Filamentous cyanobacteria are morphologically complex, with several representative species amenable to routine genetic manipulation, making them excellent model organisms for the study of development. Furthermore, two of the developmental alternatives, nitrogen-fixing heterocysts and motile hormogonia, are essential to establish nitrogen-fixing symbioses with plant partners. These symbioses are integral to global nitrogen cycles and could be artificially recreated with crop plants to serve as biofertilizers, but to achieve this goal, detailed understanding and manipulation of the hormogonium and heterocyst gene regulatory networks may be necessary. Here, using the model organism Nostoc punctiforme, we identify a previously uncharacterized hybrid histidine kinase that is confined to heterocyst-forming cyanobacteria as the earliest known participant in hormogonium development.
KEYWORDS: gliding motility, development, Nostoc punctiforme, cell motility, cyanobacteria, developmental biology, histidine kinase, hormogonia, two-component regulatory systems
ABSTRACT
Filamentous, heterocyst-forming cyanobacteria belonging to taxonomic subsections IV and V are developmentally complex multicellular organisms capable of differentiating an array of cell and filament types, including motile hormogonia. Hormogonia exhibit gliding motility that facilitates dispersal, phototaxis, and the establishment of nitrogen-fixing symbioses. The gene regulatory network (GRN) governing hormogonium development involves a hierarchical sigma factor cascade, but the factors governing the activation of this cascade are currently undefined. Here, using a forward genetic approach, we identified hrmK, a gene encoding a putative hybrid histidine kinase that functions upstream of the sigma factor cascade. The deletion of hrmK produced nonmotile filaments that failed to display hormogonium morphology or accumulate hormogonium-specific proteins or polysaccharide. Targeted transcriptional analyses using reverse transcription-quantitative PCR (RT-qPCR) demonstrated that hormogonium-specific genes both within and outside the sigma factor cascade are drastically downregulated in the absence of hrmK and that hrmK may be subject to indirect, positive autoregulation via sigJ and sigC. Orthologs of HrmK are ubiquitous among, and exclusive to, heterocyst-forming cyanobacteria. Collectively, these results indicate that hrmK functions upstream of the sigma factor cascade to initiate hormogonium development, likely by modulating the phosphorylation state of an unknown protein that may serve as the master regulator of hormogonium development in heterocyst-forming cyanobacteria.
IMPORTANCE Filamentous cyanobacteria are morphologically complex, with several representative species amenable to routine genetic manipulation, making them excellent model organisms for the study of development. Furthermore, two of the developmental alternatives, nitrogen-fixing heterocysts and motile hormogonia, are essential to establish nitrogen-fixing symbioses with plant partners. These symbioses are integral to global nitrogen cycles and could be artificially recreated with crop plants to serve as biofertilizers, but to achieve this goal, detailed understanding and manipulation of the hormogonium and heterocyst gene regulatory networks may be necessary. Here, using the model organism Nostoc punctiforme, we identify a previously uncharacterized hybrid histidine kinase that is confined to heterocyst-forming cyanobacteria as the earliest known participant in hormogonium development.
INTRODUCTION
Filamentous cyanobacteria belonging to taxonomic subsections IV and V are developmentally complex multicellular organisms (1). The vegetative filaments of these bacteria are composed of cells that actively grow and divide while performing oxygenic photosynthesis and can differentiate alternative cell or filament types. Under conditions where reduced nitrogen is limiting, approximately every 10th cell along the filament will differentiate into a nitrogen-fixing heterocyst (2). In response to other environmental factors, such as light or phosphate limitation, filaments can differentiate akinetes, metabolically dormant cells analogous to bacterial endospores, which are resistant to cold and desiccation (3). Last, filaments are capable of developing into hormogonia, motile filaments that facilitate the dispersal, phototaxis, and establishment of nitrogen-fixing symbioses (1, 4–6). Of these three developmental alternatives, heterocyst differentiation is by far the most well understood at the molecular level, having been the focus of intense study for several decades (2). In contrast, the genetic control of akinete and hormogonium development is much less defined. However, in recent years, considerable advances have been made by applying molecular approaches to define the hormogonium gene regulatory network (GRN) in the model filamentous cyanobacterium Nostoc punctiforme (7).
Initiation of hormogonium development is influenced by a complex assortment of signals that include light quality and quantity, nutrient concentrations, autogenic repressors, and signals from symbiotic partners (8–14). Once initiated, differentiating hormogonia undergo a round of synchronous cell division accompanied by architectural remodeling and filament fragmentation at heterocyst-vegetative cell junctions and necridia (apoptotic cells) to produce the distinctive hormogonium morphology, short filaments of smaller, rod-shaped cells that are devoid of heterocysts and which frequently display tapered filament termini (1). Once differentiated, subsequent cell growth and division are arrested (8), but filaments maintain metabolic activity to power gliding motility. Motility is driven by bipolar, circumferential arrays of type IV pilus motors (15) and is accompanied by the deposition of a hormogonium-specific polysaccharide (HPS) essential for movement (16).
Recent evidence indicates that the GRN promoting hormogonium development involves the activation of a hierarchical sigma factor cascade (7). At the top of this cascade is sigJ, which in turn either directly or indirectly activates the transcription of sigC and sigF, as well as a majority of the genes involved in architectural remodeling, the type 4 pilus (T4P) motor, and hormogonium-specific signal transduction systems. In turn, sigC promotes the expression of many genes associated with reductive cell division, while sigF has a remarkably limited regulon that consists primarily of pilA, which encodes the major pilin of the T4P system. Two signal transduction systems, the Hmp chemotaxis (16, 17) and Hmp partner-switching systems (18), also influence hormogonium development but appear to act downstream of the sigma factor cascade, as their transcription is influenced by sigJ and sigC, and they are dispensable for the development of morphologically distinct hormogonia. Notably, a substantial subset of hormogonium-specific genes, including several required for HPS synthesis, are still upregulated upon hormogonium induction in the absence of sigJ, implying that unidentified components of the hormogonium GRN, including a potential hormogonium master regulator, act upstream of sigJ (7).
