Manganese and zinc are essential transition metals involved in many fundamental cellular processes, including protection against external oxidative stress. In Bacillus subtilis, Zur and MntR are key transcriptional regulators of zinc and manganese homeostasis, respectively. In this work, proteome analysis of B. subtilis wild-type, ΔmntR, and Δzur strains provided new insights into bacterial adaptation to deregulation of essential metal ions. Deletions of mntR and zur genes increased bacterial sensitivity to lysozyme, beta-lactam antibiotics, and external oxidative stress and impacted the cell wall thickness. Overall, these findings highlight that Zur and MntR regulatory networks are connected to antibiotic sensitivity and cell wall plasticity.
KEYWORDS: Bacillus, metal, MntR, proteomics, Zur, stress
ABSTRACT
The Bacillus subtilis MntR and Zur transcriptional regulators control homeostasis of manganese and zinc, two essential elements required in various cellular processes. In this work, we describe the global impact of mntR and zur deletions at the protein level. Using a comprehensive proteomic approach, we showed that 33 and 55 proteins are differentially abundant in ΔmntR and Δzur cells, respectively, including proteins involved in metal acquisition, translation, central metabolism, and cell wall homeostasis. In addition, both mutants showed modifications in intracellular metal ion pools, with significant Mg2+ accumulation in the ΔmntR mutant. Phenotypic and morphological analyses of ΔmntR and Δzur mutants revealed their high sensitivity to lysozyme, beta-lactam antibiotics, and external oxidative stress. Mutant strains had a modified cell wall thickness and accumulated lower levels of intracellular reactive oxygen species (ROS) than the wild-type strain. Remarkably, our results highlight an intimate connection between MntR, Zur, antibiotic sensitivity, and cell wall structure.
IMPORTANCE Manganese and zinc are essential transition metals involved in many fundamental cellular processes, including protection against external oxidative stress. In Bacillus subtilis, Zur and MntR are key transcriptional regulators of zinc and manganese homeostasis, respectively. In this work, proteome analysis of B. subtilis wild-type, ΔmntR, and Δzur strains provided new insights into bacterial adaptation to deregulation of essential metal ions. Deletions of mntR and zur genes increased bacterial sensitivity to lysozyme, beta-lactam antibiotics, and external oxidative stress and impacted the cell wall thickness. Overall, these findings highlight that Zur and MntR regulatory networks are connected to antibiotic sensitivity and cell wall plasticity.
INTRODUCTION
Metal ions such as iron, zinc, and manganese are crucial for central cellular processes, as they are structural components of many proteins and membranes while participating in the catalysis of metabolic reactions and electron transfer. However, metals are highly toxic when in excess. The mammalian innate immune system responds to infection by combining deprivation of some metals (Fe, Zn, and Mn) with a metal poisoning strategy (Cu and Zn) to prevent bacterial replication (1–3). Restrictive access to critical transition metals is a defense known as nutritional immunity (4). Examples involve lipocalin, which binds siderophores and thereby prevents Fe acquisition by bacterial pathogens (5), and calprotectin, which restricts acquisition of Zn or Mn (6). In the macrophage phagosome, NRAMP1 removes Mn and Fe to starve intracellular pathogens (7, 8). In contrast, in macrophage antimicrobial pathways, Zn and Cu toxicity is used to combat invading microbes.
Among transition metals, Mn and Zn are important in many fundamental cellular processes, including protection against reactive oxygen species (ROS). There is now evidence that the invading microbe utilizes Mn as a key micronutrient to counteract the effects of host-mediated oxidative stress. Thus, Mn plays a significant role in adaptation of pathogenic bacteria to the human host (9). It protects bacterial cells from oxidative stress, either as a cofactor for Mn-dependent catalases and superoxide dismutases or via its inherent ability to quench free-radical-mediated reactions (9–11). Similarly, Zn is important for resistance to both hydrogen peroxide (H2O2) and the thiol-oxidizing agent diamide (12). Zn2+ may protect thiols from oxidation and even displace other redox-active metals in protein thiol-containing active sites to maintain the function of proteins (13). Pathogenic bacteria deficient in maintaining proper metal homeostasis are less virulent (2, 8, 14, 15).
Intracellular metal homeostasis in bacteria is ensured by finely tuned import and efflux systems (16). Metalloregulatory proteins act as metal-sensing regulatory transcription factors. In the Gram-positive bacterium Bacillus subtilis, MntR and Zur are key regulators of manganese and zinc homeostasis, respectively. MntR is a bifunctional regulator that binds Mn2+ as an effector (17, 18). The MntR regulon includes 7 genes involved in Mn2+ uptake or efflux (18–22). The Zur metalloprotein binds Zn2+ as a corepressor (23–26). The Zur regulon contains 11 genes, including genes for zinc transporters (12, 23) and zinc-independent alternative ribosomal proteins (27–30). However, Zur binds 80 regions on the chromosome, indicating far broader control by this regulator (31). Zinc homeostasis is also maintained via a zinc-inducible efflux pump, CzcD, which is regulated at the transcriptional level by the metalloregulator CzrA (22). Together, Zur and CzrA sense changes in the labile Zn2+ pool and modify expression of genes encoding proteins that mediate zinc homeostasis (32, 33).
The roles of the MntR and Zur proteins at the transcriptional level have been studied extensively. However, a study of the global impact of mntR and zur deletions at the protein level is still missing. Here, we performed a comprehensive study on global protein changes in B. subtilis ΔmntR and Δzur mutants by quantitative proteomic analysis and validated the obtained results by physiology tests and microscopy observation. Together, our results show that Zur and MntR regulators are necessary to ensure bacterial fitness upon environmental stress.
RESULTS
Impact of mntR and zur deletions on the B. subtilis proteome.
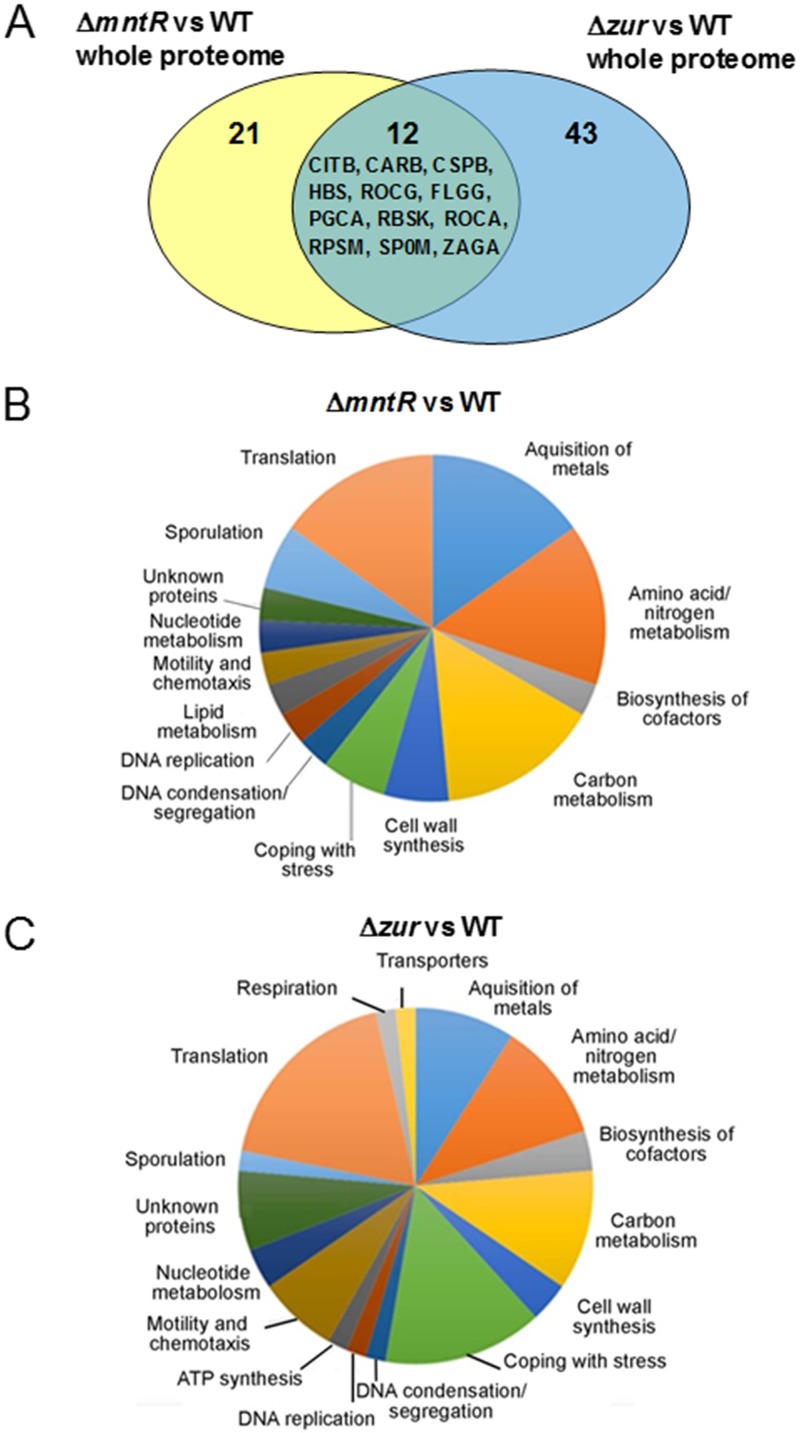
To understand the physiological state of cells with the deleted mntR or zur gene deleted, we performed a comparative analysis of the cytosolic and membrane proteome of ΔmntR, Δzur, and wild-type (WT) cells grown in exponential phase (Table 1). Optimized analyses by liquid chromatography-tandem mass spectrometry (LC-MS/MS) of four technical replicates resolved more than 1,700 proteins (see Table S1 in the supplemental material). In total, 33 and 55 proteins were statistically significantly differentially abundant (P < 0.05 by the Kruskal-Wallis test and one-way analysis of variance [ANOVA]) in ΔmntR and Δzur cells, respectively (Table 2). Twelve proteins were common between ΔmntR and Δzur proteomic data sets (Fig. 1A). Among them, CarB and Spo0M changes in regulation diverged between the two mutants. Proteins involved in metal acquisition, translation, stress response, cell wall synthesis, and amino acid/nitrogen/carbon metabolism were mainly affected in ΔmntR and Δzur mutants (Fig. 1B and C).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotypea | Reference or source |
|---|---|---|
| BSB1 | trp+ | 76 |
| BSAS45 | Δzur::aphA3 | 31 |
| BSAS46 | ΔmntR::aphA3 | This work |
aphA3, Enterococcus faecalis kanamycin resistance gene.
