Abstract
Appropriate post-translational processing of collagen requires prolyl hydroxylation, catalyzed by collagen prolyl 3-hydroxylase and collagen prolyl 4-hydroxylase, and is essential for normal cell function. Here we have investigated the expression, transcriptional regulation, and function of the collagen prolyl 3-hydroxylase and collagen prolyl 4-hydroxylase families in melanoma. We show that the collagen prolyl 3-hydroxylase family exemplified by Leprel1 and Leprel2 is subject to methylation-dependent transcriptional silencing in primary and metastatic melanoma consistent with a tumor suppressor function. In contrast, although there is transcriptional silencing of P4HA3 in a subset of melanomas, the collagen prolyl 4-hydroxylase family members P4HA1, P4HA2, and P4HA3 are often overexpressed in melanoma, expression being prognostic of worse clinical outcomes. Consistent with tumor suppressor function, ectopic expression of Leprel1 and Leprel2 inhibits melanoma proliferation, whereas P4HA2 and P4HA3 increase proliferation, and particularly invasiveness, of melanoma cells. Pharmacological inhibition with multiple selective collagen prolyl 4-hydroxylase inhibitors reduces proliferation and inhibits invasiveness of melanoma cells. Together, our data identify the collagen prolyl 3-hydroxylase and collagen prolyl 4-hydroxylase families as potentially important regulators of melanoma growth and invasiveness and suggest that selective inhibition of collagen prolyl 4-hydroxylase is an attractive strategy to reduce the invasive properties of melanoma cells.
INTRODUCTION
Melanoma is an aggressive skin cancer with a high metastatic potential. Cytotoxic drugs are rarely effective in melanoma, but targeted agents and immunotherapy have improved the prognosis (Koeblinger et al., 2017; Robert et al., 2015). Notwithstanding these developments, the prognosis for many patients with advanced melanoma remains poor, and novel therapeutic strategies continue to be urgently required.
Alterations in the properties of the basement membrane are a recognized feature of cancer cells. Collagen, the principal structural component (Shoulders and Raines, 2009), must undergo appropriate post-translational processing for efficient function, and this requires the activities of the prolyl 3-hydroxylases (C-P3Hs) and prolyl 4-hydroxylases (C-P4Hs). The C-P3H Leprecan Like 1 (Leprel1, P3H2) has been shown to participate in the processing of basement membrane collagens such as type IV collagen (Fernandes et al., 2011), but the substrate specificity of Leprel2 is unknown. Like the C-P3H proteins, the C-P4Hs also have important functions in collagen synthesis (Gorres and Raines, 2010; Myllyharju, 2008). In contrast to the C-P3Hs, the C-P4Hs are tetrameric proteins comprising 2α and 2β subunits. P4HA1, P4HA2, and P4HA3 encode the catalytic α subunits of the three C-P4H proteins, while a common β subunit is encoded by P4HB. P4HA1 and P4HA2 have been well studied, but little is known of the functions of P4HA3 since its molecular cloning (Van Den Diepstraten et al., 2003). We have previously shown that C-P3H genes are tumor suppressors, subject to methylation-dependent transcriptional silencing in breast cancer (Shah et al., 2009). In non-Hodgkin’s lymphoma, C-P3H and C-P4H genes are silenced by methylation at high frequencies, with distinct patterns of methylation in different lymphoma types (Hatzimichael et al., 2012). P4HA2 is a direct transcriptional target for p53, suggesting a tumor suppressor function (Teodoro et al., 2006). Conversely, a recent study found P4HA1 and P4HA2 to be essential for breast cancer metastasis (Gilkes et al., 2013).
Changes in the genome and epigenome of melanoma cells that drive metastasis remain incompletely described. Evidence from our own and others’ studies show that changes in CpG island methylation occur in metastatic melanoma. For example, increased methylation in the CpG island of TFPI2 is associated with metastatic melanoma (Lo Nigro et al., 2013; Tanemura et al., 2009), whereas the opposite effect is seen with NT5E (Wang et al., 2012).
Here, we have analyzed the expression, regulation, and function of the collagen prolyl hydroxylase genes in melanoma.
