Abstract
Background:
Endocrine-disrupting chemicals have been shown to have broad effects on development, but their mutagenic actions that can lead to cancer have been less clearly demonstrated. Physiological levels of estrogen have been shown to stimulate DNA damage in breast epithelial cells through mechanisms mediated by estrogen-receptor alpha (). Benzophenone-3 (BP-3) and propylparaben (PP) are xenoestrogens found in the urine of of U.S. population.
Objectives:
We investigated the effect of BP-3 and PP on estrogen receptor–dependent transactivation and DNA damage at concentrations relevant to exposures in humans.
Methods:
In human breast epithelial cells, DNA damage following treatment with (), BP-3, and PP was determined by immunostaining with antibodies against and 53BP1. Estrogenic responses were determined using luciferase reporter assays and gene expression. Formation of R-loops was determined with DNA: RNA hybrid–specific S9.6 antibody. Short-term exposure to the chemicals was also studied in ovariectomized mice. Immunostaining of mouse mammary epithelium was performed to quantify R-loops and DNA damage in vivo.
Results:
Concentrations of and BP-3 or PP increased DNA damage similar to that of treatment in a manner. However, BP-3 and PP had limited transactivation of target genes at and concentrations. BP-3 and PP exposure caused R-loop formation in a normal human breast epithelial cell line when was introduced. R-loops and DNA damage were also detected in mammary epithelial cells of mice treated with BP-3 and PP.
Conclusions:
Acute exposure to xenoestrogens (PP and BP-3) in mice induce DNA damage mediated by formation of R-loops at concentrations 10-fold lower than those required for transactivation. Exposure to these xenoestrogens may cause deleterious estrogenic responses, such as DNA damage, in susceptible individuals. https://doi.org/10.1289/EHP5221
Introduction
Endocrine-disrupting chemicals (EDCs) alter the endocrine system by binding directly to the receptors and modulating downstream signaling pathways. Xenoestrogens are structurally diverse EDCs that affect estrogen receptor (ER) signaling pathways. BP-3 (oxybenzone, or 2-Hydroxy-4-methoxybenzophenone, CAS No. 131-57-7) is a UV-filter used in personal care products, such as sunscreens, cosmetics, and lotions, with concentrations up to 0.148% (Liao and Kannan 2014) and a maximum allowed concentration of 6% by Food and Drug Administration (FDA) and European commission (EC 2017). BP-3 was detected in the urine samples of 96.8% of U.S. population in the 2003–2004 National Health and Nutrition Examination Survey (NHANES) conducted by the Centers for Disease Control and Prevention (CDC) (Calafat et al. 2008). Similarly, PP (propyl parahydroxybenzoate, CAS No. 94-13-3) is widely used as an antimicrobial agent in food and personal care products. Although the FDA limits PP to 0.1% in food, currently there is no specific limit for preservatives in personal care products. PP is banned as a food preservative, and maximum permissible levels in personal care products is 0.4% in the European Union (EU) (Snodin 2017; EC 2014). PP was detected in the urine samples of of U.S. population surveyed during 2003–2005 by the CDC (Ye et al. 2006).
Estrogenic responses are determined by the action of two distinct estrogen receptor (ER) subtypes, estrogen receptor () and estrogen receptor (). Ligand-activated ER recruits coactivators to estrogen response elements (ERE) in promoters of target genes leading to transcription initiation (Shang et al. 2000; Yi et al. 2017). In expressing breast cancer cells, proliferation is among the types of cellular responses (Henderson et al. 1988; Musgrove and Sutherland 1994). Hence, estrogenic responses to putative xenoestrogens is most often determined by transactivation of ERE-reporters, endogenous gene expression and cell proliferation in ER-expressing MCF-7 and T47D cell lines, where is the dominant subtype (Buteau-Lozano et al. 2002; Vladusic et al. 2000). These studies showed BP-3 was a weak agonist of ER at (Kerdivel et al. 2013; Schlotz et al. 2017; Schlumpf et al. 2001). BP-3 was found in the urine samples of 25 volunteers who used sunscreen containing 4% BP-3 twice a day for 5 d, suggesting it was readily absorbed through skin (Gonzalez et al. 2006). Metabolites of BP-3, such as 2,4-diOH-BP and 2,3,4-triOH BP, were shown to form by oxidation in rat and human liver microsomes (Okereke et al. 1994; Watanabe et al. 2015). 2,4-diOH-BP was detected in the urine samples of women scheduled to undergo a diagnostic and/or therapeutic laparoscopy or laparotomy as part of the ENDO study (Kunisue et al. 2012) and was shown to have higher ER transactivation potential in comparison with BP-3 (Watanabe et al. 2015). BP-3 and BP-3 metabolite 4,4′-dihydroxybenzophenone were also detected in 27 of the 79 breast milk samples from mothers who had normal pregnancy and delivery, and who participated in the Breast Milk Bank of the Blood and Tissue Bank of Catalonia (Spain) (Molins-Delgado et al. 2018). Exposure of BP-3 during pregnancy and lactation in mice resulted in altered mammary gland ductal architecture that persisted for weeks after exposures ended (LaPlante et al. 2018). Long-term exposure of MCF-7 breast cancer cells to BP-3 for increased the motility of these cells (Alamer and Darbre 2018). This increase was also observed in estrogen nonresponsive cell line MDA-MB-231, suggesting alternate pathways of BP-3 actions at this dose. Similarly, PP was shown to be an effective ER-agonist with 1.3-fold induction of gene expression using reporter assays (ERE-CAT reporter) at , increased expression of estrogen-responsive gene Trefoil Factor 1 (TFF1, also known as pS2) and increased proliferation of MCF-7 cells at (Byford et al. 2002). Proliferation induced by PP was inhibited by ER antagonist (fulvestrant) indicating dependence on . PP also increased cell motility (increased scratch closure) in both short-term (7-d) and long-term (20-wk) treatments in the MCF-7 cell line (Khanna et al. 2014).
In addition to stimulating cell proliferation and motility, estrogen also induces genotoxicity and DNA damage and is considered a major risk factor in breast cancer etiology (Roy and Liehr 1999; Yager and Davidson 2006). Estrogen has been shown to induce DNA damage by a) metabolic activation of estrogen and b) hormonal carcinogenesis (Santen et al. 2009). is metabolized to form catechol estrogens ( or and ), which can be oxidized to form reactive semiquinone (SQ) intermediates and quinone derivatives. Two such compounds, -3-4-Q and -2-3-Q form stable DNA adducts or depurinating adducts, such as -1-N7Gua and -1-N3Ade, which were associated with increased breast cancer risk, but micromolar levels of -3-4-Q and -2-3-Q were required to show DNA adduct formation in vitro (Cavalieri and Rogan 2016). The SQ and quinone derivatives can also generate ROS through redox cycling, which can be genotoxic (Fussell et al. 2011; Wang et al. 2010). Similarly, ER–independent DNA damage was shown in cell lines using the COMET assay (Rajapakse et al. 2005), cII mutagenesis assay (Zhao et al. 2006), or LOH (Huang et al. 2007; Russo et al. 2003). The concentrations of or used in these studies were , with the exception of Russo et al. 2003, who reported increased clonal efficiency of MCF10F cells at . However, the median level during pregnancy is and in normal cycling women (Table 1), and the level of circulating estradiol metabolites are 100-fold lower (Xu et al. 2007; Ziegler et al. 2010). Clinical data show that in postmenopausal women with ER-positive early breast cancer, endocrine therapy with an aromatase inhibitor was associated with significantly lower recurrence than tamoxifen (TAM) therapy (EBCTCG 2015), which could be because of either lower levels of estrogen metabolites or reduced ER activation. Epidemiological data show that for a given level of total estrogen, increased levels of , and are associated with reduced risk in breast cancer (Dallal et al. 2014; Moore et al. 2016; Sampson et al. 2017) or no independent association with risk (Sampson et al. 2017); however, an earlier study reported levels to be associated with higher breast cancer risk (Fuhrman et al. 2012). Hence, the impact of metabolic activation of estrogen at physiologically relevant concentrations on DNA damage remains to be demonstrated.
Table 1.
Estimation of estrogen and xenoestrogens concentrations of estradiol (), benzophenone-3 (BP-3), or propylparaben (PP) in urine/blood samples of women and female mice.
| Ligand | Median () | 90th or 95th percentile () | Relative transactivation activity at 90th or 95th percentile (% RTA vs. ) | References |
|---|---|---|---|---|
| BP-3 (urine) | ||||
| Non-Pregnant | 0.137 | 6.70b | Woodruff et al. 2011 | |
| Pregnant | 0.47 | 29.5b | Philippat et al. 2013 | |
| PP (urine) | ||||
| Non-Pregnant | 0.161 | 1.98b | Calafat et al. 2010 | |
| Pregnant | 0.253 | 3.26b | Philippat et al. 2013 | |
| (blood) | ||||
| Human | ||||
| Ovulatory | 0.0003–0.0018 | Clarke et al. 1997 | ||
| Luteal | 0.0002–0.0008 | O'Leary et al. 1991 | ||
| Pregnant | 0.074 | 0.118a | Schock et al. 2016 | |
| Mouse | Majewski et al. 2018 | |||
90th percentile of exposure in the given population.
95th percentile of exposure in the given population.
