Abstract
Circular RNAs have been reported to play a vital role in the development and progression of various types of cancer. However, the underlying molecular role of circular RNA CTDP1 (circCTDP1) in the tumorigenesis of nasopharyngeal carcinoma (NPC) remains unknown. In the present study, circCTDP1 expression was found to be markedly upregulated in NPC tissues and cell lines (SUNE1, SUNE2 and 6-10B cell lines). Knockdown of circCTDP1 resulted in inhibition of proliferation, migration and invasion, and promoted apoptosis of NPC cells. Moreover, circCTDP1 directly interacted with microRNA (miR)-320b based on bioinformatics prediction and dual luciferase assay, and transfection with an miR-320b inhibitor reversed the effects of circCTDP1 knockdown on NPC cells. Furthermore, circCTDP1/miR-320b promoted NPC progression by regulating the expression of homeobox A10 (HOXA10). In addition, it was demonstrated that HOXA10 may exert its oncogenic role in NPC by regulating the expression of transforming growth factor β2 (TGFβ2). Taken together, these results revealed a novel regulatory mechanism, which may provide an improved understanding of NPC tumorigenesis and be useful in the development of potential targets for NPC therapy.
Keywords: nasopharyngeal carcinoma, circular RNA CTDP1, competing endogenous RNA, microRNA-320b, homeobox A10, transforming growth factor β2
Introduction
Nasopharyngeal carcinoma (NPC) is a malignancy that occurs in nasopharyngeal epithelial tissues and has a specific ethnic and geographic distribution, with the highest incidence in Southeast Asia and North Africa (1). Despite great improvements in diagnosis and surgical treatment, the 5-year survival rate for advanced NPC is approximately 70%, which remains unsatisfactory (2,3). Currently, the main therapeutic methods for NPC are irradiation therapy, alone or combined with chemotherapy (4,5). However, the physical and psychological side effects of the treatments are severe. Therefore, an investigation of the molecular mechanisms of this disease is required to develop novel treatment strategies for patients with NPC.
Circular RNAs (circRNAs) are a class of non-coding transcripts that contain a ring structure (6). Increasing evidence indicates that the aberrant expression of circRNAs is associated with a variety of diseases, including cancer (7-9). Additionally, some circRNAs play vital roles in the development and progression of NPC. For example, circRNA ZNF609 (10), circRNA_0008450 (11) and circRNA_000543 (12) accelerate NPC tumorigenesis and metastasis. Circular RNA CTDP1 (circCTDP1) was reported to function as a competitive endogenous RNA (ceRNA) for microRNA (miRNA/miR)-29a-3p to regulate the expression of hyaluronan synthase 3, integrin subunit β1, DNA methyltransferase 3β and vascular endothelial growth factor A (VEGFA), and subsequently promote the growth and metastasis of bladder cancer (13). However, the molecular mechanisms of circCTDP1 action in the tumorigenesis of NPC are unclear.
miRNAs are a family of small non-coding RNAs of approximately 22 nucleotides in length, which regulate gene expression by complementary binding or complex mechanisms (14). In multiple studies, several miRNAs, such as miR-101 (15), miR-184 (16), miR-543 (17) and miR-449 (18), have been demonstrated to suppress proliferation, migration and invasion of NPC. miR-320b was also reported to act as a tumor suppressor during occurrence and progression in different types of cancer, including NPC. In a study by Li et al (19), the overexpression of miR-320b inhibited NPC cell proliferation and promoted apoptosis, while knockdown of miR-320b accelerated tumor growth and inhibited apoptosis. circRNAs can act as ceRNAs to regulate the development and progression of various cancers (20-22), which led to the hypothesis that circCTDP1 may promote NPC progression via miR-320b.
Homeobox A10 (HOXA10), a member of the homeobox gene family, plays a critical role in embryonic development (23). Abnormal expression of HOXA10 has been observed in several types of cancer, including endometrial carcinoma, ovarian cancer and breast cancer (24-26). Shen et al (27) also reported that HOXA10 is upregulated in NPC tissues compared to normal tissues, and promotes NPC progression by binding to the promoter of Zic family member 2. However, the precise mechanism of the regulation of HOXA10 in NPC remains unclear.
In the present study, the molecular mechanism of the circCTDP1/miR-320b/HOXA10/transforming growth factor β2 (TGFβ2) axis was investigated with regard to tumorigenesis and the progression of NPC. The aim of the study was to provide a better understanding of NPC initiation and progression, which may help in the future development of diagnostic and therapeutic targets for NPC.
Materials and methods
Clinical specimens
A total of 32 paired NPC tissues and paracarcinoma tissues were obtained from 32 NPC patients (22 males and 10 females) with a median age of 47 years (range, 27-79 years) between August, 2016 and May, 2018. The patients all provided written informed consent for the use of their samples. All experimental protocols were approved by the Ethical and Scientific Committee of the Third Affiliated Hospital of Soochow University (Jiangsu, China).
Cell culture
A normal human nasopharyngeal epithelial cell line (NP69), 3 NPC cell lines (SUNE1, SUNE2 and 6-10B) and 293T cells were obtained from the American Type Culture Collection (ATCC; Manassas). The cell lines were cultured in RPMI-1640 medium, and supplemented with 10% fetal bovine serum. The 293T cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS and 1% penicillin-streptomycin. All the cell lines were maintained at 37°C in a humidified atmosphere with 5% CO2.
Cell transfection
The short hairpin RNA (shRNAs) targeting circCTDP1 (shcircCTDP1; 5′-UCA AGA AUG CAG GCU CAA C-3′) with negative control (shNC; 5′-UCUCCG AUGCAG GCU CAA C-3′), miR-320b mimics (5′-AAA GCU GGG UUG AGA GGG CAA-3′) with negative control (miR-NC; 5′-AAU UCU CCG AAC GUG UCA CUU-3′) and miR-320b inhibitor (5′-UUG CCC UCU CAA CCC AGC UUU U-3′) with negative control (inh-miR-NC; 5′-CAG UAC UUU UGU GUA GUA CAA-3′) were synthesized by GenePharma (Shanghai, China). The full length of HOXA10 was subcloned into pcDNA3.1 to overexpress HOXA10 levels with empty pcDNA3.1 serving as control. The pcDNA3.1 vector was bought from GenePharma (Shanghai). Transfection of the cells with shcircCTDP1 (10 nM) or shNC (10 nM) and the miR-320b mimics (10 nM) or miR-NC (10 nM) and miR-320b inhibitor (10 nM) or inh-miR-NC (10 nM) was conducted with Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The efficiency of transfection was determined in each experiment using RT-qPCR 24 h post-transfection. All functional experiments were carried out 48 h post-transfection.
