Abstract
Background:
Changes in cholesterol metabolism are common hallmarks of neurodevelopmental pathologies. A diverse array of genetic disorders of cholesterol metabolism support this claim as do multiple lines of research that demonstrate chemical inhibition of cholesterol biosynthesis compromises neurodevelopment. Recent work has revealed that a number of commonly used pharmaceuticals induce changes in cholesterol metabolism that are similar to changes induced by genetic disorders with devastating neurodevelopmental deficiencies.
Objectives:
We tested the hypothesis that common environmental toxicants may also impair cholesterol metabolism and thereby possibly contribute to neurodevelopmental toxicity.
Methods:
Using high-throughput screening with a targeted lipidomic analysis and the mouse neuroblastoma cell line, Neuro-2a, the ToxCast™ chemical library was screened for compounds that impact sterol metabolism. Validation of chemical effects was conducted by assessing cholesterol biosynthesis in human induced pluripotent stem cell (hiPSC)–derived neuroprogenitors using an isotopically labeled cholesterol precursor and by monitoring product formation with UPLC-MS/MS.
Results:
Twenty-nine compounds were identified as validated lead-hits, and four were prioritized for further study (endosulfan sulfate, tributyltin chloride, fenpropimorph, and spiroxamine). All four compounds were validated to cause hypocholesterolemia in Neuro-2a cells. The morpholine-like fungicides, fenpropimorph and spiroxamine, mirrored their Neuro-2a activity in four immortalized human cell lines and in a human neuroprogenitor model derived from hiPSCs, but endosulfan sulfate and tributyltin chloride did not.
Conclusions:
These data reveal the existence of environmental compounds that interrupt cholesterol biosynthesis and that methodologically hiPSC neuroprogenitor cells provide a particularly sensitive system to monitor the effect of small molecules on de novo cholesterol formation. https://doi.org/10.1289/EHP5053
Introduction
Human exposure studies with organic solvents, metal exposures, and air pollutants have revealed the existence of developmental neurotoxicants (Costa et al. 2014; Grandjean and Landrigan 2006, 2014). To proactively address the potential for environmental contributors to neurological disease, protocols were proposed in the late 1980s to identify and screen for neurodevelopmental toxicants (Wier et al. 1989). Yet, the difficulty of translating the identification of potential neurodevelopmental toxicants into human health risk assessments has been apparent, given the need for anthropomorphized animal behavior experiments in addition to human epidemiological studies to support the adverse outcome of this class of toxicants (Vrijheid et al. 2014). An alternative method to study neurodevelopmental toxicants is through the use of developing neural tissue—most commonly from human stem cell or induced pluripotent stem cell (hiPSC) sources (Bal-Price et al. 2010; Fritsche et al. 2018a, 2018b; Kumar et al. 2012). Further, these model systems can be used to screen for developmental neurotoxicants by prioritizing chemicals that result in metabolic phenotypes of neurological disorders (Kleinstreuer et al. 2011).
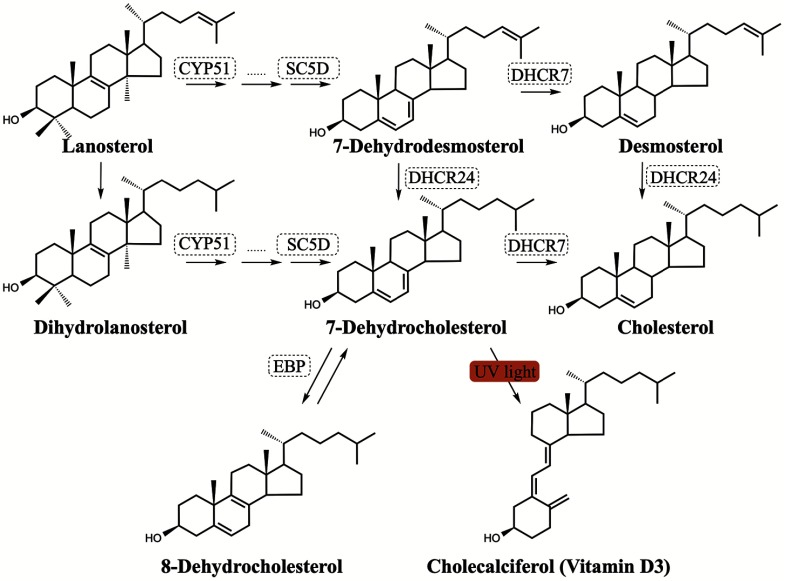
Hypocholesterolemia is a common metabolic feature of many neurodevelopmental disorders (Porter and Herman 2011) and neurodegenerative diseases, including Huntington’s disease and Alzheimer’s disease (Leoni and Caccia 2014; Martín et al. 2014; Vance 2012). Cholesterol is an indispensable lipid in the central nervous system, with more than 25% of the total cholesterol content of a human body residing in the brain (Björkhem and Meaney 2004). In addition to cholesterol serving as an important structural component for cellular membranes and myelin sheathes, a number of cholesterol metabolites, including neurosteroids, serve as critical signaling molecules (Schroepfer 2000, Zhang and Liu 2015). To maintain this indispensable pool of cholesterol, the anatomy of the central nervous system requires an active yet independent cholesterol biosynthetic pathway (Figure 1). This requirement is because plasma cholesterol from either diet or hepato-synthesis does not freely cross the blood–brain barrier (Dietschy 2009). The most convincing evidence for the necessity of cholesterol in neural biology are in rare genetic syndromic disorders intimately tied to mutations in the enzymes of cholesterol biosynthesis (Porter and Herman 2011). One such disorder is Smith-Lemli-Opitz Syndrome (SLOS). SLOS is the result of genetic defects in the enzyme 7-dehydrocholesterol reductase (DHCR7) which compromises the cell’s ability to convert 7-dehydrocholesterol (7-DHC) to cholesterol. Research has found numerous connections of SLOS with neurological deficiencies (Table 1). One of the more compelling connections between the enzyme DHCR7 and neurobehavior is the number of independent diagnoses of autism spectrum disorder (ASD) in the SLOS patient population (Sikora et al. 2006). Reports have suggested that as many as 20% of individuals on the autism spectrum have mild hypocholesterolemia likely due to defects in cholesterol metabolism (Lee and Tierney 2011; Tierney et al. 2006).
Figure 1.
Schematic of cholesterol biosynthesis. Note: Selected enzymes involved in cholesterol biosynthesis are shown in dashed boxes.
Table 1.
Smith-Lemli-Opitz syndrome (SLOS) patient characteristics and associated severity scores observed clinically [adapted from Kelley and Hennekam (2000) and Porter (2008)].
| Classification (score) | Biochemical featuresa | Physical and behavioral symptomsb,c |
|---|---|---|
| Mild () |
of normal plasma cholesterol levels 7-DHC levels |
|
| Classical (20–50) | of normal plasma cholesterol levels |
|
| Severe () |
of normal plasma cholesterol levels 7-DHC levels |
|
Biochemical values are typically graded across a spectrum of severity; shown are averaged values from multiple sources.
Craniofacial defects are characteristic of SLOS patients with anteverted nares, microcephaly, and cleft palate all common.
Dehydrocholesterol (7-DHC and 8-DHC) levels correlate with loss of cognitive function and adaptive functioning (Thurm et al. 2016) and score is correlated with increased aggression.
In addition to genetic-based evidence for the inhibition of cholesterol biosynthesis as a relevant metabolic feature for neurotoxicity, there is also chemical support for this suggestion. During the 1960s’ drug-discovery efforts to develop efficacious cholesterol-lowering pharmaceuticals, numerous lead-hit compounds failed in commercial development due to toxicity. For instance, AY-9944 was designed to inhibit DHCR7, but the compound never passed preclinical phases due to its potent teratogenicity (Dvornik and Hill 1968; Roux et al. 2000). A few decades later, AY-9944 was used to develop a rodent model of SLOS that had links to behavioral deficits found in the disorder (Jung et al. 2013; Kolf-Clauw et al. 1996). Given the history with the small molecule AY-9944, the question arises: are there environmental chemicals that affect human neural development by disrupting cholesterol metabolism?
Some environmental sterol biosynthesis disruptors have already been identified by cell culture experiments and the use of in silico predictive modeling. Namely, the widely used antiseptic benzalkonium chlorides were shown to inhibit DHCR7 (Herron et al. 2016). Another class of environmental sterol metabolism disruptors are the conazole fungicides. These agricultural pesticides are known to inhibit CYP51 by coordinating with the heme group, which halts substrate binding with a resulting increase of lanosterol (Figure 1) in cell culture (Lepesheva et al. 2008). These compounds can also disrupt normal reproductive cell function in both murine and human cell models (Roelofs et al. 2014). Indeed, screening of a library of pharmacologically active molecules identified a number of hypocholesterolemic compounds when tested in mouse neuroblastoma cells (Kim et al. 2016; Korade et al. 2016) and human fibroblasts (Korade et al. 2016); in support of this, a screen of an independent pharmacopeia-based library identified about 1 in 4 drugs as hypocholesterolemic compounds, more than 20% of which targeted DHCR7 (Wages et al. 2018). The most common drug class to treat hypercholesterolemia, statins, is contraindicated for pregnancy due to an increased risk of spontaneous pregnancy loss (McGrogan et al. 2017), yet it is controversial whether this loss is due to teratogenicity (Karalis et al. 2016). Regarding chemical-induced hypocholesterolemia, it was shown in mice that the presence of aripiprazole, a commonly prescribed antipsychotic and known DHCR7 inhibitor, in utero impaired embryonic neurodevelopment (Genaro-Mattos et al. 2019).
The knowledge that numerous pharmaceutical agents inhibit DHCR7, a gene associated with a number of fetal anomalies as well as ASD, raised the possibility of the existence of environmental xenobiotics that inhibit DHCR7. We report here the results of a study that sought to address two potential public health issues: a) to identify and characterize environmental cholesterol biosynthesis disruptors and b) to characterize whether those identified compounds have the potential to inhibit cholesterol biosynthesis in developing human neurons.
Methods
Materials
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich. HPLC grade solvents were purchased from Thermo Fisher Scientific Inc. All cell culture reagents were from Mediatech, Life Technologies, and Greiner Bio-One. All sterol standards, natural and isotopically labeled, used in this study are available from Kerafast, Inc. The CytoTox 96® NonRadioactive Cytotoxicity Assay was obtained from Promega (G1780) to assess cytotoxicity.
Immortalized Cell Lines
The neuroblastoma cell lines Neuro-2a, SK-N-SH and IMR-32 were purchased from American Type Culture Collection. The hepatocyte carcinoma cell line HepG2 and lung carcinoma cell line A549 were purchased from the European Collection of Authenticated Cell Cultures (via Sigma-Aldrich). All cell lines were maintained as recommended by distributors, subcultured every 4–5 d up to 10 passages, and culture medium was changed every 2 days. All media components were purchased from Thermo Fisher Scientific Inc., including minimum essential medium eagle with glutamine (MEM, MT100010CV), Dulbecco’s modified eagle medium with glucose and sodium pyruvate without glutamine (DMEM, MT15013CV), L-glutamine (25-030-081), penicillin-streptomycin solution (P/S, MT30002CI) and N-2 Supplement (17-502-048). Premium grade fetal bovine serum (FBS) was obtained from VWR/Seradigm (97068-085). Delipidated FBS (dFBS) was prepared by stirring of silica (S5130, Sigma-Aldrich) into of FBS for 24 h at 4°C followed by centrifugation for 1 h at at 4°C in a Thermo Scientific Sorvall RC-6 Plus superspeed centrifuge. Supernatant was collected and validated for delipidation of cholesterol using UPLC-MS/MS derivatization with 4-phenyl-1,2,4-triazoline-3,5-dione (PTAD).
Cortical Neuroprogenitors Derived from Human Induced Pluripotent Stem Cells (hiPSCs)
hiPSCs lines [(CC-3 and CD2 by Aaron Bowman’s laboratory at Vanderbilt University Medical Center, Nashville, TN) and (CX-3 by Kevin Ess’s laboratory also at Vanderbilt University Medical Center)] were reprogrammed and validated from human dermal fibroblasts that were isolated from skin biopsy of healthy human subjects after patient consent and assent under guidelines of an approved IRB protocol (Vanderbilt No. 080369, Kevin C. Ess PI) as previously reported (Tidball et al. 2016; Armstrong et al. 2017). Briefly, fibroblasts were reprogrammed into hiPSCs by electroporation with CXLE-hOCT3/4-shp53-F, pCXLE-hSK, and pCXLE-hUL (Addgene plasmid #27077, #27078, and #27079, respectively) plasmid vectors using Neon® transfection system (Life Technologies) described in Okita et al. (2011). The hiPSCs were cultured and passaged in mTeSR1 media (StemCell Technologies) on Matrigel (BD Biosciences)-coated six-well plates. hiPSCs were replated at , and differentiation was initiated when cells were 100% confluent. Cortical neural induction was performed following the dual SMAD protocol published by Chambers et al. (2009), except that LDN (Stemgent Cat. No. 04-0074) at was used instead of noggin. SB431542 () was purchased from (Stemgent Cat. No. 04-0010). Additional details and validation of this differentiation method have been previously reported (Chambers et al. 2009; Neely et al. 2012; Tidball et al. 2015). Briefly, neuralization medium consists of Knockout DMEM/F12 (Invitrogen #12660), Knockout Serum (Invitrogen #10828), glutamax (; Invitrogen #35050-061), Pen/Strep (; Mediatech #30-002-Cl), nonessential amino acids (, Invitrogen #M7145), and (Sigma #M3148) and N2 medium, which consists of DMEM/F12 (Invitrogen #10,565-018, ), D-Glucose, and N2 supplement (Thermo Fisher Scientific #17502048). Cortical neuroprogenitors were replated at on day 8 into 96-well plates for exposures that were conducted on day 10.
