Abstract
Influenza viruses often pose a serious threat to animals and human health. In an attempt to explore the potential of herbal medicine as a treatment for influenza virus infection, eleutheroside B1, a coumarin compound extracted from herba sarcandrae, was identified, which exhibited antiviral and anti-inflammatory activities against influenza A virus. In this study, high-throughput RNA sequencing and isobaric tags for relative and absolute quantification (iTRAQ) assays were performed to determine alterations in the non-coding RNA (ncRNA) transcriptome and proteomics. Bioinformatics and target prediction analyses were used to decipher the potential roles of altered ncRNAs in the function of eleutheroside B1. Furthermore, long ncRNA (lncRNA) and mRNA co-expressing networks were constructed to analyze the biological functions by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. The analysis of RNA sequencing data revealed that 5 differentially expressed ncRNAs were upregulated and 3 ncRNAs were downregulated in the A549 cells infected with A/PR8/34/H1N1, with or without eleutheroside B1 treatment (PR8+eleu and PR8, respectively). Nuclear paraspeckle assembly transcript 1 (NEAT1) was differentially expressed between the PR8 and A549 cell groups. GO and KEGG pathway analyses indicated that eleutheroside B1 took advantage of the host cell biological processes and molecular function for its antiviral and anti-inflammatory activities, as well as for regulating cytokine-cytokine receptor interaction in the immune system, consistent with previous findings. The results of the iTRAQ assays indicated that L antigen family member 3 (LAGE3) protein, essential for tRNA processing, tRNA metabolic processes and ncRNA processing, was down-regulated in the PR8+eleu compared with the PR8 group. In the present study, these comprehensive, large-scale data analysis enhanced the understanding of multiple aspects of the transcriptome and proteomics that are involved in the antiviral and anti-inflammatory activities of eleutheroside B1. These findings demonstrate the potential of eleutheroside B1 for use in the prevention and treatment of influenza A virus-mediated infections.
Keywords: eleutheroside B1, influenza virus, RNA sequencing, iTRAQ
Introduction
Influenza viruses cause worldwide outbreaks and seasonal pandemics and pose serious risks to human and animal health. Due to the lack of proofreading mechanisms in the negative sense RNA genomes during replication, the virus has a high rate of mutation (1). Thus, considerable time is required to update vaccines when novel mutated viruses appear. Thus, it is necessary to continue the development of novel and effective antiviral drugs against the influenza viruses, particularly drugs that do not cause resistance.
Current anti-influenza virus drugs include amantadine, rimantadine, oseltamivir, zanamivir and paramivir, which target the life cycle of a virus (2,3). However, worldwide resistance to these drugs due to prolonged usage and the resulting immuno-compromised status already exists (4,5). For example, the influenza strains H3N2 and pdmH1N1 have been reported to be adamantine-resistant (6) and the H7N9 virus is also resistant to oseltamivir (7). Herbal/plant-based medicine, already a global trend in the pharmaceutical industry, is a potential resource for novel therapeutic agents (8,9). Traditional Chinese Medicine (TCM) has been used in the treatment of illnesses for centuries, including influenza and influenza-like illnesses (10). TCM tends to regulate the host response and to treat disease by using multi-host targets, rendering TCM unlikely prone to drug resistance. Following these insights, there has been an increasing focus on the development of anti-influenza drugs using traditional medicines as novel prospects for influenza management (11,12).
Eleutheroside B1, a coumarin compound extracted from herba sarcandrae, demonstrated a wide spectrum of anti-human influenza virus efficacy in a previous study (13). However, the mechanisms of action of eleutheroside B1 remain elusive. More than 85% of the human genome is transcribed into RNA; however, only <3% of the genome encodes proteins. Thus, most transcripts of the human genome are non-coding RNA (ncRNAs) without protein-coding capacity. The pervasive transcription of the human genome produces thousands of previously unidentified long intergenic ncRNAs (14). Recent studies have proven that ncRNAs, including microRNAs (miRNAs or miRs) and long non-coding RNAs (lncRNAs) play an important role in a number of biological phenomena and human diseases (15-18). Some studies have demonstrated that the segment-specific non-coding sequences of influenza A virus and host non-coding RNA are crucial for influenza A virus replication (19,20). It has also been recognized that some ncRNAs are critically involved in the virus-host interaction as key regulators of transcription or post-transcription during viral infection. There is increasing evidence to indicate the functional involvement of these regulatory miRNAs, vault complex-associated RNAs (vtRNAs) and lncRNAs in influenza virus replication (21,22). For example, some studies have demonstrated that ncRNAs can regulate the activation of pattern recognition receptor (PRR)-associated signaling and transcription factors, as well as the production of interferons (IFNs) and the expression of critical IFN-stimulated genes (ISGs) (23,24). In addition to these ncRNAs, circular RNAs (circRNAs) are a novel type of RNA that, unlike linear RNAs, form a covalently closed continuous loop, and are highly represented in the eukaryotic transcriptome (25,26). Some studies have suggested that circRNAs, derived from both the host and viruses, interact with these double-stranded RNAs (dsRNAs) by binding antiviral proteins and interface with the host-virus interaction (27,28). The present study investigated whether eleutheroside B1 regulates host ncRNAs and cellular pathways for its anti-influenza virus and anti-inflammation activities.
High-throughput methods for obtaining biological data, such as RNA-sequencing and isobaric tags for relative and absolute quantification (iTRAQ) assay have undergone rapid technological advances that have led to the identification of novel molecules for the effective treatment of diseases, and the investigation of the underlying mechanisms of actions and the specific targets of interactions, such as DNA, RNA, protein and enzymes (29,30). Previous studies have investigated the mRNA expression profiles of A549 cells, following infection by the influenza virus and treatment with potential drug targets (31,32). However, these studies were limited to mRNA, and did not provide an overview of the transcriptome-wide responses to changes in host cells, nor explored their association with the differentially expressed ncRNAs and proteins. In the present study, human lung cancer cells (A549) were treated with eleutheroside B1 following influenza A virus infection, in order to assess the pathway profiles of ncRNAs and proteins. RNA-sequencing and iTRAQ assay were used to comprehensively investigate the mechanisms responsible for the eleutheroside B1 activity against influenza A viral infection. Moreover, molecular insights from target gene enrichment of altered ncRNAs and differentially expressed proteins provided critical knowledge to enhance the understanding of the anti-influenza virus mechanisms of eleutheroside B1.
Materials and methods
Compound, cells and viruses
Eleutheroside B1, extracted from Sarcandra glabra (also known as herba sarcandrae) was characterized by proton and carbon nuclear magnetic resonance (1H and 13C NMR) spectroscopy as previously described (13,32). The results of ultra-performance liquid chromatography in time-of-flight mass spectrometry indicated >89% purity for eleutheroside B1. Eleutheroside B1 was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 50 mg/ml and stored at −20°C.
Human alveolar basal epithelial adenocarcinoma cells (A549 cells) were purchased from the American Tissue Culture Collection (ATCC) and cultured in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS) under standard conditions (37°C, 5% CO2). Influenza virus A/PR/8/34 (H1N1) was also purchased from ATCC and propagated in the allantoic cavities of chicken eggs (9 days). After 48 h, these chicken eggs were broken for the collection of chicken embryo allantoic fluid with the influenza virus.
Cell culture, viral infection and sample preparation
A549 cells were trypsinized with 0.25% trypsin, containing 10 mM EDTA (pH 7.4), and seeded in 6-well culture plates (BD Bioscciences) at up to 80% confluence. Following 24 h, the A549 cells were infected with A/PR/8/34 (H1N1) (MOI=0.1), and incubated with serum-free medium at 37°C. After removing the inoculums, the cells were treated with or without eleutheroside B1 (100 µg/ml, TC50=250 µg/ml) (13). The cells were lysed in TRIzol reagent (Thermo Fisher Scientific, Inc.), following 24 h of infection, for RNA sequencing and iTRAQ assay. All samples were stored at -80°C.
RNA sequencing
The total RNA extracts from each sample were obtained following the manufacturer's instructions (Thermo Fisher Scientific, Inc.). The quality of the RNA was evaluated by electrophoresis on a 1% agarose gel. The A260/A280 ratio was determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific, Inc.) and was accepted between 1.8 and 2.0. RNA integrity was assessed by Agilent 2100 Tape Station analysis (Agilent Technologies, Inc.) and RIN >7 were found to be acceptable (33). RNA sequencing was performed on an Illumina X-ten RNA-seq sequence production system (Illumina, Inc.).
Identification of novel ncRNAs
The raw data were first filtered to eliminate low-quality reads, based on the reading mapping to the reference genome, and StringTie was used to assemble clean data. The combined transcripts were annotated using the GFFCompare program. The filtering of the RNA coding for the putative protein was performed using the minimum length and the threshold of the number of exons. ncRNAs included: i) Small ncRNAs (<200 nt), such as miRNAs, small interfering RNA (siRNA) and PIWI-interacting RNAs (piRNAs); ii) ncRNAs, such as lncRNAs (transcript lengths of >200 nt); and iii) long antisense RNAs (34). CPC/CNCI/PFAM was used to further screen the coding and non-coding genes. The coding potential score of <1 was considered as a novel ncRNA by CPC software (35).
