To the Editor — Rapid advances in DNA-sequencing and bioinformatics technologies in the past two decades have substantially improved understanding of the microbial world. This growing understanding relates to the vast diversity of microorganisms; how microbiota and microbiomes affect disease1 and medical treatment2; how microorganisms affect the health of the planet3; and the nascent exploration of the medical4, forensic5, environmental6 and agricultural7 applications of microbiome biotechnology. Much of this work has been driven by marker-gene surveys (for example, bacterial/archaeal 16S rRNA genes, fungal internal-transcribed-spacer regions and eukaryotic 18S rRNA genes), which profile microbiota with varying degrees of taxonomic specificity and phylogenetic information. The field is now transitioning to integrate other data types, such as metabolite8, metaproteome9 or metatranscriptome9,10 profiles.
The QIIME 1 microbiome bioinformatics platform has supported many microbiome studies and gained a broad user and developer community. Interactions with QIIME 1 users in our online support forum, our workshops and direct collaborations have shown the platform’s potential to serve an increasingly diverse array of microbiome researchers in academia, government and industry. Here, we present QIIME 2, a completely reengineered and rewritten system that is expected to facilitate reproducible and modular analysis of microbiome data to enable the next generation of microbiome science.
QIIME 2 was developed on the basis of a plugin architecture (Supplementary Fig. 1) that allows third parties to contribute functionality (https://library.qiime2.org). QIIME 2 plugins exist for latest-generation tools for sequence quality control from different sequencing platforms (DADA2 (ref.11) and Deblur12), taxonomy assignment13 and phylogenetic insertion14, which quantitatively improve the results over QIIME 1 and other tools (as detailed in the corresponding tool-specific publications). The plugins also support qualitatively new functionality, including microbiome paired-sample and time-series analysis15 (which are critical for studying the effects of treatments on the microbiome), and machine learning16. Trained machine learning models can be saved for application to new data and interrogated to identify important microbiome features. Several recently released plugins, including q2-cscs17, q2-metabolomics18, q2-shogun19, q2-metaphlan2 (ref.20) and q2-picrust2 (ref.21), provide initial support for analysis of metabolomics and shotgun metagenomics data. We are currently working with teams developing bioinformatics tools for metatranscriptomics and metaproteomics, and we expect to add new plugins supporting these data types to the ecosystem shortly. Additionally, many of the existing ‘downstream’ analysis tools, such as q2-sample-classifier16, can already work with these data types individually or in combination if they are provided in a feature table. Thus, QIIME 2 has the potential to serve not only as a marker-gene analysis tool but also a multidimensional and powerful data science platform that can be rapidly adapted to analyze diverse microbiome features.
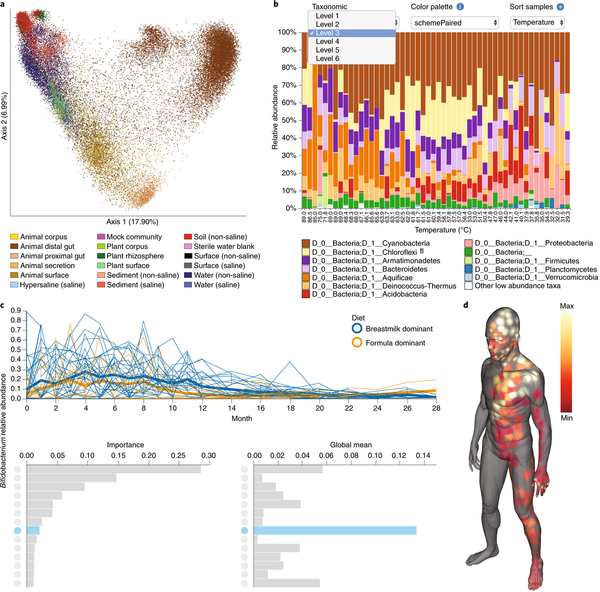
QIIME 2 provides many new interactive visualization tools facilitating exploratory analyses and result reporting. Static versions of interactive visualizations resulting from four worked examples are provided in Fig. 1. QIIME 2 View (https://view.qiime2.org) is a unique new service (Supplementary Methods) that allows users to securely share and interact with results without installing QIIME 2. The QIIME 2 visualizations presented in Fig. 1 are provided in Supplementary File 1 to allow readers to interact with QIIME 2 View. Corresponding worked QIIME 2 example code is provided in the Supplementary Methods.
Fig. 1 |. QIIME 2 provides many interactive visualization tools.