Chemical Screening and LC-MS Analysis
The 1,851 chemicals that comprise the ToxCast™ chemical library were obtained through Ann Richard [U.S. Environmental Protection Agency (U.S. EPA)]. The chemicals were supplied blinded in 384-well plates with one chemical per well at a target concentration of in DMSO. Chemical plates were shipped, registered, and stored in the Vanderbilt University High-Throughput Screening and Compound Management Facility. All screening experiments were conducted in the Vanderbilt University High-Throughput Screening Facility as previously described (Wages et al. 2018). Briefly, of each compound (at ) from the ToxCast™ Chemical Library were deposited into a separate well of a black 384-well cell culture plate (Grenier Bio-One) using a Labcyte Echo 550/555. Neuro-2a cells were trypsinized, counted by hemocytometer, resuspended in DMEM with reduced serum (0.5% FBS, L-glutamine, P/S), and of cell suspension was dispensed to 384-well plates (on top of predeposited compounds) using Multidrop Combi (Thermo Scientific) at a final plating density of Neuro-2a cells per well. In addition to the test compounds, four positive control compounds identified and validated from previous studies (Korade et al. 2016; Kim et al. 2016) were included on each plate to probe the cholesterol biosynthetic pathway. These four compounds included simvastatin, which inhibits HMG-CoA Reductase (lowers all sterols analyzed); econazole, which inhibits CYP51 (increases lanosterol levels); aripiprazole, which inhibits DHCR7 (increases 7-DHC); and clomiphene, which inhibits DHCR24 (increases desmosterol). After 24 h of incubation at 37°C and 5% , Hoechst dye (Molecular Probes) was added using Agilent Velocity 11 Bravo and incubated at room temperature for 30 min in the dark. Cells were imaged and counted based on nuclear staining using ImageXpress Micro XL (Molecular Devices) with a objective. For analysis of sterols, MeOH containing internal standards ( for , for , for , and for ) was added to each well after removing the medium to extract all endogenous lipids. Plates were centrifuged, and supernatant was transferred by an automated liquid handler Agilent Velocity 11 Bravo to a plate (Waters #186002631, flat bottomed) with of 4-phenyl-1,2,4-triazoline-3,5-dione (PTAD) predeposited in each well. The plates were immediately sealed with Easy Pierce Heat Sealing Foil (ThermoScientific AB-1720), followed by 20 min of agitation at room temperature and then centrifuged for 10 min using a Sorvall swing rotor. Similar sample preparation via PTAD-derivatization was described previously (Korade et al. 2016; Liu et al. 2014) for UPLC-MS/MS analysis. Samples were analyzed on an Acquity UPLC system equipped with ANSI-compliant well plate holder. The sterols ( injection) were analyzed on an UPLC C18 column (Acquity UPLC BEH C18, , ) with 100% MeOH (0.1% v/v acetic acid) mobile phase at a flow rate of and runtime of 1.2 min. A TSQ Quantum Ultra tandem mass spectrometer (ThermoFisher) was used for MS detections, and data were acquired with a Finnigan Xcalibur software package (version 2.2 SP1.48; Thermo Xcalibur). Selected reaction monitoring (SRM) of the PTAD derivatives was acquired in the positive ion mode using atmospheric pressure chemical ionization (APCI). The monitored transitions included: Chol , , 7-DHC , , Des , Lan , , and . MS parameters were optimized for the 7-DHC-PTAD adduct and were as follows: auxiliary nitrogen gas pressure at and sheath gas pressure at ; discharge current at and vaporizer temperature at 342°C. Collision-induced dissociation (CID) was optimized at under of argon. Endogenous sterol levels were quantified based on the known internal standard amount and then normalized to nuclear cell count, reported as nmol/million cells.
Lead-Hit Validation and in Vitro Dosing
Determination of lead-hit compounds from screening involved the calculation of screening window coefficients “Z” and “z-score” (Zhang et al. 1999); z-Scores are calculated from the following equation:
where is lead-hit screened sterol level, is averaged vehicle (0.1% DMSO) sterol level, and is standard deviation of vehicle sterol level (compound: , vehicle: ). Compounds with a were selected as lead-hits and effects validated in Neuro-2a cells in 96-well plates (Grenier Bio-One). Additionally, any chemicals that resulted in significant cell loss were selected as lead-hit compounds as well, specifically to be tested at lower doses: 0.01, 0.1, and . Neuro-2a cells were seeded at Neuro-2a cells/well in DMEM with 10% FBS, L-glutamine, P/S. After 24 h, media was aspirated by pipette and replaced with DMEM exposure media ( N-2 supplement, L-glutamine, P/S) containing lead-hit compound (0.05% DMSO) for 24 h. Neuro-2a cells were then prepped for the targeted lipidomic mass spectrometric analysis as with 384-well plates, but to each well in the 96-well plate was added the internal standards ( of stock solution in MeOH: , , for , for ) and MeOH (). All verified lead-hits were then exposed to Neuro-2a cells at 4 doses for 24 h in DMEM exposure media: 0.1, 0.4, 1, and .
Using selection criteria (Table S1 and Figure S1) remaining lead-hits were tested in four human immortalized cell lines. All cell lines were cultured and maintained in MEM with 10% FBS, P/S. For dosing experiments, cells were seeded into 96-well plates ( SK-N-SH cells/well, IMR-32 cells/well, A549 cells/well, HepG2 cells/well). After 24 h, media was aspirated by pipette and replaced with MEM exposure media (10% dFBS, P/S) containing compound at three doses for 24 h: 0.01, 0.1, and (0.05% DMSO). Each dosing experiment was conducted so that no two cell lines were cultured onto the same 96-well plate. After exposure, cells were then prepped for mass spectrometric analysis as with 384-well plates, but to each well of the black 96-well plates (Grenier Bio-One) was added the internal standards ( of stock solution in MeOH: , , , , for ), and MeOH (). Updated monitored transitions for UPLC-MS/MS included Chol , , 7-DHC , , 8-DHC , , Des , Lan , , and . The baseline sterol profile for each cell line was determined by assaying cells treated only with media in parallel to exposed cells.
For hiPSC experiments, neuroprogenitor cells were exposed to lead-hit compounds (fenpropimorph, spiroxamine) and positive control compound (haloperidol) at day 10 of differentiation. The hiPSC neuroprogenitor cells were exposed to chemicals at two doses (0.01 and , 0.05% DMSO) for four different durations of time (4, 8, 12, and 24 h) with and without to measure de novo cholesterol synthesis as previously reported (Korade et al. 2017a). Briefly, was added to the media (defined in section above) at 0.01% ethanol in order to trace the synthesis of to to using the PTAD-UPLC-MS/MS analysis method. To evaluate de novo cholesterol synthesis a residual cholesterol synthesis (RCS) was calculated as described previously (Genaro-Mattos et al. 2018; Honda et al. 1995; Wassif et al. 2005) by the following equation: []. After exposure of compound in the presence of , hiPSC neuroprogenitors were then prepped for mass spectrometric analysis as with 384-well plates, but to each well in the 96-well plate was added the internal standards ( of stock solution in MeOH: , , ), and MeOH (). Updated monitored transitions for UPLC-MS/MS included Chol , , 7-DHC , , 8-DHC , , Des , Lan , , , , , and . Traceable isotopically labeled sterol levels were quantified based on the known internal standard amount or determined response factor to for and . Values were then normalized to nuclear cell count, reported as nmol/million cells.
Detailed in vitro dosing information for all experiments is referenced in Table S2.
Chemical Synthesis and Characterization of 8-Dehydrocholesterol
Isolation of 8-DHC-PTAD adducts.
PTAD (, ) was added to a solution of 8-DHC (, ) in MeOH () and benzene ( for solubility). After 1 h, the reaction mixture was concentrated and purified on a Si SPE cartridge (hexanes:EtOAc, 1:1) to remove most of the PTAD. The reaction mixture was further purified by semipreparative HPLC (C18, MeOH) to isolate the ene; 5,7,9-PTAD; and 5,9,14-MeOH adducts. The NMR data for the isolated products matched the independently synthesized adducts, which is described below. The ene adduct could not be isolated pure, likely due to the presence of minor amounts of the other ene adducts, as well as its instability. Only the diagnostic NMR signals are reported for the ene: NMR () 7.59-7.35 (m, 4H), 7.38-7.35 (m, 1H), 5.35 (d, 1H, ), 5.13 (s, 1H), 3.59-3.50 (m, 1H), 2.39 (d, 2H, ), 1.21 (s, 3H), 0.93 (d, 3H, ), 0.86 (br s, 3H), 0.85 (br s, 3H), 0.67 (s, 3H); NMR () 151.5, 150.4, 145.8, 142.1, 131.4, 129.3, 128.2, 126.0, 125.7, 122.5, 116.4, 72.2, 54.4, 53.1, 49.8, 42.9, 42.1, 39.5, 38.7, 36.11, 36.08, 36.0, 35.4, 151.5, 150.4, 145.8, 142.1, 131.4, 129.3, 128.2, 126.0, 125.7, 122.5, 116.4, 72.2, 54.4, 53.1, 49.8, 42.9, 42.1, 39.5, 38.7, 36.11, 36.08, 36.0, 35.4, 31.9, 30.0, 29.4, 28.7, 28.0, 23.6, 23.3, 23.1, 22.8, 22.7, 22.6, 18.8, 11.6.
Synthesis of Ac-7-DHC 1.
Acetyl chloride (, ) was added to a solution of 7-DHC (, ), DMAP (, ), and (, ) in (). After 1 h, the reaction mixture was washed with and then with brine and then dried over . The product was purified by column chromatography (hexanes:EtOAc, 9:1) and isolated as a white powder (, 83%). NMR () 5.53 (dd, 1H, , ), 5.36–5.34 (m, 1H), 4.72–4.62 (m, 1H), 2.47 (ddd, 1H, , 4.9, ), 2.33 (t, 1H, ), 2.11–2.00 (m, 1H), 2.01 (s, 3H), 1.95–1.83 (m, 5H), 1.70–1.47 (m, 5H), 1.36–1.10 (m, 12H), 0.92 (s, 3H), 0.91 (d, 3H, ), 0.85 (s, 3H), 0.83 (s, 3H), 0.59 (s, 3H); NMR () 170.4, 141.5, 138.4, 120.2, 116.2, 72.7, 55.8, 54.4, 46.0, 42.8, 39.5, 39.1, 37.9, 37.0, 36.6, 36.1, 36.0, 28.1, 28.0, 23.8, 23.0, 22.8, 22.5, 21.4, 21.0, 18.8, 16.1, 11.8.
Synthesis of 2.
A solution of Ac-7-DHC (, ) in () was added to a solution of (, ) in HOAc (). After 30 min, a white precipitate formed and eventually turned yellow. After allowing the reaction to stir overnight, it was diluted with ether and filtered through a pad of silica. The filtrate was concentrated. The product was purified by column chromatography (hexanes:EtOAc, 9:1) and isolated as a yellow powder (, 50%). NMR () 5.67 (dd, 1H, , ), 5.48 (d, 1H, ), 5.37 (d, 1H, ), 4.69–4.58 (m, 1H), 2.51 (t, 1H, ), 2.42–2.31 (m, 2H), 2.22–209 (m, 2H), 2.01 (br s, 2H), 1.96–1.91 (m, 2H), 1.76–1.67 (m, 4H), 1.59–1.50 (m, 4H), 1.40–1.28 (m, 6H), 1.23 (s, 3H), 1.16-0.99 (m, 2H), 0.90 (d, 3H, ), 0.85 (d, 3H, ), 0.83 (d, 3H, ), 0.54 (s, 3H); NMR () 170.3, 143.9, 139.9, 135.6, 122.5, 119.0, 115.5, 74.0, 56.3, 50.8, 43.0, 42.2, 39.4, 39.3, 38.0, 37.4, 36.0, 35.9, 31.6, 30.2, 28.5, 28.3, 28.0, 23.9, 22.8, 22.5, 21.3, 18.3, 11.3.
Synthesis of 5,7,9-triene.
NaOH (, ) was added to solution of 2 (, ) in MeOH () and () and then heated to 80°C. After 1.5 h, the reaction mixture was cooled and diluted with EtOAc. The organics were washed with , and then with brine, and dried over . Purification by column chromatography (hexanes:EtOAc, 4:1) yielded the product as a white powder (, 75%). NMR () 5.65 (dd, 1H, ), 5.49 (d, 1H, ), 5.38 (d, 1H, ), 3.62–3.54 (m, 1H), 2.45 (t, 1H, ), 2.38–2.32 (m, 2H), 2.21–2.11 (m, 2H), 1.98-1.88 (m, 2H), 1.79–1.63 (m, 4H), 1.53–1.47 (m, 2H), 1.39–1.33 (m, 4H), 1.22 (s, 3H), 1.16–0.97 (m, 6H), 0.90 (d, 3H, ), 0.85 (d, 3H, ), 0.84 (d, 3H, ), 0.54 (s, 3H); NMR () 144.1, 141.2, 135.4, 122.5, 118.2, 115.5, 72.1, 56.3, 50.9, 43.0, 42.2, 41.5, 39.4, 39.2, 38.2, 36.0, 35.9, 32.2, 30.4, 28.4, 27.9, 23.8, 22.8, 22.5, 18.3, 11.3.
Synthesis of 5,7,9-PTAD.
