Cryo-EM structures of the undocked INX-6 hemichannels reveal a pore obstruction and N-terminal rearrangement in lipids.
Abstract
Gap junctions form intercellular conduits with a large pore size whose closed and open states regulate communication between adjacent cells. The structural basis of the mechanism by which gap junctions close, however, remains uncertain. Here, we show the cryo–electron microscopy structures of Caenorhabditis elegans innexin-6 (INX-6) gap junction proteins in an undocked hemichannel form. In the nanodisc-reconstituted structure of the wild-type INX-6 hemichannel, flat double-layer densities obstruct the channel pore. Comparison of the hemichannel structures of a wild-type INX-6 in detergent and nanodisc-reconstituted amino-terminal deletion mutant reveals that lipid-mediated amino-terminal rearrangement and pore obstruction occur upon nanodisc reconstitution. Together with molecular dynamics simulations and electrophysiology functional assays, our results provide insight into the closure of the INX-6 hemichannel in a lipid bilayer before docking of two hemichannels.
INTRODUCTION
Gap junction channels, composed of membrane proteins with four transmembrane helices, function as an oligomeric conduit that penetrates lipid bilayers of adjacent cells. A full gap junction channel is generated by the docking of two opposing hemichannels. These channels mediate intercellular communications that play substantial roles in a variety of biologic events, including development, inflammation, cell death, immune responses, and muscle contractions (1). Gap junction channel malfunction causes various human inherited diseases (2). Elucidating the structural and functional bases of gap junction channels is therefore important for the development of therapies and drugs to treat these diseases.
Gap junction channels are generally found in most multicellular organisms, but the component peptides vary. Connexins are found in chordates and innexins are widely found in pre-chordates, and the two families share almost no amino acid sequence similarity (3). The membrane topology of these two types of gap junction channels, however, is the same, i.e., four transmembrane helices (TM1 to TM4), two extracellular loops (E1 and E2), and cytoplasmic domains containing an N terminus, a cytoplasmic loop, and a C terminus, as confirmed by high-resolution structures of connexin26 (Cx26), connexin46/50 (Cx46/50), and innexin-6 (INX-6) determined by x-ray or cryo–electron microscopy (cryo-EM) (4–7). These two types of gap junction channels also have high structural similarity in terms of their helical arrangements, despite a different oligomeric number and lack of sequence similarity (6). In addition, genome sequence projects revealed that two non-gap junction channel proteins in mammals—LRRC8, which is a family of volume-regulated anion channels, and pannexin, which could be a family of adenosine 5′-triphosphate (ATP) release channels—form sister groups of innexin with weak homology (8). Most recently, structural studies using cryo-EM revealed that LRRC8A has a structural arrangement of four transmembrane domains similar to INX-6 and Cx26 (9–11). Thus, the structural study of innexin may also enhance our understanding of these non-gap junction proteins in mammals.
A common characteristic of connexins and INX-6 is the pore size, which is quite large compared with general tetrameric channel assemblies. Connexin channels may have multiple regulatory mechanisms for closing the channel (12, 13), and thus, the structural basis of the gating mechanism might be complex. There are two closure models, subunit rotation and physical blockage of the pore, but both are based on low-resolution structural studies (14, 15). The three high-resolution structures of gap junction channels show a constriction surrounded by the N-terminal funnel with a diameter greater than 10 Å, which would allow the passage of hydrated ions such as K+ and Na+ (~8 Å diameter), and are therefore thought to be in an open conformation (4, 6, 7). On the basis of recent Cx26 x-ray structures in which the N-terminal portion was not resolved, Bennett et al. (5) proposed that an electrostatic barrier in the pore pathway contributes to an ionic conduction block in larger size pores. How complete closure of a pore this large is achieved, however, remains an unanswered question due to the lack of high-resolution gap junction structures in an explicitly closed conformation, in contrast to voltage-gated ion channels for which the gating mechanism is well studied (16–18).
A possible explanation is the involvement of lipid molecules. All of the previously described high-resolution structures of gap junction proteins were determined in the absence of lipids (4, 6, 7). Here, we obtained the cryo-EM structure of the wild-type (WT) INX-6 undocked hemichannel reconstituted in a nanodisc. We also investigated the structures of undocked hemichannels of an INX-6 N-terminal deletion mutant (INX-6ΔN) in a nanodisc and WT INX-6 in detergent micelles. Combined with molecular dynamics (MD) simulations and functional analyses with electrophysiology, our observations supply a potential explanation for the N-terminal rearrangement after an INX-6 hemichannel is configured in the lipid bilayer, providing insight into how the large size pore of INX-6 is sealed before functioning as a docked junction channel.
RESULTS
Cryo-EM structure of an undocked hemichannel of WT INX-6 in a nanodisc
We previously reported the structure of WT INX-6 in a docked junction form surrounded by detergent (6). The undocked hemichannels of INX-6 were prepared by a NaCl concentration of 150 mM at all purification steps. To investigate the structure in a lipid bilayer, we reconstituted undocked WT INX-6 hemichannels in nanodiscs using the membrane scaffold protein 2N2 (MSP2N2) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), as confirmed by Coomassie brilliant blue–stained gel and negatively stained electron micrographs (fig. S1A). After collecting the cryo-EM data (fig. S1B), particles categorized as undocked hemichannels were selected for two-dimensional (2D) classification (fig. S1C). The finally obtained 3D reconstruction represented an undocked channel in a nanodisc at 3.8 Å resolution (Fig. 1A; fig. S1, D to H; and table S1).
Fig. 1. Cryo-EM structure of the undocked INX-6 hemichannel in a nanodisc.
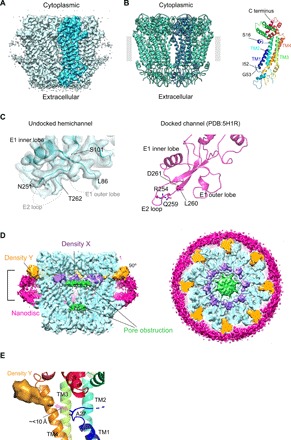
(A) The 3D reconstruction of a nanodisc-reconstituted WT INX-6 hemichannel viewed parallel to the membrane. The subunit highlighted in blue represents a monomer. The map contour level is 2σ. (B) Octameric (left) and monomeric (right) ribbon models of an undocked hemichannel of WT INX-6 in a nanodisc. A single subunit of the octamer is indicated in dark blue, the extracted monomer is in rainbow colors, and the lipid bilayer boundary is indicated by a gray band. The N-terminal residue is assigned from S16. S16, I52, and G53 are represented in stick style. The four transmembrane helices are indicated as TM1 to TM4. (C) Comparison of the extracellular docking surface of WT INX-6. (Left) Ribbon style model and density map (surface representation in gray) of the nanodisc-reconstituted undocked hemichannel are superimposed. Disordered peptides of the E1 outer lobe and E2 loop are shown as dashed lines, and the flanking residues of L86, S101, N251, and T262 are indicated. (Right) Ribbon style model of a docked junction channel (PDB: 5H1R) (6). The amino acid side chains that participate in the hydrogen bonds are depicted in stick style and are all included in the disordered regions in the undocked hemichannel. The map contour level is 2σ. (D) Sliced views of the Gaussian-filtered 3D density map of the undocked WT INX-6 hemichannel as a vertical cross section (left) and cytoplasmic top view of the slab surface (right). Slab thickness corresponds to the bracket in a side view (left). The surrounding nanodisc densities are shown in magenta, and the double-layer densities that obstruct the pore are shown in green. Newly observed densities X and Y are shown in slate and orange, respectively. The map contour level is 1.5σ. (E) Positional relationship of density Y to TM1 of WT INX-6. Density Y is located within a distance of less than 10 Å from A27, the closest residue in TM1 to density Y. The map contour level is 2σ.
