Abstract
Context
There is an unmet need for biomarkers of pancreatic beta-cell death to improve early diagnosis of type 1 diabetes, enroll subjects into clinical trials, and assess treatment response. To address this need, several groups developed assays measuring insulin deoxyribonucleic acid (DNA) with unmethylated CpG sites in cell-free DNA. Unmethylated insulin DNA should be derived predominantly from beta-cells and indicate ongoing beta-cell death.
Objective
To assess the performance of three unmethylated insulin DNA assays.
Design and Participants
Plasma or serum samples from 13 subjects undergoing total pancreatectomy and islet autotransplantation were coded and provided to investigators to measure unmethylated insulin DNA. Samples included a negative control taken post-pancreatectomy but pretransplant, and a positive control taken immediately following islet infusion. We assessed technical reproducibility, linearity, and persistence of detection of unmethylated insulin DNA for each assay.
Results
All assays discriminated between the negative sample and samples taken directly from the islet transplant bag; 2 of 3 discriminated negative samples from those taken immediately after islet infusion. When high levels of unmethylated insulin DNA were present, technical reproducibility was generally good for all assays.
Conclusions
The measurement of beta cell cell-free DNA, including insulin, is a promising approach, warranting further testing and development in those with or at-risk for type 1 diabetes, as well as in other settings where understanding the frequency or kinetics of beta cell death could be useful.
Keywords: Beta cell, type 1 diabetes, islet transplantation, cell-free DNA
Type 1 diabetes results from progressive immune-mediated loss of pancreatic β-cells, leading to a lifelong dependence on exogenous insulin (INS) therapy. The disease is recognized to progress in stages (1,2). Stage 1 is considered the start of disease and is defined by the presence of multiple islet autoantibodies and normal glucose tolerance; Stage 2 represents the progression to abnormal glucose tolerance; and Stage 3 is the onset of clinically recognized disease. To address the core pathology, a number of immunomodulatory approaches have been tested in clinical trials. A handful of drugs have shown success in delaying the loss of INS secretion when administered shortly after Stage 3 disease onset (3–7), and anti-CD3 therapy was shown recently to delay the progression from Stage 2 to Stage 3 diabetes (8). While these results are encouraging, progress is hindered due to considerable heterogeneity in type 1 diabetes disease progression and response to therapy. Subjects in the control arms of immunotherapy trials show varied rates of progression through Stages 1 and 2 (9) and differing rates of loss of INS secretion at Stage 3 (10,11), and not all subjects appear to respond to any given immunotherapy.
Robust and noninvasive biomarkers able to accurately reflect the health status of the β-cell could aid in dissecting disease-related heterogeneity, selecting patients for clinical trials, and monitoring response to therapeutic interventions. Since first described in 2011(12), several groups have developed novel assays to measure β-cell death based on the identification of differentially methylated INS DNA fragments in the serum or plasma; unmethylated INS DNA is thought to specifically emanate from dying β-cells (12–16). For this reason, the majority of cell-free deoxyribonucleic acid (cfDNA) assays proposed for the detection of β-cell death in type 1 diabetes have focused on the detection of unmethylated INS cfDNA. The development of assays seeking to measure β-cell death was driven by the principle of liquid biopsies, namely, that fragments of DNA are released from dying cells and then briefly circulate in the blood, prior to clearance by the liver or kidney. Such circulating cfDNA molecules, extracted from serum or plasma, have been successfully used in monitoring and detection of cancer (17), in prenatal diagnostics (18), and in the identification of solid organ transplant rejection (19).
To date, these assays have been developed independently by multiple groups, using a variety of technological platforms including nested polymerase chain reaction (PCR), digital droplet PCR and next-generation sequencing, and using different segments of the INS gene. Each has reported on the biological relevance of unmethylated INS DNA in different settings. In cohorts of individuals with recent-onset type 1 diabetes (Stage 3), levels of unmethylated INS DNA or the ratio of unmethylated to total INS DNA were reported to be higher than age-matched healthy controls (12–14,20). Elevated unmethylated INS DNA has been reported in some autoantibody-positive at-risk subjects (Stage 1–2) (21,22), in individuals with long-standing type 1 diabetes (23), in gestational diabetes (24), and in those with type 1 diabetes prone to ketosis (25). In addition, decreased INS cfDNA has been detected after anti-CD3 immunotherapy (20). More recently, unmethylated INS DNA has been detected after islet transplant (26–28); these experiments have also suggested that the half-life of circulating INS DNA after transplant is short, on the order of 2 hours (21), similar to the cfDNA turnover rate shown in other settings (29).
Driven by these provocative initial findings and an interest in translating the assays toward clinical utility, questions have arisen regarding the specifics of assay performance. To address these questions, we conducted a collaborative workshop to assess the performance characteristics of three unmethylated INS assays. Three participating investigators were identified to participate in the workshop, based on their willingness and the availability of an Opportunity Fund Grant from the Human Islet Research Network Consortium on Beta Cell Death and Survival. A single clinical site was engaged to prospectively collect samples from subjects undergoing Total Pancreatectomy and Islet Auto Transplant (TPIAT), using specific collection protocols that were optimized for each participating investigator. A neutral arbiter, the JDRF Core for Assay Validation, coordinated sample access, blinded and distributed the samples, and analyzed study data.
As a first step in understanding assay performance, we collaboratively investigated the reproducibility and linearity exhibited by each assay in samples collected from TPIAT subjects at times expected to show high or low levels of circulating unmethylated INS DNA. β-cell death is known to occur at the time of transplantation due to isolation stress and possibly the inflammatory reaction from intraportal infusion (27,30,31) and thus was selected as a reliable model to generate a positive signal for assay testing.
Materials and Methods
Subjects and study ethics review
Adult subjects with chronic pancreatitis undergoing TPIAT at the University of Minnesota during an enrollment period of approximately 7 months were prospectively enrolled in this study, under a protocol approved by the University of Minnesota institutional review board. At the time of enrollment, subjects did not have diabetes or chronic kidney disease. The study aimed to enroll a minimum of 10 subjects during this accrual period and ultimately enrolled 13 participants. Written informed consent was obtained for all participants. The surgical procedure and islet isolation processes were performed as previously described (27).
