild-type and variant alleles are quite common. It is remarkable that some single-nucleotide substitutions have no apparent phenotypic affect, whereas the presence of others may be lethal. Nucleotide substitutions can have a range of effects. (1) They may alter an encoded amino acid. (2) They may cause early truncation of a protein. (3) They may be silent, changing the nucleotide sequence but not the amino sequence (a “synonymous SNP”). (4) They may alter the splicing of the transcribed RNA. (5) They may alter gene expression by changing noncoding regulatory regions. (6) They may alter other DNA-encoded molecules (e.g., various forms of noncoding RNAs). There are many additional subtleties. For example, synonymous changes in coding sequence can affects splicing without altering the encoded amino acids. SNPs are widely used as genetic markers: genotyping SNPs can allow people to look at patterns of inheritance, test relatedness between individuals, pinpoint the location of phenotype-causing alleles, and explore human history. mIR, miRNA encoding DNA sequence; miRNA, microRNA.
Table 1.
Glossary of terms
| Types of Genetic Variation |
|---|
| 1. Single-nucleotide substitution: Describes any substitution of one nucleotide for another |
| Variant terminology |
| Rare: mutations |
| Common: polymorphisms or SNPs. A subset of SNPs changes restriction endonuclease sites. Such polymorphisms are referred to as RFLPs |
| Coding or noncoding |
| Coding sequence variants may alter amino acids or lead to frameshifts in translation or premature protein termination. Coding sequence variants may also leave the amino acid encoded intact or alter gene regulation in subtle ways |
| Noncoding variants may alter aspects of gene regulation and transcription (e.g., promoter activity, RNA splicing). Most noncoding variants have no identified functional effects but can be useful as genetic markers in mapping studies |
| Examples |
| (1) APOL1 G1 variant is an allele composed of two different SNPs that alter amino acids |
| (2) Many different disease-causing mutations in PKD1 alter the amino acid sequence by leading to an altered protein sequence |
| (3) Common noncoding SNPs in UMOD have been found to be associated with altered risk of CKD |
| 2. Insertions/deletions (indels): Refers to insertions or deletions of small stretches of DNA |
| Examples |
| (1) APOL1 G2 variant is a deletion of six nucleotides that removes two amino acids from the encoded APOL1 protein |
| (2) Repeat sequences (also referred to as microsatellites and minisatellites) are tracts of DNA where short stretches of DNA are repeated multiple times (from 2 to 50 or more). These are a form of variation that was very useful in earlier genetic mapping studies because of their use as genetic markers. Rarely such repeat sequences are in coding sequence. Alterations in a microsatellite in the MUC1 gene coding sequence cause a form of autosomal dominant tubulointerstitial kidney disease |
| 3. Structural variation: Refers to larger-scale changes or rearrangements in DNA, often visible by cytogenetic methods |
| Examples |
| A significant fraction of syndromic and nonsyndromic CAKUT is seen in association with structural variants |
SNP, single-nucleotide polymorphism; CAKUT, congenital anomaly of the kidney and urinary tract.
Complex versus Mendelian
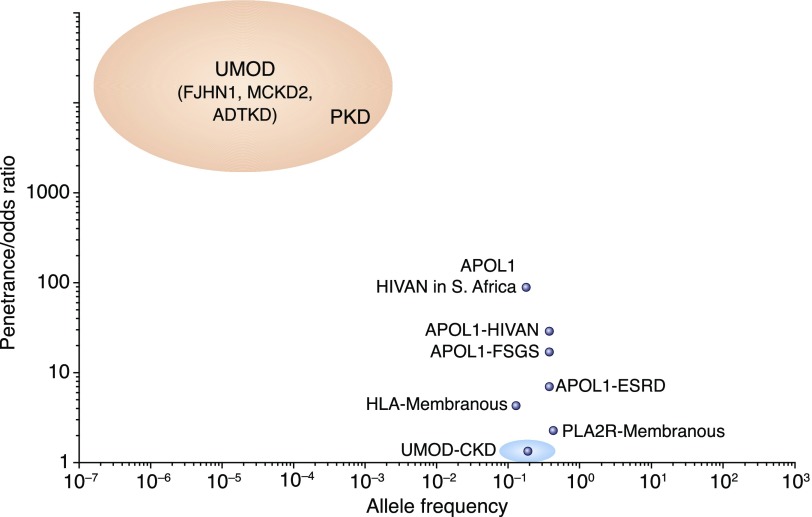
Genetically influenced diseases are often classified as either Mendelian or complex. Conceptually, Mendelian and common complex diseases fall at opposite ends of a spectrum. On one end of this spectrum, there are very rare but very highly penetrant genetic variants in a single gene, whereas on the other, there are very common variants at multiple loci in the genome with more subtle effects on phenotype (Figure 2). Evolutionary pressures can lead to variants and associated phenotypes that deviate from this pattern with atypical combinations of frequency and effect size.
