Abstract
Endothelial cell senescence is closely related to the occurrence of cardiovascular diseases and microRNAs (miRNAs/miRs) are considered as therapeutic targets for cardiovascular disease. The current study aimed to investigate the role of miR-20b in the senescence process of endothelial cells and its underlying mechanism. Cell viability, proportion of senescent cells and the cell cycle were respectively determined by Cell Counting Kit-8, SA-β-galactosidase and flow cytometry. The relative expressions of mRNA and protein were detected by reverse transcription-quantitative polymerase chain reaction and western blotting, respectively. The possible target genes and binding sites of miR-20b were predicted using Targetscan and further verified by dual luciferase reporter assay. The present study found that H2O2 inhibited cell viability, caused cell cycle arrest in G1 phase, decreased miR-20b level and induced cell senescence. Moreover, high expression of miR-20b promoted cell viability and reduced H2O2-induced cell senescence, whereas low expression of miR-20b produced the opposite effects. Thioredoxin interacting protein (TXNIP) was predicted as a target gene for miR-20b and knockdown of TXNIP increased cell viability, inhibited cell senescence, reduced the expression of p16, p21, TXNIP, NLR family pyrin domain containing 3 (NLRP3) and cleaved Caspase-1 and reversed the promoting effects of the miR-20b inhibitor and H2O2 on cell senescence. Furthermore, the knockdown of TXNIP inhibited the Wnt/β-catenin pathway. The finding reveals that high expression of miR-20b inhibits the senescence of human umbilical vein endothelial cells through regulating the Wnt/β-catenin pathway via the TXNIP/NLRP3 axis.
Keywords: microRNA-20b, cellular senescence, thioredoxin interacting protein/NLR family pyrin domain containing 3 axis, Wnt/β-catenin pathway, endothelial cell
Introduction
Population aging is one of the main problems facing the 21st century. Aging is associated with an increased incidence of age-related diseases such as Parkinson's, Alzheimer's, cardiovascular and other diseases (1). Cardiovascular disease is a chronic disease closely related to aging (2) and atherosclerosis is the primary cause of cardiovascular disease. As we age, the structure and function of artery vasculature changes, the vessel lumen expands, vascular stiffness increases, eventually affecting other tissues and organs (3). Noticeably, the damage and functional changes of vascular endothelial cells play a crucial role in the occurrence and development of atherosclerosis, and penetrate into the whole pathological process of atherosclerosis (4).
Vascular endothelial cells, which are monolayer squamous epithelial cells located between vascular endothelium subcutaneous tissues and blood, function as a physical barrier between blood and tissues. Vascular endothelial cells play an important role in maintaining normal vascular function and tissue structure and in the regulation of cells proliferation (5,6). Endothelial cell senescence, which is closely related to atherosclerosis, is accompanied by the destruction of endothelial cell integrity and functional damage of endothelial cells that in turn cause vascular dysfunction, therefore creating conditions for the occurrence of cardiovascular diseases (7,8). Thus, it is highly necessary to understand the mechanism of endothelial cell senescence, to provide patients with effective clinical treatment and reduce the occurrence of cardiovascular diseases.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs and have been regarded as therapeutic targets and biomarkers for evaluating cardiovascular disease in recent years (9). Among miRNAs, miR-222-221 can inhibit the proliferation and migration of endothelial cells and induce apoptosis (10,11). miR-126-3p and miR-126-5p are considered as potential biomarkers of atherosclerosis (12). A recent study reported that miR-20b is up-regulated in insulin-resistant skeletal muscle and is involved in glucose metabolism regulation (13). Moreover, miR-20b has been shown to play a critical role in maintaining vascular integrity, as the down-regulation of miR-20b was found to be able to down-regulate the level of a cellular senescence marker (14). However, studies are still needed to confirm the role and mechanism of miR-20b in endothelial cell senescence. In this study, the occurrence of senescence of HUVECs cells was induced by hydrogen peroxide (H2O2) and the role of miR-20b in the process of cellular senescence and its possible mechanism were investigated.
Materials and methods
Cell culture
Human umbilical vein endothelial cells (HUVECs) were purchased from Sciencell Research Laboratories, Inc., (cat. no. 8000). The cells were cultured in the endothelium cell medium (Sciencell Research Laboratories, Inc.) containing 5% fetal bovine serum (Sigma-Aldrich; Merck KGaA), 1% endothelial growth factor (Sigma-Aldrich; Merck KGaA), 100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.) in an incubator with 95% O2 and 5% CO2 at 37°C.
Induction of HUVECs cell senescence by H2O2
H2O2 was used to stimulate and induce cell senescence in HUVECs. The cells were randomly divided into 5 groups and treated with different concentrations of H2O2 (0, 10, 50, 100 and 500 µM). The blank group (0 µM H2O2) was cultured in normal medium, whereas the H2O2 group was incubated with normal medium at different concentrations of H2O2 for 24 h. The cell viability, proportion of senescent cells and cell cycle in each group were determined by Cell-Counting-Kit 8 (CCK-8; Beyotime Institute of Biotechnology), SA-β-galactosidase (SA-β-gal; Beyotime Institute of Biotechnology) and FACSCalibur Flow Cytometer (cat. no. 342973; BD Biosciences; Becton, Dickinson and Company).
Transfection
The specific groups were: Scramble group, miR-20b mimic group, miR-20b inhibitor group, scramble + H2O2 group, miR-20b mimic + H2O2 group and miR-20b inhibitor + H2O2 group. The sequences required for transfection are shown in Table I. The cells were transfected with the miR-20b mimic (100 pmol), miR-20b inhibitor (100 pmol) or 100 pmol scramble (Sigma-Aldrich; Merck KGaA) using Lipofectamine 2000 Transfection Reagent (Invitrogen; Thermo fisher Scientific, Inc.). Next, the Lipofectamine 2000 reagent was mixed with DMEM and incubated with the cells for 24 h at 37̊C with 5% CO2. After that, the medium was replaced and certain groups of cells were stimulated by H2O2 (100 µM) for 24 h, while the rest was left untreated. The Scramble served as a negative control and whether the transfection was successful was confirmed by performing reverse transcription-quantitative (RT-q)PCR.
Table I.
The sequences required for transfection.
| Sequence | 5′-3′ |
|---|---|
| miR-20b mimics | ACUGUAGUAUGGGCACUUCCAG |
| miR-20b inhibitor | UGCUCAUAGUGCAGGUAGUU |
| Scramble C | AUACAUUACCCGAAGUCUA |
| siTXNIP | |
| Forward | CUCCCUGCUAUAUGGAUGUTT |
| Reverse | ACAUCCAUAUAGCAGGGAGTT |
| siTXNIP negative control | |
| Forward | UUCUCCGAACGUGUCACGUTT |
| Reverse | ACGUGACACGUUCGGAGGAGAATT |
miR, microRNA; si, small interfering.
