Abstract
Optogenetic tools for controlling gene expression are ideal for tuning synthetic biological networks due to the exquisite spatiotemporal control available with light. Here we develop an optogenetic system for gene expression control integrated with an existing yeast toolkit allowing for rapid, modular assembly of light-controlled circuits in the important chassis organism Saccharomyces cerevisiae. We reconstitute activity of a split synthetic zinc-finger transcription factor (TF) using light-induced dimerization mediated by the proteins CRY2 and CIB1. We optimize function of this split TF and demonstrate the utility of the toolkit workflow by assembling cassettes expressing the TF activation domain and DNA-binding domain at different levels. Utilizing this TF and a synthetic promoter we demonstrate that light-intensity and duty-cycle can be used to modulate gene expression over the range currently available from natural yeast promoters. This work allows for rapid generation and prototyping of optogenetic circuits to control gene expression in Saccharomyces cerevisiae.
Keywords: optogenetics, Saccharomyces cerevisiae, yeast, light-inducible promoter, MoClo
The budding yeast Saccharomyces cerevisiae is an important chassis organism for synthetic biology and metabolic engineering (Buchholz & Collins, 2013). These disciplines integrate biological parts (e.g. coding sequences, promoters) into biological circuits with novel cellular function. This process has become more routine in Saccharomyces cerevisiae due to the creation of large libraries of well-characterized parts (Chen, et al., 2018). However, the inner workings of the cell continue to make the function and performance of engineered biological circuits unpredictable. Engineering efforts benefit greatly from the ability to tune the concentration of individual components to test and adjust circuit function. Additionally, tunability can allow circuits to be temporally and spatially adjusted to match dynamic constraints, such as the external environment or bioprocess phase (Scott, Sweeney, & McClean, 2019).
Optogenetic approaches offer a potential solution for flexible tuning. Optogenetics take advantage of light-sensitive genetically encoded proteins to actuate processes inside of the cell in a light-dependent manner. Light is a powerful actuator as it is inexpensive, easily controlled in time and space, and S. cerevisiae contains no known native photoreceptors (Salinas, Rojas, Delgado, Agosin, & Larrondo, 2017). The ability to rapidly add and remove light from cell culture or spatially target specific cells makes it particularly advantageous for applications that require spatiotemporal precision such as dynamic stimulation or real-time feedback control of cellular processes (Milias-Argeitis, Rullan, Aoki, Buchmann, & Khammash, 2016; Toettcher, Gong, Lim, & Weiner, 2011; Benzinger & Khammash, 2018; Harrigan, Madani, & El-Samad, 2018; Ng, et al., 2019; Rullan, Benzinger, Schmidt, Milias-Argeitis, & Khammash, 2019; Lugagne & Dunlop, 2019; Castillo-Hair, Igoshin, & Tabor, 2015). To continue expanding the utility of optogenetics in S. cerevisiae, here we report the construction and optimization of a light-activated transcription factor and associated components for use with an existing toolkit of yeast parts (Lee, Deloache, Cervantes, & Dueber, 2015). Addition of these optogenetic components to the toolkit allows for rapid and modular assembly of light-controlled circuits to tune gene expression dynamically and over the range defined by native yeast constitutive promoters.
