Abstract
Background
Cachexia is the direct cause of at least 20% of cancer‐associated deaths. Muscle wasting in skeletal muscle results in weakness, immobility, and death secondary to impaired respiratory muscle function. Muscle proteins are massively degraded in cachexia; nevertheless, the molecular mechanisms related to this process are poorly understood. Previous studies have reported conflicting results regarding the amino acid abundances in cachectic skeletal muscle tissues. There is a clear need to identify the molecular processes of muscle metabolism in the context of cachexia, especially how different types of molecules are involved in the muscle wasting process.
Methods
New in situ ‐omics techniques were used to produce a more comprehensive picture of amino acid metabolism in cachectic muscles by determining the quantities of amino acids, proteins, and cellular metabolites. Using matrix‐assisted laser desorption/ionization (MALDI) mass spectrometry imaging, we determined the in situ concentrations of amino acids and proteins, as well as energy and other cellular metabolites, in skeletal muscle tissues from genetic mouse cancer models (n = 21) and from patients with cancer (n = 6). Combined results from three individual MALDI mass spectrometry imaging methods were obtained and interpreted. Immunohistochemistry staining for mitochondrial proteins and myosin heavy chain expression, digital image analysis, and transmission electron microscopy complemented the MALDI mass spectrometry imaging results.
Results
Metabolic derangements in cachectic mouse muscle tissues were detected, with significantly increased quantities of lysine, arginine, proline, and tyrosine (P = 0.0037, P = 0.0048, P = 0.0430, and P = 0.0357, respectively) and significantly reduced quantities of glutamate and aspartate (P = 0.0008 and P = 0.0124). Human skeletal muscle tissues revealed similar tendencies. A majority of altered amino acids were released by the breakdown of proteins involved in oxidative phosphorylation. Decreased energy charge was observed in cachectic muscle tissues (P = 0.0101), which was related to the breakdown of specific proteins. Additionally, expression of the cationic amino acid transporter CAT1 was significantly decreased in the mitochondria of cachectic mouse muscles (P = 0.0133); this decrease may play an important role in the alterations of cationic amino acid metabolism and decreased quantity of glutamate observed in cachexia.
Conclusions
Our results suggest that mitochondrial dysfunction has a substantial influence on amino acid metabolism in cachectic skeletal muscles, which appears to be triggered by diminished CAT1 expression, as well as the degradation of mitochondrial proteins. These findings provide new insights into the pathobiochemistry of muscle wasting.
Keywords: Cancer cachexia, Mitochondrial dysfunctions, Amino acids, MALDI, Mass spectrometry imaging
Introduction
Cachexia, which is often caused by cancer, leads to massive loss of total body mass, generalized inflammation, and pronounced muscle wasting, producing a significant decrease in the quality of life. Loss of skeletal muscle results in weakness, leading to immobility and eventual death secondary to impaired respiratory muscle function.1 Muscle wasting is thereby one of the most devastating characteristics of cachexia in late‐stage cancer, for which no specific treatments are currently available. Several metabolic changes have been associated with loss of muscle mass in cachexia, including decreased protein synthesis, increased protein degradation, and deranged amino acid metabolism.2
Until now, researchers have focused on understanding the molecular mechanisms behind protein and amino acid metabolism alterations in cachexia.3 On the one hand, the activation of the ubiquitin‐dependent proteasome pathway seemed to play a major role in protein degradation in cancer cachexia.4 Specific ubiquitin ligase enzymes (E3) catalyzing the movement of the ubiquitin from the E2 enzyme to the substrate and are strongly up‐regulated in an animal model of cachexia.5 Besides, several proteasome subunits are also up‐regulated in a transcriptional manner in cachexia.6 On the other hand, protein synthesis is influenced, for example, by the activity of mechanistic (a.k.a., mammalian) target of rapamycin complex 1,7 which is progressively decreased in cachectic mice.8 All of these molecular processes are associated with a changed amino acid turnover. Meanwhile, only a few studies have reported altered quantities of amino acids in cachectic skeletal muscle,9, 10, 11, 12 and their results have been conflicting. Furthermore, amino acid quantities in the context of protein degradation and synthesis and energy metabolism is a still unexploited topic. Of note, these issues are of utmost importance, as amino acids and proteins are linked and individual amino acids contribute to the energy kinetics of skeletal muscle cells.13 In addition, a massive amino acid efflux to the circulation can also be occurring by enhanced protein degradation,2 while a significant part of amino acids is received by the circulation for skeletal muscles.14
New in situ “‐omics” approaches allow the identification of a broad and comprehensive range of possible alterations in amino acid and protein metabolism to help generate a more global picture of the molecular alterations associated with muscle wasting in cachexia. In the current study, we used matrix‐assisted laser desorption/ionization (MALDI) mass spectrometry imaging as a sophisticated label‐free, non‐targeted, in situ ‐omics method of examining skeletal muscles from cachectic and non‐cachectic mice and patients. This is the first time amino acids, energy and other cellular metabolites, and proteins have been simultaneously examined in situ in the context of cancer cachexia.
Determination of the three classes of molecules enabled the investigation of functional relationships between and within amino acids, energy metabolites, and proteins. It allowed the identification of specific proteins that were potentially degraded and contributing to amino acid release in cachexia. Because cachexia is characterized by increased resting energy expenditure and low energy intake,1 the interplay between specific amino acids and energy metabolites was examined to detect molecular alterations. The further scope of the following study was the analysis of derangements of amino acids, to identify functional relationships with proteins, and simultaneously examine energy changes in skeletal muscles related to cancer cachexia.
In mice, we found higher quantities of cationic amino acids and lower quantities of glutamate and aspartate in skeletal muscle tissues during cachexia, whereas patient tissues provide first insights in similar molecular changes. Several protein expression changes in the mitochondria of cachectic tissues were associated with alterations in energy and amino acid metabolism. To the best of our knowledge, we herein present the first in situ study describing molecular alterations in cachexia caused by functional changes in the mitochondria of skeletal muscle tissues. Furthermore, we determined that cationic amino acid transporter 1 (CAT1), a mitochondria‐associated protein, is involved in this process.
Methods
Collection of tissue samples
This study was approved by the Ethics Committee of the Medical Faculty of the Technical University of Munich (#4916/S), and written consent was obtained from all patients before surgery. The compound mutant mouse line (pVillin‐KRASV12G x Apc1638N), with the inbred C57BL/6N background, was bred at the Zentrum für Präklinische Forschung at Klinikum rechts der Isar, TUM, Munich. The animals were assessed at specific timepoints for the presence of intestinal tumours, as previously described, and their body weights were determined at the time of sacrifice (see Table 1 for baseline characteristics).15 The non‐cachectic group consisted of tumour‐free wild‐type and single transgenic littermates from the same breedings. Cachectic mice were defined as animals with at least a 10% reduction in body weight, compared with the median weight of the non‐cachectic group. All mice were maintained with a 12 hr light‐dark cycle and fed a standard diet and water ad lib. Overall, 21 mice were analysed, and 11 tumour‐bearing mice were classified as cachectic. Ten mice represented the non‐cachectic group. Mice were euthanized via cervical dislocation. The quadriceps muscles were subsequently resected and then immediately shock frozen and stored until further use in liquid nitrogen.
Table 1.
Baseline characteristics of mouse genetic cancer models
| Characteristic | Non‐cachectic | Cachectic |
|---|---|---|
| Number | 10 | 11 |
| Gender (n) | ||
| Male | 9 | 5 |
| Female | 1 | 6 |
| Age (months) | ||
| Mean | 8.7 | 6.6 |
| Range | 4–15 | 4–10 |
| Weight (g) | ||
| Mean | 31.7 | 26.7 |
| Range | 27.4–36.0 | 21.7–32.0 |
Patient tissues (samples from the musculus rectus abdominis) were obtained from patients diagnosed with pancreatic ductal adenocarcinoma (see Table 2 for baseline characteristics) during routine surgery. The patients underwent surgery at the Department of Surgery, TUM, from 2008 until 2015. Samples were obtained from 36 patients, 13 of whom exhibited a weight loss of at least 7% within the 6 months before surgery and were thereby defined as cachectic patients. The remaining 23 patients, with a weight loss of less than 5%, were considered non‐cachectic. Definition of cachexia was according to an international consensus.16 Tissues from six patients (three non‐cachectic and three cachectic patients; Supporting Information, Appendix S1 for baseline characteristics) were stored in liquid nitrogen as fresh‐frozen samples until MALDI mass spectrometry imaging analysis. Tissues from the remaining 30 patients were fixed in formalin and embedded in paraffin and then used to construct two‐tissue microarrays, which were stored at room temperature until immunohistochemistry (IHC) analysis.
Table 2.