Control serum and plasma used to produce a sample set of serial dilutions were drawn from a 26-year-old white male subject with no known health concerns and no family history of autoimmunity. The subject provided written informed consent to participate in a Benaroya Research Institute institutional review board approved protocol under the BRI Immune Mediated Disease Registry and Repository.
Sample collection and processing
Sample collection and processing procedures were approved by the participating investigators prior to study enrollment. Samples were collected prospectively from 13 subjects undergoing TPIAT for pancreatitis. Clinical and demographic characteristics of the subjects, as well as the number of islets transplanted, are presented in Table 1. Subject IDs were assigned in order of date of study enrollment. Of note, the number of islets available for transplant varied almost 10-fold between subjects. Based on previous data from 2 of the assays (26,27), timepoints pretransplant and posttransplant were selected to represent both a confirmed negative timepoint (4 hours after pancreatectomy but before islet transplantation) and a likely strong positive timepoint (15–30 minutes after islet transplant). Samples were collected and processed using protocols specified in advance by each participating investigator. Multiple aliquots were collected at each timepoint to allow assessment of reproducibility. A dilution curve, composed of serial dilutions of supernatant from the islet transplant bag of one TPIAT subject diluted into serum or plasma from a control subject, was tested to assess linearity.
Table 1.
Clinical and demographic characteristics of TPIAT subjects
| Subject ID | Gender | Age (years) | BMI (kg/m2) | No. islets transplanted |
|---|---|---|---|---|
| Summary | 7 F (53.8%)a | 50 (27–66)b | 28.0 (19.1–31.1)b | 182 800 (38 860–377 100)a |
| 01 | M | 56 | 29.2 | NA |
| 02 | F | 53 | 21.8 | 146 000 |
| 03 | F | 36 | 28.0 | 185 400 |
| 05 | M | 41 | 30.8 | 314 400 |
| 06 | M | 49 | 28.2 | 196 200 |
| 07 | F | 62 | 29.3 | 212 700 |
| 08 | F | 50 | 28.4 | 95 700 |
| 09 | F | 66 | 25.2 | 168 100 |
| 10 | M | 37 | 25.4 | 160 950 |
| 11 | M | 49 | 31.1 | 377 100 |
| 12 | F | 27 | 19.1 | 38 860 |
| 13 | F | 57 | 24.5 | 198 350 |
| 14 | M | 57 | 24.9 | 180 200 |
Abbreviation: NA, not available. TPIAT, total pancreatectomy and islet auto transplant.
an (%);
bmedian (range).
Plasma samples were collected for Assay D; serum samples were collected for assays H and M. Assays are identified by the surname of the investigator in whose lab they were developed (Dor, Herold, and Mirmira, respectively). Plasma was collected in K2-EDTA tubes and spun immediately at 1500 rcf × 10 min at 4°C. Supernatant was removed and then centrifuged again at 3000 rcf × 10 min at 4°C. Serum samples were collected in red-top vacutainers (no additive) and allowed to clot for 30 minutes before a single 15-minute room temperature spin at 1500 rcf. Islet supernatant samples were collected after islet washing in the final transplant media but prior to intravenous (IV) bag loading, and snap frozen. Mid-stream urine samples were collected using Norgen Urine Collection and Preservation Tubes (Norgen Biotek, Thorold, Ontario, Canada).
Over 900 individual aliquots were sent from the University of Minnesota to the Core for Assay Validation (BRI) for coding and later distribution to each study investigator. Each aliquot was labelled with a randomly generated 6-digit code. Coded data were returned to BRI for decoding and analysis; investigators were blinded to all sample identifiers until data were provided to BRI and all analysis for this study was complete.
Assay methods
All assays were developed independently; each laboratory selected the sites to be studied based on observations that the sites were either uniquely or preferentially unmethylated in β-cells as compared to other cell types. The original selections of sites and complete methods for each assay were described in the following references: Assay D (13,26), Assay H (21,27), and Assay M, (14,32). All assays are described here in brief. Primers for all assays are provided in Table 2. Assay D is conducted using next-generation sequencing. cfDNA was extracted from 1 mL plasma aliquots using a QIAsymphony liquid-handling robot (Qiagen) and treated with bisulfite (Zymo Research). A 150bp INS gene fragment containing 6 CpG (16) was then PCR-amplified using primers specific for the bisulfite-treated DNA, but outside of the methylation sites under study. Products were sequenced at a ~50,000 read depth on a Miseq machine (Illumina), and the fraction of fully unmethylated molecules (i.e., PCR products in which all 6 CpG sites were converted to T) was calculated as previously described (26). Values from Assay H are presented as percentage β-cell DNA, defined as fully unmethylated INS fragments in the original cfDNA template. Assay H is conducted using droplet digital PCR. cfDNA was extracted from 500 uL serum using Quick cfDNA Serum and Plasma kit (Zymo Research), bisulfite treated (Zymo Research), and amplified using primers and detection probes specific for the methylation status on the droplet digital PCR platform (BioRad). Primers amplified a 106 bp segment of the INS gene, containing 2 CpG sites. Data are presented as the ratio of unmethylated INS DNA copy number divided by the sum of the methylated plus unmethylated copy numbers. Assay M also uses droplet digital PCR. DNA was extracted from 50 uL serum using the QIAamp DNA blood kit (Qiagen) with added poly-A DNA as carrier. DNA was treated with bisulfite (Zymo Research) and amplified using digital droplet PCR (Biorad). The locus amplified was 157 bp long, and contained a single CpG site. Data are presented as copies/uL after normalization to the carrier DNA concentration.
Table 2.