Knocking down thioredoxin interacting protein (TXNIP) using small interfering RNA (siRNA)
TXNIP was knocked down by siRNA (Forward, RNA oligo: 5′-AGA GAA AAA GCC UUC U UUC CC-3′, and reverse, RNA oligo: 5′-GAA AGA AGG CUU UUU CUC UGA-3′, GE Healthcare Dharmacon Inc.) and a non-specific siRNA served as a negative control (NC sense RNA oligo: 5′-AGA AUC ACU GUA CAU CAA CUC UA-3′, antisense RNA oligo: 5′-GAG UUG AUG UAC AGU GAU UCU GC-3′). The HUVECs cells were transfected with siTXNIP (100 pmol) by Lipofectamine 2000 Transfection Reagent, at the same time, the cells incubated with Lipofectamine 2000 only were used as blank control group. Whether the transfection was successful was confirmed by performing RT-qPCR.
Next, the cells were co-transfected with miR-20b inhibitor (100 pmol) and siTXNIP (100 pmol) or NC, with NC group served as the control group. Some of the cells were stimulated by H2O2 (100 µM) for 24 h, while the rest was left untreated. The specific groups were as follows: NC group, siTXNIP miR-20b inhibitor + NC group, miR-20b inhibitor + siTXNIP group, miR-20b inhibitor + H2O2 + NC group and miR-20b inhibitor + H2O2 + siTXNIP group.
CCK-8 assay
The experimental operation of determining cell viability was performed according to the protocol of the CCK-8 (Beyotime Institute of Biotechnology). The cells were cultured at 37°C with 95% O2 and 5% CO2 in an incubator for 24 h, and CCK-8 solution was then added to further culture the cells for 2 h. The optical density (OD) value of each well was measured at 450 nm by enzyme-labeling instrument (Muliskan MK3; Thermo Fisher Scientific, Inc.).
SA-β-gal assay
The previously treated HUVECs were washed twice by PBS (Gibco; Thermo Fisher Scientific, Inc.) and fixed by cell fixative solution in β-gal staining kit (Beyotime Institute of Biotechnology) for 20 min at 37°C. Next, the cells were washed three times by PBS to remove the solution. β-gal dyeing liquid prepared in following the instruction was mixed with the cells and incubated together overnight in a regular incubator at 37̊C for 24 h without CO2. The cells with blue cytoplasm by β-gal staining were positive under a light microscope (BX41; Olympus Corporation). A total of 10 fields in each group were selected under the microscope and the total cells and SA-β-gal positive cells in each field were counted by ImageJ (version 5.0; Bio-Rad Laboratories, Inc.). The proportion of positive cells was obtained by calculating the ratio of average SA-β-gal positive cell number to average total cell number in each group.
Cell cycle assay
The cells were fixed by precooled 70% ethanol at 4°C overnight, then re-suspended in PBS and cultured with RNAase (10 mg/ml) at 4°C for 1 h. The cells were then incubated with propidium iodide (PI; 10 µg/ml) solution in the dark at 4°C for 1 h. The FACSCalibur Flow Cytometer (BD Biosciences; Becton, Dickinson and Company) was used to detect cell cycle, and data were processed by CellQuest software (version 5.1; BD Biosciences; Becton, Dickinson and Company).
Target gene prediction of miR-20b
Potential target genes for miR-20b were predicted by Targetscan (http://www.targetscan.org/). RT-qPCR was carried out to determine the effect of miR-20b on the expression of target genes and Targetscan was used to predict the binding sites between the target genes and miR-20b.
Plasmid transfection and dual luciferase reporter assay
The binding ability of miR-20b and target genes was detected by dual luciferase reporter. The wild-type or mutant (mut) 3′-untranslated region (UTR) sequences of TXNIP, or NLRP3 targeting miR-20b was amplified by RT-qPCR and then respectively inserted into the pGL3 control vectors (Promega Corporation). The HUVECs were randomly divided into eight groups, namely: Control + TXNIP-3′-UTR group, miR-20b mimic + TXNIP-3′-UTR group, Control + TXNIP-3′-UTR mut group, miR-20b mimic + TXNIP-3′-UTR mut group, Control + NLRP3-3′-UTRgroup, miR-20b mimic + NLRP3-3′-UTR group, Control + NLRP3-3′-UTR mut group and miR-20b mimic + NLRP3-3′-UTR mut group and transfected by Lipofectamine 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The recombinant vectors or miR-20b mimic was respectively transfected into the cells for 24 h. The luciferase activities were measured using the Dual-Glo Luciferase assay kits (Promega Corporation) and normalized using a Renilla luciferase reference plasmid.
RT-qPCR
Total RNA was extracted by using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and total RNA (2 µg) were reverse-transcribed into cDNA by using a Taqman MicroRNA Reverse Transcription kit (Applied Biosystems, Inc.) at 37°C for 15 min, at 85°C for 5 sec and preserved at 4°C. A SYBR-Green Master (Rox) kit (Thermo Fisher Scientific, Inc.) and RT-qPCR Detection System (ABI 7500; Life Technology; Thermo Fisher Scientific, Inc.) were used to perform RT-qPCR, the cycles were set as follows: Pretreatment at 95°C for 2 min, followed by 40 cycles at 95°C for 30 sec, at 60°C for 35 sec and at 95°C for 15 sec and finally preserved at 4°C. The 2−ΔΔCq method was used to calculate the data (15). U6 and GAPDH expression served as the internal control. The primer sequences for RT-qPCR are shown in Table II.
Table II.
The primer sequences for reverse transcription-quantitative PCR.
| Primer | Forward sequence (5′-3′) | Reverse sequence (5′-3′) |
|---|---|---|
| miR-20b | GCTCATAGTGCAGGTAGAA | TGTCAACGATACGCTACG |
| STAT3 | CCTTCCTCACCGTGTACTGG | AGCGTAGGGTAAGGTTCTTGC |
| SMAD7 | ACTCCAGATACCCGATGGATTT | CCTCCCAGTATGCCACCAC |
| TXNIP | CAGAAGCTCCTCCCTGCTATATG | GATGCAGGGATCCACCTCAG |
| NLRP3 | CTTCCTTTCCAGTTTGCTGC | TCTCGCAGTCCACTTCCTTT |
| p16 | CACGGGTCGGGTGAGAGT | CCCAACGCACCGAATAGTTAC |
| p21 | GCCTGGACTGTTTTCTCTCG | ATTCAGCATTGTGGGAGGAG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| GAPDH | AGTCAGCTCTCTCCTTTCAGG | TCCACCACCCTGTTGCTGTA |
miR, microRNA; STAT3, signal transducer and activator of transcription 3; SMAD7, mothers against decapentaplegic homolog 7; TXNIP, thioredoxin interacting protein; NLRP3, NLR family pyrin domain containing 3.
