Abstract
Background:
Sharing research data uses resources effectively; enables large, diverse data sets; and supports rigor and reproducibility. However, sharing such data increases privacy risks for participants who may be re-identified by linking study data to outside data sets. These risks have been investigated for genetic and medical records but rarely for environmental data.
Objectives:
We evaluated how data in environmental health (EH) studies may be vulnerable to linkage and we investigated, in a case study, whether environmental measurements could contribute to inferring latent categories (e.g., geographic location), which increases privacy risks.
Methods:
We identified 12 prominent EH studies, reviewed the data types collected, and evaluated the availability of outside data sets that overlap with study data. With data from the Household Exposure Study in California and Massachusetts and the Green Housing Study in Boston, Massachusetts, and Cincinnati, Ohio, we used k-means clustering and principal component analysis to investigate whether participants’ region of residence could be inferred from measurements of chemicals in household air and dust.
Results:
All 12 studies included at least two of five data types that overlap with outside data sets: geographic location (9 studies), medical data (9 studies), occupation (10 studies), housing characteristics (10 studies), and genetic data (7 studies). In our cluster analysis, participants’ region of residence could be inferred with 80%–98% accuracy using environmental measurements with original laboratory reporting limits.
Discussion:
EH studies frequently include data that are vulnerable to linkage with voter lists, tax and real estate data, professional licensing lists, and ancestry websites, and exposure measurements may be used to identify subgroup membership, increasing likelihood of linkage. Thus, unsupervised sharing of EH research data potentially raises substantial privacy risks. Empirical research can help characterize risks and evaluate technical solutions. Our findings reinforce the need for legal and policy protections to shield participants from potential harms of re-identification from data sharing. https://doi.org/10.1289/EHP4817
Introduction
The trade-off between sharing personal data and the risks to privacy has become an everyday concern for consumers as social networks, search engines, ride-sharing apps, credit cards, and smart home devices, for example, ask consumers to “allow” access to location, purchases, internet searches, and more. Privacy researchers have demonstrated that diverse types of data—for example, movie rentals (Narayanan and Shmatikov 2008), bicycle shares (Pennarola et al. 2017), electricity meter readings (Buchmann et al. 2013), and hospital visits (Sweeney 2013)—can be linked back to individuals, even when they are shared without names or other overt identifiers such as address or exact birth date. In 2013, researchers demonstrated that surnames can be identified from genetic sequencing data (Gymrek et al. 2013), and, recently, genetics data posted by consumers on genealogy websites were used to identify crime suspects (Justin 2018)—such databases may soon have sufficient coverage to facilitate re-identification (re-ID) of nearly any American of European descent (Erlich et al. 2018). Most recently, Rocher et al. (2019) estimated that nearly all Americans can be identified in any data set by using 15 demographic attributes. This process of linking “de-identified” data that lack obvious personal identifiers, such as name, birth date, or address, back to one individual or a few likely matches is referred to as re-ID. Reports of successful re-ID increased rapidly in the last decade, although a recent review found that only 8 of 55 reports were published in academic journals, limiting dissemination of these risks to the broader research community (Henriksen-Bulmer and Jeary 2016).
Re-ID is increasingly relevant to environmental health (EH) research, because of growing pressures to share data, more personalized exposure measurements, and rapidly expanding repositories of public and commercial data. Environmental exposure measurements are often individual- or home-specific, such as chemical biomonitoring data or measurements in personal spaces. The advent of wearable sensors (e.g., smartphones and devices like Fitbit®) that continuously collect data such as location, exposure, and biometrics creates added vulnerability. In addition, EH studies can include genetic, medical, or household data that are themselves vulnerable to re-ID, creating disclosure risks for the entire data set. Loss of privacy from re-ID could result in stigma for individuals and communities; affect property values, insurance, employability, and legal obligations; or reveal embarrassing or illegal activity (Goho 2016; Zarate et al. 2016). It could damage trust in research, harming the study and research more generally. Because EH studies often focus on groups with the highest exposures, privacy risks potentially compound harms faced by the most vulnerable communities. Entities that might be motivated to re-identify EH data include, for example, employers or insurance companies (who may wish to discriminate against individuals or properties on the basis of environmental exposures) and corporations affected by environmental regulations (who may wish to discredit litigants or studies demonstrating EH harms, or to discourage participation in EH research). Other parties might leverage the environmental variables to gain access to other parts of the data set, such as sensitive health information.
At the same time, researchers, funders, the public, and study participants may want to share data to maximize its value to science and health. Biological and environmental samples are expensive to collect and analyze. Sometimes they are nonrepeatable, for example, in the case of samples collected after environmental disasters. Sharing data creates new opportunities to gain knowledge from an initial and often public investment. Data sharing can also facilitate the creation of larger and geographically and demographically more diverse data sets. Research consortia, such as the Environmental Health Influences on Child Health Outcomes Program (ECHO), and, ultimately, large-scale research, such as the All of Us Study, are specifically designed around the concept of multiple researchers pooling data collected using common protocols (NIH 2017, n.d.-c).
Paradigms for data sharing span a continuum from agreements among researchers under Institutional Review Board (IRB) oversight—consistent with strong pledges of confidentiality to study participants—to data without overt identifiers shared without restriction and without IRB oversight, and, in its most open form, publicly. Public research funding is increasingly tied to open access requirements for digital data. In 2013, the White House Office of Science and Technology Policy mandated that U.S. federal agencies develop plans to make scholarly publications created with federal funds—and their underlying digital data—publicly available (Holdren 2013). Since then, 22 agencies have issued public access plans, including the National Institutes of Health (NIH) and the U.S. Environmental Protection Agency (U.S. EPA) (https://www.science.gov/publicAccess.html). The European Commission has issued similar open access recommendations for publicly funded research (European Commission 2018), leading to implementation efforts such as the Horizon 2020 Open Research Data pilot and European Open Science Cloud (European Commission 2017, n.d.). Scholarly journals also favor policies requiring access to data to ensure the rigor and reproducibility of statistical analyses [e.g., the American Association for the Advancement of Science (AAAS n.d.)].
In the United States, scientists have faced particular pressure from government officials and private interests to make data publicly available from studies used to support regulatory decisions. In the late 1990s, the tobacco industry sought to discredit scientific findings through a multipronged “sound science” campaign that included efforts to legislate data access to federally funded research (Baba et al. 2005). At the same time, industries affected by air pollution standards sought raw, individual data from the NIH-funded Six Cities Study, which was cited in regulations under the Clean Air Act (Fischer 2013). In response to these coordinated efforts, the U.S. Congress passed a law in 1999 that included a provision often referred to as the Shelby Amendment, which extended the reach of Freedom of Information Act (FOIA) requests (Fischer 2013). Previously, the U.S. Supreme Court had found that data from federal grantees were not subject to FOIA requests (in contrast to data from intramural agency research) (Fischer 2013). With the Shelby amendment, federal grantees at outside nonprofit institutions are required to provide data in response to FOIA requests when the research was used to support regulation (OMB 1999). More recently, in 2018, the U.S. EPA issued a proposed rule titled, “Strengthening Transparency in Regulatory Science,” which would require data to be publicly available as a precondition for using it to support regulatory decisions (EPA 2018). The proposed rule raised concerns among researchers about threats to participants’ privacy, violations of assurances in informed consent, and barriers to recruiting participants for future studies (Schwartz 2018). The U.S. EPA is expected to issue a supplemental proposed rule on this topic in 2020 (Science and Technology at the Environmental Protection Agency 2019).
Although all the policies discussed include stated exemptions to protect personal privacy, they do not provide guidance regarding which data constitute a risk of privacy violations. As expectations for publicly available data increase, EH researchers and decision-makers need to better understand privacy risks in order to make responsible choices that optimize data sharing while protecting privacy. These issues came to our attention through our Household Exposure Study (HES). The HES measured 87 endocrine-disrupting compounds (EDCs) in 120 people and their homes on Cape Cod in Massachusetts in the first comprehensive report on indoor exposure to these chemicals (Rudel et al. 2003), and the study later expanded to California (Brody et al. 2009; Rudel et al. 2010). The study also collected information about demographics, housing characteristics, and behaviors that might be related to the chemical measurements. These data provided a unique resource for understanding exposure and health risks from consumer product chemicals, and the U.S. EPA staff approached us about sharing the data online in ExpoCast™ (Cohen-Hubal 2009). We were uncertain whether the data might be vulnerable to re-ID, so we began an empirical investigation of that risk.
A common re-ID strategy uses linkage of two or more data sets with overlapping fields (Sweeney et al. 2017). When study data overlap with externally available data sets, the combined data can be used to re-ID participants. Using this approach, we conducted a re-ID experiment using data from the Northern California HES in Bolinas, California, and in selected neighborhoods of Richmond, California (Sweeney et al. 2017). HES researchers first redacted the data set to exclude information that cannot be shared under the Safe Harbor provision of the U.S. Health Information Portability and Accountability Act (HIPAA) and removed or aggregated other variables that we considered to be vulnerable—based on our knowledge of which data were likely to be publicly or commercially available—while maintaining the scientific utility of the data. For example, we removed data on pet ownership and aggregated dates, such as the year the home was built, into categories. The remaining data set included chemical measurements from the homes and variables such as race, gender, birth year, home ownership, square footage of living area, number and types of rooms, and decade group for when the house was built and when the participant moved in. Using information in the peer-reviewed articles from the study (Brody et al. 2009; Rudel et al. 2003, 2010), property data from local tax assessor records, and person-level information acquired from public data brokers, the re-ID team, led by L. Sweeney, was able to correctly separate records for residents of Bolinas from records for Richmond residents, and they correctly and uniquely identified 8 of 50 participants (16%) by name in a data set that met HIPAA requirements. One participant (2%) was correctly named even when the data set was further redacted to birth year by decade (Sweeney et al. 2017). Matches associate a name to a study participant’s record, which includes environmental exposure measurements in this study and could include protected health and genetic information or other personal data in other studies.
