Abstract
Biological membrane of many cell types contains large-pore channels that permeate a wide variety of ions and metabolites. Examples include connexin, innexin, and pannexin, which form gap junctions and/or bona fide cell surface channels. The most recently identified large-pore channels are the calcium homeostasis modulators (CALHMs), which permeate ions and ATP in a voltage-dependent manner to control neuronal excitability, taste signaling, and pathologies of depression and Alzheimer’s disease. Despite such critical biological roles, the structures and patterns of oligomeric assembly remain unclear. Here, we reveal the structures of two CALHMs, chicken CALHM1 and human CALHM2, by single particle cryo-electron microscopy, which show novel assembly of the four transmembrane helices into channels of 8-mers and 11-mers, respectively. Furthermore, molecular dynamics simulations suggest that lipids can favorably assemble into a bilayer within the larger CALHM2 pore, but not within CALHM1, demonstrating the potential correlation between pore-size, lipid accommodation, and channel activity.
Permeation of ions and/or substrates such as ATP by CALHM proteins is fundamental to the physiology of depression1 and cognition2 as well as the pathology of Alzheimer’s disease (AD)3. The CALHM family comprises six members, CALHM1–6 (sequence homology ~30–50%), amongst which CALHM1 has been the most extensively studied family to date. CALHM1 forms a channel that conducts ATP4 and ions including Ca2+, Na+, K+, and Cl− in a voltage-dependent manner5. A single nucleotide polymorphism (SNP) within the calhm1 gene that results in a Pro86Leu mutation was reported to be a risk factor for early onset of AD3 and at the cellular level, this mutation has been shown to promote deposition of amyloid beta, a hallmark of AD6. Later studies showed that CALHM1 proteins are expressed in type II taste bud cells to mediate ATP efflux which results in purinergic signaling for sweet, bitter, and umami taste sensations4. ATP efflux from CALHM1 was also shown to control ciliary beat frequency for mucociliary clearance in airways7. More recently, the function of CALHM2 proteins expressed in astrocytes have been linked to depression1 and implicated to play a role in glial-neuronal functions1. While CALHM3 has been shown to form heteromeric channels with CALHM18, the functions and biological roles of the remaining members, CALHM4–6, are currently unknown. The calhm genes are conserved throughout vertebrates and non-vertebrates. Furthermore, CALHM1 from Caenorhabditis elegans has been shown to possess similar ion channel properties to that of human CALHM1 (hCALHM1)9, demonstrating functional conservation throughout diverse species.
Topological prediction of the CALHM protein family has been difficult. Originally, CALHM was suggested to have similar membrane topology to N-methyl-D-aspartate receptors (NMDARs)3 and later to hemichannel and gap junction proteins including connexin and innexin based on secondary structure prediction and comparison5. The CALHM family members have no primary sequence homology with NMDARs or any of the hemichannels and gap junction protein families mentioned, thus making it difficult to design of functional experiments to address mechanisms. Overall, despite playing critical roles in human physiology and pathology, the structure and function of CALHM proteins at the molecular level remain unclear. Here, we show structures of the two CALHM family members, CALHM1 and CALHM2 in lipid nanodiscs. Protomers of both CALHM1 and CALHM2 harbor four transmembrane domains and a long cytoplasmic helix, which do not resemble the known structures of other channels including connexin, innexin, and NMDARs. Despite belonging to the same protein family, CALHM1 and CALHM2 form channels with distinct oligomeric states, 8-mer and 11-mer, respectively. Molecular dynamics simulation shows that the 11-mer assembly in CALHM2 can stably accommodate lipids in its hydrophobic channel whereas the 8-mer assembly of CALHM1 cannot suggesting a possibility for lipid accommodation as a means to regulate channel function.
Results
Preparation of CALHM proteins
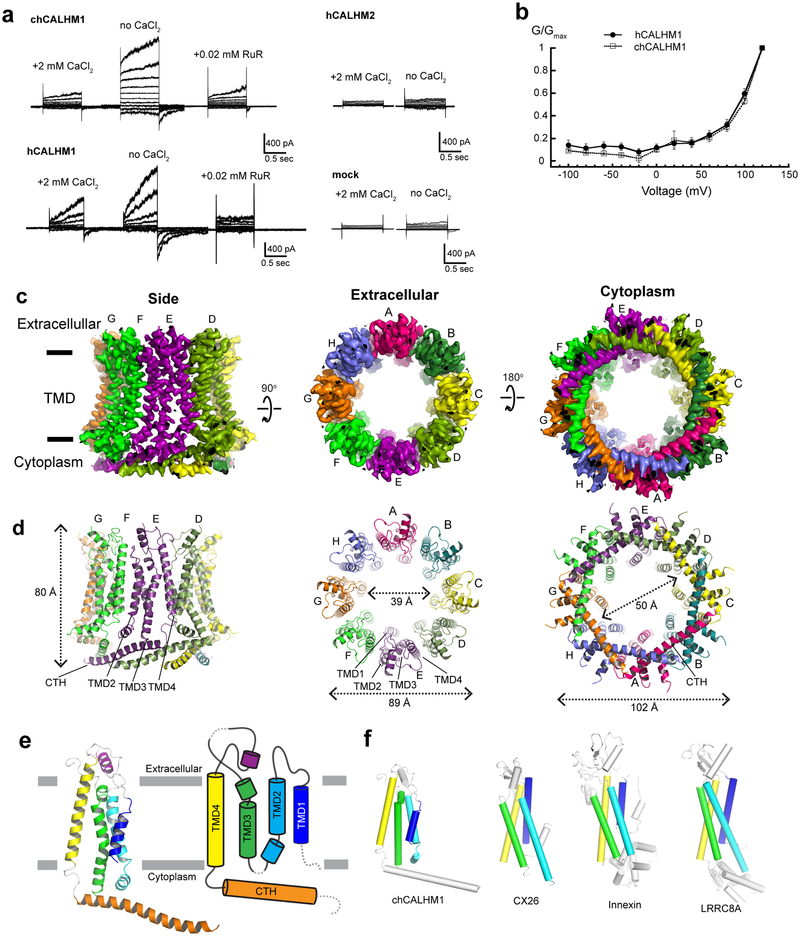
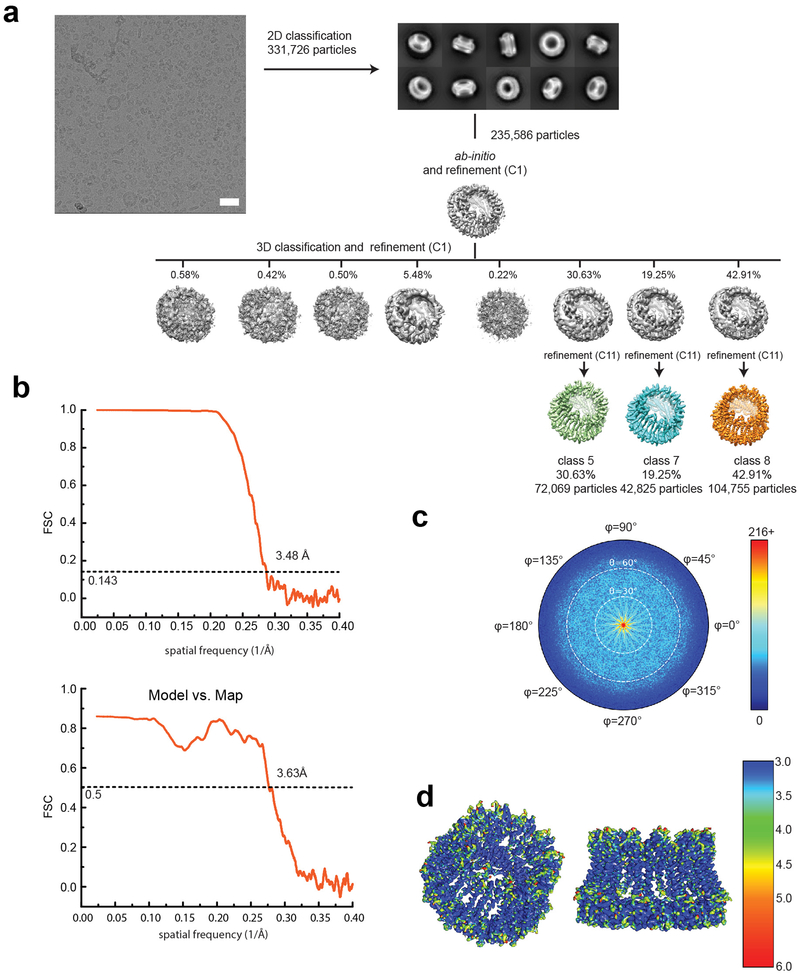
In this study, we focused on the two major family members, CALHM1 and CALHM2, which are involved in controlling excitability of neurons1,2. We first conducted expression screening of CALHM orthologues using fluorescence coupled size-exclusion chromatography (FSEC)10, which concluded that chicken CALHM1 (chCALHM1) and human CALHM2 (hCALHM2) show protein size homogeneity suitable for structural analysis. The primary sequences of chCALHM1 and human CALHM1 (hCALHM1) have 67.7% identity and 80.2% similarity overall and 81.4% identity and 93.8% similarity within the transmembrane domains (TMDs) (Supplementary Figure 1). chCALHM1 also contains Asp120, the equivalent residue of which in hCALHM1 (Asp121) has been shown to be critical for its ion channel activity5 (Supplementary Figure 1). Indeed, our patch-clamp electrophysiology shows that chCALHM1 has similar functional properties to hCALHM1 including voltage-sensitivity, calcium sensitive inhibition, and channel blockade by ruthenium red (Fig. 1a–b). Both chCALHM1 and hCALHM2 proteins were recombinantly expressed in Sf9 insect cells, infected with the respective recombinant baculoviruses11, purified, and reconstituted into lipid nanodiscs prior to vitrification for the cryo-EM study (Extended Data Fig. 1; also see Methods).