The overall architecture of the undocked WT INX-6 hemichannel was highly consistent with the octameric hemichannel part of an INX-6 docked junction channel in detergent micelles (6), as we observed no marked conformational change in the transmembrane domains or dome-like helix-rich cytoplasmic domains (C-dome) (Fig. 1B). Although the densities of I52 and G53, which connect TM1 and E1, became clearer (fig. S1G), the extracellular domains of E1 and E2 remained poorly resolved. The antiparallel E2 β sheets mostly disappeared, and the hairpin loop was disordered from R252 to D261 (Fig. 1C and fig. S2, A and B), including R254, Q259, L260, and D261, which participate in the polar interactions between the opposed INX-6 hemichannels (figs. S2B and S3) (6). The E1 outer lobe contained relaxed and disordered residues from D87 to V100, which prevent the E1 outer lobe from interfacing with the opposed hemichannel (Fig. 1C and fig. S2, A and B). As a result, the undocked INX-6 hemichannel does not have an adequate extracellular cavity to accommodate the E2 β hairpin loop of the opposed hemichannel, consistent with the observation that the docking interface of two opposing INX-6 hemichannels is maintained by the opposing E1 outer lobes and E2 β hairpins (6). The contribution of the E2 loop to the docking interface is consistent with connexin channels, in which the determinant of the docking specificity is located in the E2 loop (19, 20). On the other hand, the E1 inner lobe between E1β and E1H does not change markedly (fig. S2C). The side chains of W59 to Q67 are oriented inside, slightly closer to the pore center, but still create a wide pore diameter of 16 Å (fig. S2, E and F). While both the E1 and E2 of Cx26 and Cx46/50 are involved in the polar interactions on the docking surface (4, 7), the E1 of INX-6 does not contain any hydrogen bonds to the opposed interface (6). These findings suggest that the functional significance of the E1 of INX-6 is likely different from that of the connexin family proteins.
The 3D map of a WT INX-6 hemichannel in a nanodisc has distinctive features. One is the flat double-layer densities along the membrane plane inside the pore that can obstruct the passage (Fig. 1D, green densities), and thereby no pore funnel-like conformation is observed. Additional bulb densities are observed at the border of the cytoplasmic and transmembrane domains. The bulbs located in the pore region, referred to as density X (Fig. 1D, slate-colored densities), are near the cytoplasmic end of TM1. The distribution is close to the position of the N-terminal funnel, which is formed in the pore of the docked junction channel of INX-6 in a solubilized form (6). The other density, referred to as density Y, appears as a wedge between the adjacent subunits and orients to the outside of the pore (Fig. 1D, orange densities). The distribution is close to A27 of TM1 (<10 Å; Fig. 1E). Because assigning amino acid residues to these blob densities should be performed cautiously, the following two cryo-EM structures were determined.
Undocked WT INX-6 hemichannel in detergent reveals an N-terminal funnel
We obtained a cryo-EM structure of the undocked WT INX-6 hemichannel in detergent micelles prepared using GraDeR (21). After GraDeR, the glycerol was removed by dialysis in 50 mM NaCl, yielding mostly undocked hemichannels of INX-6 (fig. S4A). Following the cryo-EM data collection and image processing, the final resolution of the postprocessed map was estimated to be 3.8 Å using golden standard Fourier shell correlation (22) analysis (fig. S4, B to G, and table S1).
In the hemichannel structure of WT INX-6 in detergent, the extracellular E1 outer lobe and antiparallel E2 β sheets were poorly resolved, but the constriction formed by the E1 inner lobe had a diameter of 17 Å (fig. S4H). The N-terminal funnel was clearly observed in the pore (Fig. 2A), generating a wide-open pore, although the assigned residue began from N12 (Fig. 2B). In contrast to the structure reconstituted in a nanodisc, neither densities X and Y nor the double-layer pore densities are observed in detergent (Fig. 2A). I52 and G53 were disordered (Fig. 2B), suggesting that the stability of the TM1 extracellular side is linked with the N-terminal arrangement and lipid bilayer environment. Because a pore funnel is also commonly observed in the INX-6 docked junction structure (6), the INX-6 N termini generally assume a funnel configuration in detergent regardless of the form—docked or undocked. The N-terminal funnel was similarly observed in the Cx26 x-ray 3D structure crystallized with detergent micelles (4) and in the Cx46/50 structure surrounded by amphipol (7). These observations indicate that densities X and Y and double-layer pore densities found in the nanodisc-reconstituted WT INX-6 hemichannel (Fig. 1D) are not a consequence of undocking events between two opposing hemichannels but are due to the reconstitution into a lipid bilayer.
Fig. 2. Undocked hemichannel structures of WT INX-6 in detergent and INX-6ΔN in a nanodisc.
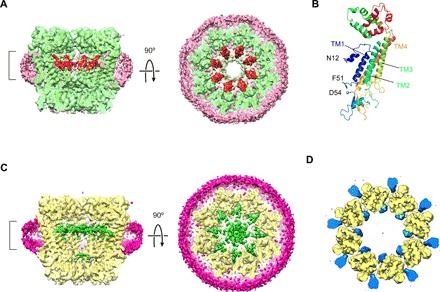
(A) Sliced views of the Gaussian-filtered 3D density map of WT INX-6 in detergent. The surrounding detergent densities are shown in pink, and the N-terminal funnels are shown in red. The ribbon style model is superimposed. The map contour level is 1.5σ. (B) Ribbon style model of a monomeric structure of WT INX-6 in detergent. The assigned residues at the N terminus begin from N12 (shown in stick style), and the I52 and G53 residues are not visible due to disorder. (C) Sliced views of the Gaussian-filtered 3D density map of INX-6ΔN in a nanodisc. The densities corresponding to the nanodisc are shown in magenta, and the ribbon style model is superimposed. The double-layer densities in the pore are shown in green. The thickness of the top view slab is indicated by the bracket in the side view. The map contour level is 1.5σ. (D) Comparison of WT INX-6 to INX-6ΔN in nanodiscs. A subtraction map of INX-6ΔN from WT INX-6 (blue) is superimposed on the structure of INX-6ΔN (yellow), where the represented slab corresponds to the bracket in (C). The map contour level is 3σ.