PTAD (, ) was added to a solution of the 5,7,9-triene (, ) in MeOH () and benzene ( for solubility). After 1 h, the reaction was concentrated and purified by column chromatography (hexanes:EtOAc, 1:1). NMR () 7.42–7.37 (m, 4H), 7.32–7.28 (m, 1H), 6.51 (d, 1H, ), 6.20 (d, 1H, ), 5.59 (dd, 1H, , ), 4.41–4.33 (m, 1H), 3.21 (dd, 1H, , ), 2.73 (dd, 1H, , ), 2.42–2.32 (m, 2H), 2.09 (d, 1H, ), 2.06–1.86 (m, 4H), 1.83–1.77 (m 2H), 1.74–1.61 (m, 3H), 1.53–1.45 (m, 1H), 1.40–1.33 (m, 5H), 1.13 (s, 3H), 1.09–1.02 (m, 3H), 0.92 (d, 3H, ), 0.85 (d, 3H, ), 0.84 (d, 3H, ), 0.67 (s, 3H); NMR () 151.1, 149.8, 141.9, 134.4, 131.6, 129.2, 128.9, 127.9, 126.0, 122.0, 67.3, 66.4, 64.3, 54.9, 47.5, 43.3, 41.5, 40.5, 39.5, 35.9, 35.4, 34.4, 32.8, 29.7, 28.0, 27.6. 25.4, 23.7, 22.9, 22.8, 22.6, 18.6, 12.5.
Synthesis of 3.
Diethyl azodicarboxylate (, ) was added to a solution of the Ac-7-DHC (, ) in benzene (), then heated to reflux. After overnight period, the reaction mixture was cooled and concentrated. The reaction resulted in a 3:1 mixture of isomers in favor of the desired product. The mixture was purified by column chromatography (hexanes:EtOAc, 4:1) to yield the product as a white powder (, 96%). The isomers could be partially separated by column chromatography using toluene:EtOAc (4:1).
Synthesis of 8-DHC.
() was condensed into a flask at . A cooled solution of 3 (, ) in THF () was added, followed by Li° (, ). The reaction mixture turned blue on completion. After 3 h, the reaction was quenched with saturated and warmed to room temperature to dissipate the . The reaction was diluted with EtOAc and washed with additional saturated and brine and dried over . The product was purified by column chromatography (hexanes:EtOAc, 4:1) and isolated as a white powder (, 42%). Product of higher purity was obtained after purification by semipreparative HPLC (C18, :MeOH, 70:30). NMR () 5.41 (br s, 1H), 3.58–3.47 (m, 1H), 2.51 (br s, 2H), 2.32–2.28 (m, 2H), 2.15–2.06 (m, 3H), 2.01–1.94 (m, 1H), 1.91-1.82 (m, 3H), 1.64–1.22 (m, 12H), 1.16 (s, 3H), 1.12–1.09 (m, 4H), 0.91 (d, 3H, ), 0.85 (d, 3H, ), 0.83 (d, 3H, ), 0.63 (s, 3H); NMR () 138.8, 132.0, 126.4, 119.5, 71.4, 54.7, 51.8, 42.2, 41.9, 39.5, 37.3, 36.8, 36.2, 36.1, 35.6, 31.9, 29.0, 28.8, 28.0, 23.9, 22.9, 22.8, 22.5, 22.2, 18.7, 11.3.
Synthesis of 5,9,14-triene.
PTAD (, ) was added to a solution of 8-DHC (, ) in (). The reaction resulted in a mixture of the desired triene and the 5,7,9-PTAD. After 2 h, the reaction mixture was concentrated and purified by column chromatography (hexanes:EtOAc, 4:1). NMR () 5.48 (br s, 1H), 5.36 (br s, 1H), 3.55-3.52 (m, 1H), 2.85 (d, 1H, ) 2.57 (dd, 1H, , ), 2.39–2.16 (m, 5H), 2.10–1.88 (m, 4H), 1.62–1.54 (m, 3H), 1.53–1.48 (m, 2H), 1.45–1.29 (m, 4H), 1.19 (s, 3H), 1.15–1.13 (m, 4H), 0.92 (d, 3H, ), 0,86 (s, 3H), 0.84 (s, 3H), 0.83 (s, 3H); NMR () 149.8, 137.7, 137.2, 120.8, 119.2, 118.2, 71.7, 57.2, 45.0, 42.2, 39.4, 37.8, 36.8, 36.0, 35.9, 35.5, 34.0, 31.9, 27.9, 27.1, 23.7, 22.8, 22.7, 22.5, 21.8, 18.8, 15.4.
Synthesis of 5,9,14-MeOH adduct.
PTAD (, ) was added to a solution of the 5,9,14-triene (, ) in MeOH () and benzene (, for solubility). After 1 h, the reaction was concentrated and purified by semipreparative HPLC (C18, MeOH). NMR () 5.34 (d, 1H, ), 4.73 (d, 1H, ), 3.66–3.61 (m, 1H), 3.20 (s, 3H), 2.88 (br s, 2H), 2.43 (d, 1H, ), 2.21–2.13 (m, 2H), 2.10–1.96 (m, 3H), 1.91–1.85 (m, 2H), 1.79–1.72 (m, 2H), 1.51–1.45 (m, 4H), 1.39–1.37 (m, 3H), 1.18–1.08 (m, 6H), 1.05 (s, 3H), 0.96 (d, 3H, ), 0.88 (d, 3H, ), 0.87 (d, 3H, ), 0.84 (s, 3H); NMR () 152.8, 137.7, 129.3, 119.4, 77.5, 70.8, 70.4, 54.6, 52.1, 43.5, 43.4, 41.4, 39.5, 38.9, 37.0, 36.0, 33.7, 31.3, 29.7, 29.2, 28.0, 27.9, 24.3, 24.1, 23.8, 22.8, 22.5, 19.0, 18.0.
Synthesis of 5.
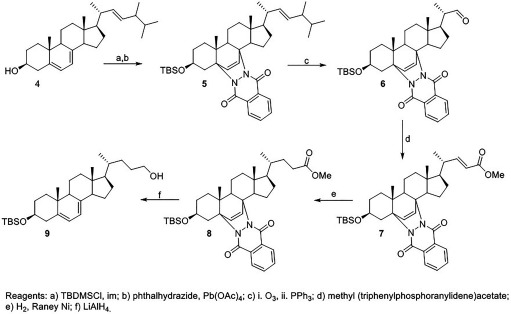
TBDMSCl (, ) and imidazole (, ) were added to a solution of ergosterol (, ) in a mixture of THF:DMF (, 1:1). After the reaction stirred overnight, it was diluted with hexanes:EtOAc (1:1) and washed with and brine. The organic layer was dried over and concentrated. The product was purified by column chromatography (hexanes:EtOAc, 9:1) and isolated as a white powder (, 91%).
(, ) was added in portions to a solution of the TBDMS-protected ergosterol (, ) and phthalhydrazide (, ) in () and HOAc (). After 1 h, the reaction mixture was carefully neutralized with saturated . The organic layer was partitioned, washed with brine, and dried over . Column chromatography (hexanes:EtOAc, 9:1) afforded the product (, 54%) as a yellow foam. NMR () 8.15-8.06 (m, 2H), 7.67–7.64 (m, 2H), 6.62 (d, 1H, ), 6.23 (d, 1H, ), 5.22–5.08 (m, 2H), 3.94–3.83 (m, 2H), 3.61-3.51 (m, 1H), 2.10–1.93 (m, 4H), 1.86–1.79 (m, 2H), 1.62–1.58 (m, 4H), 1.49–1.24 (m, 8H), 0.99 (app t, 6H, ), 0.88-0.78 (mult s, 12H), 0.83 (s, 9H), 0.06 (s, 3H), (s, 3H); NMR () 161.8, 159.5, 138.4, 135.3, 132.6, 132.4, 132.0, 130.5, 130.2, 128.6, 126.9, 126.5, 68.4, 67.4, 67.0, 56.5, 50.5, 49.0, 44.1, 42.6, 40.4, 39.8, 39.2, 35.6, 34.7, 33.0, 30.5, 28.1, 25.9, 24.4, 21.8, 20.8, 19.9, 19.6, 18.5, 18.0, 17.4, 13.3, , .
Synthesis of aldehyde 6.
Ozone was bubbled through a solution of 5 (, ) in :MeOH (, 3:1) at 0°C. After 1 h, the ozone was stopped and (, ) was added. The reaction was allowed to warm to room temperature and concentrated after 1 h. The product was purified by column chromatography (hexanes:EtOAc, 4:1) and isolated as a yellow foam (, 74%). NMR () 9.52 (d, 1H, ), 8.13–8.06 (m, 2H), 7.68–7.65 (m, 2H), 6.44 (d, 1H, ), 6.20 (d, 1H, ), 3.98 (dd, 1H, , ), 3.85 (dd, 1H, , ), 3.61–3.50 (m, 1H), 2.35–2.27 (m, 1H), 2.10–2.03 (m, 2H), 1.96–1.90 (m, 3H), 1.78–1.70 (m, 2H), 1.63–1.58 (m, 4H), 1.49–1.34 (m, 4H), 1.10 (d, 3H, ), 0.99 (s, 3H), 0.84 (s, 3H), 0.82 (s, 9H), 0.05 (s, 3H), (s, 3H); NMR () 204.5, 161.8, 159.6, 138.8, 132.7, 132.5, 130.4, 130.1, 128.1, 127.0, 126.5, 68.0, 67.3, 67.0, 51.7, 50.6, 48.9, 48.4, 44.7, 40.4, 39.1, 35.5, 34.6, 30.4, 26.5, 25.9, 24.3, 22.2, 18.5, 18.0, 13.5, 13.4, , .
Synthesis of unsaturated ester 7.
Methyl (triphenylphosphoranylidene)acetate (, ) was added to a solution of 6 (, ) in (). After overnight, the reaction mixture was concentrated. Purification by column chromatography (hexanes:EtOAc, 4:1) yielded the product (, 80%) as a yellow powder. NMR () 8.10–8.02 (m, 2H), 7.64–7.61 (m, 2H), 6.76 (dd, 1H, , ), 6.59 (d, 1H, ), 6.17 (d, 1H, ), 5.68 (d, 1H, ), 3.91 (dd, 1H, , ), 3.82 (dd, 1H, , ), 3.64 (s, 3H), 3.58–3.47 (m, 4H), 1.83–1.73 (m, 1H), 1.61–1.55 (m, 4H), 1.48–1.25 (m, 6H), 1.03 (d, 3H, ), 0.95 (s, 3H), 0.81 (s, 9H), 0.79 (s, 3H), 0.02 (s, 3H), (s, 3H); NMR () 167.1, 161.7, 159.5, 154.2, 138.6, 132.6, 132.5, 130.4, 130.1, 128.3, 126.9, 126.4, 118.9, 68.0, 67.3, 67.0, 55.5, 51.3, 50.5, 48.6, 44.4, 40.3, 39.2, 35.5, 34.6, 30.4, 27.6, 25.8, 24.3, 21.8, 19.2, 18.5, 18.1, 14.1, 13.3, , .
Synthesis of saturated ester 8.
Raney Ni (, 20%/wt) was added to a solution of 7 (, ) in THF (). The reaction mixture was sparged with for 15 min, and then left under a balloon of . After 30 min, the reaction was complete as determined by NMR. The reaction mixture was filtered through a pad of Celite® and concentrated. No purification was necessary and the product (, 98%) was isolated as a yellow foam. NMR () 8.16–8.05 (m, 2H), 7.69–7.62 (m, 2H), 6.61 (d, 1H, ), 6.22 (d, 1H, ), 3.93–3.81 (m, 2H), 3.62 (s, 3H), 3.59–3.51 (m, 1H), 2.38–2.28 (m, 1H), 2.25–2.14 (m, 1H), 2.09–1.93 (m, 4H), 1.82–1.72 (m, 2H), 1.61–1.50 (m, 4H), 1.40–1.33 (m, 8H), 0.98 (s, 3H), 0.90 (d, 3H, ), 0.82 (s, 9H), 0.79 (s, 3H), 0.05 (s, 3H), (s, 3H); NMR () 174.5, 161.8, 159.5, 138.5, 132.6, 132.5, 130.5, 130.2, 128.5, 126.9, 126.5, 68.3, 67.4, 67.0, 56.2, 5.4, 50.5, 48.8, 44.3, 40.3, 39.3, 35.5, 34.6, 30.7, 30.6, 30.4, 27.4, 25.9, 24.4, 21.8, 18.5, 18.1, 18.0, 13.1, , .
Synthesis of 7-dehydrocholenol 9.
(1M/THF, , ) was added to a solution of 8 (, ) in THF (), and then heated to reflux. After 30 min, the reaction mixture was cooled and quenched with 10% HCl. The reaction mixture was diluted with EtOAc and washed with and then with brine, and dried over . Purification by column chromatography (hexanes:EtOAc, 4:1) yielded the product as a white powder (, 65%). NMR () 5.53 (d, 1H, ), 5.37–5.34 (m, 1H), 3.61–3.52 (m, 3H), 2.31 (d, 2H, ), 2.08–2.04 (m, 1H), 1.93–1.80 (m, 4H), 1.72–1.53 (m, 6H), 1.49–1.19 (m, 10H), 0.94 (d, 3H, ), 0.91 (s, 3H), 0.87 (s, 9H), 0.60 (s, 3H), 0.05 (s, 6H); NMR () 141.0, 140.7, 119.1, 116.3, 71.2, 63.5, 55.7, 54.4, 46.2, 42.9, 41.3, 39.2, 38.5, 37.0, 35.9, 32.4, 31.7, 29.3, 28.1, 25.9, 23.0, 21.1, 18.8, 18.2, 16.3, 11.8, .