Western blotting (WB)
The cells were lysed to obtain total proteins using radio immunoprecipitation assay lysis buffer (RIPA; Sigma Aldrich; Merck KGaA). The concentration of proteins was determined by Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.) and equal amounts of total proteins (2 µg) were added to 10% SDS-PAGE and transferred to polyvinylidene difluoride membrane (PVDF; Bio-Rad Laboratories, Inc.). The PVDF membrane was blocked by 5% non-fat milk at room temperature for 30 min and first incubated with primary antibodies for 4°C overnight and then with secondary antibodies for 2 h. The antibodies used during WB were anti-TXNIP antibody (Rabbit; cat. no. ab188865; 1:1,000; Abcam), anti-GAPDH antibody (mouse; cat. no. ab8245; 1:1,000; Abcam), anti-NLRP3 antibody (Rabbit; cat. no. ab214185; 1:200; Abcam), anti-Cleaved Caspase-1 antibody (Rabbit; cat. no. 4199; 1:1,000; Cell Signaling Technology, Inc.), anti-Caspase-1 antibody (Rabbit; cat. no. 3866; 1:1,000; Cell Signaling Technology, Inc.), anti-β-catenin antibody (Rabbit, cat. no. ab32572; 1:5,000; Abcam), goat anti-mouse IgG H&L (HRP; 1:2,000; cat. no. ab205719; Abcam) and goat anti-rabbit IgG H&L (HRP; 1:2,000; cat. no. ab205718; Abcam). ECL Blotting Detection Reagents (Applygen Technologies, Inc.) was used to analyze the protein bands. The gray values of the protein bands were analyzed by ImageJ (version 5.0; Bio-Rad, Laboratories, Inc.) and the relative expressions of the target proteins were determined by the ratio of the gray scale of target protein to the internal reference. GAPDH served as internal control.
Statistical analysis
Data were analyzed by SPSS 21.0 system (IBM, Corps.) and shown as the mean ± standard deviation. Significant differences among different groups were analyzed using analysis of variance followed by the Tukey post hoc test. P<0.05 was considered to indicate a statistically significant difference. All experiments were repeated three times.
Results
H2O2 induces cell senescence and inhibits miR-20b expression
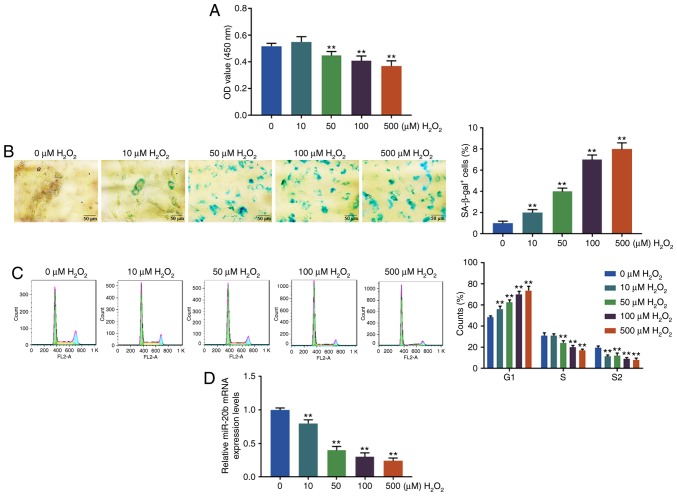
The cell viability, senescence and cycle of the HUVECs were determined by CCK-8, SA-β-gal staining and flow cytometry, respectively, after treating HUVECs at different H2O2 concentrations (0, 10, 50, 100 and 500 µM). It was found that H2O2 at 50, 100 or 500 µM significantly reduced the cell viability of HUVECs compared with 0 µM (P<0.01; Fig. 1A). As shown in Fig. 1B, the blue cytoplasm was positive and the proportion of the positive cells significantly increased as the concentration of H2O2 became higher (P<0.01; Fig. 1B). At the same time, the proportion of HUVECs in G1 phase significantly increased with the increase of H2O2 concentration, while the proportion of HUVECs in S and G2 phase significantly decreased after H2O2 treatment (P<0.01; Fig. 1C). The results suggested that H2O2 inhibited cell viability, caused cell cycle arrest in G1 phase and induced cell senescence.
Figure 1.
Cell senescence induced by oxidative stress. (A) Cell viability was determined by Cell Counting Kit-8. (B) Cell senescence was detected by SA-β-gal staining. (C) The cell cycle was determined by flow cytometry. (D) The miR-20b level was detected by reverse transcription quantitative-PCR. The human umbilical vein endothelial cells were randomly divided into 5 groups and treated with different concentrations of H2O2 (0, 10, 50, 100 and 500 µM). The blank group (0 µM H2O2) was cultured in normal medium. The H2O2 group was incubated by normal medium with different concentrations of H2O2. The experiments were repeated in triplicate. **P<0.001 vs. 0 µM H2O2. miR, microRNA; OD, optical density; SA-β-gal, SA-β-galactosidase.
Moreover, the expression level of miR-20b in HUVECs was determined by RT-qPCR and the present study found that the miR-20b level was significantly downregulated by H2O2 treatment (P<0.01; Fig. 1D). The percentage of SA-β-gal+ cells in HUVECs exposed to H2O2 at 50 µM was only ~4%, while the percentage of SA-β-gal + cells in HUVECs exposed to H2O2 at 100 µM was 7% (Fig. 1B). Based on the above results and previous studies (16,17), to obtain more senescent cells and avoid potentially excessive damage from H2O2 at higher concentration (500 µM), H2O2 at 100 µM was chosen for further experiments.
HUVECs cells are successfully transfected with an miR-20b mimic or miR-20b inhibitor
In order to investigate the effect of miR-20b expression on the senescence of HUVECs, the miR-20b mimic and miR-20b inhibitor were respectively transfected into cells. RT-qPCR was performed to confirm whether the transfection was successfully conducted. The results demonstrated that compared with the scramble group, the expression of the miR-20b in miR-20b mimic and miR-20b mimic + H2O2 group were increased, while those in miR-20b inhibitor, scramble + H2O2 and miR-20b inhibitor + H2O2 group were significantly decreased (P<0.05; Fig. 2A). Compared with the scramble + H2O2 group, the miR-20b expression was significantly increased in miR-20b mimic + H2O2 group, but significantly decreased in miR-20b inhibitor + H2O2 group (P<0.05; Fig. 2A). Therefore, the present results showed that the expression of miR-20b in HUVECs was successfully upregulated and downregulated.
Figure 2.
Effects of miR-20b expression level on cell senescence. (A) The cells were transfected with the miR-20b mimic, miR-20b inhibitor or scramble and the transfection rate was detected by reverse transcription-quantitative PCR. (B) The effect of miR-20b on cell viability was detected by Cell Counting Kit-8. (C) The effect of miR-20b on cell senescence was detected by SA-β-gal staining. The HUVECs were divided into 6 groups, namely the scramble group, miR-20b mimic group, miR-20b inhibitor group, scramble + H2O2 group, miR-20b mimic + H2O2 group and miR-20b inhibitor + H2O2 group. The cells were transfected with the miR-20b mimic, miR-20b inhibitor or scramble and then stimulated with H2O2 (100 µM), or left untreated. **P vs. scramble; ##P<0.001 vs. scramble + H2O2. HUVECs, human umbilical vein endothelial cells; miR, microRNA; SA-β-gal, SA-β-galactosidase; OD, optical density.
High expression of miR-20b inhibits cell senescence
The cell viability and senescence were determined by CCK-8 and SA-β-gal staining, respectively, and it was found that the cell viability was upregulated in miR-20b mimic group but down-regulated in miR-20b inhibitor and miR-20b inhibitor + H2O2 group compared with the scramble group (P<0.05; Fig. 2B). In comparison with the scramble + H2O2 group, the cell viability was increased in the miR-20b mimic + H2O2 group but decreased in the miR-20b inhibitor + H2O2 group (P<0.05; Fig. 2B).
