Abstract
Background:
Tributyltin (TBT) is a persistent and bioaccumulative environmental toxicant. Developmental exposure to TBT has been shown to cause fatty liver disease (steatosis), as well as increased adiposity in many species, leading to its characterization as an obesogen.
Objective:
We aimed to determine the long-term effects of developmental TBT exposure on the liver.
Methods:
C57BL/6J mice were exposed to a dose of TBT ( body weight per day; ) below the current developmental no observed adverse effect level (NOAEL) via drinking water, or drinking water alone, provided to the dam from preconception through lactation. Sires were exposed during breeding and lactation. Pups from two parity cycles were included in this study. Animals were followed longitudinally, and livers of offspring were analyzed by pathological evaluation, immunohistochemistry, immunoblotting, and RNA sequencing.
Results:
Developmental exposure to TBT led to increased adiposity and hepatic steatosis at 14 and 20 weeks of age and increased liver adenomas at 45 weeks of age in male offspring. Female offspring displayed increased adiposity as compared with males, but TBT did not lead to an increase in fatty liver or tumor development in female offspring. Liver tumors in male mice were enriched in pathways and gene signatures associated with human and rodent nonalcoholic fatty liver disease (NAFLD) and hepatocellular carcinoma (HCC). This includes down-regulation of growth hormone receptor (GHR) and of STAT5 signaling, which occurred in response to TBT exposure and preceded liver tumor development.
Conclusions:
These data reveal a previously unappreciated ability of TBT to increase risk for liver tumorigenesis in mice in a sex-specific manner. Taken together, these findings provide new insights into how early life environmental exposures contribute to liver disease in adulthood. https://doi.org/10.1289/EHP5414
Introduction
Environmental exposures during development have emerged as a significant risk factor for liver disease (Foulds et al. 2017). Some of the clearest demonstrations of the impact of early life environment on risk for liver disease are studies on the Dutch Hunger Winter and Great Chinese Famines, which have consistently reported significant associations between early life starvation and development of nonalcoholic fatty liver disease (NAFLD) in adulthood (Sandboge et al. 2013; Chen et al. 2016; N Wang et al. 2016, 2017; Z Wang et al. 2017). Similarly, in rodent models, developmental exposures to chemicals such as the xenoestrogen bisphenol A (BPA), polycyclic aromatic hydrocarbon benzo[a]pyrene (B[a]P), perfluorooctane sulfonic acid (PFOS), phthalates such as bis(2-ethylhexyl) phthalate (DEHP), and organotins such as tributyltin (TBT) have been shown to promote hepatic steatosis and/or nonalcoholic steatohepatitis (NASH) (Lee et al. 2010; Maranghi et al. 2010; Denison et al. 2011; Lv et al. 2013; Ortiz et al. 2013; Foulds et al. 2017; Shimpi et al. 2017; Treviño and Katz 2018).
Data linking early life exposures to endocrine disrupting compounds to development of liver tumors in adulthood has been reported in mice for BPA (Weinhouse et al. 2014) and arsenic (Nohara et al. 2012). However, although development of NAFLD in humans is an established risk factor for hepatocellular carcinoma (HCC) (Foulds et al. 2017), in animal studies the linkage between development of NAFLD and increased risk for liver tumors is less well established. Thus, although the underlying etiology of NAFLD and mechanisms linking this disease to increased risk for liver tumorigenesis remain ill-defined, both rodent models and human epidemiological studies clearly indicate that environmental exposures early in life are important determinants of risk for later-life development of liver disease (Foulds et al. 2017; Treviño and Katz 2018). Consequently, additional studies are needed to obtain insights into the molecular underpinnings and etiology of liver disease using model systems in which early life exposures promote both NAFLD and liver tumorigenesis.
TBT is an organotin with many industrial applications including use in polyvinyl chloride (PVC) catalysts, broad-spectrum biocides, agricultural fungicides, construction materials, wood and textile preservatives, and antifouling agents for marine vessels (Fent 1996; U.S. EPA 2003; Fromme et al. 2005; Kannan et al. 2010). The major route of human TBT exposure is reported to be via ingestion of contaminated water and food, specifically seafood (Azenha and Vasconcelos 2002; Chien et al. 2002; Antizar-Ladislao 2008; Airaksinen et al. 2010; Jadhav et al. 2011; Kucuksezgin et al. 2011; Filipkowska et al. 2016; Ashraf et al. 2017). In the 1960s, widespread use of TBT as an antifouling agent began, but after discovering detrimental effects on aquatic life including imposex in mollusks (Fent 1996; Lima et al. 2015), TBT was banned by the International Marine Organization in 2008 (Sonak 2009; Sonak et al. 2009). Despite this ban, high levels of TBT continue to be detected in recreational vessels around the Baltic sea (Lagerström et al. 2017), as well as in aquatic life and fish in more recent years (Filipkowska et al. 2016; Ashraf et al. 2017). TBT has been shown to accumulate in birds and marine mammals and is associated with reproductive and metabolic dysfunction (Coenen et al. 1992; Dorneles et al. 2008). In addition to fatty liver disease, developmental exposure to TBT has also been linked to the accumulation of fat in adipose depots in mice (Grün et al. 2006), with increased weight of adipose depots shown to be a result of both increased adipose cell size and cell number (Chamorro-García et al. 2013). These findings, as well as the ability of TBT to induce lipid accumulation in cells and promote adipogenesis in murine 3T3-L1 cell culture, have led to the characterization of TBT as a prototypical obesogen (Kanayama et al. 2005; Grün et al. 2006; Li et al. 2011).
The purpose of this study was to ascertain the long-term effects of developmental exposure to TBT. We exposed male and female mice throughout gestation and lactation to TBT or vehicle (VEH) and assessed several parameters of liver health (including gene expression) and adiposity over time. To our knowledge, these data are the first to reveal that early life TBT exposure promotes tumorigenesis in the male mouse liver and provide new insights into how early life exposures impact the adult liver to induce fatty liver and promote tumorigenesis.
Methods
Animal Exposures
Male () and female () C57BL/6J mice from the Jackson Laboratory were housed in a controlled environment with a 12-h light/dark cycle, at 68°C–72°C, and 30–70% relative humidity, in a facility known to have Murine Norovirus (MNV) and Pseudomonas sp. Mice arrived at 4 weeks of age and were allowed to acclimate for 1 week before being randomly assigned to their exposure groups. Breeding began 2 weeks after treatment at 7 weeks of age. All animals received a phytoestrogen-free diet (7% corn oil; Teklad/Envigo). Females () were exposed to TBT chloride (TBT 96% purity; Sigma) in their drinking water (made from animal facility water), whereas control females drank animal facility water () beginning 2 weeks prior to breeding, throughout gestation, and lactation. Animal facility water is carbon filtered, particulate filtered, and ultrafiltered, then rechlorinated to . Given that water bottles were situated on the top of the cage, there is potential that older weanlings could access the water. At the time of breeding, females were put into the male’s cage and both sexes were then exposed to study water for the duration of the experiment. After weaning, all pups were given nontreated water for the duration of the study. Pups from the VEH group were used to create additional mating pairs that followed the same exposure paradigm. Animals averaged a body weight (BW) of , and pregnant females became as large as during gestation. Females were bred to one male each in the male’s cage, and the male was kept in the cage until litters were weaned. Twelve litters were produced from the VEH-treated dams and 15 litters were produced from the TBT-treated dams; both VEH- and TBT-treated dams averaged two litters/dam (see Excel Table S1). Pup weights were measured only on postnatal Day 3 (PND3) in the first round of mating, which included five litters from each treatment group. Pups were chosen at random from each litter and assigned to each assay (see Excel Table S1). Animals not assigned to an assay were kept in case additional assays were warranted and euthanized at 45 weeks of age. If pups were unassigned and euthanized at 45 weeks of age, their liver tumor status was noted and included in the tumor data.
Preliminary water consumption surveys were conducted in nonpregnant females to calculate the administered dose of TBT. Consumption levels were found to spike immediately after birth at the onset of lactation for a period of 3 d. Females were found to drink of water per day; therefore, they were given of TBT each week (equivalent to ) in the drinking water provided by the animal facility in typical plastic water bottles provided by the animal facility. This dose is below the lowest dose in the current developmental no observed adverse effect level (NOAEL) range of BW as per the Concise International Chemical Assessment Documents (CICADs) guidelines for developmental toxicity (Benson 1999). Offspring were then maintained on the same phytoestrogen-free diet throughout life in ventilated cages with 3–4 animals per cage, on corncob bedding. Throughout the study, cages were changed twice per week. Animals were weighed and inspected weekly to assess general health and were euthanized if they lost 10% of their BW or appeared to be in poor health. Animals were euthanized by carbon dioxide () asphyxiation and cardiac puncture at 14, 20, or 45 weeks of age. Animal care was performed in accordance with approved institutional guidelines (IACUC protocol AN-7189) and complied with National Institutes of Health policy.
Histology
Upon study termination at 45 weeks [for male VEH-treated animals, (from 12 litters); for male TBT-treated animals, (from 11 litters); for female VEH-treated animals, (from 11 litters), for female TBT-treated animals, (from 8 litters)], tissue from the left liver lobe was immediately fixed in 10% neutral buffered formalin for 24 h, then processed, embedded in paraffin, sectioned (), and stained with hematoxylin and eosin [MD Anderson Cancer Center Research Histology, Pathology, and Imaging Core (RHPI)]. Slides were analyzed by a pathologist (M.J.F.), who was blinded to treatment, and imaged by the Digestive Diseases Core facility at Baylor College of Medicine.
Oil Red O Staining
Previously reported data has shown that lipid accumulation in the liver in response to developmental TBT exposure can occur as early as 8 weeks of age (Chamorro-García et al. 2013). We therefore investigated lipid accumulation in our subset of mice euthanized at 20 weeks of age to determine whether our mice also developed steatosis. At euthanasia (20 weeks of age), a slice from the left liver lobe was fixed in optimal cutting temperature (OCT) compound and frozen on dry ice (for VEH-treated males, ; for TBT-treated males, ; for VEH-treated females, ; and for TBT-treated females, ). Blocks were then cryosectioned () and stained with Oil Red O for 10 min (MD Anderson Cancer Center RHPI). One slide per animal was imaged using a Zeiss Axioscan.Z1 and quantitated by the Integrated Microscopy Core at Baylor College of Medicine in MATLAB™ (version 9.4) using a custom-made algorithm. Briefly, pixelated digital histologic images, which were originally stored in a red, green, and blue format, were converted into a MATLAB™ colorspace format consisting of three separate color spectrums; gray, blue, and red. Each pixel in the images was located on gray, blue, and red spectrums by calculating the smallest Euclidian distance between the pixel’s colorspace values and reference markers consisting of pure gray, blue, and red. After locating each pixel within one of three subregions corresponding with reference histologic material [i.e., nucleic material, (blue), lipid, (red), or vacant background (gray)], blue pixels and red pixels were converted to either pure blue or pure red in accordance with their predominant color. This binary conversion allowed greater distinction of lipid droplets from nucleic material for visual inspection. The number and size of lipid droplets, as well as the number of nuclei, were then measured for each liver tissue specimen (May et al. 2018).