To further understand how data linkage (also called record linkage) can contribute to privacy risks in EH data, we conducted two additional investigations reported here. We sought to evaluate how data in well-known EH studies may be vulnerable to linkage: a) We evaluated whether data types in 12 major environmental studies are currently available as part of public or commercial registries and therefore potentially pose privacy risks from linkage, and b) We used clustering methods to investigate how environmental measurements, which themselves are not currently vulnerable to data linkage, might contribute to re-ID by identifying subgroups (e.g., location, race, disease status) within a data set so that linkage is limited to smaller numbers of possible matches. For example, Gymrek et al. (2013) used U.S. state of residency as part of the strategy to match individuals to genetic data. In the Northern California HES, partitioning the data set by city (Richmond vs. Bolinas) and housing development (Liberty Village vs. Atchison Village) was crucial to the success of re-ID. Could similar partitions be achieved with the chemical measurement data alone? Answering this question is helpful for evaluating whether investigators must consider the possibility that variables such as location can be reconstructed from environmental measurements after they have been redacted.
Through these investigations, we aim to determine the availability of vulnerable data types in EH studies and assess additional risk of re-ID introduced by sharing detailed environmental sampling results. Understanding re-ID risks can contribute to realistic informed consent statements and the development of privacy-preserving research policies and practices.
Methods
EH Data Types That Are Vulnerable to Linkage
We selected 12 EH studies to evaluate for the presence of data vulnerable to linkage to existing public or commercial registries. We first selected candidate studies, based on our knowledge and experience as NIEHS-supported EH researchers, as examples of significant EH studies that have made or are expected to make important contributions to the field. Eleven studies were chosen to include a range of scenarios in EH. Because we were motivated by the HES, we included other studies of household exposures in addition to biomonitoring studies. We visited the websites of the selected studies and recorded information about the data that are publicly available or are offered for restricted sharing with other researchers. We sent our descriptions of the studies to the investigators to verify their accuracy. We categorized the data in the studies into broad categories of data types that are important for EH studies, because they represent exposures or outcomes of interest. To focus our investigation, we did not include demographic variables or survey data, which are less distinctive to EH, although these are commonly collected and could be used in re-ID. We also searched for public information about each study’s data sharing policy, which governs who can access the data outside the original research team. Because the first step in re-ID is gaining access to the data, data sharing practices are an important contributor to risk of re-ID. We convened an Advisory Council of EH researchers, privacy experts, ethicists, study-participant representatives, and public officials to consider the candidate studies and discuss data sharing and privacy issues in EH. The Advisory Council concurred with our choices of EH studies and data types. Advisory Council members are shown in Table S1. The selected studies are all U.S.-based, reflecting the expertise of the authors and Advisory Council. We later added the Green Housing Study because of its inclusion in the clustering analysis, and we revisited the websites for the studies to update information about available data.
Finally, we evaluated the availability of external public or commercial data that could be used in linkage strategies to match to the EH study data types. We searched for data sets online and drew on the experience of author L. Sweeney with obtaining commercial data sets and performing re-IDs (Sweeney 2002, 2013; Sweeney et al. 2017, 2018). We also searched for published examples of re-IDs where these data types were used as part of the linkage strategy.
Unsupervised Clustering of Environmental Chemical Measurements
The biological and individual-level environmental chemical measurements that are fundamental to EH studies are not usually expected to contribute directly to re-ID via data linkage, because currently there are very few public repositories of matching data (lead results, which may become public in some jurisdictions, are a possible example of matching data). However, as noted earlier, chemical measurements can potentially contribute to re-ID by identifying subgroups that narrow the matching task for other variables. We tested the ability to infer subgroup membership in a data set by using a data clustering approach in two studies of household exposure to environmental chemicals, each conducted in multiple geographic locations. We selected these data sets because they contained relevant exposure measurements, and we had access to the “true” group membership (location in this case) to score the success of clustering. We hypothesized that some exposures vary systematically between locations, creating an opportunity to infer likely geographic location from the chemical measurements.
Description of data.
Massachusetts and Northern California HES.
The Massachusetts Household Exposure Study was conducted in 1999–2001 (Rudel et al. 2003). Dust and indoor air samples were collected from 120 households in Cape Cod, Massachusetts, but the current analysis is restricted to 72 participants who were identified as deceased as of May 2016, because of restrictions by the Massachusetts Cancer Registry on use of the data for this analysis. The Northern California Household Exposure Study was conducted in 2006 (Rudel et al. 2010). Dust and indoor and outdoor air samples were collected from 50 households in Richmond and Bolinas. Samples in both studies were analyzed for a broad suite of chemicals, including pesticides, phthalates, polycyclic aromatic hydrocarbons (PAHs), polybrominated diphenyl ethers (PBDEs), polychlorinated biphenyls (PCBs), alkylphenols, parabens, and other phenols and biphenyls identified as EDCs.
We created a combined data set restricted to analytes measured in the same medium (dust or indoor air) in both locations in a majority of homes. The data set included 24 chemicals measured in indoor air in 122 homes and 44 chemicals measured in dust in 120 homes.
Green Housing Study.
The Green Housing Study (GHS)—a project of the Centers for Disease Control and Prevention and Department of Housing and Urban Development—is evaluating the effects of “green” housing on indoor environmental quality and children’s respiratory health. The study sites are in Boston, Massachusetts, Cincinnati, Ohio, and New Orleans, Louisiana (Coombs et al. 2016; Dodson et al. 2017). Indoor air samples were collected from households with children ages 7–12 with physician-diagnosed asthma living in subsidized housing in Boston () and Cincinnati () in 2012–2013. Air samples were analyzed for 35 semivolatile organic compounds, including flame retardants, phthalates, environmental phenols, fragrance chemicals, and PCBs. New Orleans data were not available at the time of this analysis. Some homes () had two air samples collected approximately 6 months apart; for these homes, we calculated the average exposure for each chemical. Homes that have two visits are not expected to differ systematically with respect to exposure from homes that have one visit.
Cluster analysis.
Chemicals with detection frequencies above 10% were used for cluster analysis. Detection frequencies were calculated as the percent of samples with measured masses above the method reporting limit (MRL). Samples below the MRL were reported as either not detected or as estimated values. Estimated values were used in the cluster analysis. For samples reported as not detected, concentrations were substituted with the sample-specific reporting limit (SSRL), which was calculated as the MRL divided by the sample-specific volume of air or sample-specific mass of dust. Concentration data were natural log transformed. All analyses were conducted using R (version 3.5.0; R Development Core Team).
We applied k-means clustering to the chemical exposure data. k-Means is a common method for unsupervised clustering that can be used to blindly classify groups of similar observations when no information about group membership is available. It uses an iterative process that partitions observations into k clusters based on distance to the cluster centroid. The number of clusters, k, is specified by the investigator a priori. The initial cluster centroids are assigned randomly to points in the n-dimensional space that spans all n chemicals in the analysis, and observations are assigned to the nearest centroid. Then, centroids are recalculated as the arithmetic means of the chemical measurements assigned to the cluster. Next, observations are reassigned to the new nearest centroid, and the process is repeated until no observations are reassigned to a different centroid. We used the Hartigan-Wong algorithm, which assigns observations to centroids by minimizing the within-cluster sum of squared errors (Hartigan and Wong 1979), as implemented in the “kmeans” function from the R base stats package (version 3.5.0; R Development Core Team), and we specified 100 random starts and a maximum of 10,000 iterations to converge on a stable solution.
We hypothesized that cluster analysis would partition the data by geographic region, so we specified two clusters () for each analysis (i.e., Massachusetts and California for HES, Boston and Cincinnati for GHS). This correct answer for the number of geographic centers would be known to anyone from publications about the studies. We were not aware of subgroupings in these studies other than location (such as gender or race/ethnicity) that were likely to be relevant to household exposures and did not also covary with location, so we did not explore additional numbers of clusters.
To score the results of the k-means analysis, we checked the true location associated with each data point after clustering was complete. We assigned each cluster a geographic identity (e.g., Massachusetts or California) based on the site to which the majority of records belonged. We calculated the accuracy of the clustering by counting the correctly grouped records (e.g., Massachusetts records in the Massachusetts cluster and California records in the California cluster) and dividing by the total number of records analyzed. We also calculated the adjusted Rand index (Hubert and Arabie 1985), a measure of similarity between two partitions—in this case, the k-means clustering result and the true distribution. The adjusted Rand index has an expected value of zero for random clusters and a maximum value of 1 in the case of perfect agreement.
To examine which chemicals were influential in the clustering analysis, we ran principal component analysis (PCA) on the same data sets using eigendecomposition of the covariance matrix [function “princomp” from R base stats (version 3.5.0; R Development Core Team)]. We visually examined the clustering results on a plot of PC2 vs. PC1. We assessed whether variation along either axis contributed to separation between the clusters and examined the PC loadings to determine which chemicals contributed most strongly to the separation.
Differences in data-collection protocols or analytic methods between study sites may make it easier to partition participants by study site, thus increasing the risk of re-ID. Therefore, when study data are shared, researchers may want to consider data-masking methods to obscure site differences that represent methodological artifacts. To illustrate the potential influence of data-masking approaches on our ability to partition study participants by location using chemical exposure data, we created censored data sets that eliminated systematic site differences in reporting limits (in the HES) and sample volumes (in the GHS). In the HES, MRLs systematically differed between Massachusetts and California for some chemicals. For each chemical, we calculated the most frequent MRL reported in each site (in cases of ties, we used the lower value) and defined the MRL for the censored analysis () as the higher of the two modal MRLs. We calculated censored sample-specific reporting limits () as divided by the sample-specific volume of air or sample-specific mass of dust. For all records where the original SSRL or detected or estimated concentration was lower than , the concentration was substituted with . This substitution effectively masked differences in the exposure distribution resulting from differences in reporting limits. We repeated the cluster analysis using the same procedures described above but using the censored concentration. We did not mask differences by site in sample volume or sample mass, which were small relative to differences in MRL in the HES. In GHS, MRLs did not differ by site, but the sample-specific volume of air was systematically higher in Cincinnati () than Boston () (Welch’s two sample t-test: , , ). To assess whether differences in sample volume were driving the cluster results, we repeated the cluster analysis with nondetects substituted with the MRL divided by the median volume of air across Boston, Massachusetts, and Cincinnati, Ohio (i.e., a constant value).