Structure of an undocked hemichannel of the N-terminal–deleted INX-6 in a nanodisc
We previously reported the 3D reconstruction of INX-6ΔN (referring to the N-terminal deletion of residues 2 to 19 unless otherwise noted) by electron crystallography at 10 Å resolution (23). The same construct was used in this study to further investigate the difference from WT INX-6 in nanodiscs. The undocked INX-6ΔN hemichannels were purified and reconstituted in nanodiscs, and cryo-EM analysis was used to determine the structure of the final 3D reconstruction at 3.6 Å (fig. S5, A to G, and table S1).
The map of INX-6ΔN shows flat double-layer densities in the pore (Fig. 2C), as in the nanodisc-reconstituted WT INX-6 structure (Fig. 1D). These flat densities are therefore encompassed during the nanodisc formation process but not derived from the N terminus of INX-6. Bulb densities X and Y do not appear in the INX-6ΔN map, as supported by the difference map focusing on the transmembrane domains (Fig. 2D). This observation reveals that the INX-6 N termini contribute to densities X and Y. Currently, the N-terminal helix (NTH) is simply placed on density X because the distribution is very close to the N-terminal funnel of solubilized connexin and innexin observed in the pore (Fig. 2A) (4, 6, 7). As mentioned above, the distance between TM1 and density Y is sufficiently small for the NTH to reach (Fig. 1E). These bulb densities X and Y may represent a different distribution probability of the N terminus that can assume flexible conformations in and around the pore at random when the hemichannel is embedded in a lipid bilayer.
MD simulation study of the INX-6 hemichannels
As observed in the hemichannel structures in nanodiscs, it is likely that phospholipids exist inside the pore when the channel is reconstituted in a lipid bilayer and presumably in native membranes as well. To study the accessibility of phospholipids around the INX-6 hemichannel, we performed the MD simulations of all three INX-6 structures virtually embedded in POPC phospholipids with a calculation time of 100 ns (120 ns for INX-6ΔN; Fig. 3 and fig. S6). For the WT INX-6– and INX-6ΔN–in–nanodisc models, POPC molecules were initially distributed inside and outside the channel but not between the subunits. For the WT INX-6–in–detergent model, POPC molecules were located only outside the channel because of the wide-open pore pathway. After MD simulation, we observed that the POPC molecules were located between adjacent subunits in the transmembrane region in all three structures (Fig. 3, A to C), and specifically, the lipids from outside and inside the channel contacted each other in the nanodisc models (Fig. 3, A and B). The transmembrane helix bundles of INX-6 do not have lateral interactions between adjacent subunits (23), where the unassigned densities, possibly detergent or lipid, are observed in the high-resolution structure of INX-6 (6). In the present study, these unassigned densities were consistently observed in the three structures (Fig. 3, D to F). The MD simulation suggests that phospholipids around the INX-6 hemichannel could penetrate the spaces flanked by the transmembrane helix bundles of the adjacent subunits.
Fig. 3. MD simulation of the undocked INX-6 hemichannels embedded in phospholipids and unassigned densities in cryo-EM maps.
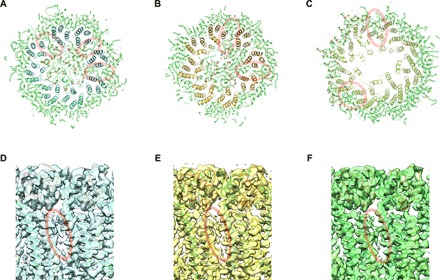
(A to C) The final models of the three atomic structures obtained by cryo-EM in this work after independently performed MD simulations in POPC for 100 ns [120 ns for (B)]. Slab sections of WT INX-6 in a nanodisc (A), INX-6ΔN in a nanodisc (B), and WT INX-6 in detergent (C) corresponding to the transmembrane domain along with POPC molecules are viewed from the cytoplasmic side. POPC models that are inserted in the space between adjacent subunits are indicated by red circles. (D to F) Unassigned densities (red circles) observed in the space between transmembrane helix bundles of adjacent subunits of WT INX-6 in a nanodisc (D), INX-6ΔN in a nanodisc (E), and WT INX-6 in detergent (F) viewed from the orientation horizontal to the membrane plane.
The N terminus is required for the normal function of INX-6 hemichannels
To investigate the functional importance of the N terminus in undocked INX-6 hemichannels and the functionality of the INX-6 constructs used for our cryo-EM study, we measured hemichannel currents in single oocytes injected with complementary RNA (cRNA) derived from WT INX-6 and a series of various INX-6ΔNs. While membrane currents were observed for WT INX-6 depending on the applied membrane potential, all other mutants except for the INX-6ΔN2-3 mutant (N-terminal deletion of residues 2 and 3) exhibited no activity compared with background levels (Fig. 4, A and B). The WT activity was inhibited by 10 μM carbenoxolone, indicating that the observed currents derived from the INX-6 hemichannels (Fig. 4A). To measure the inhibitory effect of the nonfunctional deletion mutants, the mutants of INX-6ΔN2-6 (N-terminal deletion of residues 2 to 6) and INX-6ΔN2-19 (N-terminal deletion of residues 2 to 19, used for cryo-EM) were coinjected with WT INX-6 in a single oocyte. The measured current represented a strong dominant negative effect induced by INX-6ΔN2-6 and INX-6ΔN2-19 (Fig. 4A). We also observed that oocytes injected with WT INX-6–green fluorescent protein (GFP) (C-terminal GFP fusion), INX-6ΔN2-19–GFP, and WT INX-6–GFP coinjected with INX-6ΔN2-19 without tag exhibited the fluorescence signals at the plasma membrane of Xenopus oocytes (fig. S7, A to D). These suggest that the nonfunctional mutant peptides have the potential to form heteromeric hemichannels at the oocyte plasma membrane. As the N-terminal deletion of five residues (2 to 6) is sufficient to completely suppress the WT activity (Fig. 4A), a nearly intact N terminus is required to exhibit the electrical functionality of the undocked INX-6 hemichannel. These findings suggest that the construct of WT INX-6 used for the cryo-EM study can open the pore at the oocyte plasma membrane.
Fig. 4. Functional analysis of INX-6 hemichannels with single Xenopus oocyte voltage clamp.