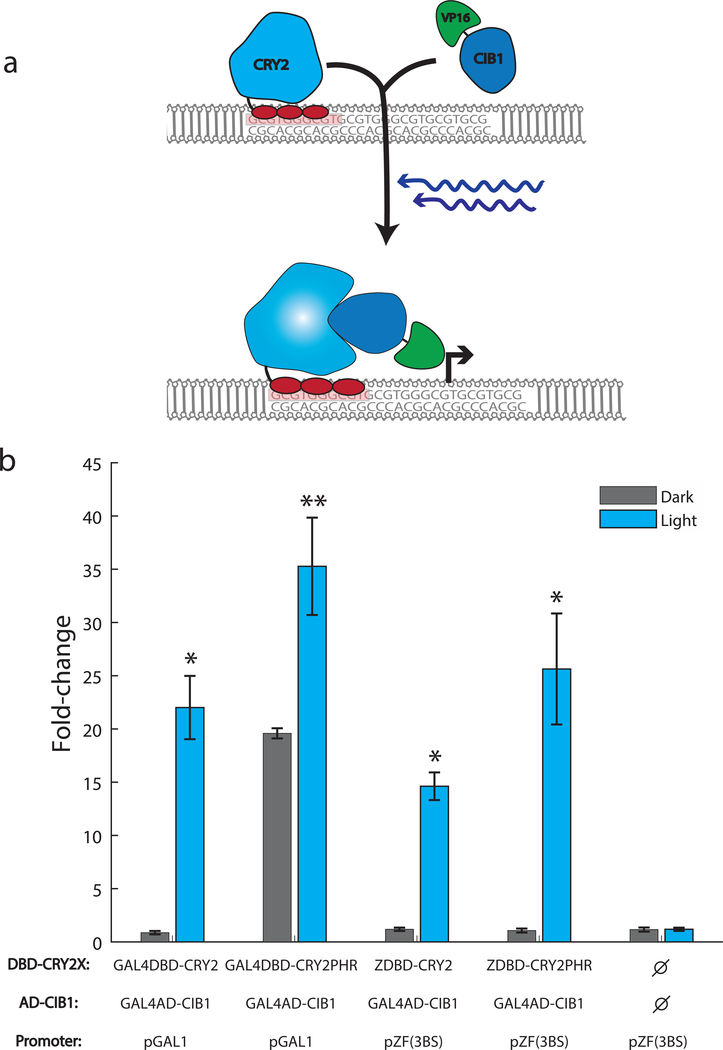
We took advantage of the naturally occurring Arabidopsis cryptochrome CRY2 and binding partner CIB1 to reconstitute the activity of a split transcription factor in a blue light-dependent manner. The feasibility of this approach was previously demonstrated by fusing CRY2 to the scGAL4 DNA-binding domain and CIB1 to the scGAL4 activation domain (Kennedy, et al., 2010). The GAL4 protein is a native S. cerevisiae transcription factor and using the GAL4 DNA-binding domain (DBD) to create synthetic transcription factors leads to crosstalk with native GAL4-inducible promoters (McIsaac, Oakes, Wang, Botstein, & Noyes, 2013). To avoid this crosstalk orthogonal DNA-binding domains from natural sources, such as those from bacterial TFs, could be used to replace the GAL4DBD as has been successfully demonstrated using the LexA DNA-binding domain (Hughes, Bolger, Tapadia, & Tucker, 2012). We chose to replace the GAL4DBD with a Cys2-His2 zinc finger (ZF) DNA-binding domain due to ZFs modularity and potential for engineered sequence specificity (Khalil, et al., 2012). We used the three-finger DNA-binding domain from the Zif268 mouse transcription factor which specifies a 9-bp sequence that occurs infrequently (<20 instances) in the S. cerevisiae genome. This domain has been shown to be a powerful, orthogonal DNA-binding domain for generating chemically-inducible transcription factors in S. cerevisiae (McIsaac, Oakes, Wang, Botstein, & Noyes, 2013). We fused the Zif268-DBD to the N-terminus of the CRY2 protein (ZDBD-CRY2) and the viral VP16 activation domain to CIB1 (VP16AD-CIB1) (Figure 1A). To create a promoter responsive to our artificial transcription factor, we removed the native GAL4 binding sites and integrated variable numbers and orientations of the Zif268 binding site (5’-GCG TGG GCG-3’) into the scGAL1 promoter (Supplemental Figure 1, 2). Utilizing a promoter with three binding sites for the Zif268 DNA-binding domain (pZF(3BS)) upstream of yEVenus and plasmids containing ZDBD-CRY2 and GAL4AD-CIB1 constructs we showed that the ZDBD-CRY2 based system induced gene expression in response to blue light as well as the original GAL4DBD-CRY2 system (Kennedy, et al., 2010) (Figure 1B). Consistent with previous reports (Kennedy, et al., 2010) the GAL4DBD-CRY2PHR construct containing only the CRY2 photolyase homology region (PHR) showed a stronger light-induced fold change as well as higher background gene expression. This background expression was greatly reduced in the ZDBD-CRY2PHR transcription factor. We verified that the native S. cerevisiae GAL machinery does not exhibit crosstalk with the pZF(3BS) promoter and that ZDBD-CRY2 did not activate expression from the scpGAL1 promoter (Supplemental Figure 3). We arrived at our final ZDBD-CRY2PHR construct and pZF(3BS) promoter by testing different design considerations (i.e. linkers, nuclear localization signals, binding site number) as detailed in the Supplemental Material (Supplemental Figures 2–5).