Results
EH Data Types That Are Vulnerable to Linkage
The 12 studies chosen for investigation included the National Health and Nutrition Examination Survey (NHANES), a cross-sectional sample designed to be representative of the U.S. population (Zipf et al. 2013); cohort studies, including occupational groups [Agricultural Health Study (Alavanja et al. 1996) and California Teachers Study (Bernstein et al. 2002)], a disease-risk group (Sister Study; Sandler et al. 2017), and children [Breast Cancer and the Environment Research Program (BCERP) (Biro et al. 2010) and Center for the Health Assessment of Mothers and Children of Salinas (CHAMACOS) Study (Eskenazi et al. 2003)]; environmental disaster response [Gulf Long Term Follow-Up (GuLF) Study (Kwok et al. 2017)]; and household chemical exposures [American Healthy Homes Survey (Stout et al. 2009), Relationships of Indoor Outdoor and Personal Air (RIOPA) (Weisel et al. 2005), Pesticide and Chemical Exposure (PACE) Study (Adamkiewicz et al. 2011), HES, and GHS]. We selected eight features of EH studies for investigation: a focus on specific locations, inclusion of multiple family members in the study, medical data, genetic data, occupation data, housing data, exposure data from biological samples, and exposure data from home or personal environment samples. We also investigated each study’s data-sharing policy. The studies and study features are identified in Table 1.
Table 1.
Study characteristics and data types that may contribute to re-identification risk in selected environmental health studies.
| Study | Study characteristics | Data types | ||||||
|---|---|---|---|---|---|---|---|---|
| Focus on specific locationsa | Family members in studyb | Medical data | Genetic data | Occupation data | Housing datac | Exposure data from biological samples | Exposure data from home/personal environment samples | |
| Agricultural Health Study. Private pesticide applicators and their spouses in Iowa and North Carolina; licensed pesticide applicators in Iowa. | x | x | x | x | x | x | x | x |
| American Healthy Homes Survey. Representative sample of U.S. homes 2005–2006. | — | — | — | — | x | x | — | x |
| Breast Cancer and the Environment Research Program (BCERP) Puberty Study. Girls recruited at 6–8 years of age in New York City, California Bay Area, and Greater Cincinnati. | x | — | x | x | — | — | x | — |
| California Teachers Study. Current and former female public school teachers or administrators. | x | — | x | x | x | — | x | — |
| CHAMACOS Study. Mothers and children in Salinas Valley, CA. | x | x | x | x | x | x | x | x |
| Green Housing Study. Children with physician-diagnosed asthma living in public housing in greater Boston, Cincinnati, and New Orleans. | x | — | x | — | — | x | x | x |
| Gulf Long Term Follow-up (GuLF) Study. Participants in the Deepwater Horizon oil spill cleanup or training. | x | — | x | x | x | x | x | x |
| National Health and Nutrition Examination Survey (NHANES). Nationally representative sample of the U.S. population, collected from specific locations in each two-year cycle. | — | — | x | x | x | x | x | — |
| Pesticide and Chemical Exposure (PACE) Study. Residents of urban MA and rural FL neighborhoods. | x | — | — | — | x | x | — | x |
| Relationships of Indoor, Outdoor, and Personal Air (RIOPA). Adults and children in Elizabeth, NH; Houston, TX; and Los Angeles, CA. | x | x | x | — | x | x | — | x |
| Silent Spring Institute Household Exposure Study (HES). Residents in Cape Cod, MA, and Bolinas, CA, and Richmond, CA. | x | — | — | — | x | x | x | x |
| Sister Study. Women (cancer-free at enrollment) with a sister diagnosed with breast cancer. | — | — | x | x | x | x | x | x |
Note: —, not a study characteristic or a data type collected in the study.
One enrollment criterion was living or working in a publicly defined geographic area. In addition, NHANES samples from 15 locations per year, although these locations are not intended to be a focus of study.
The study enrolled family members as part of its study design. Additional studies, for example NHANES and the Sister Study, allow enrollment of multiple members of the same family.
Characteristics of participants' homes, such as number or type of rooms; square footage; year built; information about heating, ventilation, and air conditioning; presence of certain furnishings or appliances, etc.
EH data types and potential for linkage.
Each study included between three and eight of the EH features we investigated. Here we review publicly available data sets that could match to each of these features in linkage-based re-ID. The data sets that follow illustrate the most apparent vulnerabilities but are not an exhaustive list of currently available data relevant to re-ID.
Focus on specific locations.
Nine studies are limited to specific geographic locations, so linkage efforts can focus on matching to data from that location. Locations may be statewide, such as the California Teachers Study and Agricultural Health Study (two states), or an environmentally defined region, such as the CHAMACOS Study in the intensive-agriculture region of the Salinas Valley and the GuLF Study of the area affected by the Deepwater Horizon oil spill. Other studies are limited to metropolitan areas (BCERP Puberty Study cohorts), cities (GHS, RIOPA, Northern California HES), or counties (PACE Study, Massachusetts HES). In the Northern California HES, specific neighborhoods were named in one city. NHANES is a nationally representative sample that does not overtly include location, but the locations and dates associated with data collection cycles can sometimes be discovered in local news stories and photographs online. However, because multiple locations are pooled in each cycle, identifying subsets of data associated with specific locations would require further work, such as data matching or clustering. Otherwise, linkage analysis must be performed between the entire NHANES data set and external data from all locations included in each cycle. The ability to identify location improves the ease and likelihood of matching demographic information, such as gender, race, and age, to voter lists or commercial lists of residents (e.g., Sweeney et al. 2017). Lists of residents by name, address, and demographic characteristics—along with countless other personal data elements—are readily available by geographic area from data brokers (Ramirez et al. 2014). Examples of major data brokers include Acxiom, Experian, Equifax, CoreLogic, and TowerData. Since January 2019, the State of Vermont has required data brokers who trade data of Vermont residents to register in a public database; as of this writing, there are 154 active registrations (Vermont Secretary of State 2019). In addition, knowing location narrows the search and improves the likelihood of matching data in other domains, such as housing and occupation, described below.
Occupation data.
Ten studies have information about occupation of the participant or the parent of the participant, providing data that potentially matches to licensing lists for pesticide applicators, teachers, nurses, and other professions, or to publicly accessible LinkedIn profiles, professional society membership lists, and institutional employer websites (e.g., Sweeney et al. 2018). In the same way that location can narrow sources of matching data, lists of licensed professionals can serve as the population registry or be cross-referenced with other population registries (e.g., voter lists) to restrict the pool of possible matches. Lists of licensed professionals may also contain linkable information, such as the year in which a professional obtained a license or allowed it to expire.
Genetic data.
Seven studies have genetic data that could potentially be matched to ancestry sites (e.g., Erlich et al. 2018); indeed, these sites are designed to facilitate record matching. Conversely, it is possible to begin from a publicly available genome on an ancestry site that also includes the individual’s name or family surname and then infer whether the individual participated in a research study reporting some genomic information. The shared research data could be as limited as statistical measures of linkage disequilibrium from a genome-wide association study (Wang et al. 2009), or a genomic data-sharing beacon that returns a yes or no answer for the presence of a single nucleotide polymorphism (SNP) in the data set (von Thenen et al. 2019). Identifying someone as a participant can associate the individual with any sensitive characteristics of the study population, such as residence in a community with environmental contamination or diagnosis of disease.
Medical data.
Nine studies collected medical data that may be linked to data disclosed in hospital discharge records, pharmacy sales, local news stories and obituaries, social media, and disease-centered online communities (Culnane et al. 2017; El Emam et al. 2011; Sweeney 2013). A 2013 survey found that 33 states release some form of publicly available—but not necessarily free—patient-level hospital discharge data (Hooley and Sweeney 2013), and we found current examples of these practices (e.g., New York State Department of Health 2019; Vermont Department of Health 2019; Virginia Health Information 2018). In addition to some demographic information, hospital discharge records may include information like admission and discharge dates, diagnoses, and cost of stays. Anonymized patient records are also being created by health information–data-mining companies that aggregate data purchased from pharmacies, insurance companies, and health care providers (Tanner 2017). Some of these data sources, such as news stories and obituaries, contain direct identifiers that could be used to re-ID participants; others, like hospital discharge records, may contain additional quasi-identifiers that could be used as part of a multistage re-ID strategy.
Housing data.
Ten studies collected housing data to characterize potential exposures, such as construction years and materials, residence years, and floor plan characteristics. These data may be linked to local records such as tax assessor data, real estate transactions, and building permits, as well as records available on real estate websites, such as Zillow (e.g., Sweeney et al. 2017).
Biological or personal environment exposure data.
All 12 EH studies contained at least one type of exposure data. Because few public repositories contain any matching exposure data, these data are less vulnerable to straightforward linkage approaches. However, biological and household samples tested for consumer product chemicals could potentially be linked to commercial data on credit card purchases. In addition, we evaluate in this article their potential use for identifying subgroup membership using cluster analysis.
Multiple family members.
Three studies included multiple family members in the same study by design (e.g., spouses, parents and children). If one member is re-identified, then it becomes trivial to identify all family members in the study.