Moreover, the SA-β-gal staining results showed that the proportion of β-gal positive cells significantly increased in the miR-20b inhibitor, scramble + H2O2, miR-20b mimic + H2O2 and miR-20b inhibitor + H2O2 group (P<0.05; Fig. 2C). However, compared with scramble + H2O2 group, the proportion of β-gal positive cells decreased in the miR-20b mimic + H2O2 group but increased in miR-20b inhibitor + H2O2 group (P<0.05; Fig. 2C).
The results indicated that miR-20b could increase cell viability and inhibit cell senescence, and that miR-20b produced protective effect on HUVECs cells from H2O2-induced cell senescence.
TXNIP is the target gene for miR-20b
Targetscan predicted that the possible target genes for miR-20b were signal transducer and activator of transcription 3 (STAT3), mothers against decapentaplegic homolog 7 (SMAD7), TXNIP and NLRP3. RT-qPCR was performed to detect the effect of miR-20b on the possible target genes and it was found that in the scramble + H2O2, miR-20b mimic + H2O2, and miR-20b inhibitor + H2O2 groups, the levels of STAT3 were significantly downregulated, while that of SMAD7 was significantly upregulated compared with the scramble group (P<0.05; Fig. 3A). The levels of TXNIP and NLRP3 were significantly decreased in the miR-20b mimic group compared with those in the scramble group, but were still increased in the miR-20b inhibitor, scramble + H2O2, miR-20b mimic + H2O2 and miR-20b inhibitor + H2O2 groups compared with those in the scramble group (P<0.05; Fig. 3A). Compared with the scramble + H2O2 group, the levels of TXNIP and NLRP3 were significantly downregulated in the miR-20b mimic + H2O2 group but were significantly upregulated in the miR-20b inhibitor + H2O2 group (P<0.05; Fig. 3A). Thus, the data indicated that the TXNIP and NLRP3 may be the target genes for miR-20b.
Figure 3.
Target gene of miR-20b was predicted. (A) The potential target genes of miR-20b were predicted by Targetscan (http://www.targetscan.org/) and RT-qPCR was performed for detection. **P<0.001 vs. scramble; ##P<0.001 vs. scramble + H2O2. (B) The binding sites of the target gene and miR-20b were predicted by Targetscan. (C) The binding abilities of miR-20b and TXNIP were detected by dual luciferase reporter. **P<0.001 vs. Control + TXNIP-3′-UTR; ##P<0.001 vs. miR-20b mimic + TXNIP-3′-UTR mut. (D) The binding abilities of miR-20b and NLRP3 were detected by dual luciferase reporter. **P<0.001 vs. Control + TXNIP-3′-UTR; ##P<0.001 vs. miR-20b mimic + TXNIP-3′-UTR mut. HUVECs were divided into 8 groups, namely, Control + TXNIP-3′-UTR group, miR-20b mimic + TXNIP-3′-UTR group, Control + TXNIP-3′-UTR mut group, miR-20b mimic + TXNIP-3′-UTR mut group, Control + NLRP3-3′-UTRgroup, miR-20b mimic + NLRP3-3′-UTR group, Control + NLRP3-3′-UTR mut group and miR-20b mimic + NLRP3-3′-UTR mut group. The pGL3 control vectors were used to generate recombinant vectors with mut sequence or a 3′UTR sequence of TXNIP or NLRP3 and the recombinant vectors and/or miR-20b mimic were transfected into cells. Mut, mutant; UTR, untranslated region; NLRP3, NLR family pyrin domain containing 3; HUVECs, human umbilical vascular cell; TXNIP, thioredoxin interacting protein.
In order to further determine the direct target gene for miR-20b, Targetscan website was used to predict the binding sites between miR-20b and TXNIP, and NLRP3 (Fig. 3B), and the results were further verified by dual luciferase reporter assay. The present study observed that in the miR-20b mimic + TXNIP-3′-UTR and miR-20b mimic + NLRP3-3′-UTR groups, the relative luciferase activities were significantly decreased, and the relative luciferase activity changes of TXNIP was the most significant (P<0.05; Fig. 3C and D). Thus, TXNIP was determined as the direct target gene for miR-20b.
siTXNIP reverses the effects of miR-20b inhibitor and H2O2
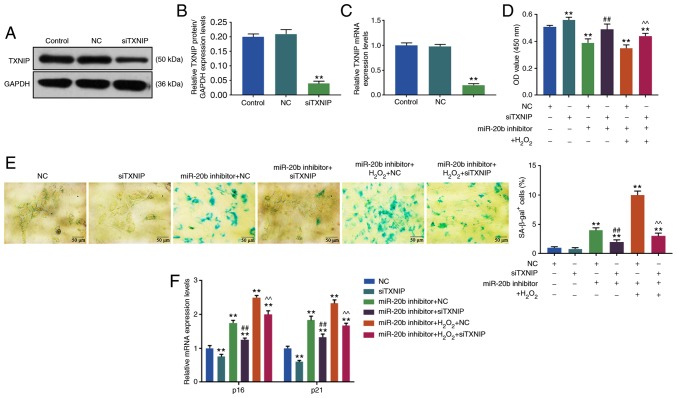
In order to investigate the mechanism of miR-20b in the cell senescence of HUVECs, TXNIP siRNA was used to transform HUVECs and the results were verified by RT-qPCR and WB. As shown in Fig. 4B and C, the expression of TXNIP decreased significantly in siTXNIP group (P<0.05), suggesting that HUVECs cells with a low expression of TXNIP were successfully constructed.
Figure 4.
Effects of siTXNIP on cell senescence. (A) Western blotting was used to determine the level of TXNIP. The knockdown of TXNIP was achieved by siRNA, the HUVECs cells were transfected with siTXNIP, the blank control group and NC group were set at the same time (Control group, NC group, and siTXNIP group). (B) The siTXNIP reduced the expression of TXNIP protein. (C) RT-qPCR was used to determine the TXNIP mRNA expression. (D) Cell viability was detected by Cell Counting Kit-8. The HUVECs were divided into 6 groups, namely, NC group, siTXNIP group, miR-20b inhibitor + NC group, miR-20b inhibitor + siTXNIP group, miR-20b inhibitor + H2O2 + NC group and miR-20b inhibitor + H2O2 + siTXNIP group. The cells were co-transfected with miR-20b inhibitor and siTXNIP or NC, and stimulated by H2O2 (100 µM), or left untreated. (E) Cell senescence was detected by SA-β-gal staining. (F) The expression of cell senescence-related genes were detected by RT-qPCR. **P<0.001 vs. NC; ##P<0.001 vs. miR-20b inhibitor + NC; ^^P<0.001 vs. miR-20b inhibitor + H2O2 + NC. RT-qPCR, reverse transcription-quantitative; miR, microRNA; si, small interfering; HHUVECs, human umbilical vascular endothelial cells; NC, negative control; TXNIP, thioredoxin interacting protein.