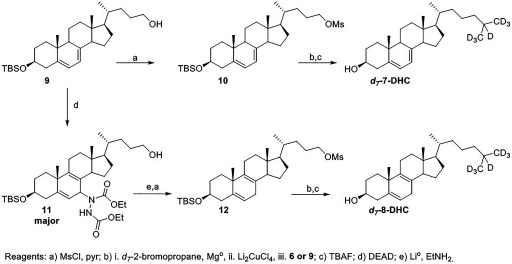
Synthesis of mesylate 10.
Methanesulfonyl chloride (, ) was added to a solution of 9 (, ) in anhydrous pyridine () at 0°C. After 30 min, the reaction mixture was concentrated under high vacuum. The residue was dissolved in EtOAc and washed with , and then with brine, and dried over . The product (, 89%) was used in the next reaction without any purification. NMR () 5.51 (d, 1H, ), 5.36-5.34 (m, 1H), 4.18 (app dt, 2H, , ), 3.61–3.51 (m, 1H), 2.98 (s, 3H), 2.30 (d, 2H, ), 2.07–2.02 (m, 1H), 1.93–1.76 (m, 5H), 1.71–1.05 (m, 4H), 0.94 (d, 3H, ), 0.90 (s, 3H), 0.86 (s, 9H), 0.59 (s, 3H), 0.04 (s, 6H); NMR () 140.8, 140.7, 119.1, 116.4, 71.2, 70.6, 55.5, 54.4, 46.2, 42.9, 41.3, 39.1, 38.5, 37.3, 37.0, 35.6, 32.3, 31.4, 28.0, 25.9, 25.8, 22.9, 21.1, 18.6, 18.2, 16.3, 11.8, .
Synthesis of .
A solution of (, ) in THF () was added in portions to Mg° (, ) in a minimal amount of THF. After 30 min, a solution of (, , ) was added, followed by the mesylate 10 (, ). The was freshly prepared from LiCl (, ) and (, ) in THF (). The reaction mixture turned from gray to dark brown. After 1 h, the reaction was quenched with saturated and extracted with EtOAc. The organics were washed with brine and dried over . The product was purified by column chromatography (hexanes:EtOAc, 19:1) and isolated as a white powder (, 77%).
TBAF (1M/THF, , ) was added to a solution of the previous compound (, ) in THF (). After overnight, the reaction mixture was diluted with EtOAc, washed with brine, and dried over . Purification by column chromatography (hexanes:EtOAc, 4:1) yielded the product as a white powder (, 77%). Higher purity was attained with purification by semipreparative HPLC (C18, MeOH). NMR () 5.55 (dd, 1H, , ), 5.38–5.34 (m, 1H), 3.66-3.56 (m, 1H), 2.45 (ddd, 1H, , 6.3, ), 2.26 (t, 1H, ), 2.10–2.03 (m, 1H), 1.98-1.85 (m, 4H), 1.72–1.53 (m, 4H), 1.40–1.09 (m, 14H), 0.92 (s, 3H), 0.91 (d, 3H, ), 0.59 (s, 3H); NMR () 141.4, 139.7, 119.6, 116.2, 70.4, 55.8, 54.4, 46.2, 42.9, 40.7, 39.1, 38.3, 37.0, 36.1, 31.9, 28.0, 23.8, 23.0, 21.1, 18.8, 16.2, 11.8.
Synthesis of 11.
Diethyl azodicarboxylate (, ) was added to a solution of the 7-dehydrocholenol 9 (, ) in benzene () and then heated to reflux. After 4 h, the reaction mixture was cooled and concentrated. The reaction resulted in a 3:1 mixture of isomers in favor of the desired product. The mixture was purified by column chromatography (hexanes:EtOAc, 1:1) to yield the product as a white powder (, 66%). The isomers could be partially separated by column chromatography using toluene:EtOAc (1:1).
Synthesis of TBS-8-dehydrocholenol.
() was condensed into a flask at . A cooled solution of 11 (, ) in THF () was added, followed by Li° (, ). The reaction mixture turned blue on completion. After 1 h, the reaction was quenched with saturated and warmed to room temperature to dissipate the . The reaction was diluted with EtOAc and washed with additional saturated and then with brine and dried over . The product was purified by column chromatography (hexanes:EtOAc, 4:1) and isolated as a white powder (, 61%). NMR () 5.38 (br s, 1H), 3.59 (t, 2H, ), 3.53–3.43 (m, 1H), 2.51 (br s, 2H), 2.32 (t, 1H, ), 2.23–21.6 (m, 1H), 2.12–2.09 (m, 2H), 2.02–1.72 (m, 4H), 1.64–1.55 (m, 4H), 1.46–1.23 (m, 10H), 1.15 (s, 3H), 0.93 (d, 3H, ), 0.86 (s, 9H), 0.63 (s, 3H), 0.03 (s, 6H); NMR () 139.7, 132.3, 126.2, 119.0, 72.2, 63.5, 54.5, 51.8, 42.7, 42.0, 37.4, 36.8, 36.0, 35.8, 32.4, 31.7, 29.4, 28.9, 28.8, 25.9, 22.9, 22.8, 22.2, 18.6, 18.2, 11.2, .
Synthesis of mesylate 12.
Methanesulfonyl chloride (, ) was added to a solution of the TBS-8-dehydrocholenol (, ) in anhydrous pyridine (). After 1 h, the reaction mixture was concentrated under high vacuum. The residue was dissolved in EtOAc and washed with and then with brine and then dried over . The product (, 100%) was used without purification in the next reaction. NMR () 5.36 (br s, 1H), 4.16 (dt, 2H, , ), 3.50–3.42 (m, 1H), 2.96 (s, 3H), 2.49 (br s, 2H), 2.30 (t, 1H, ), 2.20–2.08 (m, 3H), 1.97–1.71 (m, 5H), 1.63–1.21 (m, 12H), 1.13 (s, 3H), 0.92 (d, 3H, ), 0.85 (s, 9H), 0.61 (s, 3H), 0.01 (s, 6H); NMR () 139.6, 132.3, 126.1, 119.0, 72.2, 71.8, 70.6, 54.4, 51.7, 42.7, 42.0, 37.4, 37.3, 36.7, 35.7, 35.6, 32.4, 31.4, 28.9, 28.7, 25.9, 25.8, 22.9, 22.8, 22.2, 18.5, 18.2, 11.2, .
Synthesis of .
A solution of (, ) in THF () was added in portions to Mg° (, ) in a minimal amount of THF. After 30 min, a solution of (, , ) was added, followed by the mesylate 9 (, ). The was freshly prepared from LiCl (, ) and (, ) in THF (). The reaction mixture turned from gray to dark brown. After 1 h, the reaction was quenched with saturated and extracted with EtOAc. The organics were washed with brine and dried over . The product was purified by column chromatography (hexanes:EtOAc, 19:1) and isolated as a colorless oil (, 51%).
TBAF (1M/THF, , ) was added to a solution of the previous compound (, ) in THF (). After overnight period, the reaction mixture was diluted with EtOAc, washed with brine, and dried over . Purification by column chromatography (hexanes:EtOAc, 4:1) yielded the product as a white powder (, 71%). Higher purity was attained with purification by semipreparative HPLC (C18, :MeOH, 70:30). NMR () 5.40 (br s, 1H), 3.57–3.46 (m, 1H), 2.51 (br s, 2H), 2.31–2.27 (m, 2H), 2.11–2.05 (m, 3H), 2.01–1.95 (m, 1H), 193–1.81 (m, 3H), 1.62–1.26 (m, 11H), 1.16–1.08 (m, 4H), 1.16 (s, 3H), 0.90 (d, 3H, ), 0.62 (s, 3H); NMR () 138.8, 132.0, 126.4, 119.5, 71.4, 54.7, 51.8, 42.2, 41.9, 39.2, 37.3, 36.8, 36.2, 36.1, 35.6, 31.9, 29.0, 28.8, 23.8, 23.0, 22.8, 22.2, 18.7, 11.3.
Statistics
Statistical analyses were performed with Prism software (V5.01, GraphPad Prism 5) in combination with spreadsheet-based software including Excel® (Microsoft®). In addition, p-values were considered significant, and specific analyses used are indicated with each set of data and experiment. Unless otherwise noted, at least three replicates of all experiments were undertaken.
Results
High-Throughput Screen for Environmental Cholesterol Biosynthesis Disruptors
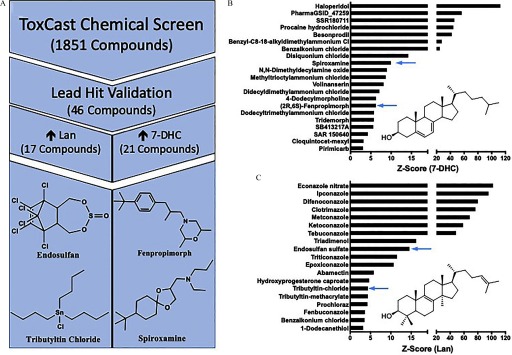
The mouse neuroblastoma cell line, Neuro-2a, was screened against the ToxCast™ Chemical Library for lead-hit identification of environmental cholesterol biosynthesis disruptors. The identities of the 1,851 chemicals tested were blinded throughout the screening process. Known bioactive compounds that affect cholesterol biosynthesis were included on each plate to assess the health and responsiveness of the cells (Figure S2). The results of these positive controls demonstrated that the cells were actively synthesizing cholesterol and that expected metabolic changes could be induced. In alignment with similar screens for cholesterol biosynthesis disruptors using the PTAD-derivatization method, an exposure of a single concentration of for 24 h was used (Korade 2016; Kim 2016; Wages 2018). Furthermore, z-scores () were calculated for each chemical tested from two independent screens and then averaged (Table S3). For the four sterol metabolites assayed (lanosterol, desmosterol, 7-dehydrocholesterol, and cholesterol), 46 lead-hit chemicals were identified as having a z-score greater than three (Figure 2). Of the 1,851 chemicals screened, 4 caused substantial cell loss in both screens, so they were included in lead-hit dose–response verification to determine whether doses at lower concentrations caused changes in sterol biology. Those compounds were colchicine, tributyltin methacrylate, tributyltin chloride, and tributyltin benzoate.
Figure 2.
Lead-hit determination of ToxCast™ Chemical Library for environmental cholesterol biosynthesis disruptors. (A) Workflow of high-throughput screen from entire library screened to the four selected lead-hit compounds, from bottom left, clockwise: tributyltin chloride, endosulfan sulfate, fenpropimorph, spiroxamine. Lead-hit compounds were identified through the results of two independent screens of the ToxCast™ library using Neuro-2a cells as an in vitro model at a screening exposure of for 24 h. Of the compounds screened, those determined as lead-hits for elevating 7-dehydrocholesterol (B) and lanosterol (C) are presented with their respective z-score values and those prioritized indicated with blue arrows.
All lead-hits were validated in a similar experimental paradigm but with conditioning the cells to an exposure media that was serum-free, and thus cholesterol-free, to reduce confounding sources of cholesterol. Although the three compounds that elevated cholesterol and five compounds that elevated desmosterol in the primary screen were not confirmed in the validation experiment (Table S1), the activity of each of the 21 7-DHC (Figure 2B) and 17 lanosterol (Figure 2C) elevating chemicals were verified at the dose. However, an additional five 7-DHC lead-hits and 4 lanosterol lead-hits were deprioritized because they could not reliably induce a dose–response effect in Neuro-2a cells. Additionally, the four compounds that were further tested due to cytotoxicity observed in the screening experiments provided two additional lead-hits (tributyltin methacrylate and tributyltin chloride) as lanosterol elevating chemicals with z-scores of 4.4 at , each (Figure 2C).
Prioritization of lead-hits for future experimentation was based on two additional factors: a) evidence of current commercial use in the environment and b) unknown impact on mammalian sterol biosynthesis (Table S1 and Figure S1). Five compounds were deprioritized because of a lack of potential human exposure and because they remain experimental compounds, and 19 compounds were deprioritized because data showing their capacity to inhibit DHCR7 or CYP51 already exist (Table S1). Tributyltin methacrylate was deprioritized due to experimental limitations. The chemicals ultimately selected for continued assessment for their potential impact on human cholesterol biosynthesis were endosulfan sulfate, tributyltin chloride, fenpropimorph, and spiroxamine (Figure 2A).
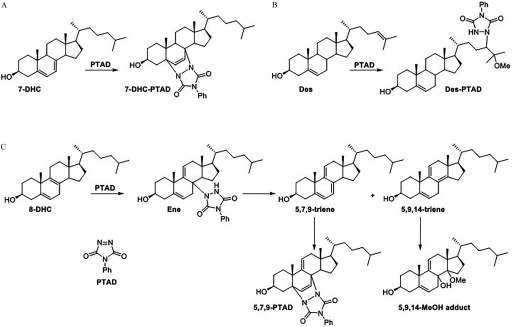
Derivatization of 8-Dehydrocholesterol with PTAD
The derivatization of 7-DHC, lanosterol, and desmosterol by PTAD occurs by either a Diels-Alder reaction of the 7-DHC diene (Liu et al. 2014) or an ene reaction of the lanosterol and desmosterol tail olefin (Korade et al. 2016). However, the reaction of 8-DHC with PTAD was more complex and involved the formation of multiple products. Figure 3 shows the products formed from the reaction of 8-DHC and PTAD. To account for these products, it seems reasonable to suggest that 8-DHC undergoes an ene reaction with PTAD to give an adduct that undergoes elimination to the 5,7,9- and 5,9,14-trienes. The ene product shown in Figure 3 is isolable and has been characterized, but there are two other possible ene intermediates (not shown), both of which could eliminate PTAD to generate the same two trienes. The 5,7,9-triene readily undergoes a Diels-Alder reaction with PTAD, and this triene is never isolated in any reaction mixtures. However, the 5,9,14-triene does not undergo a Diels-Alder reaction and instead ultimately results in the MeOH adduct.