Figure 1. DBD-CRY2 AD-CIB1 optogenetic system:
(A) Schematic of the ZDBD-CRY2 and VP16AD-CIB1 optogenetic system. In response to blue light, CRY2 undergoes a conformational change that allows CIB1 to bind CRY2. This recruits the activation domain to a promoter containing Zif268 binding sites (GCG-TGG-GCG). (B) Yeast cells were transformed with the following DBD-CRY2 or DBD-CRY2PHR, AD-CIB1, and reporter plasmids: GAL4DBD-CRY2/GAL4AD-CIB1/pGAL1-yEVenus, GAL4DBD-CRY2PHR/GAL4AD-CIB1/pGAL1-yEVenus, ZDBD-CRY2/GAL4AD-CIB1/pZF(3BS)-yEVenus, or ZDBD-CRY2PHR/GAL4AD-CIB1/pZF(3BS)-yEVenus. The control sample contains pZF(3BS)-yEVenus and empty vector controls only. Cultures were grown for 12 hours in 15 μW/cm2 470nm blue-light. No light controls were grown in identical conditions without illumination. Induction at T=12 hours is displayed as fold-change relative to the same sample at T=0 hours. Data is presented as the average ±SEM. Samples indicated with a * (p<0.05) or ** (p<0.0.1) were significantly induced at T=12 hours relative to T=0 hours (Welch’s t-test). ANOVA followed by Tukey’s-HSD indicated that the GAL4DBD split transcription factors do not induce significantly better that the ZDBD transcription factors (F(3,8)=5.4, p=0.0252, Groups: GAL4DBD-CRY2/GAL4AD-CIB1-ab, GAL4DBD-CRY2PHR/GAL4AD-CIB1-b, ZDBD-CRY2/GAL4AD-CIB1-a, and ZDBD-CRY2PHR/GAL4AD-CIB1/pZF(3BS)-ab).
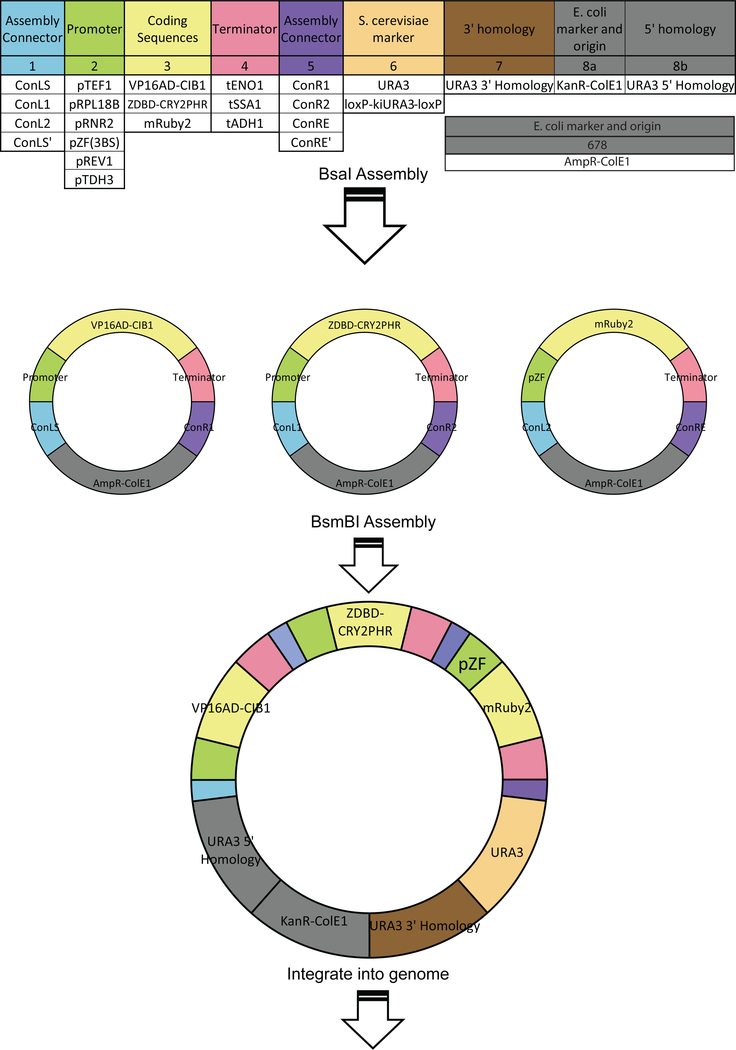
In order to allow for rapid assembly of light controlled circuits, we domesticated our optogenetic components to interface with an existing yeast toolkit (Lee, et al., 2015). This toolkit contains highly characterized yeast components (i.e. promoters, terminators) categorized as “types” based on their function and location in the completed circuit (e.g. promoter types, coding sequence types) (Figure 2). Using a Golden Gate based MoClo (modular cloning) method parts can be rapidly assembled into single or multigene cassettes and integrated into the yeast genome. To domesticate our optogenetic components we removed the restriction enzyme sites (BsaI, BsmBI, NotI) used in the MoClo assembly scheme (Lee, et al., 2015). Our ZDBD-CRY2PHR and VP16AD-CIB1 components became coding sequence (“Type 3”) parts and the pZF(3BS) promoter became a promoter (“Type 2”) part. All parts created in this study that are compatible with the MoClo scheme are shown in Supplemental Figure 6. Using the MoClo scheme with our optogenetic parts and additional parts from the yeast toolkit allows for rapid assembly of multigene integration vectors containing all the necessary components for light-induced expression of a gene of interest.