RESULTS
C-P3H and C-P4H genes are epigenetically regulated in melanoma cell lines
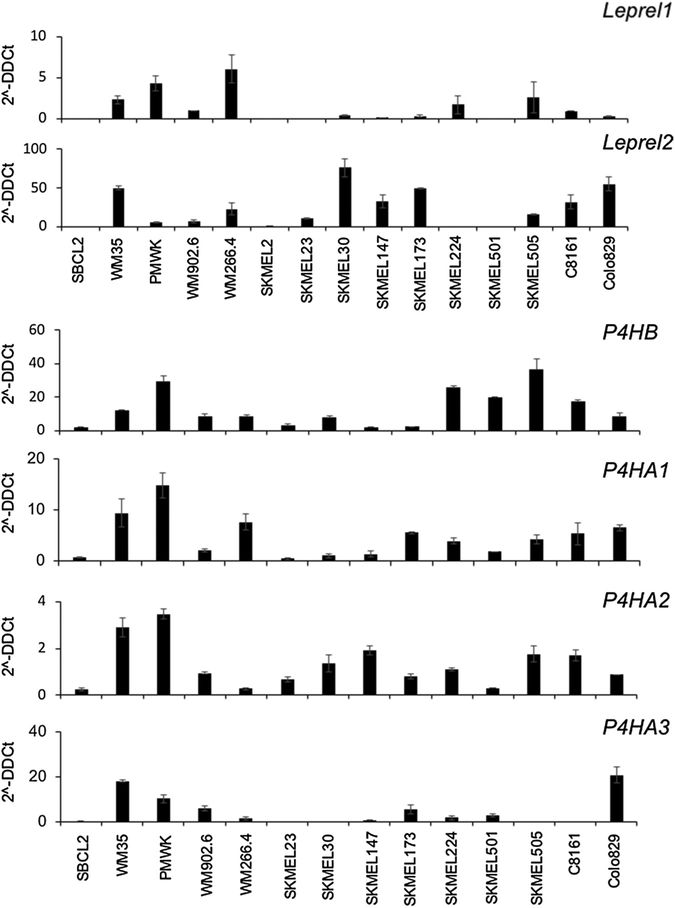
We examined mRNA expression of the C-P3H (Leprel1 and Leprel2) and C-P4H (P4HA1, P4HA2, P4HA3, and P4HB) encoding genes in a panel of melanoma cell lines (Figure 1). Leprel1 was expressed in the radial growth phase melanoma WM35 and PMWK and in WM266.4 and the metastatic melanoma cell lines SKMEL224, SKMEL505, and C8161, but was undetectable in SBCL2, SKMEL2, SKMEL23, SKMEL147, and SKMEL501 (Figure 1). Leprel2 was more abundantly expressed across the cell line panel, but not detectable in SBCL2, SKMEL224, and SKMEL501 (Figure 1). P4HB (encoding the β subunit common to all C-P4H proteins) was detectable in all cell lines (Figure 1). Similarly, P4HA1 and P4HA2 mRNA was detectable in all cell lines. In contrast to the other C-P4H encoding genes, P4HA3 was undetectable in some cell lines (SBCL2 [radial growth phase melanoma], SKMEL23, SKMEL30, SKMEL505, and C8161) and detectable only at very low levels in SKMEL147, SKMEL224, and WM266.4. Of note, in each of these cell lines with undetectable or extremely low expression of P4HA3, there was correspondingly moderate to high expression of P4HA1 and especially P4HA2 (Figure 1).
Figure 1. Expression of C-P3H and C-P4H genes in melanoma cell lines.
Quantitative PCR analysis of each C-P3H and C-P4H genes in the melanoma cell line panel. cDNA was prepared and quantitative PCR performed as described in Materials and Methods. Expression is shown as 2-ΔΔCt calculated as described in Materials and Methods. Data shown are mean ± 1 standard deviation. Each experiment was performed in triplicate and repeated at least twice.
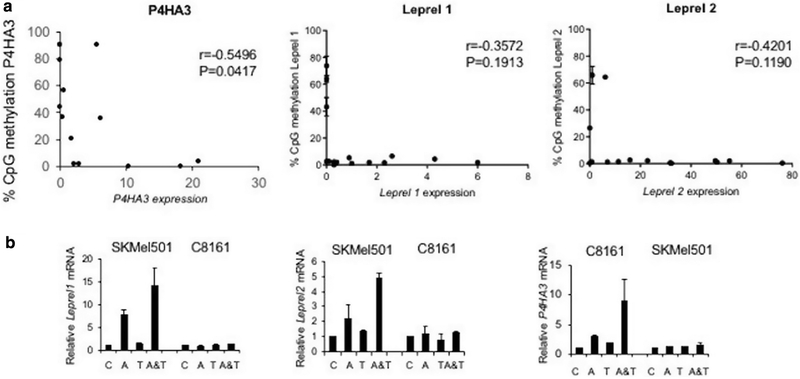
A CpG island is located in the upstream regulatory sequences of all examined genes, and we used pyrosequencing to quantitatively test for methylation. Representative pyrograms are shown in Supplementary Figure S1 online and full methylation profiles in Supplementary Figure S2 online. Each gene was unmethylated in normal melanocytes. Dense methylation in Leprel1 was detected in SKMEL2, SKMEL23, and SKMEL50 and in Leprel2 in SBCL2, PMWK, and SKMEL50. The P4HA1 CpG island was uniformly unmethylated and, similarly, P4HA2 was generally unmethylated, but we did detect methylation in SKMEL23 and SKMEL30. However, this did not correlate with reduced expression (Figure 1). There was, however, a good correlation between methylation in the P4HA3 CpG island and downregulation of expression with dense methylation in SBCL2, WM902.6, SKMEL505, SKMEL23, SKMEL30, SKMEL147, and C8161. P4HA3 expression was undetectable in all these lines with the exception of WM902.6, in which expression was reduced relative to unmethylated cell lines, such as Colo829. Correlation analysis demonstrated that expression of P4HA3 was negatively associated with methylation (r = −0.5496; P = 0.0417) (Figure 2a). Leprel1 and Leprel 2 also demonstrated a negative correlation (r = −0.3572 and r = −0.4201, respectively). Although this did not quite reach statistical significance, it is clear that the presence of dense CpG island methylation is associated with low to no expression for both genes. For example, the three cell lines that have dense CpG island methylation in Leprel1 (SKMEL2, SKMEL23, and SKMEL501), all have undetectable expression (Figure 2a). Similarly, the three cell lines with dense CpG island methylation in Leprel2 (SBCL2, SKMEL501, and PMWK) have undetectable (SBCL2 and SKMEL501) or low (PMWK) expression (Figure 2a). To further confirm that methylation downregulates expression, we performed demethylation experiments with 5′ azacytidine and trichostatin A. C8161 cells (express Leprel1, Leprel2 but do not express P4HA3 mRNA) and SKMEL501 (do not express Leprel or Leprel2 but do express P4HA3) were grown with or without 5′ azacytidine and trichostatin A. Quantitative PCR analysis showed that P4HA3, but not Leprel1 and Leprel2, was upregulated by 5′ azacytidine and 5′ azacytidine + trichostatin A in C8161 cells, whereas P4HA3 was not upregulated in SKMEL501, but both Leprel1 and Leprel2 were upregulated (Figure 2b).