Fig. 1. Structure and function of chCALHM1.
a, Currents elicited by hCALHM1 and chCALHM1 with voltage steps from −100 to +100 mV in 10 mV increment in the presence and absence of extracellular calcium. The current of chCALHM1 can be blocked by 0.02 mM ruthenium red (RuR) as previously shown for hCALHM1. No significant voltage-dependent current compared to mock transfected cells were observed for hCALHM2 consistent with the previous report8. b, G-V plot of chCALHM1 and hCALHM1 with no CaCl2 (right panel). Error bars represent SEM for six individual patches (n=6) for hCALHM1 and seven (n=7) for chCALHM1). c-d, Cryo-EM density (c) and atomic models (d) of chCALHM1 viewed from the side of the membrane, the extracellular region, and the cytoplasm. e, Ribbon (left) and schematic (right) representations of the chCALHM1 protomer. The TMDs are colored as blue, cyan, green, and yellow for TMD1, 2, 3, and 4, respectively. Dashed lines represent regions that are not visible in our structure. f, Protomers of chCALHM1, CX2620, innexin17, and LRRC8A21. The TMDs are colored as in panel e for comparison. Data for the graphs in panel b are available as source data.
Cryo-EM structure of CALHM1
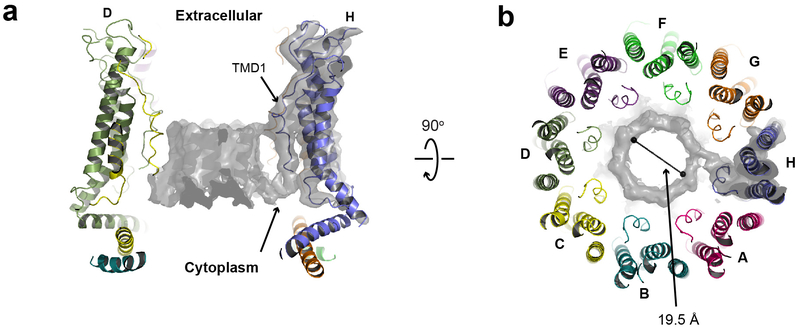
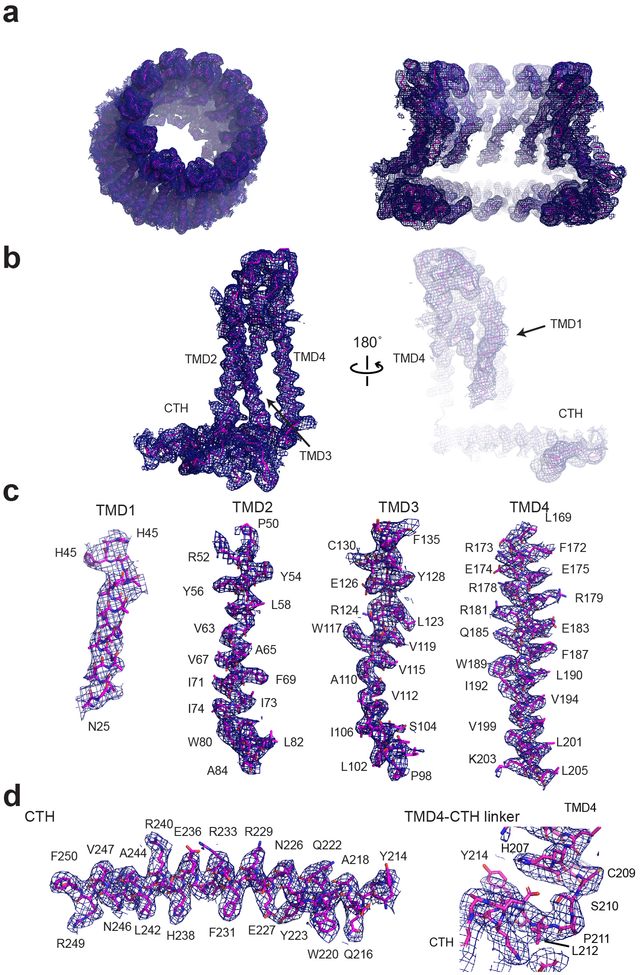
We solved the structure of chCALHM1 using single particle cryo-EM analysis at an overall resolution of 3.63 Å (Fig. 1c–e, Extended Data Figs. 2 and 3, Table 1) as assessed by Fourier Shell Correlation (FSC)12,13. The cryo-EM structure was solved in the presence of EDTA to remove free divalent ions such as calcium, and therefore likely mimic the active state. The cryo-EM density of the extracellular domain, the four TMD helices, and the cytoplasmic helices (CTHs) were of sufficient quality to conduct de novo modeling between residue numbers 26–79, 91–137 and 151–247, altogether spanning 198 out of 342 amino acids. Most of the missing density is in the carboxyl terminal region after the CTH where 72 out of 94 residues are predicted to be unstructured by a secondary structure analysis14. The structure confirms the previous prediction that CALHM1 harbors four transmembrane domains with the amino and carboxyl termini facing the cytoplasm15. The cryo-EM density for TMD1 facing the pore is weaker compared to the other three TMDs, indicating the presence of conformational flexibility. Our current structure clearly shows octameric assembly with a pore-like structure in the middle of the oligomer (Fig. 1c–d). The assembly is mediated mainly by interactions between TMD2 and TMD4, between TMD1 and TMD3, and between the forty-residue long CTHs of neighboring subunits (Fig. 2). The octameric assembly shown in our high resolution cryo-EM structure differs from a previous study suggesting hexameric assembly of CALHM1 based on Blue Native-PAGE and photobleaching of the hCALHM1-EGFP constructs15. Nevertheless, the subunit-interface residues are highly conserved between chCALHM1 and hCALHM1, strongly implying preservation of oligomeric mechanisms (Supplementary Figures 1 and 2, 88.5% identity and 100% similarity over 35 residues in TMDs and CTH). In the present study, 2D or 3D classification did not support the presence of other oligomeric species such as hexamers. Furthermore, there is clear density for TMD4, CTH, and the TMD4-CTH linker (Extended Data Fig. 3), indicating that the inter-protomer interaction mode is well defined and stable. Some unresolved density extends from TMD1 towards the middle of the channel at the cytoplasmic side, likely representing the amino terminal residues in multiple conformations (Extended Data Fig. 4). In CALHM1s from human and Caenorhabditis elegans, the first nine residues have been shown to alter voltage-sensitivity16, thus, we suggest that these voltage-sensing residues are located in the inner-pore within the membrane spanning region (Extended Data Fig. 4). The pore diameter measured based on the above cryo-EM density is 19.5 Å which is similar to the previously estimated diameter of 14 Å, based on a dye-permeation assay and electrophysiological measurement of tetralkylammonium cation permeation15.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| chCALHM1 (EMD-21143, PDB 6VAM) | hCALHM2 (EMD-21141, PDB 6VAK) | hCALHM2 gap junction (EMD-21140, PDB 6VAI) | chCALHM1 hCALHM2 chimera (EMD-21142, PDB 6VAL) | |
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 130,000 | 130,000 | 130,000 | 130,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 70 | 70 | 70 | 70 |
| Defocus range (μm) | 1.5–3.0 | 1.2–2.8 | 1.5–3.0 | 1.4–2.8 |
| Pixel size (Å) | 1.06 | 1.06 | 1.06 | 1.06 |
| Symmetry imposed | C8 | C11 | D11 | C11 |
| Initial particle images (no.) | 533,665 | 331,726 | 119,211 | 397,362 |
| Final particle images (no.) | 308,916 | 104,755 | 52,737 | 123,664 |
| Map resolution (Å) | 3.63 | 3.48 | 3.68 | 3.87 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 3.0–6.0 | 3.0–6.0 | 3.0–6.0 | 3.0–6.0 |
| Refinement | ||||
