Abstract
Bacterial mutualists generate major fitness benefits for eukaryotes, reshaping the host phenotype and its interactions with the environment. Yet, microbial mutualist populations are predicted to generate mutants that defect from providing costly services to hosts while maintaining the capacity to exploit host resources. Here, we examined the mutualist service of symbiotic nitrogen fixation in a metapopulation of root-nodulating Bradyrhizobium spp. that associate with the native legume Acmispon strigosus. We quantified mutualism traits of 85 Bradyrhizobium isolates gathered from a 700 km transect in California spanning 10 sampled A. strigosus populations. We clonally inoculated each Bradyrhizobium isolate onto A. strigosus hosts and quantified nodulation capacity and net effects of infection, including host growth and isotopic nitrogen concentration. Six Bradyrhizobium isolates from five populations were categorized as ineffective because they formed nodules but did not enhance host growth via nitrogen fixation. Six additional isolates from three populations failed to form root nodules. Phylogenetic reconstruction inferred two types of mutualism breakdown, including three to four independent losses of effectiveness and five losses of nodulation capacity on A. strigosus. The evolutionary and genomic drivers of these mutualism breakdown events remain poorly understood.
Keywords: cheating, evolutionary instability, host control, interspecific conflict, mutualism breakdown
1. Introduction
Bacterial mutualists offer an array of fitness-enhancing services to plants, animals and other multicellular hosts [1], including antibiotic protection [2], accelerated host growth [3], enhanced immune defence [4], and improved outcomes from host interactions with predators, pathogens and competitors [5]. However, bacteria have a tremendous evolutionary advantage over hosts in terms of population size and generation time [6], and natural selection is predicted to favour the evolution of mutants that defect from providing services to hosts [7]. Consistent with evolutionary instability, bacterial populations display immense diversity in mutualist effects [8], often encompassing beneficial genotypes as well as genotypes that provide negligible benefit to the host [3,8–11]. However, convincing evidence for mutualism breakdown—the evolution of uncooperative mutants from mutualist ancestors [12]—has been scant [12–14], suggesting to some biologists that mutualism instability has little ecological relevance [15,16].
The legume–rhizobia symbiosis is an ideal system to investigate the evolution of symbiotic effectiveness (i.e. microbial capacity to enhance host fitness). Rhizobia encompass soil-dwelling proteobacteria [17] that instigate nodule formation on legume roots and fix nitrogen [18]. Rhizobia vary genotypically in symbiotic effectiveness [6], ranging from beneficial genotypes that enhance host growth through nitrogen fixation to ineffective rhizobia that nodulate the host but fix no nitrogen [10]. Legumes exhibit host control traits that can constrain infection and in planta proliferation of ineffective rhizobia [19–25], and are predicted to impose selection against rhizobia that exploit hosts [26–29]. Nonetheless, ineffective rhizobia have been recovered from agricultural [30–38] and unmanaged soils [3,10,39–41], suggesting that nitrogen fixation might be recurrently lost in populations.
Here, we investigated symbiotic effectiveness in a metapopulation of Bradyrhizobium on the host plant Acmispon strigosus. Acmispon strigosus (formally Lotus strigosus) is an annual legume native to California [42] nodulated by Bradyrhizobium spp. [43–45]. Acmispon legumes regulate nodule growth dependent upon the net benefits gained from specific rhizobia strains [21,46,47] and ‘sanction’ rhizobia by arresting in planta proliferation of ineffective strains [22,23,48]. However, the degree to which these control mechanisms are independent remains unknown. Two ineffective Bradyrhizobium strains were isolated from geographically distant A. strigosus hosts, suggesting independent origins of non-fixing rhizobia [3,40]. The present study investigated 85 Bradyrhizobium isolates originating from 10 native A. strigosus populations across a 700 km transect in California, ranging from mesic coastal sites in northern California to desert sites in southeastern California [43–45]. To quantify Bradyrhizobium effectiveness, we performed clonal inoculations onto A. strigosus seedlings. We estimated the capacity of each clonal inoculum to induce nodule formation, to affect host growth and to fix nitrogen on A. strigosus. We used four loci distributed across the Bradyrhizobium genome to reconstruct phylogenetic relationships among our isolates, and also inferred relationships to Bradyrhizobium that associate with other host legumes. Our first goal was to examine the frequency and spatial distribution of ineffective Bradyrhizobium in natural populations of hosts, to assess how common ineffective rhizobia are and whether they are more prevalent in certain regions of the host range. Our second goal was to reconstruct the evolutionary history of ineffective Bradyrhizobium to resolve whether ineffective strains represent a single evolutionary origin or if they are unrelated and have evolved recurrently from independent ancestors.
2. Material and methods
(a). Bradyrhizobium isolates
Bradyrhizobium were previously isolated from the nodules of A. strigosus, its root surface and from surrounding bulk soil at 10 natural sites along a greater than 700 km transect, encompassing 1292 isolates [43–45]. For nodule and root surface isolates, whole plants were excavated, brought to the laboratory and roots were washed with tap water to remove all soil particles. Whole nodules were dissected, surface sterilized in bleach (5% sodium hypochlorite), rinsed in sterile water and individually cultured by crushing with a sterile glass rod and plating nodule contents onto plates of modified arabinose gluconate (MAG, 1.8% w/v agar) [43]. A single colony was archived from each nodule, which was assumed to contain a single genotype of Bradyrhizobium [24]. For root surface isolates, rinsed plant roots were dissected into approximately 1 cm sections, vortexed in sterile water and the wash solution was plated on a glucose-based rhizobia defined medium (GRDM) [49]). Bradyrhizobium were selected from the resultant colonies based upon growth on selective media and genotyping [43,44]. For bulk soil isolates, soils immediately adjacent to A. strigosus were collected from three southern California A. strigosus populations (electronic supplementary material, table S1) and prepared into slurries before being inoculated onto axenic A. strigosus seedlings. Soil cores were collected in August 2014, sieved to 2 mm, saturated with sterile water, filtered through eight layers of sterile cheesecloth and the supernatant was inoculated onto axenic A. strigosus seedlings originating from the same sites as the soil cores (14 August 2014). Plants were raised six weeks in a growth room, fertilized weekly with nitrogen-free Jensen's solution [50], and de-potted. Nodules were cultured onto MAG plates and a single colony from each plate was archived. Soil isolates were only cultured from white or yellow nodules (i.e. those that are lacking apparent plant expression of leghaemoglobin associated with symbiotic nitrogen fixation [51]) to improve chances of isolating ineffective rhizobial genotypes.
(b). Bradyrhizobium genotyping and phylogenetic reconstruction
Nodule and root surface isolates were previously sequenced for glnII and recA on the Bradyrhizobium chromosome (CHR), and nodZ and nolL on the symbiosis island (SI) [44,45]. The SI can be integrated on the CHR or exist as a plasmid [52] and can be transferred horizontally among CHR lineages [44,45]. Sequences from each genome region (CHR, SI) were aligned separately using Clustal Omega [53] (electronic supplementary material, table S1).