Prediction of the target and functional analysis
Many ncRNAs, particularly intergenic ncRNAs can regulate gene transcription via different mechanisms, including cis-regulatory mechanisms. A previous study demonstrated that intergenic ncRNAs were more likely to be similar to regulatory genes, and intergenic ncRNAs located within 5 kb gene-flanking regions as 'gene-proximate intergenic ncRNAs' (36). These intergenic ncRNAs are potentially cis-regulatory and often regulate regulatory genes (36). Therefore, ncRNA analyses were performed on protein-coding genes that were located in the 5 kb flanking region of the individual cDNAs. Whilst predicting the genes encoding the target protein of a trans-acting ncRNA, the sequence of each ncRNA was obtained, based on the browser of the human genome, and Gene Ontology (GO; http://www.geneontology.org/) was used to functionally classify these neighboring genes in humans (37). For GO analysis, the R package was used to separately prepare biological processes, molecular functions and cellular components. Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) pathway analysis was performed to obtain enriched pathways using the predicted target genes. In order to analyze the regulatory network of differentially expressed ncRNAs and target mRNAs, the DIANA-LncBase database (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex) was used for prediction. In addition, Circnet (38) was used to predict the association between differentially expressed ncRNA and circRNAs. The default parameter in the website was used in this study.
iTRAQ assays
A total of 1×107 cells were mixed with 1,000 µl RIPA buffer (with protease inhibitor cocktail) in 1.5 ml tubes, then sonicated on ice. The cells were treated with or without eleutheroside B1 (100 µg/ml) 24 h following infection and were used for both iTRAQ-based proteomic analysis and RNA sequencing. Following centrifugation for 15 min at 4°C at 12,000 × g the supernatant was transferred to new tubes and kept on ice. The protein in the supernatant was quantified using the BCA assay (Pierce BCA Protein Assay kit; Thermo Fisher Scientific, Inc.) and detected by SDS electrophoresis. Peptides were obtained following reduction, alkylation, acetone precipitation and trypsin digestion. Equal amounts of peptides from each sample were individually labeled with specific TMT reagents (126C, 127C, 128C, 129C, 130C). After cleaning with sodium deoxycholate, the peptides were desalted using C18 solid phase extraction. A total of 100 µg peptides were fractionated to 120 fractions with high pH RPRP-HPLC and combined to eight fractions. For each fraction, 2 µg peptide were separated and analyzed with a Nano-HPLC/EASY-nLC1200 (Thermo Fisher Scientific, Inc.) coupled to Q-Exactive mass spectrometry (Thermo Finnigan). Separation was performed using a reversed-phase column (100 µm, ID × 15 cm, Reprosil-Pur 120 C18-AQ, 1.9 µm, Dr. Math). Mobile phases contained H2O with 0.1% FA, 2% ACN (phase A) and 80% ACN, 0.1% FA (phase B). Samples were separated with a 120 min gradient at 300 nl/min flow rate. Gradient B: 5% for 3 min, 8-35% for 92 min, 35-45% for 20 min, 45-100% for 2 min, 100% for 2 min, 100-2% for 2 min and 2% for 2 min.
Data acquisition and iTRAQ analysis
Data-dependent acquisition was performed in profile and positive mode with a Orbitrap analyzer at a resolution of 70,000 (200 m/z) and m/z range of 350-1,600 for MS1. For MS2, the resolution was set to 17,500 (200 m/z) with a fixed first mass of 120 m/z. The automatic gain control (AGC) target for MS1 was set to 3.0e+06 and 1.0e+05 for MS2. The top 20 most intense ions were fragmented by HCD with normalized collision energy (NCE) of 32%, and isolation window of 2 m/z. The dynamic exclusion time window was 30 sec. Raw MS files were processed with MaxQuant (version 1.5.6.0). The Human protein sequence database (Uniprot_HUMAN_2016_09) was downloaded from UNIPROT. This database and its reverse decoy were used by MaxQuant software. The quantification type was reporter ion MS2 with 6-plex TMT specific to Lys (K) and unmodified N-term; Filter by PIF (0.75). Trypsin was set as specific enzyme with up to 2 miss cleavage; oxidation [M] and acetyl [protein N-term] were considered as variable modification, Carbamidomethyl [C] was set as fixed modification; min peptide length was 7 and max peptide mass was 4600. Both peptide and protein FDR should be <0.01. Only unmodified unique peptides were used for quantification. The iBAQ label-free quantification was also measured with log fit checked. All the other parameters were reserved as default.
All the identified proteins were annotated and classified by GO and KEGG. The differentially expressed proteins were then processed by DAVID Functional Annotation Tool 6.8 (http://david.abcc.ncifcrf.gov/) for the analysis of term enrichment. The results were filtered on the basis of a Fisher Exact statistical methodology, as previously described. The GO biological network was evaluated using the ClueGO of Cytoscape software (http://www.cytoscape.org/, Cytoscape 3.7.2). The analysis of the protein-protein interaction was performed by STRING v10.0 (http://www.string-db.org/), and a high coefficient value of 0.7 was used as a factor reduction. A cluster analysis was performed to identify the protein expression profiles differentially expressed (fold change ≥1.2 or ≤0.8 and P<0.05) using hcluster (https://pypi.python.org/pypi/hcluster/0.2.0).
Integrative analysis of proteome and transcriptome data
Transcriptome data obtained from a previous study (39) was used for integrative analysis with proteome data. In order to identify proteins that have been consistently expressed at RNA and protein levels, the differentially expressed proteins were compared with differentially expressed ncRNAs. Through previous studies and the starBase database (40), potential target genes of the differently expressed ncRNAs were predicted. The predicted potential target genes of the differently expressed ncRNAs were also compared with the differentially expressed proteins. The interaction network of the corresponding proteins and ncRNAs related genes was constructed by the STRING v10.0 software and Cytoscape software.
Reverse transcription-quantitative PCR (RT-qPCR)
The A549 cells were seeded in a 6-well plate at 37°C with 5% CO2, and then infected with influenza virus (PR8, 0.1 MOI). Following incubation for 2 h, the cells were treated with eleutheroside B1 (100 µg/ml). At 24 h post-infection, the cells were collected for the mRNA expression testing of selected genes [nuclear paraspeckle assembly transcript 1 (NEAT1) and L antigen family member 3 (LAGE3)] by RT-qPCR. The primer sequences of NEAT1 and LAGE3 are as presented in Table I. RNA was extracted with RnaExTM Total RNA Isolation Solution (GENEray, Inc.). The production of cDNA was then achieved using the Rayscript cDNA Synthesis kit (GENEray, Inc.) with 60 min at 37°C, and 5 min at 85°C. Subsequently, cDNA was used for qPCR using SYBR-Green Power qPCR PreMix (GENEray, Inc.). Primers of NEAT1 and LAGE3 were designed with Entrez Gene: 283131 and Entrez Gene: 8270. The thermocycling conditions were 1 cycle conditions including 10 min of initial denaturation at 95°C and 40 cycles of 10 sec denaturation at 95°C, 34 sec annealing at 60°C, 15 sec denaturation at 95°C, and 1 solubility curve cycle of 60 sec of annealing at 60°C, 30 sec annealing at 95°C, 15 sec annealing at 60°C. The method of quantification used was that of Livak and Schmittgen (2−ΔΔCq) (39).
Table I.
The primers of NEAT1, LAGE3 and GAPDH mRNA.
| Gene | Primers | Sequence (5′→3′) |
|---|---|---|
| NEAT1 | Forward | GTTCCGTGCTTCCTCTTCTG |
| Reverse | GTGTCCTCCGACTTTACCAG | |
| LAGE3 | Forward | AAACCGCAGCCTCTCAAC |
| Reverse | TCTCTGTGGCTCCTTCCC | |
| GAPDH | Forward | GCTGAGTATGTTGTGGAGTC |
| Reverse | GCAGAAGGAGCAGAGATGA |
NEAT1, nuclear paraspeckle assembly transcript 1; LAGE3, L antigen family member 3.
Western blot analysis
Eleutheroside B1 (100 µg/ml) was added to the A549 cells following 2 h of incubation with influenza virus (PR8) at 37°C. The samples, including A549 cells, A549 cells infected with PR8, PR8-infected A549 cells plus eleutheroside B1 (100 µg/ml) and A549 cells treated with eleutheroside B1 (100 µg/ml) were collected for protein extraction. Whole cell extracts were obtained by using cell lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, protease inhibitors, 1 mM PMSF). As determined by BCA assay, 1 µg proteins in the cell extract was loaded per lane and run on 10% SDS-PAGE gel, separated, transferred to PVDF membranes through a Trans-Blot Semi-Dry transfer machine, then blocked with PBST with milk (5% w/v milk, 0.05% v/v Tween in PBS), followed by incubation in PBST containing milk and incubation with a 1:1,000 dilution of the primary antibody to LAGE3 (PA5-46520, Thermo Fisher Scientific, Inc.) overnight at 4°C and a 1:2,000 dilution goat anti-rabbit IgG cross-adsorbed secondary antibody (G-21234, Thermo Fisher Scientific, Inc.) for 1 h at room temperature. The membrane was covered with ECL detection reagent and detected by the Bio-Rad ChemiDoc™ MP system. The gray value of the result was analyzed using ImageJ software (https://imagej.en.softonic.com/mac) and GraphPad Prism 7 (GraphPad Software Inc).