TABLE 2.
Total proteins differentially abundant in the cytosolic and membrane fractions of the B. subtilis ΔmntR and Δzur mutants
| Mutant | No. of proteins |
||||
|---|---|---|---|---|---|
| Cytosolic fraction |
Membrane fraction |
Common in cytosolic and membrane fractions | |||
| Upregulated | Downregulated | Upregulated | Downregulated | ||
| ΔmntR | 14 | 14 | 6 | 1 | 2 |
| Δzur | 20 | 16 | 18 | 10 | 9 |
FIG 1.
Functional classification of proteins with altered abundance in the B. subtilis ΔmntR and Δzur mutants. (A) Venn diagram of the 76 proteins showing significant abundance changes between ΔmntR, Δzur, and WT cells. (B and C) Proteins are classified according to their involvement in biological processes.
The proteome analysis of the ΔmntR mutant indicated a greater increase of the MntA and MntB proteins, as expected since they are components of the MntABCD Mn uptake systems (Table 3). Of note, proteins of the Mn2+ efflux systems, MneP and MneS, were not identified in the membrane or cytosolic subfraction (Table S1), suggesting that their levels were too low to be detected under our conditions. Three proteins involved in metal ion homeostasis, ZagA, DhbB, and DhbF, were increased. The ZagA zinc metallochaperone is encoded by a Zur-regulated gene (34). DhbB and DhbF are involved in biosynthesis of the catecholate siderophore bacillibactin. The dhbABCDF operon belongs to the Fur regulon (35), whose transcriptional expression has been shown to weakly decrease in an mntR mutant (19). Under our conditions, we assumed that mismetallation of Fur occurs in ΔmntR cells, which would lead to increased expression of the dhb operon. Interestingly, the SodA Mn-dependent superoxide dismutase was less abundant in ΔmntR cells, while two increased proteins, YtsJ and RbsK, participate in metabolic pathways generating NADPH (Table 3). The malic enzyme YtsJ was proposed to be important to balance the intracellular redox pool (36, 37). The ribokinase RbsK belongs to the pentose phosphate (PP) pathway, which yields two molecules of NADPH per glucose molecule (38, 39). NADPH is the unique provider of reducing equivalents to maintain or regenerate bacterial detoxifying and antioxidative defense systems (40). One can hypothesize that ΔmntR cells need NADPH to cope with internal oxidative stress and therefore need a metabolic adaptation to maintain the NADPH/NADP+ ratio. However, we observed that mntR and zur deletions were accompanied by maintained levels of the NADPH/NADP+ ratio during the exponential phase, without significant increase in the NADPH level (data not shown).
TABLE 3.
Proteins differentially abundant in the B. subtilis ΔmntR mutant
| Protein | Fold change in abundance in ΔmntR vs wild type | Gene | Function | Functional categorya |
|---|---|---|---|---|
| Proteins upregulated in cytosolic fraction | ||||
| DhbBb | 12.75 | dhbB | Isochorismatase | Acquisition of iron |
| CitB | 10.5 | citB | Aconitase | TCA cycle |
| BglH | 9.25 | bglH | Phospho-beta-glucosidase | Utilization of salicin |
| Spo0M | 9.25 | spo0M | Sporulation | Sporulation |
| PgcA | 8.25 | pgcA | Alpha-phosphoglucomutase | Biosynthesis of teichoic acid |
| RocA | 7.25 | rocA | 3-Hydroxy-1-pyrroline-5-carboxylate dehydrogenase | Arginine utilization/nitrogen metabolism |
| RbsK | 7 | rbsK | Ribokinase | Utilization of ribose |
| MntAb | 6.75 | mntA | Manganese ABC transporter | Acquisition of manganese |
| Syv | 6.5 | valS | Valyl-tRNA synthetase | Translation |
| Mao4 | 6.25 | ytsJ | NADP-dependent malate dehydrogenase | Utilization of malate |
| Syh | 5.75 | hisS | Histidyl-tRNA synthetase | Translation |
| RocGb | 5.25 | rocG | Glutamate dehydrogenase | Arginine utilization/nitrogen metabolism |
| MenB | 5.25 | menB | Naphthoate synthase | Biosynthesis of menaquinone |
| ZagA | 5.25 | zagA | Zinc metallochaperone | Acquisition of zinc |
| Proteins upregulated in membrane fraction | ||||
| DhbFb | 29 | dhbF | Involved in bacillibactin biosynthesis | Acquisition of iron |
| MntAb | 17.5 | mntA | Manganese ABC transporter | Acquisition of manganese |
| MtnK | 8.25 | mtnK | 5-Methylthioribose kinase | Methionine metabolism |
| RocGb | 7.5 | rocG | Glutamate dehydrogenase | Arginine utilization/nitrogen metabolism |
| MntB | 6.25 | mntB | Manganese ABC transporter | Acquisition of manganese |
| DnaX | 5.5 | dnaX | DNA polymerase III | DNA replication |
| Proteins downregulated in cytosolic fraction | ||||
| RplD | −25.5 | rplD | Ribosomal protein L4 | Translation |
| CarB | −22.5 | carB | Carbamoyl-phosphate transferase-arginine | Biosynthesis of arginine |
| CspB | −20.5 | cspB | RNA chaperone | Cold stress proteins |
| FabI | −20.5 | fabI | Enoyl-acyl carrier protein reductase | Fatty acid biosynthesis |
| YpfD | −11.5 | ypfD | Similar to ribosomal protein S1 | Translation |
| SodM | −11 | sodA | Superoxide dismutase | Resistance against oxidative stress |
| YkaA | −9.75 | ykaA | Unknown | Proteins of unknown function |
| PunA | −8.75 | pupG | Purine nucleoside phosphorylase | Nucleotide metabolism |
| RpsM | −8.25 | rpsM | Ribosomal protein S13 | Translation |
| OppA | −7.25 | oppA | Oligopeptide ABC transporter | Utilization of peptides |
| PtsG | −6.5 | ptsG | Glucose permease | Carbon core metabolism |
| Hbs | −6.25 | hbs | Nonspecific DNA-binding protein | DNA condensation/segregation |
| AsnB | −6 | asnB | Asparagine synthase | Control of peptidoglycan hydrolysis |
| PbpC | −6 | pbpC | Penicillin-binding protein 3 | Cell wall synthesis |
| Protein downregulated in membrane fraction: FlgG | −5.75 | flgE | Flagellar hook protein | Motility and chemotaxis |
According to the SubtiWiki database (77).
Protein detected in both cytosolic and membrane fractions.
Proteomic analysis of the Δzur mutant indicated higher levels of proteins (AdcA, AdcC, FolE2, RpmE2, YciB, ZagA, and ZinT) encoded by Zur-regulated genes, as expected (Table 4). We also observed increased amounts of the CadA efflux ATPase, which is involved in zinc and cadmium metal export (22). The Δzur strain displayed reduced levels of proteins related to stress resistance (Table 4): (i) the bacilliredoxin YqiW (renamed BrxB) promotes a redox switch in response to oxidative stress (41); (ii) the membrane protein YceD, similar to a tellurium resistance protein, is required for survival under ethanol stress (42); and (iii) LiaH is involved in resistance to envelope stress conditions (43). Therefore, the proteomic data suggest that Δzur cells could be more sensitive to environmental stresses. Interestingly, the molecular chaperone DnaK was significantly increased in Δzur cells. DnaK contributes to membrane and overall cell recovery under different stress conditions (44–46), suggesting that the Δzur strain faces internal stress conditions. Finally, the WapA tRNase toxin-WapI antitoxin system (47) was derepressed in the Δzur strain compared to the wild-type strain. Increased expression of WapA (the putative toxin) and decreased expression of WapI (the antitoxin) indicate that Δzur cells may be more susceptible to growth inhibition than wild-type cells.
TABLE 4.