Immunohistochemistry
At 45 weeks of age, mice were euthanized and liver and adenoma tissues were fixed in 10% neutral buffered formalin for 24 h, then transferred to 70% ethanol, processed, and paraffin embedded. Blocks were then sectioned at , deparaffinized and subjected to antigen retrieval. Sections were incubated with 3% hydrogen peroxide, then incubated with Ki67, a cellular marker for proliferation [1:1,000, Abcam, ab15580, and VEH-exposed liver (V), ; TBT-exposed liver (T), ; Adenoma (A), ], F4/80 (1:250, AbD Serotec, MCA497; seven adenomas were stained]; cytokeratin 19 (K19; 1:600, made in-house, and seven adenomas were stained), alpha-fetoprotein (, 1:200, Biocare CP028, and seven adenomas were stained), and major urinary protein (MUP1, 1:50, R&D Systems, MAB6560, and five adenomas from TBT-treated males and five livers from VEH-treated males were stained) antibodies by the Digestive Diseases Core, Cellular and Molecular Morphology. The Ki67 antibody was detected by incubation with biotinylated anti-rabbit secondary antibody (Vector Laboratories; BA-1000) followed by horseradish peroxidase (Vector Laboratories; PK-6100). F4/80 was detected by incubation with biotinylated anti-rat secondary antibody (Vector Laboratories; BA-9400) followed by horseradish peroxidase (Vector Laboratories; PK-6100). MUP1 was detected using Histomouse Max (Life Technologies; 879551). K19 and were detected using an IMPress anti-rabbit polymer (Vector Laboratories; MP-7401). All stains were developed using DAB (Biocare; DB801). Ki67-stained slides were imaged using a Zeiss Axioscan.Z1 and quantitated by the Integrated Microscopy Core at Baylor College of Medicine. Image analysis (Image Pro 10; Media Cybernetics) was performed via automated channel splitting (R, G, and B) and filtering of the red (R) channel for all nuclei and Ki67-positive cells on the blue (B) channel. Filters were applied to remove outliers: pixel area (5–300), roundness 1.0–20. The total percent of Ki67-positive cells of the total nuclei detected was calculated for tumor and liver samples [defined by a pathologist (M.J.F.)] using the entire section.
Body Composition Analysis
Previous reports have shown that body composition in mice is altered in response to developmental exposure to TBT as early as 19 weeks of age (Chamorro-Garcia et al. 2017). Magnetic resonance imaging (MRI) of body fat depots in mice was performed at 20 weeks of age [VEH-treated females (from one litter), for TBT-treated females (from two litters with two pups per litter), for VEH-treated males , and for TBT-treated males ] in the Small Animal MRI Core Facility at Texas Children’s Hospital. Mice were anesthetized using isoflurane, placed in the animal holder (Bruker; BioSpin) and imaged using 9.4T, Bruker Avance BioSpec Spectrometer/AVIII with a horizontal bore (Bruker; BioSpin) and a resonator. To delineate the distribution of adipose tissue in the mice, a Dixon Fat Imaging sequence was used with a , , ; , , and matrix. Saturation slabs were also incorporated. After acquisition, an in-house MATLAB™ code was utilized to separate the water and fat images, and the fat images were then quantified using Amira™ software (version AllAmira6Beta 6.5; Thermo Scientific), 3D images and tissue surface area values were exported and presented (Blesson et al. 2016).
Alanine and Aspartate Transaminase Activity
Whole blood was collected from animals [males displaying macroscopic adenomas (A, ), VEH controls (V, ), and TBT-treated males without any macroscopic adenomas (T, )] at the time of euthanasia (45 weeks of age) via cardiac puncture, and plasma was isolated by centrifugation at 4°C for 10 min at after red blood cell lysis with EL Buffer (Qiagen, Catalog No. 79217) and stored at until analysis. Alanine and aspartate transaminase (ALT/AST) activity levels were analyzed in the plasma of 45-week-old animals using an alanine transaminase activity assay (ab105134; Abcam) or an aspartate transaminase activity assay (MAK055; Sigma) as per the manufacturer’s instructions. Briefly, plasma was diluted (1:5 for ALT and 1:10 for AST) and mixed with Reaction Mix, then read on the BioTek EL808 plater reader (BioTek, Instruments) in a kinetic fashion every 3 min for 60 min while incubating at 37°C. For analysis two time points (T) were chosen where all samples fell within the standard curve. Pyruvate/glutamate levels were calculated from a standard curve and activity (in nanomoles per minute per milliliter or in milliunits per milliliter) was calculated as for ALT or for AST, where , , , and . From this data, the ALT/AST ratio was calculated.
RNA Sequencing Transcriptomics Analysis
At study termination (20 or 45 weeks of age) [VEH-treated 20-weeks, ; TBT-treated 20-weeks, ; VEH-treated 45-weeks, (from five litters); TBT-treated 45-weeks, (from nine litters) where seven animals displayed macroscopic adenomas and five animals did not)], liver or adenoma tissue was flash frozen in liquid nitrogen. RNA was isolated from of tissue (RNeasy; Qiagen, Catalog No. 74104) and treated with DNase (Qiagen, Catalog No. 79254) as per the manufacturer’s instructions. Quality was verified on the Bioanalyzer 2100 (Agilent, Catalog No. G2939BA) with . Library preparation and RNA-seq (Stark et al. 2019) analysis was conducted by the MD Anderson Science Park Next Generation Sequencing Core facility using the Illumina TruSeq Stranded Total RNA Protocol and the Hiseq 3000 SBS platform. Paired-end reads were trimmed using TrimGalore and mapped using HISAT2 (Kim et al. 2015) to the UCSC mm10 genome build, then gene expression data was quantified using featureCounts (Liao et al. 2014) and the GENCODE gene model (Harrow et al. 2012). Differential expression (DE) analysis was conducted using EdgeR (Robinson et al. 2010). Differentially expressed genes were denoted as statistically significant for the false discovery rate (FDR) adjusted with a fold change exceeding (i.e., ). We sequenced 34–50 million read pairs per sample after trimming of low-quality base pairs.
Real-Time Polymerase Chain Reaction Analysis
RNA was isolated from liver tissue of 45-week-old animals [VEH-treated, (from five litters); TBT-treated, (from nine litters) where seven animals displayed macroscopic adenomas and five animals did not)] (AllPrep DNA/RNA; Qiagen, Catalog No. 80224), and was reverse-transcribed using SuperScript III (Invitrogen). Next cyclic DNA (cDNA) was diluted 1:5 in nuclease-free water and amplified using Fast SYBR® Green Mix (Applied Biosystems; ABI) and primers (IDT DNA Technologies) listed in Table 1. Real-time polymerase chain reaction (RT-PCR) was conducted on the QuantStudio™ 7 Flex system (ABI) and QuantStudio™ Real-Time PCR Software (version 1.2; ABI) using the two-step fast PCR protocol (initial denaturation: 95°C for 20 s, followed by 40 cycles of 95°C for 1 s, 60°C for 10 s; data was collected during the 60°C step). Relative values were quantified using the using glyceraldehyde 3-phosphate dehydrogenase (Gapdh) as the housekeeping gene and VEH-exposed samples as the reference group. Statistical differences between two groups were calculated by analysis of difference between medians (Mann-Whitney U-test).
Table 1.
Primers used.
| Gene | Forward | Reverse |
|---|---|---|
| mGapdh | CCCTTAAGAGGGATGCTGCC | TACGGCCAAATCCGTTCACA |
| mUbd | TGACCTCTGTGATCCCTAAG | GTGCAGTGTGTTGTCAGAAAG |
| mLy6d | TCAGCCTGCTCACTGTTATG | ACACTGACGACTAGAAGGGA |
| mPtgds | TCCTGGACACTACACCTAC | CTTGGTGCCTCTGCTGAATA |
| mCbr3 | TTCCGATGTGACACACTTACC | CCTGTCCGCTTTCCTCTTT |
| mCyp2a4 | GAATGCTGGAGGAGAAGAAGAA | ATGGACCTTGGCCTCAATATC |
| mSlc7a11 | GTGGGAGGCTGGTAGTTAATG | CTGCTGTACCGTGGTTATGT |
| mLrtm2 | CCCTTGGGTGGATGTCATTAT | GTGTGTGTGTGTGTGACTTTG |
| mKrt20 | CCTACCAGAACCTGGAGATAGA | CTCAAGCCGGGTCTTTATGT |
| mMup1 | GAGATGAAGAGTGCTCGGAATTA | AAATCTGGTTCTCGGCCATAG |
| mMup12 | GCATACTATTATCCTGGCCTCTG | CCGAGCACTCTTCATCTCTTAC |
| mMup16 | GGCATACTATTATCCTGGCTTCT | CGGAGCACTCTTCATCTCTTAC |
| mMup11 | GCATACTATTATCCTGGCCTCTG | CCGAGCACTCTTCATCTCTTAC |
| mMup7 | CTATTATCCTGGCCTCTGACAAG | CGGAGCACTCTTCATCTCTTAC |
Growth Hormone Response Gene Set Derivation
In order to determine differential gene expression downstream of growth hormone receptor (GHR) signaling in murine livers, publicly available data [GSE93382 (Connerney et al. 2017)] from livers from control and growth hormone-treated mice were used to generate a signature (see Excel Table S2). A similar mapping and quantification approach as described above in the RNA Sequencing Transcriptomics Analysis section was used; Edger was used to assess differential genes, and significance was achieved for FDR-adjusted and a fold change exceeding . To identify direct STAT5 gene targets in mouse livers, we utilized the STAT5 ChIP sequencing (ChIP-Seq) GSE31578 (Zhang et al. 2012) peak calls reported for male mice. The STAT5 peaks were first converted from UCSC build mm9 to UCSC build mm10 using the UCSC liftOver program; next, STAT5 target genes were determined using the BEDTOOLS (Quinlan and Hall 2010) software and the GENCODE gene model. Specifically, a gene was considered a STAT5 target if a STAT5 peak was found within from the transcriptional start site (TSS) of the gene. We overlapped the GHR gene signature (from seven adenomas and either five TBT-exposed livers or six VEH-exposed livers) and the STAT5 gene targets, considering separately the GHR up-regulated gene targets and the GHR down-regulated gene targets.