Statistical analysis
Differences between 2 groups were analyzed using a Student's t-test. The differences between multiple groups were analyzed using one-way ANOVA with Fishers' Least Significant Difference test. A value of P<0.05 was considered to indicate a statistically significant difference. Genes in 2 groups, whose |logFC|>2 and q value were <0.05, were defined as differential expression genes in the present study. For RNA-seq and iTRAQ, 3 repeated samples in each group were combined for testing. For RT-qPCR and western blot analysis, 3 independent experiments were performed.
Results
Sequencing and alignment
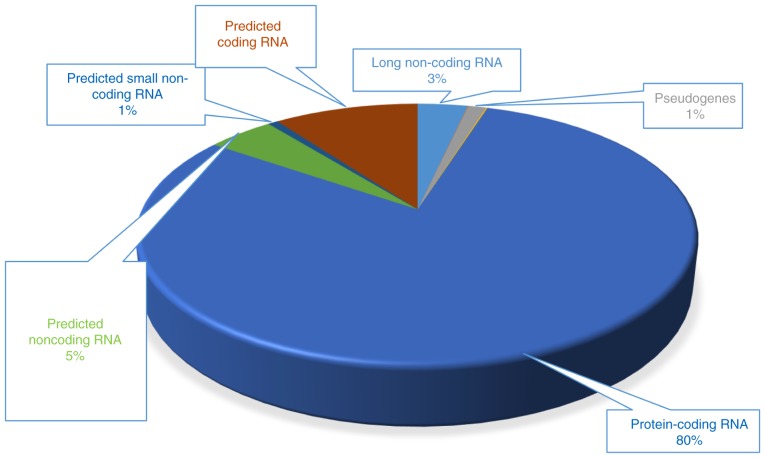
To elucidate the molecular mechanisms of action of eleutheroside B1 against influenza A virus infection, RNA-seq of influenza A virus-infected human lung epithelial (A549) cells were performed in the presence or absence of eleutheroside B1 treatment using an Illumina X-ten RNA-seq sequence production system. There were 3 experimental conditions: A549 cells without infection (A549), A549 cells infected with A/PR8/34/(H1N1) (PR8) and A549 cells infected with A/PR8/34/(H1N1) and treated with Eleutheroside B1 (100 µg/ml) (PR8+eleu). More than 13 million raw reads for each sample were generated from constructed RNA-seq libraries. After filtering, 34.75 M (97.11%, A549), 33.97 M (95.93%, PR8), 34.79 M (95.45%, PR8+eleu) sequencing reads were unambiguously mapped against the human reference genome. Furthermore, CPAT v.1.2. 2 was used to predict whether these transcripts are coding or non-coding. The results demonstrated that most of these were protein-coding RNAs, and some were novel transcripts which were not found in the database (Fig. 1). For non-coding RNAs, predicted RNAs indicate mRNAs which are near to ncRNAs and exhibit similar functions. The expression value (genes) for each sample is summarized in Table II.
Figure 1.
RNA response in A549 cells following eleutheroside B1 treatment. Following RNA sequencing, transcripts were predicted as coding or non-coding RNA. In addition, the percentage of protein-coding RNA, some novel transcripts and long non-coding RNA were calculated.
Table II.
Summary of expression value for samples.
| #SN | >0 | >1 | >2 | >3 |
|---|---|---|---|---|
| A549 | 37,994 | 35,031 | 30,627 | 24,075 |
| PR8 | 38,477 | 35,335 | 30,157 | 23,529 |
| PR8+eleu | 38,217 | 34,855 | 29,385 | 23,100 |
According to data from Illumina X-ten RNA-seq sequence production system (Illumina, Inc.), there were 37,994 genes, and their expression value was >0 (SN >0).
Non-coding RNA expression profiles in A549 cells infected with influenza A virus
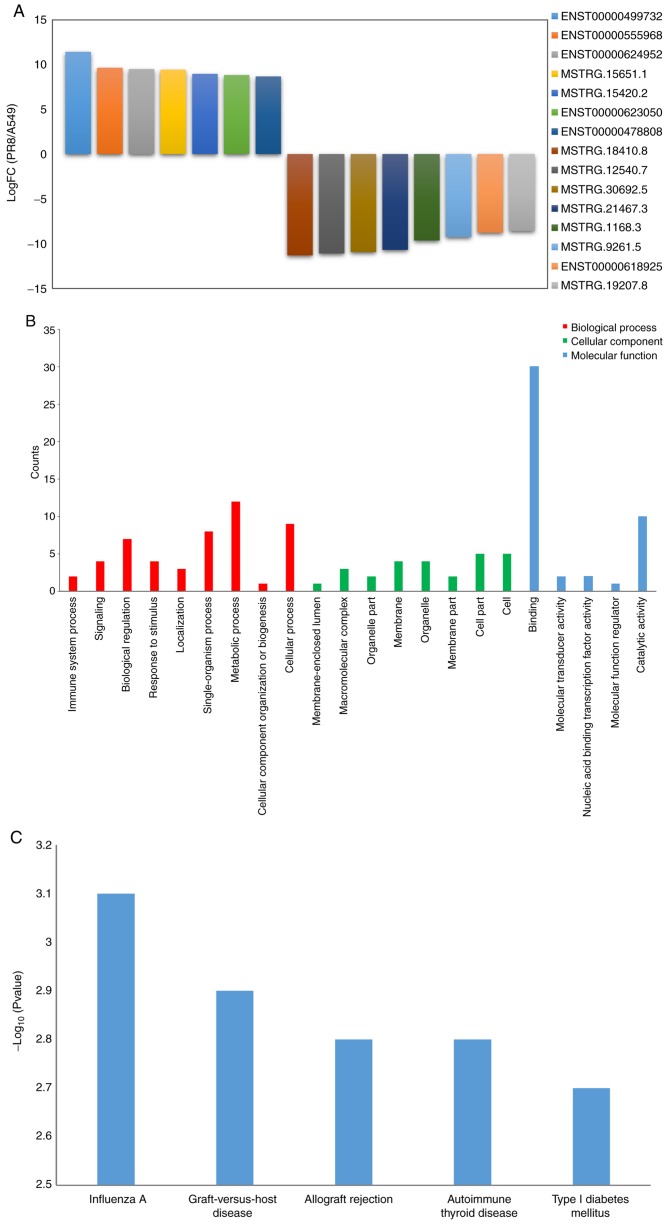
In order to determine specific ncRNA expression induced by the influenza A virus, the differentially expressed non-coding RNAs between the uninfected A549 cells and the virus-infected cells were analyzed. Compared with the control cell group, 7 ncRNAs that were upregulated and 8 ncRNAs that were downregulated were identified in the A549 cells following influenza virus infection (logFC=log2|PR8/A549|); q≤0.05). The differentially expressed ncRNAs included NEAT1, RP11-66N24.3, R P11- 609D21.3, MST RG.15651, MST RG.15420, RP11-717F1.2, USP30-AS1, MSTRG.18410, MSTRG.12540, MST RG. 3 0 69 2, MST RG. 214 67, MST RG.116 8, MSTRG.9261, MALAT1 and MSTRG19207 (alternative transcript names: ENST00000499732, ENST00000555968, ENST00000624952, MSTRG.15651.1, MSTRG.15420.2, ENST00000623050, ENST00000478808, STRG.18410.8, MSTRG.12540.7, MSTRG.30692.5, MSTRG.21467.3, MSTRG.1168.3, MSTRG.9261.5, ENST00000618925 and MSTRG.19207.8) (Fig. 2A). Of these ncRNAs, NEAT1 is a lncRNA that is upregulated in both influenza A virus and HSV-1 infections, causing larger paraspeckles. NEAT1 also modulates HIV-1 post-transcriptional expression. To predict the function of differentially expressed ncRNAs, the genes closely associated with ncRNAs were analyzed by the GO and KEGG pathways. In the present study, 4,091 of these intergenic ncRNAs were identified, and 14,969 genes were predicted to interact with these ncRNAs. Following statistical analysis (logFC=log2 (PR8/A549), q≤0.05), 7 upregulated ncRNAs and 8 downregulated ncRNAs were selected for GO classification and KEGG pathway analysis. According to the GO classification, these target genes were enriched (P<0.05) in the biological process (localization, metabolic process, cellular process and single-organism process); cellular component (cell part, membrane part and macromolecular matrix); and molecular function (binding and catalytic activity) (Fig. 2B). The KEGG pathway analysis revealed that these genes were involved in influenza A, graft-versus-host disease, allograft rejection, autoimmune thyroid disease, type I diabetes mellitus and RIG-I-like Receptor (Fig. 2C).
Figure 2.
Enrichment analysis results of differentially expressed ncRNAs regulated by influenza virus in A549 cells. (A) In total, 7 ncRNAs were upregulated and 8 ncRNAs were downregulated in A549 cells following influenza virus infection. (B) Differentially expressed ncRNAs were enriched in Gene Ontology term according to biological process, cellular component and molecular function. (C) Differentially expressed ncRNAs were analyzed through the Kyoto Encyclopedia of Genes and Genomes pathways. ncRNA, non-coding RNAs.