Figure 3.
Proposed mechanism for the reaction of (A) 7-dehydrocholesterol, (B) desmosterol (or lanosterol), and (C) 8-dehydrocholesterol with PTAD. 7-DHC undergoes a Diels-Alder reaction, whereas Des and Lan undergo an ene reaction on the tail olefin. For 8-DHC, the Diels-Alder and MeOH adducts are the major products formed through a series of ene, elimination, and Diels-Alder reactions.
To confirm the identity of the 8-DHC adducts, as well as provide evidence for the scheme shown in Figure 3, we synthesized and characterized the proposed intermediates as described below. The authentic adducts were analyzed by LC-MS and compared with an 8-DHC-PTAD reaction mixture (Figure 4). The ene intermediate was not observed in the actual sample mixture; it is likely unstable and eliminates to one of the trienes. However, the 5,7,9-PTAD and 5,9,14-MeOH adducts were found in roughly equal amounts. We chose to monitor the Diels-Alder product in sample analyses because it appears to be much more stable than the MeOH adduct and it gives characteristic fragmentation patterns.
Figure 4.
LC-MS analysis of (A) isolated and characterized products compared with (B) reaction of 8-dehydrocholesterol with PTAD under conditions used in sample analysis. The products were analyzed on an UPLC C18 column (Acquity UPLC BEH C18, , ) with 100% MeOH (0.1% v/v acetic acid) mobile phase at a flow rate of and runtime of 1.2 min. The following SRMs were monitored: 5,7,9-PTAD , ene , and 5,9,14-MeOH .
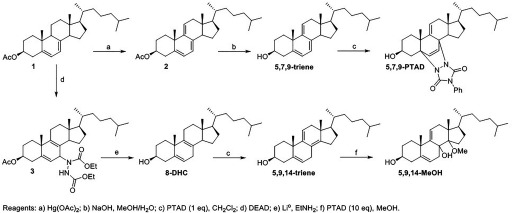
Independent Synthesis of 8-DHC PTAD Adducts
The intermediate trienes and their PTAD adducts shown in Figure 3 were independently synthesized to provide evidence for the mechanism and confirm the identity of the product used in the analysis. The 5,7,9- and 5,9,14-trienes were both synthesized from 7-DHC (Figure 5). Oxidation of the protected 7-DHC with generated the 5,7,9-triene. This triene underwent a Diels-Alder reaction with PTAD to form the 5,7,9-PTAD adduct. The Ac-7-DHC was converted to 8-DHC through an ene reaction with diethyl azodicarboxylate (DEAD) followed by reduction with Li° in . When 8-DHC was treated with one equivalent of PTAD in , it underwent an ene reaction followed by elimination to generate the 5,9,14-triene. Treatment of this triene with an excess of PTAD in MeOH, mimicking the analysis reaction conditions, resulted in the 5,9,14-MeOH adduct. Characterization of these intermediate trienes and their PTAD adducts by NMR and LC/MS confirmed the proposed products shown in Figure 3.
Figure 5.
Synthesis of 5,7,9- and 5,9,14-trienes and their PTAD adducts.
Synthesis of Isotopically Labeled Standards
The syntheses of the Des and Lan have been previously reported (Korade et al. 2016). The and were both synthesized from the 7-dehydrocholenol intermediate 9, which was prepared starting with ergosterol (Figure 6). The alcohol and diene of ergosterol were protected as the silyl ether and Diels-Alder adduct, respectively. The protected ergosterol 5 was ozonized to generate the aldehyde and subsequently underwent a Horner-Wadsworth-Emmons reaction to generate the ester 7. The ester was selectively hydrogenated using Raney nickel as the catalyst. The ester and phthalhydrazide protecting group were then reduced to generate the 7-dehydrocholenol 9.
Figure 6.
Synthesis of 7-dehydrocholenol intermediate for and .
The TBS-7-dehydrocholenol was converted to the mesylate 10. A Grignard reaction with followed by deprotection afforded the . TBS-7-dehydrocholenol was converted to TBS-8-dehydrocholenol 11 using the same route as described above for 8-DHC. An ene reaction with DEAD, followed by a Li° reduction generated the desired product. The was synthesized following the same Grignard reaction sequence as that used for the (Figure 7).
Figure 7.
Synthesis of and .
Concentration-Dependent Effects of Environmental Cholesterol Biosynthesis Disruptors
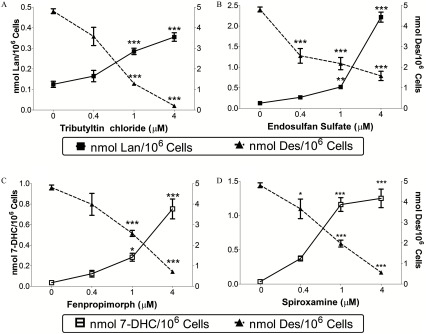
The capacity of endosulfan sulfate and tributyltin chloride to elevate lanosterol was assessed by exposing Neuro-2a cells across a range of concentrations (); fenpropimorph and spiroxamine were similarly tested for their impact on 7-DHC levels (Figure 8). values for each compound to inhibit either CYP51 or DHCR7 were determined in Neuro-2a cells: endosulfan sulfate, ; tributyltin chloride, ; fenpropimorph, ; and spiroxamine, . Concentrations of tributyltin chloride exceeding were determined to be cytotoxic via lactate-dehydrogenase release detected in the media. In addition to the monitoring of elevations in lanosterol or 7-DHC, the immediate cholesterol precursor desmosterol was shown to decrease in a concentration-dependent manner for each chemical exposure (Figure 8).
Figure 8.
Concentration-dependent response of lead-hit compounds on intracellular levels of lanosterol (A,B; solid squares), 7-dehydrocholesterol (C,D; open squares) and desmosterol (solid triangles) in Neuro-2a cells. Cells were exposed to tributyltin chloride (A), endosulfan sulfate (B), fenpropimorph (C), and spiroxamine (D) at the indicated concentration for 24 h. Data presented as nmol sterol/million cells (, ). *, **, *** as determined by post hoc Dunnett’s test following one-way analysis of variance (ANOVA), using vehicle as the control comparison.
Human cell lines from three different tissue lineages were identified to more firmly establish the potential effect that the two CYP51 inhibitors and two DHCR7 inhibitors have on human populations. Each cell line was determined to have a distinct sterol profile based on absolute values of metabolites analyzed (Table 2). For instance, the liver (Hep-G2) and neural (SK-N-SH) cell lines’ sterol profiles both consisted of less than 2% of the cholesterol precursors analyzed in this study; however, the Hep-G2 cell line had a 1.35-fold higher absolute amount of cholesterol than the neural cell line had. In contrast, the lung cell line (A549) had a substantial contribution of the sterol precursor desmosterol of the sterols analyzed in this study, whereas the other neural cell line tested, IMR-32, had a significant proportion to the total sterol content from 7-DHC.
Table 2.
Sterols ( , ) in four different cell lines of human lineage.
| Cholesterol | 7-Dehydrocholesterol | 8-Dehydrocholesterol | Desmosterol | Lanosterol | |
|---|---|---|---|---|---|
| SK-N-SH | |||||
| IMR-32 | |||||
| A549 | |||||
| Hep-G2 |
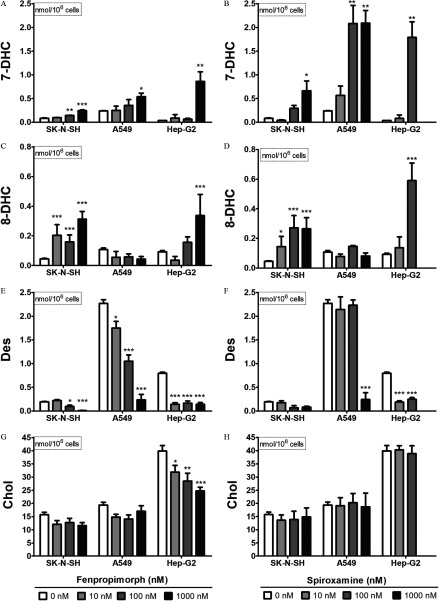
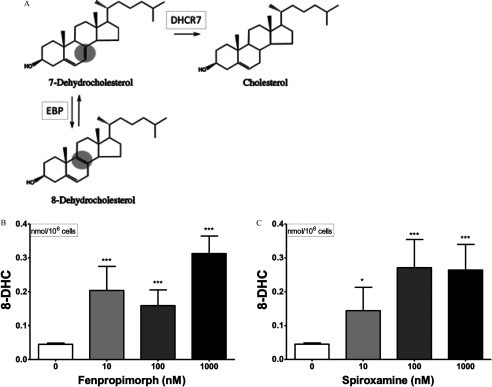
The IMR-32 cell line was particularly sensitive to fenpropimorph and spiroxamine, with concentrations as low as inducing significant cytotoxicity as determined by lactate dehydrogenase detected in the media after a 24-h exposure (Figure S3). The three less-sensitive cell lines (SK-N-SH, A549, and Hep-G2) were challenged with three concentrations of fenpropimorph and spiroxamine from to (Figure 9).
Figure 9.
Impact of fenpropimorph and spiroxamine on 7-dehydrocholesterol (7-DHC; A,B), 8-dehydrocholesterol (8-DHC; C,D), desmosterol (Des; E,F), and cholesterol levels (Chol; G,H) in three different human-derived cell lines: (from left to right) SK-N-SH, A549, and Hep-G2. Cells were exposed to compound (0, 10, 100, ) for 24 h. Omitted bars reflect significant toxicity as determined by release of lactate dehydrogenase. Data presented as nmol sterol/million cells (, ). *, **, *** as determined by post hoc Dunnett’s test following one-way analysis of variance (ANOVA), using vehicle as the control comparison.
No increase in lanosterol was observed for exposures to tributyltin chloride or endosulfan sulfate in any of the four human cell lines across the concentrations tested in this study, with the one exception that tributyltin chloride at the exposure in A549 cells elevated lanosterol levels 2.5-fold (Figure S4A). Positive control inhibition of CYP51 using econazole at for all four cell lines did show significant elevation of lanosterol levels as determined by Student’s t-test (, Figure S5). Both tributyltin chloride and endosulfan sulfate were deprioritized as compounds of interest due to the observation that neither could reliably inhibit CYP51 in human cells. In contrast to the CYP51 lead-hit compounds, 7-DHC levels were elevated in response to exposure to both fenpropimorph and spiroxamine in the three human cell lines for at least one of the concentrations tested (Figure 9A,B). It is noteworthy that the exposure of spiroxamine in the Hep-G2 cells was cytotoxic. In most cases, 7-DHC levels and desmosterol levels were inversely altered after the chemical exposures (Figure 9E,F). Unlike spiroxamine (Figure 9H), fenpropimorph did lower absolute cholesterol levels measured in all cell lines tested but showed a reliable concentration-dependent effect only with the Hep-G2 cell line (Figure 9G).
Further experimentation with fenpropimorph and spiroxamine was undertaken to evaluate levels of other 7-DHC relevant metabolites. In addition to 7-DHC serving as a precursor to both cholesterol and vitamin D, this sterol is also isomerized biosynthetically to 8-dehydrocholesterol (8-DHC) by the enzyme Emopamil Binding Protein (EBP, , ) (Figure 1). Thus, if DHCR7 is selectively inhibited, and EBP is unaffected by a compound, then 8-DHC would be elevated concomitantly with 7-DHC. Using the newly developed method to assay 8-DHC via PTAD-derivatization, the absolute levels of 8-DHC were determined for the four cells lines tested (Table 2). All concentrations tested in the neural SK-N-SH cells for both fenpropimorph (Figure 10B) and spiroxamine (Figure 10C) demonstrated a significant elevation of 8-DHC levels. Because a concomitant increase in 7-DHC and 8-DHC levels along with decreases in the DHCR7 metabolites desmosterol and cholesterol is observed following exposure to fenpropimorph and spiroxamine, it leads to the conclusion that these two pesticides inhibit DHCR7.
Figure 10.
Biological synthesis of 8-dehydrocholesterol via isomerization of 7-dehydrocholesterol by the enzyme EBP is shown (A). SK-N-SH cells were exposed to fenpropimorph (B) and spiroxamine (C) at indicated concentrations for 24 h. Data presented as nmol 8-dehydrocholesterol/million cells (, ). *, *** as determined by post hoc Dunnett’s test following one-way analysis of variance (ANOVA), using vehicle as the control comparison.