Proteins differentially abundant in the B. subtilis Δzur mutant
| Protein | Fold change in abundance in Δzur vs WT | Gene | Function | Functional categorya |
|---|---|---|---|---|
| Proteins upregulated in cytosolic fraction | ||||
| ZagAb | 243 | zagA | Zinc metallochaperone | Acquisition of zinc |
| AdcAb | 34 | znuA | ABC transporter for zinc | Acquisition of zinc |
| FolE2b | 22.25 | folE2 | GTP cyclohydrolase IB | Biosynthesis of folate |
| CarB | 18 | carB | Carbamoyl-phosphate transferase-arginine | Biosynthesis of arginine |
| DnaK | 15.5 | dnaK | Molecular chaperone | Protein quality control |
| RocA | 14.25 | rocA | 3-Hydroxy-1-pyrroline-5-carboxylate dehydrogenase | Arginine utilization/nitrogen metabolism |
| AcoN | 10.5 | citB | Aconitase | TCA cycle |
| ZinTb | 8 | zinT | Zinc-binding protein | Acquisition of zinc |
| GlmU | 7.75 | glmU | Bifunctional N-acetylglucosamine-1-phosphate | Biosynthesis of peptidoglycan |
| RbsK | 7.25 | rbsK | Ribokinase | Utilization of ribose |
| PgcA | 7.25 | pgcA | Alpha-phosphoglucomutase | Biosynthesis of teichoic acid |
| Syi | 7.25 | ileS | Isoleucyl-tRNA synthetase | Translation |
| Fbp | 7 | fbp | Fructose-1,6-bisphosphatase | Gluconeogenesis |
| GndA | 7 | gndA | NADP-dependent phosphogluconate dehydrogenase | Pentose phosphate pathway |
| Oatb | 7 | rocD | Ornithine transaminase | Ornithine utilization/nitrogen metabolism |
| PyrGb | 6.75 | pyrG | CTP synthase | Nucleotide metabolism |
| RocGb | 6.25 | rocG | Glutamate dehydrogenase | Arginine utilization/nitrogen metabolism |
| AdcCb | 5.75 | znuC | ABC transporter for zinc | Acquisition of zinc |
| Rl31B | 5.5 | rpmE2 | Accessory ribosomal protein | Translation |
| Sya | 5.5 | alaS | Alanine-tRNA synthetase | Translation |
| Proteins upregulated in membrane fraction | ||||
| ZagAb | 184.75 | zagA | Zinc metallochaperone | Acquisition of zinc |
| AdcAb | 100.75 | znuA | ABC transporter for zinc | Acquisition of zinc |
| YjlD | 29.25 | ndh | NADH dehydrogenase | Respiration |
| GlpK | 28.25 | glpK | Glycerol kinase | Utilization of glycerol |
| MetK | 22.75 | metK | S-Adenosylmethionine synthetase | Methionine metabolism |
| ZinTb | 19.25 | zinT | Zinc-binding protein | Acquisition of zinc |
| AdcCb | 16.75 | znuC | ABC transporter for zinc | Acquisition of zinc |
| WapA | 15.5 | wapA | Cell wall-associated protein precursor | Toxins, antitoxins, and immunity against toxins |
| AsnB | 14.75 | asnB | Asparagine synthase | Control of peptidoglycan hydrolysis |
| FolE2b | 12.25 | folE2 | GTP cyclohydrolase IB | Biosynthesis of folate |
| PyrGb | 11 | pyrG | CTP synthase | Nucleotide metabolism |
| CadA | 10 | cadA | Cadmium transporting ATPase | Resistance against toxic metals |
| YciB | 8.5 | yciB | Putative l,d-transpeptidase | Acquisition of zinc |
| Oatb | 7.75 | rocD | Ornithine transaminase | Ornithine utilization/nitrogen metabolism |
| MurG | 6.75 | murG | Peptidoglycan precursor biosynthesis | Biosynthesis of peptidoglycan |
| RocGb | 6.75 | rocG | Glutamate dehydrogenase | Arginine utilization/nitrogen metabolism |
| Smc | 6.25 | smc | Segregation of replication origins | DNA condensation/segregation |
| DnaN | 5.5 | dnaN | DNA polymerase III | DNA replication |
| Proteins downregulated in cytosolic fraction | ||||
| Rl7 | −46.75 | rplL | Ribosomal protein L12 | Translation |
| CspB | −37.25 | cspB | RNA chaperone | Cold stress proteins |
| Tkt | −25.75 | tkt | Transketolase | Pentose phosphate pathway |
| Rs6 | −10.5 | rpsF | Ribosomal protein S6 | Translation |
| RpsM | −10 | rpsM | Ribosomal protein S13 | Translation |
| Hbs | −9 | hbs | Nonspecific DNA-binding protein | DNA condensation/segregation |
| YqiW | −7 | yqiW | Bacilliredoxin | Resistance against oxidative stress |
| YjlCb | −7 | yjlC | Unknown | Protein of unknown function |
| YxkC | −6.5 | yxkC | Unknown | Protein of unknown function |
| WapI | −6.25 | wapI | Immunity protein | Toxins, antitoxins, and immunity against toxins |
| PthP | −6 | ptsH | Phosphotransferase system-dependent sugar transport and carbon catabolite repression | Transporters |
| Rl15 | −6 | rplO | Ribosomal protein L15 | Translation |
| YceD | −5.75 | yceD | Similar to tellurium resistance protein | Resistance against toxic metals |
| Rl10 | −5.5 | rplJ | Ribosomal protein L10 | Translation |
| Rs19 | −5.5 | rpsS | Ribosomal protein S19 | Translation |
| YqeY | −5.25 | yqeY | Unknown | Protein of unknown function |
| Proteins down-regulated in membrane fraction | ||||
| Fla | −85 | hag | Flagellin protein | Motility and chemotaxis |
| Rs8 | −18.25 | rpsH | Ribosomal protein S8 | Translation |
| CheA | −8.5 | cheA | Two-component sensor kinase | Motility and chemotaxis |
| YjlCb | −8 | yjlC | Unknown | Protein of unknown function |
| AtpF | −7.5 | atpF | ATP synthase | ATP synthesis |
| FliL | −6.5 | fliL | Flagellar protein | Motility and chemotaxis |
| LiaH | −6 | liaH | Phage shock-like protein | Resistance against oxidative stress and cell wall antibiotics |
| Spo0M | −6 | spo0M | Unknown | Sporulation |
| FlgG | −5.5 | flgE | Flagellar hook protein | Motility and chemotaxis |
| GgaB | −5 | ggaB | Membrane protein | Biosynthesis of teichoic acid |
According to the SubtiWiki database (77).
Protein detected in both cytosolic and membrane fractions.
The proteomic analysis revealed elevated levels of proteins related to the arginine utilization pathway (RocA, RocD, and RocG) in ΔmntR and Δzur cells. The rocABC, rocDEF, and rocG operons are under the transcriptional control of RocR and are induced by the presence of arginine, ornithine, or proline in the growth medium. In contrast, expression of rocE and rocD was repressed in the transcriptomic profile of a zur mutant (12). The relationship between Zur and the RocR regulon is not clear. However, our result reinforced the idea that alteration of zinc homeostasis affects expression of the RocR regulon.
Finally, we observed changes in the levels of proteins involved in peptidoglycan synthesis or hydrolysis as well as those involved in the biosynthesis of teichoic acid (Tables 3 and 4). This set includes PgcA, PbpC, and AsnB in ΔmntR cells and AsnB, GgaB, GlmU, MurG, and PgcA in Δzur cells. This suggested a possible impact of mntR and zur deletions on the cell wall structure.
Interestingly, the global proteomics analysis brought new data and revealed some unexpected regulatory effects in the ΔmntR and Δzur mutants. It is well known that mRNA and protein expression levels may differ despite being quantified in the same bacterial cells and under similar conditions (48). Previous transcriptomic studies of zur and mntR mutants in Luria-Bertani (LB) medium identified mainly genes belonging to the Zur and MntR regulons, respectively (12, 19). Our data highlighted a broader impact of mntR and zur deletions on proteins whose levels are modified in response to perturbation of cellular metal ion pools.
Disruption of metal homeostasis in ΔmntR and Δzur mutants.
To verify whether deletion of mntR or zur modifies the cellular metal ion pool, total cell-associated metal ions were quantified in the wild-type, ΔmntR and Δzur strains. For this, mid-exponential-phase cells were cultivated at the same optical density at 600 nm (OD600) in LB medium and analyzed by inductively coupled plasma mass spectrometry (ICP-MS).
The ΔmntR mutant displayed 2-, ∼1.5-, ∼1.5-, and ∼1.2-fold increases in the levels of Mg2+, Mn2+, Zn2+, and Cd2+, respectively, compared to the wild type (Table 5). In contrast, levels of Fe2+, Co2+, Cu2+, and Ni2+ were similar in both strains. Manganese accumulation in ΔmntR cells despite activation of efflux systems (21) may indicate that B. subtilis tolerates a mild Mn2+ intracellular increase without intoxication. The increased level of Cd2+ correlates with the higher level of the MntH transporter, which imports both Mn2+ and Cd2+ ions (18). Accumulation of Zn2+ and Mg2+ in a ΔmntR mutant was unexpected.
TABLE 5.
Metal content of B. subtilis wild type, ΔmntR and Δzur cells measured by ICP-MS
| Metal ion | Concn (μM)a in: |
||
|---|---|---|---|
| WT | ΔmntR mutant | Δzur mutant | |
| Fe2+ | 203 ± 29.2 | 229.4 ± 39.1 | 151.1 ± 49.6 |
| Mg2+ | 523.7 ± 85.5 | 1,155.5 ± 166.8 | 621.3 ± 70.1 |
| Mn2+ | 26.4 ± 3.1 | 42.1 ± 6.4 | 28 ± 4.5 |
| Zn2+ | 224.3 ± 25.1 | 302.7 ± 28.5 | 237.6 ± 32.7 |
| Cd2+ | 2.8 ± 0.2 | 3.6 ± 0.4 | 1.4 ± 0.3 |
| Co2+ | 0.25 ± 0.15 | 0.18 ± 0.1 | 0.18 ± 0.1 |
| Cu2+ | 6.2 ± 0.6 | 6.5 ± 0.6 | 8.8 ± 1.4 |
| Ni2+ | 6.4 ± 1.1 | 7.1 ± 1.4 | 7.5 ± 1.6 |
Results are means ± standard deviations.
The Δzur mutant displayed a 1.5-fold-increased Cu2+ level and a 2-fold-decreased Cd2+ level compared to the wild type (Table 5). The Δzur mutant had no significant differences in total Mn2+, Zn2+, Mg2+, Fe2+, Co2+, and Ni2+ levels. As the internal Zn2+ concentration was maintained in Δzur cells, B. subtilis appeared to tightly control the level of Zn2+ to avoid zinc intoxication. Copper accumulation in a Δzur mutant was intriguing. In B. subtilis, copper uptake involves a well-defined transporter, YcnJ, whose synthesis responds to copper availability (49). The lower level of Cd2+ detected in Δzur cells was in line with increased CadA, the major determinant for Cd2+ resistance (Table 4). In zinc excess, CzrA binds Zn2+ to trigger derepression of CadA (22, 50).
Differential Mg2+-dependent growth of Δzur and ΔmntR cells.