Bisulfite PCR primers and products
| Assay D | Assay H | Assay M | |
|---|---|---|---|
| Forward primer | TTTTTGGGGATTTGATTTAGT | GTGGTTTATATTTGGTGGA | GGAAATTGTAGTTTTAGTTTTTAGTTATTTGT |
| Reverse primer | ACCCTACAAATCCTCTACCTCC | ATTAACTCACCCTACAAATC | AAAACCCATCTCCCCTACCTATCA |
| Unmethylated fragment probe | ATTTAAGATTTGTTGGGAGGTAGAG | ACCCCTACCACCTAAC | |
| Methylated fragment probe | ATTTAAGATTCGTCGGGAGGTAGAG | ACCCCTACCGCCTAAC | |
| Product size | 144 base pairs | 106 base pairs | 157 base pairs |
Statistical methods
For all assays, replicate measures for each subject at each timepoint were summarized by analysis of mean values. To evaluate variability, percentage coefficients of variation were computed for each subject at all timepoints for all assays. Fold change was computed for each subject for those samples only collected at two timepoints. Within-subject paired differences were used to assess changes in β-cell death for each subject between timepoints for each of the assays.
Spearman’s correlations were used to assess the association between unmethylated INS measurements and number of islets transplanted for each of the assays at each timepoint. For serial dilution analysis, linear regression models were fit for assay comparison and were fit with a quadratic term for the log-transformed dilution factor to evaluate the dose-response curves for each assay.
All analyses were conducted at the Core for Assay Validation (BRI) and performed using JMP Pro 14 and SAS 9.4.
Results
Reproducibility of measurements of unmethylated INS DNA in samples from positive and negative control subjects
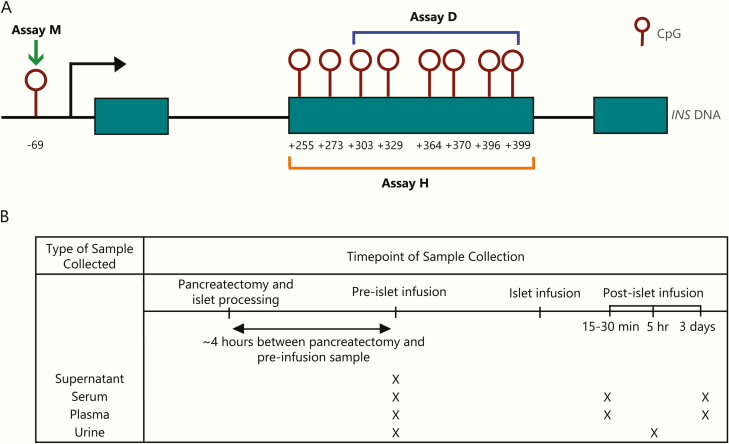
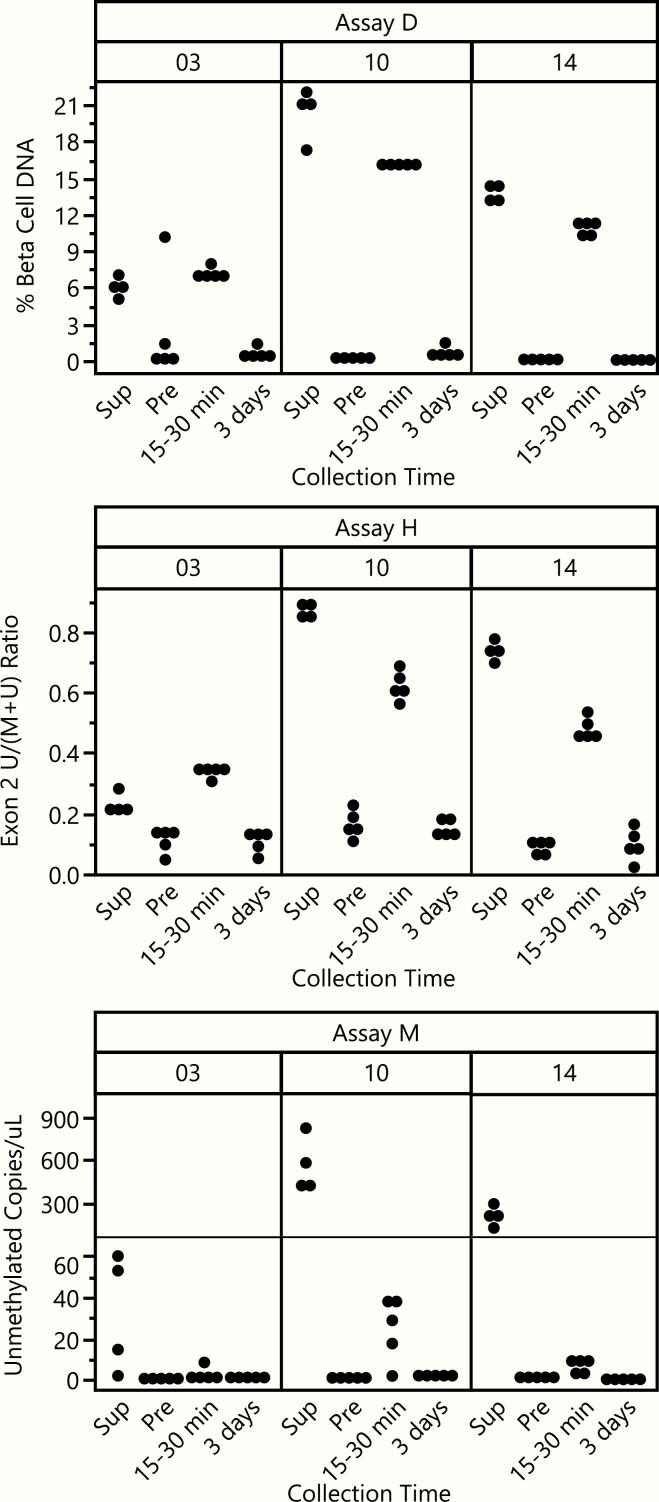
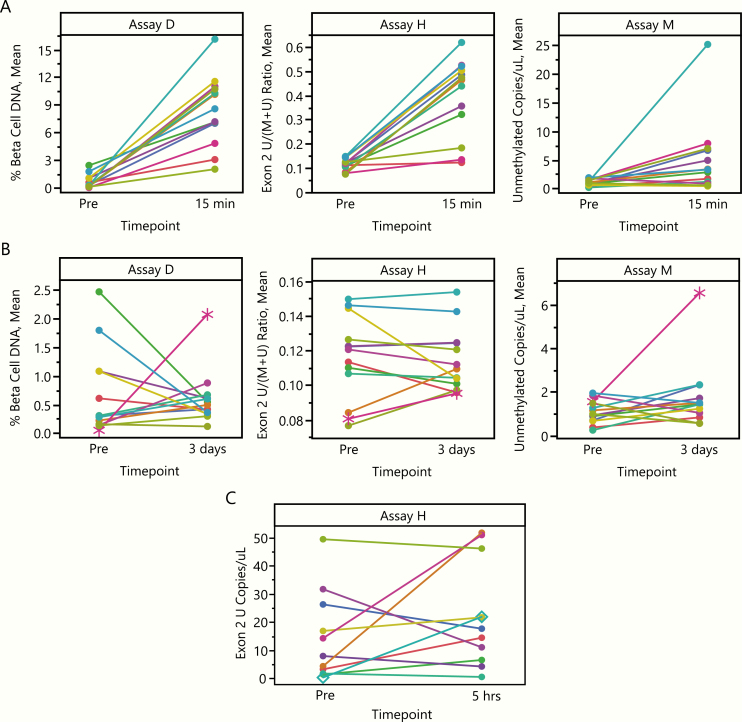
Three assays were included in this workshop; each used different methods, measured different sites in the INS gene (Fig. 1A), and used different sample volumes (50 uL Assay M; 500 uL Assay H; 1000 uL Assay D). Initially, reproducibility of all assays was tested. For each subject, five replicate aliquots of serum or plasma were tested in each assay at timepoints before, 15 to 30 minutes after, and 3 days after islet transplant (Fig. 1B). Four aliquots were also measured for each subject using supernatant samples collected from the islet transplant IV bag. Representative raw data for each aliquot tested for three randomly selected subjects are provided in Fig. 2, to highlight the consistency between replicate aliquots and the change in INS DNA detection between timepoints. The pretransplant timepoint, taken several hours after pancreatectomy, would be expected to have little β-cell DNA detected, given the half-life of cfDNA. All assays showed a consistent ability to discriminate between the pretransplant timepoint and samples of supernatant from the islet transplant IV bag; 2 of 3 assays (D and H) also showed similar discrimination between the pretransplant and 15- to 30-minute posttransplant timepoint.