Temporal Changes in Neuroprogenitor Sterol Levels Exposed to 7-DHC Elevating Pesticides
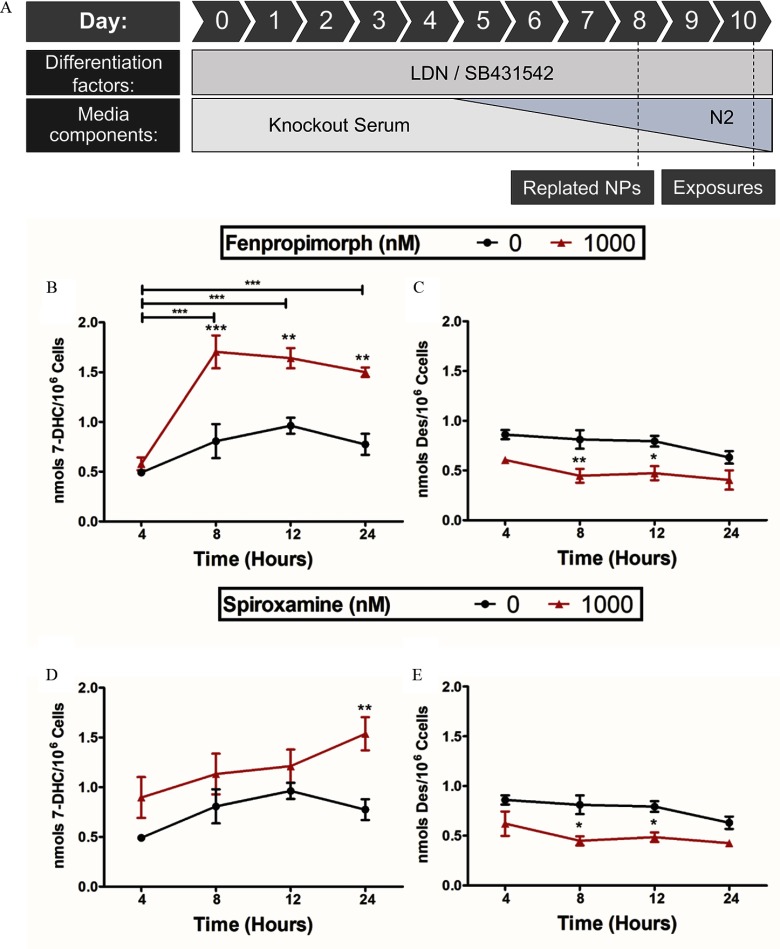
The accumulated observations for the effect of the two pesticides, fenpropimorph and spiroxamine, suggested that both compounds inhibit human DHCR7. To model the cell type most likely affected by a developmental neurotoxicant such as a DHCR7 inhibitor, hiPSCs were differentiated into neuroprogenitors of the cortical glutamatergic lineage and then exposed to chemicals (Figure 11A). To account for differentiation-induced changes in cholesterol biosynthesis, each de-identified donor cell line was used to produce three separate sets of hiPSC-derived neuroprogenitors for experiments, and the results were then averaged derived from three distinct differentiations (Figure S6). In addition to fenpropimorph and spiroxamine, the most potent 7-DHC elevating lead-hit from the original screen and validation, haloperidol, was used as a reference positive control. Haloperidol at 10 and concentration significantly elevated 7-DHC levels from cells to and nmol cells, respectively, after 8 h of exposure in hiPSC neuroprogenitors. The maximum effect of haloperidol was observed with a , 24-h exposure, plateauing at nmol cells.
Figure 11.
Human induced pluripotent stem cells (hiPSCs) were differentiated towards a neuroectoderm lineage as shown (A). These hiPSC-derived neuroprogenitor cells were exposed on day 10 of differentiation to fenpropimorph (B,C) or spiroxamine (D,E) at (red triangle) for 4, 8, 12, and 24 h. Neuroprogenitor cells were then analyzed for 7-dehydrocholesterol (B,D) or desmosterol (C,E) and compared with vehicle control (black circle; 0.01% DMSO). Three distinct differentiations were conducted for each donor and averaged; shown is the average of the three donors (). *, **, *** as determined by Bonferroni posttests following two-way analysis of variance (ANOVA).
Fenpropimorph and spiroxamine at led to elevated 7-DHC levels (Figure 11B,D) and reduced desmosterol levels in the hiPSC-derived neuroprogenitors (Figure 11C,E) in comparison with their time-matched vehicle control. Exposure to chemicals at did not significantly change sterol levels in the cells. However, beginning at 4 h, exposure to both pesticides at altered hiPSC sterol profiles as indicated by an increase in 7-DHC levels and decrease in desmosterol levels with spiroxamine (Figure 11D,E) and a decrease in desmosterol levels with fenpropimorph (Figure 11C). After 24 h, similar absolute levels of 7-DHC were observed (fenpropimorph, cells; spiroxamine, cells), but fenpropimorph reached peak elevation of 7-DHC 8 h after exposure (Figure 11B).
De Novo Cholesterol Synthesis in hiPSC Cells Treated with Fenpropimorph and Spiroxamine
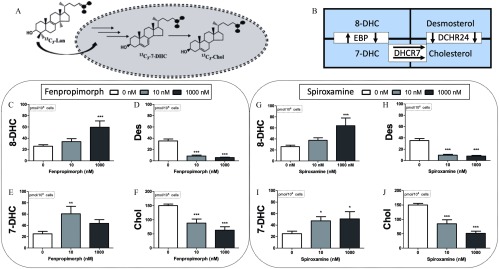
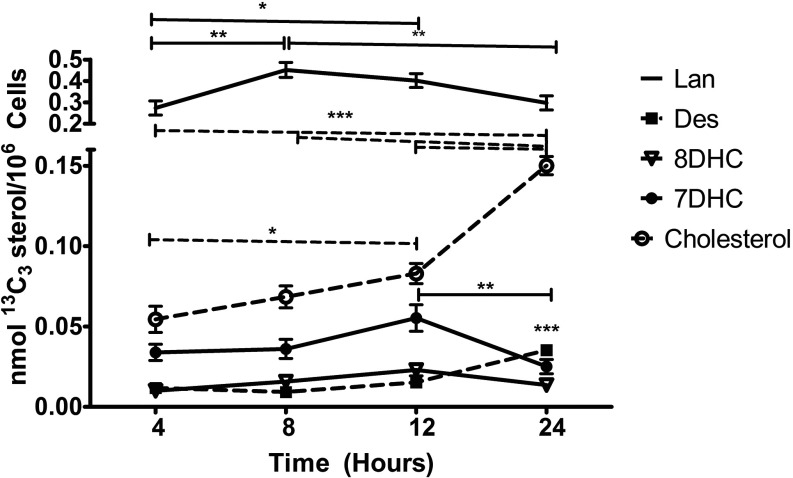
An isotopically labeled sterol precursor provides a highly sensitive means to track the enzymatic turnover of post-lanosterol metabolites during a chemical exposure. Thus, hiPSCs were incubated with a synthetic lanosterol labeled with three atoms (see Figure 12), and the isotopically labeled sterol products of this precursor—7-DHC, 8-DHC, desmosterol, and cholesterol—were analyzed by LC-MS/MS. A gradual increase in levels was observed in the neuroprogenitor cells with a doubling of the absolute amount occurring between 12 and 24 h (Figure 12). Between those same time points, a significant decrease of and an increase in were also observed (Figure 13). The dynamic change in the profile of isotopically labeled sterols at 24 h provided a useful time point to interrogate the effect of the pesticides on de novo cholesterol synthesis.
Figure 12.
De novo synthesis of cholesterol and sterol precursors was accomplished by incubating hiPSC-derived neuroprogenitor cells with for 24 h and monitoring for including and (A). Simplified cholesterol biosynthetic pathway is shown (B). Neuroprogenitor cells were exposed to fenpropimorph (C–F) or spiroxamine (G–J) at and for the same duration as the incubation. Absolute values of (C,G), (D,H), (E,I), and (F,J) were detected, quantified and normalized to cell number. Three distinct differentiations were conducted for each donor and averaged; shown is the average of the three donors (). *, **, *** as determined by post hoc Dunnett’s test following one-way analysis of variance (ANOVA).
Figure 13.
De novo synthesis of cholesterol and sterol precursors was accomplished by incubating hiPSC-derived neuroprogenitor cells with for 24 h and monitoring for at indicated time points. Isotopically labeled sterols analyzed are lanosterol (solid line), desmosterol (solid square, dashed line), 8-DHC (open triangle, solid line), 7-DHC (solid circle, solid line), and cholesterol (open circle, dashed line). Three distinct differentiations were conducted for each donor and averaged; shown is the average of the three donors (). *, **, *** as determined by repeated measures one-way analysis of variance (ANOVA).
Fenpropimorph and spiroxamine affected de novo cholesterol biosynthesis similarly. Exposures of either compound at for 24 h increased levels (Figure 12E,I) and at a higher dose of , levels were also significantly elevated (Figure 12C,G). Of importance is that both products of the enzyme DHCR7, (Figure 12D,H) and (Figure 12F,J), were significantly decreased in all exposures tested. The enzymatic capacity of DHCR7 to convert 7-DHC to cholesterol can be assessed by the ratio of []:[], also known as the RCS. Ideally, the RCS is a value of 1 in normal, functional tissues (Genaro-Mattos et al. 2018; Honda et al. 1995; Wassif et al. 2005). The hiPSC-derived neuroprogenitors exposed to the vehicle control (0.01% DMSO) provided an RCS value of , whereas our reference positive control of haloperidol at and for 24 h led to an RCS of and , respectively. Fenpropimorph compromised the RCS significantly, as determined by one-way analysis of variance (ANOVA), with the exposure causing an RCS of and exposure causing an RCS of . Spiroxamine also reduced DHCR7 activity at with an observed RCS of .
Discussion
This study expanded the chemical space known to inhibit human cholesterol biosynthesis by screening the ToxCast™ Chemical Library and demonstrated that two agricultural pesticides, fenpropimorph and spiroxamine, induced an in vitro metabolic phenotype similar to the genetic recessive disorder SLOS. The ToxCast™ library is a carefully constructed chemical toolset composed of environmentally relevant compounds maintained by the ToxCast™ program at the U.S. Environmental Protection Agency (Richard et al. 2016). A constraint of our findings is the requirement that all compounds in the screening library must be suitable for high-throughput screening (Richard et al. 2016). In this regard, many relevant environmental agents are highly reactive, gaseous, volatile, or DMSO-insoluble and are therefore not included in the ToxCast™ library. Ozone, for example, is a priority chemical for environmental health scientists and governmental regulatory bodies, and there is ample precedent for this highly reactive environmental pollutant to affect human health via oxidation of cholesterol (Pryor et al. 1992; Pulfer and Murphy 2004; Speen et al. 2016).
In comparison with other methods available to quantify sterols, the LC/MS/MS method used in this study has the advantage that it is specific for the important post-lanosterol sterols 7-DHC, 8-DHC, desmosterol, and cholesterol, and it is well-suited for a screen of a library of modest size. Although the method reliably provides quantification of these four sterols and lanosterol in a one-minute run time, we recognize the need to develop rapid high-throughput methods to decrease analysis times further and to expand screening studies to other sterols. Those efforts are underway and will be reported in due course.
Of the compounds screened against the Neuro-2a cell line, 46 were found to affect sterol levels of lanosterol, desmosterol, 7-DHC, or cholesterol. Over half of the 13 validated lead-hits that elevated lanosterol in our screen are of the azole fungicide class. These azole fungicides were designed to inhibit the CYP51 homolog in fungi and, not surprisingly, have affinity toward mammalian CYP51 as well (Rozman et al. 1996; Warrilow et al. 2013). In fact, only 3 of the 13 validated lanosterol-elevating lead-hits identified had no previous history of an effect on sterol metabolism: endosulfan sulfate, tributyltin chloride, and tributyltin methacrylate. The majority of the 16 validated lead-hit compounds that elevated 7-DHC are pharmaceuticals or experimental drugs; seven lead-hits are antiseptics of the benzalkonium chlorides, and two are pesticides. Many of the pharmaceuticals (Korade et al. 2017b) and antiseptics (Herron et al. 2016) that may be relevant to human health today had been identified previously as DHCR7 inhibitors, corroborating our screening results. The impact of pharmaceuticals and other health products found in the environment on public health is a growing concern (Boxall 2004), and it is particularly noteworthy that a number of antipsychotics, including some of the most potent DHCR7 inhibitors identified in this study, such as haloperidol, can be detected in drinking water sources as a consequence of improper medical waste disposal (Logarinho et al. 2016; Silveira et al. 2013; Van De Steene et al. 2010).
In contrast to the exposures resulting from improper medical waste disposal, the two potent commercial pesticides fenpropimorph and spiroxamine are unique in ToxCast™ because they pose a more direct primary exposure threat because few studies had yet to explore the potential impact of compounds intended for the environment on human cholesterol biosynthesis. Both fenpropimorph and spiroxamine are used in the prevention of rust disease in viticulture as well as in cereal crops such as wheat and barley. Although no current study exists identifying a relevant human exposure dose for either pesticide, assuming complete absorption, the exposures levels used in this study are in the range of the indicated acute reference dose of daily dietary intake of fenpropimorph () and spiroxamine (), as established by the U.S. EPA (2004, 2005). As is the case with most fungicides, the most common route of human exposure is through diet, with concentrated areas of exposure where cereal crops are most produced, available, and inexpensive. For instance, exposure studies of European populations near these crops reveal that metabolites of fenpropimorph can be detected in the blood of nonworker individuals, including pregnant women (Buerge et al. 2016; Jamin et al. 2014). Less is known about human exposures to spiroxamine, but evidence exists that applications of spiroxamine to Israeli orchards led to measurable concentrations of the pesticide in the surrounding air (Zivan et al. 2017), as well as evidence of seasonal elevations near German wastewater discharge sites (Beckers et al. 2018). Further, these fungicides have been suggested to have no health effects in the majority of the human population (Muri et al. 2009); therefore, the compounds have a broad global footprint. In fact, due to the current safety profile for morpholine-like pesticides such as fenpropimorph, research is being undertaken to enhance their fungicidal effectiveness by incorporating silicon moieties into their structure, which would extend the half-life of the compounds in mammalian species (Jachak et al. 2015). These efforts suggest that human exposure to fenpropimorph occurs and that exposure scenarios to spiroxamine are likely. Due to the need to cultivate and maintain cereal crops to sustain world populations, vulnerable and underrepresented groups are also more likely exposed to these pesticides than in the past.