Phylogenetic trees of the 85 isolates were reconstructed separately for the concatenated nucleotides of the CHR and SI loci with Mesorhizobium loti (MAFF303099) as an outgroup taxon. Sequences were aligned using default parameters. The GTR + I + G model of evolution was selected from the Akaike information criterion in jModeltest2 [54]. Phylogenetic trees were reconstructed with MrBayes 3.1.2 [55] using 5 × 106 generations, a heating temperature of 0.01, a ‘burnin’ of the first 10 000 trees and two parallel runs starting with random trees, each with four simultaneous chains. A plot of log-likelihood scores of sampling points (sample frequency = 500) against generation number was observed in each case to ensure that stationarity had been reached during the ‘burnin’ period. We sampled approximately 105 post-burnin trees for phylogenetic reconstruction.
To examine evolutionary relationships with Bradyrhizobium from other studies, a single gene from each locus was also used as a query sequence in BLASTN searches against the NCBI refseq_genomic database masked to Bradyrhizobium with an e-value cut-off of 10−5. Nucleotide sequences were aligned using MAFFT v. 7.402 [56] and IQ-TREE v. 1.6.12 with the options ‘-m TEST -bb 1000 -alrt 1000’ was used to select evolutionary models for each dataset and generate separate phylogenetic trees for each gene (100 tree searches, 1000 ultrafast bootstrap replicates, 1000 aLRT test replicates) [57].
(c). Selection of isolates for analysis
Eighty-five Bradyrhizobium isolates were chosen for this study following criteria of (i) sampling from the 10 A. strigosus field sites (mean = 8.5 isolates per site, range, 4–18; electronic supplementary material, table S1), (ii) including all 12 previously identified species-like clades of Bradyrhizobium isolated from A. strigosus [44], and (iii) including all isolation methods (62 nodule isolates, 8 root surface isolates, 15 bulk soil isolates).
(d). Inoculation experiments
Bradyrhizobium cultures were plated from clonal stocks and incubated until lawns formed (29°C, approx. 8 days), then washed from plates and resuspended in liquid MAG to estimate concentration via optical density [3]. Washed cells were centrifuged (4000g, 20 min) to remove media and resuspended in sterile water to a concentration of 108 cells ml−1. Inoculated plants received 5 × 108 rhizobial cells in 5 ml of sterile water and uninoculated control plants received 5 ml of sterile water.
Acmispon strigosus is a permissive host that forms nodules with diverse Bradyrhizobium spp. [44,45]. Previous inoculation studies of A. strigosus and Bradyrhizobium found relatively consistent effects of rhizobial genotypes upon different host genotypes [46,58]. Thus, a single inbred A. strigosus host line from the Claremont population was used for the experiment (AcS049.Cla.m01.g1.r02; [46]). Seeds were surface sterilized, nick scarified and germinated in sterile nitrogen-free Jensen's solution [50]. Seedlings were planted into sterilized Cone-tainers (Steuwe and Sons, Corvallis, OR) filled with sterilized quartzite sand, incubated in a growth chamber for two weeks, and moved to the greenhouse under approximately 50% shade for hardening (4 days, 1 × aily misting). One week after planting, seedlings were fertilized with 1 ml sterile nitrogen-free Jensen's solution, which was increased to 3 ml per plant at two weeks after planting. Beginning three weeks after planting (approx. 2 days before inoculation), plants were fertilized weekly with 4.5 ml Jensen's solution supplemented with a low concentration of 15N-enriched potassium nitrate (KNO3; 0.05 g l−1; 5 atm%15N). The KNO3 treatment represents approximately 10% of the nitrogen concentration needed to maximize A. strigosus shoot growth in the absence of rhizobial infection [42].
Size-matched groups of axenic seedlings were randomly assigned to inoculation treatments and blocks. Rhizobial treatments were separated into four groups to be inoculated on separate days (electronic supplementary material, table S1), each with separate uninoculated control plants. All plants that were treated with the same Bradyrhizobium isolate were inoculated on the same day. Each inoculation treatment was replicated on 10 plants separated into individual blocks, except for treatments in the last inoculation group which had five replicate plants in one separate block, due to poor germination (85 inoculation + 4 control treatments (separate controls in each block) × 10 replicates per treatment, except for inoculation group 4 which had 5 replicates = 805 plants total). Plants were harvested approximately eight weeks after inoculation.
During harvest, plants were de-potted, soil was washed from roots and plants were wrapped and stored at 4°C until dissection. Shoots were separated from roots to measure dry shoot biomass. Nodules were removed from the roots, counted and photographed. Roots, shoots and nodules were separated and oven-dried (60°C, greater than or equal to 4 days) prior to weighing. Because root dissection is time-intensive when plants are nodulated, only a subset of replicates had their roots and nodules dissected for analysis. For treatments with consistent presence of root nodules, four replicate plants per treatment were de-potted, washed and dissected. For the remaining plant replicates in each treatment, shoots were removed at the root–shoot junction and roots were not analysed. For treatments in which plants exhibited inconsistent nodulation or the absence of nodules, all replicates were dissected.
(e). Leaf atm%15N assays
Subsequent to biomass measurement of shoots, leaflets from four replicate plants per inoculation and control treatment were removed from dried shoots and ground to a fine powder. Samples were analysed for atom per cent 15N (atm%15N) at UC Santa Cruz Stable Isotope Laboratory. We compared leaf atm%15N between inoculated and control plants for each Bradyrhizobium isolate. Since we fertilized with 15N-enriched KNO3 (5%), plants infected with symbiotically effective strains are expected to exhibit significant reductions in 15N/14N relative to uninfected plants, consistent with substantial assimilation of 14N from the atmosphere via biological nitrogen fixation.
(f). Data analysis
Bradyrhizobium traits were analysed using general linear mixed models (GLMMs) in JMP Pro 13.0. Data were log-transformed as needed to improve normality. GLMMs were used to analyse variation among collection sites (fixed effect: collection site, random effect: isolate). Variation in symbiotic effectiveness among isolates and within each population was also analysed using ANOVAs. Symbiotic effectiveness was estimated as the host's growth response (HGR) to Bradyrhizobium inoculation relative to uninoculated controls (i.e. HGR = ((shoot mass of inoculated plant – shoot mass of control plant)/shoot mass of control plant) × 100 [3]). Bradyrhizobium isolates were considered effective only if they (i) consistently formed nodules on inoculated hosts, (ii) significantly improved host growth, and (iii) fixed significant amounts of nitrogen for the hosts such that they could be differentiated from uninoculated controls in terms of atm%15N (i.e. independent samples t-test, inoculated treatments compared to uninoculated controls). We quantified in planta fitness proxies for nodulating Bradyrhizobium including the mean number of nodules formed and the mean individual biomass of nodules. Nodules are typically initiated by one or a few rhizobial cells [24,59], so nodule number and size can quantify the progeny of founding cells in clonal inoculations [60]. Nodule size also takes into account the proliferation of rhizobia that occurs within the nodule [3,60] and is often positively correlated with rhizobial population sizes in nodules of A. strigosus [3], Medicago truncatula [61,62], Glycine max [19], Lotus japonicus [47] and Lupinus arboreus [24].