Clinical characteristics of patients with cancer
| Characteristic | Non‐cachectic | Cachectic |
|---|---|---|
| Number | 23 | 13 |
| Gender (n) | ||
| Male | 15 | 4 |
| Female | 8 | 9 |
| Age (years) | ||
| Mean | 67 | 65 |
| Range | 45–88 | 44–83 |
| Weight (kg) | ||
| Mean | 77 | 64 |
| Range | 40–106 | 41–98 |
| Body mass index (kg/m2) | ||
| Mean | 25.6 | 22.5 |
| Range | 16.2–34.2 | 15.8–28.4 |
| Weight loss (%) | ||
| Mean | 1.0 | 13.3 |
| Range | 0–4.8 | 7.7–25.5 |
Matrix‐assisted laser desorption/ionization mass spectrometry imaging
Frozen muscle tissue samples were cryosectioned into 12 μm sections using Microm560 (Microm International, Walldorf, Germany) and thaw mounted onto indium tin oxide‐coated conductive slides (Bruker Daltonics, Bremen, Germany). The slides were pre‐treated with 1:1 poly‐l‐lysine (Sigma‐Aldrich, Munich, Germany) and 0.1% Nonidet P‐40 (Sigma) before mounting. Briefly, the samples were covered with 10 mg/mL 9‐aminoacridine (9AA) matrix (Sigma‐Aldrich) in 70% methanol for metabolite analysis or 10 mg/mL 1,5‐diaminonaphthalene (DAN) matrix (Sigma‐Aldrich) in 70% acetonitrile for amino acid analysis, using a SunCollect sprayer (Sunchrom, Friedrichsdorf, Germany). The following preferences were used for the automatic sprayer (for both 9AA and DAN): vial distance of 0.50 mm for the X direction and 2.00 mm for the Y direction, 20 mm Z position and offset of the spray head, and medium X/Y speed. The matrix was deposited in eight layers using variable increasing spray rates, as previously reported by Ly et al..17 For metabolite analysis, data were acquired in negative ion mode using a Bruker Solarix 7.0 T Fourier‐transform ion cyclotron resonance (FTICR) mass spectrometer (Bruker Daltonics) over a mass range of 50–1000 m/z and at a lateral resolution of 100 μm. Amino acids were analysed over a mass range of 50–250 m/z. Because DAN matrix is not stable in a vacuum, a faster measurement protocol was established by changing the lateral resolution for amino acids to 150 μm. For protein analysis, 10 mg/mL sinapinic acid matrix (Sigma‐Aldrich) in 60% acetonitrile and 0.2% trifluoroacetic acid was deposited onto the sections using an ImagePrep automated sprayer (Bruker Daltonics). Linear positive ion mode over a mass range of 2000–25 000 m/z was used for protein measurements with the Bruker Ultraflex III MALDI‐TOF/TOF MS (Bruker Daltonics). The lateral resolution for examining intact proteins was predefined as 60 μm. The Smartbeam‐II Nd:YAG laser (355 nm) fired 300 times for protein ablation with a frequency of 200 Hz and a sample rate of 0.50 GS/s. External calibration was performed with Protein Calibration Standard I (Bruker Daltonics), mixed 1:1 (v/v) with the matrix solution, and spotted onto the slide.
Tissue staining and co‐registration
After acquisition of the mass spectrometry data, the matrix was removed with 70% ethanol, and the tissue sections were stained with haematoxylin and eosin. The sections were transferred to dH2O for 1 min and then haematoxylin (Carl Roth, Karlsruhe, Germany) for 1 min; after washing in tap water for approximately 5 min, they were transferred to eosin Y (Sigma‐Aldrich) for 1 min. The sections were subsequently dehydrated using an increasing alcohol solution series [70%, 90%, and 100% ethanol (Merck) and isopropanol (Merck); 30 s each], transferred to xylene (Carl Roth) for at least 2 min, coverslipped, scanned with a Mirax desk slide scanner (Zeiss, Göttingen, Germany) using a 20× magnification objective, and co‐registered with the respective mass spectrometry imaging data using flexImaging v. 4.0 (Bruker Daltonics).
Matrix‐assisted laser desorption/ionization mass spectrometry imaging data analysis
flexImaging software was used for normalization against the root mean square (FTICR) or total ion count [time of flight (TOF)] of all data points. “Virtual microdissection” was performed, defining regions of interest (ROIs) of cross‐sectioned muscle fibres and excluding non‐muscle tissue components. Specification of ROIs and exportation of spectral data for each ROI were similarly performed using flexImaging.
MATLAB script, including the bioinformatics and image processing toolboxes (v.7.10.0, MathWorks, Natick, MA, USA), was used for subsequent FTICR data processing. In this script, flexImaging‐exported spectra were processed using the LIMPIC algorithm.18 This algorithm includes a baseline subtraction (100 data points window size), resampling (0.001 m/z bin width), and smoothing (Kaiser filter with factor 3) to exclude chemical and electronic noise before peak picking. Peak picking itself was performed with a minimum peak width of 5.0E – 4 m/z, a signal‐to‐noise threshold of 2, and a minimum intensity threshold of 0.01% for the base peak of each spectrum. To enable direct comparisons between peak lists from different patients, peaks were clustered with a specific mass tolerance (5.0E – 8 * (m/z)2.023). TOF peak lists were generated using SCiLS Lab (v2019a, Bremen, Germany). Metabolites, amino acids, and proteins were identified by matching accurate mass data with Metlin (http://metlin.scripps.edu/index.php),19 the Human Metabolome Database (http://www.hmdb.ca),20 and the MaTisse database,21 allowing a window of 4 ppm for metabolites or amino acids.
Ion images were exported by flexImaging as single colour visualizations with a black background. The colour was detected entirely by image processing software and was visualized over the corresponding scanned haematoxylin and eosin‐stained tissue. Adenosine monophosphate (AMP), adenosine diphosphate (ADP), adenosine triphosphate (ATP), and energy charge visualization was performed by an in‐house developed python script based on the previously published MSIdV tool for mass spectrometry imaging.22 The remaining heatmap visualizations were received from SCiLS Lab.
Transmission electron microscopy
For transmission electron microscopy (TEM) analysis, fresh‐frozen tissues were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, and TEM fixation buffer (Electron Microscopy Sciences, Hatfield, USA) at 4 °C. The samples were subsequently post‐fixed in 2% aqueous osmium tetraoxide 44, dehydrated in increasing concentrations of ethanol (30–100%) and propylene oxide, embedded in Epon (Merck, Darmstadt, Germany), and dried for 24 h at 60 °C. Semithin sections were cut and stained with toluidine blue. Ultrathin sections of 50 nm were collected on 200‐mesh copper grids and stained with uranyl acetate and lead citrate before examination by TEM (Zeiss Libra 120 Plus, Carl Zeiss NTS GmbH, Oberkochen, Germany). Images were acquired using a Slow Scan CCD camera and iTEM software (Olympus Soft Imaging Solutions, Münster, Germany). The number of mitochondria was determined by manually counting the mitochondria per field of view in longitudinally sectioned fibres, viewed at 1600× magnification.
Immunohistochemistry staining and digital image analysis
For IHC analyses, parts of the mouse muscle tissues were fixed in 4% (v/v) neutral‐buffered formalin and embedded in paraffin by an automatic processor. CAT1 analysis of patient tissues was performed on the formalin‐fixed paraffin‐embedded tissues. Expression levels of CAT1 and voltage‐dependent anion channel (VDAC) were analysed on 3 μm consecutive sections by IHC staining with the CAT1 antibody [14195‐1‐AP (Proteintech, Manchester, UK), 1:20] and VDAC antibody [#4866 (Cell Signaling Technology, Frankfurt am Main, Germany), 1:100]. For validation purposes of mitochondrial proteins, cytochrome c oxidase subunit (COX) 7C [PA5‐51284 (Invitrogen, Carlsbad, CA, USA), 1:10] and cytochrome c [GTX108585 (GeneTex, Inc., Alton Pkwy Irvine, CA, USA), 1:25] stainings were performed on 12 μm cryosectioned mouse skeletal muscle tissues. Used tissues are representing consecutive tissue sections as analysed in MALDI imaging. The automated slide processing system Ventana DISCOVERY XT System (Ventana Medical Systems, Inc., Tucson, AZ, USA) was used in accordance with the manufacturer's instructions. All stained slides were digitalized at 20× objective magnification using the Mirax desk slide scanner. IHC staining results were quantified using the image analysis software Definiens Developer XD2 (Definiens AG, Munich, Germany). This software allows quantification of IHC staining intensities within a user‐specified ROI. ROIs were defined in longitudinally sectioned muscle fibres. Algorithms were developed, modified specifically for CAT1, VDAC, COX7C, and cytochrome c, and established in semantic and context‐based segmentation processes, which included staining intensity, shape, area, colour features, and neighbourhood. In addition, the algorithms were optimized to consider only mitochondrial staining. The quantified parameters for CAT1, VDAC, COX7C, and cytochrome c were values representing a point on a continuous spectrum of protein expression in relative units.