Figure 2.
Different kidney disease relevant gene variants have a large range of allele frequencies and effect sizes. Genetic variants that cause or predispose to kidney disease can be characterized according to their frequency (x axis) and penetrance (y axis). Mendelian disease gene variants are very rare but very powerful (red ellipse). It is hard to precisely ascertain their frequency or effect size because detection typically depends on the variant being highly penetrant and the phenotype being unambiguous. The variants that cause susceptibility to common complex kidney phenotypes, such as low GFR (CKD), have small effect sizes, and therefore, they need to be common to be detected (blue ellipse). Some variants have unusual combinations of moderately strong effect size and high frequency (APOL1 and PLA2R). Most of these variants identified to date are immunity genes where the allele that promotes kidney dysfunction may also confer some benefit to the immune system. Some genes fall into more than one category. For example, common variants in UMOD are among the strongest common contributors in studies of CKD or eGFR, whereas rare variants can cause severe Mendelian phenotypes. HIVAN, HIV-associated nephropathy.
It is not always clear when we should call an observed phenotype a “disease” or not. Some differences between individuals reflect normal variation (genetically or nongenetically mediated). For example, the GFR declines with age. Should we label a 100-year-old man with a serum creatinine level of 1.1 and an eGFR of 55 as having “CKD?” Perhaps not, but quantifiable differences in things like rate of kidney function decline, differences in kidney size, etc., are still of interest to these seeking to understand the genetic underpinnings of kidney function and dysfunction (2).
Good versus Bad Alleles
It is common in the lay literature and even among physicians to refer to particular genes and their variants are “bad” or “good.” Things are generally not so simple. People homozygous (or compound heterozygous) for loss-of-function variants in the Na-Cl cotransporter (SLC12A3) develop Gitelman syndrome, which is characterized by hypokalemia and metabolic alkalosis, and the consequent metabolic sequelae. People heterozygous for such variants have, on average, higher bone mineral density and lower BPs (3,4). Similarly, although inheritance of two copies of two specific APOL1 alleles is associated with a greatly increased susceptibility to kidney disease, inheritance of one such allele seems to provide expanded protection against certain parasitic infections (5,6). In patients with congenital nephrotic syndrome or polycystic kidney disease (PKD), there is little harm in referring to loss-of-function variants in NPHS1 or PKD1 as “deleterious” or “mutations.” Still, it is probably best to use more neutral language (e.g., “variants”) in referring to most human genetic differences.
Phenocopies
Often, we do not know whether a particular phenotype is “one thing” etiologically or a mix of many things. The case of Gitelman syndrome provides a good example. Prior to the molecular genetic elucidation of this and the various subtypes of Bartter syndrome, the distinctions between these entities were somewhat fuzzy, and they were determined on the basis of phenotypic characteristics, such as age of onset and severity of sodium and potassium loss, and the level of urine calcium excretion. Now that the molecular etiologies of these multiple forms of recessive hypokalemic alkalosis are largely understood, we can lump and split patients more rationally, with a refined understanding of the phenotype and its molecular underpinnings.
This leads to a circular problem. Without knowing the molecular etiology of disease, we may not be lumping/splitting on the basis of etiologically meaningful differences. For example, many recent studies have found type 4 collagen genes to be frequently mutated in individuals carrying the diagnosis FSGS. Should we label someone with COL4A4-associated FSGS as “having FSGS?” Having Alport syndrome? Both? Neither? Defining a phenotype precisely can aid in genetic discovery; genetic discoveries may allow a more refined understanding of phenotype variability. The variability in nomenclature used to describe kidney disease does not help. Sometimes, we describe a patient’s disease by its clinical features (e.g., nephrotic syndrome); other times, we describe it by histology (e.g., minimal change disease). Sometimes, a name really does describe a single disease entity (for example, GLA mutations and Fabry disease), but other names (such as FSGS) are just useful descriptors of shared phenotypic features that will need replacement after etiologies are better defined.