(g). Phylogenetic trait analyses
Bradyrhizobium traits were tested for phylogenetic signal (a prerequisite for ancestral state reconstruction [63]) on the CHR and SI trees, and a tree that used all four loci. The same parameters were used as stated above, except a ‘burnin’ of 12 000 trees was used in the four-locus tree. In cases where a single Bradyrhizobium genotype included multiple isolates, a representative isolate was randomly selected to include in analyses. This approach eliminates polytomies (a prerequisite to analyse phylogenetic signal). We estimated Blomberg's K, which is ideal for continuous variables (i.e. host growth response, atm%15N) using the ‘phylosignal’ function in the ‘picante’ R package [64], where K compares the observed signal in a trait to the signal under a Brownian motion model [63]. K values close to 1 indicate a Brownian motion process and suggest some degree of phylogenetic signal, whereas K values close to 0 correspond to a random pattern of trait evolution. We tested if K was significantly greater than 0 (i.e. phylogenetic signal) with 999 randomizations and report the mean ± standard error of K and average p-values calculated across 200 randomly selected post-burnin trees to account for phylogenetic uncertainty.
Ancestral states of nodulation and nitrogen fixation on A. strigosus were inferred using a consensus reconstruction of the post-burnin Bayesian trees, and were inferred with maximum likelihood and parsimony. Losses of nodulation or nitrogen fixation on A. strigosus were inferred by estimating a range of minimum to maximum values. For the minimum value of loss events, we only included monophyletic clades with Bayesian posterior support values greater than 0.80 that contain taxa that exhibit lack of nodulation or nitrogen fixation, derived from ancestors with a positive proportional likelihood of nodulation or nitrogen fixation status (i.e. greater than 0.90). For the maximum value, we used relaxed criteria, including all genetically and spatially diverged taxa that exhibit lack nodulation or nitrogen fixation derived from ancestors with a high proportional likelihood of nodulation or nitrogen fixation status (i.e. greater than 0.90). Sequences queried from NCBI were used to examine whether Bradyrhizobium from other studies are intermixed on the phylogeny with isolates from A. strigosus. Using genetic distance, we tested whether ineffective isolates were more closely related to Bradyrhizobium isolated from other legume species, suggesting adaptation to other hosts. Bradyrhizobium SI loci typically cluster phylogenetically with host species, whereas CHR loci are less informative of host origin [65,66].
3. Results
(a). Categorical analysis of Bradyrhizobium symbiotic effectiveness
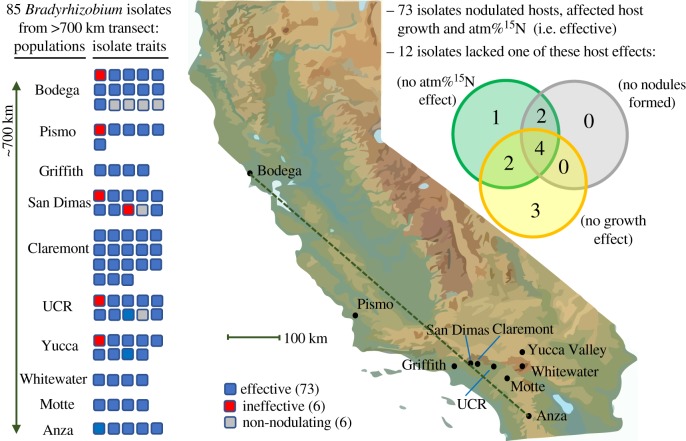
Seventy-nine of the 85 Bradyrhizobium isolates nodulated all inoculated plants. Among the remaining isolates, five failed (nos 40, 44, 53, 61, 199) to nodulate any inoculated plant and no. 149 formed a single nodule on one plant replicate. Four of these isolates, nos 40, 44, 53 and 61, were cultured from the root surface. Isolate no. 199 was originally cultured from an A. strigosus nodule, suggesting that it co-infected the original host with a nodulating isolate [67].
Seventy-three of the 85 isolates were effective on A. strigosus because they (i) consistently induced nodules, (ii) caused significant increases in host growth, and (iii) caused significant uptake of 14N from the atmosphere via biological nitrogen fixation. Conversely, six isolates (nos 2, 155, 186, 187, 200, CW1) from five collection sites were categorized as ineffective on A. strigosus because they consistently formed nodules but did not cause enhanced host growth via nitrogen fixation (figure 1).
Figure 1.
Biogeography of Bradyrhizobium effectiveness. Bradyrhizobium were sampled from A. strigosus across a 700 km transect in California (left) and categorized as effective, ineffective or non-nodulating. The map (centre) indicates locations of the collection sites with black dots and a dashed line approximating the transect. The Venn diagram (right) indicates traits from the inoculation experiment. (Online version in colour.)
Among the six isolates that did not instigate nodule formation on A. strigosus, isolates nos 44 and 61 caused modest but significant increases in host growth but did not exhibit evidence for nitrogen fixation. The remaining isolates, nos 40, 53, 149 and 199, did not have any significant effects on hosts during inoculation (electronic supplementary material, table S1).
(b). Phylogenetic reconstruction
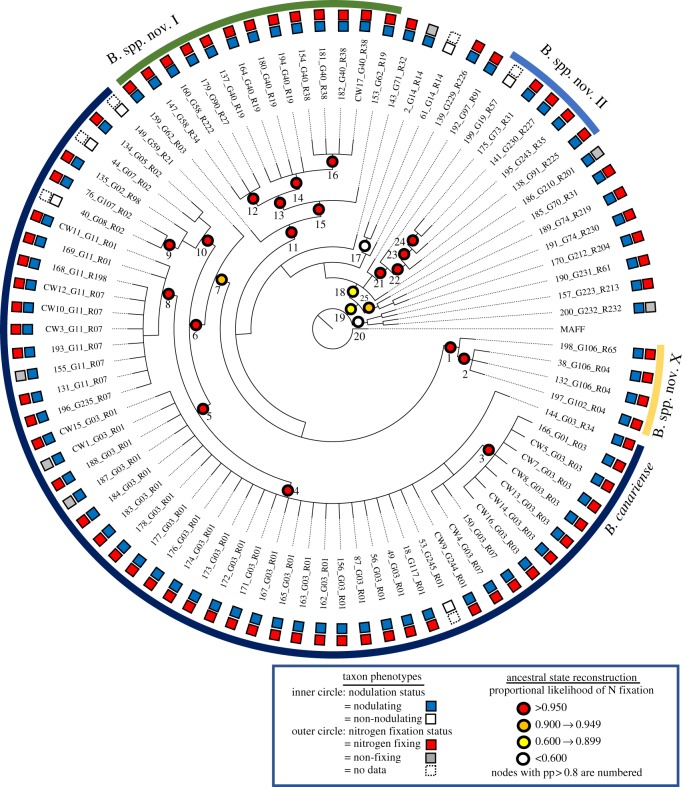
Phylogenetic reconstruction of the CHR loci recovered four species-level clades (i.e. monophyletic lineages encompassing one previously identified species, posterior support greater than or equal to 0.9, greater than or equal to 3 isolates; [44]) including B. canariense [68] and three unnamed species [44] (figure 2; electronic supplementary material, figure S2 and table S3). The remaining isolates shared genetic similarity to a diversity of reference strains and unnamed species [44] (electronic supplementary material, table S1). Reconstruction of the SI loci recovered a tree with four clades (pp > 0.80, descending from nodes 2, 11, 12 and 14) encompassing all but three isolates, which were derived on unresolved branches (i.e. nos 157, 187, 195; electronic supplementary material, figures S4 and S5, table S3). For the nodulating isolates—including the ineffective ones—we were always able to amplify and sequence at least one of the nodulation loci. We were unable to successfully amplify nodZ for isolate nos 170, 189, 190, 200 and nolL for no. 182. Conversely, we were unable to PCR amplify any SI loci on isolates that failed to nodulate A. strigosus (electronic supplementary material, table S1). Previous work in Bradyrhizobium found that no SI locus could be amplified in non-nodulating strains, suggesting the degradation or absence of the SI [44].