The cell viability and senescence of HUVECs were determined by CCK-8 and SA-β-gal staining, respectively, and it was found that the cell viability of HUVECs was increased in the siTXNIP group compared with in the NC group, moreover, the miR-20b inhibitor + NC group had a lower cell viability compared with in the miR-20b inhibitor + siTXNIP group. Furthermore, the cell viability of HUVECs was significantly increased in the miR-20b inhibitor + H2O2 + siTXNIP group compared with in miR-20b inhibitor + H2O2 + NC group (P<0.05; Fig. 4D) and the cell viability in miR-20b inhibitor + NC and miR-20b inhibitor + H2O2 + NC groups decreased compared with NC group (P<0.05; Fig. 4D).
SA-β-gal staining results showed that the proportions of β-gal positive cells in miR-20b inhibitor + NC and miR-20b inhibitor + H2O2 + NC groups were significantlu increased compared with the NC group (P<0.05; Fig. 4E), but decreased in the miR-20b inhibitor + siTXNIP group compared with in miR-20b inhibitor + NC group (P<0.05; Fig. 4E). Moreover, the number of β-gal positive cells was significantly increased in miR-20b inhibitor + H2O2 + siTXNIP group compared with that in miR-20b inhibitor + H2O2 + NC group (P<0.05; Fig. 4E).
The present data suggested that siTXNIP increased cell viability and partially reversed the inhibitory effects of miR-20b inhibitor and H2O2 on cell viability and the promoting effects of miR-20b inhibitor and H2O2 on cell senescence.
SiTXNIP reduces the expression of cell senescence-related genes
The results of WB showed that compared with NC group, the levels of p16 and p21 were decreased in siTXNIP group but were significantly increased in the miR-20b inhibitor + NC and miR-20b inhibitor + H2O2 + NC groups (P<0.05; Fig. 4F). However, the levels of p16 and p21 were decreased in the miR-20b inhibitor + siTXNIP group compared with those in the miR-20b inhibitor + NC group, and that the levels of p16 and p21 were lower in the miR-20b inhibitor + H2O2 + siTXNIP group compared with those in miR-20b inhibitor + H2O2 + NC group (P<0.05; Fig. 4F).
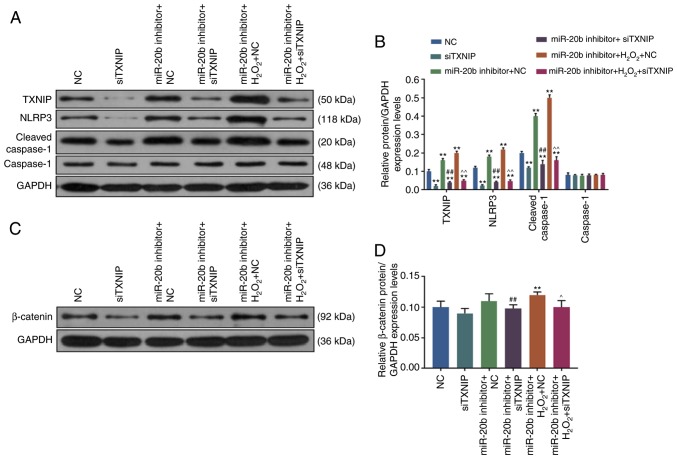
Compared with NC group, the expression of TXNIP, NLRP3 and cleaved Caspase-1 was significantly decreased in the siTXNIP group but increased in the miR-20b inhibitor + NC and miR-20b inhibitor + H2O2 + NC groups (P<0.05; Fig. 5B). Moreover, compared with the miR-20b inhibitor + NC group, the expression of TXNIP, NLRP3 and cleaved Caspase-1 in miR-20b inhibitor + siTXNIP group was decreased. However, the present study also found that the expression of TXNIP, NLRP3 and cleaved Caspase-1 was significantly decreased lower in the miR-20b inhibitor + H2O2 + siTXNIP group compared with miR-20b inhibitor + H2O2 + NC group, (P<0.05; Fig. 5B). Thus, the data showed that siTXNIP reduced the expression of senescence-related genes and inhibited cell senescence.
Figure 5.
siTXNIP inhibits the Wnt/β-catenin pathway. (A) The expression of TXNIP, NLRP3, cleaved Caspase-1 and Caspase-1 was determined by WB. (B) siTXNIP reduced TXNIP, NLRP3 and cleaved Caspase-1 expression. (C) β-catenin expression was determined by WB. (D) siTXNIP reduced β-catenin expression. **P<0.001 vs. NC; ##P<0.001 vs. miR-20b inhibitor + NC; ^P<0.001 and ^^P<0.001 vs. miR-20b inhibitor + H2O2 + NC. NC, negative control; miR, microRNA; si, small interfering; WB, western blotting; NLRP3, NLR family pyrin domain containing 3; TXNIP, thioredoxin interacting protein.
SiTXNIP inhibits the Wnt/β-catenin signaling pathway
The present study found that compared with NC group, the expression level of β-catenin was downregulated in siTXNIP group but was significantly upregulated in miR-20b inhibitor + H2O2 + NC group (P<0.05; Fig. 5D), at the same time, the expression of β-catenin in miR-20b inhibitor + siTXNIP and miR-20b inhibitor + H2O2 + siTXNIP groups was significantly decreased compared with in the miR-20b inhibitor + NC and miR-20b inhibitor + H2O2 + NC groups (P<0.05; Fig. 5D), suggesting that siTXNIP decreased β-catenin expression and inhibited the Wnt/β-catenin pathway. The working model of miR-20b is shown in Fig. 6.
Figure 6.
Working model of miR-20b in senescence of HUVECs. H2O2 induced senescence in HUVECs. The expression level of miR-20b was decreased with the increased concentration of H2O2. In normal HUVECs without H2O2 stimulation, miR-20b targeted and suppressed TXNIP and NLRP3 mRNAs, causing decreased cleaved Caspase-1. In senescent HUVECs with H2O2 stimulation, the absence of miR-20b induced upregulation of TXNIP and NLRP3, causing increased cleaved Caspase-1. HUVECs, human umbilical vascular endothelial cells; miR, microRNA; TXNIP, thioredoxin interacting protein; NLRP3, NLR family pyrin domain containing 3; SA-β-gal, SA-β-galactosidase.
Discussion
Organismal aging, which refers to the progressive decline of physiological functions, also involves a series of inevitable physiological events (18-21). Manifested as growth stagnation and changes in gene expression profiles, cell senescence is defined by the irreversible detachment of cells from the cell cycle and the loss of proliferative capacity (22,23). Vascular senescence refers to cellular senescence in the vascular system and is closely related to the occurrence of cardiovascular and metabolic disorders (24). Endothelial cell senescence, which can lead to vascular endothelial dysfunction and aging of tissues and organs, is the major risk factor for developing cardiovascular disease (6). A previous study demonstrated that aging of endothelial cells is triggered by a variety of factors such as oxidative stress, ionizing radiation and telomere dysfunction (25). Oxidative stress plays a crucial role in the occurrence of endothelial dysfunction and is characterized by increased oxygen free radical level and damage to organs and tissues (26,27).