Figure 2. Circuit construction using the Yeast Toolkit scheme:
Part plasmids contain unique upstream and downstream BsaI-generated overhangs to assemble into the appropriate position in “cassette” plasmids. Cassette plasmids are fully functional transcriptional units that are further assembled into multigene plasmids using BsmBI assembly and appropriate Assembly Connectors. This figure utilizes the color scheme and organization from Lee, et al 2015 to illustrate how the new optogenetic components integrate with an existing yeast toolkit.
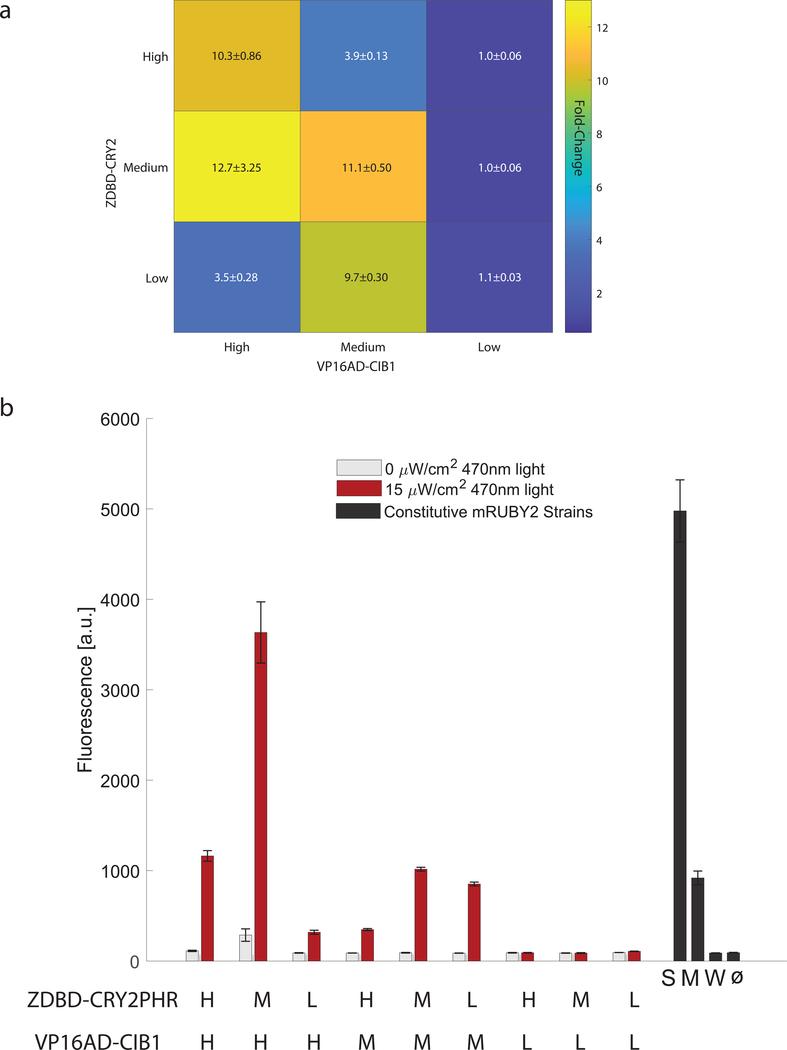
We demonstrated the utility of the toolkit workflow (Figure 2) by using it to optimize our synthetic split TF. We reasoned that the concentration and ratio of the two TF components, the DNA-binding component (ZDBD-CRY2PHR) and the activation component (VP16AD-CIB1), might be important for circuit function. We generated nine different optogenetic constructs with the two components under low (L), medium (M), and high (H) strength yeast promoters. To test gene expression control we took advantage of the red fluorescent protein mRuby2 already in the toolkit, which has an excitation spectrum further from the blue light used to excite CRY2 than yEVenus, and generated pZF(3BS)-mRuby2 reporters. We found that expression of VP16AD-CIB1 under a high strength promoter (pTDH3) and DBD-CRY2PHR under a medium strength promoter (pRPL18B) gave us maximal expression from pZF(3BS) (Figure 3A). However, this ZDBD-CRY2PHRmedium/VP16AD-CIB1high strain exhibited a growth defect. Indeed, all strains highly expressing the VP16AD-CIB1 construct exhibited a growth defect (Supplemental Figure 7). The ZDBD-CRY2PHRmedium/VP16AD-CIB1medium strain on the other hand, with both components under the control of pRPL18B, exhibited expression from pZF(3BS) equivalent to a medium strength yeast promoter without exhibiting growth defects (Figure 3B). Maximal expression was reached after approximately 6 hours (Supplemental Figure 8), which is comparable to commonly used yeast induction systems (McIssac, et al., 2011). The dosage of the ZDBD-CRY2PHR and VP16AD-CIB1 components also affected the basal (dark) induction (Figure 3B). Our results tempt us to hypothesize that too little absolute DBD component (ZDBD-CRY2PHR) reduces function due to a lack of promoter occupancy, while too much ZDBD-CRY2PHR relative to the activation domain component (VP16AD-CIB1) reduces function by decreasing the probability that a ZDBD-CRY2PHR bound at pZF(3BS) is also bound to VP16AD-CIB1. This leads to maximal fold induction along the diagonal as seen in Figure 3a. This was a crude dialing of protein concentration but suggests that one way to further optimize a split TF system would be by carefully titrating the total and relative dosage of each component, a knob not available in single-component and homogeneous two-component optogenetic systems (Benzinger & Khammash, 2018; Zhao, et al., 2018; Xu, et al., 2018; Hughes, Bolger, Tapadia, & Tucker, 2012; Kennedy, et al., 2010; Salinas, et al., 2018). It is worth noting that the variability between biological replicates decreased when the ZDBD-PHR2/VP16AD-CIB1/pZF(3BS) components were chromosomally integrated using the toolkit (Figure 3b) rather than maintained on episomal plasmids (Figure 1b). This is consistent with known variability caused by plasmid copy number (Lee, et al., 2015 Deloache, Cervantes, & Dueber, 2015) and demonstrates that the ability to easily integrate optogenetic components into the chromosome is a further advantage of the toolkit format.