RT-qPCR
According to the manufacturer's instruction, total RNA was extracted from tissues and cell lines using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was reverse transcribed to cDNA by using a Reverse Transcription Kit (Takara). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 3 min; 40 cycles of 95°C for 5 sec and 60°C for 30 sec. A melt curve step from 65-95°C was performed in increments of 0.5°C per 5 sec. The relative expression levels were calculated by comparing to the expression of GAPDH or U6 using the 2-ΔΔCq method (28). The primer sequences used for amplification were: circCTDP1 forward, 5′-TAA GAA CGG GAA GCA GCA GG3′ and reverse, 5′-TCC AAG TCC ACC ATG AGC AC3′; miR-320b forward, 5′-TCC GAA ACG GGA GAG TTG G-3′ and reverse, 5′-GTG CAG GGT CCG AGG T-3′; HOXA10 forward, 5′-GGG TAA GCG GAA TAA ACT-3′ and reverse, 5′-GCA CAG CAG CAA TAC AAT A-3′; GAPDH forward, 5′-TGC ACC ACC AAC TGC TTA GC-3′ and reverse, 5′-GGC ATG CAC TGT GGT CAT GAG-3′; and U6 forward, 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′ and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′.
Wound healing assay
The migration ability of cells was evaluated by wound healing assay. Transfected SUNE2 and 6-10B cells were cultured in RPMI-1640 supplemented with 10% FBS at a density of 8×104 cells/ml in a humidified atmosphere of 5% CO2 at 37°C and grown to a fully confluent monolayer. After 6 h, culture medium was replaced with serum-free medium, and a sterile tip was employed to generate single-line scratch and then washed twice with phosphate-buffered saline (PBS) to remove detached cells from the plates. After 24 h, the medium was replaced with PBS, and the wound gap was observed. Images of cell migration were captured with an inverted microscope (magnification, ×200; Olympus Corporation). The gap distance of each monolayer was quantitatively evaluated using ImageJ.
Cell invasion assay
Cell invasion was determined by Transwell chambers (8 µm pore size; Millipore) precoated with 100 µl of Matrigel (BD Biosciences). Transfected SUNE2 and 6-10B cells (8×104 cells) were added to the upper chamber containing 150 µl RPMI-1640 without FBS. Extra 550 µl RPMI-1640 medium was added to the lower chamber. After 24 h of incubation at 37°C with 5% CO2, the cells that did not pass through the membrane were cleared using cotton swabs and 4% paraformaldehyde was added to fix the cells at room temperature for 20 min, followed by staining the cells with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for 20 min at room temperature. The number of cells invading through the Matrigel was counted in 3 randomly selected visual fields from the central and peripheral portion of the filter using an inverted microscope (magnification, ×200; Olympus Corporation).
Flow cytometric analysis
Flow cytometric analysis was performed using a FITC Annexin V Apoptosis kit (BD Biosciences) according to the manufacturer's instructions. Briefly, transfected SUNE2 and 6-10B cells were washed twice with PBS and resuspended in 1X Annexin V binding buffer containing 10 mM HEPES/NaOH (pH 7.4) (1×106 cells/ml). Then the cells were mixed with FITC-Annexin V (5 µl) and propidium iodide for at 37°C for 20 min, and analyzed using a flow cytometer (BD Biosciences).
Bioinformatic prediction and luciferase reporter assay
StarBase (http://starbase.sysu.edu.cn) and TargetScan databases (http://www.targetscan.org) were used to predict the potential miRNAs that can bind to circCTDP1. A luciferase reporter assay was employed to investigate the regulatory relationship between circCDTP1 and miR-320b. circCTDP1 that contained the miR-320b binding site was cloned into the psiCHECK-2 vector (Promega Corporation) to construct wild-type circCTDP1. Mutant circCTDP1 that included a mutated version of the miR-320b binding site was also cloned into psiCHECK-2. Subsequently, miR-320b mimic or miR-NC and miR-320b inhibitor or miR-NC inhibitor were transfected into 293T cells that were transfected with wild-type circCTDP1 or mutant circCTDP1. The relationship between miR-320b and HOXA10 was confirmed using the same method. Luciferase activity was evaluated by Dual-Luciferase Reporter Analysis system (Promega Corporation). Firefly lucif-erase activity was normalized to Renilla (Promega Corporation) luciferase gene activity.
Western blot analysis
Following transfection for 48 h, proteins were extracted from transfected SUNE2 and 6-10B cells using radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology). Protein concentration was measured with the bicinchoninic acid assay (Beyotime Institute of Biotechnology). Following denaturation, 10 µg protein/lane was separated by 10% SDS-PAGE. Proteins were transferred onto polyvinylidene difluoride (PVDF) membranes and blocked in 5% non-fat milk for 2 h at room temperature. The membranes were incubated with primary antibodies against HOXA10 (1:1,000; mouse monoclonal antibody, sc-271139; Santa Cruz Biotechnology) and GAPDH (1:1,000; mouse monoclonal antibody, sc-47724; Santa Cruz Biotechnology) overnight at 4°C. Following primary incubation, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:1,000; goat anti-mouse IgG, ab205719 and goat anti-rabbit IgG, ab205718; Abcam) for 2 h at room temperature. Protein bands were visualized using the Pierce ECL Western Blotting kit (Pierce; Thermo Fisher Scientific, Inc.). Protein expression was quantified using Image-Pro® Plus software (version 6.0; Media Cybernetics, Inc.). GAPDH was used as endogenous control for data normalization.