Immunofluorescence analysis of myosin heavy chain expression
Immunofluorescence analysis of myosin heavy chain (MHC) expression was achieved using primary antibodies against MHCI (BA‐F8, 1:50), MHCIIa (SC‐71, 1:600), and MHCIIb (BF‐F3, 1:100) according to Bergmeister et al..23 Primary antibodies were purchased from the Developmental Studies Hybridoma Bank (University of Iowa), whereas secondary antibodies (AF633 IgG2b, AF488 IgG1, and AF555 IgM, all 1:250) were purchased from Invitrogen. Slides were digitalized at 20× objective magnification using an Axio Scan.Z1 (Zeiss). For fibre‐type analysis, all fibres within an ROI (as described for MALDI mass spectrometry imaging) were characterized. Quantification was performed by digital image analysis in Definiens Developer XD2.
Statistical analysis
To determine significant differences between cachectic and non‐cachectic tissues, the rank‐based Mann–Whitney U test was used. Further statistical testing for functional relationships between and within metabolites, amino acids, and proteins was investigated using Spearman's rank correlation analysis. Correlation plots and calculations were generated and performed using R (The R Foundation, Vienna, Austria; “corrplot” package). P values equal to or less than 0.05 were considered statistically significant.
Results
Amino acid derangements were detected by in situ metabolomics in cachexia
Sixteen amino acids were detected in situ in mouse skeletal muscle tissues. Lysine (P = 0.0037), arginine (P = 0.0048), tyrosine (P = 0.0357), and proline (P = 0.0430) were significantly more abundant in cachectic mouse muscle tissues than in non‐cachectic mice. Alanine, asparagine, glutamine, leucine/isoleucine, methionine, phenylalanine, threonine, tryptophan, and valine analysis revealed a higher relative mean intensity in cachectic mice than in non‐cachectic ones, but the differences were not statistically significant (Figure 1A). In contrast, quantities of glutamate (P = 0.0008) and aspartate (P = 0.0124) were significantly less in cachectic mouse skeletal muscle tissues. All intensities and P values of determined amino acids in non‐cachectic and cachectic mice can be obtained from Supporting Information, Appendix S2 . Figure 1B shows the results of false colour visualization of amino acids, demonstrating the differences in skeletal muscle tissues between the cachectic and non‐cachectic mice. Results in skeletal muscle tissues of cachectic patients revealed similar amino acid profiles to those observed in mice (Supporting Information, Appendix S3 ); however, the amino acid differences between cachectic and non‐cachectic patients did not reach statistical significance. As the amino acid alterations could be due to protein breakdown processes,2 our subsequent investigations examined this issue.
Figure 1.
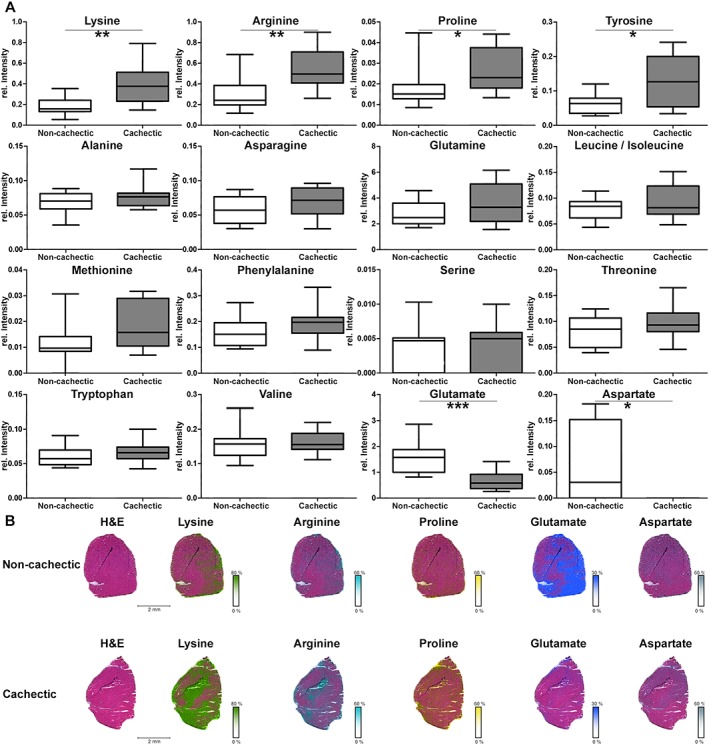
In situ analysis of amino acids in cachectic mouse skeletal muscle tissues. (A) Quantities of amino acids in skeletal muscles from cachectic and non‐cachectic mice. All intensity values were determined by mass spectrometry imaging. Quantities of lysine, arginine, proline, and tyrosine were significantly higher in cachectic mice than in non‐cachectic ones. The intensities for alanine, asparagine, glutamine, leucine/isoleucine, methionine, phenylalanine, threonine, tryptophan, and valine also revealed a higher relative mean intensity in cachectic mice than in non‐cachectic ones. Glutamate and aspartate intensities were significantly decreased in cachectic mouse muscles. Whiskers of the boxplots illustrate the minimal and maximum intensity values. (B) False colour visualization of amino acids in mouse skeletal muscle tissues. Lysine, arginine, and proline were increased, and glutamate and aspartate were decreased in cachectic mouse muscles, compared with non‐cachectic ones. * P < 0.05, ** P < 0.01, and *** P < 0.005. H&E, haematoxylin and eosin.
Amino acids are linked to protein breakdown processes
To detect possible protein breakdown targets, protein expression was determined by MALDI‐TOF and correlated with the quantities of amino acids. A negative correlation between amino acids and proteins suggests the presence of protein breakdown.
We determined the correlations between all detected proteins and the quantities of amino acids. Spearman's rank correlation analysis revealed 15 annotated proteins, which have significant negative correlations with amino acid quantities (Supporting Information, Appendix S4 ). The proteins for which significant correlations were detected included thymosin beta‐4, individual proteins for oxidative phosphorylation (OXPHOS), histone H2B, and glutathione S‐transferase P. Specifically, thymosin beta‐4 (Figure 2A) correlated negatively with the quantity of lysine (P = 0.0090), arginine (P = 0.0026), proline (P = 0.0049), and threonine (P = 0.0431). Cytochrome c oxidase subunit (COX) 6B1 correlated negatively with lysine (P = 0.0405), arginine (P = 0.0027), glutamine (P = 0.0209), and tryptophan (P = 0.0368). Figure 2B shows an example of the detected proteins: false colour visualization demonstrated lower expression of COX6B1 in cachectic mouse tissues than in non‐cachectic mice. Statistical analysis of the visualization results showed significantly decreased COX6B1 in cachexia (Figure 2C; P = 0.0008). Supporting Information, Appendix S5 shows the analyses for three OXPHOS proteins: COX7C, ATPase F6, and cytochrome c. At this, the abundances of COX7C and ATPase F6 were significantly decreased (P = 0.0127 and P = 0.0048, respectively), determined by MALDI mass spectrometry imaging. Interestingly, a majority of detected proteins that correlated negatively with amino acids were associated with the mitochondria. In addition, validation experiments were performed by IHC stainings and confirmed MALDI mass spectrometry imaging analysis results for COX7C as well as for cytochrome c (Supporting Information, Appendix S5 ). COX7C abundance detected by IHC was also significantly decreased (P = 0.0159) in cachectic mouse skeletal muscle tissues in comparison with non‐cachectic mice.
Figure 2.
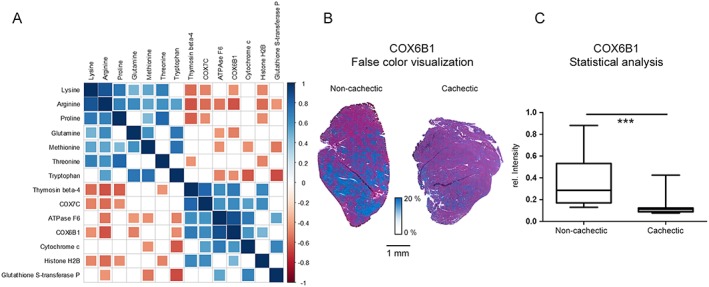
Evaluation of potential protein breakdown targets. (A) Visualization of Spearman's rank correlation analysis results examining the relationships between amino acids and proteins. Square sizes represent the magnitude of the Spearman's rank correlation coefficient. Blue squares indicate positive correlations, and red squares indicate negative correlations. Non‐significant correlations (P > 0.05) are indicated by empty squares. Many OXPHOS proteins seemed to be degraded. (B) False colour visualization of COX6B1. A reduced quantity of COX6B1 was observed in cachectic mouse tissues, compared with non‐cachectic ones. (C) Statistical analysis of the false colour visualization results depicted in (B). COX6B1 intensities were significantly lower in cachectic mouse muscle tissues than in non‐cachectic ones (P = 0.0008). Boxplot whiskers represent the minimum and maximum intensities. *** P < 0.005.