Figure 2. C-P3H and C-P4H are epigenetically regulated in melanoma.
(a) Scatter plot showing that methylation in the P4HA3, Leprel1 and Leprel2 CpG islands is negatively correlated with expression. Data are mean ± standard error of the mean (n = 15). Correlation analysis was done using Pearson’s correlation coefficient. (b) Demethylation reactivates expression of P4HA3 in melanoma cells with CpG island methylation. C8161 and SKMEL501 cells were grown in the presence of azacytidine (A), trichostatin A (T) or both agents (A&T). Expression was determined by quantitative PCR. Data shown are mean ± 1 standard deviation relative to control cells (C).
C-P3H and C-P4H methylation in primary and metastatic clinical melanoma cases
These results prompted us to analyze methylation in a series of 50 clinical melanoma cases. We used benign pigmented nevi as control tissues. Representative pyrograms are shown in Supplementary Figure S1. Methylation levels for each of the C-P3H and C-P4H genes were uniformly low in benign nevi (Supplementary Figure S3 online). In melanomas, P4HA1 was unmethylated in all cases, and P4HA2 mean methylation was <5%. For both Leprel1 and P4HA3, levels of methylation were significantly higher in melanoma than in the benign nevi (P < 0.001 for both genes) (Supplementary Figure S3), showing that CpG island methylation in Leprel1 and P4HA3 is specific to malignant melanocytes. We then analyzed Leprel1 and P4HA3 methylation in metastatic melanomas. For Leprel1, the density of methylation was similar in nodal metastatic lesions and primary lesions, but for P4HA3 methylation was higher in some lymph node metastases than in primary cases (P < 0.01; Supplementary Figure S3).
C-P3H and C-P4H have opposing effects on melanoma
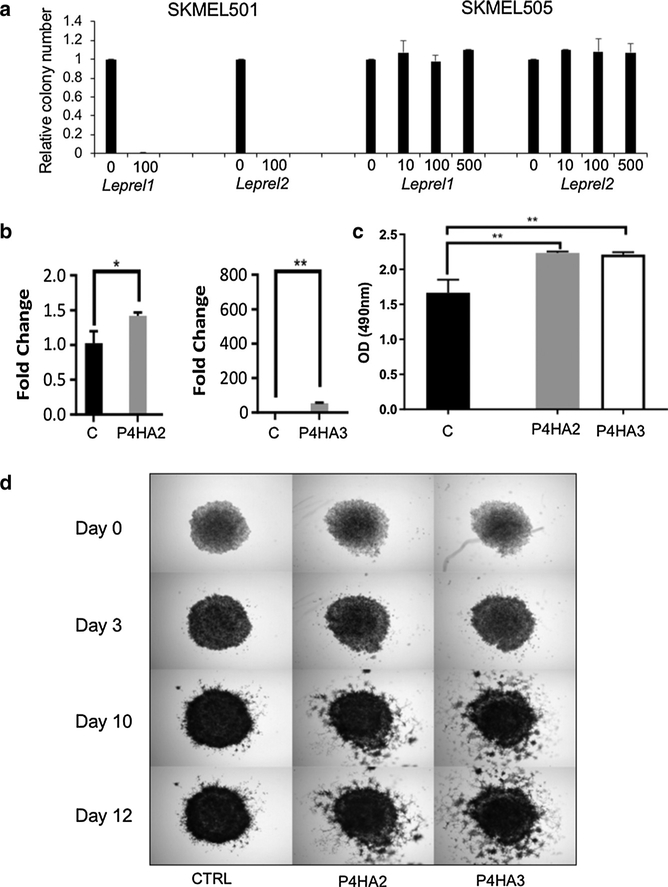
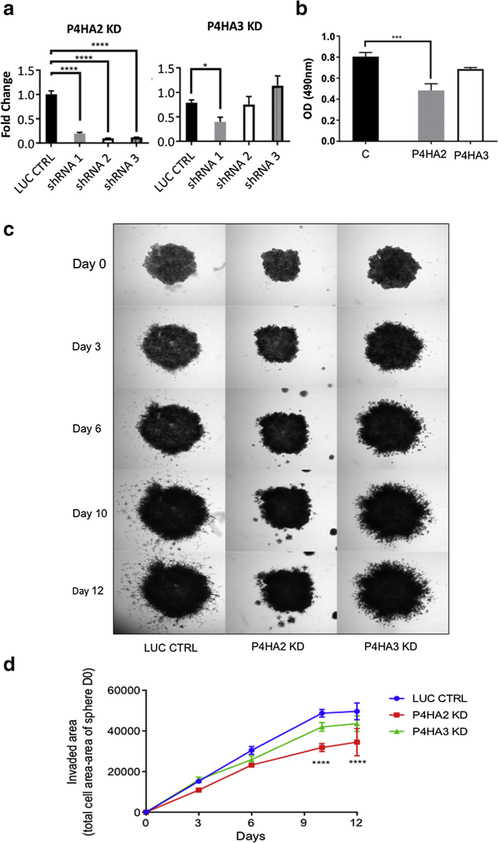
These results imply that the two major families responsible for post-translational modification of collagen have opposing effects on the properties of melanoma cell lines. In initial studies to validate this hypothesis, we transfected various melanoma cells with expression plasmids for C-P3H and C-P4H and attempted to select clones stably overexpressing the transfected sequences. Leprel1 and Leprel2 were overexpressed in SKMEL501 (methylated Leprel1 and Leprel2) and SKMEL505 cells (unmethylated Leprel1 and Leprel2), transfected cells were selected in G418 and colony formation determined after 10–14 days. Whereas colony numbers (relative to vector only controls) were essentially unaffected by ectopic expression of either gene in SKMEL505, no surviving colonies could be recovered from SKMEL501 cells transfected with either gene (Figure 3a). The results are consistent with a growth suppressor function for Leprel1 and Leprel2. Next, we tested the effect of overexpressed P4HA2 and P4HA3. We performed these studies in PMWK cells to determine whether changes in C-P4H expression are able to confer a more aggressive phenotype to an early radial growth phase melanoma. Cells were transfected with expression plasmids encoding P4HA2 and P4HA3, colonies selected and overexpression confirmed using quantitative PCR (Figure 3b). Increased expression of both genes was associated with a significant increase in basal proliferation rate in PMWK cells (Figure 3c). We then tested whether invasiveness of melanoma cells was affected by ectopic expression of P4HA2 and P4HA3 and analysis in