Xenograft experiment
Six male BALB/c nude mice (6 weeks old) were maintained under specific pathogen-free conditions and randomly divided into two groups, and were housed individually in microisolator ventilated cages (temperature, 26-28°C; 40-60% humidity and ventilation 10-15 times/h) with free access to water and food. Cells transfected with shNC or shcircCTDP1 were suspended in 100 µl PBS and injected subcutaneously into the mice. Tumors were examined every 5 days. On 30th day, the mice were sacrificed by cervical dislocation after deep anesthesia with 2% isoflurane (Baxter Healthcare Corporation) to obtain the tumors. The tumors were photographed and tumor weights were measured. Tumor length (L) and width (W) were measured. Tumor volume was calculated using the formula: V = 1/2 × L × W2. The animal experiments were approved by the Ethics Committee of the Third Affiliated Hospital of Soochow University.
Statistical analysis
Statistical analyses were performed with SPSS 18.0 software (SPSS, Inc.). Data were presented as mean ± SD. Comparisons of parameters between two groups were analyzed by a paired Student's t-test. Comparisons among multiple groups were performed using one-way ANOVA followed by Tukey's test. The correlation between gene expression levels was analyzed by Pearson's correlation coefficient. Kaplan-Meier analysis and the log-rank test were used to estimate survival curves. Cut-off values were determined using Youden's index. P<0.05 was considered to indicate a statistically significant difference.
Results
CircCTDP1 is upregulated in NPC tissues and cell lines
Using RT-qPCR, the expression levels of circCTDP1 in clinical tissues (NPC specimens and paracarcinoma tissues) was examined. As presented in Fig. 1A, circCTDP1 was significantly upregulated in NPC tissues compared with normal tissues. Similarly, circCTDP1 expression in 3 NPC cell lines (SUNE1, SUNE2 and 6-10B) was markedly upregulated compared with expression in NP69 cells, a normal nasopharyngeal epithelial cell line (Fig. 1B). Moreover, Kaplan-Meier analysis showed that patients with high circCTDP1 expression exhibited a worse prognosis compared with patients with low circCTDP1 expression (Fig. 1C). These results suggest circCTDP1 may play a pivotal role in the progression of NPC.
Figure 1.
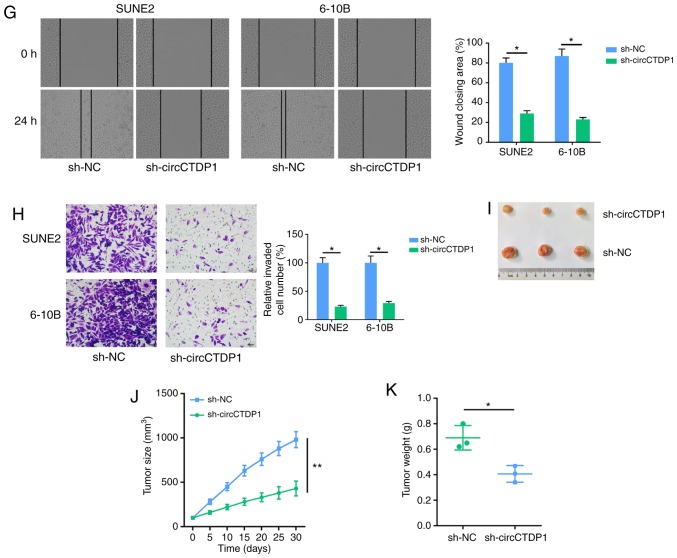
Knockdown of circCTDP1 inhibits the progression of NPC cells. (A) RT-qPCR analysis shows the relative circCTDP1 expression in NPC tissues and para-carcinoma tissues, n=32. (B) RT-qPCR analysis shows relative circCTDP1 expression in normal nasopharyngeal epithelial cell line (NP69) and NPC cell lines (SUN1, SUNE2 and 6-10B). (C) Kaplan-Meier survival analysis shows correlation between circCTDP1 expression and prognosis of NPC patients. (D) RT-qPCR analysis shows the relative circCTDP1 expression of SUNE2 and 6-10B cell lines transfected with shNC and shcircCTDP1. (E) MTT assay shows the cell growth rate of SUNE2 and 6-10B cell lines transfected with shNC and shcircCTDP1 at different time points of 0, 24, 48, and 72 h. (F) Flow cytometry assay shows the cell apoptosis of SUNE2 and 6-10B cell lines transfected with shNC and shcircCTDP1. The data were presented as mean ± SD (*P<0.05; **P<0.01). Knockdown of circCTDP1 inhibits the progression of NPC cells. (G) Wound healing assay of SUNE2 and 6-10B cell lines transfected with shNC and shcircCTDP1. (H) Cell invasion assay of SUNE2 and 6-10B cell lines transfected with shNC and shcircCTDP1. (I) Xenograft tumor assay shows tumor growth of cells transfected with shNC and shcircCTDP1. (J) Tumor volume throughout the experiment. (K) Statistical analysis of tumor weights. The data were presented as mean ± SD (*P<0.05; **P<0.01).
Knockdown of circCTDP1 suppresses NPC progression
To investigate the involvement of circCTDP1 in NPC, shRNAs targeting circCTDP1 were designed. RT-qPCR indicated that the expression of circCTDP1 in SUNE2 and 6-10B cells was decreased after cells transfected with shcircCTDP1 (Fig. 1D). Subsequently, an MTT assay revealed that cell viability was markedly reduced when NPC cells were transfected with shcircCTDP1 (Fig. 1E). Moreover, flow cytometric analysis revealed that circCTDP1 knockdown significantly promoted apoptosis in SUNE2 and 6-10B cell lines (Fig. 1F). To further explore the effect of circCTDP1 on the migration and invasion of NPC cells, wound healing and cell invasion assays were performed. As presented in Fig. 1G and H, the migration and invasion abilities of the shcircCTDP1 cells were reduced relative to shNC cells. Moreover, an in vivo xenograft experiment revealed that depletion of circCTDP1 reduced the tumor growth rate in mice (Fig. 1I-K). A statistically significant difference in tumor volume was observed between the two groups and the maximum tumor volume was 1,153 mm3. These data indicate that circCTDP1 knockdown suppresses proliferation, mobility and promotes apoptosis of NPC cells.
