Abstract
Myogenesis is controlled by a well-established transcriptional hierarchy that coordinates the activities of a set of muscle genes. Recently, roles in myogenesis have been described for non-coding RNAs, including a role of circular RNA (circRNA) to regulate muscle gene expression. However, the functions of circRNA and the underlying mechanism by which circRNAs affect myogenesis remain poorly understood. In this study, we analyzed circRNA high-throughput sequencing results of bovine skeletal muscle samples and constructed a circRNA-miRNA-mRNA network according to the competitive endogenous RNA (ceRNA) theory. The putative circHUWE1-miR-29b-AKT3 network was analyzed and its involvement in myogenesis was confirmed through a series of assays. To assess the potential function of this regulation, bovine myoblasts were infected with overexpression plasmids and small interfering RNAs (siRNAs) that target circHUWE1. Next, cell proliferation, apoptosis, and differentiation were analyzed using Cell Counting Kit-8 (CCK-8), 5-ethynyl-2′-deoxyuridine (EdU), flow cytometry, western blotting, and qRT-PCR assays. The results suggest that circHUWE1 facilitates bovine myoblast proliferation and inhibits cell apoptosis and differentiation. Next, bioinformatics, dual-luciferase reporter assay, and AGO2 RNA immunoprecipitation (RIP) approaches were used to verify the interaction between circHUWE1, miR-29b, and AKT3. Subsequently, we identified that circHUWE1 could directly interfere with the ability of miR-29b to relieve AKT3 suppression, which ultimately activates the AKT signaling pathway. These findings suggested a new regulatory pathway for bovine skeletal muscle development, and they also expand our understanding of circRNA functions in mammals.
Keywords: circHUWE1-miR-29b-AKT3, ceRNA, skeletal muscle
Introduction
Vertebrate skeletal muscle plays a crucial role in overall body metabolism, is mostly derived from the paraxial mesodermal somites, and undergoes a complex series of hyperplasia and hypertrophy processes successively.1,2 Myogenic cells proliferate to multiply cell numbers and then fuse to form multinucleated myotubes, which can then undergo further differentiation during embryogenesis, postnatal growth, and regeneration.3,4 This process is precisely coordinated by several well-established transcription factors, including the myogenic regulatory factors (MRFs) and members of the myocyte enhancer factor 2 (MEF2) family.5,6 In recent years, increasing evidence has suggested a potential role for non-coding RNAs (ncRNAs) in skeletal muscle development at the post-transcriptional level.7
MicroRNAs (miRNAs) are highly conserved endogenous ncRNAs that can target the 3′ untranslated regions (UTRs) of genes to negatively regulate gene expression. Several miRNAs, such as miR-1, miR-133, miR-206, and miR-499, have been reported to exert crucial roles in myogenesis.8 Another major class of ncRNAs is long ncRNAs (lncRNAs), which have mRNA-like transcripts longer than 200 nt. The lncRNAs usually function as competing endogenous RNAs (ceRNAs) by competitively binding to miRNA response elements (MREs) to relieve the suppression of target mRNAs.9 Acting as ceRNAs, lncRNAs such as H19,10 lncMyoD,9 linc-MD1,11 and lnc-mg12 have been shown to participate in myogenesis. Additionally, circular RNAs (circRNAs), a novel class of ncRNAs lacking 5′ caps and 3′ poly tails, have been widely detected in eukaryotes due to the improvement of RNA sequencing (RNA-seq) methodology and more sophisticated bioinformatics.13 Accumulating studies show that circRNAs can participate in various physiopathologic processes by mediating protein-RNA interactions,14,15 serving as miRNA or protein sponges,16 or modulating protein translation,17 mostly by acting as ceRNA to relieve suppression. The covalently closed-loop structure of circRNA makes these RNAs more stable than miRNAs and lncRNAs, which may make then more efficient in post-transcriptional regulation. Evidence of the effects of circRNA came from studies of ciRS-7 and Sry, acting as effective sponges of miR-7 and miR-138, respectively, to regulate target mRNA expression.18 However, there has been little characterization of the roles of circRNAs in myogenesis.
In this study, we reconstructed a circRNA-associated ceRNA network in bovine skeletal muscle development. The interactions of the circHUWE1-miR-29b-AKT3 axis were screened and the potential related functions of this axis and the underlying mechanisms affecting regulation of myogenesis were further explored. The data showed that circHUWE1 promoted myoblast proliferation and reduced cell apoptosis and differentiation by sponging miR-29b and targeting AKT3, which indirectly activates the AKT signaling pathways. Overall, these findings provide new insight into the ceRNA role of circRNA in myogenesis.