Figure 3. Optimization of the split transcription factor.
(A) Fold-induction in gene expression in response to 17 hours of 470nm 15μW/cm2 blue-light from the pZF(3BS)-mRuby2 reporter was measured in nine strains expressing different ratios of the DNA-binding domain (ZDBD-CRY2PHR) and the activation domain (VP16AD-CIB1) under High (pTEF1), Medium (pRPL18B), or Low (pRNR2) strength promoters to create ZDBD-CRY2high/med/low/VP16AD-CIB1high/med/low strains. Data is presented as mean±SEM. A one-way ANOVA (F(8,45)=30.04, p=1.35X10−15), followed by Tukey’s-HSD shows that the fold-changes are significantly different with the following groups (ZDBD-CRY2high/med/low/VP16AD-CIB1high/med/low): M/H-a; H/H, M/M, M/L-b; H/L, M/H-c; L/H, L/M, L/L-d. (B) Raw fluorescence data for strains shown in (A). Gene expression was compared to yeast strains expressing mRuby2 under constitutive promoters of different strengths (Strong-pTDH3, Medium-pRPL18B, Weak-pREV1). All constructs except those with the Low activation domain show very significant induction in the light (p<0.0001, Welch’s t-test). ANOVA followed by Tukey’s HSD (F(12,65)=248, p=2.21×10−49) shows that the following promoters or split TF combinations are significantly different: pTDH3 alone, M/H alone, the group containing H/H, M/M, L/M, pRPL18B, and the group containing L/H and H/M. All other lowly expressing promoters including the weak constitutive pREV1 promoter belong to the same group.
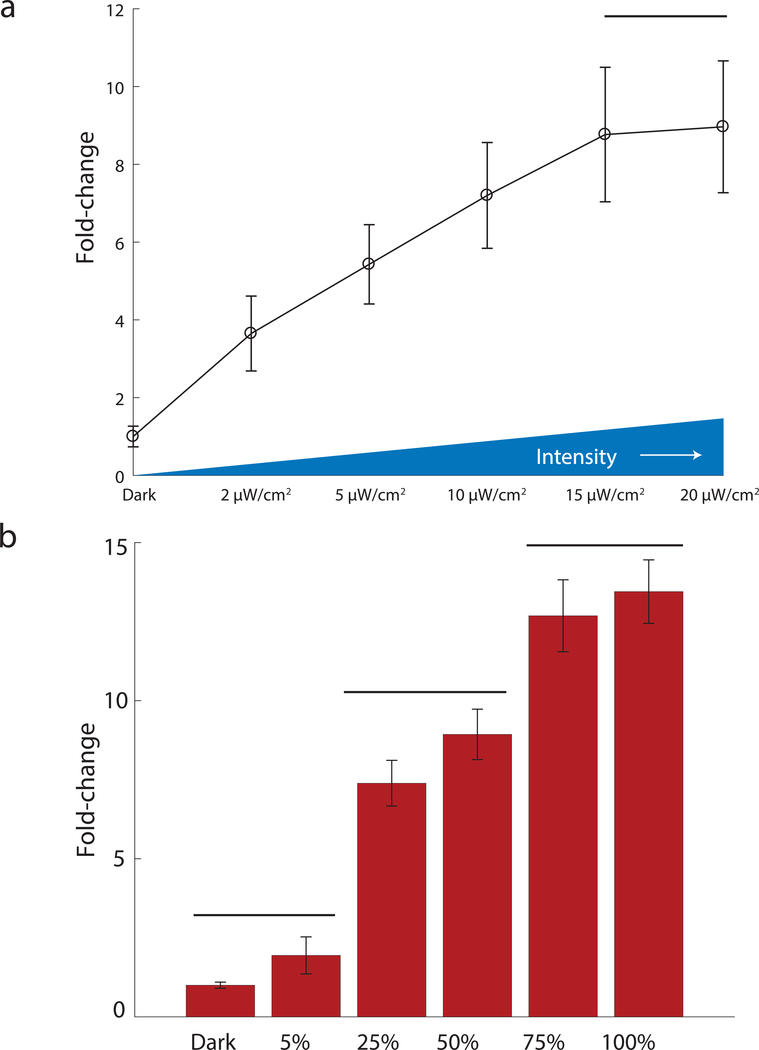
One of the advantages of light as an inducer is the ability to tune its intensity and duty cycle in cultures of cells. We examined our ability to tune output from the ZDBD-CRY2PHRmedium/VP16AD-CIB1medium strain as a function of light intensity. We found that we could tune output from the ZDBD-CRY2PHRmedium/VP16AD-CIB1medium system up to 15μW/cm2 of light, at which point output from the system saturated (Figure 4A). We also measured output from the ZDBD-CRY2PHRmedium/VP16AD-CIB1medium strain as a function of duty cycle. We varied light at 15μW/cm2 from a duty cycle of 5% (1min on/19 min off) to 100% (constant light). Gene expression output increased as a function of duty cycle. Utilizing either the ZDBD-CRY2PHRmedium/VP16AD-CIB1medium or ZDBD-CRY2PHRmedium/VP16AD-CIB1high strain we could achieve gene expression outputs equivalent to the weakest and strongest promoters in the Lee, et al yeast toolkit (Supplemental Figure 9). Thus, by putting the expression of a circuit component under the control of pZF(3BS) and using light chemostats or programmable LED plates (Melendez, et al., 2014; Gerhardt, et al., 2016) one could continuously and dynamically tune component concentration and monitor its effect on circuit function. To allow this optogenetic machinery