One likely candidate to serve as the upstream activator is a two-component or phosphorelay system, where a sensor histidine kinase, upon perception of a signal, either directly (two-component), or indirectly through a histidine phosphotransferase protein (Hpt) intermediary (phosphorelay), phosphorylates a response regulator to elicit an appropriate cellular response (19). Response regulators control a variety of functions, including, most commonly, modulation of transcription. Such systems have been shown to serve as master regulators in other bacterial developmental programs, including fruiting body development in Myxococcus xanthus (20) and sporulation in Bacillus subtilis (21).
In this study, we apply a forward genetic approach to identify HrmK, a putative hybrid histidine kinase required for hormogonium development in N. punctiforme. Phenotypic characterization and targeted transcriptional analyses of the hrmK deletion strain indicate that hrmK acts upstream of the sigma factor cascade in the hormogonium GRN to initiate the developmental program.
RESULTS
Identification of hrmK, encoding a hybrid histidine kinase regulating hormogonium development.
During an ongoing study using transposon mutagenesis to identify genes involved in hormogonium development and motility (22), three nonmotile mutants with unique transposon insertions in a gene here designated hrmK (hormogonium hybrid histidine kinase, open reading frame Npun_R3825) were identified (Fig. 1A). The gene encodes a putative hybrid histidine kinase (HHK), with the C-terminal portion containing the phosphoacceptor (pfam00512) and HATPase (pfam02518) domains of a typical histidine kinase, followed by a response regulator receiver domain (REC, pfam00072). The N-terminal portion of HrmK lacks homology to any previously characterized sensory domains (NCBI BLASTP), and based on the absence of predicted transmembrane domains (23), it is unlikely that HrmK is a transmembrane sensor. Genomic context (24) and previously published RNA sequencing (RNA-seq) data (7) indicate that hrmK is monocistronic and encodes an orphan HHK with no potential cognate Hpt or response regulator proteins encoded at the same genetic locus.
FIG 1.
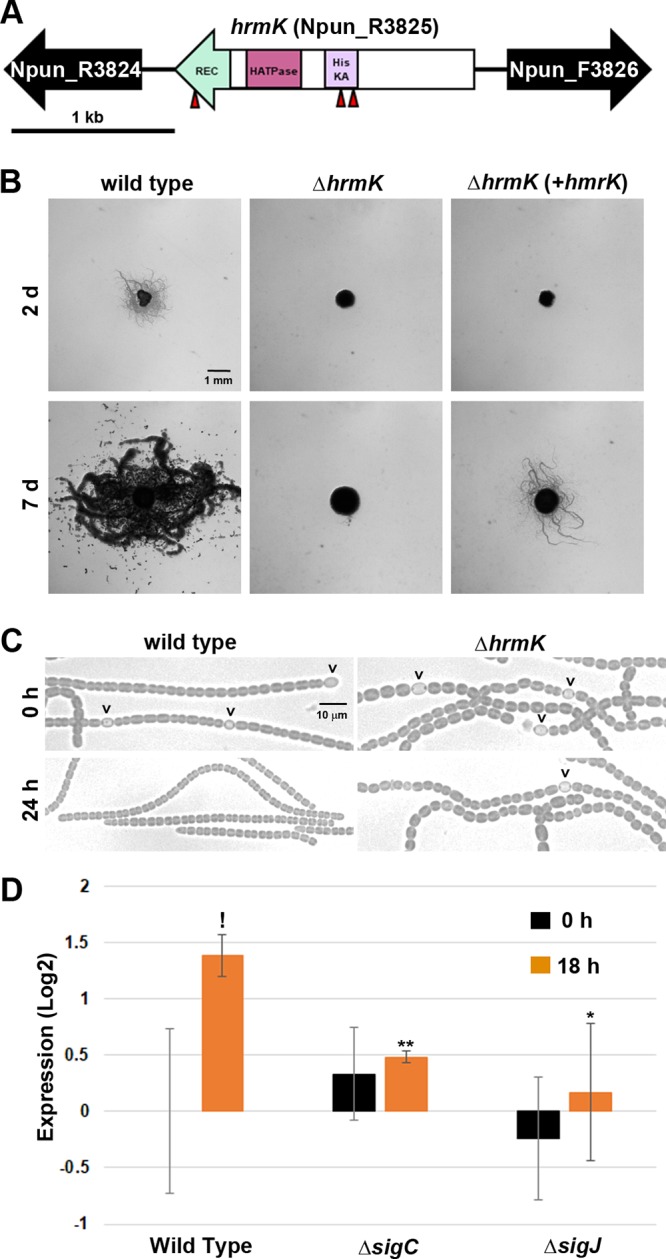
Characterization of hrmK. (A) Gene map of the hrmK locus. Triangles indicate the sites of transposon insertions. HisKA, His kinase A (phosphoacceptor) domain; HATPase, ATPase domains of histidine kinase; REC, response regulator receiver domain. (B) Plate motility assays of the wild-type strain, ΔhrmK mutant, and ΔhrmK mutant with hrmK expressed in trans from a shuttle vector (+hrmK) (as indicated). Images were taken at 2 days and 7 days postinduction. (C) Light micrographs of the filament morphology for the wild-type and ΔhrmK mutant strains (as indicated) at 0 h and 24 h post-hormogonium induction (as indicated). Carets indicate the presence of heterocysts attached to filaments. (D) RT-qPCR of hrmK in the wild-type, ΔsigJ mutant, and ΔsigC mutant strains 0 and 18 h after induction for hormogonia. *, P < 0.05; **, P < 0.01, as determined by two-tailed Student’s t test between the wild type and each deletion strain at the corresponding time point. !, P < 0.05; as determined by two-tailed Student’s t test between 0 and 18 h for the same strain.