Hormonal carcinogenesis is postulated to act through ER to initiate lesions as well as stimulate progression of tumors. treatment stimulated renal tumors in male Syrian hamsters (Liehr et al. 1988). TAM reduced tumors but did not alter levels of DNA-adducts, suggesting the primary effect of being mediated by ER. Similarly, blockage of ER activation through selective estrogen receptor modulators (SERMs), such as TAM and raloxifene, reduced the incidence of breast cancer by 50%–75% in women (Cummings et al. 1999; Cuzick and International Breast Cancer Intervention Study 2001; Martino et al. 2004). Bilateral oophorectomy and hysterectomy in women under 40 years of age reduced breast cancer later in life by 75% (Feinleib 1968). Administration of aromatase inhibitor (exemestane) for 35 months to a cohort of postmenopausal women with Gail score of 1.66 and prior atypical ductal/lobular hyperplasia or ductal carcinoma in situ treated with mastectomy but noncarriers for BRCA1/2 and no prior invasive ductal carcinoma resulted in 65% relative reduction of breast cancer (Goss et al. 2011). Mobley and Brueggemeier showed that 8-oxo-dG production with buthionine sulfoximine (BSO), () and H2O2 treatment could be reduced with TAM treatment in ER-positive MCF7 cells but not in ER-negative MDA-MB-231 cells, suggesting DNA damage was at least partially ER-mediated (Mobley and Brueggemeier 2004). Stork et al. showed lack of DNA damage marker (phosphorylated H2AX) in MCF10A cells following treatment of and for (Stork et al. 2016). In T47D cells, -mediated was diminished with treatment of ER inhibitors like TAM or fulvestrant (Periyasamy et al. 2015). ER signaling stimulates proliferation, which was causally linked to tumorigenesis, by increasing the probability of replication errors, which are propagated in daughter cells (Henderson and Feigelson 2000; Preston-Martin et al. 1990). Therefore, can be considered as a carcinogen through its actions on progression of cancer that was initiated by other factors.
The studies involving DNA damage by have used different cell lines, tissues, and end points. Therefore, there is no consistent way to discriminate the contribution of ER-dependent and ER-independent mechanisms across published studies. It is possible that both mechanisms contribute to -mediated carcinogenesis.
Recent studies have shown that ER stimulation leads to transcription-coupled DNA damage, suggesting a distinct mechanism. Interaction of with chromatin forms transcriptional coactivator/corepressor complexes to initiate transcription (Chao et al. 2002; Fullwood et al. 2009; Shang et al. 2000). The open chromatin in these complexes were susceptible to DNA damage by formation of RNA:DNA triplex structures, called R-loops (Stork et al. 2016). Therefore, estrogen can stimulate carcinogenesis by initiating direct DNA damage mediated by and proliferation that expands the population of breast cells.
Bioassays of transcriptional activities have been valuable in rapidly assessing the risk posed by xenoestrogens. However, it is unclear whether the transcriptional activities of xenoestrogens reflect their potential mutagenic activity mediated by . DNA damage by selective agonists such as diethylstilbestrol (DES) and 4,4′,4′′-(4-Propyl-[1H]-pyrazole-1,3,5-triyl) trisphenol (PPT) (Periyasamy et al. 2015) suggest that transcriptional DNA damage needs to be assessed to determine potential breast cancer risk posed by xenoestrogens. In this study, we evaluated effects of two xenoestrogens, BP-3 and PP, which differ in structure and transcriptional potency and compared these with .
Methods
Cell Culture
T47D (ATCC #HTB-133), T47DKBluc (ATCC #CRL-2865) and MCF-7 (ATCC #HTB 22) cells were passaged in growth media containing phenol red–free (PRF) DMEM-F12 (Sigma #D6434) or MEM 1X (Gibco #51200-038) with 10% heat-inactivated FBS (Omega Scientific #FB-02) and insulin (Sigma #9278), L-glutamine (Hyclone #SH30034.01), gentamycin (Gibco #15750-060), and 1X antibiotics/antimycotics (AB/AM, Gibco #15240-062) and incubated at 37°C with 5% . For experiments, cells were grown in clearing media with charcoal-stripped serum (CSS) (MEM 1x with 10% charcoal dextran–treated FBS (Omega Scientific #FB-04), insulin, and L-glutamine) for before being plated for experiments.
The 76N-Tert cell line, a human mammary epithelial cell line immortalized with expression of human telomerase reverse transcriptase (TERT), was a gift from Dr. Vimla Band (Zhao et al. 2010). These cells were grown in F-media [ DMEM (-pyruvate) (Gibco #11965-092), Ham’s F12 (Gibco #11765-054), 5% FBS, hydrocortisone (Sigma #H4001), human epidermal growth factor (Tonbo Biosciences #21-8356-U100), cholera toxin vibrio (Millipore Sigma #227035), human insulin solution, and 1X antibiotic/antimycotic] and passaged every 2–3 d.
Generation of 76N-Tert-ESR1 Cells
An inducible (ESR1) construct was generated using the pINDUCER14 vector (Meerbrey et al. 2011). Specifically, FLAG tag sequence was amplified from pFLAG-CMV-2 (Andersson et al. 1989) using forward primer 5′-ATACCGGTACCATGGACTACAAAGACGATGACGAC-3′ and reverse primer 5′-TCGACCGGTACGCGTGCGATCGCTGAATTCGCGGCAAG-3′. The amplified FLAG sequence was then cleaned using the Monarch PCR & DNA Cleanup Kit (NEB #T1030) and ligated into pINDUCER14 by digesting both plasmids with AgeI, performing dephosphorylation with shrimp alkaline phosphatase (NEB #M0371S) and gel electrophoresis and extracting from agarose gel (DNAland Scientific #GP1001). Sequencing of pINDUCER14-FLAG confirmed that the FLAG sequence was inserted.
ESR1 was amplified from a plasmid expressing ESR1 made in our lab (pIRES-hrGFPII-ESR1, unpublished data). pIRES-hrGFPII-ESR1 contained the ESR1 cDNA sequence (Open Biosystems #MHS6278-211691051) in the multiple cloning site of the pIRES-hrGFPII vector (Stratagene #240157). ESR1 was amplified from pIRES-hrGFPII-ESR1 using forward primer 5′- GCAGAAATGACCATGACCCTCCACACCAAAGC-3′ and reverse primer 5′- TAAACGCGTTCAGACCGTGGCAGGGAAACCCT-3′. Ligation of ESR1 into pINDUCER14-FLAG was done by digesting both plasmids with EcoRI and MluI and then performing dephosphorylation, cleanup, and extraction as described above. Two linker sequences (Linker A: 5′-AATTGCGCGATCGCGG-3′ and Linker B: 5′-AATTCCGCGATCGCGC-3′) between FLAG and ESR1 were added to keep the ESR1 sequence in frame. Sequencing of this final pINDUCER14-FLAG-ESR1 confirmed that all inserts were in the correct orientation relative to the vector, both FLAG and ESR1 were in frame, and the ESR1 sequence was identical to the Homo sapiens ESR1 gene (Sequencing Primers: F: 5′-CGGTGGGAGGCCTATATAAG-3′, M: 5′-GCTACCATTATGGAGTCTGG-3′, and R: 5′-ACTTATATACGGTTCTCCCC-3′). This final construct was referred to as pIND-ESR1 and expressed a constitutive GFP reporter as well as with N-terminal FLAG tag.
In addition, 293T cells were cultured in DMEM:F12 (Sigma #D8900) supplemented with 10% FBS, gentamycin (Gibco #15750-060), and 1x antibiotic/antimycotic. Cells were lifted with 0.05% trypsin and plated in tissue culture dishes at cells per dish for next-day use. 293T cells were then transfected with pIND-ESR1, psPAX2 (Addgene #12260) (gag, pol, and rev packaging vector), and pMD2.G (Addgene #12259) (vsv-g packaging vector) in antibiotic-free media using Lipofectamine 2000 (Thermo Fisher Scientific). Media was refreshed after , and viral media was collected at 48 and post initial transfection. Viral media from transfected 293T was filtered using a filter (Corning #431220) and added to 76N-Tert cells twice, apart, in a 1:1 ratio with F-media. After , viral media was removed and replaced with F-media. Following cell expansion, the cells were pooled and resuspended in 1% FBS in PBS. Selection of the stably transduced cells was performed by FACS for GFP-positive cells using FACSAria II (Becton-Dickinson). 76N-Tert uninfected cells were used as a control to set the background fluorescence. Approximately 5% cells were GFP-positive, suggesting pIND-ESR1 expression. The GFP-positive cells were collected to 90% purity. These cells were expanded and referred to as 76N-Tert-ESR1.
Luciferase Reporter Assay
T47DKBluc cells were grown in clearing media for and plated in a 24-well plate at cells/well density. After , cells were treated with (, Sigma #E2758), fulvestrant (F, ICI 182, 780, Tocris #1047), 0.5 to BP-3 (Sigma #H36206) or 0.5 to PP (Sigma #P53357). Stock solutions were prepared in DMSO (Sigma #D8418), then diluted to working concentrations in media. Luciferase assays were performed using the Promega Dual-Luciferase Reporter Assay (Promega #E1910). Cells were lysed in 1X Passive Lysis Buffer after treatment for and then stored at . Luciferase activity was determined in lysates by using the Polar Star OPTIMA plate reader (BMG Labtech) and expressed in relative light units (RLU). Treatments were compared with included on the plate, and relative transactivation activity (RTA) is defined as percent transactivation in comparison with .
RT-qPCR
RNA from T47D cells, MCF-7 cells, or flash-frozen fourth mammary gland was isolated with TRIzol (ThermoFisher Scientific #15596018) and Direct-zol RNA MiniPrep Plus (Zymo Research #R2072). cDNA was prepared from of RNA in reaction mix with Protoscript II First Strand cDNA Synthesis Kit (New England Biolabs #E6560S), following the standard protocol provided by the manufacturer. qPCR for TFF1, progesterone receptor (PGR/Pgr), and Amphiregulin (AREG/Areg) was performed using primers in Table S1 (Integrated DNA Technology) and iTaq Universal SYBR Green Supermix (Biorad, #1725121) on CFX96 Real-Time System thermocycler (Bio-Rad). Each run (96-well qPCR plate) included an inter-run calibrator to normalize across experiments. No housekeeping gene was included in the experiment to avoid possible variation due to treatments. Results represent average of three experiments. Data was analyzed with method, and relative fold change in expression of target gene was compared among control and treatments.