a 3-dimensional spheroid invasiveness assay (Figure 3d). PMWK 3-dimensional tumor spheroids stably expressing ectopic P4HA2 and P4HA3 clearly had greater invasive activity than control spheroids (Figure 3d). We next used inhibitory RNA via lentiviral vectors to selectively target gene expression of P4HA2 and P4HA3 in PMWK cells and checked knockdown by quantitative PCR (Figure 4a). Knockdown of P4HA2 caused a significant reduction in proliferation relative to control cells (Figure 4b). In the case of P4HA3 knockdown clones were obtained with short hairpin RNA1 and proliferation was reproducibly lower than control cells, although this did not reach statistical significance (possibly due to lower levels of knockdown) (Figure 4a, 4b). Next, we tested the invasive potential of the same PMWK knockdown clones (Figure 4c, 4d). Due to limitations in image thresholding, the inhibitory effect could not be quantified in its entirety. Nonetheless, there was a significant reduction in cancer cell invasion (Figure 4d), and representative images clearly show that P4HA2 knockdown almost completely abolishes invasive leader cells capable of traveling to sites distant of the sphere (Figure 4c). Similarly, P4HA3 knockdown was reproducibly associated with reduced invasiveness.
Figure 3. Collagen prolyl 3-hydroxylase and collagen prolyl 4-hydroxylase have opposing effects on melanoma progression.
(a) Ectopic expression of Leprel1 and Leprel2 in cells lacking endogenous expression blocks proliferation. (b) Quantitative PCR analysis in PMWK engineered to express of P4HA2 and P4HA3. Data are mean ± standard error of the mean (n = 3). Significance was tested using one-way analysis of variance with Dunnett’s post-hoc testing.(c) P4HA2 and P4HA3 overexpression significantly increase proliferation in PMWK cells. Graphs shows OD at 490 nm 6 days post treatment. Data are mean ± standard error of the mean over biological repeats (n = 2) performed in triplicate. Significance was tested using two-way analysis of variance with Tukey’s post-hoc testing. (d) P4HA2 and P4HA3 overexpression increase the invasiveness relative to control (CTRL) of PMWK early radial growth phase melanoma cells. Images are representative of two independent experiments performed in triplicate. OD, optical density.
Figure 4. Inhibition of collagen prolyl 4-hydroxylase gene expression reduces melanoma proliferation and invasiveness.
(a) Quantitative PCR analysis of P4HA2 and P4HA3 in PMWK cells expressing short hairpin RNA. Data are mean 2-ΔΔCt ± standard error of the mean (n = 3). Significance: one-way analysis of variance with Dunnett’s post-hoc testing. (b) P4HA2 and P4HA3 knockdown reduce proliferation of PMWK cells. Quantification was performed using sulforhodamine b as described in Materials and Methods. Data are mean ± standard error of the mean over biological repeats (n = 2) performed in triplicate. Significance shows the difference to appropriate control, using one-way analysis of variance with Sidak’s post-hoc testing.(c) Collagen prolyl 4-hydroxylase knockdown reduces invasiveness of PMWK cells. Representative images are shown. Spheroids were imaged at ×2.(d) Reduced invasion of PMWK 3-dimensionl spheroids. Data are mean ± standard error of the mean (n = 2), in triplicate. Significance: two-way analysis of variance with Tukey’s post-hoc testing. KD, knockdown; LUC CTRL, luciferase control; shRNA, short hairpin RNA.