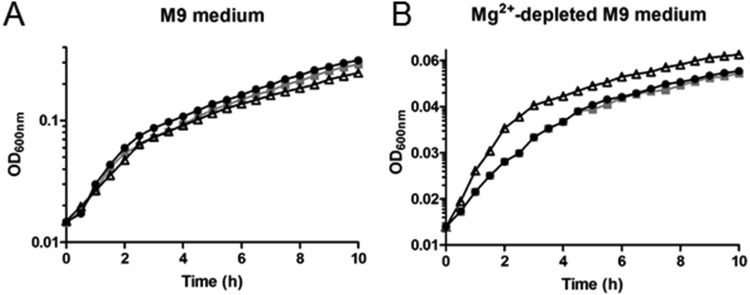
We detected an intracellular Mg2+ concentration of ∼0.5 to 1.0 mM in the three B. subtilis strains (Table 5). To gain insight into the physiological relevance of Mg2+ accumulation in the ΔmntR strain, growth assays were performed under conditions of Mg2+ starvation. Precultures of the wild-type, ΔmntR, and Δzur strains in LB medium were inoculated in M9 defined medium in the presence or absence of Mg2+. In M9 medium containing 25 mM Mg2+, all strains grew with similar growth kinetics (Fig. 2A). In Mg2+-depleted M9 medium (containing only traces of Mg2+), only the ΔmntR mutant showed a growth benefit (Fig. 2B). Therefore, Mg2+ accumulation in ΔmntR cells grown in LB medium appeared to be effective to support growth in Mg2+-depleted M9 medium. It is worth noting that supplementation of growth media with 5 to 25 mM Mg2+ may suppress morphological and vital defects of several B. subtilis mutants with mutations in cell wall-related genes (e.g., ponA, ugtP, pgcA, gtaB, or asnB) (51, 52), but the mechanisms underlying this rescuing role are still unknown. At present, we cannot explain how intracellular Mg2+ compensates for the absence of external Mg2+ ions.
FIG 2.
Effect of magnesium starvation on growth of wild-type, ΔmntR, and Δzur cells. (A) Growth curves of the wild-type (black symbols), ΔmntR (white symbols), and Δzur (gray symbols) strains grown in M9 medium in the presence of 25 mM MgCl2. A representative assay is represented. (B) Growth curves of the wild-type (black symbols), ΔmntR (white symbols), and Δzur (gray symbols) strains grown in MgCl2-depleted M9 medium. The doubling time of wild-type and Δzur cells is approximately 208 min. The doubling time of ΔmntR cells is approximately 122 min. A representative assay is represented. Note that the x axes are different in panels A and B for a better view of the data.
Sensitivity of ΔmntR and Δzur cells to environmental stresses.
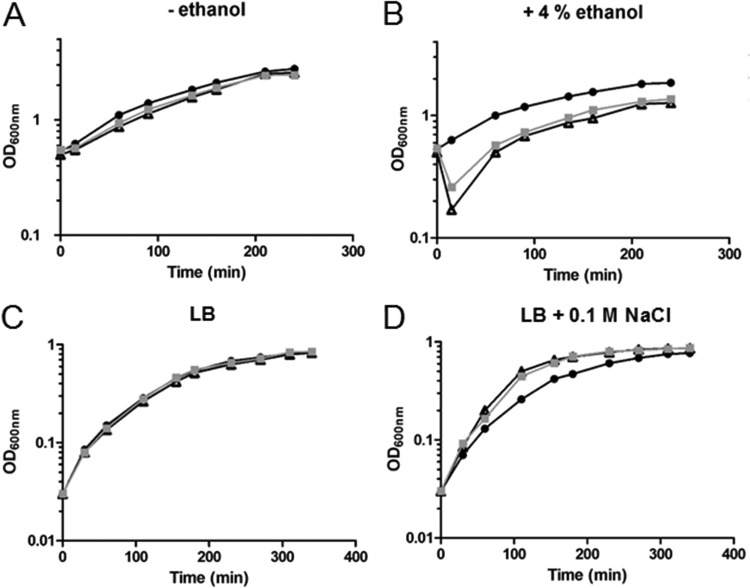
As the proteomic analysis revealed that Δzur cells displayed reduced levels of proteins related to stress resistance (BrxB, YceD, and LiaH), we tested whether environmental stresses such as organic solvent and increased salinity could affect growth of Δzur and ΔmntR cells compared to that of the wild type. Growing bacteria were exposed to a final sublethal concentration of 4% (vol/vol) ethanol. Under the conditions used, no effect on the growth rate was observed for the wild-type cells, whereas 4% ethanol transiently affected growth of the Δzur and ΔmntR cells (Fig. 3A and B). The direct target of ethanol is the membrane bilayer. A lower resistance to ethanol exposure suggests that Δzur and ΔmntR cells are less efficient in activating an early response to stress and/or that their membrane lipids and proteins differ from those in the wild-type cells. Remarkably, we observed in ΔmntR cells a 20-fold-decreased amount of the FabI protein, which is involved in fatty acid biosynthesis (Table 3).
FIG 3.
Effects of ethanol and NaCl stresses on growth of wild-type, ΔmntR, and Δzur cells. (A) Growth curves of the wild-type (black symbols), ΔmntR (white symbols), and Δzur (gray symbols) strains grown in LB medium without ethanol addition. A representative assay is represented. (B) Growth curves of the wild-type (circles), ΔmntR (triangles), and Δzur (squares) strains grown in LB medium after addition of 4% ethanol (final concentration) at an OD600 of 0.6. A representative assay is represented. (C) Growth curves of the wild-type (black symbols), ΔmntR (white symbols), and Δzur (gray symbols) strains grown in LB medium without NaCl addition. A representative assay is represented. (D) Growth curves of the wild-type (circles), ΔmntR (triangles), and Δzur (squares) strains grown in LB medium with addition of NaCl at 0.1 M. A representative assay is represented.
We further tested the effect of NaCl on growth. A moderate saline stress was imposed by incubation of the cells in the presence of 0.1 M NaCl. Under these conditions, we observed a slight but reproducible enhanced fitness of the Δzur and ΔmntR strains compared to the wild-type strain (Fig. 3C and D). No difference in resistance was observed with 0.5 M NaCl (data not shown). As bacterial responses to osmotic challenges are very complex, it is not obvious how the ΔmntR and Δzur strains are better suited than the wild type to cope with moderate saline stress.
Sensitivity of ΔmntR and Δzur cells to lysozyme and beta-lactam antibiotics.
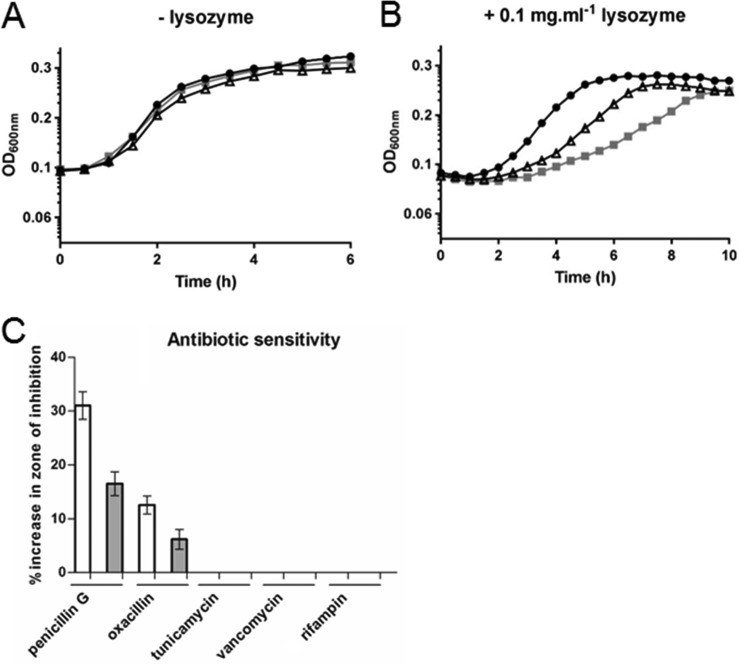
The proteomic analysis revealed modified levels of proteins (AsnB, GlmU, and MurG) involved in peptidoglycan biosynthesis in ΔmntR and Δzur cells. To test whether mntR and zur deletions modified the peptidoglycan integrity, bacteria were treated with lysozyme. Both the ΔmntR and Δzur mutants were more sensitive to 0.1 mg · ml−1 lysozyme than the wild-type strain (Fig. 4A and B). With 0.5 mg · ml−1 lysozyme, ΔmntR, Δzur, and wild-type cells showed similar growth defects (data not shown). In addition, a disk diffusion assay was performed to compare the sensitivities of the mutant and wild-type strains to several antibiotics. The ΔmntR and Δzur mutants were more sensitive than the wild-type strain to penicillin G and oxacillin, which inhibit formation of peptidoglycan cross-links in the bacterial cell wall (Fig. 4C). In contrast, the two mutants had the same sensitivity as the wild-type strain to vancomycin and to tunicamycin, which target other steps of cell wall synthesis, and to the RNA polymerase inhibitor rifampin. The decreased resistance of the ΔmntR and Δzur strains to lysozyme and beta-lactam antibiotics strongly suggests that structural modifications take place in the cell wall of mutant cells. This is in line with the (above-described) proteomic analysis, which indicated deregulation of proteins involved in cell wall plasticity.
FIG 4.
Increased sensitivity of the ΔmntR and Δzur mutants to lysozyme and antibiotics. (A) Growth curves of the wild-type (black symbols), ΔmntR (triangles), and Δzur (gray symbols) strains grown in LB medium. A representative assay is represented. (B) Growth curves of the wild-type (circles), ΔmntR (triangles), and Δzur (squares) strains grown in LB medium in the presence of 1 mg · ml−1 lysozyme. A representative assay is represented. (C) Results of disk diffusion assays with the indicated antibiotics. Bars indicate the percent increase in zone of inhibition for the ΔmntR (white bars) and Δzur (gray bars) mutants relative to the wild type. All the experiments were performed at least in three biological replicates.
Bacterial interfacial potential and cell wall thickness are altered in ΔmntR and Δzur mutants.