Cell Proliferation Assay
T47D cells grown in clearing media for was plated as of cells suspension having 5,000 or 10,000 cells per well on five 96-well plates (one for each day). The 96-well plate had 12 cell-free wells for a blank and 7 wells per treatment on each plate. After , media was changed in appropriate wells on each plate to reach the desired final concentration of (), BP-3 (5, ) or PP (1, ) in the given wells. All plates were maintained in a , 5% incubator until media were exchanged, on one plate per day, for 10% Alamar Blue in plating media. Plates were read at the same time each day at and after media exchange on a BioTek Synergy 2 plate reader (Bio-Tek) at and . Percent Alamar Blue reduction was calculated as per the Alamar Blue protocol:
Immunostaining
T47D, MCF7, 76N-Tert, or 76N-Tert-ESR1 cells were grown in clearing media for and plated on glass uncoated coverslips in 12-well plates with a density of cells/well. After of growth, cells were treated with , 1 or BP-3, and 1 or PP with or without fulvestrant for . For , cells were fixed in ice-cold methanol (100%) for 10 min and quenched with glycine for 15 min. Cells were washed with 1X PBS, blocked in 2% BSA/PBS with 0.1% Triton-X 100 for at room temperature (RT), incubated overnight with anti- antibody (Cell Signaling #9718S), antibody (Santa Cruz Biotechnology #sc-8002) or anti-53BP1 antibody (Abcam #ab36823) at 4°C, followed by with anti-rabbit AlexaFluoro 488–conjugated secondary antibody (Cell Signaling #8889S) or anti-mouse AlexaFluoro 488–conjugated (Cell Signaling #4408S) at RT. For S9.6, cells were fixed in ice-cold 100% methanol for 10 min at , permeabilized in 100% acetone for 1 min at RT, blocked for 30 min in saline sodium citrate pH 7 (SSC, 4X), 3% BSA, and 0.1% Triton-X and incubated with S9.6 antibody (Kerafast #ENH001) for at RT, followed by with anti-mouse AlexaFluoro 596–conjugated secondary antibody (Life Technologies #A11062) or antimouse AlexaFluoro 488–conjugated (Cell Signaling #4408S). For each treatment, two replicates of slides were stained with one set of replicates treated with RNase H (NEB #M0297L) for at 37°C prior to incubation with primary antibody. Stained cells were mounted with Vectashield mounting medium containing DAPI (Vector Laboratories #H-1,200). Slides were imaged at 60X (immersion oil) with Nikon A1 spectral confocal microscope. Analysis of and S9.6 intensity per nucleus or foci per nucleus was calculated using Nikon analysis software, where DAPI was used as a mask for the nucleus.
Western Blot
Cells from MCF7 grown in growth media, 76N-Tert (parental), 76N-Tert-ESR1, and 76N-Tert-ESR1 grown in F-media treated with doxycycline for and 76N-Tert-ESR1 treated with doxycycline and for were lysed with ice-cold RIPA lysis buffer [ Tris–HCl, pH 8.0; NaCl; EDTA; 1% Triton X-100; 1% Sodium deoxycholate; 0.1% SDS; 1% protease inhibitors (Sigma-Aldrich #P8340), 1% phosphatase inhibitor #2 (Sigma-Aldrich #P5726), and 1% phosphatase inhibitor #3 (Sigma-Aldrich #P0044)]. Homogenate was centrifuged at for 15 min at 4°C to remove cellular debris. Protein quantification was performed using BCA protein assay (Thermo Scientific #23225). Equal amounts of protein () were separated by SDS-PAGE on 10% acrylamide under denaturating conditions and then blotted onto PVDF membrane (Millipore #IPVH00010). Nonspecific binding was blocked with 5% nonfat dry milk in TBST (Tris-buffered saline and Tween® 20 containing Tris-HCl, pH 7.5; NaCl; 0.05% Tween® 20) for . The blot was incubated with 1:100 antibody (Abcam #ab16660) overnight at 4°C. After incubation, the blot was washed with TBST and then incubated with HRP-conjugated secondary antibody (1:5000, GE Healthcare #NA934V) for . Bands were detected using enhanced chemiluminescence solution and visualized using G-box imaging system (Syngene). The blot was washed with TBST and incubated with actin antibody (1:5000, Sigma #A1978) overnight at 4°C. After washing with TBST and HRP secondary antibody incubation for (1:5,000, GE Healthcare #NA931C), bands were detected with enhanced chemiluminescence and G-box system. Expected molecular weights were 67 kDa () and 42 kDa ( actin).
Animal Treatment
Forty mature BALB/c female mice (8 wk old) were purchased from Jackson Laboratory and housed in temperature-controlled facilities with a set temperature of and humidity of 30%–70%, alternating day/night light cycle and fed LabChow 5058 ad libitum. All procedures were in accordance with the national guidelines for the care and use of animals and approved by the University of Massachusetts Amherst’s Institutional Animal Care and Use Committee.
The mice were ovariectomized before treatment. Briefly, each mouse was anesthetized with a mix of isoflurane and oxygen. The flanks were shaved, sterilized with povidone-iodine (Betadine) and cleaned with alcohol. An incision was made to the skin on the right flank. The underlying muscle layer was nicked to reveal a small hole through which the ovary was pulled out by grasping the periovarian fat. A Serrefine clamp was used to hold the ovary. After ensuring that the blood vessels were constricted to prevent breeding, the ovary was cut from the uterine horn. The periovarian fat was restored into the peritoneum. The peritoneum was closed with one or two stiches and the skin was closed with wound clips. The procedure was repeated on the contralateral side. The mouse was monitored for a week post procedure, and wound clips were removed after 10 d. After 1 wk of recovery, the mice were randomized to four groups and began an acute oral treatment via pipette with vehicle control (tocopherol-stripped corn oil) () or one of three different compounds (), BP-3 (), and PP () for 4 d. Each mouse was administered of oil per gram of body weight to deliver , BP-3, or PP or vehicle control. For BP-3 and PP, these doses represent the toxicologically no-adverse-effect-level (NOAEL) doses for each compound based on development and reproductive toxicity assays (Scientific Committee on Consumer Products 2005, 2008; Soni et al. 2001).
Six hours prior to sacrifice, all of the mice were treated with a 5-gray (Gy) dose of gamma radiation. Then 2 h before sacrifice, all mice were injected intraperitoneally with body weight of BrdU (Sigma Aldrich; Cat. #B5002) that was previously prepared at in PBS and filter sterilized. The mice were sacrificed using carbon dioxide followed by cervical dislocation. Whole blood was collected by cardiac puncture and tissues were harvested. One of the fourth mammary gland was fixed in 10% NBF and transferred to 70% alcohol prior to paraffin embedding. The other fourth mammary gland was cleared of lymph node and stored in . The whole blood was allowed to coagulate at RT for 20 min and then spun down at for 10 min at to retrieve the serum.
Immunostaining of Mouse Mammary Gland
Freshly cut paraffin-embedded sections were deparaffinized/rehydrated with 100% xylenes 3 times for 5 min each, 2 times with 100% ethanol for 5 min each, 95% ethanol for 3 min, and 70% ethanol for 3 min. Samples were rinsed with PBS. Antigen unmasking was performed by boiling the samples in EDTA for . Samples were cooled down to RT and then treated with SSC 0.2X with gentle shaking at RT for 20 min. Samples were blocked in 3% BSA in PBS with 0.5% Tween® 20 for at RT. Primary antibody incubation was done with monoclonal S9.6 antibody (Kerafast #ENH001) or anti-H2AX antibody (Cell Signaling #9718S) for overnight at 4°C. After primary incubation, samples were washed 3 times with PBS containing 0.5% Tween® 20 and then incubated with anti-mouse AlexaFluoro 488–conjugated (Cell Signaling #4408S) or anti-rabbit AlexaFluoro 488–conjugated secondary antibody (Cell Signaling #8889S) for at RT. Samples were washed 2 times with PBS containing 0.5% Tween® 20 and 2 times with PBS and then mounted with Vectashield mounting medium containing DAPI. Slides were imaged at 60× with Nikon A1 spectral confocal microscope. Analysis of S9.6 intensity per nucleus or foci per nucleus were calculated using Nikon analysis software, where DAPI was used as a mask for the nucleus. IHC for Ki67 was performed on a DakoCytomation autostainer using 1:1,000 D2H10 primary antibody (cell signaling #9027T) and the Envision HRP detection system (Dako). Positive cells were counted using ImageJ software. A total of 1,200 cells were counted per slide to determine percent Ki67 positive.
ELISA
The serum from whole blood that was harvested from all the mice were quantified using a –specific enzyme-linked immunosorbent assay (ELISA) (Calbiotech #ES180S-100).
Statistical Analyses
Unless specified, data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) multiple-range test using GraphPad Prism 8 statistical analysis software or R program (version 3.6.0; R Development Core Team). The difference between control and fulvestrant/RNase H treated groups were evaluated with two-way ANOVA followed by Bonferroni correction. Results are presented as mean ± standard error of the mean (SEM). Data were considered statistically significant at . Growth curves were fitted to linear regression model, and slopes were compared between control and treatment conditions. Slopes and 95% confidence interval are reported in Table 2.
Table 2.