To confirm that hrmK is required for hormogonium development and/or motility, a strain with an in-frame deletion of hrmK was constructed and characterized. The deletion of hrmK completely abolished motility, as evidenced by the lack of colony spreading in plate motility assays and the absence of motility for individual filaments observed by time-lapse microscopy (Fig. 1B; see also Movie S1 in the supplemental material). The loss of motility in the ΔhrmK mutant strain could be complemented by the reintroduction of hrmK, along with its 5′ intergenic region, in trans on a replicative shuttle vector, although there was a substantial delay in the appearance of colony spreading (Fig. 1B).
We routinely employ the supplementation and subsequent removal of 4 mM sucralose from the growth medium to synchronously induce hormogonium development (25). However, higher concentrations of sucralose lead to the formation of aseriate colonies, which become encapsulated in a thick polysaccharide sheath (25). It was noted that the ΔhrmK mutant strain was more sensitive to sucralose, forming aseriate colonies in liquid cultures at 4 mM sucralose instead of 6 mM sucralose for the wild type (Fig. S1). We therefore employed supplementation and removal of 2 mM sucralose for induction of hormogonia in the ΔhrmK mutant strain to avoid complications that might arise from attempting to induce hormogonia from the aseriate colonies, where washing of the filaments to remove sucralose could be impeded by the presence of the thick polysaccharide sheath.
In liquid cultures prior to induction, filaments of the ΔhrmK mutant strain contained cells that were larger and more spherical than those of the wild type (Fig. 1C and S2). Following induction, the ΔhrmK mutant strain failed to show any of the characteristics of wild-type hormogonium morphology, which include a reduction in cell size, loss of heterocysts, and the appearance of tapered filament termini (Fig. 1C and S2). This morphological analysis could indicate that hrmK functions at an early stage in hormogonium differentiation, given that sigJ and sigC are currently the only other genes known to be required for the development of morphologically distinct hormogonia (7).
Previous transcriptomic analyses have indicated that the expression of hrmK is moderately enhanced in developing hormogonia (7) and that this induction is diminished in the absence of sigJ or sigC. To confirm this transcriptional profile, the hrmK transcript was quantified by reverse transcription-quantitative PCR (RT-qPCR) at 0 and 18 h postinduction in the wild-type, ΔsigJ mutant, and ΔsigC mutant strains. The results confirmed hormogonium-specific induction of hrmK that is both sigJ and sigC dependent (Fig. 1D).
hrmK is required for the accumulation of hormogonium-specific proteins and polysaccharide.
The absence of obvious hormogonium morphology does not necessarily preclude the activation of a substantial portion of the hormogonium GRN. For example, both sigC and sigJ are required for the development of morphologically distinct hormogonia, but only the deletion of sigJ completely prevents hormogonium-specific production of the major pilin PilA or the methyl-accepting chemotaxis-like protein HmpD (7). To further probe which portions of the hormogonium GRN may be disrupted in the ΔhrmK mutant strain, immunological and lectin-based assays were employed to measure the accumulation of PilA, HmpD, and HPS. The results from immunoblot analysis demonstrated that unlike the wild-type strain, the ΔhrmK mutant strain failed to accumulate detectable levels of either PilA or HmpD following hormogonium induction (Fig. 2A). When staining for accumulated extracellular HPS, heterocyst-specific staining with Ulex europaeus agglutinin (UEA)-fluorescein was detected in both the wild-type and ΔhrmK mutant strains prior to hormogonium induction, due to cross-reaction of UEA-fluorescein with heterocyst envelope polysaccharide, as previously reported (Fig. 2B) (26). However, upon hormogonium induction, the wild-type strain accumulated large amounts of HPS loosely associated with the filaments, while no obvious increase was observed for the ΔhrmK mutant strain (Fig. 2B).
FIG 2.