A series of structural and functional changes, for example, reduced cell division and proliferation, increased dyeing rate of SA-β-gal, expression of senescence markers and senescent-associated secretory phenotypes could also occur during the process of endothelial cell senescence (28). It has been reported that different concentrations of H2O2 treatment can induce cell senescence in a variety of types of cells (29). In the current study, the HUVECs were stimulated by H2O2 at different concentrations and it was found that the cell viability was decreased, while SA-β-gal positive cells were increased, and cell cycle arrest occurred after H2O2 stimulation intervention. Such results indicated that HUVECs cells treated by H2O2 had typical characteristics of cell senescence and that H2O2 can induce cell senescence through oxidative stress. Meanwhile, the H2O2 reduced miR-20b expression. Interestingly, H2O2 at 10 M did not decrease the cell viability, however, the level of miR-20b was downregulated, suggesting that the miR-20b expression change was more sensitive to H2O2 stimulation than cell viability change and that the reduced expression in miR-20b at 10 M H2O2 stimulation was not enough to cause decreased cell viability.
miRNAs are directly involved in the pathological process of numerous cardiovascular diseases (30). Lou et al (31) pointed out the unique role of miR-20b in controlling tuberculosis progression. Wong et al (32) showed that hsa-miR-20b is downregulated in tumor necrosis factor (TNF)-α-induced senescent microvascular endothelial cells. In addition, miR-20b is associated with aging and tends to be highly-expressed in the thymus of young mice (33) and upregulated in UVB-induced senescent diploid fibroblasts (34). However, the exact mechanisms of miR-20b in the regulation of endothelial cell senescence remains to be further studied, for such a purpose, the present study successfully constructed HUVECs cells with high and low expression of miR-20b. The results showed that the high expression of miR-20b increased cell viability and inhibited cell senescence, while the low expression of miR-20b produced the opposite effects, suggesting that a high level of miR-20b protected endothelial cells and inhibited H2O2-mediated cell senescence. These results indicated that loss of miR-20b expression might be involved in promoting senescence of HUVECs. Additionally, it would be better to perform cell cycle analysis on the miR-20b mimic or miR-20b inhibitor transfected cells. However, the present study focused on the cell senescence phenotype and cell viability, and didn't have enough resources to perform the cell cycle assay in each stage of this experiment. In addition, previous studies in animal models indicate that miR-20b is positively involved in hepatic ischaemia/reperfusion injury (35), breast cancer resistance (36), cardiac hypertrophy (37). However, whether it regulates the cardiovascular senescence in animal model remains unknown.
To study the mechanism of miR-20b in endothelial cell senescence, the potential target genes for miR-20b were predicted by Targetscan and verified by RT-qPCR and dual luciferase reporter. One recent report indicated that SMAD7 is a targeted gene for miR-20b in insulin-resistant skeletal muscle (13). Another recent study also showed that miR-20b is a circulating biomarker associated with type 2 diabetes and can target STAT3 (38). In the current study, SMAD7, STAT3, TXNIP and NLRP3 were all predicted to be the targets for miR-20b by Targetscan. However, RT-qPCR and dual lucif-erase reporter analyses showed that TXNIP and NLRP3 were the main direct target genes for miR-20b, while SMAD7, STAT3 could not be regulated by miR-20b. Nevertheless, the expression of SMAD7 and STAT3 were reduced by H2O2 stimulation. One study showed that depletion of SMAD7 causes β cell aging (39). Another study also indicated that the activation of STAT3 is necessary for TNFα-induced senescence (40). Thus, the present study inferred that SMAD7 and STAT3 may have a role in H2O2 -induced cell senescence, although it has not been confirmed in this study. Additionally, it seems that the luciferase activity of cells transfected with TXNIP-3-UTR could be more severely suppressed by the miR-20b mimic than cells transfected with NLRP3-3-UTR, thus TXNIP was chosen for further exploration. siRNA technology was applied to reduce the expression of TXNIP and detect the role of TXNIP in endothelial cell senescence. It was discovered that siTXNIP increased cell viability, but decreased SA-β-gal positive cells and partially reversed the effects of the miR-20b inhibitor and H2O2 on endothelial cells. Senescent cells are typically characterized by increased expression of cell cell-cycle inhibitors (such as p21 and/or p16), senescence-associated secretion phenotype, DNA damage and induced SA-β-gal activity (41). The senescence markers p16 and p21 play a regulatory role in the process of cell senescence, and can mediate cell cycle arrest and reduce the accumulation of damaged DNA (42,43). RT-qPCR data revealed that the levels of p16 and p21 were downregulated by siTXNIP, and that low level of TXNIP could partially reverse the senescence of endothelial cell induced by miR-20b inhibitor and H2O2.
Endothelial dysfunction is associated with the activation of the TXNIP/NLRP3 inflammasome axis (44). TXNIP plays a critical role in redox regulation, cell growth and other pathways, moreover, the increased expression of TXNIP leads to cell senescence of endothelial cells (45). The NLRP3 inflammasome is involved in the mechanism of cells senescence (46). TXNIP can bind to NLRP3 under oxidative stress, thereby activating the NLRP3 inflammasome (47). The activated NLRP3 inflammasome can further activate Caspase-1, while the activated Caspase-1 increases the expression of pro-inflammatory cytokines and promotes inflammation response and oxidative stress, therefore inducing endothelial cell senescence (48,49). In the current study, siTXNIP reduced the expression of TXNIP, NLRP3 and cleaved Caspase-1. The results reveal that siTXNIP inhibits the activation of the NLRP3 inflammasome and Caspase-1, reduces oxidative stress and reverses the cell senescence induced by oxidative stress.
The Wnt/β-catenin pathway is an important pathway that regulates the biological behavior of cells and plays different roles in diverse tissues and diseases (50). Some studies revealed that the excessive activation of the Wnt/β-catenin signaling pathway is associated with cellular senescence and that the p53/p21 pathway is the main mediator in cell senescence induced via Wnt/β-catenin pathway (51,52). In this study, siTXNIP was found to inhibit Wnt/β-catenin pathway and the present study speculated that TXNIP mainly regulate cell senescence via regulating the Wnt/β-catenin pathway. Moreover, a relatively slight decrease of cleaved Caspase-1 was observed compared with the change in TXNIP and NLRP3. A previous study indicated that the PI3K pathway is also involved in the activation of cleaved Caspase-1 and the present study inferred that TXNIP and NLRP3 regulate the activation of cleaved-Caspase 1; however, it is not the only pathway involved in activating cleaved-Caspase 1 in senescent HUVECs. In addition, as the interleukin (IL)-1β precursor is cleaved by cleaved-Caspase 1, further experiments should be performed to determine the level of IL-1β in HUVECs. Additionally, experiments in animals should be performed to confirm the present conclusion.