The products of four worked examples are presented here, and interactive versions of these screen captures are available in Supplementary File 1 and at https://github.com/qiime2/paper1. Detailed descriptions and methods, including the commands used to generate each of these visualizations, are provided in Supplementary Methods. a, Unweighted UniFrac principal coordinate analysis plot containing 37,680 samples, illustrating the scalability of QIIME 2. Colors indicate sample type, as described by the Earth Microbiome Project ontology (EMPO). b, Interactive taxonomic composition bar plot illustrating the phylum-level composition of microbial-mat samples collected along a temperature gradient in Yellowstone National Park Hot Spring outflow channels (Steep Cone Geyser). The many interactive controls available in this plot vastly decrease the burden of exploratory analysis over QIIME 1. c, Feature volatility plot (https://msystems.asm.org/content/3/6/e00219-18) illustrating the change in Bifidobacterium abundance over time in breast-fed and formula-fed infants. Temporally interesting features can be interactively discovered with this visualization. Bar charts rank the importance (predictive power for time point) and mean abundance of all microbial features. These bar charts provide an interface for visualizing volatility plots (line plots) of individual features in the context of their importance and abundance; clicking on a bar will display the volatility plot of that feature and highlight in blue that feature’s importance and abundance in the bar charts below. d, Molecular cartography of the human skin surface. Colored spots represent the abundance of the small-molecule cosmetic ingredient sodium laureth sulfate on the human skin. Sample data can be interactively visualized in three-dimensional models, thus supporting the discovery of spatial patterns.
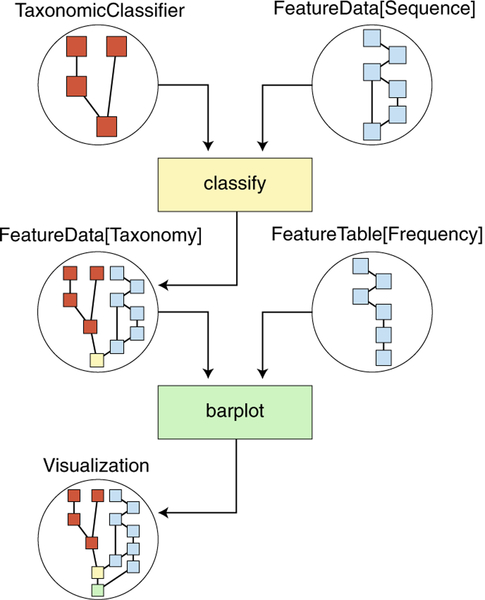
Reproducibility, transparency and clarity of microbiome data science are guiding principles in QIIME 2 design. To this end, QIIME 2 includes a decentralized data-provenance tracking system: details of all analysis steps with references to intermediate data are automatically stored in the results. Users can thus retrospectively determine exactly how any result was generated (Fig. 2 illustrates a simplified provenance graph derived from the data provenance of Fig. 1b). QIIME 2 also detects corrupted results indicating that the provenance is no longer reliable and the results no longer contain information enabling reproducibility. The provenance of the visualizations presented in Fig. 1 can be interactively reviewed by loading the contents of Supplementary File 1 with QIIME 2 View, providing far more detailed information than can typically be provided in Methods text. QIIME 2 results are also semantically typed (Fig. 2), and actions indicate acceptable input types, clarifying the data that actions should be applied to and making complex workflows less error prone. Complex workflows can be created and shared by using Jupyter Notebooks22 or Common Workflow Language (CWL)23, and support for other workflow engines is currently in development.
Fig. 2 |. QIIME 2 iteratively records data provenance, ensuring bioinformatics reproducibility.
This simplified diagram illustrates the automatically tracked information regarding the creation of the taxonomy bar plot presented in Fig. 1b. QIIME 2 results (circles) contain network diagrams illustrating the data provenance stored in the result. Actions (quadrilaterals) are applied to QIIME 2 results and generate new results. Arrows indicate the flow of QIIME 2 results through actions. TaxonomicClassifier and FeatureData[Sequence] inputs contain independent provenance (red and blue, respectively) and are provided to a classify action (yellow), which taxonomically annotates sequences. The result of the classify action, a FeatureData[Taxonomy] result, integrates the provenance of both inputs with the classify action. This result is then provided to the barplot action with a FeatureTable[Frequency] input, which shares some provenance with the FeatureData[Sequence] input, because they were generated from the same upstream analysis. The resulting visualization (Fig. 1b) has the complete data provenance and correctly identifies shared processing of inputs. This simplified representation was created manually from the complete provenance graph for the purpose of illustration. An interactive and complete version of this provenance graph (as well as those for other Fig. 1 panels) can be accessed through Supplementary File 1.