Molecular Phenotypes
Although traditionally, we use the term phenotype to refer to the clinical characterization of some disease entity, recent advances in a variety of technologies have allowed for a greatly expanded meaning. Investigators can now make measurements in humans, human-derived cells, or biologic samples that can precisely define person-to-person differences in organelle function, the behavior and abundance of proteins, metabolites, or the level and pattern expression of genes. People are now determining not just the variation in the genome that controls clinical disease but also, more refined “endophenotypes,” such as how variation throughout the genome affects the level of expression of genes in the kidney (7,8). As the fast pace of progress in the development of human-derived models, such as induced pluripotent stem cell-derived cells, kidney organoids, and kidneys on chips, continues, we will expand our ability to define phenotypes, including responses to various perturbations, in ways that we cannot with living humans (9,10). New technologies allow exploration of gene expression at the level of the single cell, allowing even more precise phenotyping and ultimately, genotype-phenotype studies (11).
Common Genetic Factors in Kidney Disease
In some instances, genes in which rare, highly penetrant variants cause rare Mendelian diseases also harbor more common variation that contributes to common forms of kidney disease. Common variants in UMOD, encoding uromodulin, are associated with CKD and eGFR in recent genome-wide association studies (12). Rare mutations in this same gene cause a set of related phenotypes described variously as autosomal dominant tubulointerstitial kidney disease, medullary cystic kidney disease, interstitial nephritis, and familial juvenile hyperuricemic nephropathy (13). Interestingly, in the genome-wide association studies, the more common UMOD allele, the version we share with our distant ancestors is associated with higher risk of CKD, whereas the newer minor allele is protective.
People of recent African ancestry have a much greater risk of kidney disease of various sorts than do other groups. Much of this disparity is attributable to common variants in the APOL1 gene. The independent APOL1 variants known as G1 and G2 confer high risk of multiple types of kidney disease under a recessive model of risk inheritance (14). In contrast to UMOD, rare, more highly penetrant mutations in APOL1 have not been reported as a cause of human kidney disease.
Some diseases are common, not because any particular disease-associated variant is common but because some genes happen to harbor a large number of highly penetrant mutations. PKD is such an example. Each individual disease-causing PKD1 or PKD2 mutation is rare, but in aggregate, mutations in these genes are fairly common (approximately 1 in 1000) and cause some degree of disease in nearly everyone who inherits one of these mutations (15).
Effect Size Matters
Linkage analyses and genome-wide association studies can identify genetic variants that significantly and reproducibly associate with disease with a wide range of effect size. We often pay too much attention to the statistical significance in contrast to the magnitude of the effect on phenotype (14). If the alleles of some SNP are associated with a phenotype of interest with P values of genome-wide significance in multiple independent studies, we can be quite certain that the association is not artifactual. However, how large of an effect does harboring these SNPs (zero, one, or two copies) have on the phenotype? If having one copy of a minor SNP allele increases risk of disease by 1.01-fold, knowing the genotype of an individual is going to have no effect on clinical decision making. A genotype that confers a 10-fold increase in risk is more likely to indicate a potential therapeutic target and help with diagnosis and prognosis than a genotype that confers a 1.001-fold increase in risk. Of course, even if a gene contributes only trivially to an individual’s risk of disease, identifying and understanding such genetic variants may inform our understanding of disease pathways and identify new targets for treatment.
Increasingly, investigators are attempting to develop “polygenic risk scores” not on the basis of individual SNPs but on the basis of the composite collection of all relevant variants in an individual (16). It remains unclear if such scores will be important clinically or even any better than just knowing whether someone has a family history of disease. The hope is that comprehensive risk scores for common phenotypes (such as AKI and CKD) will be useful in weighing the risks/benefits of preventative strategies. Heritability is only one factor that determines disease development, and for most diseases, we currently only understand a small fraction of the heritability component.
More about Evolution
As noted above, APOL1 kidney disease risk variants have both beneficial and deleterious effects on human health. This is an example of balancing selection. In the case of APOL1, natural selective pressures have driven these variants to high frequency because of a beneficial effect on innate immunity, despite a deleterious effect on the kidney’s susceptibility to disease (5). Similarly, evidence suggests that the common CKD-associated UMOD allele (which, unlike the case of APOL1, does not alter coding sequence but rather, gene expression) is associated with higher urinary excretion of uromodulin but has a protective effect against urinary tract infection (17).
IgA nephropathy (IgAN), like FSGS, is a histopathologically defined form of glomerular damage that likely represents a collection of etiologically distinct entities. Genome-wide association studies over the past decade have identified approximately 20 loci associated with IgAN risk (18). Many of these genes are also associated with risk of inflammatory bowel disease and maintenance of the intestinal barrier to pathogens. The ethnic and geographical differences in rates of IgAN, including the high prevalence in Asia, suggest that environmental factors may place different evolutionary pressures on the frequencies of IgAN-influencing alleles in different locales.