Modifications in cell wall composition are expected to affect bacterial surface charge (53, 54). We performed zeta potential measurements to determine bacterial surface potentials. The zeta potential indicates an electrochemical property of the bacterial cell surface which represents the transmembrane potential that maintains the cell wall/membrane architecture. Negative zeta potentials were measured for all three strains grown in LB medium, as expected for Gram-positive bacteria, which contain negatively charged teichoic acids. However, the ΔmntR mutant exhibited a lesser negative potential (−11.6 ± 0.4 mV) than the wild type (−13.5 ± 0.3 mV) and Δzur (−13.8 ± 0.7 mV) cells. This finding suggests that deletion of the mntR gene alters cell surface permeability, as a correlation between negative zeta potential and membrane integrity was previously shown (55). It should be noted that proteomic analysis indicated increased levels of PgcA, a protein involved in biosynthesis of teichoic acid that also may modify the surface charge of bacteria.
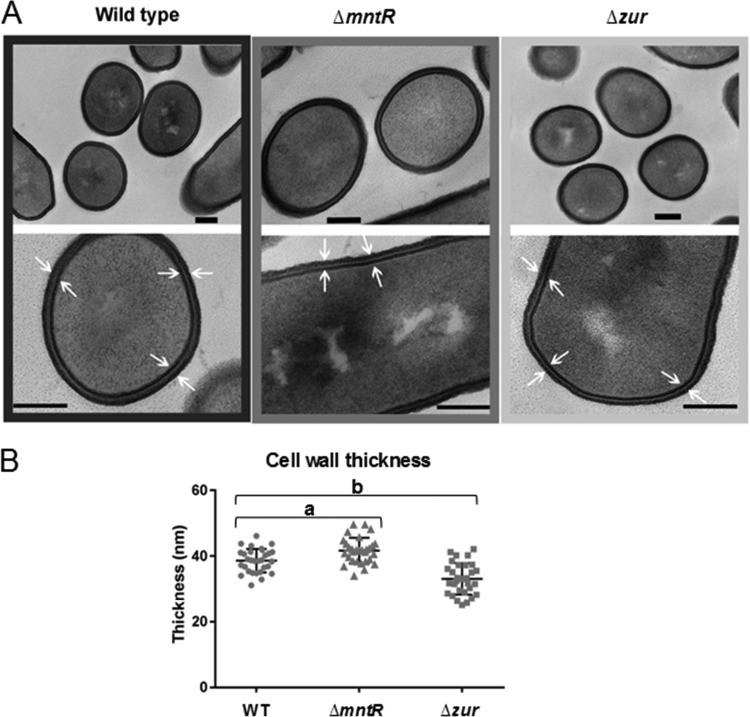
To further identify the modifications that differentiate the cell walls of ΔmntR and Δzur cells, cultures grown in LB were collected at mid-exponential phase and observed in thin cross-section by transmission electron microscopy (TEM). Bacterial cells of the 3 strains appeared as well-separated bacilli (Fig. 5A). The cell wall of the Δzur mutant was thinner (32 ± 4.5 nm) than that of the parental strain (38 ± 3.6 nm) (Fig. 5A and B). In contrast, the cell wall thickness of the ΔmntR mutant was greater (42 ± 3.5 nm) than that of the wild type. We assumed that alteration of cell wall thickness in the ΔmntR and Δzur mutants and modification of the zeta potential in ΔmntR cells resulted from changes in cell surface composition and/or structure. The interplay between manganese and zinc homeostasis and the proteins involved in cell wall plasticity identified in the proteomic approach (Tables 3 and 4) deserves future investigation.
FIG 5.
The mntR and zur deletions are associated with altered cell wall thickness. (A) Transmission electron microscopy images of representative cells of wild-type, ΔmntR, and Δzur cells during mid-log growth phase. Bars, 100 nm. (B) Dot blot graph of the cell wall thickness. Each dot represents a measurement for a single bacillus. a, P = 1.8 · 10−4 <0.05; b, P = 4.4 · 10−6 <0.05.
Low O2·− and H2O2 accumulation in ΔmntR and Δzur mutants.
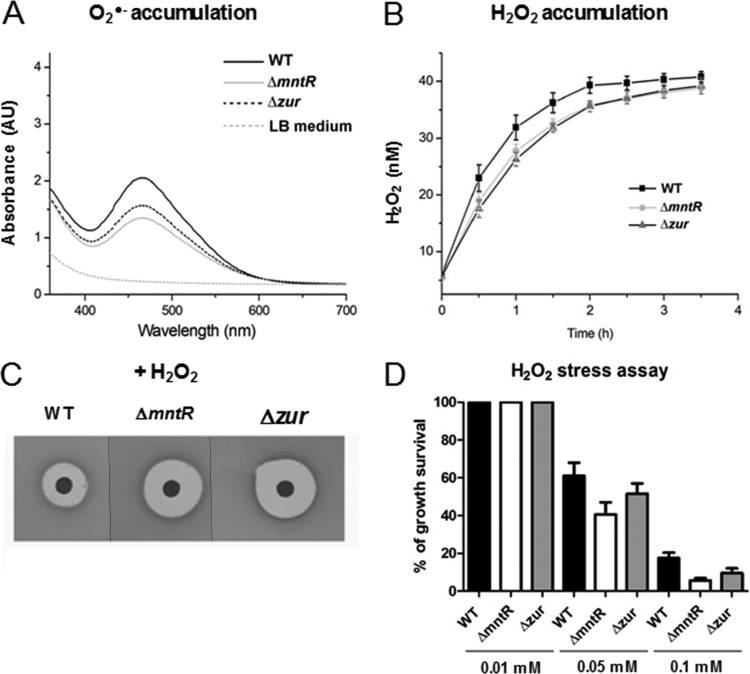
Neutralization of the surface potential of bacteria was shown to trigger the production of ROS (53, 56). Modifications in bacterial surface potential may thus result from cell wall adjustments but also from enhanced ROS production within the bacterial cells (53). However, our proteomic data were in favor of reducing ROS levels. Indeed, we observed decreased levels of BrxB and SodA in the Δzur and ΔmntR mutants, respectively. The BrxB (bacilliredoxin)- and SodA (Mn-dependent superoxide dismutase)-encoding genes can be induced by oxidative stress. Thus, a lower level of BrxB and SodA might indicate a reduction in intracellular ROS. To verify whether bacterial surface charge modifications resulted from intracellular ROS accumulation, we compared the amounts of O2·− and H2O2 production in ΔmntR and Δzur cells to those in the wild-type strain.
The 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt (XTT) assay was performed to estimate cellular production of O2·−. No light adsorption was observed for LB medium alone. XTT absorbs light at 470 nm only when reduced by O2·− and does not do so in its oxidized form. The amount of O2·− in cells in mid-exponential growth phase in LB medium was lower in ΔmntR and Δzur cells than in the wild type (Fig. 6A). The intracellular concentration of H2O2 was calculated in bacteria in mid-exponential phase based on the calibration curve obtained with pure H2O2 using the Amplex red assay. The generation of H2O2 was found to be lower in the mutants than in the wild-type strain (Fig. 6B). The ΔmntR and Δzur mutants appeared to accumulate less intracellular ROS than the wild-type strain. We thus propose that the modification in the bacterial surface potential in ΔmntR cells could result from changes in the cell wall structure rather than from intracellular ROS accumulation.
FIG 6.
ROS accumulation and oxidative stress assays in ΔmntR and Δzur mutants. (A) Quantification of O2·− produced in wild-type (WT), ΔmntR, and Δzur cells at an OD600 of 1 in LB medium. (B) Quantification of H2O2 produced in WT, ΔmntR, and Δzur cells during 4 h of growth in LB medium. (C) Plates from a disk diffusion assay with a drop of 3 M hydrogen peroxide (H2O2). (D) The histograms represent the percentage of survival of WT, ΔmntR, and Δzur cells at 15 min after addition of 0.01, 0.05, or 0.1 mM H2O2. The survival rate was determined as the ratio of the number of CFU per milliliter after 15 min to the number of CFU per milliliter before addition of H2O2.
Effect of external H2O2 on ΔmntR and Δzur cells.
To estimate bacterial sensitivity to external oxidative stress, we compared the survival of wild-type, ΔmntR, and Δzur cells when challenged with external H2O2. Disk diffusion assays indicated an increase in the zone of growth inhibition for the ΔmntR and Δzur mutants compared to that for the wild-type strain (Fig. 6C). Similarly, both the ΔmntR and Δzur mutants showed a growth defect compared to the wild-type strain when exposed to 50 or 100 μM H2O2 for 15 min in LB medium (Fig. 6D). Increased sensitivity of the ΔmntR mutant to H2O2 correlates with the role of Mn2+ as corepressor of the PerR regulator in response to peroxide stress (57–60). Increased susceptibility of the Δzur mutant to H2O2 suggests that Zur is required for expression of oxidative stress defenses, as observed in Corynebacterium diphtheriae (61).
DISCUSSION
Our results provide a new view on B. subtilis mntR and zur mutants under stress-related conditions. Proteomic analysis revealed 33 and 55 proteins whose abundance was affected in ΔmntR and Δzur mutants, respectively. The identified proteins are involved in various cellular processes, notably in translation, amino acid/nitrogen/carbon metabolism, and cell wall homeostasis. Commonly affected proteins suggest coordination between the MntR and Zur regulatory networks.
The ΔmntR and Δzur mutants displayed growth kinetics similar to those of the parental strain, although intracellular pools of some essential metal ions differed (Table 5). The 1.5-fold-greater accumulation of Mn2+ in the ΔmntR mutant versus the WT indicates that B. subtilis can tolerate an intracellular increase in Mn2+ without bacterial growth being affected. In contrast, maintenance of the Zn2+ content in a Δzur mutant provides evidence that alternative pathways can maintain intracellular Zn2+ homeostasis. Surprisingly, Mg2+ and Zn2+ metal ions also accumulate in the ΔmntR mutant (2.2-fold and 1.4-fold increased, respectively), while Cu2+ accumulates 1.5-fold in Δzur cells. It is possible that metabolic changes in ΔmntR and Δzur cells provoke secondary demands for other metal ions, leading to their increased levels. How bacteria simultaneously regulate the content of multiple metal ions merits further studies.