Finally, QIIME 2 provides a software-development kit (https://dev.qiime2.org) that can be used to integrate it as a component of other systems (such as Qiita24 or Illumina BaseSpace) and to develop interfaces targeted toward users with different levels of computational sophistication (Supplementary Fig. 2). QIIME 2 provides the QIIME 2 Studio graphical user interface and QIIME 2 View, interfaces designed for end-user biologists, clinicians and policy-makers; the QIIME 2 application programming interface, designed for data scientists who want to automate workflows or work interactively in Jupyter Notebooks22; and q2cli and q2cwl, providing a command-line interface and CWL23 wrappers for QIIME 2, designed for experts in high-performance computing. At present, computationally expensive steps support parallel computing at the individual-action level (for example, many actions including de-noising and taxonomy assignment support multiple threads). We are currently developing deeper integration with parallelism strategies available in third-party workflow engines, and workflow-level parallelism is currently possible through CWL.
There are many other powerful open-source software tools for microbiome data science, including mothur25, phyloseq26 and related tools available through Bioconductor27, and the biobakery suite20,21,28. The microbiome bioinformatics platform mothur is often compared to QIIME 1 and QIIME 2. A major difference between mothur and QIIME lies in the interactive visualizations: QIIME 2 provides many interactive visualization tools (several examples are provided in Fig. 1), whereas mothur focuses on generating data that can be easily loaded and visualized with other tools. The phyloseq tool focuses on microbiome statistical analysis and generating publication-ready visualizations but, unlike QIIME 2, begins with a feature or operational-taxonomic-unit table, leaving ‘upstream’ processing steps, such as sequence demultiplexing and quality control, to other processing pipelines, many of which (like phyloseq) are available through Bioconductor. The biobakery suite provides analytic functionality that complements that of QIIME 2, and we are actively working with biobakery developers to support interoperability by making their tools accessible as QIIME 2 plugins (for example, the q2-metaphlan2 plugin allows users to run MetaPhlAn2 through QIIME 2). QIIME 2 provides the only Python-based microbiome data-science platform that supports retrospective data-provenance tracking to ensure reproducibility, multi-omics analysis support, interfaces geared toward different user types to enhance usability and an extensibility-focused design through the plugin architecture and software-development kit. We share feedback from users of QIIME 2 on these and other features in Supplementary Methods.
The tools described in the preceding paragraph are all interoperable through plugins, exchange of files in standard formats or using multi-language environments, such as Jupyter Notebooks22. For example, the BIOM format29 is supported by all of them. A diverse ecosystem of interoperable software is beneficial for the field, because it allows both experienced users to obtain multiple perspectives on their data and novice bioinformaticians to work in the programming environments that they are most comfortable with (for example, phyloseq allows users to work in R, whereas QIIME 2 allows users to work in Python). We plan to continue working with the developers of these tools, and with organizations such as the Genomics Standards Consortium, on plugins and standards to ensure interoperability, as well as developing tools to automatically import data from microbiome data-sharing platforms such as Qiita, the European Bioinformatics Institute (EBI) European Read Archive and the National Center for Biotechnology Information (NCBI) Sequence Read Archive.
Advances in microbiome research promise to improve many aspects of health and the world, and QIIME 2 will help drive those advances by enabling accessible, community-driven microbiome data science.
Data availability
Data for the analyses presented in Fig. 1 are available as follows: Earth Microbiome Project data in Fig. 1a were obtained from ftp://ftp.microbio.me/emp/release1, and the American Gut Project (AGP) data were obtained from Qiita (http://qiita.microbio.me) study ID 10317. Sequence data in Fig. 1c are available in Qiita under study ID 10249 and the EBI under accession number ERP016173. Sequence data in Fig. 1b are available in Qiita under study ID 925 and the EBI under accession number ERP022167. Data in Fig. 1d are available in the q2-ili GitHub repository (https://github.com/biocore/q2-ili). Interactive versions of the Fig. 1 visualizations can be accessed at https://github.com/qiime2/paper1.
Code availability
QIIME 2 is open source and free for all use, including commercial. It is licensed under a BSD three-clause license. Source code is available at https://github.com/qiime2. Help for QIIME 2 is provided at https://forum.qiime2.org.