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) (Subramanian et al. 2005) was conducted on the gene rank file derived for the TBT-exposed livers or adenomas. We considered separately GHR up-regulated genes, GHR down-regulated genes, GHR up-regulated genes overlapping with STAT5 targets, and GHR down-regulated genes overlapping with STAT5 targets. GSEA was run, performing 1,000 permutations to establish statistical significance. A gene set was considered significantly enriched if it had an FDR-adjusted .
Overrepresentation Analysis
Overrepresentation analysis (ORA) was performed to detect enrichment of gene sets corresponding to pathways and biological processes or to publicly available gene signatures within the TBT transcriptomic response. Following the Molecular Signature Database methodology (MSigDB) (Subramanian et al. 2005), a hypergeometric test was used to assess the enrichment, with significance achieved at FDR-adjusted . We performed gene enrichment against the HALLMARK and Kyoto Encyclopedia of Genes and Genomes (KEGG) compendia as compiled by the Molecular Signatures Database v6.2. We further compiled publicly available NASH/NAFLD data sets curated using the National Center for Technology Information Gene Expression Omnibus (NCBI GEO) data sets GSE46300, GSE48452, GSE59930, GSE83596, GSE89632, and The Cancer Genome Atlas (TCGA) data. An additional HCC signature was derived using TCGA Liver Hepatocellular Carcinoma (LIHC) samples. Significantly different genes were determined using the Student’s t-test, as implemented in R (R Development Core Team), with significance achieved for and fold change exceeding (i.e., ). All human and rat gene signatures genes were converted to mouse gene homologs using the Ensemble Biomart repository (Durinck et al. 2009). We performed the hypergeometric test against up- and down- regulated genes from each NASH/NAFLD/HCC–derived gene signature, respectively. Significance was achieved at an FDR-adjusted .
Immunoblotting Analysis
Liver and tumor tissues from animals who were 45 weeks of age (VEH-treated, ; TBT-treated liver, ; and adenoma tissue from TBT-exposed males, ) were cryopulverized, cells were lysed in radioimmunoprecipitation assay buffer and sonicated for 7–10 30-s cycles at 4°C (Bioruptor 300; Diagenode), then protein was isolated by centrifugation at 4°C for 20 min at . Protein concentrations were assessed by bicinchoninic acid assay and of total protein was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis under reducing conditions before transfer to a polyvinylidene fluoride membrane. Membranes were blocked in Tris-buffered saline plus 0.1% Tween 20 (TBST)-5% milk and probed using antibodies against GHR (1:1,000, Santa Cruz Biotechnology; sc-137,185), or GAPDH (1:5,000, Santa Cruz Biotechnology; sc-25,778) overnight at 4°C in TBST-5% milk. Secondary antibodies conjugated to horseradish peroxidase were used at room temperature for 1 h (Santa Cruz Biotechnology), Pierce ECL (Thermo Scientific), Amersham ECL (GE Healthcare), and X-ray film exposure was used to detect and visualize primary antibodies. Bands were quantitated using ImageQuant TL software (version 7.0) and normalized to the GAPDH protein level.
Statistical Analysis
For comparisons between two groups (VEH males vs. TBT-exposed males or VEH females vs. TBT-exposed females), we used a Student’s t-test for analyzing tissue weights, fat imaging, and Oil Red O staining or a Mann-Whitney U-test for analyzing quantitative PCR (qPCR) data. For three-way comparisons, a one-way analysis of variance (ANOVA) with a Tukey’s Multiple Comparisons Test was used (ALT, AST, and Ki67 analyses). Gapdh was used as a normalization control for both qPCR and immunoblotting, and importantly, no differences were found between treatment groups in either GAPDH protein or mRNA level (see Figure S1). When analyzing contingency tables (tumor incidence) a chi-square test was utilized without Yates correction. Males and females were never directly compared to each other but, rather, each treatment group was compared to its respective control. Significance is always reported as , and tests were conducted in GraphPad Prism Software (version 8.0.0).
Data Set Availability
All RNA-seq data were submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) made publicly available upon manuscript acceptance. The accession number is GSE143304.
Results
Measurements of NAFLD in Male Mice Developmentally Exposed to TBT
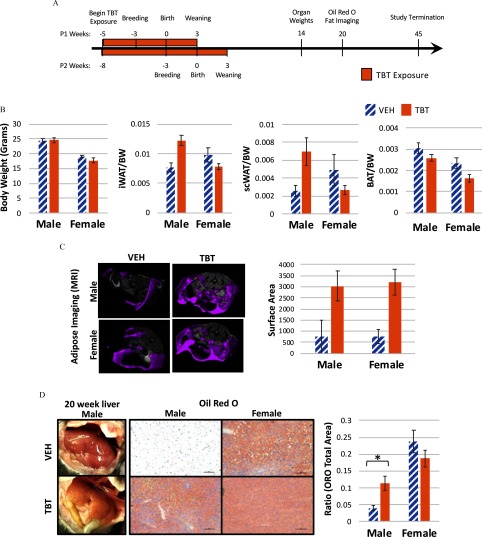
Dams were exposed to TBT () in drinking water or drinking water alone prior to breeding and during gestation through lactation, while consuming a phytoestrogen-free diet (Figure 1A). At necropsy, and throughout the study, dams and sires did not present with any signs of adverse health effects. No difference was noted in any measured parameter between pups from the first and second litters from the same TBT-exposed dams (see Figure S2). Although litter sizes were not standardized in this study, it is not thought that either group had better access to milk because the average number of pups in either treatment group was six (see Excel Table S1), with the average pup/litter weight of in the VEH-treated dams and in the TBT-treated dams. No significant difference in litter size or pup weight was observed between VEH- and TBT-exposed groups. Offspring were then weaned onto a phytoestrogen-free diet and followed longitudinally until 45 weeks of age. Body weight of offspring at 14 weeks, as well as across the life course, was unchanged between VEH- and TBT-exposed animals (Figure 1B; Figure S3). However, consistent with previous data on the obesogenic effects of this compound (Grün et al. 2006; Chamorro-García et al. 2013), MRI measurement of whole-body adiposity confirmed both male and female TBT-exposed offspring had more body fat than VEH controls (Figure 1C). In contrast, only male TBT-exposed offspring exhibited an increase in white adipose tissue accumulation in the inguinal and scapular subcutaneous depots at 14 weeks of age (Figure 1B), with the size of these fat depots that of vehicle controls, respectively. White adipose tissue was dissected away from brown adipose tissue collected from the scapular region at necropsy and both were weighed. Brown adipose tissue wet weight is shown in Figure 1B. MRI measures any adipose tissue present in the trunk of the animal, including visceral fat; suggesting that females gained adiposity in the visceral fat depots without much increase in the subcutaneous depots, whereas males gained adiposity in both depots. TBT exposure also exacerbated development of fatty liver in male offspring as assessed by hematoxylin and eosin and Oil Red O staining. As shown in Figure 1D, TBT livers presented with mixed macrovesicular and microvesicular steatosis. Quantitation of Oil Red O staining confirmed that lipid accumulation was more than twice that of VEH controls in TBT-exposed male offspring. TBT-induced development of steatosis was confirmed across multiple litters in two separate breeding cohorts of mice. Although at baseline, age-matched control female offspring had roughly four times more lipid in their livers compared with control male mice, no increase was observed in response to TBT exposure in female livers (Figure 1D).
Figure 1.
Body weight, white adipose tissue weight, adipose magnetic resonance imaging (MRI), and liver lipid of male and female mice developmentally exposed to tributyltin or vehicle. (A) Female mice were fed a phytoestrogen-free diet and treated with tributyltin (TBT) in their drinking water beginning 2 weeks prior to breeding lasting through gestation and lactation. Offspring were then weaned at 3 weeks of age and fed the same phytoestrogen-free diet until euthanasia at 14, 20, or 45 weeks of age. (B) Body weights, white adipose tissue depot [either inguinal (iWAT) or scapular (scWAT)], and brown adipose tissue (BAT) wet weights in 14-week-old animals developmentally exposed to TBT or VEH are displayed; VEH-treated females, (from two litters with two pups per litter); VEH-treated males, (from two litters); TBT-treated females, (from two litters); TBT-treated males, (from two litters, two pups per litter). (C) Representative full-body MRI scans are presented on the left and quantitation of surface area is presented on the right for 20-week-old animals: for VEH-treated females, (from one litter); for TBT-treated females, (from two litters with two pups per litter); for VEH-treated males, ; and for TBT-treated males, . (D) Macroscopic images of liver tissue at 20 weeks, and Oil Red O stained, left liver lobes are presented on the left with quantitation of Oil Red O staining on the right, for VEH-treated males, for TBT-treated males, for VEH-treated females, and for TBT-treated females. Scale bars: . In all graphs, VEH bars are hatched and TBT bars are solid, all bars represent the error of the mean (SEM) and t-tests were performed to ascertain significance (*). Excel Table S1 details information on n and litter. Note: ORO, Oil Red O.
Characterization of Tumors in Male Offspring Developmentally Exposed to TBT
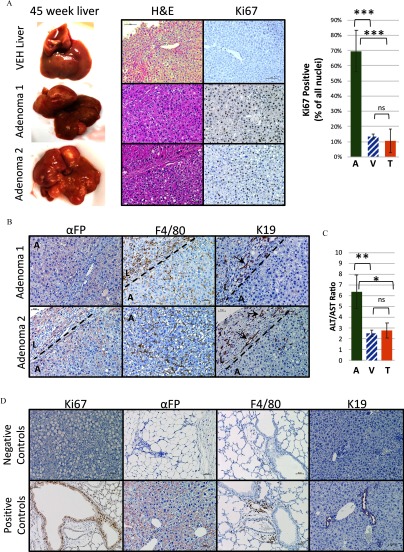
Full necropsies were conducted at 45 weeks of age when multiple large nodules were visible macroscopically (Figure 2A) throughout the liver in 46.7% (7/15 animals) of TBT-treated male offspring. This percentage was a significant increase compared with VEH-exposed male offspring (5.3%, 1/19 animals) (Table 2; chi-square ). A detailed list of tumor incidence, including the number of animals included from each litter, is presented in Excel Table S3 and remained significantly different from vehicle controls when using the litter, rather than individual, as the N (Table 2). However, no significant increase in liver tumor incidence was seen in female TBT-exposed offspring (9.1%, 1/11 animals), as compared with VEH-control females (5.9%, 1/17 animals). Thus, the absolute amount of neutral lipid in the liver (because female mice had notably more Oil Red O staining than their male counterparts; Figure 1D) did not correlate with development of liver tumors. We also assayed clinically relevant liver damage markers (AST and AST) to test whether TBT treatment increased damage. Although the individual ALT activity, AST activity, or ALT/AST activity ratio was not significantly different between VEH- (labeled V) and TBT-exposed male offspring that did not develop tumors (labeled T), as expected, the ALT, AST, and ALT/AST ratio was significantly increased in the plasma of TBT-exposed offspring presenting with liver tumors (labeled A for adenoma) (Figure 2C; Figure S4).