| Initial model used (PDB code) | de novo | de novo | 6VAK | 6VAM/6VAK |
| Model resolution (Å) | 3.92 | 3.61 | 3.79 | 4.32 |
| FSC threshold | 0.5 | 0.5 | 0.5 | 0.5 |
| Map sharpening B factor (Å2) | −150 | −90 | −90 | −90 |
| Model composition | ||||
| Nonhydrogen atoms | 13,312 | 22,781 | 45,584 | 21,615 |
| Protein residues | 1,712 | 2,292 | 5,784 | 2,827 |
| Ligands | 0 | 0 | 0 | 0 |
| B factors (Å2) | ||||
| Protein | 85.99 | 43.91 | 38.29 | 101.43 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.003 | 0.007 | 0.005 | 0.005 |
| Bond angles (°) | 0.603 | 0.746 | 0.678 | 0.728 |
| Validation | ||||
| MolProbity score | 1.70 | 2.13 | 1.99 | 1.90 |
| Clashscore | 6.08 | 8.72 | 8.94 | 9.19 |
| Poor rotamers (%) | 0 | 1.97 | 0.96 | 1.02 |
| Ramachandran plot | ||||
| Favored (%) | 94.71 | 93.28 | 91.34 | 94.02 |
| Allowed (%) | 5.29 | 6.72 | 8.66 | 5.98 |
| Disallowed (%) | 0 | 0 | 0 | 0 |
| C-beta deviation | 0 | 0 | 0 | 0 |
| EMRinger Score | 1.83 | 2.73 | 3.11 | 2.18 |
| CaBLAM outlier (%) | 3.47 | 3.79 | 2.70 | 2.04 |
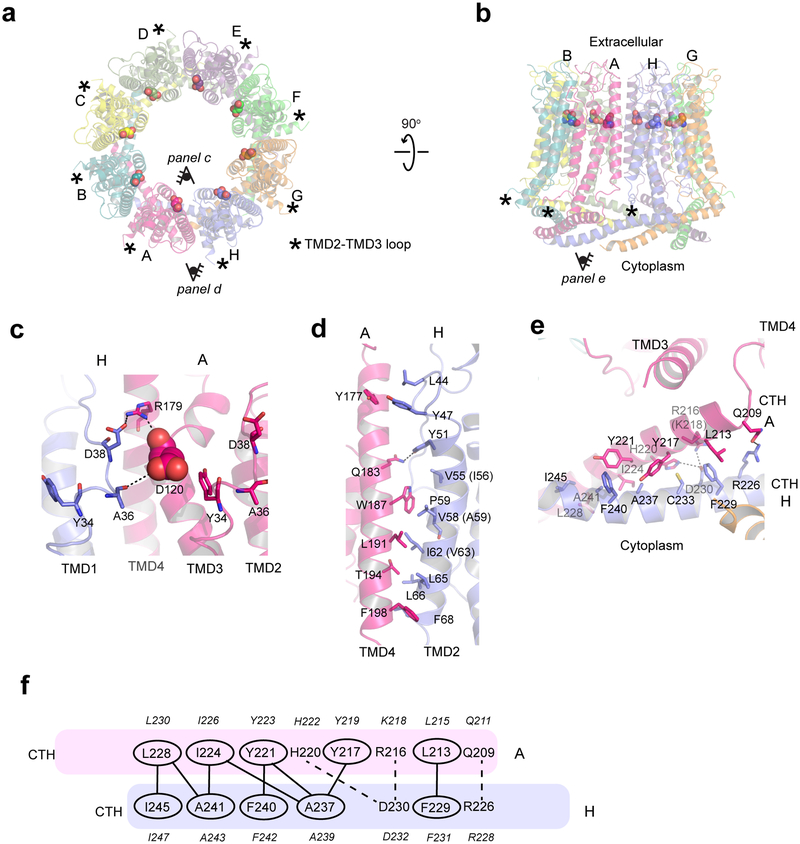
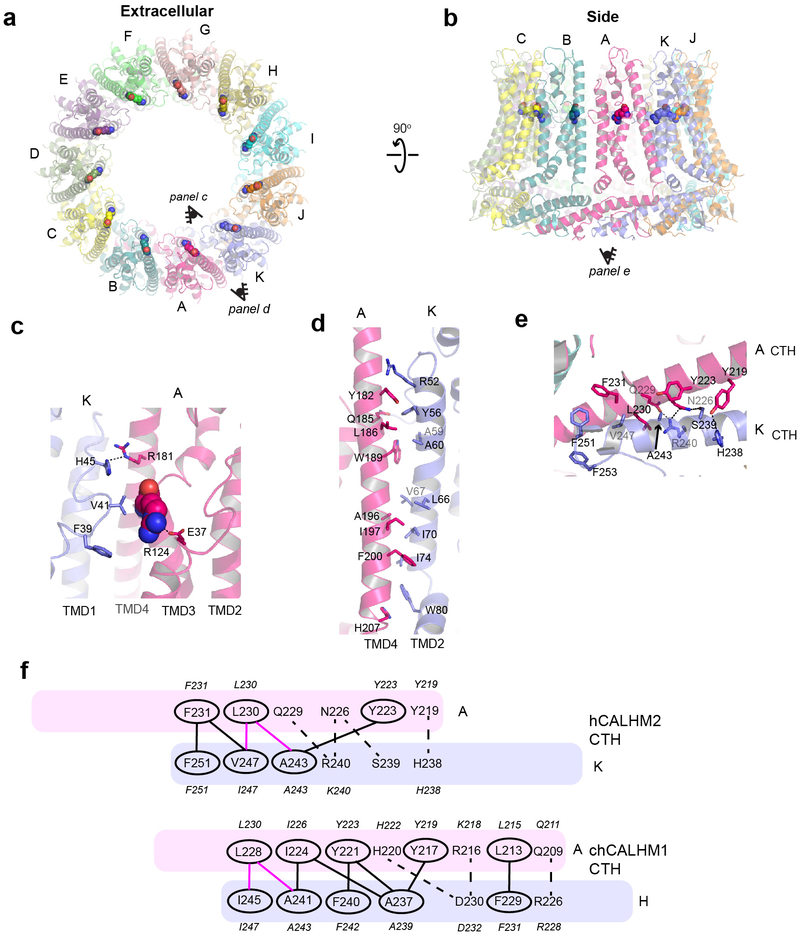
Fig. 2. Inter-subunit interface of chCALHM1.
a-b, The chCALHM1 structure viewed from the top of extracellular region (a) and the side of the membrane (b). Shown in spheres are the Asp120 residues critical for calcium sensitivity and ion permeation. c, Asp120 (sphere) and surrounding residues (sticks) form polar interactions to mediate inter-subunit interactions. d-e, The inter-subunit interactions between TMD2 and TMD4 (d) and CTHs (e). f, The schematic presentation of the interactions between two CTHs (magenta and slate blue). Polar and van der Waals interactions mediated by hydrophobic residues (ovals) are shown as dashed and solid lines, respectively. The residues in italic are the equivalent ones in hCALHM1.
The only other octameric channel reported to date is innexin17, however, it does not share similar features in the pattern of oligomeric assembly with chCALHM1. Furthermore, contrary to a previous suggestion, there is no similarity in the membrane topology, structure, and oligomeric assembly pattern of chCALHM1 to NMDARs3 which contain three TMDs and a re-entrant loop and form hetero-tetramers18,19. Importantly, the structural comparison of monomers demonstrated that chCALHM1 does not resemble other four transmembrane channel proteins including connexin (CX26)20, innexin (INX6)17, or the volume-regulated anion channel (VRAC; LRRC8)21,22 (Fig. 1f).
A key residue known to modulate CALHM1 ion permeability and calcium sensitivity, Asp120 (Asp121 in hCALHM1)5, is located in TMD3 at the interface with the neighboring subunit (Fig. 2a–c). Although in chCALHM1, each of the Asp120 residues face the inner pore, they do not appear to participate in pore formation directly as they are distant from each other (~20 Å apart between the Cαs of the neighboring Asp120 residues). Instead, Asp120 may facilitate inter-subunit interactions which stabilize channel assembly (Fig. 2c). Mutation of another key residue in hCALHM1 (Pro86Leu) has also previously been shown to be a risk factor for the age of onset of Alzheimer’s disease in selected populations3, and although the equivalent residue in chCALHM1 is Gln85, it is clear that this residue is located in the disordered loop between TMD2 and TMD3. Consistent with this location, which faces the cytoplasm and is not part of the channel-pore (Fig. 2a–b; asterisks), the Pro86Leu mutation in hCALHM1 has previously been shown not to alter channel activity5. Instead, the mechanisms underlying the association of this mutation with Alzheimer’s disease pathology may involve other factors, such as protein-protein interactions involving this loop and subsequent cell signaling events that regulate the level of amyloid beta23.
Cryo-EM structure of CALHM2
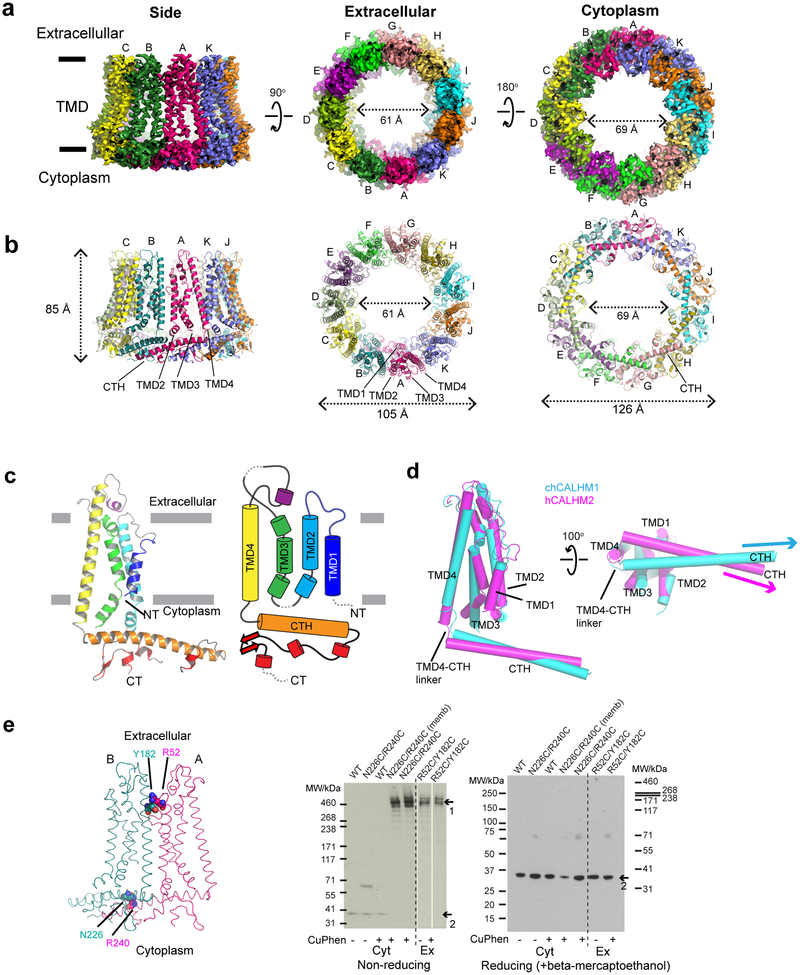
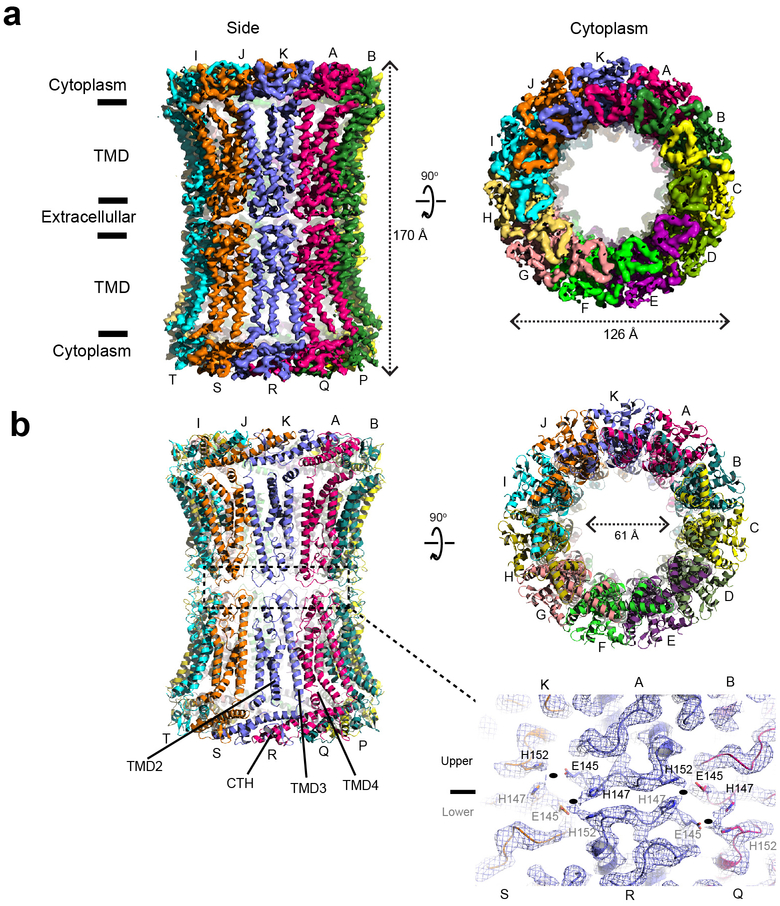
CALHM2 has a moderately high sequence similarity to CALHM1 within the predicted TMD domains (35% identity and 56% similarity between hCALHM1 and hCALHM2 and 37% identity and 55% similarity between chCALHM1 and hCALHM2; Supplementary Figure 2), yet, unlike CALHM1, does not show voltage-dependent ion channel activities (Fig. 1a)8. Thus, we wondered what structural features of CALHM1 and CALHM2 may be responsible for this functional difference. To permit an extensive comparison, we conducted a structural analysis of the human CALHM2 (hCALHM2) protein by implementing a similar protocol to that used above for chCALHM1 (Extended Data Fig. 1, also see Methods). Single particle cryo-EM analysis of the hCALHM2 sample in the absence of calcium resulted in one major 3D class at 3.48 Å resolution as assessed by FSC (Extended Data Figs. 5 and 6, Table 1). As in the case of chCALHM1, the hCALHM2 protomer contains four TMDs and the long CTH that is the signature of the CALHM family (Fig. 3a–c). The orientations of the four TMD helices in hCALHM2 are also similar to that of chCALHM1 and