Slopes of growth curve showing effect of estradiol (), benzophenone-3 (BP-3), or propylparaben (PP) on T47D cells.
| Growth Curve | Slope | 95% CI |
|---|---|---|
| Control DMSO | 0.0107 | −0.006811, 0.02821 |
| 0.08495 | 0.06604, 0.1039 | |
| PP | 0.01856 | 0.003943, 0.03318 |
| PP | 0.06387 | 0.05225, 0.07550 |
| BP-3 | 0.0202 | 0.008131, 0.03226 |
| BP-3 | 0.01581 | 0.0009721, 0.03064 |
Results
DNA Damage and TFF1 Gene Expression in Cells Treated with , BP-3, or PP
We monitored foci as a measure of DNA damage in T47D cells treated with the compounds for . A dose-dependent increase in intensity was observed following treatment (Figure 1A). Treatment with either BP-3 or PP also led to an increase in intensity. Treatment with BP-3 at 1 or increased intensity in comparison with the control () although we did not observe a dose-dependent increase ( BP-3 vs BP-3, Figure 1B). PP treatment also resulted in significantly increased intensity at 1 and in comparison with the control (). The intensity due to PP treatment was dose-dependent, similar to that of treatment ( PP vs. PP, ) (Figure 1C). We also observed a dose-dependent increase in nuclear intensity in MCF-7 with treatment of (), BP-3 () and PP () (Figure S1). We confirmed the DNA damage with immunostaining of 53BP1 (P53-binding protein 1), a DNA damage response factor, which localizes to the sites of DNA damage and forms ionization radiation–induced foci. Similar to intensity, we observed dose-dependent increases in 53BP1 nuclear intensity following treatment with () and PP () incomparison with control in both T47D and MCF-7. BP-3 treatment () showed increased nuclear 53BP1 intensity over control in both T47D and MCF-7, but only MCF-7 showed dose-dependent increase (Figure 1D and E).
Figure 1.
Evaluation of DNA damage in cells treated with (), Benzophenone-3 (BP-3), or Propylparaben (PP) for . Immunofluorescence (upper panel) and quantification (lower panel) of nuclear intensity in T47D cells treated with (A) 10 or , (B) 1 or BP-3, and (C) 1 or PP. (D) Immunofluorescence of 53BP1 staining with 10 or , 1 or BP-3, and 1 or PP in T47D (upper panel) and MCF-7 (lower panel). (E) Quantification of nuclear 53BP1 of treatments in (D) in T47D (left panel) and MCF-7 (right panel). ***, * compared with control with treatments using one-way analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) multiple-range test. biological replicates. Scale (A–C), (D). All graphs show mean ± SEM.
The effect of these compounds on was contrasted with the mRNA expression of estrogen-responsive gene TFF1. Treatment with stimulated a 13.1-fold increase in expression of the estrogen-responsive gene TFF1, whereas responses to BP-3 or PP did not differ significantly from the control (Figure 2A). The transcriptional responses to were blocked by treatment with fulvestrant (ICI 182780, ), demonstrating the dependence on ER. Blocking ER with fulvestrant also significantly reduced the effect of on intensity (Figure 2B, ) and inhibited intensity in response to BP-3 (), suggesting that the induction of DNA damage was, in part, dependent on ER. However, the foci induced by and BP-3 was incompletely blocked by fulvestrant in comparison with its inhibition of TFF1 expression.
Figure 2.
TFF1 expression and intensity in T47D cells treated with (), benzophenone-3 (BP-3), or propylparaben (PP) for with or without the ER antagonist fulvestrant. (A) Inhibition of TFF1 expression following treatment of , BP-3, and PP when cotreated with fulvestrant (ICI 182 780, ) compared with , BP-3, and PP treatments without fulvestrant. (B) Quantification of nuclear following cotreatment of fulvestrant () with (), BP-3 (), or PP () compared with (), BP-3 (), or PP () without fulvestrant treatment, respectively. *** compared control to xenoestrogens treatment and ### compared with negative fulvestrant and with positive fulvestrant using multiple comparison for 2-way ANOVA. biological replicates. All graphs show mean ± SEM.
Estrogenic Response in Cells Treated with BP-3 and PP
Reporter assays provide a sensitive means to evaluate estrogenic activity on a minimal promoter, whereas endogenous genes containing estrogen-responsive elements provide physiologically relevant targets. To saturate ER responses in these assays, is sufficient; hence, it was used as positive control that is relevant to physiologic levels () in women (Table 1). T47D-KBluc cells harbor an integrated ERE-luciferase reporter in which BP-3 showed a lowest-observed-effect at with transactivation increasing to a maximum 37% relative transactivation activity (RTA) in comparison with (Figure 3A). In contrast, PP showed 4.7% RTA at and increased to 288% at . To estimate the transactivation activity of the compounds at levels that are relevant to human exposure, we used the published urinary levels of BP-3 and PP (Table 1). At concentrations measured in the 95th percentile of pregnant women, BP-3 had , and PP had RTA (Figure S2, white and black arrows, respectively). Expression of endogenous ER target genes AREG and PGR were also quantified in T47D and MCF-7 cell lines (Figure 3B and C). Treatment of BP-3 and PP at resulted in no significant changes in mRNA expression of AREG and PGR, a concentration that led to significant increases in DNA damage in both T47D and MCF7 cells (Figure 1). Proliferation induced by these compounds was also compared with control treatment to provide an additional measure of their bioactivity (Figure 3D, Table 2). PP stimulated significant proliferation of T47D cells at , but not at PP. However, BP-3 had marginal effect at 5 or . Low concentrations of BP-3 and PP only marginally increased cell numbers in comparison with control.
Figure 3.
Evaluation of estrogen receptor transactivation and proliferation in cells treated with (), benzophenone-3 (BP-3), or propylparaben (PP) for . (A) Transactivation response (in relative light unit, RLU) of T47D-KBluc cells in response to (green), BP-3 (brown), and PP (violet) treatment. Expression of endogenous genes AREG (B) and PGR (C) with (), BP-3 (1 or ), or PP (1 or ) treatment as relative fold change over control in T47D (left panel) and MCF-7 (right panel). * and *** compared control with treatments using one-way analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) multiple-range test. biological replicates. (D) Proliferation of 47D cell as percent of Alamar Blue reduction in response to (), PP (1 or ), BP-3 (), or control. The confidence intervals of the slope are reported in Table 2. All graphs show mean ± SEM.
R-Loop Formation in T47D Cells Treated with , BP-3, or PP
R-loop formation was investigated as a possible mechanism of DNA damage using the S9.6 antibody to specifically detect DNA:RNA hybrids. Although we observed a basal level of R-loop foci in the vehicle-treated control in T47D cells, nuclear S9.6 foci were significantly increased with of BP-3 or PP treatment and comparable with responses with . Addition of RNase H to the cells treated with BP-3 or PP or abolished the S9.6 intensities, confirming the specificity of S9.6 nuclear staining (Figure 4A and B). Similarly, increase of R-loops formation was obtained with , BP-3, or PP treatment of MCF-7 cells, which was abrogated following RNase H addition post fixation (Figure 4C).
Figure 4.
R-loop formation in T47D and MCF7 cells treated with (), benzophenone-3 (BP-3), or propylparaben (PP) or vehicle with or without RNase H. (A) Immunostaining of R-loop with S9.6 antibody and DAPI in T47D cells treated with (), BP-3 () or PP () without and with RNase H treatment following fixation. Scale . (B) Quantification of the nuclear S9.6 intensity in T47D. (C) Quantification of nuclear S9.6 intensity in MCF-7. *** compared control with xenoestrogens treatment and ### compared negative RNase H and with positive RNase H using multiple comparison for 2-way ANOVA. biological replicates. All graphs show mean ± SEM.
R-Loop Formation in Normal Breast Epithelial Cell Line Treated with , BP-3, or PP
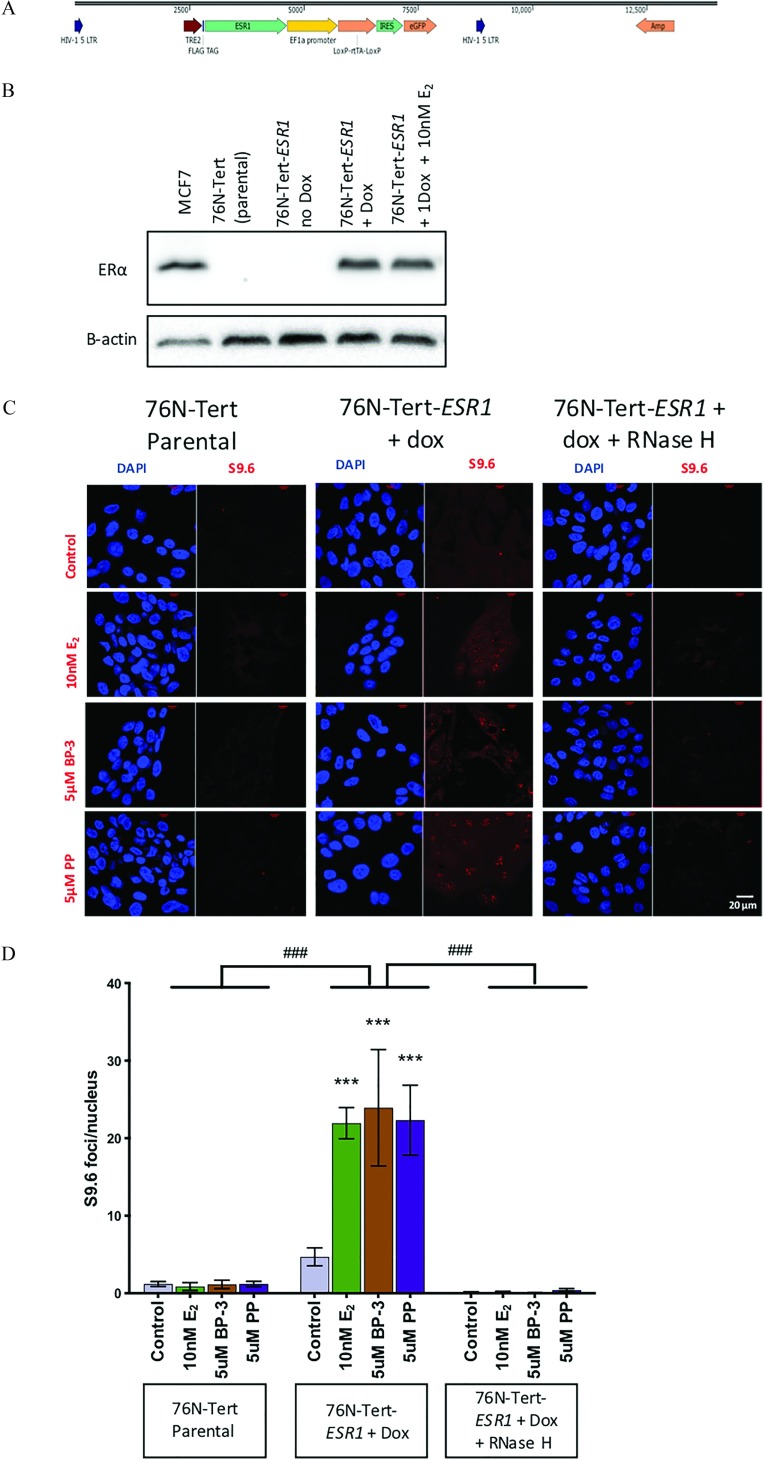
Next, we asked whether R-loops form in normal breast epithelial cells in response to exposures of BP-3 and PP (Figure 5). The 76N-Tert cells do not express endogenous ESR1, providing a null background to test R-loops. The cells were stably infected with an inducible human ESR1 (pINDUCER-ESR1) (Figure 5A) expression in 76N-Tert-ESR1 was confirmed with Western blot (Figure 5B). MCF-7 cell lysate was used as a positive control. Immunofluorescence showed 90% of the 76N-Tert-ESR1 cell population were GFP-positive ( expressing) (Figure S3).