Figure 2.
Histology of tributyltin (TBT)-treated liver adenomas in male mice. (A) At 45 weeks of age, liver adenomas were observed in many lobes of the liver in male mice. One vehicle (VEH)-exposed liver and two representative tumors are displayed with hematoxylin and eosin and Ki67 immunohistochemistry staining (left panels). The right panel shows a quantitation of Ki67 immunostaining [VEH-exposed liver (V), hatched bar, ; TBT-exposed liver (T), ; Adenoma (A), ]. Bars represent (SEM). *** by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. (B) Alpha-feto protein (), F4/80, and cytokeratin 19 (K19) immunostaining are shown for two representative tumors from male mice. A dotted line represents the tumor margin and an A marks the tumor area, and an L marks the surrounding liver area. Arrows in the K19-stained sections point to ducts. (C) Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activity levels (mU/mL) were detected in plasma from males displaying macroscopic adenomas (A, ), vehicle controls (V, hatched bar, ), and TBT-treated males without any macroscopic adenomas (T, ). The ALT/AST ratio is presented. Bars represent the and one-way ANOVA with Tukey’s multiple comparisons test were performed to ascertain significance. *; **. (D) Lung and liver tissues were used as positive and negative controls. Slides were imaged on a Nikon Ci-L microscope. Scale bars: .
Table 2.
Tumor incidence at 45 weeks.
| Sex | Treatment | Ratio | Percent | Litter | Birth cohort | Tumors per litter |
|---|---|---|---|---|---|---|
| Male | VEH | 1/19 | 5.3 | 12 | 3 | 1/12 |
| TBT | 7/15 | 46.7** | 11 | 3 | 6/11* | |
| Female | VEH | 1/17 | 5.9 | 11 | 3 | 1/11 |
| TBT | 1/11 | 9.1 | 8 | 2 | 1/8 |
Note: For source data please see Excel Table S3. Birth cohort, breeding round from which pups were born; litter, the number of litters from which the total mice were drawn; ratio, number of tumor bearing mice per total mice; TBT, tributyltin; tumors per litter, number of tumor bearing mice from distinct litters bearing at least one tumor; VEH, vehicle. ** and * by chi-square analysis comparing VEH- and TBT-exposed groups (sexes are compared separately).
Histological examination was performed by a pathologist (M.J.F.) on the macroscopic liver nodules, which were characterized as having clear demarcations compressing adjacent hepatocytes (Figure 2A). Nodules lacked normal lobular architecture but, instead, exhibited severe architectural distortion and displayed hepatocellular dysplasia. Cellular atypia was present with pleomorphic nuclei ranging from large irregular shapes to coarsely condensed chromatin. As shown in Figure 2A, nodules were highly proliferative, as evidenced by Ki67 staining, with an average of 70% of all nuclei positive for Ki67. This value is significantly higher than VEH-treated liver tissue (13%) or TBT-treated liver tissue from animals that did not develop macroscopic tumors (10%). Nodules were negative for bile ducts, demonstrated by lack of staining inside the tumors (labeled A) of the marker K19, and they displayed variable levels of , a marker for HCC, and F4/80, a macrophage marker (Figure 2B). However, both and F4/80 were higher in areas of uninvolved liver surrounding nodules (labeled L) than in the nodules themselves (labeled A). Representative images of the tumor margins (depicted by a dotted line) are presented to demonstrate the difference between staining within the nodules (labeled A) and in the surrounding liver tissue (labeled L). Liver sections in the regions outside of nodules also demonstrated striking architectural distortion, showing many irregularly situated small cytologically normal bile ducts that stained with K19 (Figure 2B). All control staining can be found in Figure 2D. Based on these observations and current guidelines (Thoolen et al. 2010), TBT-induced liver tumors were diagnosed as hepatic adenomas. After inspection of the nodule that developed in a single male VEH-exposed animal, it was determined that this was also categorized as an adenoma but with a very different pathology that included a high amount of lipid droplets and a higher degree of differentiation (see Figure S5) that distinguished it from the TBT-induced tumors.
Expression Profiling of TBT-Induced Tumors
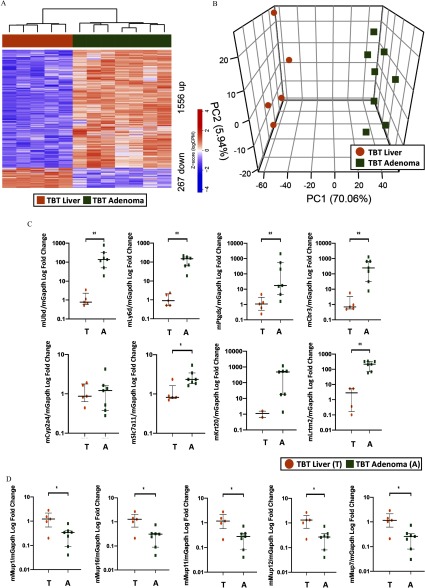
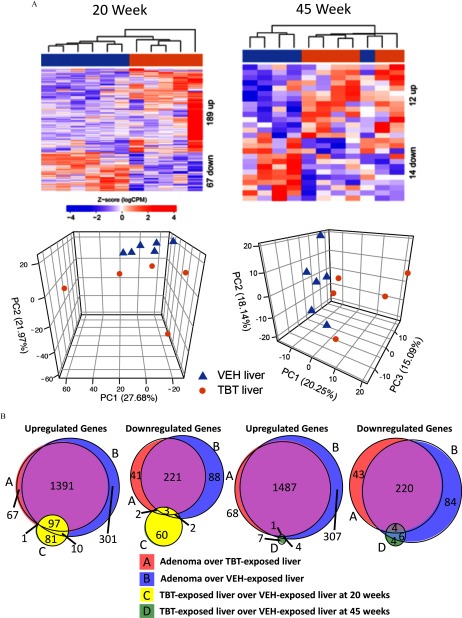
Transcriptomic analysis using RNA-seq was performed on TBT-induced adenomas, and these data were compared with RNA-seq data from livers of age-matched offspring (either VEH- or TBT-exposed) to identify genes differentially expressed between tumor and liver (Figure 3 shows adenoma vs. TBT-exposed age-matched liver; Figure S6 shows adenoma vs. VEH-exposed age-matched liver). Genes whose expression was significantly altered were identified with an FDR-adjusted and an absolute fold change exceeding . Hierarchical clustering of differentially expressed genes clustered tumors separately from TBT-exposed livers from animals that did not develop tumors (Figure 3A) as well as from VEH-exposed liver tissue (see Figure S6A). Similarly, principal component analysis (PCA) also revealed a clear separation of tumors from age-matched TBT-exposed livers (Figure 3B) or VEH-exposed livers (see Figure S6B), with PC1 separating the two groups and explaining over 70% of the overall variance in either comparison. The top 60 differentially expressed genes overexpressed in adenomas compared with TBT-exposed livers are shown in Table 3, and the 60 most repressed in adenomas are shown in Table 4 (the full list is presented in Excel Table S4). Altered expression of 8 genes comprising the top 2% of the significantly increased genes [murine ubiquitin D (mUbd), murine lymphocyte antigen 6 family member D (mLy6d), murine prostaglandin D2 synthase (mPtgds), murine carbonyl reductase 3 (mCbr3), murine cytochrome p450 family 2 subfamily a member 4 (mCyp2a4), murine solute carrier family 7 member 11 (mSlc7a11), murine keratin 20 (mKrt20), murine leucine rich repeats and transmembrane domains 2 (mLrtm2)], as well as 5 significantly decreased genes [murine major urinary protein 1 (mMup1), murine major urinary protein 16 (mMup16), murine major urinary protein 11 (mMup11), murine major urinary protein 12 (mMup12), murine major urinary protein 7 (mMup7)] were confirmed by RT-qPCR (Figure 3C,D; see also Figure S6C,D and the full list of genes presented in Excel Tables S4 and S5). Hallmark and KEGG pathway enrichment and Ingenuity Pathway Analysis (IPA) of the genes differentially expressed in tumors compared with TBT-exposed liver tissue (see Figure S7; Excel Tables S8, S9, and S10) or VEH-exposed liver tissue (see Figure S7; Excel Tables S8, S9, and S10) revealed that several pathways involved in cancer were enriched in adenomas (e.g., epithelial–mesenchymal transition, and cancer) as well as immune regulatory pathways [e.g., tumor necrosis factor-alpha () signaling via nuclear factor kappa beta ()] (see Excel Tables S8, S9, and S10). The differential gene expression signature highly overlapped between the two adenoma analyses (94% of the adenoma over TBT-exposed liver, and 81% of the adenoma over VEH-exposed liver overlap; Figure 4B). To understand TBT-induced changes that preceded adenoma development, we also examined livers from VEH- and TBT-exposed offspring at 20 weeks of age prior to tumor development. At 20 weeks of age, TBT exposure increased expression of 189 genes in the liver and decreased expression of 67 genes (Figure 4A; Excel Table S7). Interestingly, at 45 weeks, livers of offspring that did not present with adenomas were markedly more similar to age-matched VEH-exposed livers than adenomas seen at that age. This was evidenced by a much less robust change in gene expression, with only 26 genes differentially expressed [12 genes increased by TBT exposure and 14 genes decreased (Figure 4A; Excel Table S6)]. As shown in Figure 4A, PCA analysis showed little separation between livers of TBT-exposed animals that did not develop tumors and age-matched VEH-exposed livers. Together, these data suggest the interesting possibility that tumor development was not stochastic but, rather, a result in heterogeneity (and differences in susceptibility) within the population of TBT-exposed offspring. At 20 weeks of age, 108 of the 189 (57%) genes increased by TBT exposure were also increased in the adenomas (compared with either TBT- or VEH-exposed liver), whereas only 7 of the 67 (10%) genes decreased by TBT exposure are shared with the adenoma signature (Figure 4B). Only 7 of these genes exhibited altered expression at 20 weeks (i.e., were potentially directly induced by TBT), and 2 of those were perturbed in opposite directions (noted in Excel Table S6).