miR-320b inhibitor rescues the effects of circCTDP1 knock- down on NPC cells
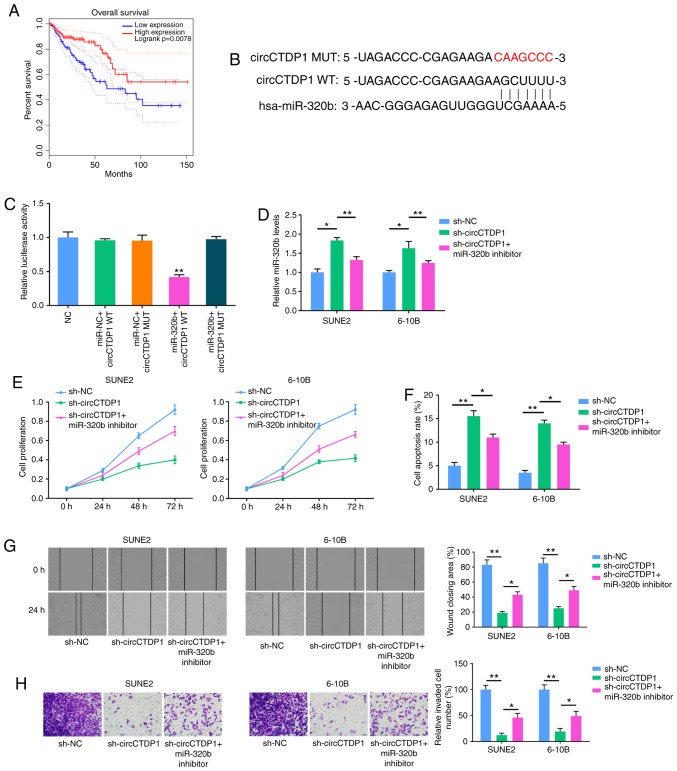
Kaplan-Meier survival analysis showed that NPC patients with low miR-320b expression had a shorter OS time compared with those patients with high miR-320b expression (Fig. 2A). Using starBase bioinformatic analysis software, circCTDP1 was suggested to bind to miR-320b via complementary base pairing (Fig. 2B). A dual luciferase reporter assay showed that a marked decrease in the luciferase activities following co-transfection of the cells with miR-320b mimics and wild-type circCTDP1 expression vector, but not with the mutant-type circCTDP1 vector (Fig. 2C). To investigate whether the effects of circCTDP1 on NPC were mediated by miR-320b, the miR-320b inhibitor was transfected into shcircCTDP1-expressing SUNE2 and 6-10B cells. RT-qPCR analysis demonstrated that the introduction of miR-320b inhibitor reduced miR-320b expression in NPC cells transfected with shcircCTDP1 (Fig. 2D). The proliferation of shcircCTDP1-transfected SUNE2 and 6-10B cells was increased when transfected with miR-320b inhibitor, as revealed by MTT assay (Fig. 2E). Furthermore, flow cytometry showed that knockdown of circCTDP1 promoted apoptosis, whereas the cell apoptotic rate was reduced by co-transfection with miR-320b inhibitor (Fig. 2F). Similarly, transfection with miR-320b inhibitor partly reversed the shcircCTDP1-mediated reduction of the migration and invasion abilities of NPC cells (Fig. 2G and H). Therefore, it was concluded that miR-320b antagonizes circCTDP1-regulated NPC progression in vitro.
Figure 2.
MiR-320b inhibitor restores the attenuated progression of shcircCTDP1-transfected NPC cell lines. (A) Kaplan-Meier survival analysis shows correlation between miR-320b expression and prognosis of NPC patients. (B) Bioinformatic prediction of binding site of miR-320b by circCTDP1. (C) Dual luciferase reporter assay shows fluorescence intensity in 293T cells transfected with NC, miR-NC+circCTDP1 WT, miR-NC+circCTDP1, MUT miR-320b+circCTDP1 WT and miR-320b+circCTDP1 MUT. (D) RT-qPCR analysis shows the relative miR-320b expression of SUNE2 and 6-10B cell lines transfected with shNC, shcircCTDP1 and shcircCTDP1 plus miR-320b inhibitor. (E) MTT assay shows cell growth rate of SUNE2 and 6-10B cell lines transfected with shNC, shcircCTDP1 and shcircCTDP1 plus miR-320b inhibitor at different time points of 0, 24, 48, and 72 h. (F) Flow cytometry assay shows the relative cell apoptosis rate of SUNE2 and 6-10B cell lines transfected with shNC, shcircCTDP1 and shcircCTDP1 plus miR-320b inhibitor. (G) Wound healing assay of SUNE2 and 6-10B cell lines transfected with shNC, shcircCTDP1 and shcircCTDP1 plus miR-320b inhibitor. (H) Cell invasion assay of SUNE2 and 6-10B cell lines transfected with shNC, shcircCTDP1 and shcircCTDP1 plus miR-320b inhibitor. The data were presented as mean ± SD (*P<0.05; **P<0.01).
miR-320b directly targets HOXA10
Using the TargetScan database, it was found that miR-320b could target HOXA10 as they have complementary DNA regions (Fig. 3A). To further examine the binding between miR-320b and HOXA10, a dual luciferase reporter assay was performed. Compared with NC cells, the results showed that the miR-320b mimic significantly reduced the luciferase activity of the wild-type HOXA10 3′UTR, whereas the miR-320b inhibitor increased luciferase activity. On the other hand, there was no effect on cells transfected with a mutant 3′-UTR of HOXA10 (Fig. 3B). Furthermore, transfection with miR-320b mimic significantly suppressed the expression of HOXA10, while miR-320b inhibitor markedly enhanced the HOXA10 expression (Fig. 3C). Taken together, these data indicate that miR-320b directly targets HOXA10 and negatively regulates its expression.
Figure 3.