Figure 2.
Bayesian cladogram inferred with glnII and recA. Four previously identified species-like clades are indicated with the outermost curved bars [44]. Symbiotic phenotypes are indicated on the tips of the tree with concentric circles, the outer indicating nitrogen fixation (red squares, significant nitrogen fixation; grey, no significant nitrogen fixation; dotted square, no data) and the inner indicating nodulation. Numbers identify clades with Bayesian posterior probabilities ≥0.80 (i.e. pp, Bayesian support value). Ancestral states for nitrogen fixation are estimated for all well-supported internal nodes using maximum likelihood. Proportional likelihood of the nitrogen fixation is reported via the colour of the node labels. In the parsimony analysis, all 20 well-supported ancestral nodes were inferred to be nitrogen fixing except for no. 17, which was ambiguous (electronic supplementary material, table S3). (Online version in colour.)
(c). Trait analysis of Bradyrhizobium isolates
Blomberg's K values for host growth response were significantly different than zero on the CHR (K = 0.031, p = 0.012) and SI trees (K = 0.070, p = 0.017), but not for the four-locus phylogeny (K = 0.044, p = 0.126; table 1; electronic supplementary material, figure S6). The same pattern was true for nitrogen fixation (CHR: K = 0.031, p = 0.007; SI: K = 0.205, p = 0.016; four-locus: K = 0.078, p = 0.070).
Table 1.
Phylogenetic signal estimated with Blomberg's K. Mean ± s.e. of K and average p-values are calculated across 200 trees to account for phylogenetic uncertainty.
|
genomea |
chromosome |
symbiosis island |
||||
|---|---|---|---|---|---|---|
| trait | K | p-value | K | p-value | K | p-value |
| HGRb | 0.04353235 | 0.126 | 0.03059187 | 0.012 | 0.06993263 | 0.017 |
| atm%15N | 0.07843186 | 0.07 | 0.03094816 | 0.007 | 0.2053341 | 0.016 |
| nodule massc | 0.2301758 | 0.203 | 0.02789672 | 0.004 | 0.0730706 | 0.005 |
aGenome refers to trees reconstructed with all four loci.
bHGR refers to host growth response.
cMean individual nodule mass.
The six isolates that were categorized as non-nodulating on A. strigosus were distributed in three independently derived clades (i.e. pp ≥ 0.80) and two long unresolved branches on the CHR phylogeny (figure 2; electronic supplementary material, figure S2, table S3). One B. canariense clade encompassed two closely related non-nodulating isolates (pp = 1.00; nos 40, 44). Both the maximum likelihood and parsimony reconstruction of ancestral states inferred five losses of nodulation on A. strigosus.
The six isolates that were categorized as ineffective on A. strigosus were independently derived in four well-supported clades (pp ≥ 0.80) and one long unresolved branch on the CHR phylogeny (pp ≥ 0.50; figure 2; electronic supplementary material, figure S2, table S3). One of the B. canariense clades encompassed two of the ineffective isolates that were closely related (i.e. pp > 0.80; no. 187, CW1, but were isolated greater than 350 km apart). Maximum likelihood and parsimony reconstructions of ancestral states using the CHR tree inferred a minimum of three or four independent losses of nitrogen fixation capacity on A. strigosus, respectively. The six isolates that were ineffective on A. strigosus were independently derived in three clades (pp ≥ 0.80) and one long unresolved branch (pp ≥ 0.50, no. 187) on the SI phylogeny (electronic supplementary material, figures S4 and S5). The maximum likelihood and parsimony reconstructions of ancestral states both inferred a minimum of three losses of nitrogen fixation on A. strigosus on the SI tree.
Bradyrhizobium isolated from Lupinus and other legume genera were intermixed with isolates from A. strigosus on the CHR trees (electronic supplementary material, figures S7 and S8, table S9). Conversely, the A. strigosus Bradyrhizobium isolates formed monophyletic clades on the SI trees that excluded isolates from other species—except for one nodule isolate from Syrmatium glabrum with a nodZ genotype shared with several of our isolates (including ineffective strains no. 155, CW1; electronic supplementary material, figures S10 and S11). Genetic distance matrices of CHR loci showed that ineffective strains were sometimes more closely related to isolates from other legume species than to beneficial strains from A. strigosus. However, this was never the case for the SI loci or when taking all four loci into account (electronic supplementary material, table S12).
(d). Variation among populations in symbiosis traits
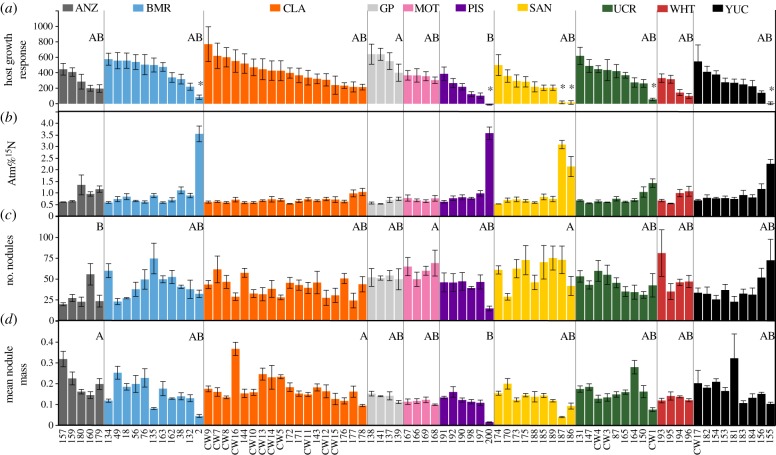
Host growth response to the Bradyrhizobium isolates varied significantly among the sampled populations (F9,66.12 = 3.0575, p = 0.0040; figure 3; electronic supplementary material, table S13) but nitrogen fixation did not (F9,69 = 1.0685, p = 0.3969). The number of nodules formed and the mean individual nodule mass of Bradyrhizobium isolates both varied significantly among the sampled populations (nodules formed: F9,77.18 = 3.1087, p = 0.0031; mean individual nodule mass: F9,70.86 = 2.0310, p = 0.0481; figure 3). Bradyrhizobium from the bulk soil isolates (which were cultured only from white or yellow nodules of plants that were inoculated with these soils) slightly increased host growth response compared to the remainder of the isolates, inconsistent with white or yellow nodules being more likely to be ineffective (F1,92.85 = 4.4699, p = 0.0372).