Figure 3.
Differential gene expression in tributyltin (TBT)-exposed adenomas vs. TBT-exposed liver samples. (A) Hierarchical clustering of RNA-seq from male mice [liver tissue from TBT-treated animals who did not develop macroscopic adenomas (T), , and adenoma tissue from TBT-treated animals (A), ] using differentially expressed genes (false discovery rate (FDR)-adjusted with a fold change exceeding ). (B) Principal component analysis of tumor and liver tissue from TBT-treated animals RNA-seq data. (C) Quantitative PCR validation of eight genes spanning the top 2% of differentially expressed genes identified in the RNA-seq analysis in panel A. (D) Quantitative PCR validation of the five most repressed major urinary protein (Mup) genes as identified in the RNA-seq analysis in panel A. In panels C and D, a T depicts TBT-exposed liver samples, and an A depicts TBT-exposed adenoma samples, the (IQ)1–IQ3 are presented, and Mann-Whitney U-tests were performed to ascertain significance. *; **. In all panels, circles depict TBT-exposed liver samples, and squares depict TBT-exposed adenoma samples. Excel Table S1 details information on n and litter. Note: A, adenoma; PC, principal component; T, TBT-treated males without any macroscopic adenomas.
Table 3.
Top 60 overexpressed genes in adenomas over tributyltin (TBT)-exposed liver.
| Gene | FC | p-Value | FDR |
|---|---|---|---|
| Lrtm2 | 8.14767385 | 1.13 | 7.89 |
| Eps8l3 | 7.94943879 | 4.58 | 3.59 |
| Krt20 | 7.94331082 | 1.38 | 1.13 |
| Lipn | 7.61854187 | 1.36 | 3.72 |
| Ucp1 | 7.52788071 | 2.94 | 7.89 |
| Aqp7 | 7.16928422 | 1.09 | 9.67 |
| Slc22a12 | 7.12755524 | 4.73 | 2.09 |
| Tekt5 | 7.11858539 | 4.72 | 4.28 |
| Spink1 | 6.95740777 | 6.69 | 4.44 |
| Timp4 | 6.86601984 | 5.61 | 8.97 |
| Gpx2 | 6.76584208 | 2.59 | 7.60 |
| Tinag | 6.75629973 | 4.14 | 4.50 |
| Syt14 | 6.70121494 | 1.49 | 1.63 |
| Slc7a11 | 6.60590914 | 1.48 | 2.34 |
| Cfap44 | 6.54600864 | 9.36 | 2.73 |
| Ly6d | 6.40078593 | 6.93 | 2.92 |
| Gipc2 | 6.37716822 | 2.05 | 4.10 |
| Styk1 | 6.33958465 | 3.85 | 8.37 |
| Muc13 | 6.3259154 | 3.23 | 1.22 |
| Inava | 6.32579732 | 1.34 | 9.80 |
| Ptprh | 6.30939473 | 4.57 | 1.14 |
| Gsta1 | 6.27773653 | 1.33 | 1.87 |
| Scn2a | 6.26264244 | 5.39 | 6.94 |
| Sult2a2 | 6.22351682 | 6.89 | 1.69 |
| Tceal5 | 6.14755118 | 4.08 | 1.37 |
| Bex4 | 6.14297967 | 8.10 | 3.53 |
| Baiap2l2 | 6.08915845 | 2.22 | 7.19 |
| Cacna1b | 6.06910411 | 9.06 | 3.75 |
| Ubd | 6.05288612 | 5.61 | 1.42 |
| Cyp2a4 | 6.0409971 | 9.62 | 1.73 |
| Gm3776 | 6.01597208 | 1.30 | 3.09 |
| Cyp2b10 | 6.00147324 | 5.28 | 6.94 |
| Tmem238l | 5.99439035 | 5.56 | 2.20 |
| Ptgds | 5.97524932 | 6.29 | 7.94 |
| Gpnmb | 5.97450643 | 2.74 | 2.16 |
| Insyn1 | 5.96205742 | 6.96 | 1.10 |
| Tubb2b | 5.95064059 | 3.80 | 1.42 |
| Elovl7 | 5.77754343 | 2.41 | 1.08 |
| Shc2 | 5.75636285 | 8.43 | 4.84 |
| Inpp5j | 5.69560793 | 4.57 | 1.47 |
| Slc1a1 | 5.66421111 | 1.96 | 1.22 |
| Rasal1 | 5.62461717 | 1.10 | 2.78 |
| Cyp2d12 | 5.58927876 | 2.97 | 1.10 |
| Tmprss4 | 5.51581358 | 2.04 | 4.69 |
| Sdcbp2 | 5.45750251 | 7.75 | 6.80 |
| Amn | 5.42274723 | 4.65 | 2.37 |
| Rbm24 | 5.42153442 | 1.62 | 1.10 |
| Cbr3 | 5.41783779 | 6.46 | 5.82 |
| Cdc25c | 5.39838395 | 1.44 | 4.36 |
| Spire2 | 5.35072302 | 6.64 | 4.96 |
| Diaph3 | 5.34680061 | 7.27 | 1.29 |
| Wfdc3 | 5.27003694 | 1.43 | 1.40 |
| Dnah1 | 5.24576415 | 6.69 | 1.02 |
| Ly6c1 | 5.22654812 | 6.56 | 3.18 |
| Fbn2 | 5.20291611 | 9.42 | 4.72 |
| Myo7b | 5.1861662 | 3.74 | 2.88 |
| Wfdc16 | 5.1642053 | 8.51 | 1.54 |
| Erich4 | 5.09937802 | 1.48 | 2.27 |
| Aplnr | 5.06456889 | 3.79 | 1.22 |
| Apela | 5.05908948 | 3.64 | 1.18 |
Note: FC, fold change; FDR, false discovery rate.
Table 4.
Top 60 repressed genes in adenoma over tributyltin (TBT)-exposed liver.
| Gene | FC | p-Value | FDR |
|---|---|---|---|
| Mup15 | 1.59 | 3.34 | |
| Mup7 | 1.03 | 3.25 | |
| Mup16 | 1.64 | 1.72 | |
| Hsd3b5 | 7.04 | 8.89 | |
| Hhip | 1.99 | 2.70 | |
| Mup12 | 6.16 | 7.06 | |
| Moxd1 | 4.80 | 3.03 | |
| Irx1 | 2.11 | 8.99 | |
| Mup1 | 4.26 | 2.69 | |
| Cd163 | 1.82 | 9.56 | |
| Mup11 | 3.51 | 3.41 | |
| Mup2 | 1.25 | 9.26 | |
| Susd4 | 1.33 | 1.12 | |
| Cxcl13 | 2.14 | 7.76 | |
| Timd4 | 3.86 | 3.56 | |
| Slc8a3 | 9.15 | 2.23 | |
| Slco1a1 | 1.07 | 5.42 | |
| Dmrta1 | 1.58 | 5.16 | |
| Mup17 | 3.04 | 1.32 | |
| Marco | 5.13 | 1.04 | |
| Mup14 | 8.18 | 3.33 | |
| Mup3 | 4.73 | 1.36 | |
| Mup22 | 5.57 | 1.96 | |
| Mup18 | 5.59 | 8.97 | |
| Mup8 | 6.56 | 9.62 | |
| Acpp | 2.51 | 6.45 | |
| Cyp2c37 | 1.48 | 5.21 | |
| Dct | 5.89 | 3.56 | |
| Mup13 | 4.87 | 1.54 | |
| Mup9 | 3.46 | 9.30 | |
| Cyp2c50 | 1.86 | 3.91 | |
| Cyp2c54 | 5.34 | 1.64 | |
| Jph1 | 1.31 | 2.76 | |
| Cyp46a1 | 1.73 | 4.96 | |
| Sult2a8 | 6.72 | 1.27 | |
| Tex12 | 1.77 | 2.90 | |
| Pnpla5 | 1.26 | 2.24 | |
| Zfp982 | 4.75 | 1.89 | |
| Cnmd | 5.73 | 1.42 | |
| Ric3 | 6.92 | 1.32 | |
| Srms | 5.85 | 9.94 | |
| Nxpe2 | 6.06 | 1.06 | |
| Mup19 | 4.14 | 6.08 | |
| Slc17a8 | 1.18 | 1.64 | |
| Gbp2b | 1.05 | 2.43 | |
| Capn8 | 2.23 | 5.00 | |
| Ripply1 | 1.53 | 1.80 | |
| Dbp | 2.23 | 2.39 | |
| Cyp8b1 | 1.95 | 2.11 | |
| Sult5a1 | 6.16 | 1.52 | |
| Colec10 | 3.25 | 4.62 | |
| Trhde | 3.14 | 8.91 | |
| Cyp2c29 | 8.65 | 8.33 | |
| Mup21 | 5.36 | 6.15 | |
| Onecut1 | 9.82 | 6.96 | |
| Efna3 | 9.01 | 1.08 | |
| Mup10 | 1.01 | 1.20 | |
| E2f8 | 1.33 | 6.25 | |
| Gna14 | 6.29 | 1.36 | |
| Cadm4 | 8.93 | 2.47 |
Note: FC, fold change; FDR, false discovery rate.
Figure 4.
Differential gene expression in tributyltin (TBT)-exposed liver samples and overlap with adenoma signatures. (A) Upper, hierarchical clustering of RNA-seq data from liver samples from 20- and 45-week-old male mice using differentially expressed genes [false discovery rate (FDR) adjusted with a fold change exceeding ]. Lower, principal component analysis of RNA-seq data at each time point. Triangles represent vehicle (VEH)-exposed liver samples, and circles represent TBT-exposed liver samples. (B) Venn diagrams of overlapping genes in all four gene expression signatures (up-regulated–left, down-regulated–right). A () over TBT-exposed liver (); B () over VEH-exposed liver (); C () over VEH-exposed liver () at 20 weeks; and D () over VEH-exposed liver (). See Excel Table S1 for detail information on n and litter.