At this stage, the combination of two different measurement techniques—high‐resolution mass spectrometry by FTICR and analysis of proteins by TOF—enabled the detection of biological relationships and thus, for example, the potential degradation of OXPHOS proteins. Four of these proteins were highlighted by correlation analysis and seemed to have enhanced breakdown in cachexia. The involvement of OXPHOS proteins suggested the possibility of alterations in energy in cachexia and thereby focused our further investigations on the mitochondria.
Energy charge is reduced in cachectic muscle tissues
Because the correlation between amino acid and protein data suggested degradation of specific OXPHOS enzymes, we conducted metabolite measurements to examine energy alterations in cachectic mouse skeletal muscle tissues. Because the quadriceps muscle is composed of mixed fibre types, we examined non‐cachectic and cachectic mouse skeletal muscle tissues for fibre‐type composition. This issue is important for regarding energy aspects, due to the adaption of specific fibre types to oxidative or glycolytic metabolism.24 No differences could be detected regarding the individual fibre type composition between non‐cachectic and cachectic mouse skeletal muscle tissues (Supporting Information, Appendix S6 ).
Adenosine monophosphate, ADP, and ATP concentrations were determined as representatives of the energy state in muscle tissues. To determine the available energy in tissues, the energy charge was calculated from the AMP, ADP, and ATP concentrations, using the formula shown in Figure 3A. The energy charge was lower in cachectic mice than in non‐cachectic mice (P = 0.0101), indicating that energy stores were reduced in cachectic mouse skeletal muscle tissues. AMP was slightly increased in the cachectic mouse skeletal muscle tissues, when compared with non‐cachectic mouse tissues (Figure 3B). ADP concentrations were similar in cachectic and non‐cachectic mice, and ATP concentration was marginally decreased in cachexia.
Figure 3.
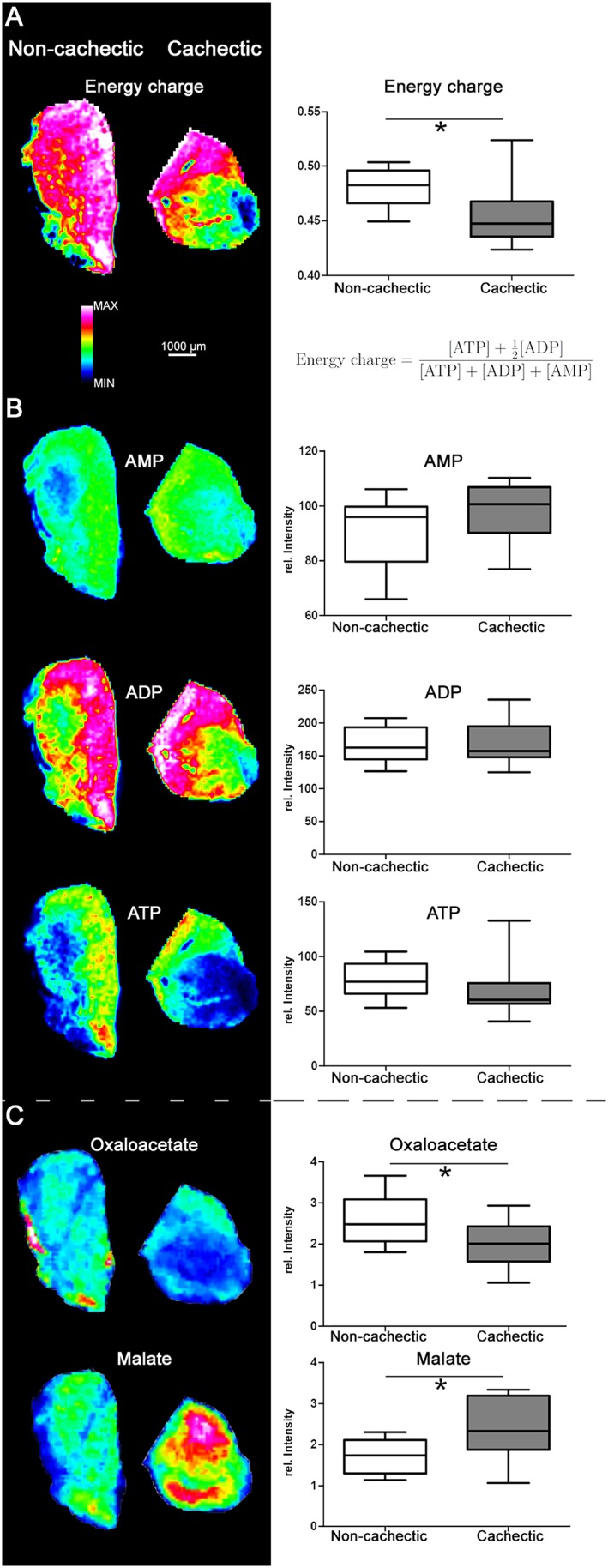
Energy changes in cancer cachexia. (A) Heatmap visualization and statistical analysis of the calculated energy charge. Calculation of the energy charge revealed a significantly lower charge in cachectic mice (P = 0.0101). (B) AMP, ADP, and ATP distribution in cachectic and non‐cachectic mouse skeletal muscle tissues. No significant differences were detected between cachectic and non‐cachectic mouse tissues. (C) Heatmap visualization and statistical analysis of changes in molecules of the tricarboxylic acid cycle. Cachectic mice exhibited significantly higher quantities of malate (P = 0.0295) and lower quantities of oxaloacetate (P = 0.0448) than non‐cachectic mice. Whiskers of the boxplots represent the lowest and highest peak intensities in each group. * P < 0.05.
Reduced energy in cachectic mouse skeletal muscle tissues could result in changed function of the tricarboxylic acid (TCA) cycle as a compensatory alteration or as a co‐influencing factor. In cell metabolomics analysis, the majority of detected TCA cycle molecules did not differ significantly between cachectic and non‐cachectic mouse muscle tissues (Supporting Information, Appendix S7 ). However, the quantity of oxaloacetate was significantly decreased (P = 0.0448) (Figure 3C) and the quantity of malate was significantly increased (P = 0.0295) in cachexia. These results indicate that cachexia was not associated with a generalized change in regulation of the TCA cycle, but it did seem to be associated with alterations of the dicarboxylic acid part.
We next used the energy charge results to examine underlying molecular mechanisms. A total of 58 proteins correlated significantly with the calculated energy charge (Figure 4). Specifically, COX7C (P = 0.0257) and COX6B1 (P = 0.0052) were all significantly and positively correlated with the energy charge. Thymosin beta‐4 (P = 0.0054), histone H2B (P ≤ 0.0001), and redox Cu/Zn superoxide dismutase (P = 0.0059) also correlated positively with the energy charge. Notably, expression of ubiquitin, which is a marker for protein degradation, correlated negatively (P = 0.0009) with the energy charge, as well as with COX7C (P = 0.0216) and other proteins. These proteins may be tagged for degradation.
Figure 4.
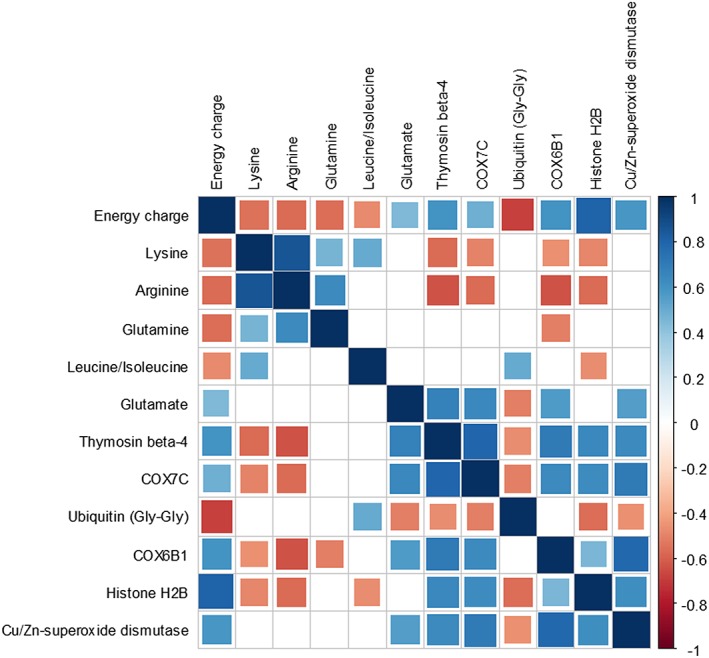
Impact of proteins and amino acids on the energy charge. Correlation plot depicting the results of Spearman's rank analysis examining the associations between amino acid and protein expression and the calculated energy charge. Square size represents the magnitude of the Spearman's rank correlation coefficient. Blue squares indicate positive correlations, and red squares indicate negative correlations. Non‐significant correlations (P > 0.05) are symbolized by empty squares. Energy charge was significantly correlated with 58 proteins and five amino acids
Five amino acids were also significantly correlated with the energy charge, suggesting a functional relationship. For example, lysine (P = 0.0122) and arginine (P = 0.0086), the quantities of which differed significantly between cachectic and non‐cachectic mouse tissues, were correlated negatively with the energy charge (Figure 4). Glutamine (P = 0.0108) and leucine/isoleucine (P = 0.0324) were likewise negatively correlated with the energy charge. Glutamate, which was significantly decreased in the cachectic state, correlated positively with the energy charge (P = 0.0437).