Figure 3.
Symbiotic traits of Bradyrhizobium isolates. Host growth response (a), atm%15N (b), mean number of nodules formed (c) and mean individual nodule mass (d) are indicated. Colours indicate different field collection sites and asterisks in the host growth response panel identify ineffective isolates. Significant differences among collection sites are indicated with capital letters (electronic supplementary material, table S13). Error bars represent 1 s.e. (Online version in colour.)
Host growth response and nitrogen fixation (atm%15N) were positively correlated (ρ = 0.6876, p < 0.0001, n = 79), consistent with nitrogen fixation being the main benefit of nodulation. Host growth response and mean individual nodule mass were also positively correlated (ρ = 0.4953, p < 0.0001, n = 79), suggesting that the plants invest more resources into nodules as the benefit of symbiotic nitrogen fixation increases [21,47]. We did not find any correlation between host growth response and the number of nodules formed (ρ = 0.0816, p = 0.4745, n = 79).
4. Discussion
Our study uncovered multiple, independent evolutionary losses of beneficial mutualism in a metapopulation of rhizobia interacting with a widespread host. Previous studies of microbial mutualist services uncovered broad genotypic variation in the magnitude of benefits that symbionts provide to hosts [8,10,11,69–71] consistent with evolutionary lability in these traits. Moreover, phylogenetic analyses have occasionally found that microbial mutualist taxa are closely related to uncooperative strains or species, allowing inference of transitions between mutualism and parasitism [12–14]. We inferred multiple transitions leading either to the loss of Bradyrhizobium nitrogen fixation or nodulation on A. strigosus in separated populations (figure 2; electronic supplementary material, figure S4). No other study that we are aware of has recovered multiple independent mutualism breakdown events occurring within a host-symbiont metapopulation. The dataset suggests that these transitions are occurring frequently, rapidly and at multiple local sites.
There is intense debate over mutualism stability. Selection for selfish traits is predicted to overcome the benefits of mutualism [72] leading its breakdown [12]; however, this might often depend on costs of cooperation or competition for partners [73,74]. We uncovered uncooperative Bradyrhizobium distributed only at the tips of the evolutionary tree, consistent with recurrent origins but no long-term fitness advantage [75]. Given the recurrent evolution of Bradyrhizobium that fail to benefit the host, what is preventing the uncooperative rhizobia from displacing beneficial strains? A null hypothesis is mutation–selection balance, wherein non-fixing rhizobia are recurrently introduced into populations via deleterious mutation and are purged at a similar rate by low fitness, either in hosts (due to host defence [6]) or in the soil environment [76]. There are conflicting data about the relative fitness of ineffective rhizobia. Our analysis here uncovered a positive correlation between symbiotic effectiveness and mean nodule mass, suggesting that cooperative strains have higher fitness in planta. However, a previous study including some of the same strains did not find a correlation [40], and instead uncovered ineffective Bradyrhizobium that achieved higher genotype frequencies than beneficial strains within populations, suggesting that cheating was favoured in those settings [40]. Evidence for rhizobial cheating has also been uncovered using nodule mass and seed mass data in the Medicago–Ensifer symbiosis [60]. It remains an open question of how often ineffective rhizobia are superior in fitness to cooperative strains, and thus can be defined as cheaters [70,77].
Bacterial mutualists can transition in their capacity to provide fitness benefits to hosts through mutation, acquisition or deletion of loci that encode symbiosis functions [3,6,13,14,78,79]. The data here are consistent with previous work, suggesting that deletion of part or all of the SI is a main driver causing rhizobia to lose capacity to nodulate hosts [3,80,81]. The evidence is less clear for rhizobia that do not fix nitrogen for a host. Losses of effectiveness on a host legume could occur if rhizobia become adapted to a novel host, and in the process lose the capacity to fix nitrogen on the initial host (i.e. G × G interactions) [10,61,80,82]. We found that some of our rhizobia were related to Bradyrhizobium isolated from other legume species, including Lupinus spp., Lablab purpureus, Syrmatium glabrum and others (electronic supplementary material, table S9), suggesting that some of these isolates might be adapted to other host species. However, for the symbiosis loci—which control host-symbiont specificity—the ineffective isolates were never more closely related to isolates from other species (electronic supplementary material, table S12). Thus, evidence is currently lacking that adaptation to a novel host drove the losses of mutualism with A. strigosus.
Ineffective rhizobia could also arise through acquisition of an SI in a genome lacking these loci [3]. A recent study of these Bradyrhizobium populations inferred recurrent evolutionary gain and loss of nodulation capacity and hypothesized that these transitions were driven by acquisition and deletion of the SI [44]. Mapping the ineffective genotypes uncovered in the current study onto a CHR tree from that larger dataset suggests that as many as three of our ineffective isolates (i.e. 2, 187, CW1) recently acquired an SI in ancestors that were non-nodulating [44]. Even with these ambiguous taxa, we still uncovered three mutualism breakdown events, independent origins of ineffective rhizobia from beneficial nodulating ancestors (i.e. 155, 186, 200). This complex evolutionary history suggests that origins of ineffective rhizobia might have multiple drivers including adaptation to other hosts, to free-living conditions in the soil, or might be due to negative epistasis caused by acquisition of novel SIs. Whole-genome datasets that deeply sample these populations are needed to better examine the mechanisms that drive these transitions and to resolve the frequency, directionality and genomic drivers of these events.
We uncovered recurrent mutualism breakdown events in a legume rhizobia metapopulation, including both the loss of nitrogen fixation and nodulation on a focal host. Parallel examples of mutualism breakdown might be expected to occur in other symbiont taxa with similar lifestyles. Vibrio fischeri is one such candidate. These marine bacteria provide the metabolically costly service of bioluminescence to diverse animal hosts, have an evolutionary advantage over hosts and also spend time in the environment between rounds of host infection [83]. However, in the well-studied bobtail squid system, host mechanisms appear to efficiently select against non-bioluminescent Vibrio [84]. Another candidate is the clade of dinoflagellate algae that provides nutrients to diverse marine hosts including corals [85], as they also share the same set of features. It would be fascinating to examine the strain and population-level variation in mutualism services in these and other taxa. More work is needed to examine the fine-scale strain-level population genomics of other microbial mutualists, to uncover shifts and losses of mutualist traits and the genomic and ecological mechanisms that drive these changes.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Data accessibility
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.3tx95x6bt [86].
Authors' contributions
K.A.G.-C. designed the study, performed the experiment, analysed the data and wrote the manuscript. C.E.W. designed the study, performed the experiment and contributed substantially to writing. A.J.W., J.H.C., P.J.S., K.A.M. and K.W.Q. helped perform the experiment and collect data. J.L.S. designed the study and wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This study was funded by US National Science Foundation (NSF, grant nos DEB 1150278 and 1738028).