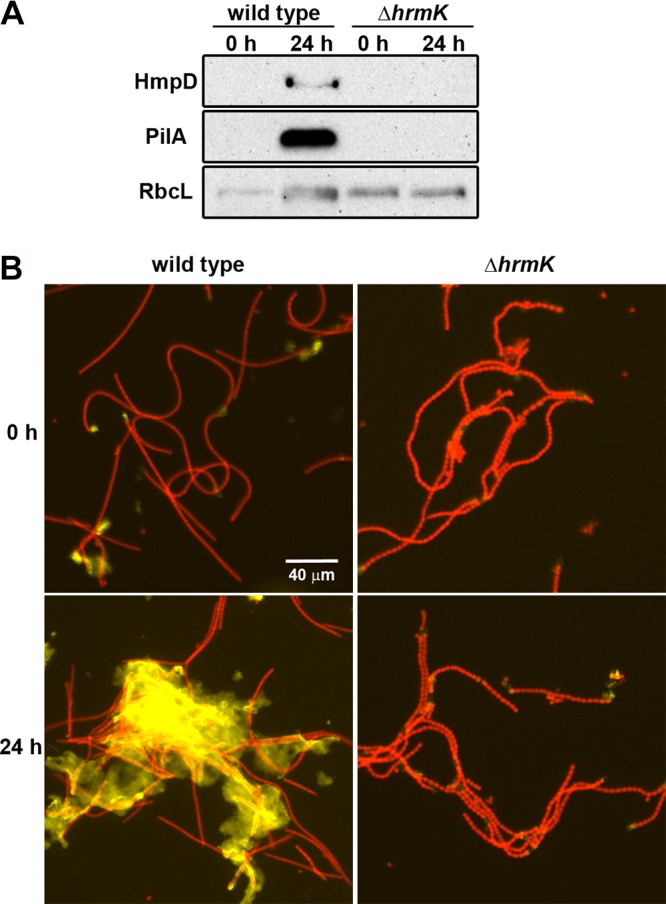
Effect of hrmK on accumulation of HmpD, PilA, and HPS. (A) Immunoblot analysis of HmpD, PilA, and RbcL in the wild-type and ΔhrmK mutant strains (as indicated) 0 and 24 h after hormogonium induction. RbcL is the large subunit of RuBisCO and serves as a protein loading control. (B) Fluorescent lectin staining of HPS in the wild-type and ΔhrmK mutant strains (as indicated). Depicted are merged images of fluorescence micrographs acquired using a 10× lens objective from cellular autofluorescence (red) and HPS (yellow) (strains as indicated) 0 and 24 h after hormogonium induction.
hrmK acts upstream of sigJ in the hormogonium GRN.
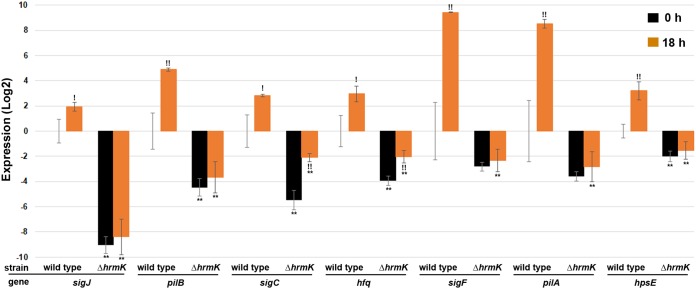
The phenotype of the ΔhrmK mutant strain is essentially indistinguishable from that of a sigJ deletion strain (7), indicating that hrmK is an early-acting factor in the hormogonium GRN, possibly either immediately upstream or downstream of sigJ. To distinguish between these two possibilities and to provide additional supporting evidence to integrate hrmK into a model of the network, RT-qPCR was used to quantify the expression of each of the three hormogonium-specific sigma factors (sigJ, sigC, and sigF), a downstream target for each (pilB, hfq, and pilA, respectively), as well as hpsE, a gene induced in hormogonia independently of the sigma factor cascade (Fig. 3). In the wild-type strain, the expression of each gene was upregulated upon hormogonium induction, as previously reported (7). In the ΔhrmK mutant strain, the expression of both sigJ and sigF and their downstream targets pilB and pilA, respectively, is drastically reduced compared to that of the wild type prior to induction and is no longer enhanced postinduction. The transcription of sigC and its downstream target hfq was also substantially reduced in the ΔhrmK mutant strain compared to that in the wild type prior to induction. A moderate increase in the expression of sigC and hfq was observed in the ΔhrmK mutant strain postinduction, but transcript levels remained well below those observed either before or following induction in the wild-type strain. Finally, hpsE expression was also reduced in the ΔhrmK mutant strain and was not differentially upregulated following induction. These data imply that hrmK acts upstream of sigJ and promotes the expression of other hormogonium-specific genes independent of the sigma factor cascade.
FIG 3.
RT-qPCR of hormogonium-specific gene expression (as indicated) in the wild-type and ΔhrmK mutant strains 0 and 18 h after induction for hormogonia. *, P < 0.05; **, P < 0.01, as determined by two-tailed Student’s t test between the wild-type and ΔhrmK mutant strains at the corresponding time point. !, P < 0.05; !!, P < 0.01, as determined by two-tailed Student’s t test between 0 and 18 h for the same strain.
Evolutionary conservation of hrmK.
Orthologs of genes essential for hormogonium development and motility in N. punctiforme are conserved in a large number of cyanobacteria. In some cases, they are widely distributed among both filamentous and unicellular species, such as those encoding the hormogonium-specific sigma factors (7), the T4P system (26), and the Hmp chemotaxis-like system (26). In others, the genes appear to be confined to filamentous cyanobacteria, as is the case for the Hmp partner-switching system (18) and several genes associated with HPS synthesis (16, 26). In contrast, hrmK is ubiquitous and exclusive to the heterocyst-forming cyanobacteria in taxonomic subsections IV and V (Fig. 4). Of the 24 heterocyst-forming cyanobacteria analyzed, only two fail to encode an ortholog of HrmK. Conversely, only 2 orthologs were detected in the 101 non-heterocyst-forming strains, one in a filamentous cyanobacterium (Leptolyngbya boryana sp. strain PCC 6306), and the other in a unicellular strain closely related to heterocyst formers (Gloeocapsa sp. strain PCC 7428). Homologous proteins identified outside the cyanobacterial lineage (NCBI BLASTP) only share sequence similarity within the C-terminal signaling domain, indicating that HrmK is unlikely to have been acquired via recent horizontal gene transfer. Therefore, it appears that HrmK is an HHK with a unique sensory domain that is confined primarily to heterocyst-forming cyanobacteria.
FIG 4.