Expression profile of ncRNAs in pulmonary epithelial cells infected with influenza A virus (A549) and treated with eleutheroside B1
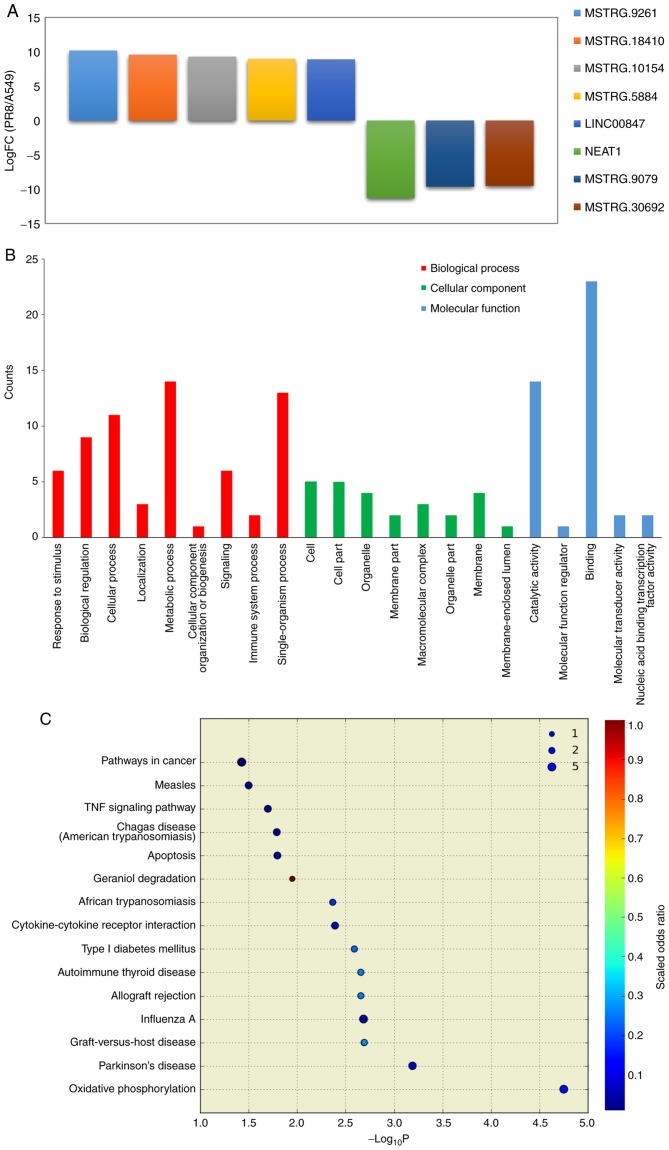
In order to identify non-coding RNA expression in influenza virus-infected cells treated with eleutheroside B1, the PR8+eleu and PR8 groups were compared and 5 upregulated and 3 downregulated ncRNAs were identified (Fig. 3A), namely NEAT1, MSTRG.9261, MSTRG.9079, MSTRG.18410, MSTRG.30692, MSTRG.10154, MSTRG.5884, LINC00847 (alternative transcript names: ENST00000499732, MSTRG.9261.5, MSTRG.9079.2, MSTRG.18410.8, MSTRG.30692.23, MSTRG.10154.6, MSTRG.5884.8, ENST00000502162). NEAT1 was downregulated in the influenza virus-infected cells treated with eleutheroside B1, although it was upregulated in the influenza virus-infected cells without drug treatment. In order to determine differences in ncRNA expression between the 2 groups, genes associated with ncRNAs were also analyzed through GO and KEGG analysis to predict their function. The GO enrichment analysis demonstrated that the majority of these ncRNAs were enriched (P<0.05) for biological process (single-organism process, biological regulation, metabolic process, cellular process); cellular component (macromolecular complex, organelle, membrane, cell part, cell); and molecular function (catalytic activity, binding) with some variations from those predicted by ncRNA (Fig. 3B). According to the KEGG pathway analysis, the genes associated with the differentially expressed ncRNAs were involved in oxidative phosphorylation, Parkinson's disease, graft-versus-host disease, influenza A, allograft rejection, autoimmune thyroid disease, type I diabetes mellitus and cytokine-cytokine receptor interaction (Fig. 3C). GO and KEGG analysis of the differentially expressed ncRNAs between the PR8 and A549 cells suggested similar pathways, such as catalytic activity, binding (GO enrichment) and influenza A (KEGG pathway analysis), which were also enriched by differentially expressed ncRNAs in the PR8+eleu/PR8 cells.
Figure 3.
Enrichment analysis results of differentially expressed ncRNAs in A549 cells with treatment of eleutheroside B1 following influenza virus infection. (A) In total, 5 upregulated and 3 downregulated ncRNAs were identified in the PR8+eleu and PR8 groups. (B) Gene Ontology analysis of ncRNAs were performed in the groups PR8+eleu vs. PR8. (C) Kyoto Encyclopedia of Genes and Genomes pathway analysis for ncRNAs in A549 cells treated with eleutheroside B1. ncRNA, non-coding RNAs.
Regulatory network of ncRNAs and mRNAs
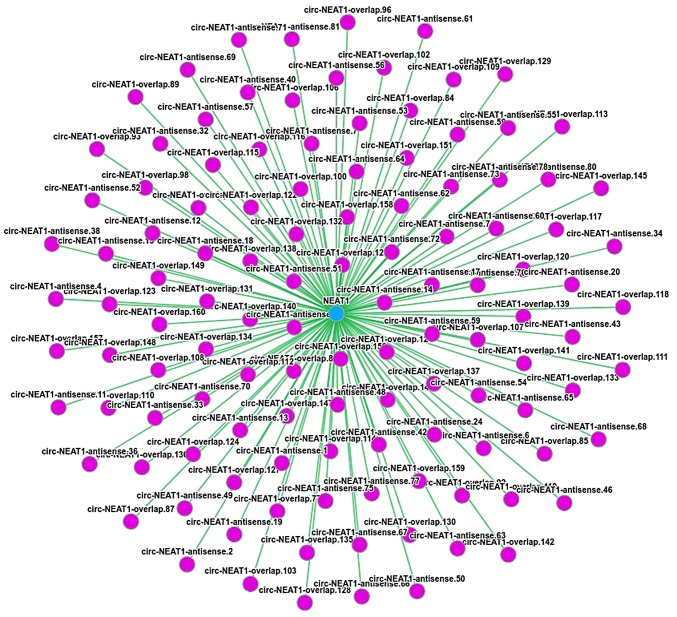
To date, previous results have demonstrated that lncRNAs have at least 6 regulatory functions, such as directly regulating the structure of DNA, transcription and translation of RNAs (41). lncRNAs also inhibit the target gene regulation of miRNAs to indirectly regulate gene expression. Conversely, ncRNAs may target different parts of an mRNA for its function. In this study, to examine the molecular mechanisms of ncRNA involvement in influenza virus infection, cells were treated with eleutheroside B1. The regulatory network analyses of differentially expressed ncRNAs and mRNAs were performed through predication on the DIANA-LncBase database (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex). In the predication analysis, NEAT1 targeted 144 mRNAs through miRNAs (Table III). According to the predication results in Circnet (38), NEAT1 exhibited an association with 120 circRNAs (Fig. 4). These results illustrated the regulatory association between ncRNAs and mRNAs in the mechanisms of eleutheroside B1 in inhibiting the influenza virus.
Table III.
Predicted miRNAs and mRNA interacting with NEAT1.
| lncRNA | miRNA | mRNA |
|---|---|---|
| NEAT1 | hsa-miR-23b-3p | ATG12, PRAP1, HIF1A, MET, BECN1, ATG12, HMGA2, TSC1, NOTCH1, RGS5, PLAU, SRC, MAP3K14, TSC1, NKX3-2, CCNG1, MAP3K14, CA2VEGFA, HIP1R, TRAF5, ATG12, TRAF5, CDH1, IL6R, PPARGC1A, MAP3K1, VEGFA, ETS1, HAS2, VHL, ZEB1, TAB2, NOTCH2, TAB3, TGIF1, KLF3, MYC, ZNF71, HMGB2, PRDX3, TAB3, MAP3K1, VEGFA, CCND1, FOXA1, ST7LFZD5, HOXB4, LPAR1 |
| NEAT1 | hsa-miR-7-5p | SNCA, RGS5, FANCG, IRS2, SMARCD1, BCL2, RAF1, TET2, EGFR, PIK3CD, REL, HOXB5, BCL2, IGF1R, PSME3, PIK3CD, FOS, GDF5, UBE2A, HOXB3, RNF183, SKP2, XIAP, XRCC2, PIK3CG, FOS, MSH3, KLF4, TET2, PAK1, BAX, CUL5, RNF183, PAX6, KMT5A, RNF183, VDAC1, IRS2,HELLS, RAF1, HOXB5, HOXB3, SRSF1, RELA, RAF1, EGFR, XIAP, TET2, PTK2, IGF1R, MSH3, HELLS, SERPINB5, IRS2, RGS5, RELA, RNF183 |
| NEAT1 | hsa-miR-30a-3p | THBS1, TMEM2, SLC7A6, BECN1, RUNX2 |
| NEAT1 | hsa-miR-146a-5p | RAD54L, TRAF6, BCLAF1, CLIP1, NUMB, IER5L, SOX2, CXCL12, LFNG, ERBB4, LFNG, HDAC7, ZDHHC13, CD86, WASF2, FANCM, NFKB1, TLR2, TGFB1, CCND1 |
| NEAT1 | hsa-miR-224-3p | RB1CC1 |
| NEAT1 | hsa-miR-3928-3p D | ICER1 |
| NEAT1 | hsa-miR-153-5p | RICTOR, TGFB2 |
| NEAT1 | hsa-miR-214-5p | RASSF5, CDK3, IGF1R, CDK3 |
| NEAT1 | hsa-miR-216a-5p | CEMIP, SMAD7, CD44, CDC42 |
| NEAT1 | hsa-miR-328-3p | PLCE1, CD44, H2AFX, CD44, PTPRJ |
| NEAT1 | hsa-miR-23a-3p | STAT3, FOXA1, HMGB2 |
| NEAT1 | hsa-miR-339-5p | BCL6 |
| NEAT1 | hsa-miR-148a-5p | CENPF, CDKN1B |
Figure 4.