Metal ions catalyze numerous metabolic processes by their roles in electron transfer. Essential pathways such as the tricarboxylic acid (TCA) cycle (CitB) and the stringent response are impacted by bacterial metabolic status (62–65). Our proteomic analyses revealed higher levels of RocA, RocD, and RocG proteins in both ΔmntR and Δzur mutants. These enzymes are involved in arginine and ornithine utilization to generate ammonium and may contribute to maintaining intracellular pH homeostasis (66).
We previously performed a genome-wide identification of Zur-binding sites by chromatin immunoprecipitation coupled with hybridization to DNA tiling arrays (ChIP-on-chip) (31). We were initially surprised that promoters identified by ChIP-on-chip did not match proteins identified in the Δzur mutant. However, the differences may be explained by the importance of posttranscriptional control of translation, as well as the effects of transcriptional regulators other than Zur in controlling gene expression.
We showed that the ΔmntR and Δzur mutants accumulate less ROS (e.g., O2·− and H2O2) than the wild-type counterpart (Fig. 6). Mn2+ and Zn2+ metal ions are intimately linked to the ability to withstand ROS. Mn2+ itself or in a complex with a cellular component is involved in cellular defense against O2·− stress and can therefore substitute for SodA (67). An elevated extracellular Zn2+ amount protects B. subtilis from peroxide stress (59). Hence, slight increases in Mn2+ and Zn2+ pools in ΔmntR cells (Table 5) may sufficiently promote the antioxidative defense mechanism to diminish the need for Mn-dependent superoxide dismutase SodA (Table 3). It is intriguing that the Δzur mutant shows a similar decrease of ROS, whereas the Mn2+ and Zn2+ levels are not modified (Table 5 and Fig. 6). This raises questions about the regulatory role of Zur in maintaining redox potential.
Remarkably, deletion of mntR or zur enhanced sensitivity to lysozyme and beta-lactam antibiotics, both of which target the cell wall (Fig. 4). Our results and previous studies suggest mechanisms that may explain these findings. First, increased sensitivity to peptidoglycan synthesis inhibitors can be due to modification of the cell wall structure and/or composition in the mutants. Our findings are in line with this, since the cell wall thickness and bacterial surface zeta potential were modified in the ΔmntR and Δzur mutants compared to the wild-type strain (Fig. 5). Second, beta-lactam antibiotics were proposed to generate ROS that contribute to bacterial death (68). We observed a greater sensitivity to external ROS of ΔmntR and Δzur cells than of the wild-type strain (Fig. 6). Third, there is an intimate connection between metal ions and antibiotic activity Some antibiotics are known to require metal ions for their activity (69). When not required to mediate target binding, interaction of metal ions with antibiotics can provide an additional mode of antibacterial action, as seen, for example, with Zn2+, which potentiates the antibiotic activity of vancomycin (70), and with ZnMgO, which increases ciprofloxacin activity (71). We showed here that ΔmntR and Δzur cells accumulate some metal ions (Table 5), which may potentiate antibiotic activity. Further investigations are needed to determine the exact role(s) of Zur and MntR in bacterial sensitivity to antibiotics.
Altogether, this work might uncover new targets for intervention to successfully combat emerging bacterial multiresistance against conventional antibiotics.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis strains used in this work are listed in Table 1. B. subtilis cells were grown in Luria-Bertani (LB) medium or in M9 minimal medium (72). In media containing NaCl, the concentrations are indicated. Antibiotics were added at the following concentrations when required: 5 μg kanamycin ml−1 and 60 μg spectinomycin ml−1. Solid media were prepared by addition of 20 g Noble agar (Difco) liter−1. Standard procedures were used to transform B. subtilis (73).
DNA manipulations.
In PCR, the Pfu DNA polymerase was used as recommended by the manufacturer (Biolabs). DNA fragments were purified from agarose gels using the QIAquick kit (Qiagen).
Construction of strains.
The mntR mutant BSAS46 was constructed by homologous replacement of the mntR coding sequence with the kanamycin resistance gene aphA3 using a joining PCR technique. The aphA3 gene was first amplified. The region upstream of the mntR gene (nucleotides 2542520 to 2543556) was amplified by PCR with a 24-bp aphA3 fragment at its 3′ end. The region downstream of mntR (nucleotides 2543796 to 2544739) was amplified with a 24-bp aphA3 fragment at its 5′ end. The three DNA fragments were combined, and then a PCR was performed with the two external oligonucleotides. The final product, corresponding to the two regions flanking mntR with the inserted aphA3 cassette in between, was purified from a gel and used to transform B. subtilis. Integration and deletion were confirmed by PCR and verified by DNA sequencing.
Intracellular metal concentration measurement by ICP-MS.
Overnight cultures of wild-type, ΔmntR, and Δzur B. subtilis strains were diluted to an optical density at 600 nm (OD600) of 0.05 in 15 ml of fresh LB medium cultured in 50-ml Falcon tubes. Bacteria were incubated at 37°C until exponential phase (OD600 of around 0.8). Cell cultures were centrifuged at 4,000 × g at 4°C for 10 min and washed three times in 5 ml of ultrapure water (Millipore) with EDTA added to 1 mM. Samples were dried overnight at 80°C and then acidified twice with Suprapur 65% nitric acid (Merck Millipore) until mineralization. Samples were further analyzed at the University of Montpellier II (Laboratoire ICP-MS, UMR5543 Géosciences). Samples were dissolved with 250 ml of nitric acid (65%) for 1 h. They were diluted 1,000-fold in double-distilled water and analyzed using an Agilent 7700x quadrupole inductively coupled plasma mass spectrometer. Concentrations were determined by analyzing standard solutions. To obtain the number of atoms of metal ions per cell, the raw data were normalized to the washing solution. The measurement for each strain was performed in three replicates.
Quantification of NADPH and NADP+.
Detection and quantification of NADPH and NADP+ content were done using the NADP/NADPH microplate assay kit (Cohesion Biosciences). Assays were performed on cells grown in LB medium until the OD600 was around 0.8. Each experiment was performed in three independent replicates.
Effects of ethanol and NaCl.
Overnight cultures were diluted in fresh LB medium to an initial OD600 of 0.1 and incubated in flasks with shaking (200 rpm) at 37°C. Bacterial growth was measured by following the optical density at 600 nm. In the ethanol stress assay, ethanol at a final concentration of 4% was added when the OD600 reached 0.6. In the saline assay, NaCl was added at final concentration of 0.1 M at the beginning of the cultures.
Effects of lysozyme and MgCl2.
Overnight cultures were diluted at least 100-fold in fresh medium. Growth curves were done in 96-well plates in a Tecan plate reader (Tecan Infinite M200PRO) with continuous agitation at 37°C. Growth conditions were as indicated elsewhere in the text. At least three independent biological replicates were performed.
Assay of sensitivity to antibiotics.
The assay of antibiotic sensitivity was performed on solid media. B. subtilis strains were grown in LB medium to an OD600 of 1. One milliliter of growing culture was spread onto petri plates containing LB medium. The plates were dried briefly in a laminar flow hood before 5-mm paper disks (Whatman) containing 25 μl of antibiotic solution were placed on the plates (penicillin G, 8 mg · ml−1; oxacillin10, μg · ml−1; tunicamycin, 375 μg · ml−1; vancomycin, 0.5 μg · ml−1). The plates were incubated at 37°C overnight. The zones of inhibition were measured by using ImageJ after scanning the plates. Standard deviations were calculated from three independent assays.
Peroxide stress assays.
To perform disk diffusion assays, the cell number was determined in exponential-phase cultures by evaluating the CFU per milliliter. One milliliter of each microbial suspension containing 1 × 108 bacterial cells ml−1 (corresponding to a 0.5 McFarland standard) was spread over the surface of the agar plates. The plates were dried briefly in a laminar flow hood. A sterile 5-mm paper disk was placed on the agar surface, and 10 μl of 3 M hydrogen peroxide (H2O2) was added to the disk. For the stress assay in liquid medium, cultures were inoculated at an OD600 of 0.05 in LB medium and incubated at 37°C with agitation at 200 rpm. At an OD600 of 0.5, H2O2 was added at 0.1 mM. After 15 min of incubation, serial dilutions were plated on LB agar. The CFU per milliliter were counted. The number of CFU per milliliter before the stress was used as a reference to measure the rate of survival.
Quantification of O2·− and H2O2 free radicals.
The bacterial production of superoxide radical ion (O2·−) was evaluated by measuring the adsorption of XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] (Sigma). XTT was dissolved in PBS (pH 7) and added to the bacterial culture to a final concentration of 0.4 mM. When reduced by O2·−, XTT forms water-soluble XTT-formazan, which adsorbs light at 470 nm. The changes in absorbance at 470 nm were monitored using an Infinite M200 luminescence reader (Tecan, Germany). XTT absorbs light at 470 nm only when reduced by O2·−.
Intracellular production of H2O2 was quantified using the Amplex red assay. B. subtilis strains were grown in LB medium to mid-exponential phase. After centrifugation, pellets were resuspended in phosphate-buffered saline (PBS) and transferred to a 96-well plate that contained 20 μl of enzymatic mix (1 μl 10-acetyl-3,7-dihydroxyphenoxazine [ADHP] reagent, 1 μl horseradish peroxidase, and 18 μl assay buffer) in each well. Resorufin fluorescence was measured using a spectrofluorometer (Tecan Infinite M200PRO) with excitation and emission wavelengths of 530 and 590 nm, respectively. H2O2 calibration was done using H2O2 standard solutions ranging from 100 to 1,500 nM. Each experiment was performed in duplicate and repeated at least twice.
Whole-cell surface potential.