C-P4H inhibition has antiproliferative effects in melanomavia induction of apoptosis
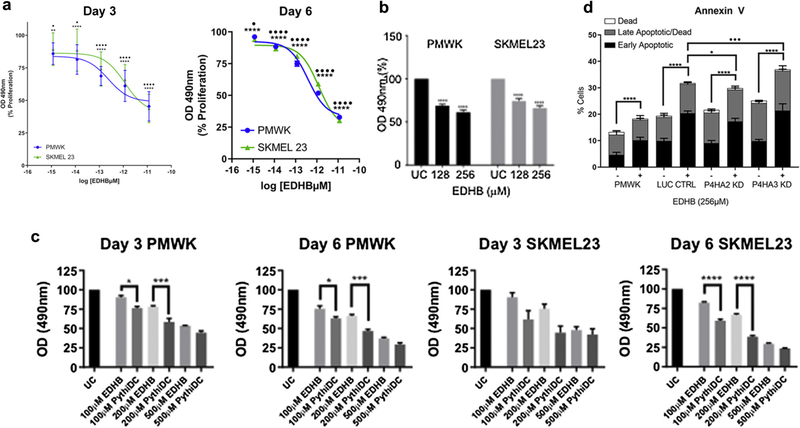
We then test whether pharmacological inhibition of C-P4H affects melanoma cell properties (Vasta and Raines, 2018). We tested two compounds, ethyl dihydroxybenzoic acid (EDHB) and the highly specific and potent C-P4H inhibitor 2-(5-carboxythiazol-2-yl)pyridine-5-carboxylic acid (Vasta et al., 2016). In initial experiments, we titrated the concentration of EDHB testing a range of concentrations from 32 μM to 512 μM in PMWK and SKMEL23 cells (Figure 5a). Concentrations of EDHB as low as 128 μM have a significant inhibitory effect on proliferation of both cell lines (Figure 5b). We then compared the potency of EDHB with 2-(5-carboxythiazol-2-yl)pyridine-5-carboxylic acid in proliferation assays (Figure 5c). 2-(5-Carboxythiazol-2-yl)pyridine-5-carboxylic acid leads to a significantly greater inhibition at various doses in both cell lines, consistent with its greater potency as a pharmacological C-P4H inhibitor (Vasta et al., 2016).
Figure 5. Inhibition of collagen prolyl 4-hydroxylase has antiproliferative effects on melanoma cells via induction of apoptosis.
(a, b) Inhibition of proliferation of PMWK (blue) and SKMEL23 (green) by EDHB. Data are mean ± standard error of the mean over biological repeats (n = 3) performed in six replicates. Significance: two-way analysis of variance with Tukey’s post-hoc testing. (c) Antiproliferative effect of EDBH and PythiDC on PMWK and SKMEL23. Data are mean ± standard error of the mean (n = 2) in triplicate. Significance: one-way analysis of variance with Sidak’s post-hoc testing. (d) EDHB induces apoptosis in PMWK cells. Apoptosis was measured via Annexin V staining on day 3. Data are mean ± standard error of the mean (n = 3). Live cell population not shown. Significance: two-way analysis of variance with Tukey’s post-hoc testing. *Differences in early-stage apoptosis. •Differences in late-stage apoptosis. EDHB, ethyl dihydroxybennzoic acid; PythiDC, 2-(5-carboxythiazol-2-yl)pyridine-5-carboxylic acid.
To determine the mechanism of cell death caused by EDHB treatment, PMWK cells were treated with EDHB for 72 hours and analyzed using the Annexin V apoptotic assay (Figure 5d). EDHB caused a significant increase in the proportion of cells in early-stage apoptosis, demonstrating that EDHB functions as an inducer of apoptosis in PMWK cells. We sought to determine whether P4HA2, P4HA3, or both, are targeted by EDHB to induce apoptosis. We therefore performed the same apoptotic assay on C-P4H knockdown cells (Figure 5d). An increase in apoptosis was seen following EDHB treatment with each sample, regardless of knockdown (P < 0.0001). However, P4HA2 knockdown cells demonstrated significantly fewer cells being induced into early-stage apoptosis compared to the luciferase control EDHB-treated control, and the same was true of P4HA3 knockdown cells with late-stage apoptotic cells. Taken together, these data suggest that both P4HA2 and P4HA3 are targeted by EDHB with resultant induction of apoptosis in PMWK cells.
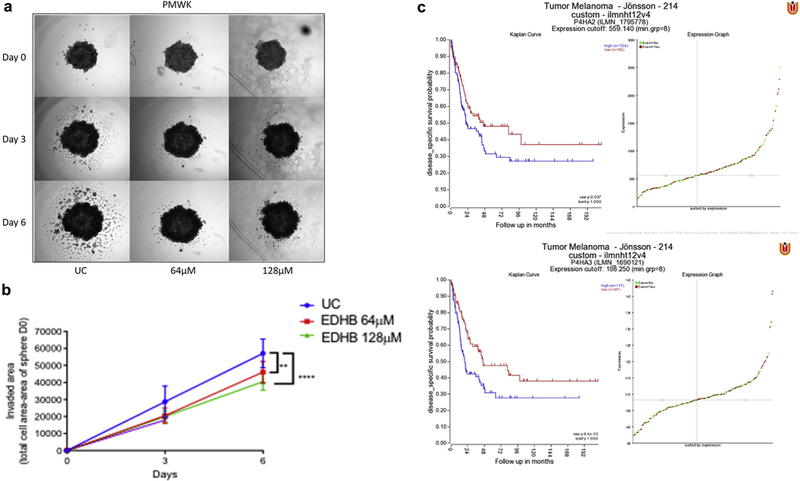
To confirm the finding that P4HA2 knockdown resulted in a significant decrease in invasion, we tested the effect of EDHB on invasion in PMWK 3-dimensional spheroids. A clear and significant decrease in invasion was demonstrated, even with low doses of EDHB (Figure 6a, 6b). Again, the limitations in image thresholding render the quantified inhibitory effect as lower than representative images suggest, although notwithstanding this issue, there was a significant inhibition of invasion by EDHB doses as low as 64 μm (Figure 6a, 6b).