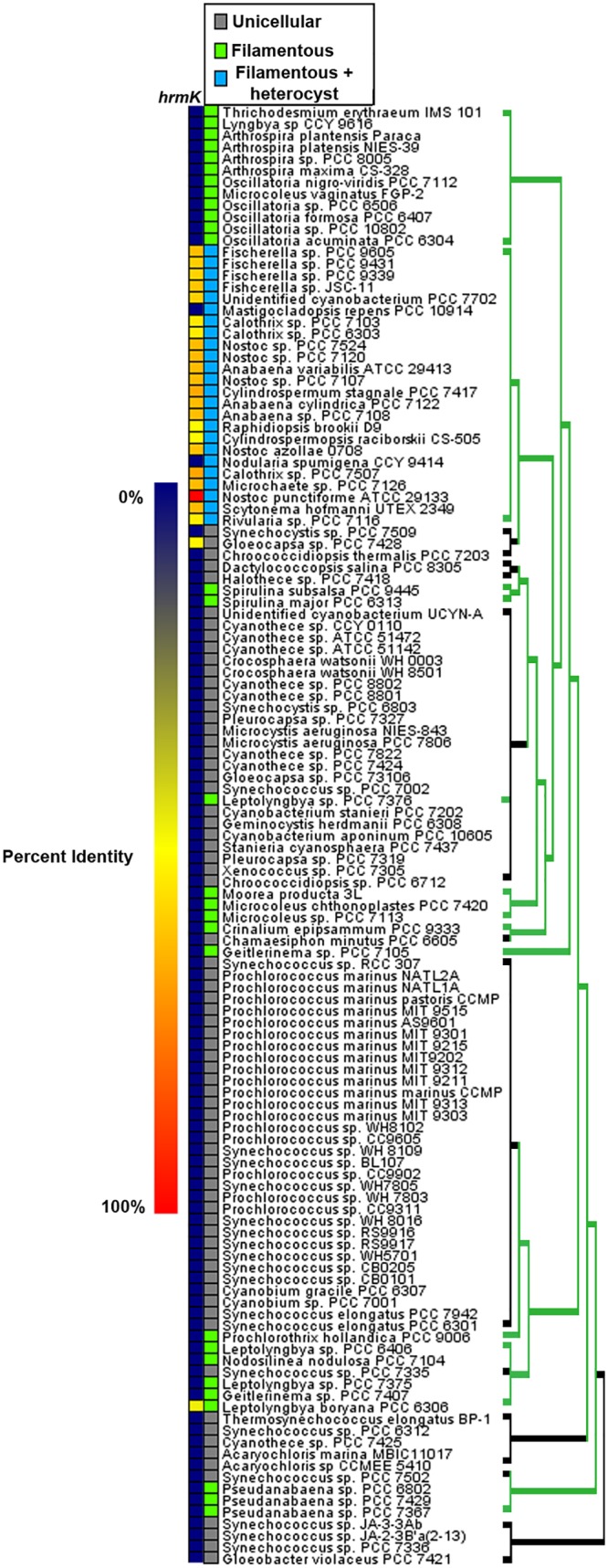
Evolutionary conservation of HrmK in cyanobacteria. Shown is a heat map depicting the percent identity for orthologs of N. punctiforme HrmK in cyanobacteria, derived from data reported by Cho et al. (26). The species organization and phylogenetic tree are based on the phylogeny reported by Shih et al. (36) but depicting the finding, as reported by Schirrmeister et al. (37), that most extant cyanobacteria are derived from a filamentous ancestor. For the phylogenetic tree, green indicates filamentous and black indicates unicellular.
DISCUSSION
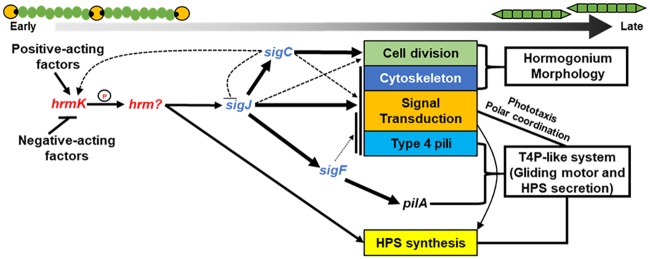
Figure 5 presents a working model of the hormogonium GRN, where hrmK indirectly promotes the transcription of both sigJ (to activate the hormogonium sigma factor cascade) and other hormogonium-specific genes independent of this cascade. This model is supported by the absence of hormogonium morphology, failure to accumulate hormogonium-specific proteins and polysaccharide, and reduced transcription of hormogonium-specific genes in the ΔhrmK mutant strain. Critically, the transcription of hpsE is diminished in the ΔhrmK mutant strain but not in any of the sigma factor deletion strains previously analyzed (7), providing key data indicating that hrmK acts upstream of the sigma factor cascade. Furthermore, the fact that transcription of hrmK is reduced in the absence of both sigJ and sigC indicates the presence of an indirect positive-feedback loop between these three genes, where sigJ promotes expression of sigC, which in turn enhances the transcription of hrmK. This model is also supported by previous experiments showing that enhanced expression of sigJ in hormogonia is sigC dependent (7). Given the presence of this positive-feedback loop, care must be taken in establishing the order of each component in the GRN; thus, while the available evidence is most consistent with hrmK as the earliest acting factor in the network, additional experimentation may be needed to definitively resolve the order.
FIG 5.
A model depicting the major findings of this report on the role of hrmK in the hormogonium GRN. Arrows indicate positive regulation, and lines with bars indicate negative regulation. The thickness of the arrows represents a combination of the number of genes regulated and the stringency of the regulation.