References
- 1.Douglas AE. 2010. The symbiotic habit. Princeton, NJ: Princeton University Press. [Google Scholar]
- 2.Currie CR, Scott JA, Summerbell RC, Malloch D. 2003. Correction: Corrigendum: Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 423, 461 ( 10.1038/nature01563) [DOI] [Google Scholar]
- 3.Sachs J, Ehinger M, Simms E. 2010. Origins of cheating and loss of symbiosis in wild Bradyrhizobium. J. Evol. Biol. 23, 1075–1089. ( 10.1111/j.1420-9101.2010.01980.x) [DOI] [PubMed] [Google Scholar]
- 4.Gerardo NM, Parker BJ. 2014. Mechanisms of symbiont-conferred protection against natural enemies: an ecological and evolutionary framework. Curr. Opin. Insect Sci. 4, 8–14. ( 10.1016/j.cois.2014.08.002) [DOI] [PubMed] [Google Scholar]
- 5.Friesen ML, Porter SS, Stark SC, von Wettberg EJ, Sachs JL, Martinez-Romero E. 2011. Microbially mediated plant functional traits. Annu. Rev. ecol. Evol. Syst. 42, 23–46. ( 10.1146/annurev-ecolsys-102710-145039) [DOI] [Google Scholar]
- 6.Sachs JL, Quides KW, Wendlandt CE. 2018. Legumes versus rhizobia: a model for ongoing conflict in symbiosis. New Phytol. 219, 1199–1206. ( 10.1111/nph.15222) [DOI] [PubMed] [Google Scholar]
- 7.Sachs JL, Mueller UG, Wilcox TP, Bull JJ. 2004. The evolution of cooperation. Q. Rev. Biol. 79, 135–160. ( 10.1086/383541) [DOI] [PubMed] [Google Scholar]
- 8.Heath KD, Stinchcombe JR. 2014. Explaining mutualism variation: a new evolutionary paradox? Evolution 68, 309–317. ( 10.1111/evo.12292) [DOI] [PubMed] [Google Scholar]
- 9.Bromfield E, Thurman N, Whitwill S, Barran L. 1987. Plasmids and symbiotic effectiveness of representative phage types from two indigenous populations of Rhizobium meliloti. Microbiology 133, 3457–3466. ( 10.1099/00221287-133-12-3457) [DOI] [Google Scholar]
- 10.Burdon JJ, Gibson AH, Searle SD, Woods MJ, Brockwell J. 1999. Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian Acacia: within-species interactions. J. Appl. Ecol. 36, 398–408. ( 10.1046/j.1365-2664.1999.00409.x) [DOI] [Google Scholar]
- 11.Hoeksema JD, et al. 2010. A meta-analysis of context-dependency in plant response to inoculation with mycorrhizal fungi. Ecol. Lett. 13, 394–407. ( 10.1111/j.1461-0248.2009.01430.x) [DOI] [PubMed] [Google Scholar]
- 12.Sachs JL, Simms EL. 2006. Pathways to mutualism breakdown. Trends Ecol. Evol. 21, 585–592. ( 10.1016/j.tree.2006.06.018) [DOI] [PubMed] [Google Scholar]
- 13.Sachs JL, Skophammer RG, Bansal N, Stajich JE. 2014. Evolutionary origins and diversification of proteobacterial mutualists. Proc. R. Soc. B 281, 20132146 ( 10.1098/rspb.2013.2146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sachs JL, Skophammer RG, Regus JU. 2011. Evolutionary transitions in bacterial symbiosis. Proc. Natl Acad. Sci. USA 108, 10 800–10 807. ( 10.1073/pnas.1100304108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frederickson ME. 2013. Rethinking mutualism stability: cheaters and the evolution of sanctions. Q. Rev. Biol. 88, 269–295. ( 10.1086/673757) [DOI] [PubMed] [Google Scholar]
- 16.Friesen ML. 2012. Widespread fitness alignment in the legume–rhizobium symbiosis. New Phytol. 194, 1096–1111. ( 10.1111/j.1469-8137.2012.04099.x) [DOI] [PubMed] [Google Scholar]
- 17.Sawada H, Kuykendall LD, Young JM. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing symbionts. J. Appl. Microbiol. 49, 155–179. ( 10.2323/jgam.49.155) [DOI] [PubMed] [Google Scholar]
- 18.Sprent JI, Sutherland J, De Faria S, Dilworth M, Corby H, Becking J, Materon LA, Drozd JW. 1987. Some aspects of the biology of nitrogen-fixing organisms and discussion. Phil. Trans. R. Soc. Lond. B 317, 111–129. ( 10.1098/rstb.1987.0051) [DOI] [Google Scholar]
- 19.Kiers ET, Rousseau RA, West SA, Denison RF. 2003. Host sanctions and the legume-rhizobium mutualism. Nature 425, 78–81. ( 10.1038/nature01931) [DOI] [PubMed] [Google Scholar]
- 20.Oono R, Anderson CG, Denison RF. 2011. Failure to fix nitrogen by non-reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc. R. Soc. B 278, 2698–2703. ( 10.1098/rspb.2010.2193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regus J, Gano K, Hollowell A, Sofish V, Sachs J. 2015. Lotus hosts delimit the mutualism–parasitism continuum of Bradyrhizobium. J. Evol. Biol. 28, 447–456. ( 10.1111/jeb.12579) [DOI] [PubMed] [Google Scholar]
- 22.Regus JU, Quides KW, O'Neill MR, Suzuki R, Savory EA, Chang JH, Sachs JL. 2017. Cell autonomous sanctions in legumes target ineffective rhizobia in nodules with mixed infections. Am. J. Bot. 104, 1299–1312. ( 10.3732/ajb.1700165) [DOI] [PubMed] [Google Scholar]
- 23.Sachs JL, Russell JE, Lii YE, Black KC, Lopez G, Patil AS. 2010. Host control over infection and proliferation of a cheater symbiont. J. Evol. Biol. 23, 1919–1927. ( 10.1111/j.1420-9101.2010.02056.x) [DOI] [PubMed] [Google Scholar]
- 24.Simms EL, Taylor DL, Povich J, Shefferson RP, Sachs JL, Urbina M, Tausczik Y. 2006. An empirical test of partner choice mechanisms in a wild legume–rhizobium interaction. Proc. R. Soc. B 273, 77–81. ( 10.1098/rspb.2005.3292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singleton PW, Stockinger KR. 1983. Compensation against ineffective nodulation in soybean. Crop Sci. 23, 69–72. ( 10.2135/cropsci1983.0011183X002300010019x) [DOI] [Google Scholar]
- 26.Akcay E, Simms EL. 2011. Negotiation, sanctions, and context dependency in the legume–rhizobium mutualism. Am. Nat. 178, 1–14. ( 10.1086/659997) [DOI] [PubMed] [Google Scholar]
- 27.Denison RF. 2000. Legume sanctions and the evolution of symbiotic cooperation by rhizobia. Am. Nat. 6, 567–576. ( 10.1086/316994) [DOI] [PubMed] [Google Scholar]
- 28.West S, Kiers ET, Pen I, Denison R. 2002. Sanctions and mutualism stability: when should less beneficial mutualists be tolerated? J. Evol. Biol. 15, 830–837. ( 10.1046/j.1420-9101.2002.00441.x) [DOI] [Google Scholar]