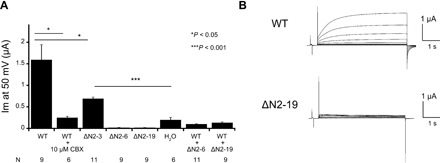
(A) Measured hemichannel currents of INX-6 and negative control (H2O) at 50 mV and steady state. Error bars correspond to SE (*P < 0.05 and ***P < 0.001). (B) Representative currents of WT INX-6 (top) and INX-6ΔN2-19 (bottom). The applied membrane potential is increased from −30 to +50 mV in 10-mV steps.
DISCUSSION
The high-resolution structures of gap junction channels so far reported are all in a solubilized form surrounded by detergent micelles or amphipol and exhibit a wide pore pathway (4–7). As the pore size of gap junction channels is much larger than that of most other ion channels, it has remained a puzzling question how such a large-sized pore can be completely closed when the channel is in a closed state. Here, we describe the first high-resolution structure of a gap junction hemichannel embedded in a lipid bilayer. Although there is no reliable way to label lipid molecules in a cryo-EM map, comparison of our three structures suggests that the double-layer densities obstructing the pore in nanodisc-reconstituted hemichannels correspond to phospholipids. It should, however, be noted that we cannot rule out the possibility that these double-layer densities appeared due to artificial effects, such as the nanodisc reaction and use of solubilized POPC without native components. It should also be considered that the N terminus of INX-6 partially contributes to these densities along with phospholipids, as the N-terminal residues are not fully resolved in the structures. On the other hand, the contribution of lipid molecules to a protein structure has often been suggested.
In a previous study using EM crystallography, an unassigned density was observed in the middle of a rotor ring of an ATP synthase (24), which was estimated to comprise phospholipids (25). A crystal structure of the K ring of bacterial V–adenosine triphosphatase (ATPase) also has lipids on the inside surface of the rotor ring (26), while a recent high-resolution cryo-EM structure of a V-ATPase Vo proton channel reconstituted in a nanodisc exhibited helices instead of lipids inside the rotor ring (27). Excluding the double-layer densities in the pore of WT INX-6 in a nanodisc, the pore diameter of 30 to 40 Å (fig. S2, E and F) is similar to the rotor ring diameters of V-ATPase (26) and FoF1-ATP synthase (28), and it is thus not surprising that lipids or helices occupy a space of this size when synthesized peptides are oligomerized in the membrane. In tetrameric ion channels, the side fenestrations of TWIK-1 are occupied by lipid alkyl chains having access to the ion-conducting pore (29). A lipid blockade gating hypothesis has alternatively been proposed for TRAAK (30). If the lipid bilayer environment is related to the INX-6 channel function, protein-lipid interactions should be further studied to clarify the mechanisms of gap junction channel closure. Pore obstruction with phospholipids may explain how closure of an undocked hemichannel is facilitated in a lipid bilayer environment (Fig. 5A).
Fig. 5. Schematic representation of the N-terminal rearrangement of the INX-6 undocked hemichannel in the lipid bilayer environment.
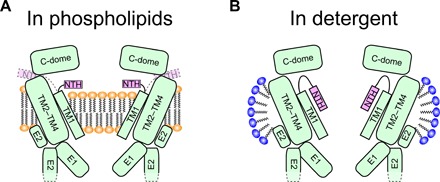
(A) INX-6 hemichannel embedded in phospholipids. Instead of the no-funnel configuration, the NTH is rearranged, and possibly the phospholipids enter the pore. The N terminus might be deflected toward the outside of the channel (dotted lines). (B) INX-6 hemichannel surrounded by detergent micelles. NTHs form a funnel configuration with a wide-open pore. The constriction is formed by the E1 inner lobes but may not be the smallest because more than 10 residues at the N terminus are still disordered.
The hemichannel structure of WT INX-6 in detergent shows a large pore pathway surrounded by the N-terminal funnel (Fig. 5B). As for connexin channels, evidence suggests that the N-terminal deletions and mutations tend to decrease connexin channel functionality (31–34), in accordance with previous and current functional findings regarding innexins (Fig. 4) (23, 35). These findings would be reasonable if intact N termini can form an appropriate open pore funnel whereby a functional conduit is generated. The N-terminal funnel has been described in three previous reports (4, 6, 7). It is intuitively interpreted as an open conformation, but the movement of the N-terminal portions has remained unclear. A low-resolution 3D reconstruction of Cx43 (36) and recent x-ray structures of Cx26 (5) do not show the N-terminal domain densities, suggesting high flexibility of the connexin N terminus. In the case of tetrameric assembly channels, no major conformational change has been observed between those in detergent and those in nanodiscs (37), but reconstitution in nanodiscs allows a detailed structural analysis in a more stable environment (38). In this study, the N-terminal arrangement of WT INX-6 in a nanodisc is distinct from a funnel configuration in detergent. Although it is possible that the N-terminal rearrangement is not shared between the innexin and connexin families, our results suggest high flexibility of the INX-6 N terminus that allows not only for assuming a funnel conformation but also an alternative conformation depending on the presence or absence of phospholipids. Considering the involvement of lipid molecules in the N-terminal rearrangement, the closure mechanism of gap junction channels is likely much more complicated than a simple plug in the pore by the N-terminal assembly (15, 39), especially when a gap junction channel is reconstituted in a lipid bilayer, such as in 2D crystals (15, 23).
Sensitivity to the transjunctional voltage gating property is observed in other innexin channels (40). For connexins, transjunctional voltage-dependent gating may be initiated by movement of an N-terminal voltage sensor (41–43). The findings of the present study should be carefully considered in this context. Although the INX-6 hemichannels exhibited voltage-dependent changes in the open probability (Fig. 4B), it is unclear in our cryo-EM structures whether the lipid-mediated N-terminal rearrangement is associated with the voltage-dependent behavior. Further studies are necessary to determine the trigger for conformational changes of the N-terminal domain of gap junction channels in vivo.
MATERIALS AND METHODS
Protein expression and purification of the undocked INX-6 hemichannels
WT and INX-6ΔN (N-terminal residues 2 to 19 are deleted) were derived from our previous studies (23, 44). In this work, INX-6ΔN corresponds to the deletion of residues 2 to 19 unless otherwise noted. All three INX-6 constructs contain GFP plus an 8×-histidine tag at the C terminus and were expressed and purified from Sf9 cells as described previously (6, 44). Briefly, Sf9 cells cultured at 27°C were infected with the recombinant virus and harvested 46 to 48 hours after infection. The following processes were all performed at 4°C. The harvested cells were sonicated for 90 s in a buffer containing 10 mM tris (pH 7.5), 150 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride and centrifuged at 12,000 rpm (22,100g) for 25 min with a JA-14 rotor. The isolated membrane was solubilized in buffer containing 10 mM tris (pH 7.5), 150 mM NaCl, and 2% (w/w) n-dodecyl-β-d-maltopyranoside (Anatrace) and mixed by rotation for 30 min. The mixture was then centrifuged at 12,000g for 10 min, and the supernatant was bound to Ni–nitrilotriacetic acid agarose (Qiagen). The protein-bound resins were washed with buffer containing 10 mM tris (pH 7.5), 150 mM NaCl, 10 mM l-histidine, and 0.1% digitonin (Wako) and eluted with 300 mM l-histidine. After digesting the C-terminal tag, gel filtration in buffer [10 mM tris (pH 7.5), 150 mM NaCl, and 0.05% n-dodecyl-β-d-maltopyranoside] with a Superose 6 Increase 10/300 GL column (GE Healthcare) was performed to separate the protein into fractions. In the gel filtration step, it is important to keep 150 mM NaCl and to use n-dodecyl-β-d-maltopyranoside to purify mostly undocked hemichannels. Peak fractions for INX-6 hemichannels were collected for further experiments.