MiR-320b directly targets HOXA10 and downregulates its expression. (A) Bioinformatic prediction of binding site of miR-320b by HOXA10. (B) Dual luciferase reporter assay shows luciferase activity in 293T cells transfected with NC, miR-320b mimic, miR-320b inhibitor. (C) RT-qPCR shows miR-320b and HOXA10 expression levels in SUNE2 cell line transfected with NC, miR-320b mimic, miR-320b inhibitor. The data were presented as mean ± SD (*P<0.05; **P<0.01).
circCTDP1/miR-320b regulates the progression of NPC via HOXA10
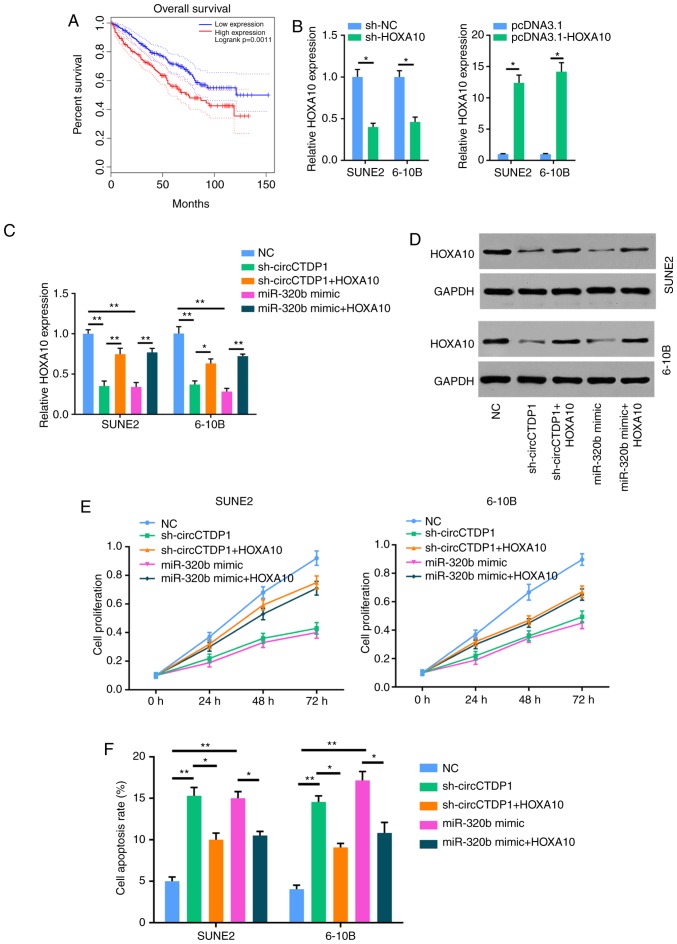
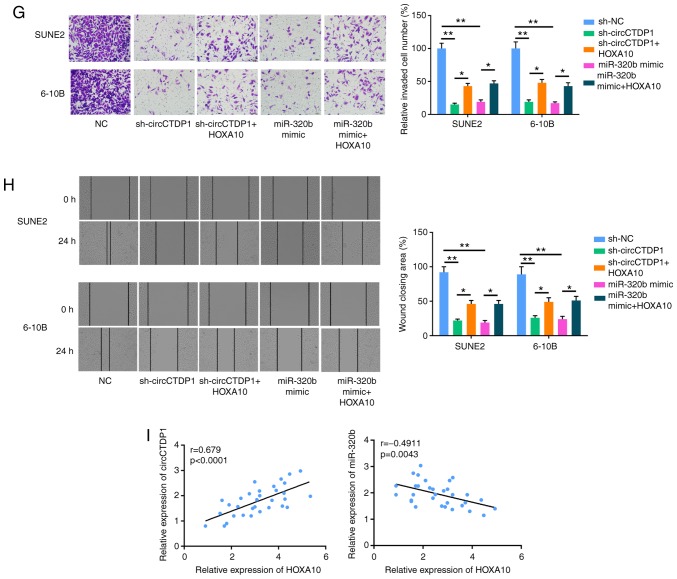
Based on the aforementioned results, miR-320b was indicated to directly target HOXA10. Therefore, it was hypothesized that HOXA10 may act as a critical factor in the circCTDP1/miR-320b axis in the development and progression of NPC. Firstly, Kaplan-Meier survival analysis showed that patients with NPC who exhibited a high HOXA10 expression had a shorter OS time compared with patients with a low HOXA10 expression (Fig. 4A). RT-qPCR analysis further demonstrated that the expression of HOXA10 was markedly reduced in NPC cells transfected with shHOXA10, but was notably increased in NPC cells transfected with HOXA10 overexpression plasmid (Fig. 4B). In addition, HOXA10 was overexpressed in shcircCTDP1- and miR-320b mimic-transfected cell lines. As presented in Fig. 4C and D, the expression level of HOXA10 was significantly increased in shcircCTDP1+HOXA10 and miR-320b+HOXA10 groups. Subsequently, an MTT assay was performed to examine cell proliferation. As shown in Fig. 4E, HOXA10 could restore the attenuated cell proliferation of shcircCTDP1- or miR-320b-trans-fected SUNE2 and 6-10B cells. Furthermore, it was found that the overexpression of HOXA10 inhibited apoptosis and enhanced cell mobility in shcircCTDP1- or miR-320b mimic-transfected SUNE2 and 6-10B cells (Fig. 4F-H). Moreover, there was a positive correlation between the expression of circCTDP1 and HOXA10 in NPC tissues, whereas there was a negative correlation between the expression of miR-320b and HOXA10 (Fig. 4I). Based on these results, the effects of shcircCTDP1 and miR-320b mimic on NPC cells were neutralized by overexpression of HOXA10, therefore suggesting that circCTDP1/mirR-320b/HOXA10 may be an important signaling pathway involved in the progression of NPC.
Figure 4.