- 29.West SA, Kiers ET, Simms EL, Denison RF. 2002. Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc. R. Soc. Lond. B 269, 685–694. ( 10.1098/rspb.2001.1878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bromfield E, Tambong J, Cloutier S, Prévost D, Laguerre G, Van Berkum P, Thi TT, Assabgui R, Barran LR. 2010. Ensifer, Phyllobacterium and Rhizobium species occupy nodules of Medicago sativa (alfalfa) and Melilotus alba (sweet clover) grown at a Canadian site without a history of cultivation. Microbiology 156, 505–520. ( 10.1099/mic.0.034058-0) [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Figueredo A, Villani H, Michajluk J, Hungria M. 2002. Diversity and symbiotic effectiveness of rhizobia isolated from field-grown soybean nodules in Paraguay. Biol. Fertil. Soils 35, 448–457. ( 10.1007/s00374-002-0493-1) [DOI] [Google Scholar]
- 32.Collins M, Thies J, Abbott L. 2002. Diversity and symbiotic effectiveness of Rhizobium leguminosarum bv. trifolii isolates from pasture soils in south-western Australia. Soil Res. 40, 1319–1329. ( 10.1071/SR01052) [DOI] [Google Scholar]
- 33.Denton M, Coventry D, Bellotti W, Howieson J. 2000. Distribution, abundance and symbiotic effectiveness of Rhizobium leguminosarum bv. trifolii from alkaline pasture soils in South Australia. Anim. Prod. Sci. 40, 25–35. ( 10.1071/EA99035) [DOI] [Google Scholar]
- 34.Fening J, Danso S. 2002. Variation in symbiotic effectiveness of cowpea bradyrhizobia indigenous to Ghanaian soils. Appl. Soil Ecol. 21, 23–29. ( 10.1016/S0929-1393(02)00042-2) [DOI] [Google Scholar]
- 35.Gibson A, Curnow B, Bergersen F, Brockwell J, Rominson A. 1975. Studies of field populations of Rhizobium: effectiveness of strains of Rhizobium trifolii associated with Trifolium subterraneum L. pastures in south-eastern Australia. Soil Biol. Biochem. 7, 95–102. ( 10.1016/0038-0717(75)90005-X) [DOI] [Google Scholar]
- 36.Moawad H, Badr El-Din SMS, Abdel-Aziz RA. 1998. Improvement of biological nitrogen fixation in Egyptian winter legumes through better management of Rhizobium. Plant Soil 204, 95–106. ( 10.1023/A:1004335112402) [DOI] [Google Scholar]
- 37.Quigley PE, Cunningham PJ, Hannah M, Ward GN, Morgan T. 1997. Symbiotic effectiveness of Rhizobium leguminosarum bv. trifolii collected from pastures in south-western Victoria. Aust. J. Exp. Agric. 37, 623–630. ( 10.1071/EA96089) [DOI] [Google Scholar]
- 38.Rangin C, Brunel B, Cleyet-Marel J-C, Perrineau M-M, Béna G. 2008. Effects of Medicago truncatula genetic diversity, rhizobial competition, and strain effectiveness on the diversity of a natural Sinorhizobium species community. Appl. Environ. Microbiol. 74, 5653–5661. ( 10.1128/AEM.01107-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehinger M, Mohr TJ, Starcevich JB, Sachs JL, Porter SS, Simms EL. 2014. Specialization-generalization trade-off in a Bradyrhizobium symbiosis with wild legume hosts. BMC Ecol. 14, 8 ( 10.1186/1472-6785-14-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gano-Cohen KA, Wendlandt CE, Stokes PJ, Blanton MA, Quides KW, Zomorrodian A, Adinata ES, Sachs JL. 2019. Interspecific conflict and the evolution of ineffective rhizobia. Ecol. Lett. 22, 914–924. ( 10.1111/ele.13247) [DOI] [PubMed] [Google Scholar]
- 41.Gaur YD, Lowther WL. 1980. Distribution, symbiotic effectiveness, and fluorescent-antibody reaction of naturalized populations of rhizobium-trifolii in otago soils. New Zeal. J. Agric. Res. 23, 529–532. ( 10.1080/00288233.1980.10417878) [DOI] [Google Scholar]
- 42.Regus JU, Wendlandt CE, Bantay RM, Gano-Cohen KA, Gleason NJ, Hollowell AC, O'Neill MR, Shahin KK, Sachs JL. 2017. Nitrogen deposition decreases the benefits of symbiosis in a native legume. Plant Soil 414, 159–170. ( 10.1007/s11104-016-3114-8) [DOI] [Google Scholar]
- 43.Sachs JL, Kembel SW, Lau AH, Simms EL. 2009. In situ phylogenetic structure and diversity of wild Bradyrhizobium communities. Appl. Environ. Microbiol. 75, 4727–4735. ( 10.1128/AEM.00667-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollowell AC, et al. 2016. Epidemic spread of symbiotic and non-symbiotic Bradyrhizobium genotypes across California. Microb. Ecol. 71, 700–710. ( 10.1007/s00248-015-0685-5) [DOI] [PubMed] [Google Scholar]
- 45.Hollowell AC, Regus JU, Turissini D, Gano-Cohen KA, Bantay R, Bernardo A, Moore D, Pham J, Sachs JL (eds). 2016. Metapopulation dominance and genomic-island acquisition of Bradyrhizobium with superior catabolic capabilities. Proc. R. Soc. B 283, 20160496 ( 10.1098/rspb.2016.0496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wendlandt CE, Regus JU, Gano-Cohen KA, Hollowell AC, Quides KW, Lyu JY, Adinata ES, Sachs JL. 2019. Host investment into symbiosis varies among genotypes of the legume Acmispon strigosus, but host sanctions are uniform. New Phytol. 221, 446–448. ( 10.1111/nph.15378) [DOI] [PubMed] [Google Scholar]
- 47.Quides KW, Stomackin GM, Lee HH, Chang JH, Sachs JL. 2017. Lotus japonicus alters in planta fitness of Mesorhizobium loti dependent on symbiotic nitrogen fixation. PLoS ONE 12, e0185568 ( 10.1371/journal.pone.0185568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regus JU, Gano KA, Hollowell AC, Sachs JL. 2014. Efficiency of partner choice and sanctions in Lotus is not altered by nitrogen fertilization. Proc. R. Soc. B 281, 20132587 ( 10.1098/rspb.2013.2587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan JT, Eardly BD, van Berkum P. 1996. Four unnamed species of nonsymbiotic rhizobia islated from the rhizosphere of Lotus corniculatus. Appl. Environ. Microbiol. 62, 2818–2925. ( 10.1128/aem.62.8.2818-2825.1996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Somasegaran P, Hoben J. 1994. Handbook for rhizobia. New York, NY: Springer-Verlag. [Google Scholar]
- 51.Lodwig E, Poole P. 2003. Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 22, 37–78. ( 10.1080/713610850) [DOI] [Google Scholar]
- 52.Okubo T, Piromyou P, Tittabutr P, Teaumroong N, Minamisawa K. 2016. Origin and evolution of nitrogen fixation genes on symbiosis islands and plasmid in Bradyrhizobium. Microb. Environ. 31, 260–267. ( 10.1264/jsme2.ME15159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 ( 10.1038/msb.2011.75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. ( 10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 56.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pahua VJ, Stokes PJN, Hollowell AC, Regus JU, Gano-Cohen KA, Wendlandt CE, Quides KW, Lyu JY, Sachs JL. 2018. Fitness variation among host species and the paradox of ineffective rhizobia. J. Evol. Biol. 31, 599–610. ( 10.1111/jeb.13249) [DOI] [PubMed] [Google Scholar]