A total of 120 circRNAs were predicted to interact with NEAT1 in the database of CircNet. Pink nodes indicate circRNAs, and green color edge illustrate the interaction between the NEAT1 and circRNA. NEAT1, nuclear paraspeckle assembly transcript 1.
Proteomic expression in A549 cells infected with the influenza virus
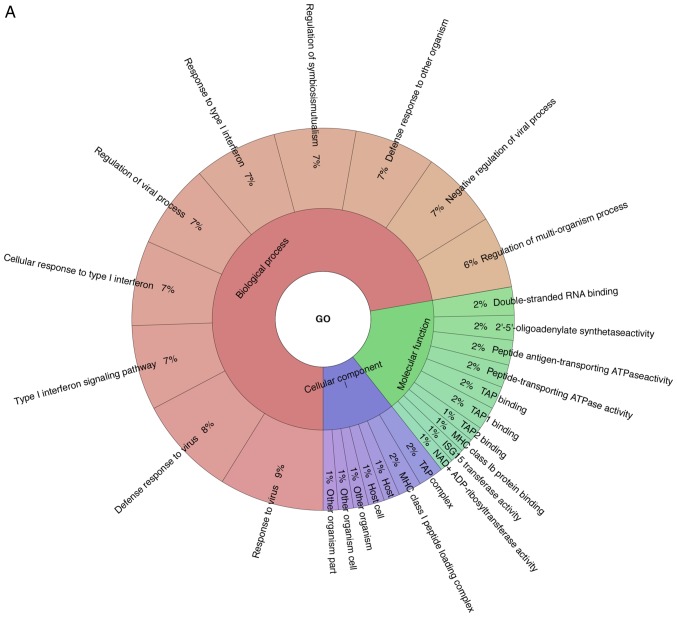
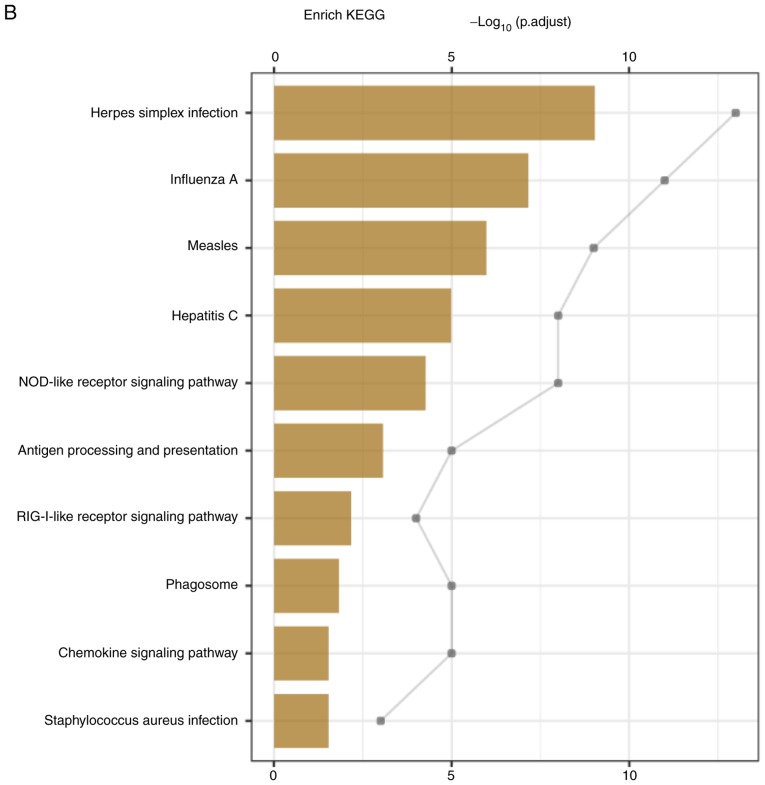
The proteomic expression levels in the A549 cells at 24 h following influenza virus infection were analyzed in order to identify the specific proteomic expression of cells treated with eleutheroside B1. The analysis of the LC-MS/MS data generated a total of 70,249 peptides. A total of 5,809 proteins were identified with at least one unique peptide with a confidence level of >95%. Through GO analysis, 71 differentially expressed proteins were enriched for the biological process, cellular component and the molecular function (Fig. 5A). The GO results demonstrated that the majority of the proteins expressed were enriched in these categories of biological processes, such as virus response, type I interferon signaling pathway, cellular response to type I interferon, regulation of viral process and response to type I interferon. The TAP complex and MHC class I peptide loading complex were the two most abundant categories in cellular component. For molecular function, double-stranded RNA binding, 2′-5′-oligoadenylate synthetase activity, peptide antigen-transporting ATPase activity, peptide-transporting ATPase activity were primary categories for proteins. As indicated by KEGG analysis, these proteins were primarily involved in Herpes simplex virus infection, influenza A virus, measles, hepatitis C virus, NOD-like receptor signaling pathway, antigen processing and presentation, RIG-I-like receptor signaling pathway, phagosome, chemokine signaling pathway, Staphylococcus aureus infection and osteoclast differentiation (Fig. 5B).
Figure 5.
Differentially expressed proteins in A549 cells infected by influenza virus. (A) Enriched in Gene Ontology term, according to biological process, cellular component and molecular function, and data was shown with Krona after enrichment analysis. (B) Proteins were analyzed through the KEGG pathways.
Differentially expressed proteins in influenza virus infection and eleutheroside B1 treatment
Proteins seldom function alone, but rather interact with other proteins to perform various functions. In this study, to explore protein interaction patterns in influenza virus infection and eleutheroside B1 treatment, differently expressed proteins identified were analyzed using STRING software and were enriched through GO and KEGG pathway. A total of 90 proteins were detected from all the test groups. Differentially expressed proteins (fold change of ≥1.5 or ≤0.666 and P<0.05) were identified between the influenza virus-infected cells treated with eleutheroside B1 and the untreated cells. Only one protein (LAGE3) was downregulated in the PR8+eleu vs. PR8 group, and 70 upregulated and 5 downregulated proteins in the PR8+eleu vs. cells group, in which no significance difference was found for the expression of LAGE3 (data not shown).
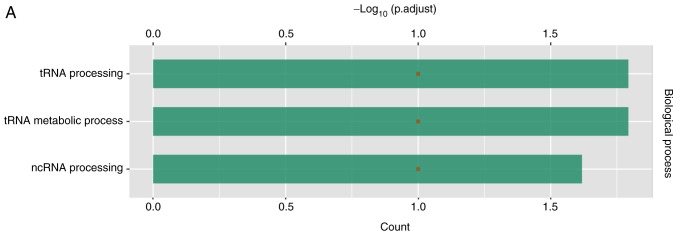
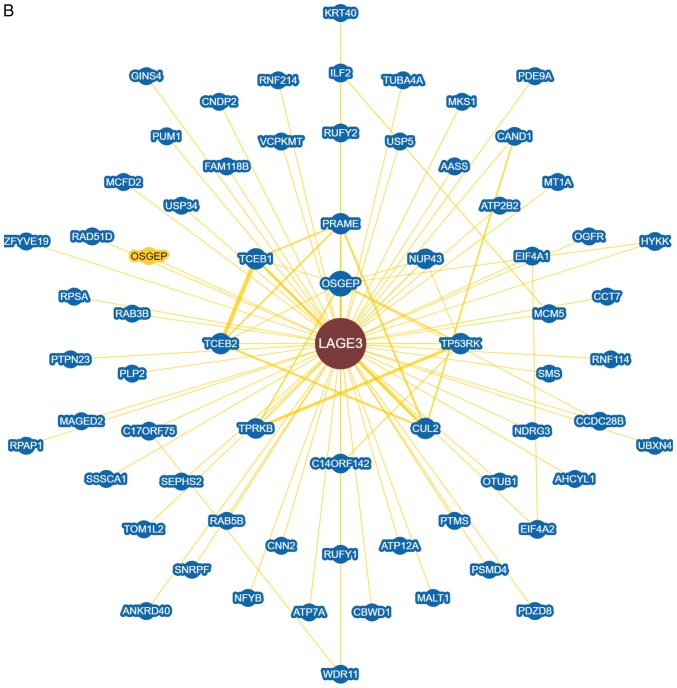
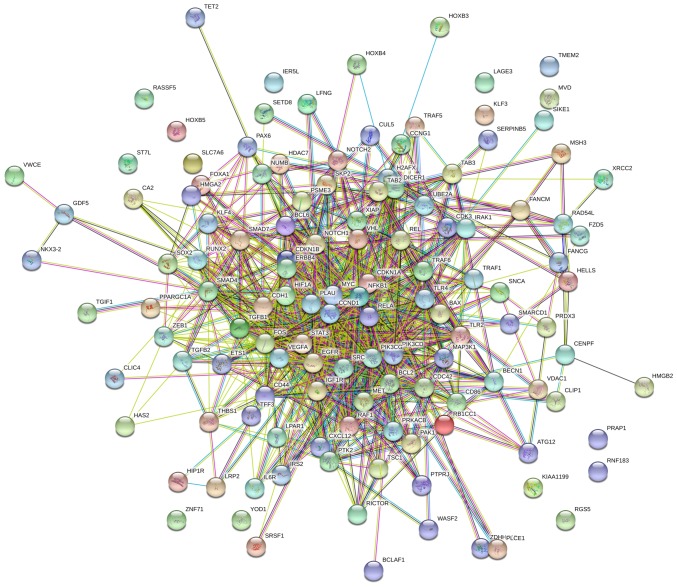
According to GO analysis, LAGE3 was involved in tRNA processing, tRNA metabolic process and lncRNA processing (Fig. 6A). Through analysis with STRING, LAGE3 was shown to interact with a series of other proteins (Fig. 6B).