Figure 1.
Methylation sites measured and timeline of sample collection. A. Each assay measures a different methylation signature in the INS gene. Maroon open circles are CpG sites. Assay M (green arrow) measures one site in the promoter region at –69. Assay H (orange bracket) measures 2 CpGs in Exon 2, +396 and +399. Assay D (blue bracket) measures 6 CpGs in Exon 2, from +303. B. Supernatant from islet transfer IV bag was collected just before islet infusion. Serum and plasma were collected at timepoints postpancreatectomy but before islet infusion, as well as 15 to 30 minutes and 3 days posttransplant. Urine was collected prior to islet transplant but after pancreatectomy and at 5 hours postpancreatectomy.
Figure 2.
Comparison of raw data from β-cell death measures for 3 TPIAT subjects. Subjects 3 (left), 10 (middle), and 14 (right) were randomly selected for presentation. Each individual data point represents a single replicate measurement; each subject had 4 to 5 replicate measurements at each timepoint. Fifteen to 30 minutes and 3 days indicates time posttransplant. Abbreviations: Sup: supernatant from islet transplant IV bag. Pre: pretransplant/postpancreatectomy timepoint. *Axis break for Assay M due to difference in unmethylated copies/uL between Sup and Pre for subjects 10 and 14.
Technical reproducibility and linearity characteristics of each assay
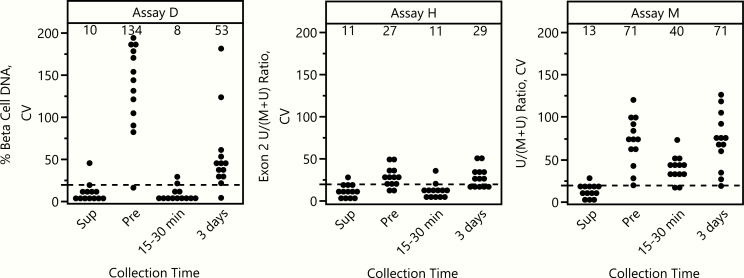
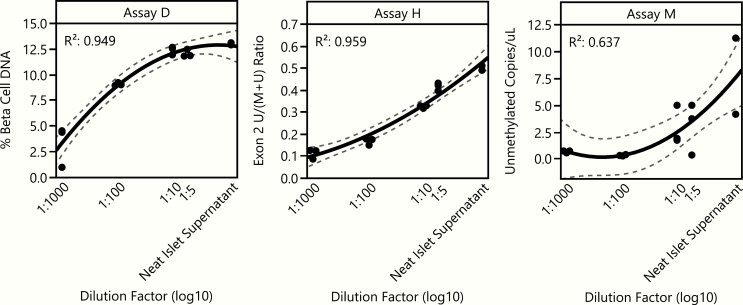
Using the replicate aliquot values, we calculated the coefficient of variation (%CV) for each subject at each timepoint for all assays (Fig. 3). %CV climbs when values measured approach zero (this is a mathematical property of the %CV); thus in this case, when little or no β-cell DNA is present, %CV values will be higher because all measurements are very small. This can be seen in data from Assay D in the pretransplant and day 3 timepoints, where little INS DNA was detected and %CVs are high. However, when ample INS DNA is present in a sample (islet bag supernatant and 15- to 30- minute timepoints), the %CV for each sample remained very low. All 3 assays showed relatively low, acceptable %CVs across all subjects for the islet transplant supernatant (Assay D: median 5.2, range 1.1–45.8; Assay H: 8.9, 2.6–28.4; Assay M: 12.7, 2.4–28.6). Two of 3 also had low %CV for the 15- to 30-minute timepoint, (Assay D: median 5.2, range 2.3–23.7; Assay H: 7.4, 3.7–36.2; Assay M: 42.0, 16.3–73.5) reflecting the detection data in Fig. 2. We also assessed the linearity of results for each assay using a dilution curve composed of islet bag supernatant at multiple dilutions (Fig. 4). All assays showed linearity based on quadratic regression analysis (R2 range of 0.64–0.96).