Figure 5.
Characterization of 76N-Tert-ESR1 and R-loop formation in 76N-Tert-ESR1 following treatment with (), benzophenone-3 (BP-3) or propylparaben (PP) with and without RNase H. (A) Map of pIN-ESR1 construct ESR1 insertion next to doxycycline(dox) inducible promoter. (B) Western blot (upper panel) with MCF-7 as positive control (lane 1), 76N-Tert parental (lane 2), 76N-Tert-ESR1 without dox (lane 3), 76N-Tert-ESR1 with dox (lane 4), and 76N-Tert-ESR1 with dox and () treatment and as loading control (lower panel). (C) Immunostaining with S9.6 antibody and DAPI with , BP-3, or PP treatment to parental 76N-Tert cells (upper panel), to 76N-Tert-ESR1 with dox induction (middle panel) without or with RNase H treatment (lower panel). Scale . (D) Quantification of nuclear S9.6 intensity in (C). *** compared control with xenoestrogens treatment and ### compared among 76N-Tert Parental, 76N-Tert with dox and () negative RNase H and 76N-Tert with dox and () positive RNase H using multiple comparison for 2-way ANOVA. biological replicates. All graphs show mean ± SEM.
In the parental 76N-Tert cell line, which does not express , treatment with , BP-3, or PP showed low nuclear S9.6 staining. After induction of with doxycycline, BP-3 or PP increased the number of nuclear S9.6 foci significantly over vehicle-treated control and was comparable with treatment. RNase H treatment reduced nuclear S9.6 foci in treated as well as BP-3 or PP treated 76N-Tert-ESR1 cell line induced with doxycycline (, Figure 5C and D).
Evaluation of R-Loop Formation and DNA Damage in Mice Treated with , BP-3, or PP
To evaluate the relevance of exposure to xenoestrogens in vivo, we treated ovariectomized BALB/c mice orally with (), BP-3 () or PP () for 4 d (Figure 6A). These doses were used in experiments evaluating effects of chronic exposures on mammary gland development (LaPlante et al. 2018). We observed 3.8-fold higher nuclear S9.6 staining in the mammary epithelium of -treated animals over control-treated animals. Exposure to BP-3 also induced 2.5-fold higher nuclear S9.6 staining in the mammary epithelial cells, whereas PP induced 3.8-fold higher in comparison with control-treated animals (Figure 6B and C). Nuclear intensity in the mammary gland of - and BP-3-treated animals was significantly higher than animals treated with vehicle control (Figure 6D). Although oral treatment of stimulated proliferation as shown by higher Ki67 straining and transcriptional activation of ER-target genes (Areg and Pgr) in the mammary gland, neither BP-3 nor PP elicited significant responses (Figure 6 E–H). Similarly, elevated serum levels of and uterine weight was observed only in -treated mice (Figure S4).
Figure 6.
Acute exposure of xenoestrogens in mice. (A) Schematic of experimental design and exposure period. (B) Immunostaining of mouse mammary epithelium with S9.6 antibody harvested from mice treated with , BP-3, or PP. Each image shows a ductal structure with luminal and myo-epithelial cell nucleus (blue) and R-loop (green) inside the nucleus. Scale . Quantification of the immunostaining data for S9.6 (C) and (D). Expression of Areg (E) and Pgr (F) from mouse mammary gland. biological replicates. Ki67 straining of luminal epithelial cells (G) and percent of Ki67 strained cells per luminal cells counted (H). Scale [Number of biological replicates (n): control (5), (8), BP-3 (12), PP (10)] ***, ** compared control with treatments using one-way analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) multiple-range test. All graphs show mean ± SEM.
Discussion
Exposure of xenoestrogens was implicated in breast cancer risk (Pastor-Barriuso et al. 2016) as well as resistance to breast cancer treatment (Goodson et al. 2011; Warth et al. 2018) due to their endocrine actions. The median urinary level of BP-3 was , and PP was in nonpregnant women in participating in the NHANES by the CDC (Calafat et al. 2010; Woodruff et al. 2011). The serum levels of BP-3 was reported to be approximately () following exposure in women (Janjua et al. 2004; Matta et al. 2019; Tarazona et al. 2013). In addition, the urinary concentrations of xenoestrogens observed in pregnant women were higher than in the general population, with median urinary concentrations of BP-3 and PP being and , respectively, and the 95th percentile concentrations in pregnant women being BP-3 and PP (Table 1). This finding raises the possibility that women may have higher exposure during pregnancy due to use of creams and lotions or that absorption and metabolism may be altered in pregnancy. These compounds were also found in normal tissues of women undergoing mastectomy for primary breast cancer (Barr et al. 2012; Barr et al. 2018) and in milk collected during the period of sunscreen use from three different cohorts of mothers of singleton children (Schlumpf et al. 2010). However, based on measures of transcriptional activity in MCF7 human breast cancer cell lines (Byford et al. 2002; Kerdivel et al. 2013), typical exposures to BP-3 and PP would appear to pose a minimal risk for breast cancer through ER-mediated transcriptional activation of target genes.
Estrogens and their metabolites have been shown to induce direct DNA damage. However, DNA damage by catechol estrogens from ER-negative cell lines requires concentrations that are 100-fold greater than the average circulating concentrations in women (Cavalieri and Rogan 2016; Savage et al. 2014; Xu et al. 2007). BP-3 and PP were shown to have the potential to cause DNA damage independent of ER transactivation, based on experiments on ER-negative cell lines. For example, treatment of BP-3 () induced foci in normal human keratin cell lines (Kim et al. 2018) and PP () showed 8-Hydroxy-2-deoxyguanosine (8-OHdG) release in Vero cells (derived from monkey kidney) (Pérez Martín et al. 2010). However, these levels exceeded typical concentrations measured in human populations (Table 1).
In the breast, epithelial cells with functional , we observed DNA damage at physiologic concentrations of . BP-3 and PP also caused DNA damage at low concentrations () (Figure 1). Both the nuclear and 53BP1 foci were diminished by fulvestrant, suggesting dependency of DNA damage. At these low concentrations ( of PP and of BP-3), we did not observe ER-mediated transcriptional response in target genes. Instead, we observed R-loop formation. We also observed increases in R-loops and in the mammary epithelial cells of ovariectomized BALB/c mice orally treated with BP-3 or PP at doses designed to model environmental exposures in humans (Figure 6D). The doses of BP-3 and PP used in mice were not sufficient to affect transcription of Areg or Pgr (Figure 6E–F) or proliferation of mammary epithelium (Figure 6G–H) in comparison with control treatments, nor were they sufficient to alter uterine weights in comparison with the control treatment in ovariectomized mice (Figure S4). This finding for BP-3 was supported by a previous study (LaPlante et al. 2018). These results with BP-3 and PP are consistent with the idea that in mammary epithelial cells of human and mice, the formation of R-loops and DNA damage is ER-dependent but is separable from gene transcription and proliferative responses.
ER-mediated DNA double-strand breaks was shown to form by collision of R-loop formed during active transcription (cotranscriptional R-loop) and replication fork in MCF7 cells (Stork et al. 2016). Alternatively, R-loop formation can occur with RNA Polymerase II pausing, which results in no increase of gene expression but leads to DNA damage (Hatchi et al. 2015; Shivji et al. 2018; Zhang et al. 2017). Indeed, our results showed that, BP-3 and PP induced formation of R-loops and DNA damage (Figure 7) but did not lead to detectable increases in full-length transcripts of TFF1, AREG, or PGR.
Figure 7.
A schematic model for ER-dependent DNA damage. or xenoestrogens binding to the ER recruit ER to the estrogen response element (ERE) in the promoter and forms R-loop. Persistence of R-loop in the promoter introduces DNA damage.
Experiments performed using a normal breast epithelial cell line 76N-Tert expressing inducible treated with , BP-3, and PP provided a) additional evidence that the R-loop formation and DNA damage were and b) that normal breast epithelial cells were susceptible to DNA damage by xenoestrogens. This finding raises the possibility that a subset of women bearing variants of R-loop processing factors may be particularly susceptible to the genotoxic effects of xenoestrogens such as BP-3 and PP. More than 300 R-loop binding proteins have been identified (Wang et al. 2018). A number of such factors were recently shown to be involved in the resolution of R-loops to limit DNA damage, including TopI (Tuduri et al. 2009), BRCA1 (Hatchi et al. 2015), BRCA2 (Shivji et al. 2018), SETX (Cohen et al. 2018; Hatchi et al. 2015), Aquaris (Sollier et al. 2014), THO/THREX complex (Bhatia et al. 2014; Gómez‐González et al. 2011), BuGZ, and Bub (Wan et al. 2015). For example, recruitment of BRCA1/SETX was important for R-loop mediated transcriptional termination. As a consequence, the mutational rate of termination regions where BRCA/SETX colocalize was higher in BRCA1-deficient tumors in comparison with BRCA1-WT tumors (Hatchi et al. 2015). Premalignant breast lesions such as atypical hyperplasia expressed higher levels of (Gregory et al. 2019) and thus may be especially sensitive to the genotoxic effects of these xenoestrogens. Therefore, limiting exposure to personal care products and foods containing these chemicals may be valuable for this subset of women.