It is interesting that many of the genes with the strongest evidence for association with kidney disease (APOL1, UMOD, PLA2R1, complement factor genes, and HLA) all seem to have important roles in defense against pathogens. This is also directly relevant to issues of modeling human kidney disease in model organisms, which are discussed more below. The environmental pressures important to human health, particularly infectious organism, are very different from those faced by rats, mice, and fish. This leads to species-specific differences in gene and gene variant behavior that depend on the complex genetic and nongenetic environments in which they are placed.
Kidney Disease?
Not all kidney diseases are diseases of the kidney. Mutations in complement factors, for example, can lead to hemolytic uremic syndrome, and mutations in the protease encoded by ADAMTS13 can cause thrombotic thrombocytopenic purpura (19). Conversely, systemic diseases, such as hypertension, may be driven by genes that primarily affect kidney function (20). Common variation in immune regulatory genes seems to affect the risk of various glomerular phenotypes (21). Diabetes, the most common cause of CKD, is the result in large part of combinations of variation in nonkidney genes; the verdict is still out on whether susceptibility to nephropathy in the setting of diabetes is driven primarily by genes directly affecting the kidney (22,23). Some people with lupus or diabetes get severe kidney disease. Others do not. Differences in the kidney’s susceptibility to systemic disease originating outside the kidney may reflect genetically determined differences in the kidney’s response to such diseases.
Modifier Genes
The number of Mendelian kidney diseases without known causal mutations continues to diminish due to the success of tools, such as next generation sequencing. The remaining unsolved diseases probably affect only an extremely small number of people. However, a new phase in the study of Mendelian kidney diseases may only be just beginning. Of special interest now are genetic variants that modify the phenotype caused by Mendelian mutations. For example, why does the age of onset and severity often vary so much between people (even within families) with the same mutation? One likely contributor is that there is other variation in the genome that can affect the severity of the phenotype caused by the primary mutation. This may be important: some Mendelian disease genes may not offer obvious therapeutic approaches because they are not “druggable targets.” However, one or more of the modifier genes (or their products) may be much better targets for small molecule therapy. Drugs for PKD, for example, may be very difficult to design, but if targeting a PKD-modifying gene can delay age of onset of GFR decline from 40 to 70, it may be just as effective as successfully targeting PKD directly. New DNA editing technologies (e.g., clustered regularly interspaced short palindromic repeats and RNA interference) may allow for therapeutic approaches that do not require a target to be easily modifiable by a traditional drug.
Proof: Going from Genotype-Phenotype Association to Causation
What constitutes proof that variations in a gene are truly causally related to disease? We can ask this from a research perspective, where we want to know (in the case of a Mendelian disease) if mutations in gene X are a cause of or a contributor to disease. We can also ask this in a clinical setting, where the question is a bit different. Is a mutation in gene X causing my patient’s disease?
In the early days of positional cloning, linkage analysis in large families (or many smaller families if they share a common genetic cause) led to the identification of a small region of the genome (usually a few million base pairs) that almost certainly contained the culprit variant. Analysis of genes in that region could then identify the most likely causal mutation. The finding of other independent mutations in other families together with an effect on the function of the gene product and perhaps, observation of a similar phenotype in a model organism engineered to have a homologous mutation along with absence of the observed or similar variants in unaffected individuals constituted compelling evidence of a causal relationship. It is important to be aware that not every gene reported as disease causing in the published literature turns out to in fact be correct after further examination.
The availability of next generation sequencing methodologies makes it relatively easy to identify the set of rare genetic variants present in an individual’s DNA. It is also, in theory, easier to evaluate the possible contribution of rare highly penetrant variants to disease, even if an accurate model of the mode of disease inheritance is lacking. Mapping of the location of a disease locus no longer typically precedes sequence analysis. We might look to see if variants meeting certain criteria are more common in patients than controls, if this difference is statistically significant (when properly adjusted for the number of hypotheses examined), and as above, if the observed changes in the gene affect the function of the gene product. Given the ease of sequencing an entire genome rather than simply a small region of the genome that we know must harbor a casual mutation, the number of candidate genes and mutations identified can be large. Nevertheless, if in repeated independent studies, variants in a gene (rare or common) are more frequent in people with than people without a phenotype, we can be reasonably confident of some sort of causal relationship. It is possible that a real and replicated relationship between a genetic variant and phenotype exists but that it is not the causal driver because associated variants may be in linkage disequilibrium with the variant(s) causally driving a relationship to phenotype.