In conclusion, the present study demonstrated high expression of miR-20b promotes cell viability and inhibits cell senescence induced by oxidative stress. TXNIP is a direct target gene for miR-20b, and silencing of TXNIP enhances cell viability, reduces the expression of cell-senescence related genes and inhibits the Wnt/β-catenin pathway. The high expression of miR-20b inhibits endothelial cell senescence, which is possibly realized through regulating the TXNIP/NLRP3 axis to inhibit the Wnt/β-catenin pathway.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
Substantial contributions to conception and design: FD and XZ, Data acquisition, data analysis and interpretation: SD, YL, KW and YQ, Drafting the article or critically revising it for important intellectual content: FD and XZ. Final approval of the version to be published: All authors. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved: All authors.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mitchell SJ, Scheibye-Knudsen M, Longo DL, de Cabo R. Animal models of aging research: Implications for human aging and age-related diseases. Annu Rev Anim Biosci. 2015;3:283–303. doi: 10.1146/annurev-animal-022114-110829. [DOI] [PubMed] [Google Scholar]
- 2.Wu IC, Lin CC, Hsiung CA. Emerging roles of frailty and inflammaging in risk assessment of age-related chronic diseases in older adults: The intersection between aging biology and personalized medicine. Biomedicine (Taipei) 2015;5:1. doi: 10.7603/s40681-015-0001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thijssen DH, Carter SE, Green DJ. Arterial structure and function in vascular ageing: Are you as old as your arteries? J Physiol. 2016;594:2275–2284. doi: 10.1113/JP270597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeda M, Hayashi T, Mizuno N, Hattori Y, Kuzuya M. Intermittent high glucose implements stress-induced senescence in human vascular endothelial cells: Role of superoxide production by NADPH oxidase. PLoS One. 2015;10:e0123169. doi: 10.1371/journal.pone.0123169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su JB. Vascular endothelial dysfunction and pharmacological treatment. World J Cardiol. 2015;7:719–741. doi: 10.4330/wjc.v7.i11.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donato AJ, Morgan RG, Walker AE, Lesniewski LA. Cellular and molecular biology of aging endothelial cells. J Mol Cell Cardiol. 2015;89:122–135. doi: 10.1016/j.yjmcc.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang S, Mi X, Chen Y, Feng C, Hou Z, Hui R, Zhang W. MicroRNA-216a induces endothelial senescence and inflammation via Smad3/IKBα pathway. J Cell Mol Med. 2018;22:2739–2749. doi: 10.1111/jcmm.13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Q, Chen K, Gao L, Zheng Y, Yang YG. Thrombospondin-1 signaling through CD47 inhibits cell cycle progression and induces senescence in endothelial cells. Cell Death Dis. 2016;7:e2368. doi: 10.1038/cddis.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dangwal S, Thum T. microRNA therapeutics in cardiovascular disease models. Annu Rev Pharmacol Toxicol. 2014;54:185–203. doi: 10.1146/annurev-pharmtox-011613-135957. [DOI] [PubMed] [Google Scholar]
- 10.Xue Y, Wei Z, Ding H, Wang Q, Zhou Z, Zheng S, Zhang Y, Hou D, Liu Y, Zen K, et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1α in the progression of atherosclerosis. Atherosclerosis. 2015;241:671–681. doi: 10.1016/j.atherosclerosis.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 11.Liu CW, Sung HC, Lin SR, Wu CW, Lee CW, Lee IT, Yang YF, Yu IS, Lin SW, Chiang MH, et al. Resveratrol attenuates ICAM-1 expression and monocyte adhesiveness to TNF-α-treated endothelial cells: Evidence for an anti-inflammatory cascade mediated by the miR-221/222/AMPK/p38/NF-κB pathway. Sci Rep. 2017;7:44689. doi: 10.1038/srep44689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med. 2014;20:368–376. doi: 10.1038/nm.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao D, Hu Y, Fu Y, Wang R, Zhang H, Li M, Li Z, Zhang Y, Xuan L, Li X, et al. Emodin improves glucose metabolism by targeting microRNA-20b in insulin-resistant skeletal muscle. Phytomedicine. 2019;59:152758. doi: 10.1016/j.phymed.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 14.Yamakuchi M, Hashiguchi T. Endothelial cell aging: How miRNAs contribute? J Clin Med. 2018;7 doi: 10.3390/jcm7070170. pii: E170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Lin YJ, Zhen YZ, Wei J, Liu B, Yu ZY, Hu G. Effects of Rhein lysinate on H2O2-induced cellular senescence of human umbilical vascular endothelial cells. Acta Pharmacol Sin. 2011;32:1246–1252. doi: 10.1038/aps.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitsumoto A, Takanezawa Y, Okawa K, Iwamatsu A, Nakagawa Y. Variants of peroxiredoxins expression in response to hydroperoxide stress. Free Radic Biol Med. 2001;30:625–635. doi: 10.1016/S0891-5849(00)00503-7. [DOI] [PubMed] [Google Scholar]
- 18.Bulterijs S, Hull RS, Björk VC, Roy AG. It is time to classify biological aging as a disease. Front Genet. 2015;6:205. doi: 10.3389/fgene.2015.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gems D. The aging-disease false dichotomy: Understanding senescence as pathology. Front Genet. 2015;6:212. doi: 10.3389/fgene.2015.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogrodnik M, Miwa S, Tchkonia T, Tiniakos D, Wilson CL, Lahat A, Day CP, Burt A, Palmer A, Anstee QM, et al. Cellular senescence drives age-dependent hepatic steatosis. Nat Commun. 2017;8:15691. doi: 10.1038/ncomms15691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat Med. 2015;21:1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrido AM, Bennett M. Assessment and consequences of cell senescence in atherosclerosis. Curr Opin Lipidol. 2016;27:431–438. doi: 10.1097/MOL.0000000000000327. [DOI] [PubMed] [Google Scholar]
- 23.Fan LM, Douglas G, Bendall JK, McNeill E, Crabtree MJ, Hale AB, Mai A, Li JM, McAteer MA, Schneider JE, et al. Endothelial cell-specific reactive oxygen species production increases susceptibility to aortic dissection. Circulation. 2014;129:2661–2672. doi: 10.1161/CIRCULATIONAHA.113.005062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katsuumi G, Shimizu I, Yoshida Y, Minamino T. Vascular senescence in cardiovascular and metabolic diseases. Front Cardiovasc Med. 2018;5:18. doi: 10.3389/fcvm.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Bloom SI, Donato AJ. The role of senescence, telomere dysfunction and shelterin in vascular aging. Microcirculation. 2019;26:e12487. doi: 10.1111/micc.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conti V, Corbi G, Simeon V, Russomanno G, Manzo V, Ferrara N, Filippelli A. Aging-related changes in oxidative stress response of human endothelial cells. Aging Clin Exp Res. 2015;27:547–553. doi: 10.1007/s40520-015-0357-9. [DOI] [PubMed] [Google Scholar]