Notably, despite the static expression of sigJ in the absence of sigC, the sigJ-dependent regulon is still rapidly upregulated upon hormogonium induction (7), an observation that could be accounted for by posttranscriptional regulation of sigJ. In contrast to the deletion of sigC, the deletion of hrmK drastically reduced sigJ expression to levels well below that of wild-type vegetative filaments and also abolished the upregulation of sigJ-dependent genes, indicating that hrmK regulates sigJ at the transcriptional level and is essential for its expression. These findings demonstrate that the hormogonium GRN is similar to those that control other developmental processes in bacteria, perhaps most notably sporulation in the Bacillus genus, where a phosphorelay system controls the initiation of a sigma factor cascade (21). It is also notable that the ΔhrmK mutant strain displayed increased sensitivity to sucralose with respect to the formation of aseriate colonies. Currently, little is known about the genetic regulation of this morphology, but it appears to be negatively influenced by the activity of hrmK. Thus, it seems likely that hrmK may function as a switch that simultaneously promotes the hormogonium GRN while suppressing the mutually exclusive aseriate morphology.
Presumably, HrmK is not directly affecting transcription but is modulating the phosphorylation state of a downstream response regulator, possibly a transcriptional activator or repressor, to control its activity. Typically, HHKs participate in phosphorelays where the phosphate is passed from the REC domain of the HHK to a standalone Hpt protein and subsequently to a response regulator (19). However, HrmK is an orphan HHK with no obvious partner Hpt or response regulator proteins encoded at the same genetic locus. This makes identifying the additional components of the putative phosphorelay more challenging. Moreover, the genomes of N. punctiforme and several other cyanobacteria investigated in a previous study are largely devoid of obvious Hpt proteins that would serve as intermediaries between HrmK and its cognate response regulator (27). Perhaps, cyanobacteria possess cryptic Hpt proteins that are not easily identified based on sequence similarity. It is also possible that HrmK participates in a less common type of phosphorelay similar to that which controls type III chromatic acclimation in the cyanobacterium Fremyella diplosiphon (28). In this system, the histidine kinase RcaE passes phosphate to a standalone REC domain protein, RcaF, which in turn phosphorylates an Hpt domain on RcaC, a transcriptional regulator that contains both an Hpt and multiple REC domains (29). Notably, there are several RcaC homologs encoded in the N. punctiforme genome (24). Alternatively, HrmK may directly phosphorylate a response regulator. Identifying the additional components of the HrmK signal transduction system is a critical area for future work.
Another aspect of HrmK that requires further investigation is the role of the N-terminal sensory domain. This domain is found exclusively in heterocyst-forming cyanobacteria and has not been previously characterized, making the prediction of its sensory capacity difficult. Given the absence of any detectable transmembrane domains within HrmK, it is likely to sense an intracellular, rather than extracellular, signal. It is clear, based on its conservation, that HrmK probably plays a critical role in promoting hormogonium development in all heterocyst-forming cyanobacteria. Further studies will be required to answer these outstanding questions.
MATERIALS AND METHODS
Strains and culture conditions.
For a detailed description of the strains used in this study, refer to Table S1 in the supplemental material. N. punctiforme ATCC 29133 and its derivatives were cultured in Allen and Arnon medium diluted 4-fold (AA/4), without supplementation of fixed nitrogen, as previously described (30), with the exception that liquid cultures were supplemented with 4 mM (for the wild-type strain) or 2 mM (for the ΔhrmK mutant strain) sucralose, and solid medium was supplemented with 10 mM sucralose, to inhibit hormogonium formation (25). For small-scale hormogonium induction for phenotypic analysis, the equivalent of 30 μg ml−1 chlorophyll a (Chl a) of cell material from cultures at a Chl a concentration of 10 to 20 μg ml−1 was harvested at 2,000 × g for 3 min, washed two times with AA/4, and resuspended in 2 ml of fresh AA/4 without sucralose. For large-scale hormogonium induction for RNA extraction and subsequent RT-qPCR analysis, this process was repeated but starting with the equivalent of 300 μg ml−1 Chl a of cell material and resuspension in 50 ml of fresh AA/4. For selective growth, the medium was supplemented with 50 μg ml−1 neomycin. Escherichia coli cultures were grown in lysogeny broth (LB) for liquid cultures or LB supplemented with 1.5% (wt/vol) agar for plates. Selective growth medium was supplemented with 50 μg ml−1 kanamycin, 50 μg ml−1 ampicillin, and 15 μg ml−1 chloramphenicol.
Plasmid and strain construction.
For a detailed description of the plasmids, strains, and oligonucleotides used in this study, refer to Tables S1 and S2. All constructs were sequenced to ensure fidelity.
To construct a plasmid for in-frame deletion of hrmK, approximately 900 bp of flanking DNA on either side of the gene and several codons at the beginning and end of the gene were amplified via overlap extension PCR (see Tables S1 and S2 for details) and cloned into pRL278 (31) as BamHI-SacI fragments using restriction sites introduced on the primers.
To construct a mobilizable shuttle vector containing hrmK and its respective promoter region, the coding region and 5′ intergenic region were amplified via PCR (see Tables S1 and S2 for details) and subsequently cloned into pAM504 (32) as a BamHI‐SacI fragment using restriction sites introduced on the primers.
The generation of transposon mutants and identification of transposon insertion sites were performed as previously described (22) using plasmid pRL1063a (33). Gene deletions were performed as previously described (16) with N. punctiforme cultures supplemented with 4 mM sucralose to inhibit hormogonium development and enhance conjugation efficiency (22, 25). To construct the ΔhrmK mutant strain, pDDR420 was introduced into wild-type N. punctiforme, creating strain UOP139.