Figure 3.
%CV values are acceptable and low when considerable unmethylated INS DNA is expected to be present. Coefficient of variation (%CV, SD/mean * 100) plotted for each subject at each timepoint in each of the assays. Each point represents the %CV for a single subject at a given timepoint, calculated from 4 to 5 aliquots per timepoint. Mean %CV is annotated above each timepoint within each assay. Data are paneled by assay.
Figure 4.
Linearity of results from serially diluted samples. β-cell death after serial dilutions of islet supernatant from a single TPIAT subject, diluted with plasma (Assay D) or with serum (Assay H, M) from a healthy adult. Points represent a single aliquot (n = 2 to 3 aliquots for each dilution). Black lines are quadratic regression lines and dashed lines represent 95% confidence intervals. Data are paneled by assay. R2 goodness-of-fit values for each assay are embedded in each panel.
Comparisons between assays
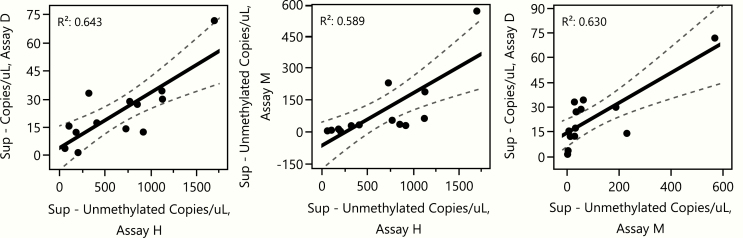
We next assessed whether each assay gave similar results for each subject by comparing the data from the islet supernatant bags across assays (Fig. 5). All values were converted to copies/uL for this assessment. While the absolute number of copies differed across assays, the correlations were relatively high and similar regardless of which pairwise assay comparison was conducted (R2 range of 0.59–0.64).
Figure 5.
Comparison across assays of detection of unmethylated INS DNA in islet bag supernatant samples. Linear regression was used to compare β-cell death, quantified as copies/uL of unmethylated INS DNA in islet supernatant, for each 2-way comparison of assays. Each point represents a single subject. Black lines are linear regressions for each comparison and dashed lines indicate the 95% confidence intervals.
Kinetics of INS DNA detection in TPIAT samples
All 3 assays showed a stronger signal of unmethylated INS at 15- to 30-minutes posttransplant, as compared to the pretransplant timepoint (Fig. 6A). Assays D and H detected an increase in INS DNA in all subjects studied; Assay M detected an increase in 10/13 subjects tested. Of note, the 2 subjects with the lowest number of transplanted islets were among the 3 with no difference between the pretransplant and 15- to 30-minute posttransplant timepoints for Assay M.
Figure 6.
Detection of unmethylated INS DNA at pretransplant and posttransplant timepoints. Each dot in all panels represents a single subject at either the pretransplant (postpancreatectomy) timepoint or at the noted posttransplant timepoint (A: 15–30 minutes; B: 3 days; C: 5 hours). Data are paired by subject (lines). A and B: Each data point represents the mean of 4 to 5 aliquots measured at the indicated timepoint. B: Star indicates subject 11. C: Urine data were collected for only one assay. Data are from a single aliquot of urine. Diamond indicates subject 11.
We also assessed the difference between the pre-transplant timepoint and the day 3 posttransplant timepoint (Fig. 6B). There was no persistent signal above pretransplant baseline observed in any assay. Subject 11, however, showed a considerably increased value at day 3 posttransplant by 2 assays (D and M) and an increasing trend for the third (H), though these values were well below the 15- to 30-minute value for this subject. Subject 11 also had the highest number of transplanted islets. Of note, this subject also had a 45-fold increase in INS DNA detection in urine at 5 hours posttransplant (urine tested only by Assay H; Fig. 6C). These findings suggest that at 3 days post auto-transplantation the extent of β-cell death is much reduced, and that cfDNA assays are capable in principle of detecting a signal in patients in which β-cell death does occur.
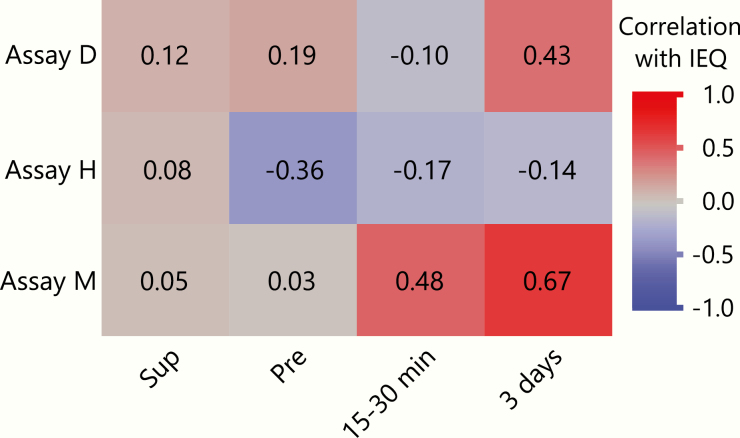
Correlations between the number of islets transplanted and assay measurements
To assess the relationship between the number of islets transplanted per subject and each assay measurement, we tested the correlations between each of these values at all timepoints (Fig. 7). As expected, limited/no correlations between the number of islets transplanted and unmethylated INS DNA levels at the pretransplant timepoint were observed. Some previous studies have detected a correlation between values in the islet IV bag supernatant and the number of islets transplanted (21); others have not (26). No correlation between values in the islet IV bag supernatant and the number of islets transplanted were observed for any assay in this study. Assay M showed a moderate correlation between the 15- to 30-minute timepoint and the number of islets transplanted (Spearman’s r = 0.48). While the absolute values of unmethylated INS DNA detected were low at day 3, both assays D and M showed positive correlations between number of islets transplanted and INS DNA at the 3-day timepoint (Spearman’s r of 0.43 and 0.67 respectively).