Figure 6. Blocking collagen prolyl 4-hydroxylase inhibits melanoma invasion.
(a) EDHB inhibits invasiveness of PMWK 3-dimensional tumor spheroids. Representative images from two independent experiments performed in triplicate. Spheroids were imaged at ×2 on days 0, 3, and 6 post treatment.(b) Quantification of the effect of EDHB on PMWK 3-dimensional spheroid invasion. Spheroids were imaged at ×2 on days 0, 3, and 6 post treatment. Data are mean ± standard error of the mean (n = 2) performed in triplicate. Significance was compared to the appropriate control using two-way analysis of variance with Tukey’s post-hoc testing. (c) Overexpression of C-P4H genes is associated with worse disease-free survival in melanoma. Kaplane–Meier plots by gene expression were created using the “Tumor Melanoma-Jönsson-214-custom-ilmnht12v4” data set (n = 214) (Cirenajwis et al., 2015). EDHB, ethyl dihydroxybennzoic acid.
Expression of C-P4H is prognostic in melanoma
Together, these data are consistent with an oncogenic function for the C-P4H family in melanoma and we were therefore interested to determine whether expression has prognostic utility in melanoma. We interrogated the Jonsson data set via the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) (Cirenajwis et al., 2015). Diseasefree survival was significantly worse in cases overexpressing both P4HA2 (P = 0.037) and P4HA3 (P = 0.0064) (Figure 6c).
DISCUSSION
We present evidence that although the C-P3H encoding genes, Leprel1 (P3H2) and Leprel2 (P3H3), are subject to transcriptional silencing in both melanoma cell lines and clinical cases of melanoma, methylation was observed only in P4HA3 among the C-P4H genes. To our knowledge, C-P4H (P4HA3) gene silencing has not been reported previously in any human non-hematologic cancer, and Leprel silencing in solid tumors has only been reported in one previous study (Shah et al., 2009). Of note, the majority of melanomas we analyzed demonstrated CpG island methylation in at least one of Leprel1 and Leprel2, but very rarely were both genes silenced. This suggests that loss of the post-translational collagen modifying functions of the C-P3H family may be important in malignant development in melanoma, but also implies that there may be functional redundancy between P3H family members to preserve one or more critical functions. We also noted that in melanoma cell lines with undetectable or extremely low expression of P4HA3, there was usually moderate to high expression of P4HA1 and especially P4HA2, again implying functional redundancy between P4H family members.
There are, however, no mechanistic studies of P4HA3 in human cancer and no previous reports of the function of either the C-P3H or C-P4H family in melanoma. Although at face value the methylation data imply a tumor suppressor function for P4HA3, our detailed mechanistic studies do not support this, as clones stably expressing P4HA3 are more invasive and proliferative than parental controls and, conversely, knockdown clones are less invasive. Methylation-dependent silencing of genes that have potentially oncogenic functions in melanoma may appear counterintuitive. There are, however, precedents in melanoma and indeed in other cancer types. For example, NT5E has definite oncogenic properties and yet is subject to methylation-dependent transcriptional silencing in both breast cancer and metastatic melanoma, with better clinical outcomes in cases with methylation (Lo Nigro et al., 2013; Wang et al., 2012).
To gain mechanistic insight, we modulated expression of Leprel1, Leprel2, P4HA2, and P4HA3 in melanoma cell lines using ectopic expression and (for P4HA2 and P4HA3) knockdown. Ectopic expression of Leprel1 and Leprel2 efficiently abrogated proliferation and colony-forming ability in cell lines in which the endogenous gene is silenced by CpG island methylation. We observed similar effects in breast cancer (Shah et al., 2009). However, clones of the cell line PMWK engineered to stably express P4HA2 and P4HA3 (expression confirmed by quantitative PCR) exhibited a higher basal proliferation rate and invasiveness than control and parental PMWK cells, and this finding was further confirmed by knocking down P4HA2 and P4HA3 expression and observing the reverse effect. These observations prompted investigation of pharmacologic inhibition of the C-P4H proteins using the C-P4H selective agents EDHB and the highly potent and selective 2-(5-carboxythiazol-2-yl)pyridine-5-carboxylic acid (Vasta et al., 2016). Both agents demonstrated efficient anti-proliferative activity in PMWK and the metastatic melanoma cell line SKMEL23 and we have further shown that EDHB induces apoptosis, which is at least partially dependent on P4HA2 and P4HA3 (knockdown of P4HA2/3 reduced EDHB-induced apoptosis). EDHB also has an anti-invasion action, completely inhibiting migratory leader cells from traveling from the tumor spheroid.
Bioinformatic analysis of a well-validated set of clinical data demonstrated a clear negative correlation between P4HA2 and P4HA3 overexpression and time-dependent clinical outcomes, such as disease-free survival.