Preparation of undocked INX-6 hemichannels for cryo-EM using nanodiscs and GraDeR
The INX-6 hemichannels were reconstituted into nanodiscs following the method described previously (38). Briefly, fractions of purified INX-6 containing hemichannels were mixed with POPC (Avanti Polar Lipids) at 10 mg ml−1 and MSP2N2 at 9.5 mg ml−1. The reconstitution was obtained at a molar ratio of INX-6:MSP2N2:POPC = 1:0.75:100 for WT and 1:0.5:30 for INX-6ΔN. Bio-beads SM-2 resin (Bio-Rad) was added to the mixture to initiate the nanodisc reconstitution reaction by removing the detergents. The mixture was incubated overnight at 4°C with constant rotation. The bio-beads were removed, and the mixture was cleared of debris by ultracentrifugation. The supernatant was loaded onto a Superose 6 Increase 10/300 GL column (GE Healthcare) in buffer without detergent [10 mM tris and 150 mM NaCl (pH 7.5)]. Reconstitution was assessed by fluorescence-detection size exclusion chromatography (45), SDS–polyacrylamide gel electrophoresis, and negative-stain EM. Peak fractions corresponding to reconstituted INX-6 hemichannels in lipid nanodiscs were collected and submitted to cryo-EM.
GraDeR (21) was performed according to a previous study (6), with minor modifications. Lauryl maltose neopentyl glycol (Anatrace) was added to the gel filtration fraction of INX-6 at a final concentration of 0.02%. Buffer A [10 mM tris (pH 7.5), 150 mM NaCl, 5% glycerol, and 0.003% lauryl maltose neopentyl glycol] was used for the top layer, and buffer B [10 mM tris (pH 7.5), 150 mM NaCl, and 25% glycerol] was used for the bottom layer. After stacking the buffers, the gradient was generated with Gradient Master 108 (BioComp Instruments). The solution containing the WT INX-6 hemichannels was loaded on the top of the gradient and centrifuged at 35,000 rpm (209,678g) for 18 hours at 4°C with an SW41Ti rotor. The solution was recovered as fractions using a peristaltic pump and analyzed by fluorescence-detection size exclusion chromatography. The INX-6 solutions were dialyzed against a 10 mM tris (pH 7.5), 50 mM NaCl buffer for 1 hour at 4°C and pooled for cryo-EM. The condition of 50 mM NaCl is also important to facilitate undocked hemichannels.
Cryo-EM for undocked INX-6 hemichannels
All cryo-EM grids were preirradiated overnight with an electron beam and glow-discharged. The proteins for cryo-EM were concentrated at 2 to 4 mg ml−1. For WT INX-6 in nanodiscs, 2 μl of the protein solution was placed on 200-mesh Quantifoil R1.2/1.3 molybdenum grids and plunged into liquid ethane with Vitrobot Mark IV (Thermo Fisher Scientific). The applied parameters were as follows: blotting force, 3; blotting time, 4 s; and 100% humidity at 4°C. For INX-6ΔN in nanodiscs and WT INX-6 in detergent, 200-mesh Quantifoil R2/2 molybdenum grids were used. The protein solution (1.5 μl) was placed on a grid, excess solution was manually blotted for 5 s at room temperature, and the grid was plunged into liquid ethane with a manual plunger on KF-80 (formerly Reichert-Jung).
The data were collected using a JEM-3000SFF (JEOL) electron microscope at 300 kV equipped with a K2 summit direct electron detector camera (Gatan). The images were recorded at a magnification of ×40,600 at a temperature of 80 to 100 K. The superresolution mode (7420 × 7676 pixels) with a pixel size of 0.616 Å at the specimen level was used for WT INX-6 in nanodiscs, and the electron counting mode (3710 × 3838 pixels) with a pixel size of 1.232 Å was used for INX-6ΔN in nanodiscs and WT INX-6 in detergent. The dose rate was 10.5 e− per physical pixel per second, corresponding to 6.9 e− per Å2 at the specimen level. The exposure time was 6.0 s, resulting in an accumulated dose of 41.5 e− per Å2. Each image includes 20 fractioned frames for the superresolution mode and 30 frames for the electron counting mode, and the last few frames were removed to adjust the total dose to 35 e− per Å2.
For image processing of WT INX-6 in nanodiscs, the dose-fractioned images in the superresolution mode were binned 2 × 2 by Fourier cropping, resulting in 3710 × 3838 pixels with a pixel size of 1.232 Å. All of the stacked frames were subjected to motion correction with MotionCorr2 (46). Defocus was estimated using CTFFIND4 (47). A total of 258,673 particles were selected using EMAN2 (48) with a box size of 180 × 180 pixels from 928 micrographs, which also generated the initial 3D model with 43,810 particles. The particles were transmitted to RELION2.0 (49), 121,699 particles were selected by 2D classification, and the particles were divided into four classes by 3D classification, resulting in only one good class containing 40,359 particles. The 3D autorefinement with C8 symmetry produced a map at 4.0 Å resolution, and the final resolution after masking and postprocessing was 3.8 Å, based on the gold standard Fourier shell correlation using a criterion of 0.143 (22). For INX-6ΔN in nanodiscs and WT INX-6 in detergent, the micrographs were processed similarly but modified for electron counting mode recording. The stacked frames were aligned by MotionCorr2 (46) without binning, followed by contrast transfer function estimation with CTFFIND4 (47). For INX-6ΔN in nanodiscs, the total number of picked particles was 138,502 from 300 micrographs. 2D classification selected 88,379 particles, and finally 54,536 particles were fractioned in one of the three classes by 3D classification. 3D autorefinement produced a map at 4.0 Å resolution followed by postprocessing with masking, resulting in a map with 3.6 Å resolution. For WT INX-6 in detergent, a total of 192,550 particles were picked from 497 micrographs, and 93,860 particles were selected by 2D classification and were fractionated into four classes, one of which contained 66,811 particles. 3D autorefinement produced a map at 4.2 Å resolution, and the postprocessed map had a resolution of 3.8 Å. The local resolution maps were calculated on ResMap (50).