Motility assays.
Both plate and time-lapse motility assays were performed as previously described (15).
RT-qPCR.
Total RNA was extracted from the equivalent of 300 μg ml−1 Chl a of cell material for each of 3 biological replicates from each strain at 0 and 18 h following hormogonium induction, using previously published methods (30). Five hundred nanograms of total RNA was used to synthesize cDNA with the ProtoScript first-strand cDNA synthesis kit and random hexamer primers (New England BioLabs, Inc.), following the specifications of the manufacturer, after which 2 μl of cDNA was used as the template for quantitative PCR (qPCR). Transcripts were amplified with the primer sets indicated in Table S2, using a StepOnePlus real-time PCR system (Applied Biosystems) and SensiFAST SYBR No-ROX kit (Bioline), following the manufacturer’s specifications. Quantification of transcript abundance was calculated from the average of two technical replicates from each of the three biological replicates using the 2−ΔΔCT method (34), with expression normalized relative to rnpB. The primer efficiencies for each primer pair were all greater than 90%.
Immunological and lectin-based analyses.
Preparation of cell material, protein extraction, and detection of PilA, RbcL, and HmpD by immunoblot analysis were performed as previously described (26). Detection of HPS by fluorescent lectin staining was performed as previously described (26).
Microscopy.
Light microscopy of filament morphology was performed using a Leica DM E light microscope with a 40× lens objective and equipped with a Leica DFC290 digital camera controlled by micromanager imaging software (35). Quantification of cell length and the percentage of filaments with attached heterocysts were determined as previously described (22).
Fluorescence microscopy was performed with an Evos FL fluorescence microscope (Life Technologies) equipped with a 10× lens objective. Excitation and emission were as follows: EVOS light cube, green fluorescent protein (GFP; AMEP4651), excitation at 470 ± 22 nm and emission at 525 ± 50 nm) for UEA-fluorescein labeled HPS, and Evos light cube, red fluorescent protein (RFP; AMEP4652), excitation at 531 ± 40 nm and emission at 593 ± 40 nm for cellular autofluorescence.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carrie Kozina and the Microbiology (BIOL 145) laboratory course teaching assistants and students for purification of chromosomal DNA from nonmotile transposon mutants of N. punctiforme.
This work was supported by NSF award number 1753690 to D.D.R.
D.D.R. and E.G.Z. designed the experiments, D.D.R., E.G.Z., N.M.F., A.G., and A.P.P. performed the experiments, and D.D.R. and E.G.Z. prepared the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Meeks JC, Campbell EL, Summers ML, Wong FC. 2002. Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch Microbiol 178:395–403. doi: 10.1007/s00203-002-0476-5. [DOI] [PubMed] [Google Scholar]
- 2.Kumar K, Mella-Herrera RA, Golden JW. 2010. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol 2:a000315. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez R, Forchhammer K, Salerno G, Maldener I. 2016. Clear differences in metabolic and morphological adaptations of akinetes of two Nostocales living in different habitats. Microbiology 162:214–223. doi: 10.1099/mic.0.000230. [DOI] [PubMed] [Google Scholar]
- 4.Meeks JC. 2006. Molecular mechanisms in the nitrogen-fixing Nostoc-bryophyte symbiosis. Prog Mol Subcell Biol 41:165–196. doi: 10.1007/3-540-28221-1_9. [DOI] [PubMed] [Google Scholar]
- 5.Wong FC, Meeks JC. 2002. Establishment of a functional symbiosis between the cyanobacterium Nostoc punctiforme and the bryophyte Anthoceros punctatus requires genes involved in nitrogen control and initiation of heterocyst differentiation. Microbiology 148:315–323. doi: 10.1099/00221287-148-1-315. [DOI] [PubMed] [Google Scholar]
- 6.Kluge M, Mollenhauer D, Wolf E. 2003. The Nostoc-Geosiphon endocytobiosis, p 1–30. In Rai AR, Bergman B, Rasmussen U (ed), Cyanobacteria in symbiosis. Springer, Dordrecht, the Netherlands. [Google Scholar]
- 7.Gonzalez A, Riley KW, Harwood TV, Zuniga EG, Risser DD. 2019. A tripartite, hierarchical sigma factor cascade promotes hormogonium development in the filamentous cyanobacterium Nostoc punctiforme. mSphere 4:e00231-19. doi: 10.1128/mSphere.0023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell EL, Meeks JC. 1989. Characteristics of hormogonia formation by symbiotic Nostoc spp. in response to the presence of Anthoceros punctatus or its extracellular products. Appl Environ Microbiol 55:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell EL, Wong FC, Meeks JC. 2003. DNA binding properties of the HrmR protein of Nostoc punctiforme responsible for transcriptional regulation of genes involved in the differentiation of hormogonia. Mol Microbiol 47:573–582. doi: 10.1046/j.1365-2958.2003.03320.x. [DOI] [PubMed] [Google Scholar]
- 10.Cohen MF, Meeks JC. 1997. A hormogonium regulating locus, hrmUA, of the cyanobacterium Nostoc punctiforme strain ATCC 29133 and its response to an extract of a symbiotic plant partner Anthoceros punctatus. Mol Plant Microbe Interact 10:280–289. doi: 10.1094/MPMI.1997.10.2.280. [DOI] [PubMed] [Google Scholar]