- 27.Liu R, Liu H, Ha Y, Tilton RG, Zhang W. Oxidative stress induces endothelial cell senescence via downregulation of Sirt6. Biomed Res Int. 2014;2014:902842. doi: 10.1155/2014/902842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mistriotis P, Andreadis ST. Vascular aging: Molecular mechanisms and potential treatments for vascular rejuvenation. Ageing Res Rev. 2017;37:94–116. doi: 10.1016/j.arr.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Crowe EP, Tuzer F, Gregory BD, Donahue G, Gosai SJ, Cohen J, Leung YY, Yetkin E, Nativio R, Wang LS, et al. Changes in the transcriptome of human astrocytes accompanying oxidative stress-induced senescence. Front Aging Neurosci. 2016;8:208. doi: 10.3389/fnagi.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bu H, Wedel S, Cavinato M, Jansen-Dürr P. MicroRNA regulation of oxidative stress-induced cellular senescence. Oxid Med Cell Longev. 2017;2017:2398696. doi: 10.1155/2017/2398696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou J, Wang Y, Zhang Z, Qiu W. MiR-20b inhibits myco-bacterium tuberculosis induced inflammation in the lung of mice through targeting NLRP3. Exp Cell Res. 2017;358:120–128. doi: 10.1016/j.yexcr.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Wong PF, Jamal J, Tong KL, Khor ES, Yeap CE, Jong HL, Lee ST, Mustafa MR, Abubakar S. Deregulation of hsa-miR-20b expression in TNF-α-induced premature senescence of human pulmonary microvascular endothelial cells. Microvasc Res. 2017;114:26–33. doi: 10.1016/j.mvr.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Guo D, Ye Y, Qi J, Tan X, Zhang Y, Ma Y, Li Y. Age and sex differences in microRNAs expression during the process of thymus aging. Acta Biochim Biophys Sin (Shanghai) 2017;49:409–419. doi: 10.1093/abbs/gmx029. [DOI] [PubMed] [Google Scholar]
- 34.Greussing R, Hackl M, Charoentong P, Pauck A, Monteforte R, Cavinato M, Hofer E, Scheideler M, Neuhaus M, Micutkova L, et al. Identification of microRNA-mRNA functional interactions in UVB-induced senescence of human diploid fibroblasts. BMC Genomic. 2013;14:224. doi: 10.1186/1471-2164-14-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang B, Bao N, He G, Wang J. Long noncoding RNA HOTAIR regulates autophagy via the miR-20b-5p/ATG7 axis in hepatic ischemia/reperfusion injury. Gene. 2019;686:56–62. doi: 10.1016/j.gene.2018.10.059. [DOI] [PubMed] [Google Scholar]
- 36.Ao X, Nie P, Wu B, Xu W, Zhang T, Wang S, Chang H, Zou Z. Decreased expression of microRNA-17 and microRNA-20b promotes breast cancer resistance to taxol therapy by upregulation of NCOA3. Cell Death Dis. 2016;7:e2463. doi: 10.1038/cddis.2016.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M, Jiang Y, Guo X, Zhang B, Wu J, Sun J, Liang H, Shan H, Zhang Y, Liu J, et al. Long non-coding RNA cardiac hypertrophy-associated regulator governs cardiac hypertrophy via regulating miR-20b and the downstream PTEN/AKT pathway. J Cell Mol Med. 2019;23:7685–7698. doi: 10.1111/jcmm.14641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katayama M, Wiklander OPB, Fritz T, Caidahl K, El-Andaloussi S, Zierath JR, Krook A. Circulating exosomal miR-20b-5p is elevated in type 2 diabetes and could impair insulin action in human skeletal muscle. Diabetes. 2019;68:515–526. doi: 10.2337/db18-0470. [DOI] [PubMed] [Google Scholar]
- 39.Xiao X, Chen C, Guo P, Zhang T, Fischbach S, Fusco J, Shiota C, Prasadan K, Dong H, Gittes GK. Forkhead box protein 1 (FoxO1) inhibits accelerated β cell aging in pancreas-specific SMAD7 mutant mice. J Biol Chem. 2017;292:3456–3465. doi: 10.1074/jbc.M116.770032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kandhaya-Pillai R, Miro-Mur F, Alijotas-Reig J, Tchkonia T, Kirkland JL, Schwartz S. TNFα-senescence initiates a STAT-dependent positive feedback loop, leading to a sustained interferon signature, DNA damage, and cytokine secretion. Aging (Albany NY) 2017;9:2411–2435. doi: 10.18632/aging.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q, Jiang D, Matsuda JL, Ternyak K, Zhang B, Chu HW. Cigarette smoke induces human airway epithelial senescence via growth differentiation factor 15 production. Am J Respir Cell Mol Biol. 2016;55:429–438. doi: 10.1165/rcmb.2015-0143OC. [DOI] [PubMed] [Google Scholar]
- 42.Jin H, Lian N, Bian M, Zhang C, Chen X, Shao J, Wu L, Chen A, Guo Q, Zhang F, Zheng S. Oroxylin A inhibits ethanol-induced hepatocyte senescence via YAP pathway. Cell Prolif. 2018;51:e12431. doi: 10.1111/cpr.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan SY, Awad EM, Oszwald A, Mayr M, Yin X, Waltenberger B, Stuppner H, Lipovac M, Uhrin P, Breuss JM. Premature senescence of endothelial cells upon chronic exposure to TNFα can be prevented by N-acetyl cysteine and plumericin. Sci Rep. 2017;7:39501. doi: 10.1038/srep39501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Yang J, Chen MH, Wang Q, Qin MJ, Zhang T, Chen XQ, Liu BL, Wen XD. Ilexgenin A inhibits endoplasmic reticulum stress and ameliorates endothelial dysfunction via suppression of TXNIP/NLRP3 inflammasome activation in an AMPK dependent manner. Pharmacol Res. 2015;99:101–115. doi: 10.1016/j.phrs.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Riahi Y, Kaiser N, Cohen G, Abd-Elrahman I, Blum G, Shapira OM, Koler T, Simionescu M, Sima AV, Zarkovic N, et al. Foam cell-derived 4-hydroxynonenal induces endothelial cell senescence in a TXNIP-dependent manner. J Cell Mol Med. 2015;19:1887–1899. doi: 10.1111/jcmm.12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin Y, Zhou Z, Liu W, Chang Q, Sun G, Dai Y. Vascular endothelial cells senescence is associated with NOD-like receptor family pyrin domain-containing 3 (NLRP3) inflammasome activation via reactive oxygen species (ROS)/thioredoxin-interacting protein (TXNIP) pathway. Int J Biochem Cell Biol. 2017;84:22–34. doi: 10.1016/j.biocel.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Sun X, Jiao X, Ma Y, Liu Y, Zhang L, He Y, Chen Y. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS-TXNIP-NLRP3 inflammasome. Biochem Biophys Res Commun. 2016;481:63–70. doi: 10.1016/j.bbrc.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 48.Spadaro O, Goldberg EL, Camell CD, Youm YH, Kopchick JJ, Nguyen KY, Bartke A, Sun LY, Dixit VD. Growth hormone receptor deficiency protects against age-related NLRP3 inflam-masome activation and immune senescence. Cell Rep. 2016;14:1571–1580. doi: 10.1016/j.celrep.2016.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He B, Zhang B, Wu F, Wang L, Shi X, Qin W, Lin Y, Ma S, Liang J. Homoplantaginin inhibits palmitic acid-induced endothelial cells inflammation by suppressing TLR4 and NLRP3 inflammasome. J Cardiovasc Pharmacol. 2016;67:93–101. doi: 10.1097/FJC.0000000000000318. [DOI] [PubMed] [Google Scholar]
- 50.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 51.Zhang DY, Wang HJ, Tan YZ. Wnt/β-catenin signaling induces the aging of mesenchymal stem cells through the DNA damage response and the p53/p21 pathway. PLoS One. 2011;6:e21397. doi: 10.1371/journal.pone.0021397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Jia YS, Liu GZ, Sun Q, Zhang F, Ma S, Wang YJ. Role of LncRNA TUG1 in intervertebral disc degeneration and nucleus pulp-osus cells via regulating Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2017;491:668–674. doi: 10.1016/j.bbrc.2017.07.146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author upon reasonable request.