Model building of the undocked INX-6 hemichannels
The atomic model of Protein Data Bank (PDB) code 5H1Q that we reported previously was used as an initial model of the undocked INX-6 hemichannels. The disordered regions in E1 and E2 were eliminated, and the N-terminal regions were manually modified on COOT (51). For the WT INX-6 in nanodiscs, the N-terminal helices were placed on density X inside the pore as a representative. As mentioned in the text, however, the strong density Y between the adjacent subunits may represent the probability of the N terminus based on the clear difference from the INX-6ΔN map. The models were refined on PHENIX (52). The real-space refine function was used in combination with restraint about the secondary structure and noncrystallographic symmetry, and the COOT/PHENIX refinement was iterated until the refinements converged. Last, the statistics calculated using MolProbity (53) were checked. The pore size for WT INX-6 in detergent was calculated using HOLE (54). The statistical surface potential map was generated using APBS (55). Figures were drawn with the PyMOL Molecular Graphics System (Schrödinger) and UCSF Chimera (56).
MD simulations of the undocked INX-6 hemichannels
The structures of the undocked INX-6 hemichannels (WT in detergent, WT in a nanodisc, and INX-6ΔN in a nanodisc) were embedded with VMD 1.9.3 (57) in solvated lipid bilayers comprising 447 to 490 POPC molecules, ~100 mM KCl, and 72,000 to 76,000 water molecules. The spatial arrangement of the proteins in the lipid bilayer was determined by referring to the data of the docked INX-6 gap junction channel (PDB ID: 5H1R) in the OPM (orientation of protein in membrane) database. In the WT-in-detergent model, lipid molecules were removed from the pore and water molecules were placed there instead. In the WT-in-nanodisc and INX-6ΔN–in–nanodisc models, lipid molecules were retained in the pores. For the WT-in-detergent model, the system was first energy-minimized and then equilibrated at 300 K and 1 × 105 bar in three consecutive runs of 500-ps MD simulations. In the first run, only non-protein atoms were allowed to move. In the second and third runs, position restraints with a force constant of 1 kcal mol−1 Å−2 were imposed on all the protein atoms and on only the Cα atoms, respectively. Last, a 100-ns MD simulation was performed without restraints. For the WT-in-nanodisc model, only the atoms of the fatty acid parts of the lipid were allowed to move in the first equilibration run. In the second run, the position restraints were imposed on all the protein atoms and the phosphorus atoms of the lipids in the pore. In the third run, they were imposed on only the Cα atoms. After the equilibration MD runs, a space was formed in the lipid bilayer in the pore because the lipid molecules moved to form closer interactions with the protein or with other lipid molecules. Lipid molecules were inserted into the space from a preequilibrated lipid bilayer model, and water molecules that overlapped with the inserted lipid molecules were removed. The equilibration-insertion process was repeated three times. The system was equilibrated again, and a 100-ns unrestrained MD simulation was performed. For the INX-6ΔN–in–nanodisc model, the equilibration-insertion process was repeated four times. The system was equilibrated again, and an MD simulation was performed without restraints. We found, however, that a space was formed in the lipid bilayer in the pore in 20 ns. Therefore, we inserted lipid molecules into the space and equilibrated the system. Then, a 100-ns unrestrained MD simulation was performed.
The CHARMM36m force-field parameters (58) were used for the protein, lipid, and ions. The TIP3P model (59) was used for water. The temperature was controlled with the Langevin dynamics method (60, 61). The pressure was regulated with the Langevin piston method (62). Bond lengths involving hydrogen atoms were constrained using the SHAKE algorithm (63, 64) to allow the use of a large time step (2 fs). Electrostatic interactions were calculated with the particle mesh Ewald method (65, 66). All MD simulations were performed with NAMD 2.12 (67), with coordinates recorded every 10 ps.
Voltage-clamp recording with a single Xenopus oocyte for INX-6 hemichannels
A series of N-terminal deletion mutants (ΔN2-3, ΔN2-6, and ΔN2-19) of INX-6 was generated according to the PrimeSTAR Mutagenesis Basal Kit protocol (Takara). Complementary DNAs (cDNAs) for INX-6 and its mutants were cloned into pGEM-HeFx plasmids (68). The plasmids were linearized using restriction enzymes and then applied to an RNA preparation kit (T7 mMESSAGE mMACHINE; Ambion) according to the manufacturer’s protocol. An adult Xenopus laevis female was anesthetized with MS-222 (Merck), and the ovarian lobes were collected using a surgical knife and forceps. The eggs were treated with collagenase solution (1 mg ml−1 collagenase type I, Life Technologies) in OR2 buffer (82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM Hepes, adjusted to pH 7.5 using NaOH) at 18°C for 1.5 hours. Stage V and VI oocytes were collected manually and used for cRNA injection. cRNA (10 ng) was coinjected with antisense oligonucleotide DNA for Xenopus cx38 (10 ng) into Xenopus oocytes. For the negative control, only antisense oligonucleotide was injected. Oocytes injected with cRNA were incubated for 3 days at 18°C in ND96 buffer (93.5 mM NaCl, 2 mM KCl, 1.8 mM MgCl2, 2 mM MgCl2, and 5 mM Hepes, adjusted to pH 7.5 using NaOH). Hemichannel currents were recorded from a single oocyte using Multi-Electrode Clamp Amplifier iTEV90. Current and voltage electrodes were prepared with a micropipette puller (P-1000, Sutter Instrument) to obtain a resistance of 0.5 to 1.0 megohm when filled with an internal solution containing 3 M KCl, 10 mM ethylene glycol tetraacetic acid (EGTA), and 10 mM Hepes, adjusted to pH 7.4 using KOH. Modified ND96 solution without CaCl2 was used as the bath solution. To obtain the hemichannel current, the cells were initially clamped at −40 mV and then subjected to 5-s voltage steps from −30 to +50 mV in 10-mV increments.
Fluorescent microscopy of GFP-tagged INX-6 in Xenopus oocytes
WT and ΔN2-19 INX-6 containing GFP plus an 8×-histidine tag at the C terminus were subcloned into pGEM-HeFx and expressed in Xenopus oocytes by cRNA injection as described above. Uninjected oocytes were used for negative control. To examine whether ΔN2-19 blocks trafficking of WT, INX-6ΔN2-19 without tag was coexpressed with WT INX-6-GFP. Oocytes expressing INX6 proteins were washed with phosphate-buffered saline (PBS) and embedded with optimal cutting temperature (OCT) compound, and the frozen sections were prepared. Fluorescent micrographs were recorded using a BZ-X700 microscope (KEYENCE). To distinguish GFP signals at the plasma membrane from yolk autofluorescence, we used a color camera along with a GFP longpass filter.