Cationic amino acid transporter 1 is changed in cachexia
We next focused on examining changes in CAT1, which was previously identified as an essential factor for l‐arginine transport into mitochondria.25 The potential role of this transport protein in cachexia was supported by our findings of prominent alterations in arginine and lysine, both of which are cationic amino acids.
The quantity of mitochondrial CAT1 in skeletal muscle tissues was lower in cachectic mice than in non‐cachectic ones (Figure 5A). Using digital image analysis of stained mouse skeletal muscle tissues, significantly less CAT1 was observed in the cachectic state (Figure 5B; P = 0.0133). There was a similar tendency towards decreased CAT1 in muscle tissues of cachectic patients, compared with non‐cachectic patients, but the difference did not reach statistical significance (P = 0.5498; Figure 5C). For comparing an equal group size of patients without gender influence, we also calculated CAT1 expression for female patients alone, which is marginally improving the statistics (P = 0.2261). CAT1 expression was significantly correlated with the quantity of lysine (P = 0.0122; negative correlation), proline (P = 0.0266; negative correlation), and glutamate (P = 0.0106; positive correlation), suggesting an important influence of CAT1 on amino acid quantities in cachectic tissues.
Figure 5.
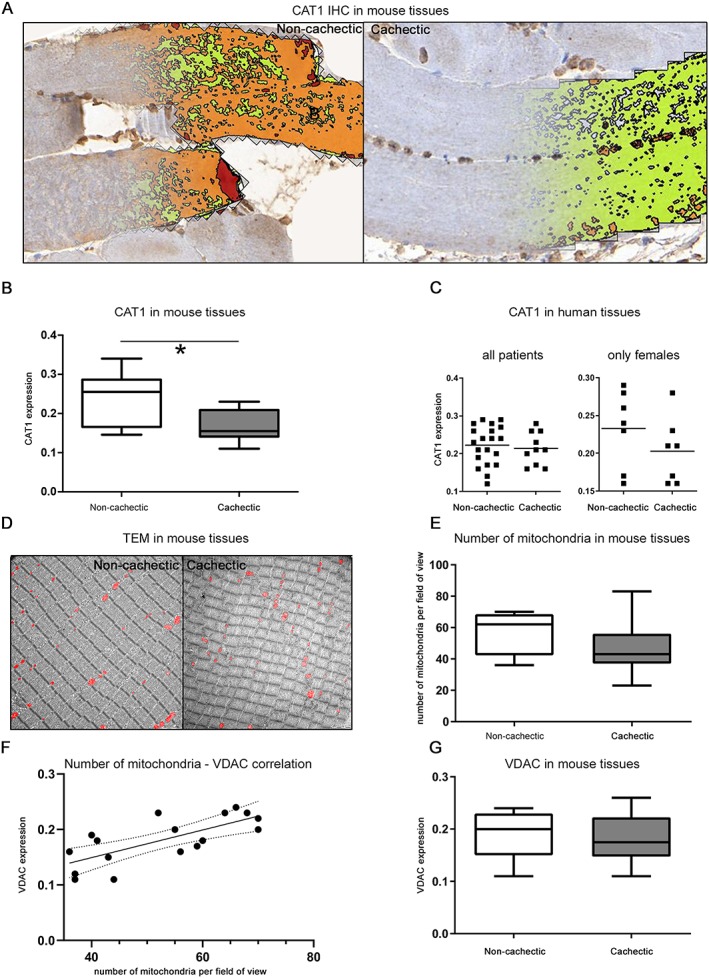
Changes in CAT1 expression in cachexia. (A) Digital image analysis of the CAT1 immunohistochemistry (IHC) results. Definiens Software Developer XD2 was used to detect the CAT1 expression in all tissue sections. (B) The detected CAT1 abundance was significantly lower in skeletal muscles of cachectic mice than in non‐cachectic ones (P = 0.0133). Whiskers of the boxplots represent the lowest and highest CAT1 expression in each group. (C) CAT1 expression in skeletal muscle tissues from humans with cancer for all and only female patients. CAT1 was slightly lower in patients with cachexia, compared with non‐cachectic patients, but the difference did not reach statistical significance. Horizontal lines represent the mean intensity of each group. (D) Number of mitochondria in mouse skeletal muscle tissues determined by transmission electron microscopy (TEM) at 1600× magnification. The red colour represents the mitochondria. Intact mitochondria were observed in tissues from both cachectic and non‐cachectic mice. (E) Boxplots illustrating the number of mitochondria in mouse skeletal muscle tissues. No significant difference was detected between cachectic and non‐cachectic mice. (F) Correlation analysis between the number of mitochondria determined by electron microscopy and the intensity of voltage‐dependent anion channel (VDAC) staining detected by IHC. The number of mitochondria was significantly correlated with VDAC expression (P = 0.0033). (G) Statistical analysis of VDAC staining in muscle tissues from cachectic and non‐cachectic mice. No significant difference was detected. * P < 0.05.
The CAT1 expression results could have been influenced by a change in the number of mitochondria, as fewer mitochondria would lead to reduced quantities of mitochondrial proteins. Therefore, TEM was performed to determine the number and possible morphological alterations of mitochondria in mouse skeletal muscle tissues. No differences in mitochondrial morphology were observed between mitochondria in cachectic and non‐cachectic mice; we did not observe electron‐lucent areas, swelling, and fragmentation of cristae, which were previously reported in the muscles of mice with cancer cachexia.26 Similarly, the number of mitochondria was not significantly decreased in the cachectic state (Figure 5D and 5E, P = 0.1051). Additionally, there were no systematic ultrastructural changes between cachectic and non‐cachectic mice.
To provide further evidence of a comparable number of mitochondria in cachectic and non‐cachectic mice, VDAC expression was determined by IHC. The quantity of VDAC correlated significantly with the number of mitochondria determined by electron microscopy (Figure 5F; P = 0.0033) and revealed no significant differences between cachectic and non‐cachectic mouse skeletal muscle tissues (Figure 5G; P = 0.5648). Thus, TEM and VDAC expression confirmed the presence of a similar number of mitochondria in cachectic and non‐cachectic mouse skeletal muscle tissues, thereby suggesting a functional—not quantitative—change in mitochondria in cachectic tissues. CAT1 also seemed to be essential for specific aspects of amino acid metabolism in skeletal muscle cells, with significantly lower expression in cachectic mouse skeletal muscle tissues.
Discussion
In this study, intact skeletal muscle tissues from a murine genetic cancer model were examined for molecular alterations associated with cancer cachexia. For the first time, a combination of three different MALDI mass spectrometry imaging methods was used to explore in situ associations between amino acids, proteins, and energy and other cell metabolites in cachectic skeletal muscle tissues. Significant amino acid alterations were detected in cachectic mice, with higher quantities of lysine, arginine, proline, and tyrosine and lower quantities of glutamate and aspartate (Figure 1A).
Changes in the quantities of glutamine, isoleucine, leucine, and valine in cachectic skeletal muscle tissues were reported in previous studies.9, 10, 11, 12 However, the specific results were conflicting, as individual studies found lower and/or higher quantities of the same amino acids in cachectic skeletal muscle tissues. Changes of detected abundances could occur because of different analytic platforms, resulting in specific sample preparations, and the individual selected skeletal muscles. Our results in quadriceps muscle tissue and MALDI analysis showed no significant changes towards a higher quantity of all of these amino acids in cachectic skeletal muscle tissues (Figure 1A). Furthermore, among all amino acids detected in our study, significant changes were especially noted with cationic amino acids and glutamate. Because lysine, arginine, and glutamate are connected by a biochemical pathway related to mitochondria,27, 28 we focused on mitochondria to understand the detected amino acid alterations. Of note, mitochondrial enzymes catalyze the synthesis of glutamate by transferring the amino group from amino acids such as lysine and arginine to α‐ketoglutarate.27, 28 Nonetheless, lysine and arginine must first enter the mitochondria.