The clinical situation is often different, especially when we are dealing with individuals rather than families (where we can track whether variants segregate with disease phenotypes). We now know, with certainty, that mutations in PKD1 cause autosomal dominant PKD. However, what if clinical genetic testing identifies a previously undescribed rare variant in PKD1 in a patient with some atypical cystic disease? A variety of software tools can assign the likelihood of being deleterious on the basis of a number of factors (e.g., evolutionary conservation and predicted effect of a specific amino acid change). Assigning a causal relationship in this individual patient is less clear. In research studies, we typically study individuals with a well defined phenotype to help us identify the genetic underpinnings of that phenotype. In the clinical setting, we often want genetics to help us care for patients when there is lack of diagnostic clarity from the observed phenotype alone: we want the genetics to help make the phenotype clearer.
Can we use model organisms to confirm causality? This is usually impractical in the clinical setting. However, even in the research setting, this is often complicated. Rats, mice, fish, and flies are all similar to humans at a genetic level, but the differences, even in the case of mammals like mice, can be large. Genes that have clearly been demonstrated to be responsible for highly penetrant kidney phenotypes in humans may produce no overt phenotype when homologous mutations are introduced into mice (for example, INF2) (24). Conversely, there are multiple genes that, when mutated in model organisms, alter kidney structure or function but have not been associated with human phenotypes.
In some cases, a model organism does not have a homologous gene. APOL1, as an example, is a gene found only in humans and a few primate species. Not even our closest relative, the chimpanzee, has a functional APOL1. Investigators have used yeast, flies, fish, and mice to model APOL1-associated disease, but these studies, although often highly informative about mechanism, do not recapitulate the activity of a human gene in a human context. Even in cases where a human gene has close homologs on other organisms, the development of engineered organisms to model and study disease is often difficult. Despite the fact that, for example, mutations in TRPC6 are a well established cause of autosomal dominant FSGS, translation into an animal model has been difficult. The human glomerulus (in particular, the podocyte) is a highly differentiated structure that must endure many physical stresses and toxic insults over many decades, with little apparent capacity for replacement. The stresses facing mouse glomeruli or drosophila nephrocytes are different, thus leading to different evolutionary pressures that have fine tuned their function.
Genetics in the Clinic
It is still not routine to perform genetic testing in people with kidney disease. Increasingly, we are seeing that unbiased assessment of variants in the genome leads to a clear genetic diagnosis in a nontrivial subset of people and in some, reclassification of the disease category (25). Genetic testing can be a nuisance: most institutions do not do this routinely, it can be costly (and not covered by insurance), and sorting out the appropriate laboratory for a particular indication may not be simple. In the not too distant future, whole-genome and/or whole-exome sequencing may replace disease-specific panels in the clinic. The choice of appropriate test can be tricky. On the one hand, we know from many recent studies that the genetic cause of disease may not have been on the original list of top suspects on the basis of clinical phenotype. On the other hand, sequencing the entire genome in a patient with, for example, cystic kidney disease may give us too much information that we do not know what to do with, akin to obtaining a whole-body magnetic resonance imaging scan routinely in the clinic. We will likely learn things that we may not want to know and do not know how to deal with. Although the American College of Medical Genetics has published guildelines on such incidental findings, this remains an unresolved and debated topic.
Unlike most diagnostic tests used in clinical care, an optimally done test of a person’s germline DNA does not change with time. Prednisone may reverse a patient’s nephrosis, but it will not change her NPHS2 genotype. This is a strong counterargument to the notion that genetic tests are too expensive: these tests do not typically need to be repeated unlike, for example, serum creatinine levels or kidney ultrasound studies.
People often ask, “will doing this test change my care for this patient?” Until genetic testing becomes more commonplace in nephrology care, we will not know the answer. Although we do not advocate routine whole-genome sequencing, we do think that well chosen, well executed genetic tests with high prior probabilities of clarifying patient diagnoses should be a much more common part of clinical care than they are at present. Commercial laboratories will of course be more than happy to do any genetic test requested in exchange for payment. However, it makes no sense, for example, to use one of the available “nephrotic syndrome” gene panels in a black patient with FSGS unless it includes APOL1 (and in fact, APOL1 is absent from several of these panels). The prior probability that such a patient has a high-risk APOL1 genotype is about 75%. The probability that causal alterations will be found in any of the Mendelian FSGS and nephrotic syndrome genes is minuscule in comparison.