Figure 7.
Correlations between islet equivalents (IEQ) transplanted and unmethylated INS DNA levels for each assay. Heatmap shows Spearman r values of the correlations between number of islets transplanted (IEQ) and β-cell death measurements for each assay at each timepoint. Y axis: assays. X axis: timepoints. Fields are colored by strength of correlation.
Discussion
Noninvasive monitoring of β-cell death in type 1 diabetes could aid in dissecting disease-related heterogeneity in type 1 diabetes and help guide the timing of immunomodulatory interventions in high risk individuals. In addition, assays that reliably detect β-cell stress may have application outside of type 1 diabetes, including in type 2 and gestational diabetes or in the monitoring of individuals undergoing islet or pancreatic transplantation. Here, we compared three assays that aim to measure β-cell death by quantification of differentially methylated fragments of the INS gene in serum, plasma, or urine. The underlying premise of this approach is that the INS gene is unmethylated in the β-cell, where it is actively transcribed, and is methylated in other cell types, as shown previously (33,34). Since first described in 2011 (12), distinct methodologies have been developed to quantitate circulating levels of INS DNA, generating questions regarding how these approaches compare to one another.
We selected the TPIAT setting as a “positive control” for this assay comparison workshop because β-cell death is known to occur immediately posttransplant (26,27). An intriguing, as yet unanswered, question is precisely what each of these assays measures in the TPIAT setting. That is, the assays may primarily measure β-cell DNA infused along with the islet preparation due to islet apoptosis prior to transfusion (30,31), or they may detect de novo β-cell death after the islets have been transplanted. In this study, we are not able to assess de novo β-cell death specifically, as no assay detected any signal at the day 3 timepoint (when de novo β-cell death may or may not occur), and 15 minutes is too early to distinguish between possible de novo cell death in vivo and DNA infused from dead or dying cells. Because we selected an extreme “positive control,” this workshop also did not test any assay’s performance in the context of cells dying at any stage of type 1 diabetes.
The levels of unmethylated INS DNA in islet supernatant were highly correlated across all three assays, and all assays showed the ability to detect a difference between those samples and the negative control sample from subjects after pancreatectomy but before islet transplant. However, the serial dilution experiment revealed marked differences in assay performance. While Assay D clearly discriminated between the 1:100 and 1:1000 dilutions, assays H and M performed better at lower dilutions (higher substrate levels). Regardless of assay, the degree to which the levels of INS DNA detected at any of these dilutions corresponds to biological settings, such as samples from early stage type 1 diabetes, is not known.
The 3 assays compared here differ structurally along several important axes. First, the volume of starting material for analysis was substantially different. Assay D used 1 mL volumes, Assay H 500 uL, and Assay M 50 uL (although a larger volume was available for testing). This difference in volume may contribute to observed assay differences in both %CV and the ability to consistently detect unmethylated INS DNA immediately after islet infusion. We note that in the context of cancer cfDNA biomarkers, much larger volumes than were used for any of these assays have been required to reliably detect cancer at a clinically meaningful stage. The assays also use different optimized sample types, collection methods, and measurement techniques. DNA isolation techniques used also differ across the assays. DNA isolation (35) and pre-analytical variable optimization (36) are active areas of investigation, which may impact techniques for these assays going forward. Finally, each assay measures a different CpG site or sites in the INS gene.
It remains unknown whether the unmethylated DNA detected by each assay is precisely diagnostic of β-cell death, as INS DNA can be unmethylated (although in a lower frequency of cells) in other cell types. For example, islet α-cells can exhibit unmethylated INS DNA, which may specifically be detectable in these TPIAT samples (37). More comprehensive analysis of the methylation patterns of the INS gene across multiple tissues, including the exocrine pancreas and endocrine cells elsewhere in the gastrointestinal tract will be required to fully define tissue specificity of these assays. The finding that all assays detected nontrivial amounts of unmethylated cfDNA in some samples obtained 4 hours after total pancreatectomy could be explained by the presence of ectopic pancreatic tissue, unmethylated INS DNA originating from nonpancreatic tissue, or the clearance rate of cfDNA being longer than previously thought. Indeed, cfDNA clearance rates could vary between TPIAT subjects who may have some degree of kidney dysfunction; this could change cfDNA levels in urine, serum/plasma, or both, although the kidney is not the sole method of cfDNA clearance. cfDNA generation and subsequent clearance remains an active area of investigation in the field of cfDNA biomarkers more globally (38). Further studies are needed to address this issue, both regarding the presence of cfDNA postpancreatectomy and the possibilities of cfDNA clearance differences between subjects.
It remains an open question as to how to optimally apply these assays to measure β-cell death in type 1 diabetes, where the frequency of β-cell death is expected to be much lower in magnitude than following TPIAT. In addition, β-cell death in type 1 diabetes may be intermittent, which represents another detection challenge due to the short half-life of cfDNA in blood, estimated to be between 15 minutes and 2 hours (21). Because unmethylated INS DNA may not be specific to β-cells in total cfDNA, other genomic loci that may contribute to a composite β-cell death signature are currently being explored (16,39). The death of other cell types may also be relevant to prediction of outcomes in type 1 diabetes or in predicting response to therapy; incorporating methylation markers for immune cell cfDNA, for example, could be useful in future iterations of these assays. In addition to the use of larger starting blood volumes, some in the cancer field have applied hundreds of independent markers measured in parallel to enhance sensitivity (40). Both of these method modifications are likely to be needed for cfDNA methylation markers to move toward clinical utility in subjects throughout the stages of type 1 diabetes. Understanding how consistently and robustly β-cell death can be detected in those with or at-risk for type 1 diabetes is a key next step for future investigation, possibly by other workshop efforts.