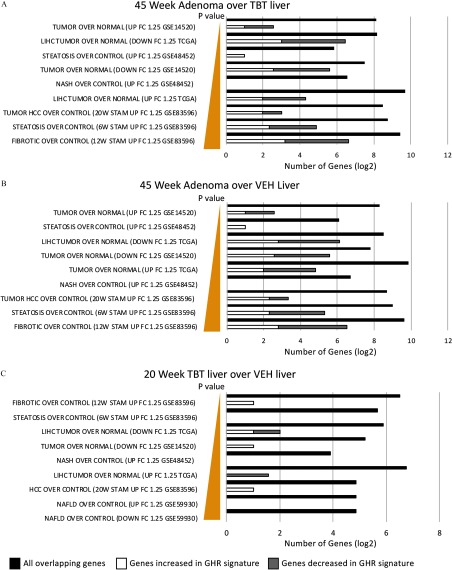
To understand the linkage(s) between tumor etiology in this model and well-characterized sequela for liver disease, we performed an ORA, following the MSigDB methodology, to ask if genes known to be involved in NASH/NAFLD/HCC in both human and mouse studies were enriched in adenomas of TBT-exposed animals or in TBT-exposed liver tissue prior to adenoma development. As shown in Figure 5, we found that a significant percentage of genes differentially expressed in TBT-induced adenomas compared with TBT-exposed or VEH-exposed liver tissue have been previously linked to steatosis, NASH, NAFLD, and/or HCC: these genes are listed in Excel Table S11. Interestingly, even in TBT-exposed liver tissue from 20-week-old animals, we observed an enrichment for expression of genes linked to liver disease, including tumorigenesis (Figure 5; Excel Table S11).
Figure 5.
Overrepresentation analysis (ORA) using publicly available mouse and human liver disease data sets. An ORA was conducted using the genes differentially expressed in the tumors from TBT-exposed males with publicly available mouse and human RNA-seq data sets in distinct liver disease states (GSE14520, LIHC TCGA, GSE48452, GSE83596, GSE59930). ORA for (A) 45 week adenoma over TBT liver, (B) 45 week adenoma over VEH liver, and (C) 20 week TBT liver over VEH liver. Significant enrichment was detected in all data sets. Closed bars represent all overlapping genes in scale, whereas open bars and gray bars represent the subset of all genes that are also present in the growth hormone receptor (GHR) signature. Note: HCC, hepatocellular carcinoma; LIHC, liver hepatocellular carcinoma; NASH, nonalcoholic steatohepatitis.
mRNA Expression of Members of the GHR/STAT5 Signaling Pathway in Male Mice Developmentally Exposed to TBT
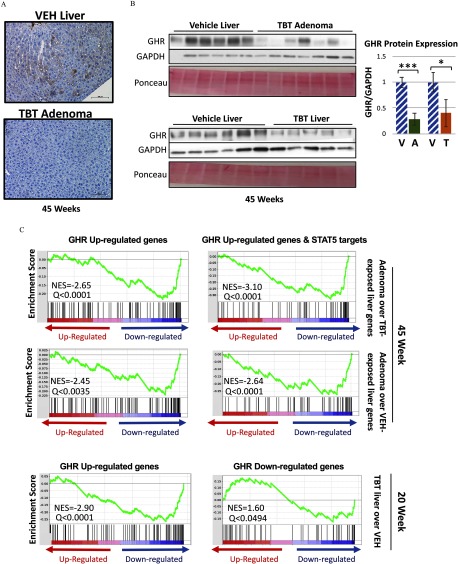
The mouse urinary proteins (Mups), members of the lipocalin gene family, are encoded by a contiguous gene cluster on mouse chromosome 4. These secreted proteins play a key role in chemical communication between animals and can trigger several types of behavioral responses (Zhou and Rui 2010). Interestingly, 5 of the 10 most repressed genes in TBT-induced adenomas were Mups 1, 7, 15, 12, and 16, and many more were included in the most reduced gene list (Table 4; Figure 3; Figure S6). Decreased Mup gene expression in adenomas resulted in decreased MUP protein expression, as confirmed via immunohistochemistry using an antibody specific for MUP1 (Figure 6A).
Figure 6.
Expression of members of the growth hormone receptor (GHR)/STAT5 signaling pathway in male mice developmentally exposed to tributyltin. (A) Representative images of immunohistochemical staining of major urinary protein 1 (MUP1) protein in vehicle (VEH)-treated liver and adenomas from tributyltin (TBT)-treated 45-week-old male mice (; scale bars:). (B) Immunoblot analysis of growth hormone receptor (GHR) protein levels in liver tissue from VEH- () and TBT-exposed () males and adenoma tissue from TBT-exposed males () at 45 weeks of age (left panels) with quantitation of the immunoblots (right panels). GHR level was normalized to GAPDH protein level in each lysate. Ponceau stain is displayed beneath each blot. Bars represent the . *, **, *** by t-test. (C) Gene Set Enrichment Analysis (GSEA) based on the transcriptomic footprint of TBT-induced adenomas vs. TBT- or VEH-exposed age-matched liver at 45 weeks using a publicly available GHR gene signature (GSE93382), either for all up-regulated genes (left) or the up-regulated genes that are also direct STAT5 targets (identified by STAT5 ChIP-Seq GSE31578, right). The 20-week transcriptome of male liver tissue was also analyzed for both up-regulated and down-regulated GHR genes (bottom panels). Significance was achieved at an FDR-adjusted Note: A, adenoma; T, TBT-treated males without any macroscopic adenomas; V, hatched, VEH-exposed liver.
In the rodent liver, Mup gene transcription is regulated by GHR signaling, which when activated results in phosphorylation and activation of the STAT5 transcription factor (Herrington et al. 2000; Clodfelter et al. 2006; Waxman and O’Connor 2006). Reduction in Mup gene expression has been linked to decreased GHR signaling, and Mup genes have previously been reported to be down-regulated during murine hepatocellular carcinogenesis (Arellanes-Robledo et al. 2010). We found that GHR protein expression was significantly reduced in TBT-induced liver adenomas compared with VEH-exposed liver (Figure 6B). Importantly, GHR expression was also significantly lower in TBT-exposed liver relative to VEH-exposed liver (Figure 6B), indicating that decreased GHR expression was a result of TBT-exposure rather than a consequence of pathophysiological changes associated with tumor development.
To evaluate perturbations in GHR signaling during tumorigenesis, we analyzed the transcriptional output of the GHR pathway in our gene expression data. We derived a growth hormone response gene signature by identifying differentially expressed genes in publicly available RNA-seq data sets of liver tissue from hypophysectomized male mice treated with growth hormone (GSE93382; Excel Table S2), which comprised 174 genes (105 that increased and 69 that decreased in response to growth hormone) (see Excel Table S12, column C). We then conducted a GSEA using this growth hormone-response signature and found a significant inverse enrichment of growth hormone-responsive genes in the genes altered in TBT-induced adenomas compared with either TBT-exposed or VEH-exposed liver tissue at 45 weeks of age (Figure 6C) consistent with decreased GHR signaling in TBT-induced adenomas. The genes that are differentially expressed in the TBT adenoma signature and overlap with the GHR gene signature, thus contributing to this enrichment, are shown in Figure S8 and Excel Table S13. Because GHR is known to signal through the transcription factor STAT5, we also tested for possible overlap between this signature and publicly available STAT5 ChIP-seq data in the murine liver treated with growth hormone. This analysis yielded a similar result (Figure 6C), and STAT5 targets are annotated in Figure S8, confirming that the TBT-induced decrease in GHR expression is consistent with a defect in GHR-mediated downstream STAT5 signaling. The negative Normalized Enrichment Score (NES) we observed from these analyses ( to , ) indicates that GHR up-regulated genes, under additional STAT5 regulation or not, were down-regulated in the TBT-treated adenomas. At 20 weeks of age, we observed a negative NES in up-regulated GHR genes [(, ) and a positive NES in down-regulated GHR genes (1.60, ), consistent with decreased GHR signaling occurring in TBT-exposed livers prior to tumor development (Figure 6C, bottom panel)].
Discussion
We report here that early life TBT exposure promoted fatty liver and development of liver adenomas in male offspring. Tumor development in this setting was preceded by decreased expression of GHR and STAT5 signaling, which in tumors resulted in down-regulation of GHR target genes such as the Mup genes. Developmental or prenatal exposure to TBT was previously shown to promote fatty liver in mice (Chamorro-García et al. 2013), and this finding was reproduced in male mice in our study. The sex bias toward males we observed in this study is consistent with the human NAFLD etiology, where males are four times more likely to develop this disease than females (Hoenerhoff et al. 2011). The TBT-induced adenoma phenotype is even more striking given that development of adenomas in the liver at 45 weeks is uncommon in mouse models in any strain or sex (Maronpot 2009), and to the best of our knowledge, developmental TBT exposure has not previously been reported to promote liver tumorigenesis.
A notable strength of our study is that it is the first to report the very long-term effect of developmental exposure to TBT leading to adenoma formation in the liver. In addition, we assessed both sexes, enabling us to discover that this is a sex-specific phenomenon. Although exciting, there were some limitations to this study, including an evaluation comparing adenomas to adjacent liver tissue from the same animal. This assessment would better describe alterations occurring in the liver prior to tumor development that are possibly contributory and will be addressed in future experiments. In some experiments, we did not have enough animals to conduct a thorough statistical evaluation, partially due to including more than one animal per litter, potentially losing the ability to properly control for dam/litter effects. These preliminary findings call for additional confirmatory experiments in the future.
In TBT-induced adenomas, Mup expression was dramatically repressed compared with livers from TBT- or VEH-exposed animals. Although MUPs are known to be excreted into the urine and bind and stabilize pheromones for communication between animals, this does not preclude them from being involved in other physiological functions, and MUP1, one of the most repressed of the major urinary proteins in our study, has been shown to be involved in several metabolic processes in mice (Hui et al. 2009; Zhou et al. 2009; Zhou and Rui 2010). The Mup genes are known to be regulated by the GHR/STAT5 pathway (Clodfelter et al. 2006; Waxman and O’Connor 2006), and decreased Mup expression in the TBT-induced adenomas is entirely consistent with reduced GHR protein and signaling observed in the tumors. Interestingly, in human studies GHR has been previously reported to be down-regulated in HCC. In an integrative analysis of gene expression alterations in seven data sets that included 135 HCC samples and 110 control liver samples, GHR was identified to be down-regulated in HCC compared with healthy liver controls (Choi et al. 2004). Another study evaluated GHR protein expression across a spectrum of liver diseases in 36 diseased patients and 9 healthy controls (Vespasiani Gentilucci et al. 2006). GHR protein expression in the cytoplasm decreased with liver disease progression, whereas GHR protein expression in the nuclear compartment increased with increasing stages of liver disease. GHR protein expression in HCC patients did not differ from levels in healthy control patients in either cellular compartment, although the study included only five patients with HCC (Vespasiani Gentilucci et al. 2006).