Data sharing practices of the studies.
Among the selected studies, RIOPA and NHANES post selected data publicly online (CDC/NCHS 2018; Health Effects Institute n.d.), and exposure data from the American Healthy Homes Survey are intended for inclusion in the public U.S. EPA ExpoCast™ database but are not yet available online (NCCT 2017). For NHANES, re-ID is prohibited by law and online instructions state that use of the data signifies agreement to use it “only for the purpose of health statistical reporting and analysis” (CDC 2015a). Additional NHANES data beyond the public-use files are available by application at a restricted data center; researchers must receive approval in advance for their analysis plans and code, and they enter the controlled area without laptops, smartphones, or other electronic communications devices (CDC 2015b). Four studies—the Agricultural Health Study, California Teacher’s Study, GuLF Study, and Sister Study—have established data access procedures that consider requests for specific variables for specific research purposes under agreements to protect the confidentiality of individual participants (California Teachers Study n.d.; Freeman et al. 2017; NIH n.d.-d; Sister Study n.d.). The Agricultural Health Study has shared selected data in response to FOIA requests (e.g., see Supplemental Material of Goodman et al. 2017), and other federally funded studies must also follow the requirements set forth in that Act and in the Shelby amendment. For the other studies we considered, data-sharing policies were not publicly specified; however, they are required to follow policies set by IRBS and funding agencies, such as NIH.
Unsupervised Clustering of Environmental Chemical Measurements
Massachusetts and Northern California HES.
Air.
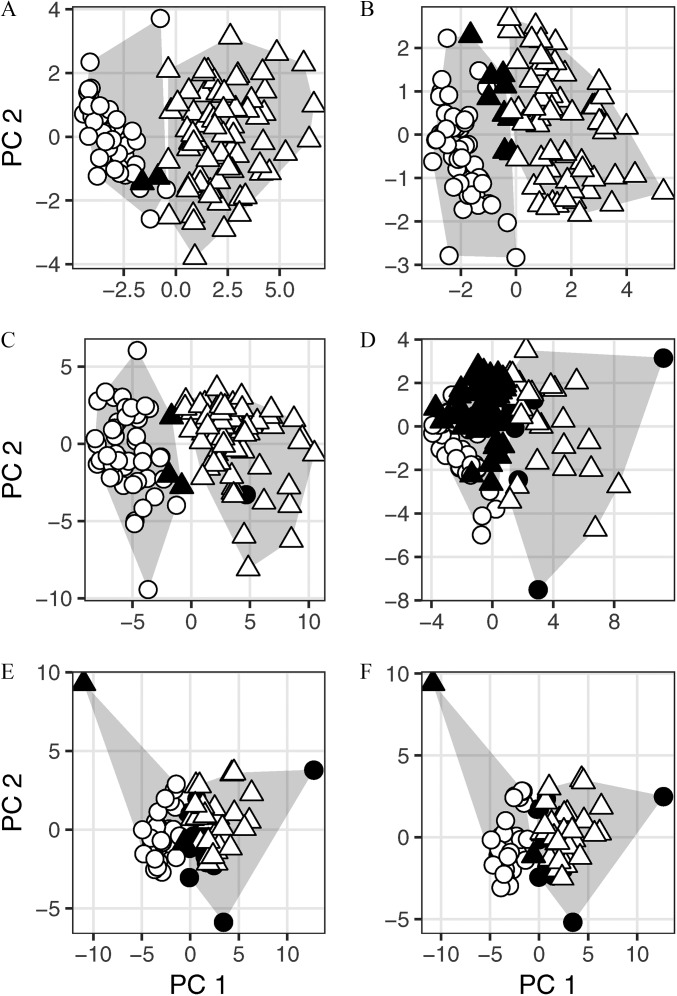
A total of 13 chemicals were detected in residential indoor air samples in at least 10% of the 122 homes in the data set using the original reporting limits. k-Means clustering of these uncensored chemical measurements grouped 70 of 72 Massachusetts homes into one cluster with positive scores for the first principal component (PC1 in Figure 1A) and grouped all 50 California homes (and 2 misclassified Massachusetts homes) into a second cluster with negative scores for PC1, resulting in a partition accuracy of 98.4% (Table 2). When differences in MRLs between the Massachusetts and California sites were masked by substituting censored sample-specific reporting limits for some observations, all 13 chemicals were retained for analysis based on detection frequency. k-Means analysis of the data set with censored detection limits grouped 63 of 72 Massachusetts homes into one cluster with positive scores on PC1, and grouped all 50 California homes (and 9 misclassified Massachusetts homes) into a second cluster with negative scores for PC1, for a partition accuracy of 92.6% (Table 2; Figure 1B). The chemicals with the highest loadings on PC1 in both analyses include banned organochlorine pesticides (e.g., heptachlor and chlordane) and the disinfectant o-phenylphenol (Table S2). Massachusetts homes had positive scores for PC1, reflecting higher levels of these chemicals—which is consistent with published findings from the HES study (Rudel et al. 2010).
Figure 1.
Individual homes plotted by principal component scores (PC1 and PC2) of residential chemical concentration data and overlaid on gray convex hulls indicating the bounds of two clusters generated using unsupervised k-means cluster analysis of the same data. All panels show homes from two regions. Homes were classified as correctly clustered (white symbols) if they were grouped in the cluster containing the majority of homes from their region; otherwise, they were classified as incorrectly clustered (black symbols). The shape of the symbol indicates the home’s true location. (A) and (B): k-means classification of 122 homes in the Household Exposure Study (72 from Massachusetts, triangles; 50 from California, circles) based on chemical concentrations in indoor air using original (A) or censored (B) reporting limits. (C) and (D): k-means classification of 120 homes in the Household Exposure Study (71 from Massachusetts, triangles; 49 from California, circles) based on chemical concentrations in indoor dust using original (C) or censored (D) reporting limits. (E) and (F): k-means classification of 77 homes in the Green Housing Study (33 from Cincinnati, Ohio, triangles, and 44 from Boston, Massachusetts, circles) based on chemical concentrations in indoor air using original (E) or constant (F) reporting limits.
Table 2.
Accuracy of k-means cluster analysis for subgrouping homes by region in the household exposure study (HES; Massachusetts and California) and Green Housing Study (GHS; Boston, Massachusetts, and Cincinnati, Ohio) using concentrations of chemicals detected in at least 10 percent of residential indoor air or dust samples.
| Study | Homes () | Sample matrix | Chemicals in studya () | Chemicals in cluster analysisb () | Reporting limits | Accuracyc (%) | Adjusted Rand indexd |
|---|---|---|---|---|---|---|---|
| HES | 122 | Air | 24 | 13 | Originale | 98.4 | 0.93 |
| HES | 122 | Air | 24 | 13 | Censoredf | 92.6 | 0.72 |
| HES | 120 | Dust | 44 | 25 | Original | 96.7 | 0.87 |
| HES | 120 | Dust | 44 | 18 | Censored | 55.8 | 0 |
| GHS | 77g | Air | 35 | 28 | Original | 80.5 | 0.36 |
| GHS | 77g | Air | 35 | 28 | Constanth | 80.5 | 0.36 |
Number of chemicals measured in the same medium in all homes in each cluster analysis.
Number of chemicals detected in at least 10% of homes given the reporting limits used in each analysis.
Number of homes correctly grouped by region using k-means clustering divided by the total number of homes analyzed.
The adjusted Rand index measures similarity between the two clusters identified by k-means analysis and the two true regional subgroups in the data. It has an expected value of zero for random clusters and a maximum value of 1 in the case of perfect agreement.
In analyses using the original reporting limits, concentrations that were not detected were substituted with the sample-specific reporting limit (SSRL). We calculated the SSRL as the method reporting limit (MRL) divided by the sample-specific volume of air or sample-specific mass of dust.
In analyses with censored reporting limits, we calculated the most frequent MRL reported in each site (in cases of ties we used the lower value). We defined as the higher of the two modal MRLs and calculated censored sample-specific reporting limits () as divided by the sample-specific volume of air or sample-specific mass of dust. For all records where the original SSRL or detected or estimated concentration was lower than , the concentration was substituted with .
Cluster analysis was performed on 77 homes comprising 105 samples. A total of 49 homes were sampled once, and 28 homes were sampled twice approximately six months apart. For homes sampled twice, we used the average exposure for each chemical.
Nondetects were substituted with the MRL divided by the median volume of air across Boston, Massachusetts, and Cincinnati, Ohio.
Dust.
A total of 25 chemicals were detected in residential indoor dust samples in at least 10% of the 120 homes in the data set using original reporting limits. k-Means clustering of these uncensored chemical measurements grouped 68 of 71 Massachusetts homes (and 1 misclassified California home) into one cluster with positive scores for PC1, and grouped 48 of 49 California homes (and 3 misclassified Massachusetts homes) into a second cluster with negative scores for PC1, resulting in a partition accuracy of 96.7% (Table 2, Figure 1C). When differences in MRLs between the Massachusetts and California sites were masked by substituting censored sample-specific reporting limits for some observations, only 18 chemicals were detected in at least 10% of homes. k-Means analysis of the data set with censored detection limits grouped 24 of 71 Massachusetts homes (and 6 misclassified California homes) into one cluster with higher scores on PC1, and grouped 43 of 49 California homes (and 47 misclassified Massachusetts homes) into a second cluster with lower scores for PC1, for a partition accuracy of 55.8% (Table 2, Figure 1D). Four of the top six chemicals that contributed to separation along PC1 (diazinon, PCB 105, PCB 153, and PCB 52) were among those excluded from analysis when reporting limits were censored (Table S2), which likely contributed to the reduction in accuracy of the unsupervised clustering.
Green Housing Study.
Air.