CircCTDP1 upregulates the expression of HOXA10 via targeting miR-320b. (A) Kaplan-Meier survival analysis shows correlation between HOXA10 expression and prognosis of NPC patients. (B) RT-qPCR analysis shows relative HOXA10 expression of SUNE2 and 6-10B cell lines (shNC or shHOXA10 and pcDNA3.1 or pcDNA3.1-HOXA10). (C) RT-qPCR analysis shows relative HOXA10 expression of SUNE2 and 6-10B cell lines (NC, shcircCTDP1, shcircCTDP1 plus HOXA10, miR-320b mimic, and miR-320b mimic plus HOXA10). (D) Western blot analysis shows the relative HOXA10 protein level of SUNE2 and CINE1 cell lines (NC, shcircCTDP1, shcircCTDP1 plus HOXA10, miR-320b mimic, and miR-320b mimic plus HOXA10). (E) MTT assay shows the cell growth rate of SUNE2 and 6-10B cell lines (NC, shcircCTDP1, shcircCTDP1 plus HOXA10, miR-320b mimic, and miR-320b mimic plus HOXA10) at different time points of 0, 24, 48, and 72 h. (F) Flow cytometry assay shows the relative cell apoptosis rate of SUNE2 and 6-10B cell lines (NC, shcircCTDP1, shcircCTDP1 plus HOXA10, miR-320b mimic, and miR-320b mimic plus HOXA10). The data were presented as mean ± SD (*P<0.05; **P<0.01). CircCTDP1 upregulates the expression of HOXA10 via targeting miR-320b. (G) Cell invasion assay of SUNE2 and 6-10B cell lines (NC, shcircCTDP1, shcircCTDP1 plus HOXA10, miR-320b mimic, and miR-320b mimic plus HOXA10). (H) Wound healing assay of SUNE2 and 6-10B cell lines (NC, shcircCTDP1, shcircCTDP1 plus HOXA10, miR-320b mimic, and miR-320b mimic plus HOXA10). (I) The circCTDP1 expressions were positively correlated with HOXA10 expressions within included NPC tissues. The miR-320b expressions were negatively correlated with HOXA10 expressions within included NPC tissues. The data were presented as mean ± SD (*P<0.05; **P<0.01).
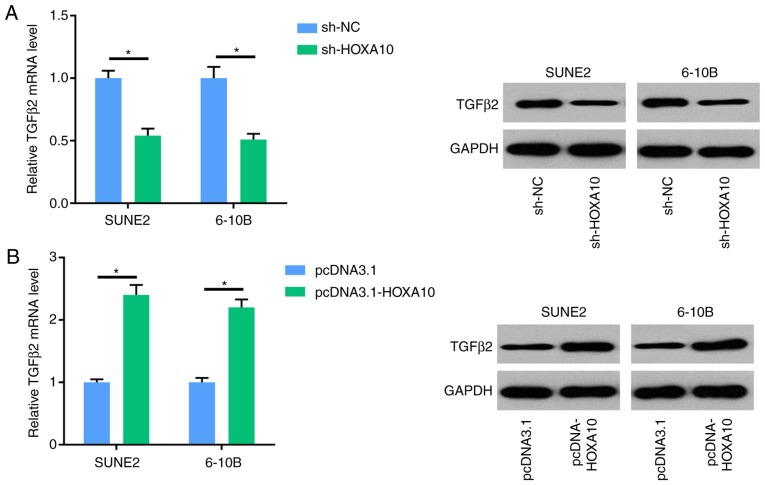
HOXA10 regulates the expression of TGFβ2 in NPC cells
TGFβ2 is a vital modulator of tumor invasion and motility. RT-qPCR and western blot analysis showed that knockdown of HOXA10 decreased the expression of TGFβ2 in SUNE2 and 6-10B cells, whereas overexpression of HOXA10 increased the expression of TGFβ2. These results demonstrated that HOXA10 could regulate the expression of TGFβ2 in NPC cells (Fig. 5A and B).
Figure 5.
HOXA10 regulates the expression of TGFβ2 in NPC cells. (A) RT-qPCR and western blot analysis shows the expression levels of TGFβ2 in SUNE2 and 6-10B cell lines transfected with shNC and shHOXA10. (B) RT-qPCR and western blot analysis shows the expression levels of TGFβ2 in SUNE2 and 6-10B cell lines transfected with pcDNA3.1 and pcDNA3.1-HOXA10. The data were presented as mean ± SD (*P<0.05).
Discussion
NPC is one of the most life-threatening tumors worldwide which is mainly ascribed to tumor recurrence and distant metastasis (29,30). Thus, developing novel prognostic markers for NPC is crucial. To the best of our knowledge, in the present study, it was demonstrated for the first time that the circCTDP1/miR-320b/HOXA10 axis may contribute to the development and progression of NPC.
Previous findings have demonstrated that aberrant expression of circRNA is involved in various human cancers (31-33). For instance, circ_0067934 overexpression is associated with poor prognosis and facilitates the development and progression of thyroid carcinoma (34). Shuai et al (35) reported that circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma. However, the biological role and potential molecular mechanism of circRNAs in NPC remain to be elucidated. In the current study, the association between the expression level of circCTDP1 and the prognosis of patients with NPC was analyzed. circCTDP1 expression was also determined in in NPC tissues and cell lines, and found to be upregulated. Knockdown of circCTDP1 suppressed the proliferation, migration and invasion of NPC cells, suggesting for the first time that circCTDP1 is associated with the progression of NPC.
Previous studies have demonstrated that circRNAs interact with miRNAs by acting as ceRNA to regulate the proliferation and migration of tumor cells (36-38). Recently, Cao et al (39) reported that circ0001429 promoted the progression of bladder cancer through sponging miR-205-3p and upregulating expression of VEGFA. Regarding NPC, Ke et al reported that circHIPK3 acts as a ceRNA to upregulate E74 like ETS transcription factor 3 by sponging miR-4288 (40). In the present study, circCTDP1 was found to directly interact with miR-320b, and to inhibit its expression through bioinformatic analysis and luciferase reporter assay. Furthermore, miR-320b inhibitor could significantly abolish the inhibitory effect of shcircCTDP1 on NPC phenotypes.
HOX genes are divided into 4 groups (HOXA, HOXB, HOXC and HOXD) encode transcription factors involved in the control of cell growth (41,42). It has been reported that HOXA10 is associated with cell migration, proliferation and survival in various types of cancer (43,44). In the current study, it was demonstrated that HOXA10 was a direct downstream target of miR-320b through bioinformatic prediction and in vitro experiments, and overexpression of HOXA10 was found to promote the progression and development of NPC cells. Moreover, expression of HOXA10 was positively correlated with that of circCTDP1, but negatively correlated with miR-320b expression. These results indicated that miR-320b may inhibit the proliferation, migration and invasion of NPC cells by downregulating HOXA10 expression.