However, the present studies do not show a direct risk of exposure to these compounds on subsequent breast cancer. Although chronic exposure to low levels of DNA damage have the potential to induce mutations that either initiate or promote carcinogenesis, the experiments were not designed to demonstrate a causal effect of BP-3 or PP on mammary tumors or breast carcinogenesis. The DNA damage observed was associated with the formation of R-loops, but it is unclear whether also contributes to the formation of R-loops or may mitigate this. Although many tissues express ERs, they vary in the levels of and as well as expression of DNA repair factors and proficiency of resolving R-loops. Therefore, this mechanism of DNA damage may be limited to the breast epithelium of a subset of individuals. Also unclear is how combinations of environmental xenoestrogens may interact to augment or dissipate the genotoxicity through competing actions on ER. Nonetheless, the data presented here reveal a need to consider the unique potential for genotoxicity of environmental xenoestrogens in tissues expressing ERs.
These studies demonstrated that xenoestrogens possessed the potential for genotoxic activity that was mediated by through the formation of R-loops and DNA double-strand breaks. These genotoxic effects were observed at concentrations well below those necessary for detectable transcriptional activation. Therefore, R-loop forming capacity provides a valuable end point to consider when evaluating the safety and activity of environmental chemicals. The inducible expression of in normal breast cells provides a tool with which to quantify the variation in sensitivity to these compounds among individuals and to determine whether a subset of individuals is preferentially susceptible to the genotoxic activities.
Supplementary Material
Acknowledgments
Research reported in this publication was supported, in part, by the National Institute of Environmental Health Sciences of the National Institutes of Health under award number U01ES026140 (D.J.J., S.S.S.), the Department of Defense under contract #W81XWH-15-1-0217 (D.J.J.), the Rays of Hope Center for Breast Cancer Research (D.J.J.), and University Grants Commission (India) for Raman Fellowship for postdoctoral research (P.D.M.). Protocol for S9.6 immunofluorescence in cells was kindly provided by G. Capranico, Professor of Molecular Biology, University of Bologna, Italy. The microscopy data was gathered in the Light Microscopy Facility and Nikon Center of Excellence at the Institute for Applied Life Sciences, UMass Amherst, with support from the Massachusetts Life Sciences Center and help from J. Chambers. The authors thank J. Kane and the Applied Molecular Biotechnology Program for assistance in preparing expression vectors. The authors also thank A. Symington for valuable discussions throughout the project.
Footnotes
Supplemental Material is available online (https://doi.org/10.1289/EHP5221).
Both authors contributed equally to this work.
L.N.V. has received funding from the Cornell Douglas Foundation and Paul G. Allen Foundation. She has been reimbursed for travel expenses by several organizations, including SweTox, the European Commission, the Mexican Endocrine Society, Advancing Green Chemistry, CropLife America, Beautycounter (Counter Brands, LLC), and many universities, to speak about endocrine disruptors. All other authors declare they have no actual or potential competing financial interests.
Note to readers with disabilities: EHP strives to ensure that all journal content is accessible to all readers. However, some figures and Supplemental Material published in EHP articles may not conform to 508 standards due to the complexity of the information being presented. If you need assistance accessing journal content, please contact ehponline@niehs.nih.gov. Our staff will work with you to assess and meet your accessibility needs within 3 working days.
References
- Alamer M, Darbre PD. 2018. Effects of exposure to six chemical ultraviolet filters commonly used in personal care products on motility of MCF‐7 and MDA‐MB‐231 human breast cancer cells in vitro. J Appl Toxicol 38(2):148–159-159, PMID: 28990245, 10.1002/jat.3525. [DOI] [PubMed] [Google Scholar]
- Andersson S, Davis D, Dahlbäck H, Jörnvall H, Russell DW. 1989. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem 264(14):8222–8229, PMID: 2722778. [PubMed] [Google Scholar]
- Barr L, Alamer M, Darbre PD. 2018. Measurement of concentrations of four chemical ultraviolet filters in human breast tissue at serial locations across the breast. J Appl Toxicol 38(8):1112–1120, PMID: 29658634, 10.1002/jat.3621. [DOI] [PubMed] [Google Scholar]
- Barr L, Metaxas G, Harbach CA, Savoy LA, Darbre PD. 2012. Measurement of paraben concentrations in human breast tissue at serial locations across the breast from axilla to sternum. J Appl Toxicol 32(3):219–232, PMID: 22237600, 10.1002/jat.1786. [DOI] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. 2014. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511(7509):362–365, PMID: 24896180, 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- Buteau-Lozano H, Ancelin M, Lardeux B, Milanini J, Perrot-Applanat M. 2002. Transcriptional regulation of vascular endothelial growth factor by estradiol and tamoxifen in breast cancer cells. Cancer Res 62(17):4977–4984, PMID: 12208749. [PubMed] [Google Scholar]
- Byford JR, Shaw LE, Drew MG, Pope GS, Sauer MJ, Darbre PD. 2002. Oestrogenic activity of parabens in MCF7 human breast cancer cells. J Steroid Biochem Mol Biol 80(1):49–60, PMID: 11867263, 10.1016/S0960-0760(01)00174-1. [DOI] [PubMed] [Google Scholar]
- Calafat AM, Wong LY, Ye X, Reidy JA, Needham LL. 2008. Concentrations of the sunscreen agent benzophenone-3 in residents of the United States: National Health and Nutrition Examination Survey 2003–2004. Environ Health Perspect 116(7):893–897, PMID: 18629311, 10.1289/ehp.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafat AM, Ye X, Wong LY, Bishop AM, Needham LL. 2010. Urinary concentrations of four parabens in the U.S. population: NHANES 2005–2006. Environ Health Perspect 118(5):679–685, PMID: 20056562, 10.1289/ehp.0901560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri EL, Rogan EG. 2016. Depurinating estrogen-DNA adducts, generators of cancer initiation: their minimization leads to cancer prevention. Clin Transl Med 5(1):12, PMID: 26979321, 10.1186/s40169-016-0088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Jeffrey C, Yiwei Z, Anjana VY, Sambasiva MR, Yi-Jun JZ. 2002. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem 277(32):28624–30, PMID: 12039952, 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- Clarke RB, Howell A, Anderson E. 1997. Estrogen sensitivity of normal human breast tissue in vivo and implanted into athymic nude mice: analysis of the relationship between estrogen-induced proliferation and progesterone receptor expression. Breast Cancer Res Treat 45(2):121–133, PMID: 9342437, 10.1023/a:1005805831460. [DOI] [PubMed] [Google Scholar]
- Cohen S, Puget N, Lin YL, Clouaire T, Aguirrebengoa M, Rocher V. 2018. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat Commun 9(1):533, PMID: 29416069, 10.1038/s41467-018-02894-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings SR, Eckert S, Krueger KA, Grady D, Powles TJ, Cauley JA. 1999. The effect of raloxifene on risk of breast cancer in postmenopausal women: results from the MORE randomized trial. Multiple Outcomes of Raloxifene Evaluation. JAMA 281:2189–2197, PMID: 10376571, 10.1001/jama.281.23.2189. [DOI] [PubMed] [Google Scholar]
- Cuzick J, International Breast Cancer Intervention Study . 2001. A brief review of the International Breast Cancer Intervention Study (IBIS), the other current breast cancer prevention trials, and proposals for future trials. Ann NY Acad Sci 949:123–133, PMID: 11795344, 10.1111/j.1749-6632.2001.tb04010.x. [DOI] [PubMed] [Google Scholar]
- Dallal CM, Tice JA, Buist DS, Bauer DC, Lacey JV Jr, Cauley JA, et al. 2014. Estrogen metabolism and breast cancer risk among postmenopausal women: a case-cohort study within B∼FIT. Carcinogenesis 35(2):346–355, PMID: 24213602, 10.1093/carcin/bgt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EBCTCG (Early Breast Cancer Trialists' Collaborative Group). 2015. Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet 386:1341–1352, PMID: 26211827, 10.1016/S0140-6736(15)61074-1. [DOI] [PubMed] [Google Scholar]
- EC (European Commission). 2014. Commission Regulation (EU) No. 1004/2014 of 18 September 2014 amending Annex V to Regulation (EC) No. 1223/2009 of the European Parliament and of the Council on cosmetic products Text with EEA relevance. https://op.europa.eu/en/publication-detail/-/publication/a22d3948-4545-11e4-a0cb-01aa75ed71a1/language-en [accessed 5 February 2019].
- EC. 2017. Commission Regulation (EU) 2017/238 of 10 February 2017 amending Annex VI to Regulation (EC) No. 1223/2009 of the European Parliament and of the Council on cosmetic products. http://data.europa.eu/eli/reg/2017/238/oj [accessed 5 February 2019].