Multiple studies in rodent models have shown that exposure to TBT during development can promote obesity and fatty liver disease later in life (Grün and Blumberg 2006; Grün et al. 2006; Grün and Blumberg 2009a, 2009b; Janesick and Blumberg 2011, 2012, 2016; Heindel et al. 2017). Our data reveal a new consequence of early life TBT exposure: promotion of tumorigenesis in a sex-dependent manner. In addition, our data adds to the accumulating knowledge of sex bias in response to early life environmental exposures. Sex bias in liver pathology has been seen in other rodent models of developmental exposures to endocrine disruptors (Maranghi et al. 2010; Strakovsky et al. 2015). However, the basis for this bias, or the sex-specific promotion of liver tumorigenesis we observed in response to TBT, is unknown. Regardless, the data presented here adds to the accumulating knowledge about how early life exposures impact target tissues such as the liver by identifying genes and signaling pathways that become dysregulated in adulthood as a result of early life TBT exposure. Furthermore, our demonstration that TBT can promote liver tumorigenesis and provides a new model system for studying how environmental exposures may contribute to the increase in liver disease that is currently paralleling the obesity epidemic worldwide.
Supplementary Material
Acknowledgments
This work was part of the Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TaRGET) II Consortium. Oil Red O staining was conducted at the MD Anderson Cancer Center Research Histology, Pathology, and Imaging Core (RHPI) (supported by P30 CA16672 DHHS/NCI Cancer Center Support Grant). Oil Red O imaging and quantification was supported by the Integrated Microscopy Core at Baylor College of Medicine with funding from the National Institutes of Health (NIH; DK56338 and CA125123), the Cancer Prevention and Research Institute of Texas (CPRIT; RP150578), the Dan L. Duncan Comprehensive Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomics. Magnetic resonance imaging was conducted by the Small Animal Imaging Facility at Texas Children’s Hospital. Immunohistochemical staining and slide scanning was conducted by the Texas Medical Center, Digestive Diseases Center, Cellular and Molecular Morphology and RNA In Situ Hybridization core facilities supported in part by NIH Public Health Service (PHS) grant P30DK056338 and a Shared Instrumentation grant from the NIH (1S10OD016167). RNA sequencing was conducted at the MD Anderson Cancer Center Science Park Next Generation Sequencing Core (supported by RP170002). C.C., S.L.G., and M.J.R. were partially supported by CPRIT (RP170005). C.E.F. was partially supported by a subaward from NIH grant 4 U24 DK097748-05. J.-P.B. was supported by an award from the Ligue Nationale Contre le Cancer and the Phillippe Foundation. This work was supported in part by NIH grants U01ES026719 and R01ES023206 to C.L.W.
Footnotes
Supplemental Material is available online (https://doi.org/10.1289/EHP5414).
The authors declare they have no actual or potential competing financial interests.
Note to readers with disabilities: EHP strives to ensure that all journal content is accessible to all readers. However, some figures and Supplemental Material published in EHP articles may not conform to 508 standards due to the complexity of the information being presented. If you need assistance accessing journal content, please contact ehponline@niehs.nih.gov. Our staff will work with you to assess and meet your accessibility needs within 3 working days.
References
- Airaksinen R, Rantakokko P, Turunen AW, Vartiainen T, Vuorinen PJ, Lappalainen A, et al. 2010. Organotin intake through fish consumption in Finland. Environ Res 110(6):544–547, PMID: 20573344, 10.1016/j.envres.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Antizar-Ladislao B. 2008. Environmental levels, toxicity and human exposure to tributyltin (TBT)-contaminated marine environment. a review. Environ Int 34(2):292–308, PMID: 17959247, 10.1016/j.envint.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Arellanes-Robledo J, Salcido-Neyoy ME, Márquez-Quiñones A, García-Román R, Beltrán-Ramírez O, Le Berre V, et al. 2010. Celecoxib activates Stat5 and restores or increases the expression of growth hormone-regulated genes in hepatocarcinogenesis. Anticancer Drugs 21(4):411–422, PMID: 20145537, 10.1097/CAD.0b013e328336e907. [DOI] [PubMed] [Google Scholar]
- Ashraf MW, Salam A, Mian A. 2017. Levels of organotin compounds in selected fish species from the Arabian Gulf. Bull Environ Contam Toxicol 98(6):811–816, PMID: 28405694, 10.1007/s00128-017-2083-9. [DOI] [PubMed] [Google Scholar]
- Azenha M, Vasconcelos MT. 2002. Butyltin compounds in Portuguese wines. J Agric Food Chem 50(9):2713–2716, PMID: 11958646, 10.1021/jf0115544. [DOI] [PubMed] [Google Scholar]
- Benson R. 1999. Concise International Chemical Assessment Documents, Document 14; Tributyltin Oxide. Geneva, Switzerland: World Health Organization; https://www.who.int/ipcs/publications/cicad/en/cicad14.pdf?ua=1 [accessed 8 December 2019]. [Google Scholar]
- Blesson CS, Schutt AK, Balakrishnan MP, Pautler RG, Pedersen SE, Sarkar P, et al. 2016. Novel lean type 2 diabetic rat model using gestational low-protein programming. Am J Obstet Gynecol 214(4):540.e541–540.e547, PMID: 26874300, 10.1016/j.ajog.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-Garcia R, Diaz-Castillo C, Shoucri BM, Käch H, Leavitt R, Shioda T, et al. 2017. Ancestral perinatal obesogen exposure results in a transgenerational thrifty phenotype in mice. Nat Commun 8(1):2012, PMID: 29222412, 10.1038/s41467-017-01944-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-García R, Sahu M, Abbey RJ, Laude J, Pham N, Blumberg B. 2013. Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice. Environ Health Perspect 121(3):359–366, PMID: 23322813, 10.1289/ehp.1205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JP, Peng B, Tang L, Sun R, Hu S, Wen XY, et al. 2016. Fetal and infant exposure to the Chinese famine increases the risk of fatty liver disease in Chongqing, China. J Gastroenterol Hepatol 31(1):200–205, PMID: 26201820, 10.1111/jgh.13044. [DOI] [PubMed] [Google Scholar]
- Chien LC, Hung TC, Choang KY, Yeh CY, Meng PJ, Shieh MJ, et al. 2002. Daily intake of TBT, Cu, Zn, Cd and As for fishermen in Taiwan. Sci Total Environ 285(1–3):177–185, PMID: 11874040, 10.1016/s0048-9697(01)00916-0. [DOI] [PubMed] [Google Scholar]
- Choi JK, Choi JY, Kim DG, Choi DW, Kim BY, Lee KH, et al. 2004. Integrative analysis of multiple gene expression profiles applied to liver cancer study. FEBS Lett 565(1–3):93–100, PMID: 15135059, 10.1016/j.febslet.2004.03.081. [DOI] [PubMed] [Google Scholar]
- Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. 2006. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol 20(6):1333–1351, PMID: 16469768, 10.1210/me.2005-0489. [DOI] [PubMed] [Google Scholar]
- Coenen TM, Brouwer A, Enninga IC, Koeman JH. 1992. Subchronic toxicity and reproduction effects of tri-n-butyltin oxide in Japanese quail. Arch Environ Contam Toxicol 23(4):457–463, PMID: 1444589, 10.1007/bf00203809. [DOI] [PubMed] [Google Scholar]
- Connerney J, Lau-Corona D, Rampersaud A, Waxman DJ. 2017. Activation of male liver chromatin accessibility and STAT5-dependent gene transcription by plasma growth hormone pulses. Endocrinology 158(5):1386–1405, PMID: 28323953, 10.1210/en.2017-00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. 2011. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci 124(1):1–22, PMID: 21908767, 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorneles PR, Lailson-Brito J, Fernandez MA, Vidal LG, Barbosa LA, Azevedo AF, et al. 2008. Evaluation of cetacean exposure to organotin compounds in Brazilian waters through hepatic total tin concentrations. Environ Pollut 156(3):1268–1276, PMID: 18440109, 10.1016/j.envpol.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W. 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4(8):1184–1191, PMID: 19617889, 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fent K. 1996. Ecotoxicology of organotin compounds. Crit Rev Toxicol 26(1):1–117, PMID: 8833456, 10.3109/10408449609089891. [DOI] [PubMed] [Google Scholar]
- Filipkowska A, Złoch I, Wawrzyniak-Wydrowska B, Kowalewska G. 2016. Organotins in fish muscle and liver from the Polish coast of the Baltic Sea: is the total ban successful? Mar Pollut Bull 111(1–2):493–499, PMID: 27345706, 10.1016/j.marpolbul.2016.06.052. [DOI] [PubMed] [Google Scholar]
- Foulds CE, Treviño LS, York B, Walker CL. 2017. Endocrine-disrupting chemicals and fatty liver disease. Nat Rev Endocrinol 13(8):445–457, PMID: 28524171, 10.1038/nrendo.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme H, Mattulat A, Lahrz T, Rüden H. 2005. Occurrence of organotin compounds in house dust in Berlin (Germany). Chemosphere 58(10):1377–1383, PMID: 15686755, 10.1016/j.chemosphere.2004.09.092. [DOI] [PubMed] [Google Scholar]
- Grün F, Blumberg B. 2006. Environmental obesogens: organotins and endocrine disruption via nuclear receptor signaling. Endocrinology 147(suppl 6):S50–S55, PMID: 16690801, 10.1210/en.2005-1129. [DOI] [PubMed] [Google Scholar]
- Grün F, Blumberg B. 2009a. Endocrine disrupters as obesogens. Mol Cell Endocrinol 304(1–2):19–29, PMID: 19433244, 10.1016/j.mce.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün F, Blumberg B. 2009b. Minireview: the case for obesogens. Mol Endocrinol 23(8):1127–1134, PMID: 19372238, 10.1210/me.2008-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün F, Watanabe H, Zamanian Z, Maeda L, Arima K, Cubacha R, et al. 2006. Endocrine-disrupting organotin compounds are potent inducers of adipogenesis in vertebrates. Mol Endocrinol 20(9):2141–2155, PMID: 16613991, 10.1210/me.2005-0367. [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22(9):1760–1774, PMID: 22955987, 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindel JJ, Blumberg B, Cave M, Machtinger R, Mantovani A, Mendez MA, et al. 2017. Metabolism disrupting chemicals and metabolic disorders. Reprod Toxicol 68:3–33, PMID: 27760374, 10.1016/j.reprotox.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J, Smit LS, Schwartz J, Carter-Su C. 2000. The role of STAT proteins in growth hormone signaling. Oncogene 19(21):2585–2597, PMID: 10851057, 10.1038/sj.onc.1203526. [DOI] [PubMed] [Google Scholar]