Fenpropimorph and spiroxamine are known to affect fungal sterol biosynthesis by inhibiting either reductase and , isomerase or both (Debieu et al. 2000); however, our data suggest that these compounds have a higher affinity to inhibit DHCR7 in human cells. If indeed fenpropimorph or spiroxamine inhibited the human homologs to reductase or , isomerase, we would expect decreases in 7-DHC and 8-DHC, not increases. One other study to date reported that fenpropimorph did indeed affect mammalian cholesterol biosynthesis, albeit in a mouse cell line (Corio-Costet et al. 1988). The CYP51 inhibitors, tributyltin chloride and endosulfan sulfate, increased lanosterol levels only in mouse cells. Even though cholesterol biosynthesis is highly conserved across the animal kingdom as determined by gene similarity of all cholesterol biosynthetic enzymes, recent studies have demonstrated that for CYP51, the difference between mouse and human expression is significant with regard to intron regulation and the existence of different isoforms of the gene (Nelson et al. 2004; Rozman et al. 1996). These interspecies differences likely explain the discordance with the deprioritization of tributyltin chloride and endosulfan sulfate. Because we were able to show that human CYP51 could still be inhibited with econazole, our results suggest that tributyltin chloride and endosulfan sulfate inhibit CYP51 via an unknown mechanism that is likely not relevant in human cells.
The primary screen data presented in Figure 2 were obtained in a murine neuroblastoma cell line (Neuro-2a), and verification of the effect in human cell lines derived from different tissues was explored. The cells examined in this study—SK-N-SH, IMR-32, A549, and Hep-G2—have distinct sterol profiles and differential responses to small molecule modifiers of cholesterol biosynthesis (Fon Tacer et al. 2010; Mitsche et al. 2015). In fact, the sterol metabolisms of each cell line and their tissue lineages are of some interest. Unlike most tissues that have high turnover rates of cholesterol, the central nervous system is effective at sequestering and storing cholesterol (Dietschy 2009). Thus, the lack of significant change in cholesterol in the human neuroblastoma SK-N-SH cell line in response to fenpropimorph or spiroxamine could be due to a biological adaptation of these cells to store cholesterol during acute exposure to small molecule inhibitors of cholesterol biosynthesis. These distinct sterolomic profiles across cell lines that match closely with their tissues of origin have the potential to provide additional insights. Even though our in vitro data suggested that the lead-hits were most potent in neural cells, the results also demonstrated that the lead-hits could impair cholesterol biosynthesis in other tissues as well. When extrapolating this study’s findings to an actual human, it would be important to consider the impact of the exposure beyond the brain, because liver and lung sterol profiles will likely be disrupted as well.
The cell lines used in this study are derived from oncogenic tissue and for this reason may be criticized as models for developmental toxicity studies. However, hiPSCs are a better model to examine developmental toxicity because they are not transformed cells, and they have the ability to differentiate into any cell type, including neuroprogenitors (Fritsche et al. 2018a, 2018b; Kumar et al. 2012). This period of differentiation allows for modeling exposure conditions with the most vulnerable human tissues to better understand neurotoxicant toxicity. Most stem cell–based developmental toxicity testing has been performed using mouse embryonic stem cells (mESCs) due to ethical concerns with human ESCs; but determining toxicity in the analogous hiPSC-based in vitro human system has advantages because toxicokinetics and toxicodynamics likely differ in a species-dependent manner, human genetic and environmental factors contributing to disease are not always conserved in mouse or other species, and hiPSCs have fewer ethical concerns relative to human ESCs given their postnatal origin (Fritsche et al. 2018a, 2018b; Luz and Tokar 2018). Indeed, of the dozens of assays carried out under different exposure conditions and in different cell types, the most sensitive system found in our study was the de novo assay of biosynthesis in neuroprogenitors using an isotopically labeled sterol precursor (see Figure 12). Although increases in 7- and 8-DHC are observed in these assays under most conditions, the decreases observed for desmosterol and cholesterol are particularly telling. Inhibition of DHCR7 should decrease de novo biosynthesis of both desmosterol and cholesterol (see Figure 1), and the decrease observed for these sterols at concentrations as low as seems particularly important.
In conclusion, the results of this study show that about one percent of the ToxCast™ library of environmental agents affected DHCR7, the enzyme that promotes the last critical step in cholesterol biosynthesis, in select mouse and human cell lines. Many of these ToxCast™ hits are found in medical waste, but fenpropimorph and spiroxamine are used extensively in cereal crops and may represent a novel human exposure threat. These two fungicides increased levels of 7- and 8-DHC and reduced levels of desmosterol and cholesterol in several human cell lines and in developing human neural tissue. Even though this work is founded solely on in vitro experiments, this research does suggest that these compounds may be developmental neurotoxicants like numerous other pesticides (Gunier et al. 2017; Roncati et al. 2017; van Wendel de Joode et al. 2016). Additionally, adverse-outcome pathway analyses may benefit from the inclusion of cholesterol biosynthesis as a possible key event or even molecular initiating event for frameworks describing environmental neurodevelopmental toxicity (Sorhus et al. 2017).
Supplementary Material
Acknowledgments
This work was supported in part by the National Institute of Environmental Health Sciences Grants ES024133 (NAP, PAW), ES000267 (PAW, ABB), and ES016931 (ABB). The authors thank A. Richard and K. Houck [U.S. EPA, National Center for Computational Toxicology (NCCT)] for assistance in obtaining the ToxCast™ Chemical Library. We thank K. Ess (Vanderbilt University Medical Center) for the CX-3 line and human dermal fibroblasts. The ToxCast™ Chemical Library was distributed by the Vanderbilt High-Throughput Screening Core Facility with assistance from P. Vinson and C. Whitwell. The Vanderbilt High-throughput Screening Core Facility receives support from the Vanderbilt Institute of Chemical Biology and The Vanderbilt Ingram Cancer Center.
Footnotes
Supplemental Material is available online (https://doi.org/10.1289/EHP5053).
The authors declare they have no actual or potential competing financial interests.
Note to readers with disabilities: EHP strives to ensure that all journal content is accessible to all readers. However, some figures and Supplemental Material published in EHP articles may not conform to 508 standards due to the complexity of the information being presented. If you need assistance accessing journal content, please contact ehponline@niehs.nih.gov. Our staff will work with you to assess and meet your accessibility needs within 3 working days.
References
- Armstrong LC, Westlake G, Snow JP, Cawthon B, Armour E, Bowman AB, et al. . 2017. Heterozygous loss of TSC2 alters p53 signaling and human stem cell reprogramming. Hum Mol Genet 26(23):4629–4641, PMID: 28973543, 10.1093/hmg/ddx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal-Price AK, Hogberg HT, Buzanska L, Lenas P, van Vliet E, Hartung T. 2010. In vitro developmental neurotoxicity (DNT) testing: relevant models and endpoints. Neurotoxicology 31(5):545–554, PMID: 19969020, 10.1016/j.neuro.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Beckers LM, Busch W, Krauss M, Schulze T, Brack W. 2018. Characterization and risk assessment of seasonal and weather dynamics in organic pollutant mixtures from discharge of a separate sewer system. Water Res 135:122–133, PMID: 29466716, 10.1016/j.watres.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Björkhem I, Meaney S. 2004. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol 24(5):806–815, PMID: 14764421, 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- Boxall AB. 2004. The environmental side effects of medication. EMBO Rep 5(12):1110–1116, PMID: 15577922, 10.1038/sj.embor.7400307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerge IJ, Krauss J, López-Cabeza R, Siegfried W, Stüssi M, Wettstein FE, et al. . 2016. Stereoselective metabolism of the sterol biosynthesis inhibitor fungicides fenpropidin, fenpropimorph, and spiroxamine in grapes, sugar beets, and wheat. J Agric Food Chem 64(26):5301–5309, PMID: 27248479, 10.1021/acs.jafc.6b00919. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27(3):275–280, PMID: 19252484, 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corio-Costet MF, Gerst N, Benveniste P, Schuber F. 1988. Inhibition by the fungicide fenpropimorph of cholesterol biosynthesis in 3T3 fibroblasts. Biochem J 256(3):829–834, PMID: 3223956, 10.1042/bj2560829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LG, Cole TB, Coburn J, Chang YC, Dao K, Roque P. 2014. Neurotoxicants are in the air: convergence of human, animal, and in vitro studies on the effects of air pollution on the brain. BioMed Res Int 2014:736385, PMID: 24524086, 10.1155/2014/736385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debieu D, Bach J, Arnold A, Brousset S, Gredt M, Taton M, et al. . 2000. Inhibition of ergosterol biosynthesis by morpholine, piperidine, and spiroketalamine fungicides in Microdochium nivale: effect on sterol composition and sterol Δ8→Δ7-isomerase activity. Pestic Biochem Physiol 67(2):85–94, 10.1006/pest.2000.2485. [DOI] [Google Scholar]
- Dietschy JM. 2009. Central nervous system: cholesterol turnover, brain development and neurodegeneration. Biol Chem 390(4):287–293, PMID: 19166320, 10.1515/BC.2009.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvornik D, Hill P. 1968. Effect of long-term administration of AY-9944, an inhibitor of 7-dehydrocholesterol delta 7-reductase, on serum and tissue lipids in the rat. J Lipid Res 9(5):587–595, PMID: 5726317. [PubMed] [Google Scholar]
- Fon Tacer K, Pompon D, Rozman D. 2010. Adaptation of cholesterol synthesis to fasting and TNF-alpha: profiling cholesterol intermediates in the liver, brain, and testis. J Steroid Biochem Mol Biol 121(3–5):619–625, PMID: 20206258, 10.1016/j.jsbmb.2010.02.026. [DOI] [PubMed] [Google Scholar]
- Fritsche E, Barenys M, Klose J, Masjosthusmann S, Nimtz L, Schmuck M, et al. . 2018a. Development of the concept for stem cell-based developmental neurotoxicity (DNT) evaluation. Toxicol Sci 165(1):14–20, PMID: 29982725, 10.1093/toxsci/kfy175. [DOI] [PubMed] [Google Scholar]
- Fritsche E, Barenys M, Klose J, Masjosthusmann S, Nimtz L, Schmuck M, et al. . 2018b. Current availability of stem cell-based in vitro methods for developmental neurotoxicity (DNT) testing. Toxicol Sci 165(1):21–30, PMID: 29982830, 10.1093/toxsci/kfy178. [DOI] [PubMed] [Google Scholar]
- Genaro-Mattos TC, Allen LB, Anderson A, Tallman KA, Porter NA, Korade Z, et al. . 2019. Maternal aripiprazole exposure interacts with 7-dehydrocholesterol reductase mutations and alters embryonic neurodevelopment. Mol Psychiatry 24(4):491–500, PMID: 30742019, 10.1038/s41380-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genaro-Mattos TC, Tallman KA, Allen LB, Anderson A, Mirnics K, Korade Z, et al. . 2018. Dichlorophenyl piperazines, including a recently-approved atypical antipsychotic, are potent inhibitors of DHCR7, the last enzyme in cholesterol biosynthesis. Toxicol Appl Pharmacol 349:21–28, PMID: 29698737, 10.1016/j.taap.2018.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean P, Landrigan PJ. 2006. Developmental neurotoxicity of industrial chemicals. Lancet 368(9553):2167–2178, PMID: 17174709, 10.1016/S0140-6736(06)69665-7. [DOI] [PubMed] [Google Scholar]
- Grandjean P, Landrigan PJ. 2014. Neurobehavioural effects of developmental toxicity. Lancet Neurol 13(3):330–338, PMID: 24556010, 10.1016/S1474-4422(13)70278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunier RB, Bradman A, Harley KG, Kogut K, Eskenazi B. 2017. Prenatal residential proximity to agricultural pesticide use and IQ in 7-year-old children. Environ Health Perspect 125(5):057002, PMID: 28557711, 10.1289/EHP504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron J, Reese RC, Tallman KA, Narayanaswamy R, Porter NA, Xu L. 2016. Identification of environmental quaternary ammonium compounds as direct inhibitors of cholesterol biosynthesis. Toxicol Sci 151(2):261–270, PMID: 26919959, 10.1093/toxsci/kfw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Tint GS, Salen G, Batta AK, Chen TS, Shefer S. 1995. Defective conversion of 7-dehydrocholesterol to cholesterol in cultured skin fibroblasts from Smith-Lemli-Opitz syndrome homozygotes. J Lipid Res 36(7):1595–1601, PMID: 7595082. [PubMed] [Google Scholar]
- Jachak GR, Ramesh R, Sant DG, Jorwekar SU, Jadhav MR, Tupe SG, et al. . 2015. Silicon incorporated morpholine antifungals: design, synthesis, and biological evaluation. ACS Med Chem Lett 6(11):1111–1116, PMID: 26617963, 10.1021/acsmedchemlett.5b00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamin EL, Bonvallot N, Tremblay-Franco M, Cravedi JP, Chevrier C, Cordier S, et al. . 2014. Untargeted profiling of pesticide metabolites by LC-HRMS: an exposomics tool for human exposure evaluation. Anal Bioanal Chem 406(4):1149–1161, PMID: 23892877, 10.1007/s00216-013-7136-2. [DOI] [PubMed] [Google Scholar]