HOXA10 has been reported to increase the levels of TGFβ2 in pancreatic cancer cells (45). Therefore, it was hypothesized that HOXA10 may exert its role in NPC by regulating the expression of TGFβ2. The findings of the present study demonstrated that upregulation of HOXA10 increased the expression of TGFβ2 and downregulation of HOXA10 decreased the expression of TGFβ2.
In the current study, it was demonstrated that circCTDP1 promotes proliferation, migration and invasion of NPC cells through a miR-320b/HOXA10 axis. This study offers an improved understanding of the pathogenesis of NPC, and the circCTDP1, miR-320b, HOXA10 and TGFβ2 may have potential as therapeutic targets for NPC.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
HL, CC and XT designed the study. JY and HX performed experiments. HL and CC analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Third Affiliated Hospital of Soochow University. Written informed consent was obtained from all patients prior to the study start. All animal experiments were conducted with the approval of the Third Affiliated Hospital of Soochow University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no that they have no competing interests.
References
- 1.Lee AW, Lin JC, Ng WT. Current management of nasopharyngeal cancer. Semin Radiat Oncol. 2012;22:233–244. doi: 10.1016/j.semradonc.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Pan F, Ruan Z, Li J, Pang X, Zhang Y, Zou L, Liang H. Radiotherapy combined docetaxel and oxaliplatin chemotherapy is effective in patients with locally advanced nasopharyngeal carcinoma. Med Oncol. 2015;32:252. doi: 10.1007/s12032-015-0698-4. [DOI] [PubMed] [Google Scholar]
- 3.Colaco RJ, Betts G, Donne A, Swindell R, Yap BK, Sykes AJ, Slevin NJ, Homer JJ, Lee LW. Nasopharyngeal carcinoma: A retrospective review of demographics, treatment and patient outcome in a single centre. Clin Oncol (R Coll Radiol) 2013;25:171–177. doi: 10.1016/j.clon.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Ma DD, Yuan LL, Lin LQ. LncRNA HOTAIR contributes to the tumorigenesis of nasopharyngeal carcinoma via up-regulating FASN. Eur Rev Med Pharmacol Sci. 2017;21:5143–5152. doi: 10.26355/eurrev_201711_13831. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang M, Zhao M, Qiu H, Shi D, Wang J, Tian Y, Lin L, Deng W. Effusanin E suppresses nasopharyngeal carcinoma cell growth by inhibiting NF-κB and COX-2 signaling. PLoS One. 2014;9:e109951. doi: 10.1371/journal.pone.0109951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pei W, Tao L, Zhang LW, Zhang S, Cao J, Jiao Y, Tong J, Nie J. Circular RNA profiles in mouse lung tissue induced by radon. Environ Health Prev Med. 2017;22:36. doi: 10.1186/s12199-017-0627-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li R, Wu B, Xia J, Ye L, Yang X. Circular RNA hsa_ circRNA_102958 promotes tumorigenesis of colorectal cancer via miR-585/CDC25B axis. Cancer Manag Res. 2019;11:6887–6893. doi: 10.2147/CMAR.S212180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, Dai J, Zhang S, Yang Q, Yi Y, Jiang Y. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18:13. doi: 10.1186/s12943-019-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52:743–754. doi: 10.3892/ijo.2018.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu L, Liu Y, Yang Y, Mao XM, Yin ZD. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci. 2019;23:2817–2826. doi: 10.26355/eurrev_201904_17558. [DOI] [PubMed] [Google Scholar]
- 11.Wei H, Liu D, Sun J, Mao Y, Zhao L, Zhu W, Xu G, Gao Z. Circular RNA circ_0008450 upregulates CXCL9 expression by targeting miR-577 to regulate cell proliferation and invasion in nasopharyngeal carcinoma. Exp Mol Pathol. 2019;110:104288. doi: 10.1016/j.yexmp.2019.104288. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Zhou H, Guan Z. CircRNA_000543 knockdown sensitizes nasopharyngeal carcinoma to irradiation by targeting miR-9/platelet-derived growth factor receptor B axis. Biochem Biophys Res Commun. 2019;512:786–792. doi: 10.1016/j.bbrc.2019.03.126. [DOI] [PubMed] [Google Scholar]
- 13.Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Q, Liu T, Zhang T, Du S, Xie GX, Lin X, Chen L, Yuan Y. MiR-101 sensitizes human nasopharyngeal carcinoma cells to radiation by targeting stathmin 1. Mol Med Rep. 2015;11:3330–3336. doi: 10.3892/mmr.2015.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu RS, Qiu EH, Zhu JJ, Wang JR, Lin HL. MiR-101 promotes nasopharyngeal carcinoma cell apoptosis through inhibiting Ras/Raf/MEK/ERK signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:150–157. doi: 10.26355/eurrev_201801_14112. [DOI] [PubMed] [Google Scholar]
- 16.Zhu HM, Jiang XS, Li HZ, Qian LX, Du MY, Lu ZW, Wu J, Tian XK, Fei Q, He X, Yin L. miR-184 inhibits tumor invasion, migration and metastasis in nasopharyngeal carcinoma by targeting Notch2. Cell Physiol Biochem. 2018;49:1564–1576. doi: 10.1159/000493459. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Dai B, Feng L. miR-543 promoted the cell proliferation and invasion of nasopharyngeal carcinoma by targeting the JAM-A. Hum Cell. 2019;32:477–486. doi: 10.1007/s13577-019-00274-0. [DOI] [PubMed] [Google Scholar]
- 18.Yin W, Shi L, Mao Y. MicroRNA-449b-5p suppresses cell proliferation, migration and invasion by targeting TPD52 in nasopharyngeal carcinoma. J Biochem. 2019;166:433–440. doi: 10.1093/jb/mvz057. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Tang X, He Q, Yang X, Ren X, Wen X, Zhang J, Wang Y, Liu N, Ma J. Overexpression of mitochondria mediator gene TRIAP1 by miR-320b loss is associated with progression in nasopharyngeal carcinoma. PLoS Genet. 2016;12:e1006183. doi: 10.1371/journal.pgen.1006183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Zhang PY, Li P, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, Huang CM, Zheng CH. Circular RNA hsa_circ_0001368 suppresses the progression of gastric cancer by regulating miR-6506-5p/FOXO3 axis. Biochem Biophys Res Commun. 2019;512:29–33. doi: 10.1016/j.bbrc.2019.02.111. [DOI] [PubMed] [Google Scholar]