- Hoenerhoff MJ, Pandiri AR, Lahousse SA, Hong HH, Ton TV, Masinde T, et al. 2011. Global gene profiling of spontaneous hepatocellular carcinoma in B6C3F1 mice: similarities in the molecular landscape with human liver cancer. Toxicol Pathol 39(4):678–699, PMID: 21571946, 10.1177/0192623311407213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui X, Zhu W, Wang Y, Lam KS, Zhang J, Wu D, et al. 2009. Major urinary protein-1 increases energy expenditure and improves glucose intolerance through enhancing mitochondrial function in skeletal muscle of diabetic mice. J Biol Chem 284(21):14050–14057, PMID: 19336396, 10.1074/jbc.M109.001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav S, Bhosale D, Bhosle N. 2011. Baseline of organotin pollution in fishes, clams, shrimps, squids and crabs collected from the west coast of India. Mar Pollut Bull 62(10):2213–2219, PMID: 21820681, 10.1016/j.marpolbul.2011.06.023. [DOI] [PubMed] [Google Scholar]
- Janesick A, Blumberg B. 2011. Endocrine disrupting chemicals and the developmental programming of adipogenesis and obesity. Birth Defects Res C Embryo Today 93(1):34–50, PMID: 21425440, 10.1002/bdrc.20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesick A, Blumberg B. 2012. Obesogens, stem cells and the developmental programming of obesity. Int J Androl 35(3):437–448, PMID: 22372658, 10.1111/j.1365-2605.2012.01247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesick AS, Blumberg B. 2016. Obesogens: an emerging threat to public health. Am J Obstet Gynecol 214(5):559–565, PMID: 26829510, 10.1016/j.ajog.2016.01.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama T, Kobayashi N, Mamiya S, Nakanishi T, Nishikawa J. 2005. Organotin compounds promote adipocyte differentiation as agonists of the peroxisome proliferator-activated receptor γ/retinoid X receptor pathway. Mol Pharmacol 67(3):766–774, PMID: 15611480, 10.1124/mol.104.008409. [DOI] [PubMed] [Google Scholar]
- Kannan K, Takahashi S, Fujiwara N, Mizukawa H, Tanabe S. 2010. Organotin compounds, including butyltins and octyltins, in house dust from Albany, New York, USA. Arch Environ Contam Toxicol 58(4):901–907, PMID: 20379706, 10.1007/s00244-010-9513-6. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360, PMID: 25751142, 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucuksezgin F, Aydin-Onen S, Gonul LT, Pazi I, Kocak F. 2011. Assessment of organotin (butyltin species) contamination in marine biota from the Eastern Aegean Sea, Turkey. Mar Pollut Bull 62(9):1984–1988, PMID: 21764084, 10.1016/j.marpolbul.2011.06.020. [DOI] [PubMed] [Google Scholar]
- Lagerström M, Strand J, Eklund B, Ytreberg E. 2017. Total tin and organotin speciation in historic layers of antifouling paint on leisure boat hulls. Environ Pollut 220(Pt B):1333–1341, PMID: 27836476, 10.1016/j.envpol.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Lee JH, Wada T, Febbraio M, He J, Matsubara T, Lee MJ, et al. 2010. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology 139(2):653–663, PMID: 20303349, 10.1053/j.gastro.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ycaza J, Blumberg B. 2011. The environmental obesogen tributyltin chloride acts via peroxisome proliferator activated receptor gamma to induce adipogenesis in murine 3T3-L1 preadipocytes. J Steroid Biochem Mol Biol 127(1–2):9–15, PMID: 21397693, 10.1016/j.jsbmb.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930, PMID: 24227677, 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lima D, Castro LF, Coelho I, Lacerda R, Gesto M, Soares J, et al. 2015. Effects of tributyltin and other retinoid receptor agonists in reproductive-related endpoints in the zebrafish (Danio rerio). J Toxicol Environ Health A 78(12):747–760, PMID: 26090559, 10.1080/15287394.2015.1028301. [DOI] [PubMed] [Google Scholar]
- Lv Z, Li G, Li Y, Ying C, Chen J, Chen T, et al. 2013. Glucose and lipid homeostasis in adult rat is impaired by early-life exposure to perfluorooctane sulfonate. Environ Toxicol 28(9):532–542, PMID: 23983163, 10.1002/tox.20747. [DOI] [PubMed] [Google Scholar]
- Maranghi F, Lorenzetti S, Tassinari R, Moracci G, Tassinari V, Marcoccia D, et al. 2010. In utero exposure to di-(2-ethylhexyl) phthalate affects liver morphology and metabolism in post-natal CD-1 mice. Reprod Toxicol 29(4):427–432, PMID: 20307648, 10.1016/j.reprotox.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Maronpot RR. 2009. Biological basis of differential susceptibility to hepatocarcinogenesis among mouse strains. J Toxicol Pathol 22(1):11–33, PMID: 22271974, 10.1293/tox.22.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T, Klatt KC, Smith J, Castro E, Manary M, Caudill MA, et al. 2018. Choline supplementation prevents a hallmark disturbance of kwashiorkor in weanling mice fed a maize vegetable diet: hepatic steatosis of undernutrition. Nutrients 10(5):653, PMID: 29786674, 10.3390/nu10050653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohara K, Tateishi Y, Suzuki T, Okamura K, Murai H, Takumi S, et al. 2012. Late-onset increases in oxidative stress and other tumorigenic activities and tumors with a Ha-ras mutation in the liver of adult male C3H mice gestationally exposed to arsenic. Toxicol Sci 129(2):293–304, PMID: 22700541, 10.1093/toxsci/kfs203. [DOI] [PubMed] [Google Scholar]
- Ortiz L, Nakamura B, Li X, Blumberg B, Luderer U. 2013. In utero exposure to benzo[a]pyrene increases adiposity and causes hepatic steatosis in female mice, and glutathione deficiency is protective. Toxicol Lett 223(2):260–267, PMID: 24107266, 10.1016/j.toxlet.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842, PMID: 20110278, 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140, PMID: 19910308, 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandboge S, Perälä MM, Salonen MK, Blomstedt PA, Osmond C, Kajantie E, et al. 2013. Early growth and non-alcoholic fatty liver disease in adulthood—the NAFLD liver fat score and equation applied on the Helsinki Birth Cohort Study. Ann Med 45(5–6):430–437, PMID: 23767967, 10.3109/07853890.2013.801275. [DOI] [PubMed] [Google Scholar]
- Shimpi PC, More VR, Paranjpe M, Donepudi AC, Goodrich JM, Dolinoy DC, et al. 2017. Hepatic lipid accumulation and Nrf2 expression following perinatal and peripubertal exposure to bisphenol A in a mouse model of nonalcoholic liver disease. Environ Health Perspect 125(8):087005, PMID: 28796629, 10.1289/EHP664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonak S. 2009. Implications of organotins in the marine environment and their prohibition. J Environ Manage 90(suppl 1):S1–S3, PMID: 18973976, 10.1016/j.jenvman.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Sonak S, Pangam P, Giriyan A, Hawaldar K. 2009. Implications of the ban on organotins for protection of global coastal and marine ecology. J Environ Manage 90(suppl 1):S96–S108, PMID: 18977581, 10.1016/j.jenvman.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Stark R, Grzelak M, Hadfield J. 2019. RNA sequencing: the teenage years. Nat Rev Genet 20(11):631–656, PMID: 31341269, 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- Strakovsky RS, Wang H, Engeseth NJ, Flaws JA, Helferich WG, Pan YX, et al. 2015. Developmental bisphenol A (BPA) exposure leads to sex-specific modification of hepatic gene expression and epigenome at birth that may exacerbate high-fat diet-induced hepatic steatosis. Toxicol Appl Pharmacol 284(2):101–112, PMID: 25748669, 10.1016/j.taap.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550, PMID: 16199517, 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoolen B, Maronpot RR, Harada T, Nyska A, Rousseaux C, Nolte T, et al. 2010. Proliferative and nonproliferative lesions of the rat and mouse hepatobiliary system. Toxicol Pathol 38(suppl 7):5S–81S, PMID: 21191096, 10.1177/0192623310386499. [DOI] [PubMed] [Google Scholar]
- Treviño LS, Katz TA. 2018. Endocrine disruptors and developmental origins of nonalcoholic fatty liver disease. Endocrinology 159(1):20–31, PMID: 29126168, 10.1210/en.2017-00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2003. Ambient Aquatic Life Water Quality Criteria for Tributyltin (TBT)—Final. EPA 822-R-03-031. Washington, DC: U.S. Environmental Protection Agency. [Google Scholar]
- Vespasiani Gentilucci U, Perrone G, Galati G, D’Avola D, Zardi EM, Rabitti C, et al. 2006. Subcellular shift of the hepatic growth hormone receptor with progression of hepatitis C virus-related chronic liver disease. Histopathology 48(7):822–830, PMID: 16722931, 10.1111/j.1365-2559.2006.02429.x. [DOI] [PubMed] [Google Scholar]
- Wang N, Chen Y, Ning Z, Li Q, Han B, Zhu C, et al. 2016. Exposure to famine in early life and nonalcoholic fatty liver disease in adulthood. J Clin Endocrinol Metab 101(5):2218–2225, PMID: 27003304, 10.1210/jc.2016-1076. [DOI] [PubMed] [Google Scholar]
- Wang N, Wang X, Li Q, Han B, Chen Y, Zhu C, et al. 2017. The famine exposure in early life and metabolic syndrome in adulthood. Clin Nutr 36(1):253–259, PMID: 26646357, 10.1016/j.clnu.2015.11.010. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li C, Yang Z, Ma J, Zou Z. 2017. Fetal and infant exposure to severe Chinese famine increases the risk of adult dyslipidemia: results from the China Health and Retirement Longitudinal Study. BMC Public Health 17(1):488, PMID: 28610568, 10.1186/s12889-017-4421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman DJ, O’Connor C. 2006. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol 20(11):2613–2629, PMID: 16543404, 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- Weinhouse C, Anderson OS, Bergin IL, Vandenbergh DJ, Gyekis JP, Dingman MA, et al. 2014. Dose-dependent incidence of hepatic tumors in adult mice following perinatal exposure to bisphenol A. Environ Health Perspect 122(5):485–491, PMID: 24487385, 10.1289/ehp.1307449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Laz EV, Waxman DJ. 2012. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol Cell Biol 32(4):880–896, PMID: 22158971, 10.1128/MCB.06312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Jiang L, Rui L. 2009. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem 284(17):11152–11159, PMID: 19258313, 10.1074/jbc.M900754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Rui L. 2010. Major urinary protein regulation of chemical communication and nutrient metabolism. Vitam Horm 83:151–163, PMID: 20831945, 10.1016/S0083-6729(10)83006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.