to be easily integrated into a yeast strain of interest we created a yeast marker (“Type 6”) part containing KlURA3 flanked by loxP sites to allow for marker recycling. We created a pre-assembled integration vector (Supplemental Figure 6) and used it to integrate ZDBD-CRY2PHRmedium/VP16AD-CIB1medium and pZF(3BS)-mRuby2. We verified that integration of ZDBD-CRY2PHRmedium/VP16AD-CIB1medium with the KlURA3 marker, followed by subsequent Cre-recombinase mediated excision of the marker, did not affect light-induced expression of mRuby2 (Supplemental Figure 10).
Figure 4. Tuning of gene expression.
(A) Gene expression in the ZDBD-CRY2PHRmedium/VP16AD-CIB1medium optogenetic strain is tunable as a function of blue-light intensity. Strains were induced for 17 hours at the indicated intensity of 470nm light. ANOVA followed by Tukey’s-HSD shows that the expression at each light intensity is significantly different except for 15μW/cm2 and 20μW/cm2, which are in the same group (F(5,12)=209.93, p-value=3.01×10−11). (B) Gene expression in the ZDBD-CRY2PHRmedium/VP16AD-CIB1medium optogenetic strain is tunable as a function of light duty cycle. Strains were induced with 20 minute periods with the indicated duty-cycle of 470nm blue light at 15μW/cm2. Fluorescence was measured as fold-change relative to the dark control. Bars distinguish significantly different groups determined by one-way ANOVA followed by Tukey’s HSD (F(5,12)=325, p=2.2×10−12).
Optogenetic approaches for controlling natural and synthetic biological networks are garnering attention as the toolkit expands and more powerful applications are demonstrated (Melendez, et al., 2014; Zhao, et al., 2018; Harrigan, Madani, & El-Samad, 2018; Ng, et al., 2019; Rullan, Benzinger, Schmidt, Milias-Argeitis, & Khammash, 2019). Here we report an orthogonal light-inducible transcriptional activator for gene expression control in S. cerevisiae. We have engineered this transcription factor to be compatible with an existing yeast toolkit which allows circuits for light-controlled gene expression to be assembled, integrated into the yeast genome, and tested in less than two weeks. We utilized this rapid prototyping to optimize the ratio and concentration of the two halves of our split transcription factor for maximal light-inducible gene expression with minimal growth defects. Both light intensity and duty cycle can be used to tune output from this gene expression system. We anticipate that this expansion of the yeast toolkit will be very useful to the community, as it will allow for rapid assembly of synthetic circuits with one or more components that can be dynamically tuned with light. This will allow for circuit optimization as well as real-time light-based control of circuit output.