Supplementary Material
Acknowledgments
We thank K. Kobayashi and H. Iwao (JEOL) for maintaining the electron microscopes and G. Christoph (Osaka University) for critical reading. We thank H. Hirano (Nagoya University) for cell culture and assistance for the biochemical experiments. Funding: This work was supported by Grants-in-Aid for Scientific Research (S), the Japan New Energy and Industrial Technology Development Organization (NEDO), and the Japan Agency for Medical Research and Development (AMED) (Y.F.); Grants-in-Aid for Scientific Research (B) under grant number JP17H03683 (M.W.), Challenging Research (Exploratory) (K.T.); the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED under grant numbers JP19am0101118 (K.T.), JP19am0101107 (support number 1380) (T.T.), and JP19ae0101050s0202 and JP19am0101074 (A.O.); Grants-in-Aid for Scientific Research (C) under grant number JP16K07266 (A.O.); and Grants-in-Aid for Scientific Research (B) under grant number JP19H03165 (A.O.). Author contributions: B.B. and R.S. expressed and purified the proteins and generated the nanodisc reconstitution; B.B., R.S., and A.O. collected cryo-EM data; A.O. and K.T. processed cryo-EM images; K.T. performed model building and refinement; M.W. measured channel activity with electrophysiology; T.T. performed the MD simulations; A.O. designed and supervised the research; and all authors contributed to writing the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Requests for the MSP2N2 plasmid should be submitted to Addgene. Cryo-EM density maps for undocked INX-6 hemichannels have been deposited in the EMDataBank (http://emdatabank.org/) under accession codes EMD-9971 (WT INX-6 in a nanodisc), EMD-9972 (WT INX-6 in detergent), and EMD-9973 (INX-6ΔN in a nanodisc). Atomic coordinates were deposited in the PDB under accession numbers 6KFF, 6KFG, and 6KFH. Cryo-EM images were deposited in the Electron Microscopy Pilot Image Archive (EMPIAR; www.ebi.ac.uk/pdbe/emdb/empiar) with accession codes EMPIAR-10289, EMPIAR-10290, and EMPIAR-10291.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/7/eaax3157/DC1
Fig. S1. Cryo-EM of the undocked WT INX-6 hemichannel in a nanodisc.
Fig. S2. Comparison of INX-6 channels between docked and undocked forms focusing on extracellular domains and the pore pathway.
Fig. S3. Sequence alignment of INX-6 and docking interface residues.
Fig. S4. Cryo-EM of the undocked WT INX-6 hemichannel in detergent prepared by GraDeR.
Fig. S5. Cryo-EM of INX-6ΔN in an undocked hemichannel form in a nanodisc.
Fig. S6. MD simulations of the undocked INX-6 hemichannels.
Fig. S7. Fluorescent micrographs of INX-6 distribution on Xenopus oocytes.
Table S1. Summary of data and statistics of the undocked INX-6 hemichannel structure determination.
Reference (69)
REFERENCES AND NOTES
- 1.Neijssen J., Pang B., Neefjes J., Gap junction-mediated intercellular communication in the immune system. Prog. Biophys. Mol. Biol. 94, 207–218 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Lai-Cheong J. E., Arita K., McGrath J. A., Genetic diseases of junctions. J. Invest. Dermatol. 127, 2713–2725 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Phelan P., Innexins: Members of an evolutionarily conserved family of gap-junction proteins. Biochim. Biophys. Acta 1711, 225–245 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Maeda S., Nakagawa S., Suga M., Yamashita E., Oshima A., Fujiyoshi Y., Tsukihara T., Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 458, 597–602 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Bennett B. C., Purdy M. D., Baker K. A., Acharya C., McIntire W. E., Stevens R. C., Zhang Q., Harris A. L., Abagyan R., Yeager M., An electrostatic mechanism for Ca2+-mediated regulation of gap junction channels. Nat. Commun. 7, 8770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oshima A., Tani K., Fujiyoshi Y., Atomic structure of the innexin-6 gap junction channel determined by cryo-EM. Nat. Commun. 7, 13681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myers J. B., Haddad B. G., O'Neill S. E., Chorev D. S., Yoshioka C. C., Robinson C. V., Zuckerman D. M., Reichow S. L., Structure of native lens connexin 46/50 intercellular channels by cryo-EM. Nature 564, 372–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abascal F., Zardoya R., LRRC8 proteins share a common ancestor with pannexins, and may form hexameric channels involved in cell-cell communication. Bioessays 34, 551–560 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Deneka D., Sawicka M., Lam A. K. M., Paulino C., Dutzler R., Structure of a volume-regulated anion channel of the LRRC8 family. Nature 558, 254–259 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Kefauver J. M., Saotome K., Dubin A. E., Pallesen J., Cottrell C. A., Cahalan S. M., Qiu Z., Hong G., Crowley C. S., Whitwam T., Lee W.-H., Ward A. B., Patapoutian A., Structure of the human volume regulated anion channel. eLife 7, e38461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasuya G., Nakane T., Yokoyama T., Jia Y., Inoue M., Watanabe K., Nakamura R., Nishizawa T., Kusakizako T., Tsutsumi A., Yanagisawa H., Dohmae N., Hattori M., Ichijo H., Yan Z., Kikkawa M., Shirouzu M., Ishitani R., Nureki O., Cryo-EM structures of the human volume-regulated anion channel LRRC8. Nat. Struct. Mol. Biol. 25, 797–804 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Peracchia C., Chemical gating of gap junction channels: Roles of calcium, pH and calmodulin. Biochim. Biophys. Acta 1662, 61–80 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Saez J. C., Retamal M. A., Basilio D., Bukauskas F. F., Bennett M. V. L., Connexin-based gap junction hemichannels: Gating mechanisms. Biochim. Biophys. Acta 1711, 215–224 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unwin P. N. T., Ennis P. D., Two configurations of a channel-forming membrane protein. Nature 307, 609–613 (1984). [DOI] [PubMed] [Google Scholar]
- 15.Oshima A., Tani K., Hiroaki Y., Fujiyoshi Y., Sosinsky G. E., Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc. Natl. Acad. Sci. U.S.A. 104, 10034–10039 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catterall W. A., Wisedchaisri G., Zheng N., The chemical basis for electrical signaling. Nat. Chem. Biol. 13, 455–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niemeyer M. I., Cid L. P., González W., Sepúlveda F. V., Gating, regulation, and structure in K2P K+ channels: In varietate concordia? Mol. Pharmacol. 90, 309–317 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Okamura Y., Fujiwara Y., Sakata S., Gating mechanisms of voltage-gated proton channels. Annu. Rev. Biochem. 84, 685–709 (2015). [DOI] [PubMed] [Google Scholar]