We have therefore shown, through mechanistic studies and bioinformatics analysis, that overexpression of P4HA2 and P4HA3 is capable of promoting malignant progression in melanoma and that this effect is reversible via inhibition of P4HA2 and/or P4HA3. It is likely that this effect is related to the role of P4HA2 and P4HA3 in regulating collagen deposition (Xiong et al., 2014). Collagens form the main structural component of the extracellular matrix and are known to control cellular processes, such as proliferation, invasion, and migration in both cancer progression and in health (Pozzi et al., 1998; Zhang et al., 2013, Provenzano et al., 2008). P4H catalyzes the formation of 4-hydroxyproline, which supports the folding of newly synthesized collagen polypeptides into stable triple helix structures (Nokelainen et al., 2001). Blocking the P4H enzyme prevents maturation of collagens, and their deposition in the extracellular matrix (Sasaki et al., 2012) and it has been shown in breast cancer models that inhibition of P4H leads to a reduction in extracellular collagen deposition along with reduced invasiveness (Xiong et al., 2014). Therefore, the reduction in phenotypic aggression we observed in melanoma cells when P4H is inhibited could be driven by a reduced level of mature collagens with resulting inhibition of invasiveness and potentially angiogenesis. Taken together, our results suggest that P4H is a viable therapeutic target for reducing growth and invasion in melanoma.
MATERIALS AND METHODS
Cell lines, plasmids, and short hairpin RNA
Melanoma cell lines and primary human melanocytes were grown as described previously (Lo Nigro et al., 2013; Wang et al., 2012). Plasmids encoding Leprel1 and Leprel2 were described previously (Shah et al., 2009). To generate a P4HA2 expression vector, the ORF was purchased from Origene and subcloned into pcDNA3-DEST40 using Invitrogen Gateway Technology to generate pcDNA3.2/V5/GW/P4HA2. pcDNA3.2/V5/GW/CAT served as the control plasmid. The sequence of both plasmids was verified by sequencing. pcDNA3P4HA3 was described previously (Van Den Diepstraten et al., 2003). Short hairpin RNA sequences were obtained from the RNAi Consortium (Moffat et al., 2006). These were inserted into EcoRI and AgeI cut pLKO.1–TRC cloning vector (Addgene #10878) and subsequently these plasmids were used for lentiviral particle production and target cell transduction following the Addgene protocol for lentiviral transduction of mammalian cells (https://www.addgene.org/tools/protocols/plko/#A). Cell proliferation was measured using the sulforhodamine b colorimetric assay.
Gene expression analysis
The expression of C-P4H genes was determined by quantitative PCR. cDNA was generated from 1 μg RNA using Moloney Murine Leukemia Virus reverse transcriptase (Sigma, St Louis, MO), and 0.5 μg random primer. PCR was performed in 96-well plates using SYBER Select Master Mix, and 0.5 μM primer pairs targeting each mRNA transcript of interest (ThermoFisher Scientific, Waltham, MA). cDNA (50 ng) was added to each well, containing 10 μl SYBER Select, 3 μl nuclease-free water, and the primer pairs. mRNA levels were determined by normalizing to the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase and using the relative quantification method (2-ΔΔCt). Data are presented as fold-change compared to housekeeping gene.
Annexin V assay
Two hundred thousand cells were plated in six-well plates and allowed to attach overnight. The following day, cells were treated with appropriate drug or vehicle control. On day 3, wells were harvested via trypsinization and stained using the Annexin V Apoptosis Detection kit, according to manufactures direction (ThermoFisher). Cells were analyzed on a Muse Cell Analyzer (Millipore Sigma, Burlington, MA).
Spheroid 3-dimensional invasion assay
Invasion was measured by generating 3-dimensional tumor spheroids and measuring their invasion into surrounding growth factor reduced Matrigel (Corning, Tewksbury, MA). Five thousand cells per well were plated in a low attachment 96-well plate with 200 μl media. Following 4 days growth, cells were visually analyzed to confirm spheroid formation.
Colony-forming assays
The 2 × 105 cells/6-cm dish were transfected after 24 hours with varying amounts of expression plasmids for Leprel1, Leprel2, P4HA2, and P4HA3 or control vector using Metafectine (Biontex, München, Germany) according to the manufacturer’s instructions. Forty-eight hours after transfection, G418 (400 ng/ml) was added and cells were monitored daily by light microscopy for the appearance of colonies, which were counted by staining with Coomassie Brilliant Blue. Cell clusters of 50 cells were defined as colonies.
Clinical material
The study was approved by The Tayside Tissue Bank, under delegated authority from the Tayside Local Research Ethics Committee, with written informed consent from patients. We analyzed 50 clinical cases of melanoma, including primary and metastatic lesions. In all cases, microdissection of tissue sections was performed to enrich for melanoma cells prior to isolation of nucleic acids. Benign pigmented nevi from sun-exposed skin were control tissues. We used proteinase K digestion to isolate genomic DNA from tissue sections.
Methylation analysis
Methylation in the CpG islands of the C-P3H and C-P4H gene families was determined using pyrosequencing with the primers and reaction conditions described previously (Hatzimichael et al., 2012).