Figure 6.
Differentially expressed proteins in A549 cells treated with eleutheroside B1 following influenza virus infection. (A) Enriched in Gene Ontology terms. (B) LAGE3 interacted with other proteins through string analysis. LAGE3, L antigen family member 3.
Interaction network of integrative proteome and transcriptome analysis
Integrative analysis of the proteome and transcriptomes were performed to determine the mechanisms through which proteins are regulated by eleutheroside B1 at the translational and transcriptional level. LncRNAs can regulate gene expression by acting on miRNAs. Based on prediction, the differentially expressed ncRNA NEAT1 in the PR8+eleu vs. PR8 groups exhibited an interaction with 144 proteins through miRNAs. The differential expression of LAGE3 in the 2 groups was regulated in the same manner as its mRNAs in the RNA sequences (data not shown). The potential ncRNA targets were used with proteomics data to construct an interaction network, which indicated that the potential ncRNA targets, such as ATG12, EGFR, CD44, CXCL12 and CDC42 were interacting, whereas there was no direct interaction between these targets and LAGE3 in the database (Fig. 7). The interaction between these proteins was also intricated in the GO terms and KEGG pathways, such as the positive regulation of cellular metabolic process (GO:0031325), regulation of cell proliferation (GO:0042127), protein binding (GO:0005515), enzyme binding (GO:0019899), nuclear part (GO:0044428), microRNAs in cancer (05206), FoxO signaling pathway (04068), indicating that eleutheroside B1 can affect cell function by regulating both ncRNA and protein. Combining the RNA-seq and iTRAQ results, it was demonstrated that eleutheroside B1 regulated some pathways, which were associated with cell proliferation and gene expression at different levels to inhibit the replication of influenza virus.
Figure 7.
Interaction network between NEAT1 potential targets and LAGE3 was built by string. NEAT1, nuclear paraspeckle assembly transcript 1; LAGE3, L antigen family member 3.
Validation of NEAT1 and LAGE3
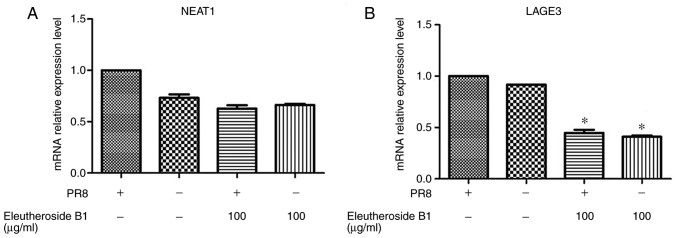
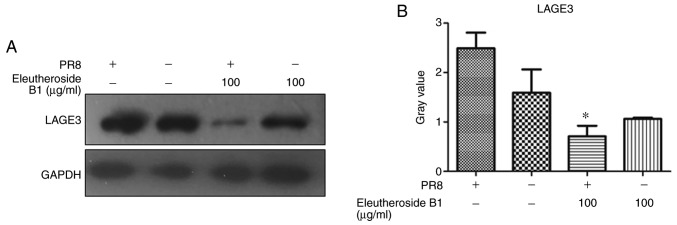
The mRNA expression levels of ncRNA NEAT1 and LAGE3 were decreased by eleutheroside B1 treatment at concentrations of 100 µg/ml (Fig. 8). However, the mRNA expression level of LAGE3 was significantly downregulated by eleutheroside B1 (P<0.05). The effect of eleutheroside B1 on the expression of LAGE3 protein was also confirmed by western blot analysis, and the result revealed a decreased protein expression of LAGE3 in the eleutheroside B1 treatment group (Fig. 9).
Figure 8.
Effects of eleutheroside B1 on the expression of NEAT1 and LAGE3 in the A/PR/8 (MOI=0.1)-infected A549 cells. (A) NEAT1 expression levels. (B) LAGE3 mRNA expression levels. *P<0.05. NEAT1, nuclear paraspeckle assembly transcript 1; LAGE3, L antigen family member 3.
Figure 9.
(A and B) Results of western blot analysis of LAGE3 protein from different samples. The signature for LAGE3 was decreased following treatment with eleutheroside B1 in PR8-infected A549 cells, compared with PR8-infected groups. *P<0.05. LAGE3, L antigen family member 3.
Discussion
The underlying mechanisms of TCM against the influenza virus at the molecular level are complex. High-throughput RNA sequencing and iTRAQ assays are good methods which provide comprehensive analysis of the mRNA, non-coding RNA and proteins induced by disease pathogenesis and drugs (31,42,43). Utilizing these methods, multiple aspects of mRNA, ncRNA and protein expression profiles were profiled and characterized in A549 cells in response to eleutheroside B1, following influenza virus infection. The result of mRNA expression profiles have been previously published (39); thus, only ncRNA and protein expression profiles are shown in this study. The transcriptome analysis revealed 8 differentially expressed ncRNAs in the PR8+eleu vs. PR8 groups (5 upregulated ncRNAs and 3 downregulated RNAs) and 15 differentially expressed ncRNAs in the PR8 vs. A549 cell groups (7 upregulated ncRNAs and 8 downregulated RNAs). In these group comparisons, the same differentially expressed ncRNA (NEAT1) was found to regulate the expression of IL-8 through sequestering splicing factor proline-glutamine rich (SFPQ/PSF) in paraspeckles, and is deregulated by stress, IAV and HSV infection, or poly I:C treatment (22). NEAT1 was also essential in the expression of various genes associated with innate immunity (44). The formation of subnuclear paraspeckle structures depend on transcription and RNA polymerase II in response to stress or cellular metabolic changes (45). The mRNA data analysis also indicated that the effect of eleutheroside B1 on RNA polymerase and RNA synthesis was required for the anti-influenza activity (46). Previous studies have proven that NEAT1 is a crucial innate immune molecule with dual functions (44). On the one hand, it is a critical part of the antiviral response to increase the expression of immune cytokines, which on the other hand, could be 'hijacked' by viruses to facilitate viral gene expression and viral replication (44,47,48). The results of the present study demonstrated that NEAT1 was downregulated by eleutheroside B1 in A549 cells infected with the influenza virus, while it is upregulated in the PR8 vs. the control A549 cells. The induction of NEAT1 by the influenza virus is necessary for the successful replication of the virus. The downregulation of NEAT1 by eleutheroside B1 suggested that eleutheroside B1 may inhibit viral replication and may decrease the expression of immune cytokines through NEAT1; this finding is consistent the findings of a previous study (13). In addition, NEAT1 also regulates some pathways, such as the Wnt/β-catenin signaling pathway (44). In the future, the authors of this study aim to construct NEAT1 overexpression and knockout cells in order to investigate the effects on the anti-influenza virus activity of eleutheroside B1.
The GO and pathway analyses for predicted target genes of altered ncRNAs in the PR8+eleu/PR8 groups revealed that the majority of the genes that were enriched (P<0.05) belong to biological process (single-organism process, biological regulation, metabolic process, cellular process), cellular component (macromolecular complex, organelle, membrane, cell part, cell, and molecular function (catalytic activity, binding). According to the KEGG pathway analysis, these genes associated with the differentially expressed ncRNAs were involved in oxidative phosphorylation, Parkinson's disease, graft-versus-host disease, influenza A, allograft rejection, autoimmune thyroid disease, type I diabetes mellitus, cytokine-cytokine receptor interaction (49). The influenza virus recruits variable and complementary host cellular pathways during its replication cycle (50-52). Current anti-influenza virus medication often targets one part of the virus life cycle, which potentially influences the development of drug-resistance (2). The data suggest that the involvement of eleutheroside B1 in altering cellular pathways is necessary for the virus life cycle, such as binding and cytokine-cytokine receptor interaction, consistent with previous studies (13,53). Cytokine-receptor complexes are ideal drug targets. However, the protein-protein interactions are difficult to block using small molecules (53,54).