- Feinleib M. 1968. Breast cancer and artificial menopause: a cohort study. J Natl Cancer Inst 41(2):315–329, PMID: 5671283. [PubMed] [Google Scholar]
- Fuhrman BJ, Schairer C, Gail MH, Boyd-Morin J, Xu X, Sue LY, et al. 2012. Estrogen metabolism and risk of breast cancer in postmenopausal women. J Natl Cancer Inst 104(4):326–339, PMID: 22232133, 10.1093/jnci/djr531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. 2009. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462(7269):58–64-64, PMID: 19890323, 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussell KC, Udasin RG, Smith PJ, Gallo MA, Laskin JD. 2011. Catechol metabolites of endogenous estrogens induce redox cycling and generate reactive oxygen species in breast epithelial cells. Carcinogenesis 32(8):1285–1293, PMID: 21665890, 10.1093/carcin/bgr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-González B, García-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marín A, et al. 2011. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J 30(15):3106–3119, PMID: 21701562, 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez H, Farbrot A, Larkö O, Wennberg AM. 2006. Percutaneous absorption of the sunscreen benzophenone-3 after repeated whole-body applications, with and without ultraviolet irradiation. Br J Dermatol 154(2):337–340, PMID: 16433806, 10.1111/j.1365-2133.2005.07007.x. [DOI] [PubMed] [Google Scholar]
- Goodson WH 3rd, Luciani MG, Sayeed SA, Jaffee IM, Moore DH 2nd, Dairkee SH. 2011. Activation of the mTOR pathway by low levels of xenoestrogens in breast epithelial cells from high-risk women. Carcinogenesis 32(11):1724–1733, PMID: 21890461, 10.1093/carcin/bgr196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss PE, Ingle JN, Alés-Martínez JE, Cheung AM, Chlebowski RT, Wactawski-Wende J, et al. 2011. Exemestane for breast-cancer prevention in postmenopausal women. N Engl J Med 364(25):2381–2391, PMID: 21639806, 10.1056/NEJMoa1103507. [DOI] [PubMed] [Google Scholar]
- Gregory KJ, Roberts AL, Conlon EM, Mayfield JA, Hagen MJ, Crisi GM, et al. 2019. Gene expression signature of atypical breast hyperplasia and regulation by SFRP1. Breast Cancer Res 21(1):76, PMID: 31248446, 10.1186/s13058-019-1157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, et al. 2015. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven dna damage repair. Mol Cell 57(4):636–647, PMID: 25699710, 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson BE, Feigelson HS. 2000. Hormonal carcinogenesis. Carcinogenesis 21(3):427–433, PMID: 10688862, 10.1093/carcin/21.3.427. [DOI] [PubMed] [Google Scholar]
- Henderson BE, Ross R, Bernstein L. 1988. Estrogens as a cause of human cancer: the Richard and Hinda Rosenthal Foundation award lecture. Cancer Res 48(2):246–253, PMID: 2825969. [PubMed] [Google Scholar]
- Huang Y, Fernandez SV, Goodwin S, Russo PA, Russo IH, Sutter TR, et al. 2007. Epithelial to mesenchymal transition in human breast epithelial cells transformed by 17beta-estradiol. Cancer Res 67(23):11147–11157, PMID: 18056439, 10.1158/0008-5472.CAN-07-1371. [DOI] [PubMed] [Google Scholar]
- Janjua NR, Mogensen B, Andersson A-M, Petersen JHH, Henriksen M, Skakkebaek NE, et al. 2004. Systemic absorption of the sunscreens benzophenone-3, octyl-methoxycinnamate, and 3-(4-methyl-benzylidene) camphor after whole-body topical application and reproductive hormone levels in humans. J Invest Dermatol 123(1):57–61, PMID: 15191542, 10.1111/j.0022-202X.2004.22725.x. [DOI] [PubMed] [Google Scholar]
- Kerdivel G, Guevel R, Habauzit D, Brion F, Ait-Aissa S, Pakdel F. 2013. Estrogenic potency of benzophenone UV filters in breast cancer cells: proliferative and transcriptional activity substantiated by docking analysis. PLoS One 8(4):e60567, PMID: 23593250, 10.1371/journal.pone.0060567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S, Dash PR, Darbre PD. 2014. Exposure to parabens at the concentration of maximal proliferative response increases migratory and invasive activity of human breast cancer cells in vitro. J Appl Toxicol 34(9):1051–1059, PMID: 24652746, 10.1002/jat.3003. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Lee E, Lee M, Ahn S, Kim J, Liu J, et al. 2018. Phosphodiesterase 4B plays a role in benzophenone-3-induced phototoxicity in normal human keratinocytes. Toxicol Appl Pharmacol 338:174–181, PMID: 29183759, 10.1016/j.taap.2017.11.021. [DOI] [PubMed] [Google Scholar]
- Kunisue T, Chen Z, Louis GM, Sundaram R, Hediger ML, Sun L, et al. 2012. Urinary concentrations of benzophenone-type UV filters in U.S. women and their association with endometriosis. Environ Sci Technol 46(8):4624–4632, PMID: 22417702, 10.1021/es204415a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlante CD, Bansal R, Dunphy KA, Jerry DJ, Vandenberg LN. 2018. Oxybenzone alters mammary gland morphology in mice exposed during pregnancy and lactation. J Endocr Soc 2(8):903–921, PMID: 30057971, 10.1210/js.2018-00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C, Kannan K. 2014. Widespread occurrence of benzophenone-type UV light filters in personal care products from China and the United States: an assessment of human exposure. Environ Sci Technol 48(7):4103–4109, PMID: 24588714, 10.1021/es405450n. [DOI] [PubMed] [Google Scholar]
- Liehr JG, Sirbasku DA, Jurka E, Randerath K, Randerath E. 1988. Inhibition of estrogen-induced renal carcinogenesis in male Syrian hamsters by tamoxifen without decrease in DNA adduct levels. Cancer Res 48(4):779–783, PMID: 3338075. [PubMed] [Google Scholar]
- Majewski AR, Chuong LM, Neill HM, Roberts AL, Jerry DJ, Dunphy KA. 2018. Sterilization of silastic capsules containing 17β-estradiol for effective hormone delivery in Mus musculus. J Am Assoc Lab Anim Sci 57(6):679–685, PMID: 30314533, 10.30802/AALAS-JAALAS-18-000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino S, Cauley JA, Barrett-Connor E, Powles TJ, Mershon J, Disch D, et al. 2004. Continuing outcomes relevant to Evista: breast cancer incidence in postmenopausal osteoporotic women in a randomized trial of raloxifene. J Natl Cancer Inst 96(23):1751–1761, PMID: 15572757, 10.1093/jnci/djh319. [DOI] [PubMed] [Google Scholar]
- Matta MK, Zusterzeel R, Pilli NR, Patel V, Volpe DA, Florian J, et al. 2019. Effect of sunscreen application under maximal use conditions on plasma concentration of sunscreen active ingredients: a randomized clinical trial. JAMA 321(21):2082–2091, PMID: 31058986, 10.1001/jama.2019.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, et al. 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA 108(9):3665–3670, PMID: 21307310, 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley JA, Brueggemeier RW. 2004. Estrogen receptor-mediated regulation of oxidative stress and DNA damage in breast cancer. Carcinogenesis 25(1):3–9, PMID: 14514655, 10.1093/carcin/bgg175. [DOI] [PubMed] [Google Scholar]
- Molins-Delgado D, Olmo-Campos MDM, Valeta-Juan G, Pleguezuelos-Hernández V, Barceló D, Díaz-Cruz MS. 2018. Determination of UV filters in human breast milk using turbulent flow chromatography and babies' daily intake estimation. Environ Res 161:532–539, PMID: 29232646, 10.1016/j.envres.2017.11.033. [DOI] [PubMed] [Google Scholar]
- Moore SC, Matthews CE, Ou Shu X, Yu K, Gail MH, Xu X. 2016. Endogenous estrogens, estrogen metabolites, and breast cancer risk in postmenopausal Chinese women. J Natl Cancer Inst 108(10), PMID: 27193440, 10.1093/jnci/djw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove EA, Sutherland RL. 1994. Cell cycle control by steroid hormones. Semin Cancer Biol 5(5):381–389, PMID: 7849266. [PubMed] [Google Scholar]
- Okereke CS, Abdel-Rhaman MS, Friedman MA. 1994. Disposition of benzophenone-3 after dermal administration in male rats. Toxicol Lett 73(2):113–122, PMID: 8048080, 10.1016/0378-4274(94)90101-5. [DOI] [PubMed] [Google Scholar]
- O'Leary P, Boyne P, Flett P, Beilby J, James I. 1991. Longitudinal assessment of changes in reproductive hormones during normal pregnancy. Clin Chem 37(5):667–672, PMID: 1827758. [PubMed] [Google Scholar]
- Pastor-Barriuso R, Fernández MF, Castaño-Vinyals G, Whelan D, Pérez-Gómez B, Llorca J, et al. 2016. Total effective xenoestrogen burden in serum samples and risk for breast cancer in a population-based multicase-control study in Spain. Environ Health Perspect 124(10):1575–1582, PMID: 27203080, 10.1289/EHP157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Martín J, Peropadre A, Herrero Ó, Freire P, Labrador V, Hazen M. 2010. Oxidative DNA damage contributes to the toxic activity of propylparaben in mammalian cells. Mutat Res/Genetic Toxicol Environ Mutagenesis 702(1):86–91, PMID: 20682357, 10.1016/j.mrgentox.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Periyasamy M, Patel H, Lai CF, Nguyen V, Nevedomskaya E, Harrod A, et al. 2015. APOBEC3B-mediated cytidine deamination is required for estrogen receptor action in breast cancer. Cell Rep 13(1):108–121, PMID: 26411678, 10.1016/j.celrep.2015.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippat C, Wolff MS, Calafat AM, Ye X, Bausell R, Meadows M, et al. 2013. Prenatal exposure to environmental phenols: concentrations in amniotic fluid and variability in urinary concentrations during pregnancy. Environ Health Perspect 121(10):1225–1231, PMID: 23942273, 10.1289/ehp.1206335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston-Martin S, Pike MC, Ross RK, Jones PA, Henderson BE. 1990. Increased cell division as a cause of human cancer. Cancer Res 50(23):7415–7421, PMID: 2174724. [PubMed] [Google Scholar]