Future Directions
People will continue to perform genome-wide association studies and sequencing studies, and they will continue to find genes and alleles that contribute to kidney disease. It is difficult to believe that there are many more genes to identify responsible for highly penetrant and moderately common Mendelian diseased, such as PKD, or common variants with large effect size, such as with APOL1. Still, the spectrum of rare variants that contribute to kidney diseases has not been fully defined. It remains unclear why widely expressed genes (such as ACTN4) can be responsible for such kidney-limited phenotypes. Understanding how genetic variation affects phenotype at the level of the individual cell and why these phenotypes are often cell-type specific is an important challenge. New technologies (e.g., single-cell RNA sequencing) enable looking at genetic questions at the level of individual cells. For genes and their variants that contribute to disease, we need to understand all of the factors (genetic, epigenetic, and environmental) that explain why people with the same variant may have differences in phenotype. Many of the modern genomics tools that can be applied to these problems are discussed in subsequent reviews in this series.
Other “Omes”
The term “genome” is thought to have been invented as a portmanteau of gene and chromosome (26). We have now come to speak of other “omes”: the metabolome, the proteome, the lipidome, the glycome, the phenome, and so on. Still, the genome will likely remain the most fundamental “ome” in understanding human biology. An organism’s DNA is essentially digital, making it unique among big biologic datasets. At the level of the individual organism, the genome is the most proximal component of the complex path from information to biology. Ultimately, all of the omes are controlled by the genome. Although other omes may come and go, it is hard to imagine a future in which the science of DNA and its variation are not a central part of biology and its applications to medicine.
Disclosures
Dr. Pollak and Dr. Friedman are coinventors of patents filed by Beth Israel Deaconess Medical Center related to APOL1 diagnostics and therapeutics. They both report receiving grant funding and consultant fees from Vertex and owning equity in Apolo1Bio.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Naidoo N, Pawitan Y, Soong R, Cooper DN, Ku C-S: Human genetics and genomics a decade after the release of the draft sequence of the human genome. Hum. Genomics 5: 577–622, 2011. 10.1186/1479-7364-5-6-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, Tin A, Wang L, Chu AY, Hoppmann A, Kirsten H, Giri A, Chai JF, Sveinbjornsson G, Tayo BO, Nutile T, Fuchsberger C, Marten J, Cocca M, Ghasemi S, Xu Y, Horn K, Noce D, van der Most PJ, Sedaghat S, Yu Z, Akiyama M, Afaq S, Ahluwalia TS, Almgren P, Amin N, Ärnlöv J, Bakker SJL, Bansal N, Baptista D, Bergmann S, Biggs ML, Biino G, Boehnke M, Boerwinkle E, Boissel M, Bottinger EP, Boutin TS, Brenner H, Brumat M, Burkhardt R, Butterworth AS, Campana E, Campbell A, Campbell H, Canouil M, Carroll RJ, Catamo E, Chambers JC, Chee ML, Chee ML, Chen X, Cheng CY, Cheng Y, Christensen K, Cifkova R, Ciullo M, Concas MP, Cook JP, Coresh J, Corre T, Sala CF, Cusi D, Danesh J, Daw EW, de Borst MH, De Grandi A, de Mutsert R, de Vries APJ, Degenhardt F, Delgado G, Demirkan A, Di Angelantonio E, Dittrich K, Divers J, Dorajoo R, Eckardt KU, Ehret G, Elliott P, Endlich K, Evans MK, Felix JF, Foo VHX, Franco OH, Franke A, Freedman BI, Freitag-Wolf S, Friedlander Y, Froguel P, Gansevoort RT, Gao H, Gasparini P, Gaziano JM, Giedraitis V, Gieger C, Girotto G, Giulianini F, Gögele M, Gordon SD, Gudbjartsson DF, Gudnason V, Haller T, Hamet P, Harris TB, Hartman CA, Hayward C, Hellwege JN, Heng CK, Hicks AA, Hofer E, Huang W, Hutri-Kähönen N, Hwang SJ, Ikram MA, Indridason