A total of 28 chemicals were detected in residential indoor air samples in at least 10% of the 105 samples collected, before duplicate samples were averaged for 28 homes. k-Means clustering of the data set with original detection limits grouped 31 of 33 Cincinnati homes (and 13 misclassified Boston homes) into one cluster with positive scores for PC1, and grouped 31 of 44 Boston homes (and 2 misclassified Cincinnati homes) into a second cluster with negative scores for PC1, resulting in a partition accuracy of 80.5% (Table 2; Figure 1E). When differences in sample volumes between Boston and Cincinnati were masked by using the median volume of air across sites to calculate reporting limits, rather than a sample-specific value, k-means analysis produced an identical partition to the analysis using original reporting limits (Table 2; Figure 1F). The chemicals with the highest loadings on PC1 in both analyses include fragrance chemicals (musk ketone, musk xylene, tonalide, galaxolide) and flame retardants (TDCIPP, BDE 47, BDE 100, BDE99, BDE 28) (Table S3). Cincinnati homes had positive scores for PC1, reflecting higher levels of these chemicals.
These results show that region of residence of a particular study participant can be inferred with substantial confidence from their cluster membership. Thus, the ability to partition data sets using differences in chemical levels can narrow the pool of potential matches for re-ID for each participant.
Discussion
This investigation considered two types of vulnerabilities that increase risk of re-ID in EH data: a) study variables that overlap with public or commercial data sets and b) environmental measurements that vary by subgroup, such as location. Our results show that EH studies collect several types of data that could facilitate re-ID, suggesting that public sharing of study data will often create privacy threats to study participants. Researchers must consider vulnerabilities in their data when deciding which data to share, with whom, and with which restrictions. Additional legal protections are also needed, parallel to protections for genetic data.
Linking EH Data in Practice
We found that information collected in EH studies (Table 1) extensively overlaps with publicly available data sets that could result in re-ID using linkage strategies. For example, occupational data may be linked to lists of licensed professionals, housing characteristics may be linked to tax and real estate data, genetics data are stored on ancestry websites, and medical information may be linked to hospital release data or news stories about accidents, illnesses, or deaths. Studies limited to well-defined geographic areas constrain the population of prospective matches, especially in combination with other data fields (e.g., registered pesticide applicators in Iowa). The data types we identified in environmental studies also are commonly collected in other types of cohort studies, posing similar risks.
As noted earlier, we do not explicitly discuss demographic information (e.g., age, gender, race/ethnicity); however, these data are nearly always collected in EH studies and are known to be vulnerable in linkage-based re-ID strategies. We also did not consider mobility traces (i.e., detailed location information) in our analysis. At the time the 12 EH studies were identified, personal sensors and mobile health technologies were not as widely and cheaply available. Recent EH studies, however, are precisely tracking individual location, for example, to infer colocated environmental data such as weather, traffic, and air quality (Habre et al. 2018). Mobility traces are extremely vulnerable to re-ID (de Montjoye et al. 2013; Douriez et al. 2016; Siddle 2014). Finally, we did not consider linkage to data sets that are not intended to be public but enter the public domain (such as through unintentional or intentional leaks); data sets obtained through malicious or illegal activities (such as hacking); or privately held data sets accessible only to the holder (such as data held by a health insurer or bank) (Culnane et al. 2017). Linkage with privately held data could be used by the holder to inform decisions without necessitating disclosure.
The fact that environmental data sets include vulnerable fields does not necessarily mean that re-ID will occur, however. One factor in real-world vulnerability is ease of access to data with the potential for linkage. Data available online have the greatest ease of access, and many public government records have migrated online in recent years. However, for older studies, or in less well-resourced municipalities, records of interest for linkage may not be available electronically, and attackers may be less likely to seek out original print records housed in government offices. Another consideration is for what years the data of interest are available. Some data may exist only historically, whereas other data may have been available at the time the study was conducted but were not archived. For example, in our previous re-ID experiment using HES data, historical tax data were available for purchase in Bolinas, California, but these data did not include detailed housing characteristics. Rather, current housing characteristics were mined from the Marin County (California) Tax Assessor’s office website up to 10 y after the study data were collected. This factor likely contributed to not obtaining any correct re-IDs in Bolinas (Sweeney et al. 2017). Because the availability of property records on Cape Cod created a similar situation, we predicted that a re-ID with the Massachusetts HES data would not provide meaningful results. Other types of data may have started to be collected only in recent years. For instance, as services have migrated to the internet, data sets containing personal data have proliferated. In addition, even when data are available, completeness and accuracy of the data will affect their ultimate utility. However, even incomplete data can create risk, and completeness and quality of data are likely to increase over time. Cost of the data is a final potential barrier. Although cost—especially of data obtained through commercial brokers—could deter an individual attacker, better-resourced entities are unlikely to be deterred, and costs may be offset by financial or other gains, such as identifying individuals who would be costly to insure or learning information relevant to a legal case. All three of these factors (cost, availability, and ease of access) can change rapidly as old files are digitized, electronic data storage becomes cheaper, and regulations about personal data evolve. Therefore, linkage strategies that are not currently possible may become possible in the future, for example, as new data become available, and current strategies could expire as data are lost to the public domain. A final consideration is the availability of EH research data. As investigators face mounting pressure from funding agencies and regulators to release data (EPA 2018; Holdren 2013), EH data could become increasingly accessible.
Inferring Group Membership Using Environmental Measurements
Even when EH data sets do not contain certain fields explicitly, they can often be inferred and therefore help triangulate re-ID approaches. In NHANES, geographic location is not specified in the data set, but the counties sampled in 2-y time periods can be identified using multiple strategies—for example, by locating news articles that announce NHANES’s presence in a community (e.g., Bawab 2018; Kowalick 2018) or by examining search engine data for locations where searches for “NHANES” or related terms have experienced spikes. We illustrated in the experiment described here that another approach could use environmental chemical measurements to facilitate re-ID by uncovering latent variables (perhaps intentionally removed to protect privacy). We found that unsupervised clustering of chemical exposure measurements alone successfully discriminated geographic location in five out of six test cases (with accuracy rates ranging from 80% to 98%). Although this study was limited to inferring geographic region, the same approach could be used with other variables. For example, behavioral correlates of gender and race/ethnicity (e.g., frequency of use of makeup or fragrance) can influence exposure levels, as can biological sex (e.g., sex differences in elimination rates contributes to sex differences in levels of per- and polyfluoroalkyl substances).
In this experiment we were able to use the true identity of the homes to assign cluster identity, but an attacker would not have access to the true identities. However, an attacker could use the cluster means, or loadings from a principal component analysis, to infer the identity of the cluster. For example, after clustering the HES, we observed one cluster with higher organochlorine pesticide levels than the other. An attacker could easily look to the published findings from the study to find this result. In this case, Rudel et al. (2010) note that “Indoor air concentrations in Cape Cod were higher than those in this California study for banned organochlorine pesticides (but not contemporary pesticides) and PCBs, and for the commercial chemicals nonylphenol and o-phenylphenol.” In addition, both papers describing the HES include a supplement with summary statistics of all chemicals measured in the study, so an attacker can learn that the 95th percentile of heptachlor in indoor air in the Northern California HES was (Rudel et al. 2010), whereas the 90th percentile in the Cape Cod HES was (Rudel et al. 2003). If these published data were not available, an attacker might logically infer that Cape Cod’s older housing and humid climate would be associated with higher residues of chlordane used for termites and that additional pesticides would be found from the historical proximity to agriculture, protecting trees from gypsy moths, and manicured greenery for the tourist economy. In contrast, a primary difference observed in the GHS data—higher fragrance levels in Cincinnati, Ohio, than in Boston, Massachusetts—would be difficult to infer without the published study findings. It is possible that an attacker would be more likely to attribute the differences to another subgroup categorization, such as ethnicity, rather than geography. The accuracy of the partition is also lower, likely because fragrance levels reflect current rather than historical use, and fragrance use has a strong personal component in addition to reflecting larger cultural trends.
A limitation of this experiment is that we used a relatively simple clustering algorithm, k-means, that is easy to implement. Other more sophisticated unsupervised methods, such as using the Expectation-Maximization algorithm to estimate Gaussian mixture models, could produce even more accurate clusters than those observed here or be used to assess the optimal number of clusters without having an a priori prediction. The failed test case (HES dust data with censored reporting limits) demonstrates that artifacts in the data set—in this case, laboratory reporting limits—can contribute to identifiability, but also that researchers can use data-masking approaches to reduce identifiability while retaining sufficient data for some analyses. In the HES, the differing reporting limits between sites resulted from advances in laboratory analytical capabilities in the years between when the data were collected. In other data sets, differences could result from samples being analyzed by different laboratories—for example, as might occur in a large consortium. By censoring the data to the same reporting limits at both sites, we masked this simple discriminator. However, we also lost some information about low levels of these chemicals at one site, which could slightly reduce the utility to other researchers using the shared data set. A final limitation is that this technique is likely only sensitive enough to reveal high-level group membership (e.g., sex, location, disease status), variables that may most often be critical to the utility of the data set and therefore unlikely to be masked in the first place.
To establish the generalizability—and limits—of using k-means or other clustering approaches for inferring subgroups from environmental data, future research should test techniques on data sets for which the true group memberships are not initially known to the study team, and on larger data sets that have more participants, contributing sites, or both. Results will help to evaluate the likely magnitude of risk associated with chemical measurement data alone in different types of studies. The k-means technique relies on every site having a unique multidimensional “exposure signature” with minimal overlap with other sites. Therefore, the combination of intrasite and intersite variation will determine cluster accuracy (i.e., subgroup identifiability). We expect that larger numbers of participants per site would decrease identifiability only if the additional participants increase intrasite variation. Similarly, we expect that additional sites would decrease identifiability only if the new sites decrease intersite variation. Greater numbers of measured analytes may increase identifiability insofar as the analyte set is more likely to contain one or more distinguishing chemicals that increase intersite variation. In this study, for example, the HES Dust censored analysis had reduced identifiability because seven chemicals were excluded that no longer met minimum detection requirements. In addition, clustering techniques should be evaluated for efficacy at uncovering latent variables other than geographic location in isolation (i.e., distinguishing sex in a single geographic location) and in combination with location. Empirical research in a variety of studies will build knowledge about factors associated with higher or lower subgroup identifiability. This knowledge will inform decisions about whether to share data, and it can also spur subsequent research to evaluate and optimize data-masking strategies that reduce identifiability while retaining the utility of the data for other analyses. Results will help researchers understand risks and consider solutions when sharing exposure measurements.