- Rajapakse N, Butterworth M, Kortenkamp A. 2005. Detection of DNA strand breaks and oxidized DNA bases at the single-cell level resulting from exposure to estradiol and hydroxylated metabolites. Environ Mol Mutagen 45(4):397–404, PMID: 15662657, 10.1002/em.20104. [DOI] [PubMed] [Google Scholar]
- Roy D, Liehr JG. 1999. Estrogen, DNA damage and mutations. Mutat Res/Fundamental and Mol Mech Mutagenesis 424(1–2):107–115, PMID: 10064854, 10.1016/S0027-5107(99)00012-3. [DOI] [PubMed] [Google Scholar]
- Russo J, Hasan Lareef M, Balogh G, Guo S, Russo IH. 2003. Estrogen and its metabolites are carcinogenic agents in human breast epithelial cells. J Steroid Biochem Mol Biol 87(1):1–25, PMID: 14630087, 10.1016/s0960-0760(03)00390-x. [DOI] [PubMed] [Google Scholar]
- Sampson JN, Falk RT, Schairer C, Moore SC, Fuhrman BJ, Dallal CM, et al. 2017. Association of estrogen metabolism with breast cancer risk in different cohorts of postmenopausal women. Cancer Res 77(4):918–925, PMID: 28011624, 10.1158/0008-5472.CAN-16-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen R, Cavalieri E, Rogan E, Russo J, Guttenplan J, Ingle J, et al. 2009. Estrogen mediation of breast tumor formation involves estrogen receptor-dependent, as well as independent, genotoxic effects. Ann NY Acad Sci 1155:132–140, PMID: 19250200, 10.1111/j.1749-6632.2008.03685.x. [DOI] [PubMed] [Google Scholar]
- Savage KI, Matchett KB, Barros EM, Cooper KM, Irwin GW, Gorski JJ, et al. 2014. BRCA1 deficiency exacerbates estrogen-induced DNA damage and genomic instability. Cancer Res 74(10):2773–2784, PMID: 24638981, 10.1158/0008-5472.CAN-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotz N, Kim G-J, Jäger S, Günther S, Lamy E. 2017. In vitro observations and in silico predictions of xenoestrogen mixture effects in T47D-based receptor transactivation and proliferation assays. Toxicol In Vitro 45(Pt 1):146–157, PMID: 28855101, 10.1016/j.tiv.2017.08.017. [DOI] [PubMed] [Google Scholar]
- Schlumpf M, Cotton B, Conscience M, Haller V, Steinmann B, Lichtensteiger W. 2001. In vitro and in vivo estrogenicity of UV screens. Environ Health Perspect 109(3):239–244, PMID: 11333184, 10.1289/ehp.01109239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumpf M, Kypke K, Wittassek M, Angerer J, Mascher H, Mascher D, et al. 2010. Exposure patterns of UV filters, fragrances, parabens, phthalates, organochlor pesticides, PBDEs, and PCBs in human milk: correlation of UV filters with use of cosmetics. Chemosphere 81(10):1171–1183, PMID: 21030064, 10.1016/j.chemosphere.2010.09.079. [DOI] [PubMed] [Google Scholar]
- Schock H, Zeleniuch-Jacquotte A, Lundin E, Grankvist K, Lakso HÅ, Idahl A, et al. 2016. Hormone concentrations throughout uncomplicated pregnancies: a longitudinal study. BMC Pregnancy Childbirth 16(1):146, PMID: 27377060, 10.1186/s12884-016-0937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scientific Committee on Consumer Products. 2005. Extended Opinion on the Safety Evaluation of Parabens. https://ec.europa.eu/health/ph_risk/committees/04_sccp/docs/sccp_o_019.pdf [accessed 11 August 2019].
- Scientific Committee on Consumer Products. 2008. Opinion on benzophenone-3. https://ec.europa.eu/health/ph_risk/committees/04_sccp/docs/sccp_o_159.pdf [accessed 11 August 2019].
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. 2000. Cofactor dynamics and sufficiency in estrogen receptor–regulated transcription. Cell 103(6):843–852, PMID: 11136970, 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Shivji MKK, Renaudin X, Williams CH, Venkitaraman AR. 2018. BRCA2 regulates transcription elongation by RNA polymerase II to prevent R-loop accumulation. Cell Rep 22(4):1031–1039, PMID: 29386125, 10.1016/j.celrep.2017.12.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodin D. 2017. Regulatory risk assessments: is there a need to reduce uncertainty and enhance robustness? Update on propylparaben in relation to its EU regulatory status. Hum Exp Toxicol 36(10):1007–1014, PMID: 28695774, 10.1177/0960327117718042. [DOI] [PubMed] [Google Scholar]
- Sollier J, Stork C, García-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. 2014. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 56(6):777–785, PMID: 25435140, 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni MG, Burdock GA, Taylor SL, Greenberg NA. 2001. Safety assessment of propyl paraben: a review of the published literature. Food Chem Toxicol 39(6):513–532, PMID: 11346481, 10.1016/s0278-6915(00)00162-9. [DOI] [PubMed] [Google Scholar]
- Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chédin F, et al. 2016. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. eLife 5:e17548, PMID: 27552054, 10.7554/eLife.17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona I, Chisvert A, Salvador A. 2013. Determination of benzophenone-3 and its main metabolites in human serum by dispersive liquid–liquid microextraction followed by liquid chromatography tandem mass spectrometry. Talanta 116:388–395, PMID: 24148420, 10.1016/j.talanta.2013.05.075. [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbé L, Conti C, Tourrière H, Holtgreve-Grez H, Jauch A, et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11(11):1315–1324, PMID: 19838172, 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladusic EA, Hornby AE, Guerra-Vladusic FK, Lakins J, Lupu R. 2000. Expression and regulation of estrogen receptor beta in human breast tumors and cell lines. Oncol Rep 7(1):157–167, PMID: 10601611, 10.3892/or.7.1.157. [DOI] [PubMed] [Google Scholar]
- Wan Y, Zheng X, Chen H, Guo Y, Jiang H, He X, et al. 2015. Splicing function of mitotic regulators links R-loop–mediated DNA damage to tumor cell killing. J Cell Biol 209(2):235–246, PMID: 25918225, 10.1083/jcb.201409073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chandrasena ER, Yuan Y, Peng KW, van Breemen RB, Thatcher GR, et al. 2010. Redox cycling of catechol estrogens generating apurinic/apyrimidinic sites and 8-oxo-deoxyguanosine via reactive oxygen species differentiates equine and human estrogens. Chem Res Toxicol 23(8):1365–1373, PMID: 20509668, 10.1021/tx1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IX, Grunseich C, Fox J, Burdick J, Zhu Z, Ravazian N, et al. 2018. Human proteins that interact with RNA/DNA hybrids. Genome Res 28(9):1405–1414, PMID: 30108179, 10.1101/gr.237362.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth B, Raffeiner P, Granados A, Huan T, Fang M, Forsberg EM, et al. 2018. Metabolomics reveals that dietary xenoestrogens alter cellular metabolism induced by palbociclib/letrozole combination cancer therapy. Cell Chem Biol 25(3):291–300, PMID: 29337187, 10.1016/j.chembiol.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Kojima H, Takeuchi S, Uramaru N, Sanoh S, Sugihara K, et al. 2015. Metabolism of UV-filter benzophenone-3 by rat and human liver microsomes and its effect on endocrine-disrupting activity. Toxicol Appl Pharmacol 282(2):119–128, PMID: 25528284, 10.1016/j.taap.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Woodruff TJ, Zota AR, Schwartz JM. 2011. Environmental chemicals in pregnant women in the United States: NHANES 2003–2004. Environ Health Perspect 119(6):878–885, PMID: 21233055, 10.1289/ehp.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Roman JM, Issaq HJ, Keefer LK, Veenstra TD, Ziegler RG. 2007. Quantitative measurement of endogenous estrogens and estrogen metabolites in human serum by liquid chromatography-tandem mass spectrometry. Anal Chem 79(20):7813–7821, PMID: 17848096, 10.1021/ac070494j. [DOI] [PubMed] [Google Scholar]
- Yager JD, Davidson NE. 2006. Estrogen carcinogenesis in breast cancer. N Engl J Med 354(3):270–282, PMID: 16421368, 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- Ye X, Bishop AM, Reidy JA, Needham LL, Calafat AM. 2006. Parabens as urinary biomarkers of exposure in humans. Environ Health Perspect 114(12):1843–1846, PMID: 17185273, 10.1289/ehp.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Wang Z, Feng Q, Chou C-K, Pintilie GD, Shen H, et al. 2017. Structural and functional impacts of ER coactivator sequential recruitment. Mol Cell 67(5):733–743, PMID: 28844863, 10.1016/j.molcel.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chiang H-C, Wang Y, Zhang C, Smith S, Zhao X, et al. 2017. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat Commun 8:15908, PMID: 28649985, 10.1038/ncomms15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Kosinska W, Khmelnitsky M, Cavalieri EL, Rogan EG, Chakravarti D, et al. 2006. Mutagenic activity of 4-hydroxyestradiol, but not 2-hydroxyestradiol, in BB rat2 embryonic cells, and the mutational spectrum of 4-hydroxyestradiol. Chem Res Toxicol 19(3):475–479, PMID: 16544955, 10.1021/tx0502645. [DOI] [PubMed] [Google Scholar]
- Zhao X, Malhotra GK, Lele SM, Lele MS, West WW, Eudy JD, et al. 2010. Telomerase-immortalized human mammary stem/progenitor cells with ability to self-renew and differentiate. Proc Natl Acad Sci USA 107(32):14146–14151, PMID: 20660721, 10.1073/pnas.1009030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler RG, Faupel-Badger JM, Sue LY, Fuhrman BJ, Falk RT, Boyd-Morin J, et al. 2010. A new approach to measuring estrogen exposure and metabolism in epidemiologic studies. J Steroid Biochem Mol Biol 121(3–5):538–545, PMID: 20382222, 10.1016/j.jsbmb.2010.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.