- Jung S, Seo JS, Kim BS, Lee D, Jung KH, Chu K, et al. . 2013. Social deficits in the AY-9944 mouse model of atypical absence epilepsy. Behav Brain Res 236(1):23–29, PMID: 22944514, 10.1016/j.bbr.2012.08.029. [DOI] [PubMed] [Google Scholar]
- Kelley RI, Hennekam RC. 2000. The Smith-Lemli-Opitz syndrome. J Med Genet 37(5):321–335, PMID: 10807690, 10.1136/jmg.37.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalis DG, Hill AN, Clifton S, Wild RA. 2016. The risks of statin use in pregnancy: a systematic review. J Clin Lipidol 10(5):1081–1090, PMID: 27678424, 10.1016/j.jacl.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Kim HY, Korade Z, Tallman KA, Liu W, Weaver CD, Mirnics K, et al. . 2016. Inhibitors of 7-dehydrocholesterol reductase: screening of a collection of pharmacologically active compounds in Neuro2a cells. Chem Res Toxicol 29(5):892–900, PMID: 27097157, 10.1021/acs.chemrestox.6b00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer NC, Smith AM, West PR, Conard KR, Fontaine BR, Weir-Hauptman AM, et al. . 2011. Identifying developmental toxicity pathways for a subset of ToxCast chemicals using human embryonic stem cells and metabolomics. Toxicol Appl Pharmacol 257(1):111–121, PMID: 21925528, 10.1016/j.taap.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Kolf-Clauw M, Chevy F, Wolf C, Siliart B, Citadelle D, Roux C. 1996. Inhibition of 7-dehydrocholesterol reductase by the teratogen AY-9944: a rat model for Smith-Lemli-Opitz syndrome. Teratology 54(3):115–125, PMID: 8987154, . [DOI] [PubMed] [Google Scholar]
- Korade Z, Genaro-Mattos TC, Tallman KA, Liu W, Garbett KA, Koczok K, et al. . 2017a. Vulnerability of DHCR7+/− mutation carriers to aripiprazole and trazodone exposure. J Lipid Res 58(11):2139–2146, PMID: 28972118, 10.1194/jlr.M079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z, Kim HY, Tallman KA, Liu W, Koczok K, Balogh I, et al. . 2016. The effect of small molecules on sterol homeostasis: Measuring 7-dehydrocholesterol in dhcr7-deficient neuro2a cells and human fibroblasts. J Med Chem 59(3):1102–1115, PMID: 26789657, 10.1021/acs.jmedchem.5b01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z, Liu W, Warren EB, Armstrong K, Porter NA, Konradi C. 2017b. Effect of psychotropic drug treatment on sterol metabolism. Schizophr Res 187:74, PMID: 28202290, 10.1016/j.schres.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KK, Aboud AA, Bowman AB. 2012. The potential of induced pluripotent stem cells as a translational model for neurotoxicological risk. Neurotoxicology 33(3):518–529, PMID: 22330734, 10.1016/j.neuro.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RW, Tierney E. 2011. Hypothesis: the role of sterols in autism spectrum disorder. Autism Res Treat 2011:653570, PMID: 22937253, 10.1155/2011/653570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni V, Caccia C. 2014. Study of cholesterol in Huntington’s disease. Biochem Biophys Res Commun 446(3):697–701, PMID: 24525128, 10.1016/j.bbrc.2014.01.188. [DOI] [PubMed] [Google Scholar]
- Lepesheva GI, Hargrove TY, Kleshchenko Y, Nes WD, Villalta F, Waterman MR. 2008. CYP51: a major drug target in the cytochrome P450 superfamily. Lipids 43(12):1117–1125, PMID: 18769951, 10.1007/s11745-008-3225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xu L, Lamberson C, Haas D, Korade Z, Porter NA. 2014. A highly sensitive method for analysis of 7-dehydrocholesterol for the study of Smith-Lemli-Opitz syndrome. J Lipid Res 55(2):329–337, PMID: 24259532, 10.1194/jlr.D043877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho F, Rosado T, Lourenço C, Barroso M, Araujo ARTS, Gallardo E, et al. . 2016. Determination of antipsychotic drugs in hospital and wastewater treatment plant samples by gas chromatography/tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 1038:127, PMID: 28029546, 10.1016/j.jchromb.2016.10.031. [DOI] [PubMed] [Google Scholar]
- Luz AL, Tokar EJ. 2018. Pluripotent stem cells in developmental toxicity testing: a review of methodological advances. Toxicol Sci 165(1):31–39, PMID: 30169765, 10.1093/toxsci/kfy174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín M, Pfrieger F, Dotti C. 2014. Cholesterol in brain disease: sometimes determinant and frequently implicated. EMBO Rep 15(10):1036–1052, PMID: 25223281, 10.15252/embr.201439225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrogan A, Snowball J, Charlton RA. 2017. Statins during pregnancy: a cohort study using the General Practice Research Database to investigate pregnancy loss. Pharmacoepidemiol Drug Saf 26(7):843–852, PMID: 28176447, 10.1002/pds.4176. [DOI] [PubMed] [Google Scholar]
- Mitsche MA, McDonald JG, Hobbs HH, Cohen JC. 2015. Flux analysis of cholesterol biosynthesis in vivo reveals multiple tissue and cell-type specific pathways. eLife 4:e07999, PMID: 26114596, 10.7554/eLife.07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muri SD, van der Voet H, Boon PE, van Klaveren JD, Brüschweiler BJ. 2009. Comparison of human health risks resulting from exposure to fungicides and mycotoxins via food. Food Chem Toxicol 47(12):2963–2974, PMID: 19345717, 10.1016/j.fct.2009.03.035. [DOI] [PubMed] [Google Scholar]
- Neely DM, Litt MJ, Tidball AM, Li GG, Aboud AA, Hopkins CR, et al. . 2012. DMH1, a highly selective small molecule BMP inhibitor promotes neurogenesis of hiPSCs: comparison of PAX6 and SOX1 expression during neural induction. ACS Chem Neurosci 3(6):482–491, PMID: 22860217, 10.1021/cn300029t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. 2004. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 14(1):1–18, PMID: 15128046, 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. . 2011. A more efficient method to generate integration-free human iPS cells. Nat Methods 8(5):409–412, PMID: 21460823, 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Porter FD. 2008. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet 16(5):535–541, PMID: 18285838, 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- Porter FD, Herman GE. 2011. Malformation syndromes caused by disorders of cholesterol synthesis. J Lipid Res 52(1):6–34, PMID: 20929975, 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor WA, Wang K, Bermúdez E. 1992. Cholesterol ozonation products as biomarkers for ozone exposure in rats. Biochem Biophys Res Commun 188(2):618–623, PMID: 1445306, 10.1016/0006-291x(92)91101-u. [DOI] [PubMed] [Google Scholar]
- Pulfer MK, Murphy RC. 2004. Formation of biologically active oxysterols during ozonolysis of cholesterol present in lung surfactant. J Biol Chem 279(25):26331–26338, PMID: 15096493, 10.1074/jbc.M403581200. [DOI] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, et al. . 2016. ToxCast chemical landscape: paving the road to 21st century toxicology. Chem Res Toxicol 29(8):1225–1251, PMID: 27367298, 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- Roelofs MJE, Temming AR, Piersma AH, van den Berg M, van Duursen MBM. 2014. Conazole fungicides inhibit Leydig cell testosterone secretion and androgen receptor activation in vitro. Toxicol Rep 1:271–283, PMID: 5598417, 10.1016/j.toxrep.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncati L, Pusiol T, Piscioli F, Lavezzi AM. 2017. Neurodevelopmental disorders and pesticide exposure: the northeastern Italian experience. Arch Toxicol 91(2):603–604, PMID: 28032145, 10.1007/s00204-016-1920-7. [DOI] [PubMed] [Google Scholar]
- Roux C, Wolf C, Mulliez N, Gaoua W, Cormier V, Chevy F, et al. . 2000. Role of cholesterol in embryonic development. Am J Clin Nutr 71(suppl 5):1270s–1279s, PMID: 10799401, 10.1093/ajcn/71.5.1270s. [DOI] [PubMed] [Google Scholar]
- Rozman D, Strömstedt M, Tsui LC, Scherer SW, Waterman MR. 1996. Structure and mapping of the human lanosterol 14alpha-demethylase gene (CYP51) encoding the cytochrome P450 involved in cholesterol biosynthesis; comparison of exon/intron organization with other mammalian and fungal CYP genes. Genomics 38(3):371–381, PMID: 8975714, 10.1006/geno.1996.0640. [DOI] [PubMed] [Google Scholar]
- Schroepfer G. 2000. Oxysterols: modulators of cholesterol metabolism and other processes. Physiol Rev 80(1):361–554, PMID: 10617772, 10.1152/physrev.2000.80.1.361. [DOI] [PubMed] [Google Scholar]
- Sikora DM, Pettit-Kekel K, Penfield J, Merkens LS, Steiner RD. 2006. The near universal presence of autism spectrum disorders in children with Smith-Lemli-Opitz syndrome. Am J Med Genet A 140(14):1511–1518, PMID: 16761297, 10.1002/ajmg.a.31294. [DOI] [PubMed] [Google Scholar]
- Silveira MAK, Caldas SS, Guilherme JR, Costa FP, BdS G, Cerqueira MBR, et al. . 2013. Quantification of pharmaceuticals and personal care product residues in surface and drinking water samples by SPE and LC-ESI-MS/MS. J Brazilian Chem Soc 24:1385–1395, 10.5935/0103-5053.20130176. [DOI] [Google Scholar]
- Sorhus E, Incardona JP, Furmanek T, Goetz GW, Scholz NL, Meier S, et al. . 2017. Novel adverse outcome pathways revealed by chemical genetics in a developing marine fish. eLife 6: e20707, PMID: 28117666, 10.7554/eLife.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speen AM, Kim HH, Bauer RN, Meyer M, Gowdy KM, Fessler MB, et al. . 2016. Ozone-derived oxysterols affect liver x receptor (LXR) signaling: a potential role for lipid-protein adducts. J Biol Chem 291(48):25192–25206, PMID: 27703007, 10.1074/jbc.M116.732362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurm A, Tierney E, Farmer C, Albert P, Joseph L, Swedo S, et al. . 2016. Development, behavior, and biomarker characterization of Smith-Lemli-Opitz syndrome: an update. J Neurodev Disord 8:12, PMID: 27053961, 10.1186/s11689-016-9145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball AM, Bryan MR, Uhouse MA, Kumar KK, Aboud AA, Feist JE, et al. . 2015. A novel manganese-dependent ATM-p53 signaling pathway is selectively impaired in patient-based neuroprogenitor and murine striatal models of Huntington‘s disease. Hum Mol Genet 24(7):1929–1944, PMID: 25489053, 10.1093/hmg/ddu609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball AM, Neely MD, Chamberlin R, Aboud AA, Kuma KK, Han B, et al. . 2016. Genomic instability associated with p53 knockdown in the generation of Huntington’s disease human induced pluripotent stem cells. PLoS One 11(3):e0150372, PMID: 26982737, 10.1371/journal.pone.0150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney E, Bukelis I, Thompson RE, Ahmed K, Aneja A, Kratz L, et al. . 2006. Abnormalities of cholesterol metabolism in autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet 141B(6):666–668, 10.1002/ajmg.b.30368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2004. Health risk assessment for spiroxamine use in/on grapes, hops, and imported bananas. https://www3.epa.gov/pesticides/chem_search/hhbp/R100183.pdf [accessed 17 June 2018]. [Google Scholar]
- U.S. EPA. 2005. Fenpropimorph hed human health risk assessment to support tolerance on imported bananas. https://www3.epa.gov/pesticides/chem_search/hhbp/R116369.pdf [accessed 17 June 2018]. [Google Scholar]
- Van De Steene JC, Stove CP, Lambert WE. 2010. A field study on 8 pharmaceuticals and 1 pesticide in Belgium: removal rates in waste water treatment plants and occurrence in surface water. Sci Total Environ 408(16):3448–3453, PMID: 20471061, 10.1016/j.scitotenv.2010.04.037. [DOI] [PubMed] [Google Scholar]
- van Wendel de Joode B, Mora AM, Lindh CH, Hernández-Bonilla D, Córdoba L, Wesseling C, et al. . 2016. Pesticide exposure and neurodevelopment in children aged 6-9 years from Talamanca, Costa Rica. Cortex 85:137–150, PMID: 27773359, 10.1016/j.cortex.2016.09.003. [DOI] [PubMed] [Google Scholar]
- Vance J. 2012. Dysregulation of cholesterol balance in the brain: contribution to neurodegenerative diseases. Dis Model Mech 5(6):746–755, PMID: 23065638, 10.1242/dmm.010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijheid M, Slama R, Robinson O, Chatzi L, Coen M, van den Hazel P, et al. . 2014. The human early-life exposome (HELIX): project rationale and design. Environ Health Perspect 122(6):535–544, PMID: 24610234, 10.1289/ehp.1307204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wages PA, Kim HH, Korade Z, Porter NA. 2018. Identification and characterization of prescription drugs that change levels of the cholesterol precursors 7-dehydrocholesterol and desmosterol. J Lipid Res 59(10):1916, PMID: 30087204, 10.1194/jlr.M086991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow AG, Parker JE, Kelly DE, Kelly SL. 2013. Azole affinity of sterol 14α-demethylase (CYP51) enzymes from Candida albicans and Homo sapiens. Antimicrob Agents Chemother 57(3):1352–1360, PMID: 23274672, 10.1128/AAC.02067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassif CA, Krakowiak PA, Wright BS, Gewandter JS, Sterner AL, Javitt N, et al. . 2005. Residual cholesterol synthesis and simvastatin induction of cholesterol synthesis in Smith-Lemli-Opitz syndrome fibroblasts. Mol Genet Metab 85(2):96–107, PMID: 15896653, 10.1016/j.ymgme.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Wier PJ, Guerriero FJ, Walker RF. 1989. Implementation of a primary screen for developmental neurotoxicity. Toxicol Sci 13(1):118–136, PMID: 2767352, 10.1093/toxsci/13.1.118. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4(2):67–73, PMID: 10838414, 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhang J, Qiang L. 2015. Cholesterol metabolism and homeostasis in the brain. Protein Cell 6(4):254–264, PMID: 25682154, 10.1007/s13238-014-0131-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivan O, Bohbot-Raviv Y, Dubowski Y. 2017. Primary and secondary pesticide drift profiles from a peach orchard. Chemosphere 177:303–310, PMID: 28314235, 10.1016/j.chemosphere.2017.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.