- 11.Khamar HJ, Breathwaite EK, Prasse CE, Fraley ER, Secor CR, Chibane FL, Elhai J, Chiu WL. 2010. Multiple roles of soluble sugars in the establishment of Gunnera-Nostoc endosymbiosis. Plant Physiol 154:1381–1389. doi: 10.1104/pp.110.162529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liaimer A, Helfrich EJN, Hinrichs K, Guljamow A, Ishida K, Hertweck C, Dittmann E. 2015. Nostopeptolide plays a governing role during cellular differentiation of the symbiotic cyanobacterium Nostoc punctiforme. Proc Natl Acad Sci U S A 112:1862–1867. doi: 10.1073/pnas.1419543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meeks JC, Elhai J. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol Mol Biol Rev 66:9–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashidoko Y, Nishizuka H, Tanaka M, Murata K, Murai Y, Hashimoto M. 2019. Isolation and characterization of 1-palmitoyl-2-linoleoyl-sn-glycerol as a hormogonium-inducing factor (HIF) from the coralloid roots of Cycas revoluta (Cycadaceae). Sci Rep 9:475–478. doi: 10.1038/s41598-019-39647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khayatan B, Meeks JC, Risser DD. 2015. Evidence that a modified type IV pilus-like system powers gliding motility and polysaccharide secretion in filamentous cyanobacteria. Mol Microbiol 98:1021–1036. doi: 10.1111/mmi.13205. [DOI] [PubMed] [Google Scholar]
- 16.Risser DD, Meeks JC. 2013. Comparative transcriptomics with a motility-deficient mutant leads to identification of a novel polysaccharide secretion system in Nostoc punctiforme. Mol Microbiol 87:884–893. doi: 10.1111/mmi.12138. [DOI] [PubMed] [Google Scholar]
- 17.Risser DD, Chew WG, Meeks JC. 2014. Genetic characterization of the hmp locus, a chemotaxis-like gene cluster that regulates hormogonium development and motility in Nostoc punctiforme. Mol Microbiol 92:222–233. doi: 10.1111/mmi.12552. [DOI] [PubMed] [Google Scholar]
- 18.Riley KW, Gonzalez A, Risser DD. 2018. A partner-switching regulatory system controls hormogonium development in the filamentous cyanobacterium Nostoc punctiforme. Mol Microbiol 109:555–569. doi: 10.1111/mmi.14061. [DOI] [PubMed] [Google Scholar]
- 19.Jung K, Fried L, Behr S, Heermann R. 2012. Histidine kinases and response regulators in networks. Curr Opin Microbiol 15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Sun H, Shi W. 2001. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J Bacteriol 183:4786–4795. doi: 10.1128/JB.183.16.4786-4795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoch JA. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol 47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 22.Khayatan B, Bains DK, Cheng MH, Cho YW, Huynh J, Kim R, Omoruyi OH, Pantoja AP, Park JS, Peng JK, Splitt SD, Tian MY, Risser DD. 2017. A putative O-linked β-N-acetylglucosamine transferase is essential for hormogonium development and motility in the filamentous cyanobacterium Nostoc punctiforme. J Bacteriol 199:e00075-17. doi: 10.1128/JB.00075-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Möller S, Croning MD, Apweiler R. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 24.Meeks JC, Elhai J, Thiel T, Potts M, Larimer F, Lamerdin J, Predki P, Atlas R. 2001. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosynth Res 70:85–106. doi: 10.1023/A:1013840025518. [DOI] [PubMed] [Google Scholar]
- 25.Splitt S, Risser D. 2015. The non-metabolizable sucrose analog sucralose is a potent inhibitor of hormogonium differentiation in the filamentous cyanobacterium Nostoc punctiforme. Arch Microbiol doi: 10.1007/s00203-015-1171-7. [DOI] [PubMed] [Google Scholar]
- 26.Cho YW, Gonzales A, Harwood TV, Huynh J, Hwang Y, Park JS, Trieu AQ, Italia P, Pallipuram VK, Risser DD. 2017. Dynamic localization of HmpF regulates type IV pilus activity and directional motility in the filamentous cyanobacterium Nostoc punctiforme. Mol Microbiol 106:252–265. doi: 10.1111/mmi.13761. [DOI] [PubMed] [Google Scholar]
- 27.Ashby MK, Houmard J. 2006. Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution. Microbiol Mol Biol Rev 70:472–509. doi: 10.1128/MMBR.00046-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery BL. 2017. Seeing new light: recent insights into the occurrence and regulation of chromatic acclimation in cyanobacteria. Curr Opin Plant Biol 37:18–23. doi: 10.1016/j.pbi.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Kehoe DM, Grossman AR. 1997. New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J Bacteriol 179:3914–3921. doi: 10.1128/jb.179.12.3914-3921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell EL, Summers ML, Christman H, Martin ME, Meeks JC. 2007. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J Bacteriol 189:5247–5256. doi: 10.1128/JB.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai YP, Wolk CP. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol 172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei TF, Ramasubramanian TS, Golden JW. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol 176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolk CP, Cai Y, Panoff JM. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci U S A 88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. 2014. Advanced methods of microscope control using μManager software. J Biol Methods 1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, Calteau A, Cai F, Tandeau de Marsac N, Rippka R, Herdman M, Sivonen K, Coursin T, Laurent T, Goodwin L, Nolan M, Davenport KW, Han CS, Rubin EM, Eisen JA, Woyke T, Gugger M, Kerfeld CA, 2013. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci U S A 110:1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schirrmeister BE, de Vos JM, Antonelli A, Bagheri HC. 2013. Evolution of multicellularity coincided with increased diversification of cyanobacteria and the Great Oxidation Event. Proc Natl Acad Sci U S A 110:1791–1796. doi: 10.1073/pnas.1209927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.