OS, Ingelsson E, Ising M, Jaddoe VWV, Jakobsdottir J, Jonas JB, Joshi PK, Josyula NS, Jung B, Kähönen M, Kamatani Y, Kammerer CM, Kanai M, Kastarinen M, Kerr SM, Khor CC, Kiess W, Kleber ME, Koenig W, Kooner JS, Körner A, Kovacs P, Kraja AT, Krajcoviechova A, Kramer H, Krämer BK, Kronenberg F, Kubo M, Kühnel B, Kuokkanen M, Kuusisto J, La Bianca M, Laakso M, Lange LA, Langefeld CD, Lee JJ, Lehne B, Lehtimäki T, Lieb W, Lim SC, Lind L, Lindgren CM, Liu J, Liu J, Loeffler M, Loos RJF, Lucae S, Lukas MA, Lyytikäinen LP, Mägi R, Magnusson PKE, Mahajan A, Martin NG, Martins J, März W, Mascalzoni D, Matsuda K, Meisinger C, Meitinger T, Melander O, Metspalu A, Mikaelsdottir EK, Milaneschi Y, Miliku K, Mishra PP, Mohlke KL, Mononen N, Montgomery GW, Mook-Kanamori DO, Mychaleckyj JC, Nadkarni GN, Nalls MA, Nauck M, Nikus K, Ning B, Nolte IM, Noordam R, O’Connell J, O’Donoghue ML, Olafsson I, Oldehinkel AJ, Orho-Melander M, Ouwehand WH, Padmanabhan S, Palmer ND, Palsson R, Penninx BWJH, Perls T, Perola M, Pirastu M, Pirastu N, Pistis G, Podgornaia AI, Polasek O, Ponte B, Porteous DJ, Poulain T, Pramstaller PP, Preuss MH, Prins BP, Province MA, Rabelink TJ, Raffield LM, Raitakari OT, Reilly DF, Rettig R, Rheinberger M, Rice KM, Ridker PM, Rivadeneira F, Rizzi F, Roberts DJ, Robino A, Rossing P, Rudan I, Rueedi R, Ruggiero D, Ryan KA, Saba Y, Sabanayagam C, Salomaa V, Salvi E, Saum KU, Schmidt H, Schmidt R, Schöttker B, Schulz CA, Schupf N, Shaffer CM, Shi Y, Smith AV, Smith BH, Soranzo N, Spracklen CN, Strauch K, Stringham HM, Stumvoll M, Svensson PO, Szymczak S, Tai ES, Tajuddin SM, Tan NYQ, Taylor KD, Teren A, Tham YC, Thiery J, Thio CHL, Thomsen H, Thorleifsson G, Toniolo D, Tönjes A, Tremblay J, Tzoulaki I, Uitterlinden AG, Vaccargiu S, van Dam RM, van der Harst P, van Duijn CM, Velez Edward DR, Verweij N, Vogelezang S, Völker U, Vollenweider P, Waeber G, Waldenberger M, Wallentin L, Wang YX, Wang C, Waterworth DM, Bin Wei W, White H, Whitfield JB; Lifelines Cohort Study; V. A. Million Veteran Program : A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 51: 957–972, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon DB, Karet FE, Hamdan JM, DiPietro A, Sanjad SA, Lifton RP: Bartter’s syndrome, hypokalaemic alkalosis with hypercalciuria, is caused by mutations in the Na-K-2Cl cotransporter NKCC2. Nat Genet 13: 183–188, 1996. [DOI] [PubMed] [Google Scholar]
- 4.Hsu YJ, Yang SS, Cheng CJ, Liu ST, Huang SM, Chau T, Chu P, Salter DM, Lee HS, Lin SH: Thiazide-sensitive Na+ -Cl- cotransporter (NCC) gene inactivation results in increased duodenal Ca2+ absorption, enhanced osteoblast differentiation and elevated bone mineral density. J Bone Miner Res 30: 116–127, 2015. [DOI] [PubMed] [Google Scholar]
- 5.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K: Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, Suszták K: Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360: 758–763, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillies CE, Putler R, Menon R, Otto E, Yasutake K, Nair V, Hoover P, Lieb D, Li S, Eddy S, Fermin D, McNulty MT, Hacohen N, Kiryluk K, Kretzler M, Wen X, Sampson MG; Nephrotic Syndrome Study Network (NEPTUNE) : An eQTL landscape of kidney tissue in human nephrotic syndrome. Am J Hum Genet 103: 232–244, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nieskens TTG, Sjögren AK: Emerging in vitro systems to screen and predict drug-induced kidney toxicity. Semin Nephrol 39: 215–226, 2019. [DOI] [PubMed] [Google Scholar]
- 10.Little MH, Hale LJ, Howden SE, Kumar SV: Generating kidney from stem cells. Annu Rev Physiol 81: 335–357, 2019. [DOI] [PubMed] [Google Scholar]