Further proteomics analysis of the function of eleuthero-side B1 was performed through iTRAQ assays. Only one protein (LAGE3) was downregulated in the PR8+eleu vs. PR8 groups, whereas 70 were upregulated and 5 proteins were downregulated in the PR8+eleu vs. cell groups. LAGE3 belongs to the NY-ESO gene family, a component of the EKC/KEOPS complex. This complex is required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t6A37) in tRNAs that read codons beginning with adenine (55,56). Some researchers have reported that the EKC/KEOPS complex is linked with the protein synthesis machinery and cell growth (57). According to GO analysis, LAGE3 was involved in tRNA processing, tRNA metabolic processes and lncRNA processing. These differentially expressed proteins could be potential targets of eleutheroside B1 in regulating the ncRNAs of host cells. Transfer RNAs (tRNAs) are substrates for protein synthesis and a central part of the translation machinery in all living organisms, which impact translational speed and fidelity by their abundance. A 'channeled tRNA cycle' includes the following: tRNAs are shuttled directly from the ribosome to their cognate tRNA synthetase and back to the ribosome for another cycle of translation without reentering the cytosolic pool (58,59). The polysome-associated tRNA population changes dramatically, following influenza virus infection; however, the total cellular tRNA population remains unaltered (60). This proves that the changes in polysome-associated tRNA levels reflect the codon usage of viral genes, suggesting the existence of local tRNA pools optimized for viral translation and enhanced translational efficiency of viral genes. Over the past decade, ncRNAs (microRNAs, lncRNAs and vtRNAs) have been identified as an important class of regulators involved in virus-host interaction, particularly in anti-viral immune response (20). Focus on ncRNAs has recently been increased. Advances in the high-throughput sequencing techniques are generating increasing numbers of newly discovered ncRNAs that are involved in infections and immunological processes (61). In some reports, ncRNAs have been shown to regulate histone modifications in influenza virus-infected cells, interact with transcription factors during virus infection and induce specific protein-coding gene expression, affect virus replication in an interferon-independent manner and control protein synthesis during influenza virus infection (62-66). The LAGE3 gene involved in lncRNA processing warrants further investigation to determine the role of eleuthero-side B1 as a lncRNA regulator for its anti-influenza virus.
Overall, the present study demonstrates that ncRNAs are involved in the interaction between host and influenza virus. Although research on non-coding RNA is currently unpopular in Chinese medicine, it is worth noting that ncRNAs, such as NEAT1, the protein LAGE3, and their pathways identified in the present study indicate the need to investigate further the details of the ability of eleutheroside B1 to inhibit influenza virus. One of the limitations of the present study was that it was only performed using A549 cells. It would be ideal to include animal or human samples to profile the RNA and protein expression, particularly in influenza virus-infected patients treated with this drug to compare whether the observed changes are comparable in a clinical setting. Unfortunately, the existing method for extracting eleutheroside B1 does not yield sufficient compounds to allow animal experiments and clinical tests. However, in the future the authors aim to study clinical samples treated with herba sarcandrae. In addition, another limitation is that ncRNA expression was not examined at different time points. Previous research (46) has often selected 24 h after infection for RNA-seq; therefore, this time point was used for detection in this study. In the future, the authors aim to perform further studies using different time points. Another limitation of the present study is that only the iTRAQ assay was used to investigate protein expression, which yields incomplete data due to the use of data-dependent acquisition that only captures one differentially expressed protein. Thus, it will be interesting to examine protein expression profiles induced by eleutheroside B1 by other proteomics methods, being cautious that ncRNAs and proteins may function in a coordinated fashion, and therefore, the overall anti-influenza virus response induced by eleutheroside B1 may be due to the effect of a cumulative response to overall changes in ncRNAs or proteins rather than individual ncRNAs or proteins. Furthermore, certain studies open novel avenues to further explore the molecular pathways based on ncRNA or protein functions. Therefore, whether these networks were induced by eleutheroside B1 in the future should be investigated.
In the validation assays, the RT-qPCR results revealed that the expression levels of ncRNA NEAT1 and mRNA LAGE3 were decreased by eleutheroside B1 (100 µg/ml) treatment. The mRNA expression level of LAGE3 was significantly downregulated by eleutheroside B1 (P<0.05). Western blot analysis also demonstrated that the protein expression of LAGE3 was decreased with eleutheroside B1 treatment. For NEAT1, the ncRNA and not the mRNA, which cannot be investigated in protein level using recent techniques. In this study, only RT-qPCR and western blot analysis were used to examine these genes of interest. However, further molecular assays, such as immunofluorescence assay, should be adopted to examine these genes in the future. In addition, the authors aim to perform further studies in which NEAT1- and LAGE3-overexpressing cells and knockout cells will be constructed to investigate their roles in the anti-influenza virus activity of eleutheroside B1.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (U1502226), the Engineering technology research center (development) of Guangdong general universities (GCZX-A1408), Natural Science Basic Research Program of Shaanxi (Program No. 2019JM-513) and Shijiazhuang Yiling Pharmaceutical Co.,Ltd (Shijiazhuang, China).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
WY wrote the manuscript and performed experiments. JC drafted the manuscript and performed experiments. ZW revised the manuscript and performed experiments. XiaohuW designed, edited and modified the manuscript. ZZ performed experiments and modified the manuscript. DT designed this study and reviewed the manuscript. YutaoW and XinhuaW obtained funds and designed this study. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yoon SW, Webby RJ, Webster RG. Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol. 2014;385:359–375. doi: 10.1007/82_2014_396. [DOI] [PubMed] [Google Scholar]
- 2.Amarelle L, Lecuona E, Sznajder JI. Anti-influenza treatment: Drugs currently used and under development. Arch Bronconeumol. 2017;53:19–26. doi: 10.1016/j.arbres.2016.07.004. In English, Spanish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takamatsu K, Marumo S, Fukui M, Hata A. Safety and efficacy of anti-influenza drugs, intravenous peramivir against influenza virus infection in elderly patients with underlying disease. J Microbiol Immunol Infect. 2017;50:541–544. doi: 10.1016/j.jmii.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Moss RB, Davey RT, Steigbigel RT, Fang F. Targeting pandemic influenza: A primer on influenza antivirals and drug resistance. J Antimicrob Chemother. 2010;65:1086–1093. doi: 10.1093/jac/dkq100. [DOI] [PubMed] [Google Scholar]
- 5.Oh DY, Panozzo J, Vitesnik S, Farrukee R, Piedrafita D, Mosse J, Hurt AC. Selection of multi-drug resistant influenza A and B viruses under zanamivir pressure and their replication fitness in ferrets. Antivir Ther. 2018;23:295–306. doi: 10.3851/IMP3135. [DOI] [PubMed] [Google Scholar]
- 6.Deyde VM, Xu X, Bright RA, Shaw M, Smith CB, Zhang Y, Shu Y, Gubareva LV, Cox NJ, Klimov AI. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis. 2007;196:249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- 7.Yen HL, McKimm-Breschkin JL, Choy KT, Wong DD, Cheung PP, Zhou J, Ng IH, Zhu H, Webby RJ, Guan Y, et al. Resistance to neuraminidase inhibitors conferred by an R292K mutation in a human influenza virus H7N9 isolate can be masked by a mixed R/K viral population. mBio. 2013;4 doi: 10.1128/mBio.00396-13. pii: e00396-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurib-Fakim A. Medicinal plants: Traditions of yesterday and drugs of tomorrow. Mol Aspects Med. 2006;27:1–93. doi: 10.1016/j.mam.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Pan SY, Zhou SF, Gao SH, Yu ZL, Zhang SF, Tang MK, Sun JN, Ma DL, Han YF, Fong WF, Ko KM. New perspectives on how to discover drugs from herbal medicines: CAM's outstanding contribution to modern therapeutics. Evid Based Complement Alternat Med. 2013;2013:627375. doi: 10.1155/2013/627375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Zhang RM, Liu GY, Wei BL, Wang Y, Cai HY, Li FS, Xu YL, Zheng SP, Wang G. Chinese herbs in treatment of influenza: A randomized, double-blind, placebo-controlled trial. Respir Med. 2010;104:1362–1369. doi: 10.1016/j.rmed.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin TJ, Lin CF, Chiu CH, Lee MC, Horng JT. Inhibition of endosomal fusion activity of influenza virus by Rheum tanguticum (da-huang) Sci Rep. 2016;6:27768. doi: 10.1038/srep27768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu W, Li R, Li X, He J, Jiang S, Liu S, Yang J. Quercetin as an antiviral agent inhibits influenza a virus (IAV) entry. Viruses. 2015;8 doi: 10.3390/v8010006. pii: E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Yan W, Chen Q, Huang W, Yang Z, Li X, Wang X. Inhibition viral RNP and anti-inflammatory activity of coumarins against influenza virus. Biomed Pharmacother. 2017;87:583–588. doi: 10.1016/j.biopha.2016.12.117. [DOI] [PubMed] [Google Scholar]