unrelated to connexin (CX26), innexin, or VRAC (Fig. 3d). However, the profound structural difference between chCALHM1 and hCALHM2 is that the oligomeric state of hCALHM2 is 11-mer (Fig. 3a–b). As in chCALHM1, the oligomeric assembly of hCALHM2 is mediated by interactions between TMD2 and TMD4, between TMD1 and TMD3, and between the CTHs of the neighboring subunits (Extended Data Fig. 7). However, the fundamental difference is in the angle between TMD4 and the CTH, which is controlled by the linker sequence that tethers these two helices together (TMD-CTH linker; Fig. 3d). Consequently, the sites of inter-CTH interactions are different between chCALHM1 and hCALHM2. This linker sequence differs between CALHM family members, suggesting that CALHM4–6 may also have distinct oligomeric states (Supplementary Figure 2). It is interesting to note that CALHM1 and CALHM3 have similar linker sequences and are known to form heteromers8. Nevertheless, the 11-mer channel assembly observed in hCALHM2 is unprecedented. Therefore, to validate the physiological relevance of this 11-mer assembly in the membrane environment, we conducted a series of disulfide-based inter-subunit crosslinking experiments (Fig. 3e). Based on the structure, we substituted residues in the inter-subunit interface at the extracellular side of TMD2 and TMD4 (Arg52/Tyr182) and at the CTHs (Asn226/Arg240) with cysteines (Fig. 3e), subjected the mutants to SDS-PAGE, and detected bands by Western blot in the presence and absence of a reducing agent. In the case of the CTH mutants located in the cytoplasm, we facilitated disulfide bond formation by addition of copper phenanthroline to the membrane fraction. In the non-reducing condition, we observed a high molecular weight band around 460 kDa. Monomer bands were exclusively observed in the reducing condition, indicating that the band shift in the non-reducing condition is mediated by disulfide bonding between the engineered cysteines (Fig. 3e). The result above shows that the oligomeric assembly observed in the cryo-EM structure is consistent with its physiological state in the membrane.
Fig. 3. Structure and function of hCALHM2.
a-b, Cryo-EM density (a) and atomic models (b) of hCALHM2 viewed from the side of the membrane, the extracellular region, and the cytoplasm. c, Ribbon (left) and schematic (right) representations of the hCALHM2 protomer. The TMDs are colored as blue, cyan, green, and yellow for TMD1, 2, 3, and 4, respectively. Dashed lines represent regions that are not visible in our structure. d, Superposition of the TMDs of the chCALHM1 (in cyan) and the hCALHM2 (in magenta) viewed from the side of the membrane (left) and the cytoplasm (right). RMSD of superposition is 1.1 Å over 105 Cα positions. e, Cysteine mutations, Tyr182Cys/Arg52Cys or Asn226Cys/Arg240Cys (spheres), were introduced at the subunit interfaces to assess formation of inter-subunit disulfide crosslinking (left). Anti-1D4 Western blots (right) of SDS-PAGE show band shifts for Tyr182Cys/Arg52Cys and Asn226Cys/Arg240Cys (arrow 1). Tyr182Cys/Arg52Cys forms disulfide bonds independent of copper phenanthroline. Formation of disulfide bonds for Asn226Cys/Arg240Cys requires copper phenanthroline treatment before (Asn226Cys/Arg240Cys memb) or after detergent solubilization. The wild-type hCALHM2 protein runs as a monomer under these conditions (arrow 2). Under reducing conditions (beta mercaptoethanol), all constructs run as monomers (arrow 2 on the right gel). Uncropped images for panel e are available as source data.
Cytoplasmic domain governs oligomeric state
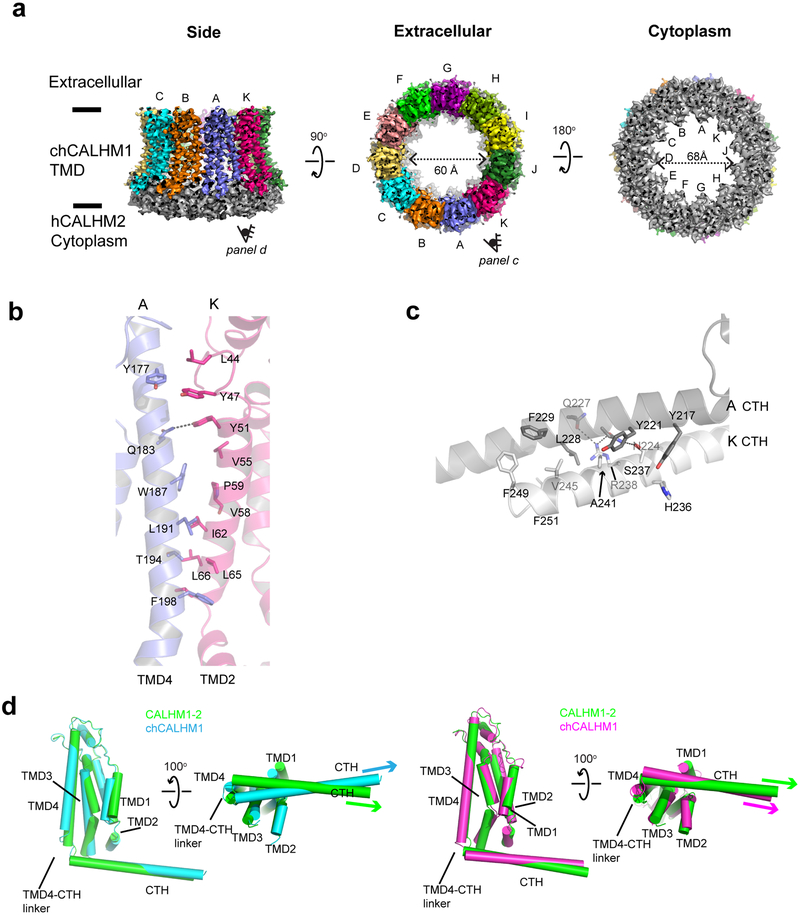
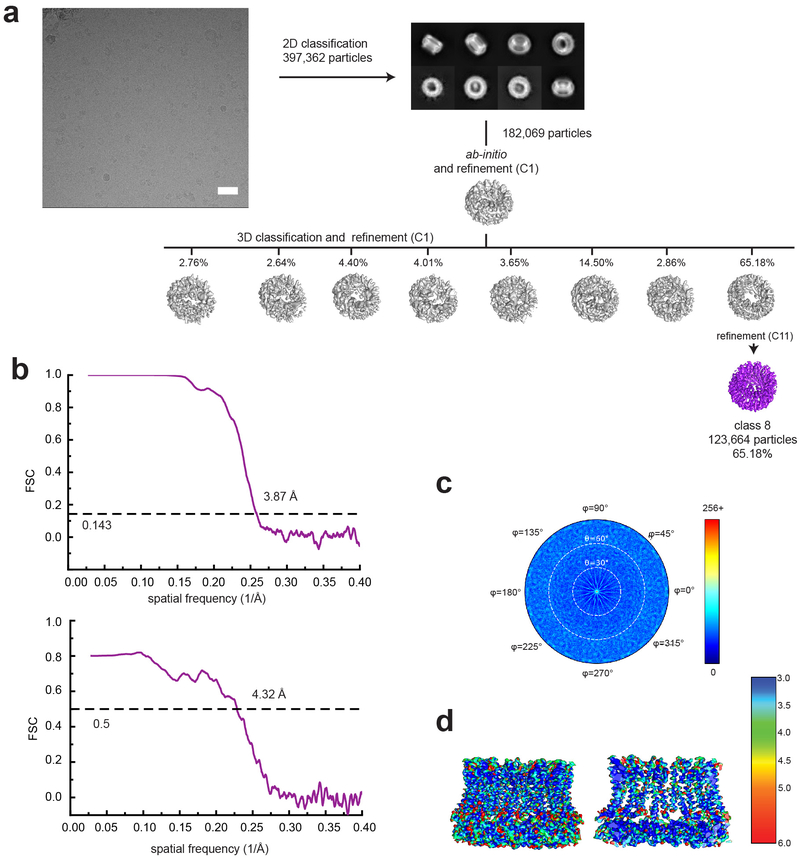
To test our structure-based hypothesis that the interactions between the CTHs control the oligomeric state of CALHMs, we made a chimeric construct of chCALHM1 TMDs and hCALHM2 cytoplasmic domains (CALHM1–2) and analyzed the oligomeric state by solving the cryo-EM structure in lipid nanodiscs as done in chCALHM1 and hCALHM2. Our cryo-EM structure at 3.9 Å resolution unambiguously showed an 11-mer assembly similar in principle to the one observed in hCALHM2 (RMSD = 2.83 Å over 2386 aligned residues) (Fig. 4a, Extended Data Fig. 8). This demonstrates that replacing the cytoplasmic domain of chCALHM1 with that of hCALHM2 converts the oligomeric state of chCALHM1 from 8-mer to 11-mer. The major structural determinants for the inter-protomer interactions are TMD2-TMD4 and CTHs as in the case of chCALHM1 and hCALHM2 (Fig. 4b–c). Consistent with the 11-mer assembly, the angle between TMD4 and CTH of CALHM1–2 is similar to that of hCALHM2 but not chCALHM1 (Fig. 4d). Moreover, the mode of inter-CTH interaction is similar between CALHM1–2 (Fig. 4c) and hCALHM2 (Extended Data Fig. 7e–f) implying that the TMD4-CTH linker and inter-CTH interactions govern the oligomeric state. Overall, our result demonstrates a proof-of-principle that the oligomeric state of CALHM1 can be manipulated by altering the cytoplasmic domain.