Technical and Policy Solutions
To evaluate data sharing plans, researchers and IRBs would benefit from empirically based methods to quantitatively evaluate re-ID potential and inform solutions. For example, the Privacert computational model (developed by Privacert, Inc., owned by coauthor L. Sweeney) predicts re-ID risk in relation to HIPAA standards, but like HIPAA itself, the model does not provide a quantitative estimate of risk (Privacert n.d.; Sweeney 2011). New models could be developed for other types of data and measures of privacy. Other benchmarks for privacy are k-anonymity (Sweeney 2002) and unicity (de Montjoye et al. 2013, 2015). K-anonymity requires that every combination of fields that could be used for linkage occurs at least k times, such that linking on these fields never reveals a set of individuals smaller than k. However, it is not clear what value of k is acceptable, because small groups can be equally harmed by re-ID. In a similar vein, unicity measures the proportion of a data set that can be uniquely re-identified given some number of outside pieces of information. Higher values of k and lower values of unicity (i.e., less uniquely identifiable data) can be achieved by redacting fields or coarsening the data into larger categories. For example, we used this method in the previous HES re-ID study by aggregating year a house was built into decade or longer groups, rather than exact year (Sweeney et al. 2017). However, decisions of how to redact and coarsen data often are not guided by a formal definition of privacy, in part because producing an optimal k-anonymous data set (one that minimizes data loss) is unlikely to be achievable with the technologies and computational methods available in the present and near future (Meyerson and Williams 2004). In addition, coarsening or suppressing data may do little to lessen identifiability (de Montjoye et al. 2013, 2015) and can completely negate the utility of the data (Aggarwal 2005; Brickell and Shmatikov 2008). More quantitative research and translational guidance is needed before researchers rely on redaction or coarsening to protect their data sets. However, as the data landscape constantly expands, new “big data” analytical techniques are developed, and computing power increases, researchers who wish to protect their data using technical solutions face an unending arms race against those parties seeking to re-identify data.
In comparison with these technical approaches, the more protective strategy is to restrict access to potentially vulnerable data sets and enact public policies or laws that protect study participants from harm due to privacy loss. Restricted access refers to any access limitation beyond open, publicly available data. Some forms of restricted access include sharing identifiable data under IRB oversight, typically with other researchers; restricted-access data centers (e.g., similar to that used by NHANES to give researchers limited access to more sensitive data fields); and legally binding data-use agreements that establish allowed uses of the data and penalties for misuse. Although NHANES data are subject to the general data-use agreement for public-use files from the National Center for Health Statistics (NCHS), which has the force of law and expressly prohibits re-ID, the agreement does not state penalties for violations, nor is it readily accessible from the NHANES website at the time of this writing (CDC 2015a). Rather, the agreement is housed in the data access section of the NCHS website. A more proactive strategy would be to require users to actively accept the data-use agreement (perhaps revised to include penalties) at the time when NHANES or other NCHS data are downloaded. Data access can also be mediated by a fixed query interface (i.e., raw data are not available). Simple fixed query systems are vulnerable to attack when results of repeated queries are combined, but a query system that uses differentially private algorithms would be less vulnerable (NASEM 2017). Differentially private algorithms, which rely on noise, provide a formal guarantee that no individual private data can be inferred from the output of a statistical query. Differentially private algorithms are promising but not ready for practical implementation, because more work is needed to understand the relationships between amount of noise introduced, accuracy of the algorithm, complexity of the statistical task, and size of the data set. The All of Us Study is confronting these issues with a two-tiered system of data access to an online research hub. The public tier contains only anonymized, aggregate data. To access the registered tier, which includes more robust data, researchers will have to register, complete research ethics training, and sign a data-use agreement. All activity on the research hub will be tracked (NIH n.d.-a, n.d.-b).
Federal policies that govern medical records research, however imperfect, can stimulate the development of parallel policies for EH. The HIPAA Privacy Rule sets limits on the use and disclosure of personal medical information (DHHS 2002), and the Genetic Information Nondiscrimination Act of 2008 protects people from insurance and employment discrimination based on genetic data (United States Congress 2008). Although a prescriptive approach to privacy protection like HIPAA Safe Harbor would be ineffective for EH data (as evidenced by Sweeney et al. 2017), legal recognition of the sensitivity of EH information is needed.
Privacy-protective policies for data sharing are necessary to fulfill researchers’ preexisting pledges of confidentiality to study participants in the informed consent. Researchers who violate these pledges, or who do not offer privacy protections in future studies, will risk suppressing research participation among people who fear loss of privacy. Some people are comfortable with open data sharing (Zarate et al. 2016), and others may be willing to accept low to moderate privacy risks for the benefit of public health. However, requiring consent for permissive data sharing could negatively affect participation of racial and ethnic minorities, populations that are already underrepresented in health research (Konkel 2015), and overburdened by diseases with environmental triggers, such as asthma (Forno and Celedón 2012). In multiple studies, African Americans have shown significantly less acceptance of broad consent than white participants (Ewing et al. 2015; Platt et al. 2014; Sanderson et al. 2017). In addition, environmental justice communities are also less able to cope with the economic harms of privacy loss. The new and growing “data justice” movement has pointed to many abuses of data for surveillance and discrimination and has called for combatting such abuses and using data for rights, justice, and fairness (Taylor 2017). Our research reflects such concerns.
Our survey of prominent EH studies and case study of clustering of environmental measurements illuminate features of EH studies that increase vulnerability to re-ID. Researchers and institutions face large knowledge gaps about the nature and magnitude of these risks. In addition, they lack technical and policy guidance, and legal protections have not been developed for potential harms from release of EH data. Our work represents a beginning effort to stimulate further consideration of a fast-moving landscape of increasing vulnerability to re-ID as more data becomes accessible online. Empirical assessments of privacy risks in EH data can contribute to decisions about when and how to share data, accurate descriptions of risk in informed consent documents, and discussions about whether new legal protections are needed to shield study participants from harm. During this time when privacy risks and solutions remain substantially underinvestigated, researchers and agencies should be cautious about sharing EH study data outside IRB protections or other explicit privacy agreements.
Supplementary Material
Acknowledgments
The authors thank O. Zarate for contributions to the analysis of vulnerable data in EH studies, including the first draft of Table 1, and R. Dodson for access to selected environmental chemicals data from the GHS. This work was supported by the National Institute of Environmental Health Sciences (R01ES021726).
Footnotes
Supplemental Material is available online (https://doi.org/10.1289/EHP4817).
L. Sweeney owns Privacert, a for-profit company that issues determinations about whether data are sufficiently deidentified in accordance with specific legal and regulatory requirements. The other authors declare they have no actual or potential competing financial interests.
Note to readers with disabilities: EHP strives to ensure that all journal content is accessible to all readers. However, some figures and Supplemental Material published in EHP articles may not conform to 508 standards due to the complexity of the information being presented. If you need assistance accessing journal content, please contact ehponline@niehs.nih.gov. Our staff will work with you to assess and meet your accessibility needs within 3 working days.
References
- AAAS (American Association for the Advancement of Science). N.d Science Journals: editorial policies. https://www.sciencemag.org/authors/science-journals-editorial-policies [accessed 3 June 2019].
- Adamkiewicz G, Dodson R, Zota A, Perovich L, Brody J, Rudel R, et al. 2011. Semi-volatile organic compounds distributions in residential dust samples from 5 US communities: key lessons for improving residential exposure assessment. Epidemiology 22(1):S160–S161, 10.1097/01.ede.0000392166.33641.22. [DOI] [Google Scholar]
- Aggarwal CC. 2005. On k-anonymity and the curse of dimensionality. In: Proceedings of the 31st International Conference on Very Large Data Bases. 30 August–2 September 2005, Trondheim, Norway. Trondheim: VLDB Endowment, 901–909. [Google Scholar]
- Alavanja MC, Sandler DP, McMaster SB, Zahm SH, McDonnell CJ, Lynch CF, et al. 1996. The Agricultural Health Study. Environ Health Perspect 104(4):362–369, PMID: 8732939, 10.1289/ehp.96104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba A, Cook DM, McGarity TO, Bero LA. 2005. Legislating “Sound Science”: the role of the tobacco industry. Am J Public Health 95(S1):S20–S27, PMID: 16030333, 10.2105/AJPH.2004.050963. [DOI] [PubMed] [Google Scholar]
- Bawab N. 2018. CDC is conducting a national health survey in Dallas. Dallas Observer, 2 November 2018. https://www.dallasobserver.com/news/the-cdc-is-conducting-a-national-health-survey-in-dallas-11320061 [accessed 21 November 2018]. [Google Scholar]
- Bernstein L, Allen M, Anton-Culver H, Deapen D, Horn-Ross PL, Peel D, et al. 2002. High breast cancer incidence rates among California teachers: results from the California Teachers Study (United States). Cancer Causes Control 13(7):625–635, PMID: 12296510, 10.1023/a:1019552126105. [DOI] [PubMed] [Google Scholar]
- Biro FM, Galvez MP, Greenspan LC, Succop PA, Vangeepuram N, Pinney SM, et al. 2010. Pubertal assessment method and baseline characteristics in a mixed longitudinal study of girls. Pediatrics 126(3):e583, PMID: 20696727, 10.1542/peds.2009-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickell J, Shmatikov V. 2008. The cost of privacy: destruction of data-mining utility in anonymized data publishing. In: Proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, August 2008, Las Vegas, NV. New York, NY: ACM, 70–78. [Google Scholar]
- Brody JG, Morello-Frosch R, Zota A, Brown P, Pérez C, Rudel RA. 2009. Linking exposure assessment science with policy objectives for environmental justice and breast cancer advocacy: the Northern California Household Exposure Study. Am J Public Health 99 (suppl 3):S600–609, PMID: 19890164, 10.2105/AJPH.2008.149088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann E, Böhm K, Burghardt T, Kessler S. 2013. Re-identification of smart meter data. Pers Ubiquit Comput 17(4):653–662, 10.1007/s00779-012-0513-6. [DOI] [Google Scholar]
- California Teachers Study. n.d California Teachers Study: for researchers. https://www.calteachersstudy.org/for-researchers [accessed 18 November 2019].