Supplementary Material
Acknowledgements
QIIME 2 development was primarily funded by NSF Awards 1565100 to J.G.C. and 1565057 to R.K. Partial support was also provided by the following: grants NIH U54CA143925 (J.G.C. and T.P.) and U54MD012388 (J.G.C. and T.P.); grants from the Alfred P. Sloan Foundation (J.G.C. and R.K.); ERCSTG project MetaPG (N.S.); the Strategic Priority Research Program of the Chinese Academy of Sciences QYZDB-SSW-SMC021 (Y.B.); the Australian National Health and Medical Research Council APP1085372 (G.A.H., J.G.C., Von Bing Yap and R.K.); the Natural Sciences and Engineering Research Council (NSERC) to D.L.G.; and the State of Arizona Technology and Research Initiative Fund (TRIF), administered by the Arizona Board of Regents, through Northern Arizona University. All NCI coauthors were supported by the Intramural Research Program of the National Cancer Institute. S.M.G. and C. Diener were supported by the Washington Research Foundation Distinguished Investigator Award. Thanks to the Yellowstone Center for Resources for research permit no. 5664 to J.R.S. for Yellowstone access and sample collection. We thank P. J. McMurdie for helpful discussion on the relationships between QIIME 2 and phyloseq. We would like to thank the users of QIIME 1 and 2, whose invaluable feedback has shaped QIIME 2. In particular, we would like to thank A. Abdelfattah (Stockholm University, Sweden), R. C. T. Boutin (University of British Columbia, Canada), D. J. Bradshaw II (Florida Atlantic University Harbor Branch Oceanographic Institute, USA), L. Bullington (MPG Ranch, USA), J. W. Debelius (Karolinska Institutet, Sweden), C. Duvallet (Massachusetts Institute of Technology, USA), E. Korzune Ganda (Cornell University, USA), A. Mahnert (Medical University of Graz, Austria), M. C. Melendrez (St. Cloud State University, USA), D. O’Rourke (University of New Hampshire, USA), A. R. Rivers (USDA ARS, USA), B. Sen (Tianjin University, China), S. Tangedal (Haukeland University Hospital and University of Bergen, Norway), P. J. Torres (San Diego State University, USA) and J. Warren (National Laboratory Service, UK) for writing end-user reviews included in the Supplementary Methods.
Footnotes
Supplementary information is available for this paper at https://doi.org/10.1038/s41587-019-0209-9.
References
- 1.Smith MI et al. Science 339, 548–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gopalakrishnan V et al. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gehring CA, Sthultz CM, Flores-Rentería L, Whipple AV & Whitham TG Proc. Natl Acad. Sci. USA 114, 11169–11174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee K, Pletcher SD, Lynch SV, Goldberg AN & Cope EK Front. Cell. Infect. Microbiol 8, 168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metcalf JL et al. Science 351, 158–162 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Rubin RL et al. Ecol. Appl 28, 1594–1605 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Pineda A, Kaplan I & Bezemer TM Trends Plant Sci. 22, 770–778 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Kapono CA et al. Sci. Rep 8, 3669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verberkmoes NC et al. ISME J. 3, 179–189 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Barr T et al. Gut Microbes 9, 338–356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callahan BJ et al. Nat. Methods 13, 581–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amir A et al. mSystems 2, e00191–16 (2017).28289731 [Google Scholar]
- 13.Bokulich NA et al. Microbiome 6, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen S et al. mSystems 3, e00021–18 (2018).29719869 [Google Scholar]
- 15.Bokulich NA et al. mSystems 3, e00219–18 (2018). [Google Scholar]
- 16.Bokulich N et al. J. Open Source Softw 3, 934 (2018). [Google Scholar]
- 17.Sedio BE, Rojas Echeverri JC, Boya PCA & Wright SJ Ecology 98, 616–623 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Wang M et al. Nat. Biotechnol 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillmann B et al. mSystems 3, e00069–18 (2018).30443602 [Google Scholar]
- 20.Truong DT et al. Nat. Methods 12, 902–903 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Langille MGI et al. Nat. Biotechnol 31, 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kluyver T et al. Positioning and power in academic publishing: players, agents and agendas. in Proc 20th International Conference on Electronic Publishing (eds Loizides F & Schmidt B) 87–90 (IOS Press, 2016). [Google Scholar]
- 23.Amstutz P et al. 10.6084/m9.figshare.3115156.v2 (2016). [DOI]
- 24.Gonzalez A et al. Nat. Methods 15, 796–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schloss PD et al. Appl. Environ. Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMurdie PJ & Holmes S PLoS One 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber W et al. Nat. Methods 12, 115–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franzosa EA et al. Nat. Methods 15, 962–968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald D et al. Gigascience 1, 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for the analyses presented in Fig. 1 are available as follows: Earth Microbiome Project data in Fig. 1a were obtained from ftp://ftp.microbio.me/emp/release1, and the American Gut Project (AGP) data were obtained from Qiita (http://qiita.microbio.me) study ID 10317. Sequence data in Fig. 1c are available in Qiita under study ID 10249 and the EBI under accession number ERP016173. Sequence data in Fig. 1b are available in Qiita under study ID 925 and the EBI under accession number ERP022167. Data in Fig. 1d are available in the q2-ili GitHub repository (https://github.com/biocore/q2-ili). Interactive versions of the Fig. 1 visualizations can be accessed at https://github.com/qiime2/paper1.