Fig. 4. Structure of the CALHM1–2 chimera.
a, Cryo-EM density of CALHM1–2 chimera viewed from the side of the TMDs, the extracellular region, and the cytoplasm. The TMDs of each subunit is colored differently and the cytoplasmic domains are in gray. CALHM1–2 assembles as 11-mer similar to hCALHM2. b-c, The inter-subunit interactions between TMD2 and TMD4 (b) and between CTHs (c). d, Comparison of protomers of the CALHM1–2 (green), chCALHM1 (cyan), and hCALHM2 (magenta) demonstrating the similar and distinct TMD4-CTH orientations between CALHM1–2 and hCALHM2 and between CALHM1–2 and chCALHM1, respectively.
Cryo-EM structure of CALHM2 gap junction
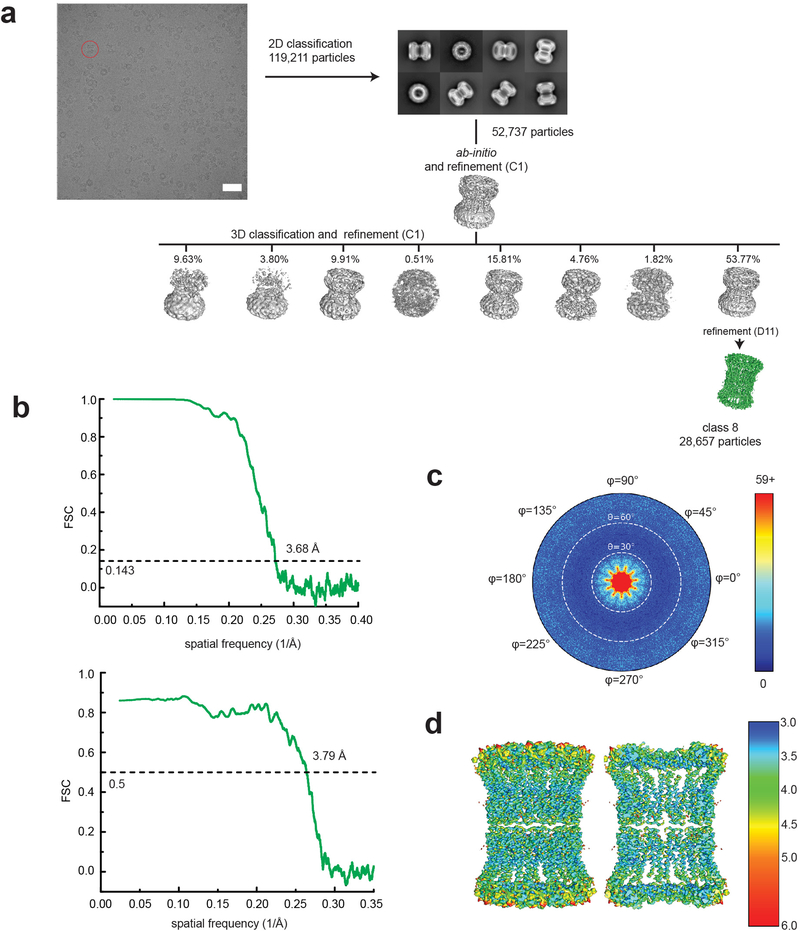
One noteworthy observation is that the hCALMH2 11-mer dimerizes to form a 22-mer reminiscent of a gap junction in the presence of 1 mM CaCl2 under cryo-EM conditions (Extended Data Figs. 9 and 10). Our 22-mer structure solved at 3.68 Å showed a high structural similarity to the 11-mer structure (RMSD = 0.853 Å over 265 residues), indicating that the calcium did not alter the overall protein architecture. The gap junction formation is mediated by His147 and His152 in the apposed hemichannels and possibly Glu145, which are located in the extracellular loop between TMD3 and TMD4 (Extended Data Fig. 10b, lower panel). The cryo-EM density between His147 and His152 is continuous potentially implying that a divalent cation may be present to mediate an interaction between these two histidines along with Glu145. In this condition, EDTA is not included, thus, a trace amount of divalent ions such as zinc ions may be present in the sample. However, inclusion of 0.1 mM ZnCl2 does not oligomerize hCALHM2 to a 22-mer assembly in solution as observed in the identical peak retention time in size-exclusion chromatography (data not shown). It is therefore unclear if the 22-mer observed by cryo-EM exists under physiological conditions or if the gap junction formation of hCALHM2 requires additional factors in solution.
Properties of channel pores
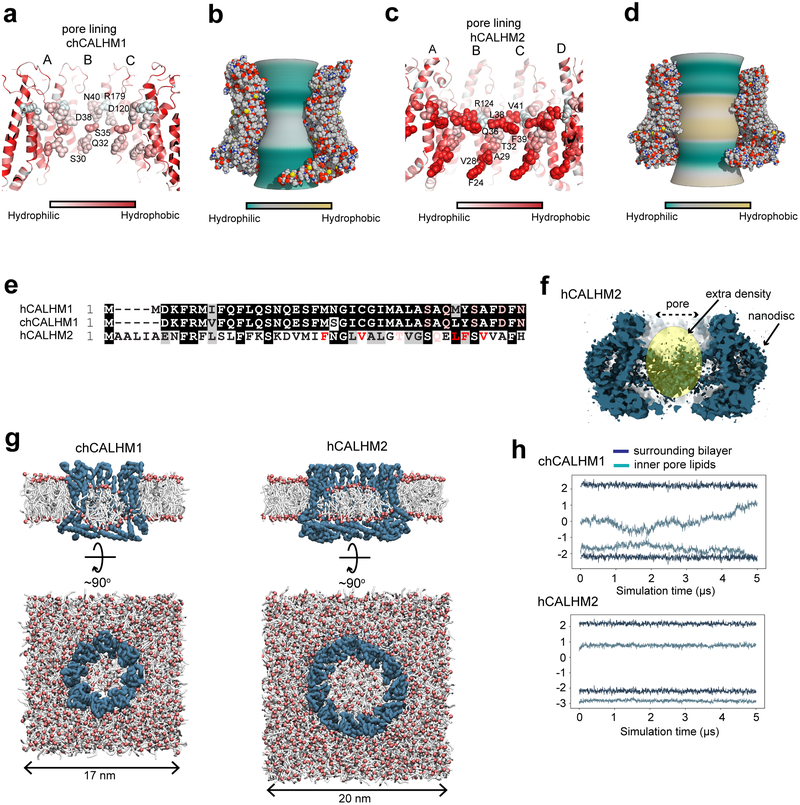
Further structural inspection revealed that the majority of the channel-lining residues (TMD1 and 3) in chCALHM1 are hydrophilic (Fig. 5a–b) whereas those in similar positions in hCALHM2 are hydrophobic (Fig. 5c–e). Thus, we speculated that hydrophobic molecules, for example lipids, may be favorably placed in this hydrophobic channel structure of hCALHM2. In hCALHM2, we observe amorphous density in the middle of the 11-mer assembly, which is discontinuous from the density for proteins (Fig. 5f). In chCALHM1, the density in the middle of the octamer is stronger, more ordered, and continuous from TMD1 (Extended Data Fig. 4). Therefore, the features of the extra cryo-EM density in the middle of the oligomeric assemblies appear to be distinct between chCALHM1 and hCALHM2. To assess if the 11-mer channel structure in hCALHM2 can accommodate lipids in the middle, we conducted molecular dynamics simulations of chCALHM1 and hCALHM2 in the presence of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Fig. 5g–h). Coarse-grained structural models24 of chCALHM1 and hCALHM2 were mixed with POPC lipids and allowed to self-assemble into bilayers25. During replicates of 500 ns simulations, a membrane bilayer formed around both proteins within the first 100 ns, with a number of lipids inside the channel pore. In hCALHM2, the central pore lipids were oriented with a bilayer-like configuration that remained stable, both in simulations of the protein-membrane systems upon conversion into their corresponding atomistic representation as well as in further, extended coarse-grained simulations (up to 5 μs per replicate) (Fig. 5h). By contrast, although phospholipids could also be accommodated within the smaller chCALHM1 pore, they did not assemble into clearly defined or stable inner and upper leaflets (Fig. 5h). These simulations imply that the hydrophobicity and larger 11-mer assembly of hCALHM2 may favor the accommodation of lipid molecules, which prevent ion permeation.
Fig. 5. Comparison of pore properties between chCALHM1 and hCALHM2.
a-d, Channel lining residues (a and c)28 and inner pore surface (b and d; calculated by CHAP29) of chCALHM1 (a-b) and hCALHM2 (c-d) colored based on relative hydrophobicity. These parameters are calculated based on the modeled residues, thus, the amino terminal ends are not taken into account. e, Sequence alignment of the N-terminal residues showing hydrophobic and hydrophilic residues facing the pore with the same color code as in panel a and c. f, Cross-section of the hCALHM2 showing an extra cryo-EM density in the middle of the pore. g, Coarse-grained MD simulations of chCALHM1 (left) and hCALHM2 (right) embedded in POPC membranes. Side (cutaway) and top views of the final frame of one 5 μs replicate are shown in each case, with the protein backbone particles in blue, phospholipid headgroups in red, and acyl tails in white. Water and ions present in the simulation systems are omitted for clarity. h, Headgroup positions of lipids inside each channel pore and in the surrounding bilayer membrane. The average headgroup z-coordinates of lipids constituting the upper- and lower-leaflets in the final frame of each simulation are respectively tracked through the 5 μs simulated duration; results from one replicate are shown for each protein.