- CDC/NCHS (Centers for Disease Control and Prevention/National Center for Health Statistics). 2015a. Data User Agreement. https://www.cdc.gov/nchs/data_access/restrictions.htm [accessed 2 July 2019].
- CDC/NCHS. 2015b. On Site at an NCHS RDC. https://www.cdc.gov/rdc/b2accessmod/acs210.htm [accessed 30 July 2018].
- CDC/NCHS. 2018. Public-Use Data Files and Documentation. https://www.cdc.gov/nchs/data_access/ftp_data.htm [accessed 18 November 2019].
- Cohen-Hubal EA. 2009. ExpoCast: exposure science for prioritization and toxicity testing. In: International Society of Exposure Science, Annual Meeting Computational Toxicology Board of Scientific Counselors Review, Research Triangle Park, NC, 29–30 September 2009. Minneapolis, MN. [Google Scholar]
- Coombs KC, Chew GL, Schaffer C, Ryan PH, Brokamp C, Grinshpun SA, et al. 2016. Indoor air quality in green-renovated vs. non-green low-income homes of children living in a temperate region of US (Ohio). Sci Total Environ 554-555:178–185, PMID: 26950631, 10.1016/j.scitotenv.2016.02.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culnane C, Rubinstein BI, Teague V. 2017. Health data in an open world. arXiv: 1712:05627. [Google Scholar]
- de Montjoye YA, Hidalgo CA, Verleysen M, Blondel VD. 2013. Unique in the crowd: the privacy bounds of human mobility. Sci Rep 3:1376, PMID: 23524645, 10.1038/srep01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Montjoye YA, Radaelli L, Singh VK, Pentland AS. 2015. Unique in the shopping mall: on the reidentifiability of credit card metadata. Science 347(6221):536–539, PMID: 25635097, 10.1126/science.1256297. [DOI] [PubMed] [Google Scholar]
- DHHS (Department of Health and Human Services). 2002. Standards for Privacy of Individually Identifiable Health Information; Final Rule. Fed Reg 67(157):53181–53273, PMID: 12180470. [PubMed] [Google Scholar]
- Dodson RE, Udesky JO, Colton MD, McCauley M, Camann DE, Yau AY, et al. 2017. Chemical exposures in recently renovated low-income housing: Influence of building materials and occupant activities. Environ Int 109:114–127, PMID: 28916131, 10.1016/j.envint.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Douriez M, Doraiswamy H, Freire J, Silva CT. 2016. Anonymizing NYC taxi data: does it matter? In: Proceedings of the 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA) October 2016 Montreal, Canada: IEEE, 140–148. [Google Scholar]
- El Emam K, Jonker E, Arbuckle L, Malin B. 2011. A systematic review of re-identification attacks on health data. PLoS One 6(12):e28071, PMID: 22164229, 10.1371/journal.pone.0028071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA (U.S. Environmental Protection Agency). 2018. Strengthening transparency in regulatory science. Fed Reg 83:18768–18774. [Google Scholar]
- Erlich Y, Shor T, Pe’er I, Carmi S. 2018. Identity inference of genomic data using long-range familial searches. Science 362(6415):690–694, PMID: 30309907, 10.1126/science.aau4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskenazi B, Bradman A, Gladstone EA, Jaramillo S, Birch K, Holland N. 2003. CHAMACOS, a longitudinal birth cohort study: lessons from the fields. J Children’s Health 1(1):3–27, 10.3109/713610244. [DOI] [Google Scholar]
- European Commission. 2017. Guidelines to the Rules on Open Access to Scientific Publications and Open Access to Research Data in Horizon 2020. Version 3.2. Brussels, Belgium: European Commission Directorate-General for Research & Innovation. [Google Scholar]
- European Commission. 2018. Commission Recommendation of 25.4.2018 on access to and preservation of scientific information. 2375 final Brussels, Belgium: European Commission. [Google Scholar]
- European Commission. N.d European Open Science Cloud (EOSC). https://ec.europa.eu/research/openscience/index.cfm?pg=open-science-cloud [accessed 2 July 2019].
- Ewing AT, Erby LA, Bollinger J, Tetteyfio E, Ricks-Santi LJ, Kaufman D. 2015. Demographic differences in willingness to provide broad and narrow consent for biobank research. Biopreserv Biobank 13(2):98–106, PMID: 25825819, 10.1089/bio.2014.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer EA. 2013. Public Access to Data from Federally Funded Research: Provisions in OMB Circular A-110. Washington, DC: Congressional Research Service, Library of Congress. [Google Scholar]
- Forno E, Celedón JC. 2012. Health disparities in asthma. Am J Respir Crit Care Med 185(10):1033–1035, PMID: 22589306, 10.1164/rccm.201202-0350ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman LB, Blair A, Hofmann J, Sandler DP, Parks CG, Thomas K. 2017. Agricultural Health Study Policy 2-4: Guidelines for Collaboration. https://aghealth.nih.gov/collaboration/AHS%20Policy%202-4%20Guidelines%20for%20Collaboration_2017.1.pdf [accessed 18 November 2019].
- Goho SA. 2016. The legal implications of report back in household exposure studies. Environ Health Perspect 124(11):1662–1670, PMID: 27153111, 10.1289/EHP187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JE, Loftus CT, Zu K. 2017. 2,4-Dichlorophenoxyacetic acid and non-Hodgkin’s lymphoma: results from the Agricultural Health Study and an updated meta-analysis. Ann Epidemiol 27(4):290–292, PMID: 28292638, 10.1016/j.annepidem.2017.01.008. [DOI] [PubMed] [Google Scholar]
- Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. 2013. Identifying personal genomes by surname inference. Science 339(6117):321–324, PMID: 23329047, 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
- Habre R, Rocchio R, Hosseini A, van Vliet E, Eckel S, Valencia L. 2018. An mHealth platform for predicting risk of pediatric asthma exacerbation using personal sensor monitoring systems: The Los Angeles Prisms Center. In: Proceedings of the ISEE Conference Abstracts 2018. [Google Scholar]
- Hartigan JA, Wong MA. 1979. Algorithm AS 136: a k-means clustering algorithm. J Royal Stat Soc Series C (Appl Stat) 28(1):100–108, 10.2307/2346830. [DOI] [Google Scholar]
- Health Effects Institute. N. D Databases. https://www.healtheffects.org/research/databases [accessed 18 November 2019].
- Henriksen-Bulmer J, Jeary S. 2016. Re-identification attacks—A systematic literature review. Int J Inform Manage 36(6, part B):1184–1192, 10.1016/j.ijinfomgt.2016.08.002. [DOI] [Google Scholar]
- Holdren JP. 2013. Increasing Access to the Results of Federally Funded Scientific Research. Washington, D.C: Office of Science and Technology Policy; https://obamawhitehouse.archives.gov/sites/default/files/microsites/ostp/ostp_public_access_memo_2013.pdf [accessed 13 May 2019]. [Google Scholar]
- Hooley S, Sweeney L. 2013. Survey of publicly available state health databases. arXiv 1306:2564. [Google Scholar]
- Hubert L, Arabie P. 1985. Comparing partitions. J Classification 2(1):193–218, 10.1007/BF01908075. [DOI] [Google Scholar]
- Justin J. 2018. To find alleged Golden State Killer, investigators first found his great-great-great-grandparents. Washington Post Washington, D.C. 30 April 2018. https://www.washingtonpost.com/local/public-safety/to-find-alleged-golden-state-killer-investigators-first-found-his-great-great-great-grandparents/2018/04/30/3c865fe7-dfcc-4a0e-b6b2-0bec548d501f_story.html [accessed 31 October 2018].
- Konkel L. 2015. Racial and ethnic disparities in research studies: the challenge of creating more diverse cohorts. Environ Health Perspect 123(12):A297–302, PMID: 26625444, 10.1289/ehp.123-A297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalick C. 2018. Wichita County selected to participate in national health survey. Times Record News Wichita Falls, KS. 8 September 2018. https://www.timesrecordnews.com/story/news/local/2018/09/08/wichita-county-selected-participate-national-health-survey/1212460002/ [accessed 11 September 2018].
- Kwok RK, Engel LS, Miller AK, Blair A, Curry MD, Jackson WB, et al. 2017. The GuLF STUDY: a prospective study of persons involved in the Deepwater Horizon oil spill response and clean-up. Environ Health Perspect 125(4):570–578, PMID: 28362265, 10.1289/EHP715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson A, Williams R. 2004. On the complexity of optimal K-anonymity. In: Proceedings of the Twenty-third ACM SIGMOD-SIGACT-SIGART Symposium on Principles of Database Systems June 2004, Paris, France: 23:223–228, 10.1145/1055558.1055591. [DOI] [Google Scholar]
- Narayanan A, Shmatikov V. 2008. Robust de-anonymization of large sparse datasets. In: Proceedings of the 2008 IEEE Symposium on Security and Privacy, 2008, Oakland, CA: 111–125, 10.1109/SP.2008.33. [DOI] [Google Scholar]
- NASEM (National Academies of Sciences Engineering and Medicine). 2017. Federal Statistics, Multiple Data Sources, and Privacy Protection: Next Steps. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- NCCT (United States Environmental Protection Agency National Center for Computational Toxicology). 2017. Exposure Forecaster. https://catalog.data.gov/dataset/exposure-forecaster-a16a1 [accessed 10 July 2019].