The zeta potential of the bacterial cell growth in exponential phase in LB medium was measured using a Zetasizer Nano ZS90 instrument (Malvern, UK). The results for zeta potential are presented as the average value from three independent cultures (10 measurements per culture).
TEM.
For transmission electron microscopy (TEM), bacterial pellets were collected at an OD600 of 1 and fixed with 2% glutaraldehyde in 0.1 m sodium cacodylate buffer, pH 7.2, at room temperature for 3 h. Samples were then contrasted with 0.5% oolong tea extract in sodium cacodylate buffer (74), postfixed with 1% osmium tetroxide containing 1.5% potassium cyanoferrate, gradually dehydrated in ethanol (30% to 100%), substituted gradually in a mixture of propylene oxide and Epon, and embedded in Epon (Delta Microscopy, Labège, France). Thin (70-nm) sections were collected onto 200-mesh copper grids and counterstained with lead citrate. Grids were examined using a Hitachi HT7700 electron microscope operated at 80 kV (Elexience, France). Images were acquired with a charge-coupled device camera (Advanced Microscopy Techniques Corp., Japan). The thickness of the cell wall was measured on TEM micrographs of at least five cells at a magnification of ×70,000, taking at least six measurements on each cell. Statistical analysis was performed with the unpaired Student t test, and a P value of <0.05 was considered significant. Analyses were performed using Prism 7 (GraphPad Software, San Diego, CA).
Sample preparation for proteomic analysis.
To analyze the proteomic profiles of the wild-type, ΔmntR, and Δzur cells, protein extraction and tryptic digestion in gel were performed as described in detail previously (75). Briefly, four independent cultures of each B. subtilis strain were grown in LB medium at an OD600 of 0.8. Cells were harvested by centrifugation and disrupted by a passage through a One Shot cell disrupter (Constant Systems Ltd., Warwickshire, UK). After centrifugation, the resulting supernatants were ultracentrifuged (100,000 × g, 1 h, 4°C). Cytoplasmic fractions were designated the soluble parts after a single ultracentrifugation step. The remaining pellets were considered the crude membrane fraction. The total protein concentration was measured using a NanoDrop instrument. All of the samples were loaded on 10% NuPAGE Bis-Tris gels (Invitrogen). In-gel digestion of the proteins was performed on bands excised from one-dimensional SDS-PAGE gel. The quantity of modified trypsin (Promega, sequencing grade) was 0.5 μg per sample. In the final step, tryptic peptides were resuspended in 25 μl of precolumn loading buffer containing 0.05% (vol/vol) trifluoroacetic acid (TFA) and 5% (vol/vol) acetonitrile prior to LC-MS/MS analysis.
LC-MS.
Mass spectrometry was performed on the Plateforme d'Analyse Protéomique de Paris Sud Ouest (PAPPSO) platform as described in detail previously (75). A NanoLC-Ultra Eksigent (Sciex) system connected to a Q-Exactive mass spectrometer (Thermo Fisher) by a nanoelectrospray ion source was used. The protein identification was performed with X!TandemPipeline (open-source software developed by PAPPSO, version 3.3.1) against a Bacillus subtilis 168 protein database (4,253 entries). The X!Tandem search parameters were as follows: trypsin specificity with two missed cleavages, fixed alkylation of cysteine (+57.0215), and variable oxidation of methionine (+15.9949). The protein identification was run with a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.02 Da. For all proteins identified with a protein E value of <0.01 in the first step, we searched for additional peptides to reinforce identification using similar parameters except that semitryptic peptides and protein N-terminal acetylations were accepted. The final search results were filtered as follows: (i) peptide E value of <0.01 with a minimum of 2 peptides per protein and (ii) protein E value of <0.05.
Relative quantification of peptides and proteins.
Peptide were analyzed by spectral counting (SC). SC takes into account the number of assigned spectra for each protein and is correlated to relative protein abundance. For control quality of data, normalization, filtration, and statistical analysis, MassChroqR (http://pappso.inra.fr/bioinfo/masschroqr/) was used. Proteins with fewer than two peptides were removed. Peptides with a variation ratio of <1.5 were eliminated, as were peptides with a standard deviation from the retention time of 20 s and higher. Repeatable peptides were those which were present in at least 7 of 8 samples. The data set was normalized based on the median retention time, and missing peptide intensities and protein abundances were imputed. The proteins whose numbers of peaks were significantly different (with a minimum difference of 5 peaks between the mutant and the wild type) were determined by using the Kruskal-Wallis test. A one-way ANOVA model was used to analyze changes, with the genotype as a fixed effect. A protein was considered significantly variable when the P value was <0.05 (see Table S1 in the supplemental material).
Availability of data.
All supporting data are included in the main article and in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We benefited from the facilities of the MIMA2 MET (GABI, INRA) and PAPSSO (Micalis, INRA) platforms, Jouy en Josas, France. We thank David Portehault (LCMCP, France) for helping with zeta potential measurements. We are grateful to Philippe Noirot for the support to initiate this project. We gratefully acknowledge Alexandra Gruss and Stéphane Aymerich for critical reading of the manuscript.
This research received funding from the European Union, Marie Curie ITN AMBER, 317338.
P.R., J.V., and S.A. conceived and designed the experiments. P.R., J.A.-M., A.G., C.P., J.V., and S.A. performed the experiments. P.R., A.A.-F., J.V., and S.A. carried out analysis and interpretation of data. J.V. and S.A. drafted the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Djoko KY, Ong CL, Walker MJ, McEwan AG. 2015. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem 290:18954–18961. doi: 10.1074/jbc.R115.647099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neyrolles O, Wolschendorf F, Mitra A, Niederweis M. 2015. Mycobacteria, metals, and the macrophage. Immunol Rev 264:249–263. doi: 10.1111/imr.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stafford SL, Bokil NJ, Achard ME, Kapetanovic R, Schembri MA, McEwan AG, Sweet MJ. 2013. Metal ions in macrophage antimicrobial pathways: emerging roles for zinc and copper. Biosci Rep 33:e00049. doi: 10.1042/BSR20130014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 6.Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol 14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cellier MF, Courville P, Campion C. 2007. Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect 9:1662–1670. doi: 10.1016/j.micinf.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Juttukonda LJ, Skaar EP. 2015. Manganese homeostasis and utilization in pathogenic bacteria. Mol Microbiol 97:216–228. doi: 10.1111/mmi.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lisher JP, Giedroc DP. 2013. Manganese acquisition and homeostasis at the host-pathogen interface. Front Cell Infect Microbiol 3:91. doi: 10.3389/fcimb.2013.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguirre JD, Culotta VC. 2012. Battles with iron: manganese in oxidative stress protection. J Biol Chem 287:13541–13548. doi: 10.1074/jbc.R111.312181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anjem A, Varghese S, Imlay JA. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol 72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaballa A, Wang T, Ye RW, Helmann JD. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J Bacteriol 184:6508–6514. doi: 10.1128/jb.184.23.6508-6514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oteiza PI. 2012. Zinc and the modulation of redox homeostasis. Free Radic Biol Med 53:1748–1759. doi: 10.1016/j.freeradbiomed.2012.08.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janulczyk R, Ricci S, Bjorck L. 2003. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect Immun 71:2656–2664. doi: 10.1128/iai.71.5.2656-2664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner AG, Ong C-LY, Gillen CM, Davies MR, West NP, McEwan AG, Walker MJ. 2015. Manganese homeostasis in group A Streptococcus is critical for resistance to oxidative stress and virulence. mBio 6:e00278-15. doi: 10.1128/mBio.00278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandrangsu P, Rensing C, Helmann JD. 2017. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol 15:338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glasfeld A, Guedon E, Helmann JD, Brennan RG. 2003. Structure of the manganese-bound manganese transport regulator of Bacillus subtilis. Nat Struct Biol 10:652–657. doi: 10.1038/nsb951. [DOI] [PubMed] [Google Scholar]
- 18.Que Q, Helmann JD. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 19.Guedon E, Moore CM, Que Q, Wang T, Ye RW, Helmann JD. 2003. The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and sigmaB regulons. Mol Microbiol 49:1477–1491. doi: 10.1046/j.1365-2958.2003.03648.x. [DOI] [PubMed] [Google Scholar]
- 20.Helmann JD. 2014. Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J Biol Chem 289:28112–28120. doi: 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, Shin JH, Pinochet-Barros A, Su TT, Helmann JD. 2017. Bacillus subtilis MntR coordinates the transcriptional regulation of manganese uptake and efflux systems. Mol Microbiol 103:253–268. doi: 10.1111/mmi.13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore CM, Helmann JD. 2005. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol 8:188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Gaballa A, Helmann JD. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol 180:5815–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 25.Outten CE, Tobin DA, Penner-Hahn JE, O'Halloran TV. 2001. Characterization of the metal receptor sites in Escherichia coli Zur, an ultrasensitive zinc(II) metalloregulatory protein. Biochemistry 40:10417–10423. doi: 10.1021/bi0155448. [DOI] [PubMed] [Google Scholar]
- 26.Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol 28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 27.Akanuma G, Nanamiya H, Natori Y, Nomura N, Kawamura F. 2006. Liberation of zinc-containing L31 (RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J Bacteriol 188:2715–2720. doi: 10.1128/JB.188.7.2715-2720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriel SE, Helmann JD. 2009. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J Bacteriol 191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanamiya H, Kawamura F. 2010. Towards an elucidation of the roles of the ribosome during different growth phases in Bacillus subtilis. Biosci Biotechnol Biochem 74:451–461. doi: 10.1271/bbb.90859. [DOI] [PubMed] [Google Scholar]