Statistics
Statistical analyses were performed using Prism 7 software (Graph-Pad, La Jolla, CA). Significance was as follows: *P < 0.05, **P <0.01, ***P < 0.001, ****P < 0.0001. Kaplane–Meier plots by gene expression were created on the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) using the “Tumor Melanoma-Jönsson-214-custom-ilmnht12v4” data set (n = 214) (Cirenajwis et al., 2015). The expression cutoff was determined using the Kaplan scan modus, which generates a Kaplane–Meier Plot based on the most optimal mRNA cut-off expression level to discriminate between a good and bad prognosis cohort.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by Melanoma Focus, The Leng Charitable Foundation, The Medical Research Council, The Tayside Tissue Bank, Barts and the London Charity, and the Grant Simpson Trust. Work in RTR’s lab is supported by grant R01 AR044276 (National Institutes of Health). TC is Scottish Senior Clinical Fellow in Medical Oncology. Work in TC’s lab is supported by the Chief Scientist’s Office of Scotland, The Anonymous Trust, Tenovus Scotland, and The Translational Medicine Research Fund of Dundee Cancer Centre. Work in NS’s lab is supported by The Brain Tumour Research Charity. CP and LW are supported by Cancer Research UK. JGP holds the Heart and Stroke Foundation of Ontario/Barnett-Ivey Chair. AA is supported by the Sharon Dunster Fund.
Abbreviations:
- C-P3H
collagen prolyl 3-hydroxylase
- C-P4H
collagen prolyl 4-hydroxylase
- EDHB
ethyl dihydroxybennzoic acid
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2018.10.038.
REFERENCES
- Cirenajwis H, Ekedahl H, Lauss M, Harbst K, Carneiro A, Enoksson J, et al. Molecular stratification of metastatic melanoma using gene expression profiling: prediction of survival outcome and benefit from molecular targeted therapy. Oncotarget 2015;6:12297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes RJ, Farnand AW, Traeger GR, Weis MA, Eyre DR. A role for prolyl 3-hydroxylase in post translational modification of fibril-forming collagens. J Biol Chem 2011;286:30662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Chatuverdi P, Bajpai S, Wong CC, Wei HH, Pitcairn S, et al. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res 2013;73:3285–96. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gorres KL, Raines RT. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol 2010;45:106–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzimichael E, Lo Nigro C, Lattanzio L, Syed N, Shah R, Dasoula A, et al. The collagen prolyl hydroxylases are novel transcriptionally silenced genes in lymphoma. Br J Cancer 2012;107:1423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelblinger P, Dornbierer J, Dummer R. A review of binimetinib for the treatment of mutant cutaneous melanoma. Future Oncol 2017;13: 1755–66. [DOI] [PubMed] [Google Scholar]
- Lo Nigro C, Wang H, McHugh A, Lattanzio L, Matin R, Harwood C, et al. Methylated tissue factor pathway inhibitor 2 (TFPI2) in serum is a novel circulating DNA biomarker of metastatic melanoma. J Invest Dermatol 2013;133:1278–85. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 2006;124:1283–98. [DOI] [PubMed] [Google Scholar]
- Prolyl Myllyharju J. 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia and their roles as treatment targets. Ann Med 2008;40:402–17. [DOI] [PubMed] [Google Scholar]
- Nokelainen M, Nissi R, Kukkola L, Helaakoski T, Myllyharju J. Characterization of the human and mouse genes for the αsubunit of type II prolyl 4-hydroxylase: Identification of a previously unknown alternatively spliced exon and its expression in various tissues. Eur J Biochem 2001;268: 5300–9. [DOI] [PubMed] [Google Scholar]
- Pozzi A, Wary KK, Giancotti FG, Gardner HA. Integrin a1b1 mediates a unique collagen-dependent proliferation pathway in vivo. J Cell Biol 1998;142:587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med 2008;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 2015;372:30–9. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio IIC, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev 2012;26:2038–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R, Smith P, Purdie C, Quinlan P, Baker L, Aman P, et al. The prolyl 3-hydroxylases P3H2 and P3H3 are novel targets for epigenetic silencing in breast cancer. Br J Cancer 2009;100:1687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem 2009;78:929–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanemura A, Terando AM, Sim MS, van Hoesel AQ, de Maat MF, Morton DL, et al. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res 2009;15:1801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro JG, Parker AE, Zhu X, Green MR. p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science 2006;313:968–71. [DOI] [PubMed] [Google Scholar]
- Van Den Diepstraten C, Papay K, Bolender Z, Brown A, Pickering JG. Cloning of a novel prolyl 4-hydroxylase subunit expressed in the fibrous cap of human atherosclerotic plaque. Circulation 2003;108:508–11. [DOI] [PubMed] [Google Scholar]
- Vasta JD, Andersen KA, Deck KM, Nizzi CP, Eisenstein RS, Raines RT. Selective inhibition of collagen prolyl 4-hydroxylase in human cells. ACS Chem Biol 2016;11:193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasta JD, Raines RT. Collagen prolyl 4-hydroxylase as a therapeutic target.J Med Chem 2018. July 23 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lee S, Nigro CL, Lattanzio L, Merlano M, Monteverde M, et al. NT5E (CD73) is epigenetically regulated in malignant melanoma and associated with metastatic site specificity. Br J Cancer 2012;106:1446–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G, Deng L, Zhu J, Rychahou PG, Xu R. Prolyl-4-hydroxylase a subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer 2014;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Corsa CA, Ponik SM, Prior JL, Piwnica-Worms D, Eliceiri KW, et al. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat Cell Biol 2013; 15:677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.