- New York State Department of Health. 2019. Hospital Inpatient Discharges (SPARCS De-Identified): 2017. https://health.data.ny.gov/dataset/Hospital-Inpatient-Discharges-SPARCS-De-Identified/22g3-z7e7 [accessed 21 November 2019].
- NIH (National Institutes of Health). 2017. About ECHO. https://www.nih.gov/echo/about-echo [accessed 26 July 2018].
- NIH. N. D.-a Privacy Safeguards. https://www.joinallofus.org/en/privacy-safeguards [accessed 21 November 2018].
- NIH. N. D.-B Workbench. https://www.researchallofus.org/data/workbench/ [accessed 5 June 2019].
- NIH. N. D.-C Program Overview–All of Us. https://allofus.nih.gov/about/about-all-us-research-program [accessed 1 November].
- NIH. N. D.-D GuLF STUDY: For Researchers. https://gulfstudy.nih.gov/en/forresearchers.html [accessed 18 November 2019].
- OMB (Office of Management and Budget). 1999. OMB Circular A-110, “Uniform Administrative Requirements for Grants and Agreements with Institutions of Higher Education, Hospitals, and Other Non-Profit Organizations.” Fed Reg 64(195):54926–54930. [Google Scholar]
- Pennarola F, Pistilli L, Chau M. 2017. Angels and daemons: is more knowledge better than less privacy? an empirical study on a k-anonymized openly available dataset. 38th International Conference on Information Systems ICIS 2017 Proceedings. December 2017 Seoul, South Korea: 1318. [Google Scholar]
- Platt J, Bollinger J, Dvoskin R, Kardia SL, Kaufman D. 2014. Public preferences regarding informed consent models for participation in population-based genomic research. Genet Med 16(1):11–18, PMID: 23660530, 10.1038/gim.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privacert. n.d HIPAA Solutions. http://privacert.com/hipaa/index.html [accessed 1 November 2018].
- Ramirez E, Brill J, Ohlhausen MK, Wright JD, McSweeny T. 2014. Data brokers: a call for transparency and accountability. Washington, DC: Federal Trade Commission; https://www.ftc.gov/reports/data-brokers-call-transparency-accountability-report-federal-trade-commission-may-2014 [accessed 18 November 2019]. [Google Scholar]
- Rocher L, Hendrickx JM, de Montjoye YA. 2019. Estimating the success of re-identifications in incomplete datasets using generative models. Nat Commun 10(1):3069, PMID: 31337762, 10.1038/s41467-019-10933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel RA, Camann DE, Spengler JD, Korn LR, Brody JG. 2003. Phthalates, alkylphenols, pesticides, polybrominated diphenyl ethers, and other endocrine-disrupting compounds in indoor air and dust. Environ Sci Technol 37(20):4543–4553, PMID: 14594359, 10.1021/es0264596. [DOI] [PubMed] [Google Scholar]
- Rudel RA, Dodson RE, Perovich LJ, Morello-Frosch R, Camann DE, Zuniga MM, et al. 2010. Semivolatile endocrine-disrupting compounds in paired indoor and outdoor air in two northern California communities. Environ Sci Technol 44(17):6583–6590, PMID: 20681565, 10.1021/es100159c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson SC, Brothers KB, Mercaldo ND, Clayton EW, Antommaria AHM, Aufox SA, et al. 2017. Public attitudes toward consent and data sharing in biobank research: a large multi-site experimental survey in the US. Am J Hum Genet 100(3):414–427, PMID: 28190457, 10.1016/j.ajhg.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler DP, Hodgson ME, Deming-Halverson SL, Juras PS, D’Aloisio AA, Suarez LM, et al. 2017. The Sister Study Cohort: baseline methods and participant characteristics. Environ Health Perspect 125(12):127003, PMID: 29373861, 10.1289/EHP1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J. 2018. Transparency” as mask? The EPA’s proposed rule on scientific data. N Engl J Med 379(16):1496–1497, PMID: 30156992, 10.1056/NEJMp1807751. [DOI] [PubMed] [Google Scholar]
- Science and Technology at the Environmental Protection Agency. 2019. Hearing Before the Committee on Science, Space, and Technology, U.S. House of Representatives, 116th Congress (September 19, 2019) (testimony of Andrew R. Wheeler, Administrator U.S. Environmental Protection Agency). https://science.house.gov/imo/media/doc/9.19.19%20Wheeler%20Testimony.pdf [accessed 20 November 2019].
- Siddle J. 2014. I know where you were last summer: London’s public bike data is telling everyone where you’ve been. https://vartree.blogspot.com/2014/04/i-know-where-you-were-last-summer.html [accessed 22 August 2019].
- Sister Study. N.d The Sister Study data sharing policy. https://www.sisterstudystars.org/Public/Sister/Documents/Data%20Access%20Policies%20and%20Procedures.pdf [accessed 18 November 2019].
- Stout DMI 2nd, Bradham KD, Egeghy PP, Jones PA, Croghan CW, Ashley PA, et al. 2009. American Healthy Homes Survey: a national study of residential pesticides measured from floor wipes. Environ Sci Technol 43(12):4294–4300, PMID: 19603637, 10.1021/es8030243. [DOI] [PubMed] [Google Scholar]
- Sweeney L. 2002. k-Anonymity: a model for protecting privacy. Int J Unc Fuzz Knowl Based Syst 10(5):557–570, 10.1142/S0218488502001648. [DOI] [Google Scholar]
- Sweeney L. 2011. Patient Identifiability in Pharmaceutical Marketing Data. Data Privacy Lab Working Paper 1015. https://dataprivacylab.org/projects/identifiability/pharma1.pdf [accessed 1 November 2018].
- Sweeney L. 2013. Matching Known Patients to Health Records in Washington State Data. ID 2289850. Rochester, NY: Social Science Research Network. [Google Scholar]
- Sweeney L, Von Loewenfeldt M, Perry M. 2018. Saying it’s anonymous doesn’t make it so: re-identifications of “anonymized” law school data. Technol Sci. 2018111301, PMID: 25078429, https://techscience.org/a/2017082801 [accessed 17 January 2018].25078429
- Sweeney L, Yoo J, Perovich L, Boronow K, Brown P, Brody J. 2017. Re-identification Risks in HIPAA Safe Harbor Data: a study of data from one environmental health study. Technol Sci. 2017082801, PMID: 30687852. [PMC free article] [PubMed] [Google Scholar]
- Tanner A. 2017. Our Bodies, Our Data: How Companies Make Billions Selling Our Medical Records. Boston: Beacon Press. [Google Scholar]
- Taylor L. 2017. What is data justice? The case for connecting digital rights and freedoms globally. Big Data & Society 4(2):2053951717736335, 10.1177/2053951717736335. [DOI] [Google Scholar]
- United States Congress. 2008. The Genetic Information Nondiscrimination Act of 2008 (GINA). Pub L No. 110–233. H.R. 493 (110th Congress 28 May 2008), https://www.gpo.gov/fdsys/pkg/PLAW-110publ233/html/PLAW-110publ233.htm [accessed 1 November 2018].
- Vermont Department of Health. 2019. Vermont Uniform Hospital Discharge Data System. https://www.healthvermont.gov/health-statistics-vital-records/health-care-systems-reporting/hospital-discharge-data [accessed 21 November 2019].
- Vermont Secretary of State. 2019. Data Broker Search. https://www.vtsosonline.com/online/DataBrokerInquire/ [accessed 18 November 2019].
- Virginia Health Information. 2018. Data Directory: Health Care Decision Support Data (updated 2018-01-23). https://vhi.org/files/PDFs_to_download_from_web/Data_Directory_2017.pdf [accessed 21 November 2019].
- von Thenen N, Ayday E, Cicek AE. 2019. Re-identification of individuals in genomic data-sharing beacons via allele inference. Bioinformatics 35(3):365–371, PMID: 30052749, 10.1093/bioinformatics/bty643. [DOI] [PubMed] [Google Scholar]
- Wang R, Li YF, Wang X, Tang H, Zhou X. 2009. Learning your identity and disease from research papers: information leaks in genome wide association study. In: Proceedings of the 16th ACM Conference on Computer and Communications Security (CCS ’09) CCS 2009. 9–13 November 2009 Chicago, IL. New York, NY: ACM, 534–544, 10.1145/1653662.1653726. [DOI] [Google Scholar]
- Weisel CP, Zhang J, Turpin BJ, Morandi MT, Colome S, Stock TH, et al. 2005. Relationship of Indoor, Outdoor and Personal Air (RIOPA) study: study design, methods and quality assurance/control results. J Expo Anal Environ Epidemiol 15(2):123–137, PMID: 15213705, 10.1038/sj.jea.7500379. [DOI] [PubMed] [Google Scholar]
- Zarate OA, Brody JG, Brown P, Ramirez-Andreotta MD, Perovich L, Matz J. 2016. Balancing benefits and risks of immortal data: participants’ views of open consent in the Personal Genome Project. Hastings Cent Rep 46(1):36–45, PMID: 26678513, 10.1002/hast.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipf G, Chiappa M, Porter K, Ostchega Y, Lewis B, Dostal J. 2013. National Health and Nutrition Examination Survey: plan and operations, 1999–2010. Vital and Health Statistics 1(56): PMID: 25078429. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.