Materials and Methods
Strains and Culture Methods
Yeast strains used in this study are shown in Supplemental Table 1. Yeast strains were grown in standard yeast media, as described in the Supporting Information. Yeast transformation was accomplished using standard lithium-acetate transformation (Gietz & Schiestl, 2007). Primers used for validating integrations are listed in Supplemental Table 2. Escherichia coli strain DH5α was used for all transformation and plasmid maintenance in this study. E. coli were grown on standard media as described in the Supporting Information with appropriate antibiotics to select for and maintain plasmids.
Blue light induction
Blue-light induction was accomplished by either (1) illuminating cultures grown in glass culture tubes on a roller drum or (2) in a Light Plate Apparatus (LPA) (Gerhardt, et al., 2016). Light intensity was measured and validated using a standard photodiode power sensor and power meter (Thorlabs #S120VC, Thorlabs #PM100D). The LPA was calibrated as described in Sweeney, et al (Sweeney, Moreno Morales, Burmeister, Nimunkar, & McClean, 2019) so that consistent light doses could be delivered between LPAs and between experiments.
Flow Cytometry
Gene expression in response to blue light was assayed using fluorescent reporters and flow cytometry. Flow cytometry was performed on either a BD Biosciences LRSII Flow Cytometer (488nm laser and 505LP dichroic filter) or an Attune NxT Flow Cytometer (ThermoFisher Scientific) with 561nm excitation laser and 620/15nm filter cube. For assaying mRuby2 fluorescence on the Attune, the voltages of the flow cytometer were calibrated using rainbow beads so that the medians of FSS, SSC, and mRuby2 fluorescence of the rainbow beads were within 10% difference. The flow cytometry data was then analyzed using FlowJo software. All samples were prepared for flow cytometry by diluting yeast cell culture 1:3 into ice-cold PBS + 0.1% Tween-20. Samples were kept on ice or at 4°C until being analyzed. Samples run on the LPA were measured without sonication. Samples grown in glass culture tubes were sonicated with 10 bursts of 0.5 seconds each once diluted in PBS and prior to flow cytometry.
Construction and Optimization of the DBD-CRY2/AD-CIB1 Optogenetic Split Transcription Factor
Various plasmids were created and tested to determine an optimal DBD-CRY2/AD-CIB1 system (Supplemental Table 3, Supplemental Methods). Function of these plasmid combinations was tested by assaying for blue-light induced gene expression in glass culture tubes or LPAs as outlined above.
Domestication for the Yeast Toolkit
The parts added to the Yeast Toolkit (Lee, et al., 2015) are shown in Supplemental Figure 6. Domestication of parts for the Yeast Toolkit (Lee, et al 2015) requires removal of BsaI, BsmBI, and NotI restriction sites. This was accomplished using the Q5 site-directed mutagenesis kit (New England Biolabs E0554S) and appropriate primers as indicated in Supplemental Table 2 to introduce a synonymous mutation to remove the undesirable restriction enzyme site. Domestication was verified by Sanger sequencing. Domesticated parts were inserted into the part-plasmid backbone (yTK001) as described in Lee, et al using primers in Supplemental Table 2.
Golden Gate Assembly of Cassette and Multigene Plasmids
Cassette plasmids (consisting of transcriptional units, i.e. promoter-coding sequence-terminator) and multigene plasmids (consisting of multiple transcriptional units linked together through assembly connectors with appropriate homology to integrate into the yeast genome) were assembled using BsaI assembly or BsmBI assembly as outlined in Lee, et al (Lee, et al 2015). Details and small adjustments are outlined in the Supplemental Methods.
Recycling of loxP-flanked markers
In order to allow for marker recycling utilizing the Cre-loxP system we created a loxP-KlURA3-loxP part plasmid (pMM0519). Cre-mediated recombination to recycle the KlURA3 marker was accomplished by adapting the Cre recombinase-mediated excision protocol from Carter and Delneri (Carter & Delneri, 2010).
Statistical Analysis
All statistical analysis was carried out as described using Matlab (Mathworks, Natick, MA).
Supplementary Material
Acknowledgements
The authors would like to acknowledge discussion and helpful comments from the members of the McClean lab throughout the project. We thank K. Sweeney and T. Scott for assistance with Matlab and Python code for analyzing flow cytometry and growth curve data. We thank Renee Szakaly for assistance with strain construction. This work was supported by the National Institutes of Health [1R35GM128873, R21AI135166] and a Lewis-Sigler Fellowship from Princeton University (M.N.M). Flow cytometry was enabled by the University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520 as well as the Princeton Flow Cytometry Resource Facility. Megan Nicole McClean, Ph.D., holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
Associated Content
Supporting Information is available online.