Discussion
Our high-resolution structures of CALHM1 and CALHM2 and structural comparison offer novel insights into assembly patterns and function of the CALHM protein family. The protomers of CALHM1 and CALHM2 have similar structural architectures including four transmembrane domains and a long CTH in the cytoplasmic face, which are unique to this protein family. However, CALHM1 and CALHM2 protomers assemble as an 8-mer and 11-mer, respectively, which marks an unusual case where different members of the same channel family form assembly with distinct oligomeric states. These distinct patterns of channel assembly appear to be mediated by the unique modes of interactions between the CTHs. The structures in the current study were solved in the absence of voltage and in the presence of EDTA to remove divalent cations which have been previously shown to inhibit the CALHM1 ion channel. Therefore, the chCALHM1 structure likely represent the active state, however, one cannot exclude a possibility that in the presence of voltage, the channel may undergo further conformational transitions.
The oligomeric states of the CALHM1 and CALHM2 structures in our study were different from the hexameric assembly previously suggested for CALHM1 by Blue Native PAGE and a fluorescence quenching assay of the EGFP-fused CALHM115. While our cryo-EM structures show 8-mer and 11-mer assemblies for CALHM1 and CALHM2, respectively, we do not rule out the possibility that they can also exist in different oligomeric states. The inter-promoter interfaces involve only neighboring promoters and thus, there is a chance that large conformational changes within protomers may trigger different oligomeric assemblies. An example of such a case is calcium/calmodulin-dependent kinase II (CaMKII) where active and inactive protomers can shuffle and form either 12-mers or 14-mers interchangeably26. Under the experimental conditions in our current study (no divalent cations and samples in lipid nanodiscs), we did not observe any evidence of other oligomeric assembly by 2D or 3D classification. Furthermore, there is clear density for the linker between TMD4 and CTH in both CALHM1 and CALHM2 (Extended Data Figs. 3 and 6), thus, indicating that the intrinsic structure of the linker is well-ordered and specific interactions between the CTHs are the major determinant for the observed oligomeric states. Buttressing this hypothesis is our result that the oligomeric state of chCALHM1 can be converted from 8-mer to 11-mer by substituting its cytoplasmic domain with that of hCALHM2 (Fig. 4). The structure of this chimeric construct, which we named CALHM1–2, shows the similar TMD4-CTH angle and mode of inter-CTH interactions to the ones observed in hCALHM2 (Fig. 3d, Fig. 4c–d, and Extended Data Fig. 7e–f). While we have not converted hCALHM2 from 11-mer to 8-mer by the reverse approach at this point, our experimental results strongly support the role of the TMD4-CTH linker and the inter-CTH interactions in defining oligomeric states.
The oligomeric states of CALHMs are also the major determinants for pore-diameters. We measured the pore-diameter of CALHM1 to be approximately 19.5 Å, which is similar to the previously reported diameter, 14 Å, estimated based on a dye permeation assay15. In our CALHM1 structure, the extra cryo-EM density extending from Ile22 does not account for the remainder of the amino-terminal residues, thus, the actual pore diameter may be slightly smaller than 19.5 Å. Nevertheless, one of the important function of CALHM1 is its capability to permeate ATP and the pore size observed here is well suited to mediate this. The 11-mer assembly of CALHM2 results in a much greater pore diameter (~60 Å) compared to that of CALHM1 and weak and amorphous cryo-EM density discontinuous from the CALHM2 proteins was observed in the middle of the channel. While some of the density may be derived from the amino-terminal residues, it is possible that hydrophobic compounds such as lipids exist in the middle of the channel since the channel lining residues of the TMD1 helices are hydrophobic in CALHM2 and are not favored to be exposed to solvent. Such presence of hydrophobic compounds may account for a lack of voltage-gated ion conductance in CALHM2 as reported previously8 and as we confirmed in this study (Fig. 1a). Existence of lipids in an oligomeric membrane protein assembly was previously observed in the structure of the bacteriorhodopin trimers in 2D crystals27. These lipids likely strengthen the trimeric assembly and also prevent ions and metabolites from permeating through the oligomeric center.
In conclusion, our study demonstrates that CAHLM1 and CALHM2 assemble as 8-mers and 11-mers, respectively, and that these different oligomeric states of CALHMs correlate with channel functions, with only the smaller 8-mer assembly displaying ion channel activity. The structural information presented here will serve as the foundation to study mechanistic questions and reagents development for CALHMs.
Methods
Cell lines
HEK293 (ATCC, catalog no. CRL-1573) are adherent cells. Spodoptera frugiperda (Sf9) cells (ThermoFisher) are suspension cells. Cell lines were not authenticated or tested for mycoplasma contamination.
Expression, purification, and nanodisc reconstitution of CALHM1 and CALHM2
The chCALHM1 or CALHM1–2 construct with N-terminally fused Strep-II, 8xHis tag, EGFP, and the 3C protease site (StrepII2-His8-GFP) and the hCALHM2 construct with a C-terminally fused Strep-II tag were expressed in the baculovirus (BV)/Sf9 system under the Drosophila Hsp70 promoter as previously described30. In brief, Sf9 cells were cultured in CCM3 (Invitrogen) supplemented with 1% non-heat inactivated FBS at 27°C, infected with BV at a cell density of 4 × 106 cell/ml, and harvested 48–52 hours after infection. The harvested cell pellets were resuspended in 20 mM Hepes-NaOH (pH 7.5), 200 mM NaCl, 1 mM EDTA and 1 mM PMSF and lysed under high-pressure homogenization (Avestin). The lysate was spun at 4,550g for 20 min and the supernatant was ultracentrifuged at 186,000g for 1 hour at 4°C. The pellet was solubilized in 20 mM Hepes-NaOH pH 7.5, 200 mM NaCl, 1 mM EDTA, and 1% C12E8 (Anatrace) for 2 hours at 4°C and ultracentrifuged at 186,000g for 1 hour at 4°C. The clarified supernatant was loaded onto a Strep-Tactin Sepharose column followed by 20 column volumes (CV) of washing with 20 mM Hepes (pH 7.5), 200 mM NaCl, 1 mM EDTA, 0.01% C12E8 (wash buffer) and elution using the wash buffer supplemented with 3 mM desthiobiotin. The purified hCALHM2 was concentrated to ~2.5 mg/ml at 4°C using 100-kDa MWCO Amicon concentrators (Millipore) before reconstitution into nanodiscs. Purified chCALHM1 or CALHM1–2 was concentrated to 1 mg/ml, digested by trypsin at a weight-to-weight ratio of 1:20 for 1 hour at 18°C to remove StrepII2-His8-GFP, and purified further by size exclusion chromatography using a Superose 6 10/300 column (GE Healthcare) in 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 0.01% C12E8. Peak fractions were pooled and concentrated prior to reconstitution into nanodiscs. For reconstitution into nanodiscs, soybean polar extract, MSP2N2, and the purified CALHM proteins, at final concentrations of 0.75, 0.3 and 0.3 mg/ml, respectively, were mixed for 1 hour at 4°C, followed by detergent removal by SM2 Bio-Beads (BioRad) overnight (~12 hours). The beads were removed and the solution was further purified by size exclusion chromatography using a Superose 6 10/300 column (GE Healthcare) in 20 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA. Peak fractions were pooled and concentrated to ~2.5 mg/ml (hCALHM2) or ~0.6 mg/ml (chCALHM1) for cryo-EM grid preparation. MSP2N2 protein was expressed and purified as previously described31.
Cryo-EM sample preparation, image collection and single particle analysis
3–4 μl of the CALHM-nanodisc complex was applied to glow-discharged 1.2/1.3 400 mesh C-flat carbon coated copper grids (Protochips). The grids were blotted for 4 s with blot force 7 at 85% humidity and 15°C prior to plunge freezing into liquid ethane using a Vitrobot Mark IV (Thermo Fisher). Here, it was critical to use blotting paper which was prewashed with 1 mM EDTA and dried in order to remove contaminations such as divalent cations. Usage of unwashed paper resulted in gap junction formation. Datasets were collected using a Titan Krios operated at an acceleration voltage of 300 keV and the GATAN K2 Summit direct electron detector coupled with the GIF quantum energy filter (Gatan Inc.) controlled by SerialEM software32. Movies were recorded with a pixel size of 1.06 Å, an exposure time of 10 s over 50 frames, and a dose rate of 1.4 e/Å2/frame. For chCALHM1, hCALHM2, and CALHM1–2, the program Warp was used to align movies, estimate the CTF and pick particles33. 2D classification, ab-initio 3D map generation, 3D refinement, 3D classification, per particle CTF refinement and B-factor sharpening were performed using the program cisTEM34. For hCALHM2 gap junction, movie alignment and CTF estimation were carried out using the program Unblur35 and CTFFind436, respectively, within the cisTEM package. Particle picking and the rest of the procedures above were performed using cisTEM. The highest resolution of 3D refinement used was 6 Å for all of the models in this study. The workflows of single particle analyses for chCALHM1, hCALHM2, and hCALHM2 gap junction are outlined in Extended Data Figs. 2, 5, 8, and 9. De novo modeling was done manually using the program Coot37. The final models were refined against the cryo-EM maps using PHENIX real space refinement38 with secondary structure and Ramachandran restraints. The FSCs were calculated by phenix.mtriage. Data collection and refinement statistics are summarized in Table 1. Local resolutions were estimated using the program ResMap39.
Cysteine crosslinking and Western blot analysis
The cysteine substituted Asn226Cys/Arg240Cys hCALHM2 and Arg52Cys/Tyr182Cys hCALHM2, as well as wild-type hCALHM2, were all C-terminally 1D4 tagged and expressed in the BV/Sf9 expression system under the CMV promoter. The membrane fraction were isolated and solubilized as above. The solubilized fraction was subjected to affinity purification by 1D4 antibody conjugated to CNBr-activated agarose (GE Healthcare). The resin was extensively washed and the protein eluted with wash buffer supplemented with 0.2 mg/ml 1D4 peptide. Samples were then either reduced with β-mercaptoethanol, left untreated, or treated with the oxidizing agent (1,10-phenanthroline) copper(II). After 30 min incubation on ice, 1 mM final concentration iodoacetamide was added to samples treated with (1,10-phenanthroline) copper(II). Samples were subjected to Western blot using anti-1D4 monoclonal antibodies (University of British Columbia) and anti-mouse Horseradish peroxidase-conjugated antibodies (GE Healthcare). Protein bands were detected by enhanced chemiluminescence on X-ray film (ECL kit; GE Healthcare). To verify that the in-membrane hCALHM2 assembly corresponded in size to detergent extracted hCALHM2, the membrane fractions containing Asn226Cys/Arg240Cys hCALHM2–1D4 were also oxidized with (1,10-phenanthroline) copper(II) prior to detergent solubilization.