CAT1, a transporter molecule in mammalian mitochondria,25 carries arginine and lysine through membranes.29 The presence of CAT1 in mitochondria was confirmed by western blot analysis of isolated cardiac mitochondria and by confocal microscopy, and it seemed to be involved in cationic amino acid transport, mitochondrial stress, survival, and diminished reactive oxygen species production.25 In our study, we detected higher quantities of the cationic amino acids lysine and arginine and a lower quantity of glutamate in the muscles of cachectic mice, compared with non‐cachectic ones. We also observed lower expression of CAT1 in cachectic mice. Correlation analysis confirmed significant correlations between CAT1 and lysine, as well as glutamate: low CAT1 expression was associated with more lysine and less glutamate in muscle tissues. These findings may be due to enhanced protein breakdown in cachexia,30 leading to increased levels of certain amino acids, such as arginine and lysine (Figure 6A). Reduced metabolic conversion because of diminished CAT1 expression could also account for the significantly higher quantities of arginine and lysine [Figure 6B, (1)]. Of note, our findings are consistent with the results of previous studies, which showed that arginine and lysine were increased in the gastrocnemius of cachectic mice12 and glutamate was reduced in quadriceps muscles of cachectic C26 hosts.11 Decreased production of glutamate from arginine and lysine because of reduced transamination reactions could account for the significantly lower quantity of glutamate in cachectic mouse tissues [Figure 6B, (2)]. Glutamate production could also be hampered by a low NADH/NAD+ ratio in mitochondria, which would favour oxidative decarboxylation of α‐ketoglutarate to succinyl‐CoA.31 An alteration in an electron shuttle system through the inner mitochondrial membrane, for example, could be an explanation for an altered ratio. This may lead to a lower amount of NADH inside mitochondria.
Figure 6.
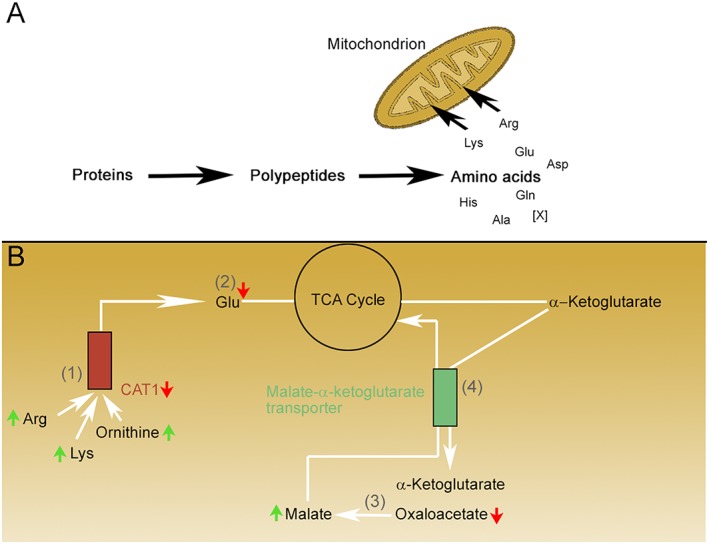
Hypothesis regarding the molecular changes in skeletal muscles during cachexia, focusing on mitochondrial dysfunction. The small red and green arrows indicate molecules for which significant changes were detected in the current study. (A) Proteins in muscle tissues of cachectic mice are degraded and subsequently processed to amino acids. Individual amino acids are then transported into the mitochondria for further metabolism. (B) (1) Lysine (Lys), arginine (Arg), and ornithine (data not shown) are transported via CAT1 into the mitochondria. (2) Specific transaminase proteins metabolize Lys, Arg, ornithine, and other amino acids and produce glutamate. Glutamate is decreased in cachexia because of reduced CAT1 expression. (3) Cytosolic NADH reduces oxaloacetate to malate; it is hypothesized that an increased quantity of cytosolic NADH in cachexia increases the quantity of malate. (4) Malate is exchanged with α‐ketoglutarate in the mitochondrial matrix by the malate‐α‐ketoglutarate transporter.
Reduced expression of CAT1 in the muscles of cachectic mice and patients could arise from several mechanisms. For instance, a lower number of mitochondria would reduce mitochondrial CAT1 expression. However, examination of the tissues with TEM (the gold standard for measuring mitochondrial content32) revealed no evidence of a reduced number or altered structural integrity of mitochondria in cachexia (Figure 5E). Likewise, examination of mitochondrial content by staining for VDAC (the most abundant protein in the outer membrane of mitochondria33) did not show significant differences between cachectic and non‐cachectic mice (Figure 5G). These results indicate that decreased CAT1 expression is not the result of an altered number of mitochondria; instead, it is due to a specific decrease in expression of the CAT1 protein itself (Figure 5B). Thus, decreased mitochondrial CAT1 expression has been described for the first time in cancer cachexia, which may be highly relevant to the amino acid metabolism changes observed in this setting, as discussed earlier.
Manifestations of cachexia can differ according to the type of tumour.34 In the current study, colorectal and pancreatic cancer revealed similar molecular changes during cachexia. Lysine, arginine, and proline quantities were increased, and glutamate and aspartate were decreased in skeletal muscle tissues of both types of tumours during cachexia. Notably, human material offered only preliminary results in the current study due to the small patient number and further studies, including more patients, are necessary for confirming these molecular alterations. A recent study comparing lung carcinoma and colon adenocarcinoma models under cachectic conditions also found molecular similarities between the two types of malignancies.35 Despite different food intake in the two models, hypothalamic expression of the orexigenic neuropeptide Y was increased in both tumour types, compared with controls. At this point, our data suggest tumour type‐independent molecular alterations, at least for colorectal and pancreatic cancers. Additional studies are necessary to further explore the influence of tumour type on the molecular manifestations of cachexia.
Protein breakdown is highly discussed in the context of cachexia.2 In the present study, simultaneous examination of proteins and amino acids enabled the identification of proteins that may be selectively degraded in cachectic skeletal muscle tissues. A majority of detected and degraded proteins in this study were OXPHOS proteins, which were negatively correlated with an overabundance of certain amino acids (Figure 2A), thereby suggesting the presence of a degradation process. Of note, only small undigested proteins (<25 kDa) are determined by MALDI imaging protein measurement and released amino acids from muscle can not be set into relation with proteins and metabolites in the current study. A recent study of rat gastrocnemius muscles revealed an accumulation of oxidatively modified mitochondrial proteins in cachexia.36 Oxidative modifications can be repaired enzymatically or lead to protein degradation. Both processes are essential for maintaining homeostasis and survival of cells.37 Oxidative modifications might also initiate degradation of the OXPHOS proteins detected in the current study. A previous study found reduced quantity of COX6B1 protein in cachectic mouse tissues, compared with controls,38 which is consistent with our results (Figure 2C). COX6B1 could be altered by oxidative modification and therefore degraded in cachectic skeletal muscle tissues. This degradation could account for the lower quantity of COX6B1 in cachectic skeletal muscle tissues (Figure 2C) and the negative correlation between COX6B1 and individual amino acids (Figure 2A). Of interest, CAT1 is also involved in reducing oxidative stress.25 Normal CAT1 expression could help maintain normal protein signatures in mitochondria by preventing oxidative damage and thereby protecting proteins from degradation.
Mitochondria, as well as OXPHOS proteins, are associated with ATP production and, therefore, energy generation in muscle tissues. Given our findings of OXPHOS protein degradation and altered mitochondrial protein signatures during cachexia, we examined the energy status of cachectic skeletal muscle tissues. The energy charge, defined as the mole fraction of ATP plus half the mole fraction of ADP, is linearly related to the amount of temporarily stored metabolically available energy.39 We found that energy charge was significantly decreased in cachexia. Furthermore, correlation analysis indicated that higher energy charge was associated with a lower quantity of individual amino acids, higher expression of OXPHOS proteins, and lower expression of ubiquitin. Nonetheless, glutamate correlated positively with the energy charge (Figure 4). A reduced rate of ATP synthesis (determined by 31P nuclear magnetic resonance spectroscopy) has been previously described in cancer cachexia.40 Degradation of mitochondrial OXPHOS proteins could at least partially explain the reduced energy stores in cachectic muscles. As ubiquitin, a marker for protein degradation, can initiate proteasomal degradation,41 lower ubiquitin tagging could reduce degradation of OXPHOS proteins and the quantity of amino acids but increase the energy charge. The positive correlation between glutamate and energy charge could underlie the anaplerotic processes of glutamate in cancer cachexia.13
Reduced energy availability in muscle tissues could lead to higher glycolytic activity, because two NADH and two ATP molecules are produced from one glucose molecule, and the NADH can be utilized for ATP synthesis in mitochondria. However, the inner mitochondrial membrane is impermeable to NADH, so cytosolic NADH must be transported into the mitochondrial matrix by transport systems, such as the malate‐aspartate shuttle.42 High glycolytic activity has been described in cachexia,12, 43 which could lead to high quantities of cytosolic NADH [Figure 6B, (3)]. Pin et al.11 formally described an increased reliance on glycolytic metabolism and a reduced flux of glycolysis‐derived pyruvate into the TCA cycle, which might also be followed by reducing energy in cachectic skeletal muscle tissues. Malate dehydrogenase 1 enzyme can utilize NADH to reduce oxaloacetate to malate44 for transporting electrons from NADH across the inner membrane via the malate‐α‐ketoglutarate transporter [Figure 6B, (4)]. This could account for the low oxaloacetate and high malate quantities found in the current study (Figure 3C). High levels of cytosolic NADH may reduce a high amount of oxaloacetate to malate, available for carrying electrons across the inner mitochondrial membrane for ATP synthesis.