In summary, we designed and executed a collaborative workshop for the comparison of three β-cell death assays, resulting in a better understanding of the reproducibility and performance of these assays in the context of positive and negative control samples. All 3 assays are still under development with an aim to further improve their performance. We propose that a workshop approach should be applied to samples from antibody-positive at-risk (Stage 1–2) and clinically diagnosed (Stage 3) type 1 diabetes subjects to help understand the reproducibility and utility of these assays in the setting where they may ultimately be the most useful. It is important to note that comparative studies like this one would likely not be competitive for many traditional funding streams, but are critical to understand the utility of proposed biomarker assays when the ultimate aim is translation to the clinic. Previous collaborative efforts to validate autoantibodies for clinical utility in type 1 diabetes were highly fruitful and resulted in widely used tests; however, they required sustained efforts to conduct and complete (41) and, in fact, continue to today as autoantibody assays improve over time (42). Thus, we encourage government and foundation funders to develop nontraditional funding sources to support future comparison efforts, not only to further our understanding of assay utility, but also to improve understanding of the rigor and reproducibility of currently available assays.
Acknowledgments
We wish to thank Peggy Ptacek (University of Minnesota) for sample collection and cataloging efforts and Rachel Hartley (BRI) for blinding, preparing, and shipping the samples to each laboratory. Anne Hocking (BRI) provided a helpful review of the manuscript. Members of the BRI Translational Research Program were involved in patient recruitment work to provide additional serum samples.
Funding Statement: This research was performed using funding provided by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)-supported Human Islet Research Network Opportunity Pool Fund (HIRN, RRID:SCR_014393; https://hirnetwork.org; U01 DK104162 to CEM), and by JDRF under the Core for Assay Validation grants #3-SRA-2016-209-Q-R to S. Alice Long and 3-SRA-2019-791-S-B to CS. KCH received support from R01 DK057846, UC4 DK104205-01, and R21 AI135562. KCH and SUB have support from R43 DK116577. Dr. Bellin is supported by R01 DKI09914.
Author contributions: CS designed the study, provided samples, coordinated sample blinding and distribution, analyzed data, and wrote the first draft of and edited the manuscript. AY designed analyses, analyzed data, wrote sections of the manuscript, and reviewed the manuscript. DN and RS acquired and analyzed data and reviewed the manuscript. SU-B and PC acquired and analyzed data and reviewed the manuscript. SAT acquired and analyzed data and reviewed the manuscript. JJW processed and provided samples and edited the manuscript. MDB enrolled subjects, provided samples, and edited the manuscript. KCH designed the study, oversaw data generation, and edited the manuscript. RGM designed the study, oversaw data generation, and edited the manuscript. YD, RS, and BG designed the study, oversaw data generation, and edited the manuscript. CEM conceived of and designed the study, oversaw data generation, and helped write early drafts of and edited the manuscript.
Additional Information
Present Affiliation: SAT and RGM’s current affiliation is Kovler Diabetes Center, University of Chicago, Chicago, IL, US.
Disclosure Summary: DN, BG, RS and YD hold a patent for Assay D; SU-B holds a patent for Assay H; and SAT and RGM have filed a patent application for Assay M. SU-B receives a salary from L2 Diagnostics. CS, AY, PC, KCH, and CEM have nothing to disclose.
Data availability: The datasets generated and analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Bingley PJ, Wherrett DK, Shultz A, Rafkin LE, Atkinson MA, Greenbaum CJ. Type 1 diabetes TrialNet: a multifaceted approach to bringing disease-modifying therapy to clinical use in type 1 diabetes. Diabetes Care. 2018;41(4):653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Insel RA, Dunne JL, Atkinson MA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care. 2015;38(10):1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haller MJ, Schatz DA, Skyler JS, et al. ; Type 1 Diabetes TrialNet ATG-GCSF Study Group Low-dose anti-thymocyte globulin (ATG) preserves β-cell function and improves HbA1c in new-onset type 1 diabetes. Diabetes Care. 2018;41(9):1917–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Orban T, Bundy B, Becker DJ, et al. ; Type 1 Diabetes TrialNet Abatacept Study Group Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet. 2011;378(9789):412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Herold KC, Gitelman SE, Ehlers MR, et al. ; AbATE Study Team Teplizumab (anti-CD3 mAb) treatment preserves C-peptide responses in patients with new-onset type 1 diabetes in a randomized controlled trial: metabolic and immunologic features at baseline identify a subgroup of responders. Diabetes. 2013;62(11):3766–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, et al. ; Type 1 Diabetes TrialNet Anti-CD20 Study Group Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N Engl J Med. 2009;361(22):2143–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rigby MR, DiMeglio LA, Rendell MS, et al. ; T1DAL Study Team Targeting of memory T cells with alefacept in new-onset type 1 diabetes (T1DAL study): 12 month results of a randomised, double-blind, placebo-controlled phase 2 trial. Lancet Diabetes Endocrinol. 2013;1(4):284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herold KC, Bundy BN, Long SA, et al. ; Type 1 Diabetes TrialNet Study Group An anti-CD3 antibody, teplizumab, in relatives at risk for type 1 diabetes. N Engl J Med. 2019;381(7):603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Writing Committee for the Type 1 Diabetes TrialNet Oral Insulin Study Group, Krischer JP, Schatz DA, Bundy B, Skyler JS, Greenbaum CJ. Effect of oral insulin on prevention of diabetes in relatives of patients with type 1 diabetes. JAMA. 2017;318(19):1891–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dufort MJ, Greenbaum CJ, Speake C, Linsley PS. Cell type-specific immune phenotypes predict loss of insulin secretion in new-onset type 1 diabetes. JCI Insight. 