Molecular dynamics
Molecular structures of chCALHM1 and hCALHM2 were separately embedded within POPC bilayer membranes that were solvated on either side at 0.2 M NaCl concentration. Simulation cells were of approximate dimensions 17 × 17 × 15 nm3 (CALHM1) and 20 × 20 × 15 nm3 (CALHM2). Each protein-membrane system was assembled and equilibrated via a previously established protocol40. Simulations were performed with GROMACS 5.141. The MARTINI 2.2 force field42 was used for coarse-grained simulations, with a time-step of 20 fs and an elastic network used to harmonically restrain Cα particles and stabilize the protein structure. Atomistic simulations were run using the OPLS all-atom protein force field with united-atom lipids43 and the TIP4P/2005 water model44. The integration time-step was 2 fs. Temperature and pressure were maintained at 37°C and 1 bar during simulations, using the velocity-rescaling thermostat45 in combination with a semi-isotropic Parrinello and Rahman barostat46, with coupling constants of τT = 0.1 ps and τP = 1 ps, respectively. Bonds were constrained through the LINCS algorithm47. A Verlet cut-off scheme was applied, and long-range electrostatic interactions were calculated using the Particle Mesh Ewald method48.
Electrophysiology
CALHM proteins were expressed in HEK293T cells infected by the recombinant BV harboring chCALHM1 or hCALHM under the CMV promoter. Recordings were obtained ~48 h post infection using borosilicate glass pipettes (Sutter Instruments) pulled and polished to a final resistance of 2–6 MΩ and backfilled with (in mM) 147 NaCl, 10 EGTA, and 10 HEPES pH 7.0 with NaOH. The bath solution contained (in mM) 147 NaCl, 13 glucose, 10 HEPES pH 7.3 with NaOH, 2 KCl, 2 CaCl2, and 1 MgCl2. Recordings performed in the absence of Ca2+ used a similar solution but with no CaCl2 added. A rapid solution exchanger (RSC-200; Bio-logic) was used to perfuse cells with various solutions. All of the recordings were done at 22°C. Data was collected on an AxoPatch 200B patch-clamp amplifier (Axon Instruments), filtered at 2 kHz (Frequency Devices), and digitized with a Digidata 1550B digitizer (Axon Instruments) using a sampling frequency of 10 kHz. Recordings were analyzed using the Clampex 11.0 software (Axon Instruments). Patches were held at −60 mV and stepped between −100 mV and +100 mV in 20 mV increments for 1 s.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
Cryo-EM maps and structural coordinates generated during this study will be deposited in the Electron Microscopy Data Bank with accession codes: EMD-21143 and PDB 6VAM (chCALHM1), EMD-21141 and PDB 6VAK (hCALHM2), EMD-21140 and PDB 6VAI (hCALHM2 gap junction) and EMD-21142 and PDB 6VAL (chCALHM1-hCALHM2 chimera). Source data for Figure 1a are available with the paper online.
Extended Data
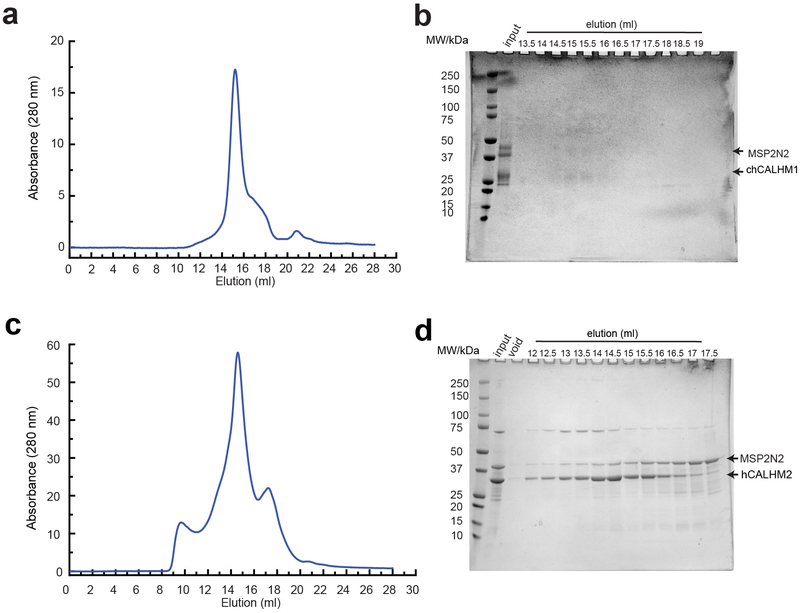
Extended Data Fig. 1. Reconstitution of chCALHM1 and hCALHM2 into lipid nanodiscs.
a, Representative Superose-6 SEC chromatograph of chCALHM1 in MSP2N2 nanodiscs with soy polar extract. b, SDS-PAGE of the fractions collected from SEC. The band for chCALHM1 has a tendency to spread out in SDS-PAGE. Fractions that eluted between 13.5–15.5 ml were pooled, concentrated and subjected to cryo-EM. c, Representative Superose 6 SEC chromatograph of hCALHM2 in MSP2N2 nanodiscs with soy polar extract. d, SDS-PAGE of the fractions collected from SEC. Fractions that eluted between 14.5–16.5 ml were pooled, concentrated and subjected to cryo-EM.
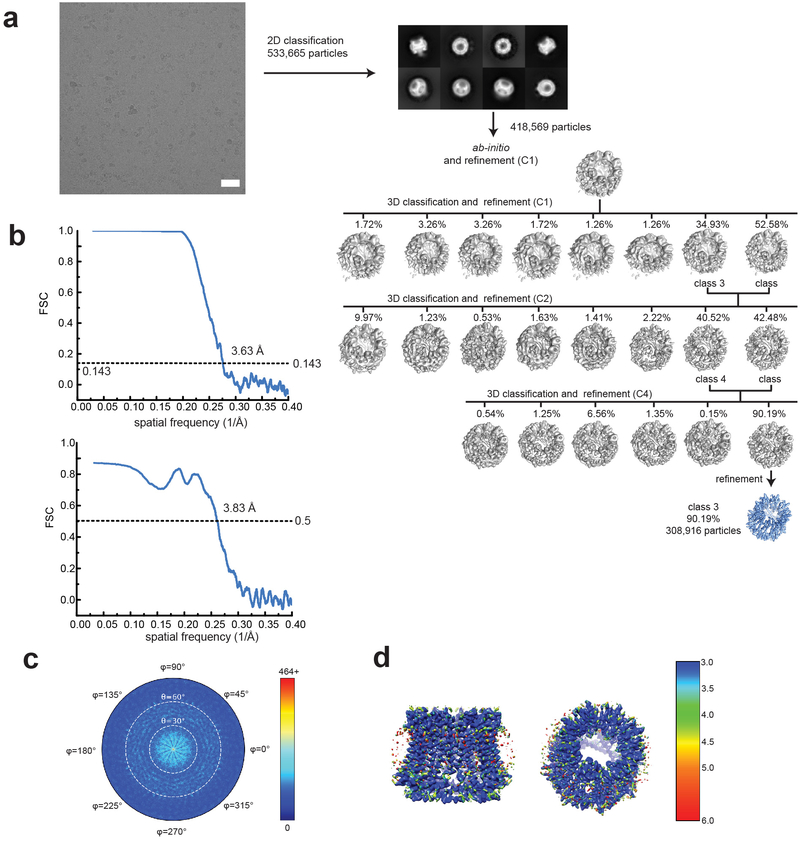
Extended Data Fig. 2. Single particle analysis of chCALHM1.
a. A representative micrograph (scale bar = 38.8 nm), representative 2D class averages, and the 3D classification workflow are shown. b. The FSC plots of the two half maps (top) and the map vs model (bottom) are shown. c. The angular distribution plot for class 3. d. Local resolutions of class 3 were calculated using ResMap.
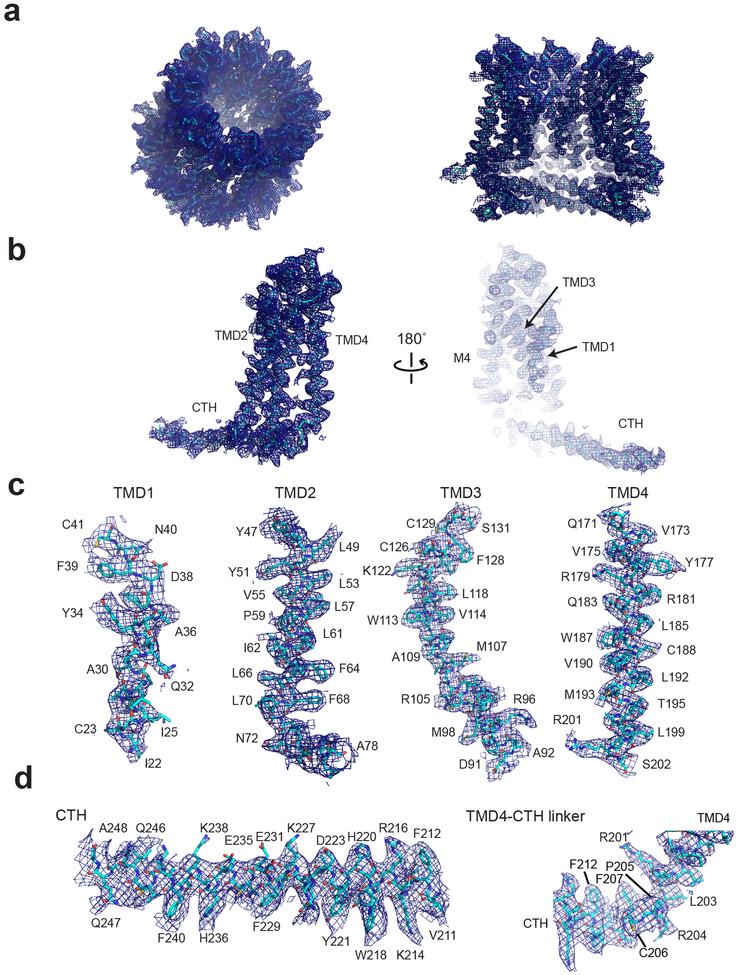
Extended Data Fig. 3. Representative cryo-EM density of chCALHM1.
a, Cryo-EM density of the overall octameric assembly (left) and the cross sectional view of the central cavity (right). b-c, Representative density for a monomer (b), and individual TMDs (c), and a CTH and a TMD4-CTH linker.