To conclude, our data revealed mitochondrial dysfunction in cachectic skeletal muscle tissue, which appeared to have a substantial influence on amino acid metabolism and protein breakdown, based on several lines of evidence. Firstly, we observed significant accumulation of lysine and arginine in cachexia, which can be metabolized in mitochondria to glutamate.27, 28 Secondly, indices of degradation suggested the presence of degradation of mitochondrial OXPHOS proteins. Thirdly, the energy charge was reduced in cachexia, which may be at least partly attributed to mitochondrial dysfunction. Finally, cachectic muscle tissues exhibited significantly reduced expression of the mitochondrial protein CAT1, which may play an important role in the observed molecular changes associated with cachexia.
Conflict of interest
None declared.
Funding
The study was funded by the Ministry of Education and Research of the Federal Republic of Germany (Bundesministerium für Bildung und Forschung; 01ZX1610B and 01KT1615), the Deutsche Forschungsgemeinschaft (SFB 824 C4 and CRC/Transregio 205/1), and the Deutsche Krebshilfe (70112617) to A.W.
Supporting information
Appendix S1. Advanced baseline characteristics for patients, which is separating patient material in individual fixation methods. FFPE tissues were used for immunohistochemical staining, and fresh frozen tissues were used for the determination of amino acids.
Appendix S2. Intensities of amino acids in skeletal muscle tissues of cachectic and non‐cachectic mice. P values were calculated by Mann–Whitney U test.
Appendix S3. In situ amino acid quantities in skeletal muscle tissues from cachectic patients with cancer. Changes in lysine, arginine, proline, glutamate, and aspartate with cachexia in humans revealed similarities to the changes observed in cachectic mouse skeletal muscle tissues. The horizontal lines represent the mean intensity of each group.
Appendix S4. P values (Spearman's rank correlation analysis) of significant negative correlations between proteins and amino acids. Included proteins were may degraded in skeletal muscle tissues of cachectic mice.
Appendix S5. Analysis of mitochondrial proteins in mouse skeletal muscle tissues (A) Statistical analysis for the OXPHOS‐related proteins COX7C, cytochrome c, and ATPase F6 determined by MALDI mass spectrometry imaging. COX7C (P = 0.0127) and ATPase F6 (P = 0.0048) expression was significantly decreased in cachexia, compared with non‐cachectic skeletal muscle tissues. Cytochrome c was also decreased in cachectic mouse skeletal muscle tissues, but the differences did not reach statistical significance (P = 0.1145). (B) Immunohistochemistry (IHC) results confirmed changes of mitochondrial proteins detected by MALDI mass spectrometry imaging. Quantification of the IHC, performed by digital image analysis, revealed a lower expression of COX7C in cachectic mouse skeletal muscle tissues (P = 0.0159) and a similar change of cytochrome c without reaching significance level (P = 0.2512). * P < 0.05, ** P < 0.01.
Appendix S6. Immunofluorescence analysis of myosin heavy chain (MHC) expressions. (A) Statistical analysis of the expression of myosin heavy chains in cachectic and non‐cachectic mouse skeletal muscle tissues. MHC1 expressions were in both comparison groups 0.02%. The Mann–Whitney U test was performed for all fibre types and revealed no significant change between non‐cachectic and cachectic mouse comparison groups. (B) Exemplary pictures for immunofluorescence stained cross‐sectioned mouse skeletal muscle tissues. The left tissue section is representing a non‐cachectic mouse, whereby the right tissue is belonging to a cachectic mouse. Shown are type I (blue), type IIA (green), type IIB (red), and type IIX (unstained) fibres.
Appendix S7. Statistical analysis regarding changes of molecules in the tricarboxylic acid (TCA) cycle. Peak intensity was significantly higher for malate (P = 0.0295) and lower for oxaloacetate (P = 0.0448) in tissues of cachectic mice, compared with non‐cachectic ones. Boxplot whiskers represent the lowest and highest peak intensities in each group. * P < 0.05.
Acknowledgements
The authors would like to thank Ulrike Buchholz, Claudia‐Mareike Pflüger, Gabriele Mettenleiter, Cristina Huebner Freitas, Elenore Samson, and Andreas Voss for their excellent technical assistance. Primary antibodies for analysis of myosin heavy chain expression were developed by Schiaffino, S., obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH, and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. The authors certify that they comply with the ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle.45
Kunzke T., Buck A., Prade V. M., Feuchtinger A., Prokopchuk O., Martignoni M. E., Heisz S., Hauner H., Janssen K.‐P., Walch A., and Aichler M. (2020) Derangements of amino acids in cachectic skeletal muscle are caused by mitochondrial dysfunction, Journal of Cachexia, Sarcopenia and Muscle, 11: 226–240. 10.1002/jcsm.12498.
References
- 1. Tisdale MJ. Cachexia in cancer patients. Nat Rev Cancer 2002;2:862–871. [DOI] [PubMed] [Google Scholar]
- 2. Argiles JM, Busquets S, Stemmler B, Lopez‐Soriano FJ. Cancer cachexia: understanding the molecular basis. Nat Rev Cancer 2014;14:754–762. [DOI] [PubMed] [Google Scholar]
- 3. Pisters PW, Pearlstone DB. Protein and amino acid metabolism in cancer cachexia: investigative techniques and therapeutic interventions. Crit Rev Clin Lab Sci 1993;30:223–272. [DOI] [PubMed] [Google Scholar]
- 4. Khal J, Wyke SM, Russell ST, Hine AV, Tisdale MJ. Expression of the ubiquitin–proteasome pathway and muscle loss in experimental cancer cachexia. Br J Cancer 2005;93:774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sandri M. Protein breakdown in cancer cachexia. Semin Cell Dev Biol 2016;54:11–19. [DOI] [PubMed] [Google Scholar]
- 6. Cornwell EW, Mirbod A, Wu CL, Kandarian SC, Jackman RW. C26 cancer‐induced muscle wasting is Iκκβ‐dependent and NF‐κβ‐independent. PLoS ONE 2014;9:e87776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duval AP, Jeanneret C, Santoro T, Dormond O. mTOR and Tumor Cachexia. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. White JP, Baynes JW, Welle SL, Kostek MC, Matesic LE, Sato S, et al. The regulation of skeletal muscle protein turnover during the progression of cancer cachexia in the Apc (Min/+) mouse. PLoS ONE 2011;6:e24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tseng YC, Kulp SK, Lai IL, Hsu EC, He WA, Frankhouser DE, et al. Preclinical investigation of the novel histone deacetylase inhibitor AR‐42 in the treatment of cancer‐induced cachexia. J Natl Cancer Inst 2015;107:djv274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. QuanJun Y, GenJin Y, LiLi W, Yan H, YongLong H, Jin L, et al. Integrated analysis of serum and intact muscle metabonomics identify metabolic profiles of cancer cachexia in a dynamic mouse model. RSC Adv 2015;5:92438–92448. [Google Scholar]
- 11. Pin F, Barreto R, Couch ME, Bonetto A, O'Connell TM. Cachexia induced by cancer and chemotherapy yield distinct perturbations to energy metabolism. J Cachexia Sarcopenia Muscle 2019;10:140–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Der‐Torossian H, Wysong A, Shadfar S, Willis MS, McDunn J, Couch ME. Metabolic derangements in the gastrocnemius and the effect of compound A therapy in a murine model of cancer cachexia. J Cachexia Sarcopenia Muscle 2013;4:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rutten EP, Engelen MP, Schols AM, Deutz NE. Skeletal muscle glutamate metabolism in health and disease: state of the art. Curr Opin Clin Nutr Metab Care 2005;8:41–51. [DOI] [PubMed] [Google Scholar]
- 14. Dickinson JM, Rasmussen BB. Amino acid transporters in the regulation of human skeletal muscle protein metabolism. Curr Opin Clin Nutr Metab Care 2013;16:638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Janssen KP, Alberici P, Fsihi H, Gaspar C, Breukel C, Franken P, et al. APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology 2006;131:1096–1109. [DOI] [PubMed] [Google Scholar]
- 16. Fearon K, Strasser F, Anker SD, Bosaeus I, Bruera E, Fainsinger RL, et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol 2011;12:489–495. [DOI] [PubMed] [Google Scholar]
- 17. Ly A, Buck A, Balluff B, Sun N, Gorzolka K, Feuchtinger A, et al. High‐mass‐resolution MALDI mass spectrometry imaging of metabolites from formalin‐fixed paraffin‐embedded tissue. Nat Protoc 2016;11:1428–1443. [DOI] [PubMed] [Google Scholar]