- 21.An J, Shi H, Zhang N, Song S. Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;511:921–925. doi: 10.1016/j.bbrc.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zanatta A, Rocha AM, Carvalho FM, Pereira RM, Taylor HS, Motta EL, Baracat EC, Serafini PC. The role of the Hoxa10/HOXA10 gene in the etiology of endometriosis and its related infertility: A review. J Assist Reprod Genet. 2010;27:701–710. doi: 10.1007/s10815-010-9471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Wang DL, Yu P. LncRNA H19 regulates the expression of its target gene HOXA10 in endometrial carcinoma through competing with miR-612. Eur Rev Med Pharmacol Sci. 2018;22:4820–4827. doi: 10.26355/eurrev_201808_15617. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Jiang Y, Wan Y, Zhou S, Thapa S, Cheng W. MicroRNA-665 suppresses the growth and migration of ovarian cancer cells by targeting HOXA10. Mol Med Rep. 2018;18:2661–2668. doi: 10.3892/mmr.2018.9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SM, Choi EY, Bae M, Choi JK, Kim YJ. A long-range interactive DNA methylation marker panel for the promoters of HOXA9 and HOXA10 predicts survival in breast cancer patients. Clin Epigenetics. 2017;9:73. doi: 10.1186/s13148-017-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen ZH, Zhao KM, Du T. HOXA10 promotes nasopharyngeal carcinoma cell proliferation and invasion via inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci. 2017;21:945–952. [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Lee AW, Poon YF, Foo W, Law SC, Cheung FK, Chan DK, Tung SY, Thaw M, Ho JH. Retrospective analysis of 5037 patients with nasopharyngeal carcinoma treated during 1976-1985: Overall survival and patterns of failure. Int J Radiat Oncol Biol Phys. 1992;23:261–270. doi: 10.1016/0360-3016(92)90740-9. [DOI] [PubMed] [Google Scholar]
- 30.Vokes EE, Liebowitz DN, Weichselbaum RR. Nasopharyngeal carcinoma. Lancet. 1997;350:1087–1091. doi: 10.1016/S0140-6736(97)07269-3. [DOI] [PubMed] [Google Scholar]
- 31.Sheng M, Wei N, Yang HY, Yan M, Zhao QX, Jing LJ. CircRNA UBAP2 promotes the progression of ovarian cancer by sponging microRNA-144. Eur Rev Med Pharmacol Sci. 2019;23:7283–7294. doi: 10.26355/eurrev_201909_18833. [DOI] [PubMed] [Google Scholar]
- 32.Jin C, Shi L, Li Z, Liu W, Zhao B, Qiu Y, Zhao Y, Li K, Li Y, Zhu Q. Circ_0039569 promotes renal cell carcinoma growth and metastasis by regulating miR-34a-5p/CCL22. Am J Transl Res. 2019;11:4935–4945. [PMC free article] [PubMed] [Google Scholar]
- 33.Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, Shu Y. CircRNAs in cancer metabolism: A review. J Hematol Oncol. 2019;12:90. doi: 10.1186/s13045-019-0776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Yan X, Zhang H, Zhan X. CircRNA circ_0067934 overexpression correlates with poor prognosis and promotes thyroid carcinoma progression. Med Sci Monit. 2019;25:1342–1349. doi: 10.12659/MSM.913463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shuai M, Hong J, Huang D, Zhang X, Tian Y. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett. 2018;16:6495–6501. doi: 10.3892/ol.2018.9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L, Yang J, Zhu X, Li D, Lv Z, Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283:2326–2339. doi: 10.1111/febs.13741. [DOI] [PubMed] [Google Scholar]
- 37.Hou Z, Xu X, Zhou L, Fu X, Tao S, Zhou J, Tan D, Liu S. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol. 2017;39:1010428317718135. doi: 10.1177/1010428317718135. [DOI] [PubMed] [Google Scholar]
- 38.Wang SH, Ma F, Tang ZH, Wu XC, Cai Q, Zhang MD, Weng MZ, Zhou D, Wang JD, Quan ZW. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. J Exp Clin Cancer Res. 2016;35:160. doi: 10.1186/s13046-016-0436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao W, Zhao Y, Wang L, Huang X. Circ0001429 regulates progression of bladder cancer through binding miR-205-5p and promoting VEGFA expression. Cancer Biomark. 2019;25:101–113. doi: 10.3233/CBM-182380. [DOI] [PubMed] [Google Scholar]
- 40.Ke Z, Xie F, Zheng C, Chen D. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol. 2019;234:1699–1706. doi: 10.1002/jcp.27041. [DOI] [PubMed] [Google Scholar]
- 41.Chen KN, Gu ZD, Ke Y, Li JY, Shi XT, Xu GW. Expression of 11 HOX genes is deregulated in esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1044–1049. [PubMed] [Google Scholar]
- 42.Carrera M, Bitu CC, de Oliveira CE, Cervigne NK, Graner E, Manninen A, Salo T, Coletta RD. HOXA10 controls proliferation, migration and invasion in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:3613–3623. [PMC free article] [PubMed] [Google Scholar]
- 43.Shao L, Chen Z, Peng D, Soutto M, Zhu S, Bates A, Zhang S, El-Rifai W. Methylation of the HOXA10 promoter directs miR-196b-5p-dependent cell proliferation and invasion of gastric cancer cells. Mol Cancer Res. 2018;16:696–706. doi: 10.1158/1541-7786.MCR-17-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Li C, Jiang Y, Wan Y, Zhou S, Cheng W. Tumor-suppressor role of miR-139-5p in endometrial cancer. Cancer Cell Int. 2018;18:51. doi: 10.1186/s12935-018-0545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui XP, Qin CK, Zhang ZH, Su ZX, Liu X, Wang SK, Tian XS. HOXA10 promotes cell invasion and MMP-3 expression via TGFβ2-mediated activation of the p38 MAPK pathway in pancreatic cancer cells. Dig Dis Sci. 2014;59:1442–1451. doi: 10.1007/s10620-014-3033-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.