Extended Data Fig. 4. Presence of extra cryo-EM density in the chCALHM1 pore.
a, Extra cryo-EM density is observed in the middle of pore-like structure of the chCALHM1. Here the pore-density and the density for only subunit H are shown for clarity. TMD1 and the pore-density are continuous (arrow). b, The density observed from the top of the extracellular region. The diameter of the pore is 19.5 Å.
Extended Data Fig. 5. Single particle analysis of hCALHM2.
a, A representative micrograph (scale bar = 38.8 nm), representative 2D class averages, and the 3D classification workflow are shown. b, The FSC plots of the two half maps (top) and the map vs. model (bottom) are shown for class 8. c, The angular distribution plot for class 8. d, Local resolutions of class 8 were calculated using ResMap.
Extended Data Fig. 6. Representative cryo-EM density of hCALHM2.
a, Cryo-EM density of the overall 11-mer assembly (left) and the cross-sectional view of the central cavity (right). b-c, Representative density for a monomer (b), and individual TMDs (c) and a CTH and a TMD4-CTH linker.
Extended Data Fig. 7. Interaction of hCALHM2 subunits.
a-b, The hCALHM2 structure viewed from the top of extracellular region (a) and the side of the membrane (b). Shown in spheres are the Arg124 residues at the equivalent position to chCALHM1 Asp120 or hCALHM1 Asp121. c, Arg124 (sphere) and surrounding residues (sticks) form polar and hydrophobic interactions to mediate inter-subunit interactions. d-e, The inter-subunit interactions between TMD2 and TMD4 (d) and CTHs (e). f, The schematic presentation of the interactions between two CTHs (magenta and slate blue) in hCALHM2 (top) and chCALHM1 (bottom). Polar and van der Waals interactions mediated by hydrophobic residues (ovals) are shown as dashed and solid lines, respectively. The lines in magenta are the conserved interactions between chCALHM1 and hCALHM2. The residues in italic are the equivalent ones in hCALHM1.
Extended Data Fig. 8. Single particle analysis of CALHM1–2.
a, A representative micrograph (scale bar = 40.5 nm), representative 2D class averages, and the 3D classification workflow are shown. b, The FSC plots of the two half maps (top) and the map vs. model (bottom) are shown for class 8. c, The angular distribution plot for class 8. d, Local resolutions of class 8 were calculated using ResMap.
Extended Data Fig. 9. Single particle analysis of 22-meric hCALHM2.
a, A representative micrograph (scale bar = 38.8 nm), representative 2D class averages, and the 3D classification workflow are shown. b, The FSC plots of the two half maps (top) and the map vs. model (bottom) are shown for class 8. c, The angular distribution plot for class 8. d, Local resolutions of class 8 were calculated using ResMap.
Extended Data Fig. 10. Structure of hCALHM2 gap junction.
a, Cryo-EM density of the 22-meric hCALHM2 viewed from the side of the membrane and from the cytoplasm. b, The structural models in the same orientation as the cryo-EM density in (a), showing locations of the TMD2–4 and the CTH. There is little or no structural change between the 22-mer and 11-mer structures except for the extracellular region (due to the inter-11-mer interaction). The interaction between the two hemichannels is mediated by His147, His152, and Glu145 in the extracellular loop between TMD3 and TMD4. Density between His147 and His152 is continuous implying the potential presence of a divalent cation. Residues from the upper and lower hemichannels are annotated with black and gray fonts. Ovals are placed at the interaction sites.
Supplementary Material
Acknowledgements
We would like to thank D. Thomas and M. Wang for managing the cryo-EM facility and the computing facility at Cold Spring Harbor Laboratory, respectively. A. Hoffmann and T. Malinauskas were involved in the early phase of the research related to hCALHM2. This work was supported by NIH (NS113632 and MH085926), Robertson funds at Cold Spring Harbor Laboratory, Doug Fox Alzheimer’s fund, Austin’s purpose, and Heartfelt Wing Alzheimer’s fund (to H.F.), Howard Hughes Medical Institute (to N.G.), and the Biotechnology and Biological Sciences Research Council (to S.J.T.). J.L.S. is supported by the Charles H. Revson Senior Fellowship in Biomedical Science.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
References
- 1.Jun M et al. Calhm2 governs astrocytic ATP releasing in the development of depression-like behaviors. Mol Psychiatry 23, 1091 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Vingtdeux V et al. CALHM1 deficiency impairs cerebral neuron activity and memory flexibility in mice. Sci Rep 6, 24250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreses-Werringloer U et al. A polymorphism in CALHM1 influences Ca2+ homeostasis, Abeta levels, and Alzheimer’s disease risk. Cell 133, 1149–61 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taruno A et al. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature 495, 223–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Z et al. Calcium homeostasis modulator 1 (CALHM1) is the pore-forming subunit of an ion channel that mediates extracellular Ca2+ regulation of neuronal excitability. Proc Natl Acad Sci U S A 109, E1963–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vingtdeux V et al. Effect of the CALHM1 G330D and R154H human variants on the control of cytosolic Ca2+ and Abeta levels. PLoS One 9, e112484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Workman AD et al. CALHM1-Mediated ATP Release and Ciliary Beat Frequency Modulation in Nasal Epithelial Cells. Sci Rep 7, 6687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Z et al. CALHM3 Is Essential for Rapid Ion Channel-Mediated Purinergic Neurotransmission of GPCR-Mediated Tastes. Neuron 98, 547–561 e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanis JE et al. CLHM-1 is a functionally conserved and conditionally toxic Ca2+-permeable ion channel in Caenorhabditis elegans. Journal of Neuroscience 33, 12275–12286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawate T & Gouaux E Fluorescence-Detection Size-Exclusion Chromatography for Precrystallization Screening of Integral Membrane Proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Regan MC et al. Structural Mechanism of Functional Modulation by Gene Splicing in NMDA Receptors. Neuron 98, 521–529 e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harauz G & van Heel M Exact Filters for General Geometry Three Dimensional Reconstruction. Optik 73, 146–156 (1986). [Google Scholar]
- 13.Rosenthal PB & Henderson R Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–45 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Slabinski L et al. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics 23, 3403–5 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Siebert AP et al. Structural and functional similarities of calcium homeostasis modulator 1 (CALHM1) ion channel with connexins, pannexins, and innexins. Journal of Biological Chemistry 288, 6140–6153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanis JE, Ma ZM & Foskett JK The NH2 terminus regulates voltage-dependent gating of CALHM ion channels. American Journal of Physiology-Cell Physiology 313, C173–C186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oshima A et al. Atomic structure of the innexin-6 gap junction channel determined by cryo-EM. Nature Communications 7, 13681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karakas E & Furukawa H Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344, 992–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CH et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 511, 191–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda S et al. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 458, 597–602 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Deneka D, Sawicka M, Lam AKM, Paulino C & Dutzler R Structure of a volume-regulated anion channel of the LRRC8 family. Nature 558, 254–259 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Kasuya G et al. Cryo-EM structures of the human volume-regulated anion channel LRRC8. Nat Struct Mol Biol 25, 797–804 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Vingtdeux V et al. CALHM1 ion channel elicits amyloid-beta clearance by insulin-degrading enzyme in cell lines and in vivo in the mouse brain. J Cell Sci 128, 2330–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Jong DH et al. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J Chem Theory Comput 9, 687–97 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Stansfeld PJ et al. MemProtMD: Automated Insertion of Membrane Protein Structures into Explicit Lipid Membranes. Structure 23, 1350–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharyya M et al. Molecular mechanism of activation-triggered subunit exchange in Ca2+/calmodulin-dependent protein kinase II. eLife 5, e13405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grigorieff N, Ceska TA, Downing KH, Baldwin JM & Henderson R Electron-crystallographic Refinement of the Structure of Bacteriorhodopsin. Journal of Molecular Biology 259, 393–421 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Eisenberg D, Schwarz E, Komaromy M & Wall R Analysis of membrane and surface protein sequences with the hydrophobic moment plot. Journal of Molecular Biology 179, 125–142 (1984). [DOI] [PubMed] [Google Scholar]
- 29.Klesse G, Rao S, Sansom MSP & Tucker SJ CHAP: A Versatile Tool for the Structural and Functional Annotation of Ion Channel Pores. J Mol Biol 431, 3353–3365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regan MC et al. Structural Mechanism of Functional Modulation by Gene Splicing in NMDA Receptors. Neuron 98, 521–529 e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie TK et al. Chapter 11 Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs. Methods in Enzymology 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schorb M, Haberbosch I, Hagen WJH, Schwab Y & Mastronarde DN Software tools for automated transmission electron microscopy. Nat Methods 16, 471–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tegunov D & Cramer P Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grant T, Rohou A & Grigorieff N cisTEM, user-friendly software for single-particle image processing. Elife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant T & Grigorieff N Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. eLife 4, e06980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of Structural Biology 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stansfeld PJ et al. MemProtMD: Automated Insertion of Membrane Protein Structures into Explicit Lipid Membranes. Structure 23, 1350–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abraham MJ et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 42.de Jong DH et al. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J Chem Theory Comput 9, 687–97 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Jorgensen WL, Maxwell DS & Tirado-Rives J Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. Journal of the American Chemical Society 118, 11225–11236 (1996). [Google Scholar]
- 44.Abascal JLF & Vega C A general purpose model for the condensed phases of water: TIP4P/2005. The Journal of Chemical Physics 123, 234505 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Bussi G, Donadio D & Parrinello M Canonical sampling through velocity rescaling. The Journal of Chemical Physics 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Parrinello M & Rahman A Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics 52, 7182–7190 (1981). [Google Scholar]
- 47.Hess B, Bekker H, Berendsen HJC & Fraaije JGEM LINCS: A Linear Constraint Solver for molecular simulations. Journal of Computational Chemistry 18, 1463–1472 (1997). [Google Scholar]
- 48.Darden T, York D & Pedersen L Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. The Journal of Chemical Physics 98, 10089–10092 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps and structural coordinates generated during this study will be deposited in the Electron Microscopy Data Bank with accession codes: EMD-21143 and PDB 6VAM (chCALHM1), EMD-21141 and PDB 6VAK (hCALHM2), EMD-21140 and PDB 6VAI (hCALHM2 gap junction) and EMD-21142 and PDB 6VAL (chCALHM1-hCALHM2 chimera). Source data for Figure 1a are available with the paper online.