2019;4(4):125556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bundy BN, Krischer JP; Type 1 Diabetes TrialNet Study Group A model-based approach to sample size estimation in recent onset type 1 diabetes. Diabetes Metab Res Rev. 2016;32(8):827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akirav EM, Lebastchi J, Galvan EM, et al. Detection of beta cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci U S A. 2011;108(47):19018–19023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lehmann-Werman R, Neiman D, Zemmour H, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A. 2016;113(13):E1826–E1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fisher MM, Watkins RA, Blum J, et al. Elevations in circulating methylated and unmethylated preproinsulin DNA in new-onset type 1 diabetes. Diabetes. 2015;64(11):3867–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Husseiny MI, Kaye A, Zebadua E, Kandeel F, Ferreri K. Tissue-specific methylation of human insulin gene and PCR assay for monitoring beta cell death. PloS One. 2014;9(4):e94591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moss J, Magenheim J, Neiman D, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. [DOI] [PubMed] [Google Scholar]
- 18. Bianchi DW, Chiu RWK. Sequencing of circulating cell-free DNA during pregnancy. N Engl J Med. 2018;379(5):464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burnham P, Khush K, De Vlaminck I. Myriad applications of circulating cell-free DNA in precision organ transplant monitoring. Ann Am Thorac Soc. 2017;14(Supplement_3):S237–S241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lebastchi J, Deng S, Lebastchi AH, et al. Immune therapy and β-cell death in type 1 diabetes. Diabetes. 2013;62(5):1676–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herold KC, Usmani-Brown S, Ghazi T, et al. , Type 1 Diabetes TrialNet Study Group Beta cell death and dysfunction during type 1 diabetes development in at-risk individuals. J Clin Invest. 2015;125(3):1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simmons KM, Fouts A, Pyle L, et al. , Unmethylated insulin as an adjunctive marker of beta cell death and progression to type 1 diabetes in participants at risk for diabetes. Int J Mol Sci. 2019;20(16):E3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neyman A, Nelson J, Tersey SA, Mirmira RG, Evans-Molina C, Sims EK. Persistent elevations in circulating INS DNA among subjects with longstanding type 1 diabetes. Diabetes Obes Metab. 2019;21(1):95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kenna LA, Olsen JA, Spelios MG, Radin MS, Akirav EM. β-Cell death is decreased in women with gestational diabetes mellitus. Diabetol Metab Syndr. 2016;8(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mulukutla SN, Tersey SA, Hampe CS, Mirmira RG, Balasubramanyam A. Elevated unmethylated and methylated insulin DNA are unique markers of A+β+ ketosis prone diabetes. J Diabetes Complications. 2018;32(2):193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gala-Lopez BL, Neiman D, Kin T, et al. Beta cell death by cell-free DNA and outcome after clinical islet transplantation. Transplantation. 2018;102(6):978–985. [DOI] [PubMed] [Google Scholar]
- 27. Bellin MD, Clark P, Usmani-Brown S, et al. Unmethylated insulin DNA is elevated after total pancreatectomy with islet autotransplantation: assessment of a novel beta cell marker. Am J Transplant. 2017;17(4):1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roels S, Costa OR, Tersey SA, et al. Combined analysis of GAD65, miR-375, and unmethylated insulin DNA following islet transplantation in patients with T1D. J Clin Endocrinol Metab. 2019;104(2):451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64(1):218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hanley S, Liu S, Lipsett M, et al. Tumor necrosis factor-alpha production by human islets leads to postisolation cell death. Transplantation. 2006;82(6):813–818. [DOI] [PubMed] [Google Scholar]
- 31. Paraskevas S, Maysinger D, Wang R, Duguid TP, Rosenberg L. Cell loss in isolated human islets occurs by apoptosis. Pancreas. 2000;20(3):270–276. [DOI] [PubMed] [Google Scholar]
- 32. Tersey SA, Nelson JB, Fisher MM, Mirmira RG. Measurement of differentially methylated INS DNA species in human serum samples as a biomarker of islet beta cell death. J Vis Exp. 2016(118). doi:10.3791/54838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cate RL, Chick W, Gilbert W. Comparison of the methylation patterns of the two rat insulin genes. J Biol Chem. 1983;258(10):6645–6652. [PubMed] [Google Scholar]
- 34. Kuroda A, Rauch TA, Todorov I, et al. Insulin gene expression is regulated by DNA methylation. Plos One. 2009;4(9):e6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diefenbach RJ, Lee JH, Kefford RF, Rizos H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 2018;228-229:21–27. [DOI] [PubMed] [Google Scholar]
- 36. Meddeb R, Pisareva E, Thierry AR. Guidelines for the preanalytical conditions for analyzing circulating cell-free DNA. Clin Chem. 2019;65(5):623–633. [DOI] [PubMed] [Google Scholar]
- 37. Neiman D, Moss J, Hecht M, et al. Islet cells share promoter hypomethylation independently of expression, but exhibit cell-type-specific methylation in enhancers. Proc Natl Acad Sci U S A. 2017;114(51):13525–13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biol Ther. 2019;20(8):1057–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Olsen JA, Kenna LA, Spelios MG, Hessner MJ, Akirav EM. Circulating differentially methylated amylin DNA as a biomarker of β-cell loss in type 1 diabetes. PloS One. 2016;11(4):e0152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan KCA, Woo JKS, King A, et al. Analysis of plasma epstein-barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377(6):513–522. [DOI] [PubMed] [Google Scholar]
- 41. Bonifacio E, Achenbach P. Birth and coming of age of islet autoantibodies. Clin Exp Immunol. 2019;198(3):294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lampasona V, Pittman DL, Williams AJ, et al. ; Participating Laboratories Islet autoantibody standardization program 2018 workshop: interlaboratory comparison of glutamic acid decarboxylase autoantibody assay performance. Clin Chem. 2019;65(9):1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]