- 19.White T. W., Bruzzone R., Wolfram S., Paul D. L., Goodenough D. A., Selective interactions among multiple connexin proteins expressed in the vertebrate lens: The second extracellular domain is a determinant of compatibility between connexins. J. Cell Biol. 125, 879–892 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White T. W., Paul D. L., Goodenough D. A., Bruzzone R., Functional analysis of selective interactions among rodent connexins. Mol. Biol. Cell 6, 459–470 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauer F., Gerle C., Fischer N., Oshima A., Shinzawa-Itoh K., Shimada S., Yokoyama K., Fujiyoshi Y., Stark H., GraDeR: Membrane protein complex preparation for single-particle cryo-EM. Structure 23, 1769–1775 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Rosental P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Oshima A., Matsuzawa T., Murata K., Tani K., Fujiyoshi Y., Hexadecameric structure of an invertebrate gap junction channel. J. Mol. Biol. 428, 1227–1236 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Meier T., Matthey U., von Ballmoos C., Vonck J., Krug von Nidda T., Kühlbrandt W., Dimroth P., Evidence for structural integrity in the undecameric c-rings isolated from sodium ATP synthases. J. Mol. Biol. 325, 389–397 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Meier T., Matthey U., Henzen F., Dimroth P., Müller D. J., The central plug in the reconstituted undecameric c cylinder of a bacterial ATP synthase consists of phospholipids. FEBS Lett. 505, 353–356 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Murata T., Yamato I., Kakinuma Y., Leslie A. G. W., Walker J. E., Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae. Science 308, 654–659 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Roh S.-H., Stam N. J., Hryc C. F., Couoh-Cardel S., Pintilie G., Chiu W., Wilkens S., The 3.5-Å CryoEM structure of nanodisc-reconstituted yeast vacuolar ATPase Vo proton channel. Mol. Cell 69, 993–1004.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klusch N., Murphy B. J., Mills D. J., Yildiz Ö., Kühlbrandt W., Structural basis of proton translocation and force generation in mitochondrial ATP synthase. eLife 6, e33274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller A. N., Long S. B., Crystal structure of the human two–pore domain potassium channel K2P1. Science 335, 432–436 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Brohawn S. G., Campbell E. B., MacKinnon R., Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature 516, 126–130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshima A., Tani K., Toloue M. M., Hiroaki Y., Smock A., Inukai S., Cone A., Nicholson B. J., Sosinsky G. E., Fujiyoshi Y., Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels. J. Mol. Biol. 405, 724–735 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyle J. W., Minogue P. J., Thomas B. C., Domowicz D. A. L., Berthoud V. M., Hanck D. A., Beyer E. C., An intact connexin N-terminus is required for function but not gap junction formation. J. Cell Sci. 121, 2744–2750 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyle J. W., Berthoud V. M., Kurutz J., Minogue P. J., Greenspan M., Hanck D. A., Beyer E. C., The N terminus of connexin37 contains an α-helix that is required for channel function. J. Biol. Chem. 284, 20418–20427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao Q., Liu Q., Lorentz R., Gong X.-Q., Bai D., Shaw G. S., Laird D. W., Structure and functional studies of N-terminal Cx43 mutants linked to oculodentodigital dysplasia. Mol. Biol. Cell 23, 3312–3321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marks W. D., Skerrett I. M., Role of amino terminus in voltage gating and unctional rectification of Shaking B innexins. J. Neurophysiol. 111, 1383–1395 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Unger V. M., Kumar N. M., Gilula N. B., Yeager M., Three-dimensional structure of a recombinant gap junction membrane channel. Science 283, 1176–1180 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Matthies D., Bae C., Toombes G. E. S., Fox T., Bartesaghi A., Subramaniam S., Swartz K. J., Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs. eLife 7, e37558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Y., Cao E., Julius D., Cheng Y., TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oshima A., Structure and closure of connexin gap junction channels. FEBS Lett. 588, 1230–1237 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Depriest A., Phelan P., Skerrett I. M., Tryptophan scanning mutagenesis of the first transmembrane domain of the innexin Shaking-B(Lethal). Biophys. J. 101, 2408–2416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verselis V. K., Ginter C. S., Bargiello T. A., Opposite voltage gating polarities of two closely related connexins. Nature 368, 348–351 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Purnick P. E., Oh S., Abrams C. K., Verselis V. K., Bargiello T. A., Reversal of the gating polarity of gap junctions by negative charge substitutions in the N-terminus of connexin 32. Biophys. J. 79, 2403–2415 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh S., Rivkin S., Tang Q., Verselis V. K., Bargiello T. A., Determinants of gating polarity of a connexin 32 hemichannel. Biophys. J. 87, 912–928 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oshima A., Matsuzawa T., Nishikawa K., Fujiyoshi Y., Oligomeric structure and functional characterization of Caenorhabditis elegans innexin-6 gap junction protein. J. Biol. Chem. 288, 10513–10521 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawate T., Gouaux E., Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Kimanius D., Forsberg B. O., Scheres S. H. W., Lindahl E., Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. P., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A., Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B. L., Grubmüller H., MacKerell A. D. Jr., CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 60.Pastor R. W., Brooks B. R., Szabo A., An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 65, 1409–1419 (1988). [Google Scholar]
- 61.Loncharich R. J., Brooks B. R., Pastor R. W., Langevin dynamics of peptides: The frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers 32, 523–535 (1992). [DOI] [PubMed] [Google Scholar]
- 62.Feller S. E., Zhang Y., Pastor R. W., Brooks B. R., Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995). [Google Scholar]
- 63.Miyamoto S., Kollman P. A., Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992). [Google Scholar]
- 64.Ryckaert J.-P., Ciccotti G., Berendsen H. J. C., Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977). [Google Scholar]
- 65.Darden T., York D., Pedersen L., Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 66.Essmann U., Perela L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G., A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995). [Google Scholar]
- 67.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K., Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furutani K., Ohno Y., Inanobe A., Hibino H., Kurachi Y., Mutational and in silico analyses for antidepressant block of astroglial inward-rectifier Kir4.1 channel. Mol. Pharmacol. 75, 1287–1295 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Thompson J. D., Higgins D. G., Gibson T. J., CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/7/eaax3157/DC1
Fig. S1. Cryo-EM of the undocked WT INX-6 hemichannel in a nanodisc.
Fig. S2. Comparison of INX-6 channels between docked and undocked forms focusing on extracellular domains and the pore pathway.
Fig. S3. Sequence alignment of INX-6 and docking interface residues.
Fig. S4. Cryo-EM of the undocked WT INX-6 hemichannel in detergent prepared by GraDeR.
Fig. S5. Cryo-EM of INX-6ΔN in an undocked hemichannel form in a nanodisc.
Fig. S6. MD simulations of the undocked INX-6 hemichannels.
Fig. S7. Fluorescent micrographs of INX-6 distribution on Xenopus oocytes.
Table S1. Summary of data and statistics of the undocked INX-6 hemichannel structure determination.
Reference (69)