- 11.Wilson PC, Humphreys BD: Kidney and organoid single-cell transcriptomics: The end of the beginning [published online ahead of print January 4, 2019]. Pediatr Nephrol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, Smith AV, Arking DE, Astor BC, Boerwinkle E, Ehret GB, Ruczinski I, Scharpf RB, Chen YD, de Boer IH, Haritunians T, Lumley T, Sarnak M, Siscovick D, Benjamin EJ, Levy D, Upadhyay A, Aulchenko YS, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, Chasman DI, Paré G, Ridker PM, Kao WH, Witteman JC, Coresh J, Shlipak MG, Fox CS: Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 41: 712–717, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bleyer AJ, Hart PS, Kmoch S: Autosomal dominant tubulointerstitial kidney disease, UMOD-related In: GeneReviews, edited by Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, Seattle, WA, University of Washington, 1993. Available at: https://www.ncbi.nlm.nih.gov/books/NBK1356/. Accessed August 10, 2019 [PubMed] [Google Scholar]
- 14.Friedman DJ, Pollak MR: Apolipoprotein L1 and kidney disease in African Americans. Trends Endocrinol Metab 27: 204–215, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergmann C, Guay-Woodford LM, Harris PC, Horie S, Peters DJM, Torres VE: Polycystic kidney disease. Nat Rev Dis Primers 4: 50, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torkamani A, Wineinger NE, Topol EJ: The personal and clinical utility of polygenic risk scores. Nat Rev Genet 19: 581–590, 2018. [DOI] [PubMed] [Google Scholar]
- 17.Ghirotto S, Tassi F, Barbujani G, Pattini L, Hayward C, Vollenweider P, Bochud M, Rampoldi L, Devuyst O: The uromodulin gene locus shows evidence of pathogen adaptation through human evolution. J Am Soc Nephrol 27: 2983–2996, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neugut YD, Kiryluk K: Genetic determinants of IgA nephropathy: Western perspective. Semin Nephrol 38: 443–454, 2018. [DOI] [PubMed] [Google Scholar]
- 19.Kavanagh D, Goodship T: Genetics and complement in atypical HUS. Pediatr Nephrol 25: 2431–2442, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toka HR, Koshy JM, Hariri A: The molecular basis of blood pressure variation. Pediatr Nephrol 28: 387–399, 2013. [DOI] [PubMed] [Google Scholar]
- 21.Debiec H, Dossier C, Letouzé E, Gillies CE, Vivarelli M, Putler RK, Ars E, Jacqz-Aigrain E, Elie V, Colucci M, Debette S, Amouyel P, Elalaoui SC, Sefiani A, Dubois V, Simon T, Kretzler M, Ballarin J, Emma F, Sampson MG, Deschênes G, Ronco P: Transethnic, genome-wide analysis reveals immune-related risk alleles and phenotypic correlates in pediatric steroid-sensitive nephrotic syndrome. J Am Soc Nephrol 29: 2000–2013, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad RB, Groop L: Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 6: 87–123, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robertson CC, Rich SS: Genetics of type 1 diabetes. Curr Opin Genet Dev 50: 7–16, 2018. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian B, Sun H, Yan P, Charoonratana VT, Higgs HN, Wang F, Lai KV, Valenzuela DM, Brown EJ, Schlöndorff JS, Pollak MR: Mice with mutant Inf2 show impaired podocyte and slit diaphragm integrity in response to protamine-induced kidney injury. Kidney Int 90: 363–372, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groopman EE, Marasa M, Cameron-Christie S, Petrovski S, Aggarwal VS, Milo-Rasouly H, Li Y, Zhang J, Nestor J, Krithivasan P, Lam WY, Mitrotti A, Piva S, Kil BH, Chatterjee D, Reingold R, Bradbury D, DiVecchia M, Snyder H, Mu X, Mehl K, Balderes O, Fasel DA, Weng C, Radhakrishnan J, Canetta P, Appel GB, Bomback AS, Ahn W, Uy NS, Alam S, Cohen DJ, Crew RJ, Dube GK, Rao MK, Kamalakaran S, Copeland B, Ren Z, Bridgers J, Malone CD, Mebane CM, Dagaonkar N, Fellström BC, Haefliger C, Mohan S, Sanna-Cherchi S, Kiryluk K, Fleckner J, March R, Platt A, Goldstein DB, Gharavi AG: Diagnostic utility of exome sequencing for kidney disease. N Engl J Med 380: 142–151, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler H: Verbreitung und Ursache der Parthenogenesis im Pflanzen- und Tierreiche, Jena, Germany, G. Fischer, 1920