- 59.Gage DJ. 2002. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 184, 7042–7046. ( 10.1128/JB.184.24.7042-7046.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Porter SS, Simms EL. 2014. Selection for cheating across disparate environments in the legume–rhizobium mutualism. Ecol. Lett. 17, 1121–1129. ( 10.1111/ele.12318) [DOI] [PubMed] [Google Scholar]
- 61.Heath KD, Tiffin P. 2007. Context dependence in the coevolution of plant and rhizobial mutualists. Proc. R. Soc. B 274, 1905–1912. ( 10.1098/rspb.2007.0495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heath KD, Tiffin P. 2009. Stabilizing mechanisms in a legume–rhizobium mutualism. Evolution 63, 652–662. ( 10.1111/j.1558-5646.2008.00582.x) [DOI] [PubMed] [Google Scholar]
- 63.Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745. ( 10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 64.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. ( 10.1093/bioinformatics/btq166) [DOI] [PubMed] [Google Scholar]
- 65.Parker MA. 2012. Legumes select symbiosis island sequence variants in Bradyrhizobium. Mol. Ecol. 21, 1769–1778. ( 10.1111/j.1365-294X.2012.05497.x) [DOI] [PubMed] [Google Scholar]
- 66.Parker MA. 2015. The spread of Bradyrhizobium lineages across host legume clades: from Abarema to Zygia. Microb. Ecol. 69, 630–640. ( 10.1007/s00248-014-0503-5) [DOI] [PubMed] [Google Scholar]
- 67.Gano-Cohen KA, et al. 2016. Nonnodulating Bradyrhizobium spp. modulate the benefits of legume-rhizobium mutualism. Appl. Environ. Microbiol. 82, 5259–5268. ( 10.1128/AEM.01116-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vinuesa P, Silva C, Werner D, Martinez-Romero E. 2005. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 34, 29–54. ( 10.1016/j.ympev.2004.08.020) [DOI] [PubMed] [Google Scholar]
- 69.Douglas AE. 2008. Conflict, cheats and the persistence of symbioses. New Phytol. 177, 849–858. ( 10.1111/j.1469-8137.2007.02326.x) [DOI] [PubMed] [Google Scholar]
- 70.Sachs J. 2015. The exploitation of mutualisms. In Mutualism (ed. JL Bronstein), pp. 93–106 Oxford, UK: Oxford University Press. [Google Scholar]
- 71.Sachs JL, Wilcox TP. 2006. A shift to parasitism in the jellyfish symbiont Symbiodinium microadriaticum. Proc. R. Soc. B 273, 425–429. ( 10.1098/rspb.2005.3346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Axelrod R, Hamilton WD. 1981. The evolution of cooperation. Science 211, 1390–1396. ( 10.1126/science.7466396) [DOI] [PubMed] [Google Scholar]
- 73.Doebeli M, Knowlton N. 1998. The evolution of interspecific mutualisms. Proc. Natl Acad. Sci. USA 95, 8676–8680. ( 10.1073/pnas.95.15.8676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferriere R, Gauduchon M, Bronstein JL. 2007. Evolution and persistence of obligate mutualists and exploiters: competition for partners and evolutionary immunization. Ecol. Lett. 10, 115–126. ( 10.1111/j.1461-0248.2006.01008.x) [DOI] [PubMed] [Google Scholar]
- 75.Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igic B. 2010. Species selection maintains self-incompatibility. Science 330, 493–495. ( 10.1126/science.1194513) [DOI] [PubMed] [Google Scholar]
- 76.Van Dyken JD, Linksvayer TA, Wade MJ. 2011. Kin selection–mutation balance: a model for the origin, maintenance, and consequences of social cheating. Am. Nat. 177, 288–300. ( 10.1086/658365) [DOI] [PubMed] [Google Scholar]
- 77.Jones EI, et al. 2015. Cheaters must prosper: reconciling theoretical and empirical perspectives on cheating in mutualism. Ecol. Lett. 18, 1270–1284. ( 10.1111/ele.12507) [DOI] [PubMed] [Google Scholar]
- 78.Price PA, Tanner HR, Dillon BA, Shabab M, Walker GC, Griffitts JS. 2015. Rhizobial peptidase HrrP cleaves host-encoded signaling peptides and mediates symbiotic compatibility. Proc. Natl Acad. Sci. USA 112, 15 244–15 249. ( 10.1073/pnas.1417797112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sullivan JT, Patrick HN, Lowther WL, Scott DB, Ronson CW. 1995. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl Acad. Sci. USA 92, 8985–8989. ( 10.1073/pnas.92.19.8985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sachs JL, Russell JE, Hollowell AC. 2011. Evolutionary instability of symbiotic function in Bradyrhizobium japonicum. PLoS Biol. 6, e26370 ( 10.1371/journal.pone.0026370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Porter SS, Faber-Hammond J, Montoya AP, Friesen ML, Sackos C. 2019. Dynamic genomic architecture of mutualistic cooperation in a wild population of Mesorhizobium. ISME J. 13, 301–315. ( 10.1038/s41396-018-0266-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heath KD. 2010. Intergenomic epistasis and coevolutionary constraint in plants and rhizobia. Evolution 64, 1446–1458. ( 10.1111/j.1558-5646.2009.00913.x0) [DOI] [PubMed] [Google Scholar]
- 83.Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid–Vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642. ( 10.1038/nrmicro957) [DOI] [PubMed] [Google Scholar]
- 84.McFall-Ngai M. 2014. Divining the essence of symbiosis: insights from the squid–Vibrio model. PLoS Biol. 12, e1001783 ( 10.1371/journal.pbio.1001783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stat M, Morris E, Gates RD. 2008. Functional diversity in coral–dinoflagellate symbiosis. Proc. Natl Acad. Sci. USA 105, 9256–9261. ( 10.1073/pnas.0801328105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gano-Cohen KA, Wendlandt CE, Al Moussawi K, Stokes PJ, Quides KW, Weisberg AJ, Chang JH, Sachs JL. 2020. Data from: Recurrent mutualism breakdown events in a legume rhizobia metapopulation Dryad Digital Repository. ( 10.5061/dryad.3tx95x6bt) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Gano-Cohen KA, Wendlandt CE, Al Moussawi K, Stokes PJ, Quides KW, Weisberg AJ, Chang JH, Sachs JL. 2020. Data from: Recurrent mutualism breakdown events in a legume rhizobia metapopulation Dryad Digital Repository. ( 10.5061/dryad.3tx95x6bt) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.3tx95x6bt [86].