- 18. Mantini D, Petrucci F, Pieragostino D, Del Boccio P, Di Nicola M, Di Ilio C, et al. LIMPIC: a computational method for the separation of protein MALDI‐TOF‐MS signals from noise. BMC Bioinformatics 2007;8:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guijas C, Montenegro‐Burke JR, Domingo‐Almenara X, Palermo A, Warth B, Hermann G, et al. METLIN: a technology platform for identifying knowns and unknowns. Anal Chem 2018;90:3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the human metabolome database. Nucleic Acids Res 2007;35:D521–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maier SK, Hahne H, Gholami AM, Balluff B, Meding S, Schoene C, et al. Comprehensive identification of proteins from MALDI imaging. Mol Cell Proteomics 2013;12:2901–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayakawa E, Fujimura Y, Miura D. MSIdV: a versatile tool to visualize biological indices from mass spectrometry imaging data. Bioinformatics 2016;32:3852–3854. [DOI] [PubMed] [Google Scholar]
- 23. Bergmeister KD, Groger M, Aman M, Willensdorfer A, Manzano‐Szalai K, Salminger S, et al. A rapid automated protocol for muscle fiber population analysis in rat muscle cross sections using myosin heavy chain immunohistochemistry. J Vis Exp 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev 2011;91:1447–1531. [DOI] [PubMed] [Google Scholar]
- 25. Williams D, Venardos KM, Byrne M, Joshi M, Horlock D, Lam NT, et al. Abnormal mitochondrial l‐arginine transport contributes to the pathogenesis of heart failure and rexoygenation injury. PLoS ONE 2014;9:e104643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shum AM, Mahendradatta T, Taylor RJ, Painter AB, Moore MM, Tsoli M, et al. Disruption of MEF2C signaling and loss of sarcomeric and mitochondrial integrity in cancer‐induced skeletal muscle wasting. Aging (Albany NY) 2012;4:133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Papes F, Surpili MJ, Langone F, Trigo JR, Arruda P. The essential amino acid lysine acts as precursor of glutamate in the mammalian central nervous system. FEBS Lett 2001;488:34–38. [DOI] [PubMed] [Google Scholar]
- 28. Morris SM Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annu Rev Nutr 2002;22:87–105. [DOI] [PubMed] [Google Scholar]
- 29. Hatzoglou M, Fernandez J, Yaman I, Closs E. Regulation of cationic amino acid transport: the story of the CAT‐1 transporter. Annu Rev Nutr 2004;24:377–399. [DOI] [PubMed] [Google Scholar]
- 30. Argiles JM, Lopez‐Soriano FJ. The ubiquitin‐dependent proteolytic pathway in skeletal muscle: its role in pathological states. Trends Pharmacol Sci 1996;17:223–226. [DOI] [PubMed] [Google Scholar]
- 31. Frigerio F, Casimir M, Carobbio S, Maechler P. Tissue specificity of mitochondrial glutamate pathways and the control of metabolic homeostasis. Biochim Biophys Acta 2008;1777:965–972. [DOI] [PubMed] [Google Scholar]
- 32. Larsen S, Nielsen J, Hansen CN, Nielsen LB, Wibrand F, Stride N, et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J Physiol 2012;590:3349–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kroemer G, Reed JC. Mitochondrial control of cell death. Nat Med 2000;6:513–519. [DOI] [PubMed] [Google Scholar]
- 34. Tisdale MJ. Mechanisms of cancer cachexia. Physiol Rev 2009;89:381–410. [DOI] [PubMed] [Google Scholar]
- 35. Dwarkasing JT, Boekschoten MV, Argiles JM, van Dijk M, Busquets S, Penna F, et al. Differences in food intake of tumour‐bearing cachectic mice are associated with hypothalamic serotonin signalling. J Cachexia Sarcopenia Muscle 2015;6:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Padrao AI, Oliveira P, Vitorino R, Colaco B, Pires MJ, Marquez M, et al. Bladder cancer‐induced skeletal muscle wasting: disclosing the role of mitochondria plasticity. Int J Biochem Cell Biol 2013;45:1399–1409. [DOI] [PubMed] [Google Scholar]
- 37. Bulteau AL, Szweda LI, Friguet B. Mitochondrial protein oxidation and degradation in response to oxidative stress and aging. Exp Gerontol 2006;41:653–657. [DOI] [PubMed] [Google Scholar]
- 38. Shum AMY, Poljak A, Bentley NL, Turner N, Tan TC, Polly P. Proteomic profiling of skeletal and cardiac muscle in cancer cachexia: alterations in sarcomeric and mitochondrial protein expression. Oncotarget 2018;9:22001–22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Atkinson DE. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry 1968;7:4030–4034. [DOI] [PubMed] [Google Scholar]
- 40. Constantinou C, Fontes de Oliveira CC, Mintzopoulos D, Busquets S, He J, Kesarwani M, et al. Nuclear magnetic resonance in conjunction with functional genomics suggests mitochondrial dysfunction in a murine model of cancer cachexia. Int J Mol Med 2011;27:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lecker SH, Goldberg AL, Mitch WE. Protein Degradation by the ubiquitin–proteasome pathway in normal and disease states. J Am Soc Nephrol 2006;17:1807–1819. [DOI] [PubMed] [Google Scholar]
- 42. Kovacevic Z. Possibility for the transfer of reducing equivalents from the cytosol to the mitochondrial compartment in Ehrlich ascites tumor cells by the malate‐aspartate shuttle. Eur J Biochem 1972;25:372–378. [DOI] [PubMed] [Google Scholar]
- 43. Mantovani G. Cachexia and wasting a modern approach. Milan: Springer; 2006. [Google Scholar]
- 44. Mitchell M, Cashman KS, Gardner DK, Thompson JG, Lane M. Disruption of mitochondrial malate‐aspartate shuttle activity in mouse blastocysts impairs viability and fetal growth. Biol Reprod 2009;80:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2017. J Cachexia Sarcopenia Muscle 2017;8:1081–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Advanced baseline characteristics for patients, which is separating patient material in individual fixation methods. FFPE tissues were used for immunohistochemical staining, and fresh frozen tissues were used for the determination of amino acids.
Appendix S2. Intensities of amino acids in skeletal muscle tissues of cachectic and non‐cachectic mice. P values were calculated by Mann–Whitney U test.
Appendix S3. In situ amino acid quantities in skeletal muscle tissues from cachectic patients with cancer. Changes in lysine, arginine, proline, glutamate, and aspartate with cachexia in humans revealed similarities to the changes observed in cachectic mouse skeletal muscle tissues. The horizontal lines represent the mean intensity of each group.
Appendix S4. P values (Spearman's rank correlation analysis) of significant negative correlations between proteins and amino acids. Included proteins were may degraded in skeletal muscle tissues of cachectic mice.
Appendix S5. Analysis of mitochondrial proteins in mouse skeletal muscle tissues (A) Statistical analysis for the OXPHOS‐related proteins COX7C, cytochrome c, and ATPase F6 determined by MALDI mass spectrometry imaging. COX7C (P = 0.0127) and ATPase F6 (P = 0.0048) expression was significantly decreased in cachexia, compared with non‐cachectic skeletal muscle tissues. Cytochrome c was also decreased in cachectic mouse skeletal muscle tissues, but the differences did not reach statistical significance (P = 0.1145). (B) Immunohistochemistry (IHC) results confirmed changes of mitochondrial proteins detected by MALDI mass spectrometry imaging. Quantification of the IHC, performed by digital image analysis, revealed a lower expression of COX7C in cachectic mouse skeletal muscle tissues (P = 0.0159) and a similar change of cytochrome c without reaching significance level (P = 0.2512). * P < 0.05, ** P < 0.01.
Appendix S6. Immunofluorescence analysis of myosin heavy chain (MHC) expressions. (A) Statistical analysis of the expression of myosin heavy chains in cachectic and non‐cachectic mouse skeletal muscle tissues. MHC1 expressions were in both comparison groups 0.02%. The Mann–Whitney U test was performed for all fibre types and revealed no significant change between non‐cachectic and cachectic mouse comparison groups. (B) Exemplary pictures for immunofluorescence stained cross‐sectioned mouse skeletal muscle tissues. The left tissue section is representing a non‐cachectic mouse, whereby the right tissue is belonging to a cachectic mouse. Shown are type I (blue), type IIA (green), type IIB (red), and type IIX (unstained) fibres.
Appendix S7. Statistical analysis regarding changes of molecules in the tricarboxylic acid (TCA) cycle. Peak intensity was significantly higher for malate (P = 0.0295) and lower for oxaloacetate (P = 0.0448) in tissues of cachectic mice, compared with non‐cachectic ones. Boxplot whiskers represent the lowest and highest peak intensities in each group. * P < 0.05.