References
- Benzinger D, & Khammash M. (2018). Pulsatile inputs achieve tunable attenuation of gene expression variability and graded multi-gene regulation. Nature communications, 9(1), 3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz K, & Collins J. (2013). The roots-a short history of industrial microbiology and biotechnology. Applied Microbiology and Biotechnology, 97, 3747–3762. [DOI] [PubMed] [Google Scholar]
- Carter Z, & Delneri D. (2010). New generation of loxP-mutated deletion cassettes for the genetic manipulation of yeast natural isolates. Yeast, 27, 765–775. [DOI] [PubMed] [Google Scholar]
- Castillo-Hair S, Igoshin O, & Tabor J. (2015). How to train your microbe: methods for dynamically characterizing gene networks. Current opinion in microbiology, 24, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Lee H, Heng Y, Chua N, Teo W, Choi W, … Chang M. (2018). Synthetic biology toolkits and applications in Saccharomyces cerevisiae. Biotechnology Advances, 36(7), 1870–1881. [DOI] [PubMed] [Google Scholar]
- Gerhardt K, Olson E, Castillo-Hair S, Hartsough L, Landry B, Ekness F, … Tabor J (2016). An open-hardware platform for optogenetics and photobiology. Scientific Reports, 6(35363). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R, & Schiestl R. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols, 2, 31–34. [DOI] [PubMed] [Google Scholar]
- Harrigan P, Madani H, & El-Samad H. (2018). Real time genetic compensation operationally defines the dynamic demands of feedback control. Cell, 175, 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R, Bolger S, Tapadia H, & Tucker C. (2012). Light-mediated control of DNA transcription in yeast. Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M, Hughes R, Peteys L, Schwartz J, Ehlers M, & Tucker C. (2010). Rapid blue-light-mediated induction of protein interactions in living cells. Nature Methods, 7(12), 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A, Lu T, Bashor C, Ramirez C, Pyenson N, Joung J, & Collins J. (2012). A synthetic biology framework for programming eukaryotic transcription functions. Cell, 150, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ME, Deloache W, Cervantes D, & Dueber J. (2015). A highly characterized yeast toolkit for modular, multipart assembly. ACS Synthetic Biology, 4(9), 975–986. [DOI] [PubMed] [Google Scholar]
- Lugagne J-B, & Dunlop MJ (2019). Cell-machine interfaces for characterizing gene regulatory network dynamics. Current opinion in systems biology, 14, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac R, Oakes B, Wang XD, Botstein D, & Noyes M. (2013). Synthetic gene expression systems with rapid, tunable single-gene specificity in yeast. Nucleic Acids Research, 41(4), e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIssac R, Silverman S, McClean M, Gibney P, Macinskas J, Hickman M, … Botstein D. (2011). Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Molecular Biology of the Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez J, Patel M, Oakes B, Xu P, Morton P, & McClean M. (2014). Real-time optogenetic control of intracellular protein concentration in microbial cell cultures. Integrative Biology, 6, 366–372. [DOI] [PubMed] [Google Scholar]
- Milias-Argeitis A, Rullan M, Aoki SK, Buchmann P, & Khammash M. (2016). Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nature Communications, 7, 12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng A, Nguyen T, Gomez-Schiavon M, Dods G, Langa R, Boyken S, … El-Samad H (2019). Modular and tunable biological feedback control using a de novo protein switch. Nature, 572, 265–269. [DOI] [PubMed] [Google Scholar]
- Rullan M, Benzinger D, Schmidt G, Milias-Argeitis A, & Khammash M. (2019). An optogenetic platform for real-time, single-cell interrogation of stochastic transcriptional regulation. Molecular Cell, 70, 745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas F, Rojas V, Delgado V, Agosin E, & Larrondo L. (2017). Optogenetic switches for light-controlled gene expression in yeast. Applied microbiology and biotechnology, 101, 2629–2640. [DOI] [PubMed] [Google Scholar]
- Salinas F, Rojas V, Delgado V, Lopez J, Agosin E, & Larrondo L. (2018). Fungal Light-Oxygen-Voltage Domains for optogenetic control of gene expression and flocculation in yeast. mBio, 9(4), e00626–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TD, Sweeney K, & McClean MN. (2019). Biological signal generators: integrating synthetic biology tools and in silico control. Current Opinion in Systems Biology, 14, 48–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney K, Moreno Morales N, Burmeister Z, Nimunkar A, & McClean M. (2019). Easy calibration of the Light Plate Apparatus for optogenetic experiments. MethodsX, 1480–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher J, Gong D, Lim W, & Weiner O. (2011). Light-based feedback for controlling intracellular signaling dynamics. Nature Methods, 8(10), 837–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Du Z, Liu R, Li T, Zhao Y, Chen X, & Yang Y. (2018). A single-component optogenetic system allows stringent switch of gene expression in yeast cells. ACS Synthetic Biology. [DOI] [PubMed] [Google Scholar]
- Zhao E, Zhang Y, Mehl J, Park H, Lalwani M, Toettcher J, & Avalos J. (2018). Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature, 555(7698), 683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.