- 30.Natori Y, Nanamiya H, Akanuma G, Kosono S, Kudo T, Ochi K, Kawamura F. 2007. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol Microbiol 63:294–307. doi: 10.1111/j.1365-2958.2006.05513.x. [DOI] [PubMed] [Google Scholar]
- 31.Prestel E, Noirot P, Auger S. 2015. Genome-wide identification of Bacillus subtilis Zur-binding sites associated with a Zur box expands its known regulatory network. BMC Microbiol 15:13. doi: 10.1186/s12866-015-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Z, Gabriel SE, Helmann JD. 2011. Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res 39:9130–9138. doi: 10.1093/nar/gkr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Z, Helmann JD. 2013. Metal homeostasis and oxidative stress in Bacillus subtilis, eibc2129 In Scott RA. (ed), Encyclopedia of inorganic and bioinorganic chemistry. John Wiley & Sons, Chichester, UK. [Google Scholar]
- 34.Chandrangsu P, Huang X, Gaballa A, Helmann JD. 2019. Bacillus subtilis FolE is sustained by the ZagA zinc metallochaperone and the alarmone ZTP under conditions of zinc deficiency. Mol Microbiol 112:751. doi: 10.1111/mmi.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baichoo N, Wang T, Ye R, Helmann JD. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol 45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- 36.Kleijn RJ, Buescher JM, Le Chat L, Jules M, Aymerich S, Sauer U. 2010. Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. J Biol Chem 285:1587–1596. doi: 10.1074/jbc.M109.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lerondel G, Doan T, Zamboni N, Sauer U, Aymerich S. 2006. YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis. J Bacteriol 188:4727–4736. doi: 10.1128/JB.00167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauer U, Hatzimanikatis V, Bailey JE, Hochuli M, Szyperski T, Wuthrich K. 1997. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat Biotechnol 15:448–452. doi: 10.1038/nbt0597-448. [DOI] [PubMed] [Google Scholar]
- 39.Schilling O, Frick O, Herzberg C, Ehrenreich A, Heinzle E, Wittmann C, Stulke J. 2007. Transcriptional and metabolic responses of Bacillus subtilis to the availability of organic acids: transcription regulation is important but not sufficient to account for metabolic adaptation. Appl Environ Microbiol 73:499–507. doi: 10.1128/AEM.02084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agledal L, Niere M, Ziegler M. 2010. The phosphate makes a difference: cellular functions of NADP. Redox Rep 15:2–10. doi: 10.1179/174329210X12650506623122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaballa A, Chi BK, Roberts AA, Becher D, Hamilton CJ, Antelmann H, Helmann JD. 2014. Redox regulation in Bacillus subtilis: the bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid Redox Signal 21:357–367. doi: 10.1089/ars.2013.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoper D, Volker U, Hecker M. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J Bacteriol 187:2810–2826. doi: 10.1128/JB.187.8.2810-2826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mascher T, Zimmer SL, Smith TA, Helmann JD. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seydlová G, Halada P, Fišer R, Toman O, Ulrych A, Svobodová J. 2012. DnaK and GroEL chaperones are recruited to the Bacillus subtilis membrane after short‐term ethanol stress. J Appl Microbiol 112:765–774. doi: 10.1111/j.1365-2672.2012.05238.x. [DOI] [PubMed] [Google Scholar]
- 45.Susin MF, Baldini RL, Gueiros-Filho F, Gomes SL. 2006. GroES/GroEL and DnaK/DnaJ have distinct roles in stress responses and during cell cycle progression in Caulobacter crescentus. J Bacteriol 188:8044–8053. doi: 10.1128/JB.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugimoto S, Higashi C, Matsumoto S, Sonomoto K. 2010. Improvement of multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 under conditions of thermal stress by heterologous expression of Escherichia coli dnaK. Appl Environ Microbiol 76:4277–4285. doi: 10.1128/AEM.02878-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. 2013. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haider S, Pal R. 2013. Integrated analysis of transcriptomic and proteomic data. Curr Genomics 14:91–110. doi: 10.2174/1389202911314020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chillappagari S, Miethke M, Trip H, Kuipers OP, Marahiel MA. 2009. Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J Bacteriol 191:2362–2370. doi: 10.1128/JB.01616-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solovieva IM, Entian KD. 2002. Investigation of the yvgW Bacillus subtilis chromosomal gene involved in Cd(2+) ion resistance. FEMS Microbiol Lett 208:105–109. doi: 10.1111/j.1574-6968.2002.tb11068.x. [DOI] [PubMed] [Google Scholar]
- 51.Carballido-López R. 2019. Rod width under control. Nat Microbiol 4:1246. doi: 10.1038/s41564-019-0528-0. [DOI] [PubMed] [Google Scholar]
- 52.Dajkovic A, Tesson B, Chauhan S, Courtin P, Keary R, Flores P, Marliere C, Filipe SR, Chapot-Chartier MP, Carballido-Lopez R. 2017. Hydrolysis of peptidoglycan is modulated by amidation of meso‐diaminopimelic acid and M g2+ in B acillus subtilis. Mol Microbiol 104:972–988. doi: 10.1111/mmi.13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arakha M, Saleem M, Mallick BC, Jha S. 2015. The effects of interfacial potential on antimicrobial propensity of ZnO nanoparticle. Sci Rep 5:9578. doi: 10.1038/srep09578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solopova A, Formosa-Dague C, Courtin P, Furlan S, Veiga P, Péchoux C, Armalyte J, Sadauskas M, Kok J, Hols P, Dufrêne YF, Kuipers OP, Chapot-Chartier M-P, Kulakauskas S. 2016. Regulation of cell wall plasticity by nucleotide metabolism in Lactococcus lactis. J Biol Chem 291:11323–11336. doi: 10.1074/jbc.M116.714303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halder S, Yadav KK, Sarkar R, Mukherjee S, Saha P, Haldar S, Karmakar S, Sen T. 2015. Alteration of Zeta potential and membrane permeability in bacteria: a study with cationic agents. SpringerPlus 4:672. doi: 10.1186/s40064-015-1476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tavares AFN, Teixeira M, Romão CC, Seixas JD, Nobre LS, Saraiva LM. 2011. Reactive oxygen species mediate bactericidal killing elicited by carbon monoxide-releasing molecules. J Biol Chem 286:26708–26717. doi: 10.1074/jbc.M111.255752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L, James LP, Helmann JD. 1993. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J Bacteriol 175:5428–5437. doi: 10.1128/jb.175.17.5428-5437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuangthong M, Herbig AF, Bsat N, Helmann JD. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J Bacteriol 184:3276–3286. doi: 10.1128/jb.184.12.3276-3286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaballa A, Helmann JD. 2002. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol Microbiol 45:997–1005. doi: 10.1046/j.1365-2958.2002.03068.x. [DOI] [PubMed] [Google Scholar]
- 60.Herbig AF, Helmann JD. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol 41:849–859. doi: 10.1046/j.1365-2958.2001.02543.x. [DOI] [PubMed] [Google Scholar]
- 61.Smith KF, Bibb LA, Schmitt MP, Oram DM. 2009. Regulation and activity of a zinc uptake regulator, Zur, in Corynebacterium diphtheriae. J Bacteriol 191:1595–1603. doi: 10.1128/JB.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer E, Sauer U. 2005. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat Genet 37:636–640. doi: 10.1038/ng1555. [DOI] [PubMed] [Google Scholar]
- 63.Luche S, Eymard-Vernain E, Diemer H, Van Dorsselaer A, Rabilloud T, Lelong C. 2016. Zinc oxide induces the stringent response and major reorientations in the central metabolism of Bacillus subtilis. J Proteomics 135:170–180. doi: 10.1016/j.jprot.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 64.Ong CL, Walker MJ, McEwan AG. 2015. Zinc disrupts central carbon metabolism and capsule biosynthesis in Streptococcus pyogenes. Sci Rep 5:10799. doi: 10.1038/srep10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smaldone GT, Revelles O, Gaballa A, Sauer U, Antelmann H, Helmann JD. 2012. A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism. J Bacteriol 194:2594–2605. doi: 10.1128/JB.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilks JC, Kitko RD, Cleeton SH, Lee GE, Ugwu CS, Jones BD, BonDurant SS, Slonczewski JL. 2009. Acid and base stress and transcriptomic responses in Bacillus subtilis. Appl Environ Microbiol 75:981–990. doi: 10.1128/AEM.01652-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Inaoka T, Matsumura Y, Tsuchido T. 1999. SodA and manganese are essential for resistance to oxidative stress in growing and sporulating cells of Bacillus subtilis. J Bacteriol 181:1939–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ming LJ. 2003. Structure and function of “metalloantibiotics”. Med Res Rev 23:697–762. doi: 10.1002/med.10052. [DOI] [PubMed] [Google Scholar]
- 70.Zarkan A, Macklyne HR, Chirgadze DY, Bond AD, Hesketh AR, Hong HJ. 2017. Zn(II) mediates vancomycin polymerization and potentiates its antibiotic activity against resistant bacteria. Sci Rep 7:4893. doi: 10.1038/s41598-017-04868-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Auger S, Henry C, Pechoux C, Suman S, Lejal N, Bertho N, Larcher T, Stankic S, Vidic J. 2018. Exploring multiple effects of Zn0.15Mg0.85O nanoparticles on Bacillus subtilis and macrophages. Sci Rep 8:12276. doi: 10.1038/s41598-018-30719-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harwood CR, Cutting SM (ed). 1990. Molecular biological methods for Bacillus. Wiley, Chichester, United Kingdom. [Google Scholar]
- 73.Kunst F, Rapoport G. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol 177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He X, Liu B. 2017. Oolong tea extract as a substitute for uranyl acetate in staining of ultrathin sections based on examples of animal tissues for transmission electron microscopy. J Microsc 267:27–33. doi: 10.1111/jmi.12544. [DOI] [PubMed] [Google Scholar]
- 75.Randazzo P, Aubert-Frambourg A, Guillot A, Auger S. 2016. The MarR-like protein PchR (YvmB) regulates expression of genes involved in pulcherriminic acid biosynthesis and in the initiation of sporulation in Bacillus subtilis. BMC Microbiol 16:190. doi: 10.1186/s12866-016-0807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RAT, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 77.Michna RH, Commichau FM, Tödter D, Zschiedrich CP, Stülke J. 2014. Subti wiki—a database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res 42:D692–D698. doi: 10.1093/nar/gkt1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.