- 14.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He JH, Han ZP, Li YG. Association between long non-coding RNA and human rare diseases (Review) Biomed Rep. 2014;2:19–23. doi: 10.3892/br.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Li N, Shi K, Chen Q. Systematic review and meta-analysis of the utility of long non-coding RNA GAS5 as a diagnostic and prognostic cancer biomarker. Oncotarget. 2017;8:66414–66425. doi: 10.18632/oncotarget.19040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin SY, Jeong JS, Lim JY, Kim T, Park JH, Kim JK, Shin C. Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genomics. 2018;19:532. doi: 10.1186/s12864-018-4897-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie H, Ma B, Gao Q, Zhan H, Liu Y, Chen Z, Ye S, Li J, Yao L, Huang W. Long non-coding RNA CRNDE in cancer prognosis: Review and meta-analysis. Clin Chim Acta. 2018;485:262–271. doi: 10.1016/j.cca.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Ng SS, Li OT, Cheung TK, Malik Peiris JS, Poon LL. Heterologous influenza vRNA segments with identical non-coding sequences stimulate viral RNA replication in trans. Virol J. 2008;5:2. doi: 10.1186/1743-422X-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winterling C, Koch M, Koeppel M, Garcia-Alcalde F, Karlas A, Meyer TF. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol. 2014;11:66–75. doi: 10.4161/rna.27504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, Chen Y, Zhang Z, Ouyang J, Wang Y, Yan R, Huang S, Gao GF, Guo G, Chen JL. Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic Acids Res. 2015;43:10321–10337. doi: 10.1093/nar/gkv1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Ouyang J, Wei J, Maarouf M, Chen JL. Involvement of host non-coding RNAs in the pathogenesis of the influenza virus. Int J Mol Sci. 2016;18 doi: 10.3390/ijms18010039. pii: E39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valadkhan S, Plasek LM. Long non-coding RNA-mediated regulation of the interferon response: A new perspective on a familiar theme. Pathog Immun. 2018;3:126–148. doi: 10.20411/pai.v3i1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Zhang J, Huo C, Ding N, Li J, Xiao J, Lin X, Cai B, Zhang Y, Xu J. Dynamic organization of lncRNA and circular RNA regulators collectively controlled cardiac differentiation in humans. EBioMedicine. 2017;24:137–146. doi: 10.1016/j.ebiom.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okholm TLH, Nielsen MM, Hamilton MP, Christensen LL, Vang S, Hedegaard J, Hansen TB, Kjems J, Dyrskjøt L, Pedersen JS. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom Med. 2017;2:36. doi: 10.1038/s41525-017-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67:228–238.e5. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–227.e7. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Brahmachari G. In: Discovery and development of antidiabetic agents from natural products. 1st edition. Waltham MA, editor. Elsevier Inc; 2016. [Google Scholar]
- 29.Luo R, Zhao H. Protein quantitation using iTRAQ: Review on the sources of variations and analysis of nonrandom missingness. Stat Interface. 2012;5:99–107. doi: 10.4310/SII.2012.v5.n1.a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith SB, Dampier W, Tozeren A, Brown JR, Magid-Slav M. Identification of common biological pathways and drug targets across multiple respiratory viruses based on human host gene expression analysis. PLoS One. 2012;7:e33174. doi: 10.1371/journal.pone.0033174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B, Li J, Liang X, Yang Z, Jiang Z. Transcriptome profiling of influenza A virus-infected lung epithelial (A549) cells with lariciresinol-4-β-D-glucopyranoside treatment. PLoS One. 2017;12:e0173058. doi: 10.1371/journal.pone.0173058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Zhang Y, Zeng X, Yang L, Deng Y. Chemical profiling of bioactive constituents in Sarcandra glabra and its preparations using ultra-high-pressure liquid chromatography coupled with LTQ Orbitrap mass spectrometry. Rapid Commun Mass Spectrom. 2011;25:2439–2447. doi: 10.1002/rcm.5123. [DOI] [PubMed] [Google Scholar]
- 33.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Li C, Chen Y. Small and long non-coding RNAs: Novel targets in perspective cancer therapy. Curr Genomics. 2015;16:319–326. doi: 10.2174/1389202916666150707155851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang YJ, Yang DC, Kong L, Hou M, Meng YQ, Wei L, Gao G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu Z, Adelson DL. Bovine ncRNAs are abundant, primarily intergenic, conserved and associated with regulatory genes. PLoS One. 2012;7:e42638. doi: 10.1371/journal.pone.0042638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu Z, Adelson DL. Identification and comparative analysis of ncRNAs in human, mouse and zebrafish indicate a conserved role in regulation of genes expressed in brain. PLoS One. 2012;7:e52275. doi: 10.1371/journal.pone.0052275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al. CircNet: A database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44(D1):D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database Issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinn JL. lncRNAs: Linking RNA to chromatin. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a018614. pii: a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, Gao Z, Wang X, Gu M, Liang Y, Liu X, Hu S, Liu H, Liu W, Chen S, et al. iTRAQ-based quantitative proteomics reveals important host factors involved in the high pathogenicity of the H5N1 avian influenza virus in mice. Med Microbiol Immunol. 2017;206:125–147. doi: 10.1007/s00430-016-0489-3. [DOI] [PubMed] [Google Scholar]
- 43.Xiao YL, Kash JC, Beres SB, Sheng ZM, Musser JM, Taubenberger JK. High-throughput RNA sequencing of a formalin-fixed, paraffin-embedded autopsy lung tissue sample from the 1918 influenza pandemic. J Pathol. 2013;229:535–545. doi: 10.1002/path.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan W, Zheng C, He J, Zhang W, Huang XA, Li X, Wang Y, Wang X. Eleutheroside B1 mediates its anti-influenza activity through POLR2A and N-glycosylation. Int J Mol Med. 2018;42:2776–2792. doi: 10.3892/ijmm.2018.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Bénard M, Fox AH, Pierron G. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi: 10.1091/mbc.e13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landeras-Bueno S, Jorba N, Pérez-Cidoncha M, Ortin J. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog. 2011;7:e1002397. doi: 10.1371/journal.ppat.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samji T. Influenza A: Understanding the viral life cycle. Yale J Biol Med. 2009;82:153–159. [PMC free article] [PubMed] [Google Scholar]
- 50.Li C, Bankhead A, III, Eisfeld AJ, Hatta Y, Jeng S, Chang JH, Aicher LD, Proll S, Ellis AL, Law GL, et al. Host regulatory network response to infection with highly pathogenic H5N1 avian influenza virus. J Virol. 2011;85:10955–10967. doi: 10.1128/JVI.05792-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo M. Influenza virus entry. Adv Exp Med Biol. 2012;726:201–221. doi: 10.1007/978-1-4614-0980-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudnicka A, Yamauchi Y. Ubiquitin in influenza virus entry and innate immunity. Viruses. 2016;8 doi: 10.3390/v8100293. pii: E293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schreiber G, Walter MR. Cytokine-receptor interactions as drug targets. Curr Opin Chem Biol. 2010;14:511–519. doi: 10.1016/j.cbpa.2010.06.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spangler JB, Moraga I, Mendoza JL, Garcia KC. Insights into cytokine-receptor interactions from cytokine engineering. Annu Rev Immunol. 2015;33:139–167. doi: 10.1146/annurev-immunol-032713-120211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long M, Wang Y, Chen D, Wang Y, Wang R, Gong D, He H, Rock DL, Hao W, Luo S. Identification of host cellular proteins LAGE3 and IGFBP6 that interact with orf virus protein ORFV024. Gene. 2018;661:60–67. doi: 10.1016/j.gene.2018.03.089. [DOI] [PubMed] [Google Scholar]
- 56.Wan LC, Maisonneuve P, Szilard RK, Lambert JP, Ng TF, Manczyk N, Huang H, Laister R, Caudy AA, Gingras AC, et al. Proteomic analysis of the human KEOPS complex identifies C14ORF142 as a core subunit homologous to yeast Gon7. Nucleic Acids Res. 2017;45:805–817. doi: 10.1093/nar/gkw1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rojas-Benitez D, Ibar C, Glavic Á. The Drosophila EKC/KEOPS complex: Roles in protein synthesis homeostasis and animal growth. Fly (Austin) 2013;7:168–172. doi: 10.4161/fly.25227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrushenko ZM, Budkevich TV, Shalak VF, Negrutskii BS, El'skaya AV. Novel complexes of mammalian translation elongation factor eEF1A.GDP with uncharged tRNA and aminoacyl-tRNA synthetase. Implications for tRNA channeling. Eur J Biochem. 2002;269:4811–4818. doi: 10.1046/j.1432-1033.2002.03178.x. [DOI] [PubMed] [Google Scholar]
- 59.Stapulionis R, Deutscher MP. A channeled tRNA cycle during mammalian protein synthesis. Proc Natl Acad Sci USA. 1995;92:7158–7161. doi: 10.1073/pnas.92.16.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pavon-Eternod M, David A, Dittmar K, Berglund P, Pan T, Bennink JR. Vaccinia and influenza A viruses select rather than adjust tRNAs to optimize translation. Nucleic Acids Res. 2013;41:1914–1921. doi: 10.1093/nar/gks986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Landeras-Bueno S, Ortin J. Regulation of influenza virus infection by long non-coding RNAs. Virus Res. 2016;212:78–84. doi: 10.1016/j.virusres.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 62.Barriocanal M, Carnero E, Segura V, Fortes P. Long non-coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor tetherin. Front Immunol. 2015;5:655. doi: 10.3389/fimmu.2014.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D'Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol. 2010;17:815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, Carnero E, Barriocanal M, Fortes P, Anthony DD, Valadkhan S. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014;42:10668–10680. doi: 10.1093/nar/gku713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article or are available from the corresponding author on reasonable request.