Results
Identification and Analysis of the circRNA-miRNA-mRNA Network
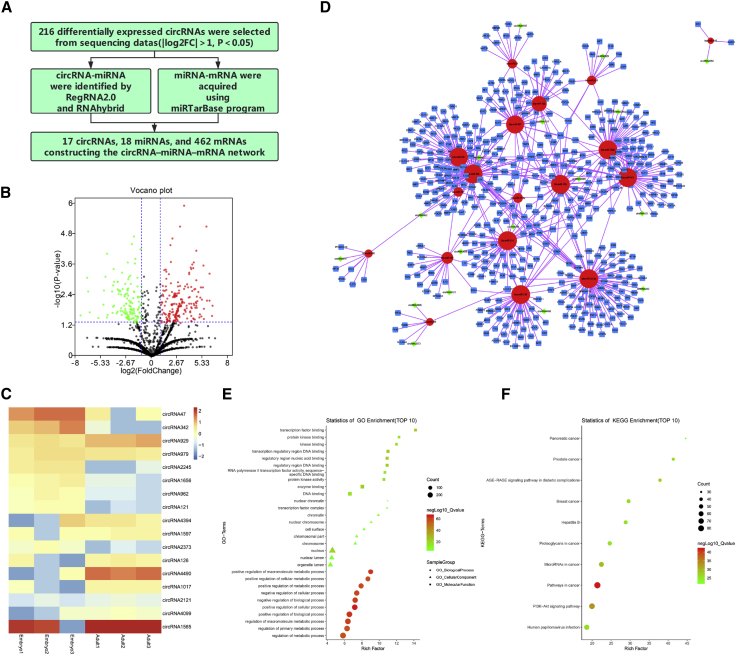
After processing (Figure 1A), 216 differentially expressed circRNAs were selected from our sequencing data (Figure 1B). As described in Materials and Methods, we identified 17 co-expressed circRNAs (Figure 1C), 18 co-expressed miRNAs, and 462 co-expressed mRNAs, which were selected to construct the circRNA-miRNA-mRNA network (Figure 1D). In this network, each node represents a biological molecule, and the edges represent the interactions between nodes. The node degree typifies the number of edges linked to a given node, where the higher the degree, the bigger is the size, which corresponds to biological functions. Interestingly, Gene Ontology (GO) analysis results revealed that these co-expressed RNAs were mainly involved in cell metabolism regulation (Table S1; Figure 1E), and pathway analysis manifested that the included mRNAs were enriched in the phosphatidylinositol 3-kinase (PI3K)-AKT signaling pathway (Table S2; Figure 1F). These results suggested close links between the circRNA-miRNA-mRNA network and myogenesis.
Figure 1.
circRNA-miRNA-mRNA Network Construction and Analysis
(A) Schematic of data processing. (B) Volcano plot of 216 differentially expressed circRNAs for embryo versus adult. Criteria are based on |log2 FC| >1 and p values <0.05. (C) Heatmap of 17 co-expressed circRNAs identified by comparison of embryonic and adult stages. (D) Global view of the circRNA-miRNA-mRNA network in bovine skeletal muscle development, including 17 circRNAs (green diamonds), 18 miRNAs (red circles), and 462 mRNAs (blue squares). (E and F) Advanced bubble chart of GO (E) and KEGG (F) analysis.
Identification of the Circular Structure and Expression Levels of circHUWE1
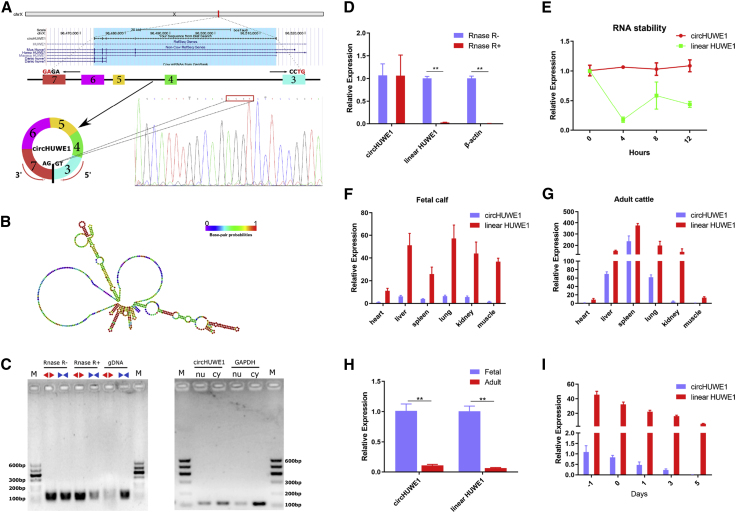
The circRNA47 was termed circHUWE1 because it derives from exons 3–7 of the gene encoding HECT, UBA, and WWE domain containing E3 ubiquitin protein ligase 1 (HUWE1), which is located on chromosome X (Table S3; Figure 2A). The circHUWE1 back-splicing junction was verified by Sanger sequencing, and the secondary structure of circHUWE1 was predicted using the RNAfold web server (Figure 2B). To confirm the circular structure of circHUWE1, convergent primers for the HUWE1 mRNA and divergent primers to amplify circHUWE1 were designed. We detected circHUWE1 expression using the two sets of primers in PCR reactions with cDNA, RNase R-treated cDNA, or genomic DNA (gDNA) isolated from bovine skeletal muscle. The PCR reactions were visualized by agarose gel electrophoresis. The results showed the successful amplification of circular transcripts by divergent primers using the cDNA or the RNase R-treated cDNA as template, but not when gDNA was used as the template. Linear transcripts were amplified by convergent primers in PCR reactions containing cDNA or gDNA, but were only weakly detected when RNase R-treated cDNA was used as template (Figure 2C). To further investigate the circHUWE1, nuclear and cytoplasmic localization experiments were performed and revealed that circHUWE1 was mainly localized in the cytoplasm (Figure 2C). Next, resistance to the RNase R exonuclease was assayed by a qRT-PCR assay. As shown in Figure 2D, the expression levels of linear HUWE1 and β-actin were obviously decreased by RNase R, while the circHUWE1 transcripts were resistant to RNase R treatment. To assess the stability of circHUWE1, myoblasts were treated with actinomycin D to inhibit transcription and then the half-lives of circHUWE1 and linear HUWE1 were measured. The results showed that circHUWE1 was more stable than linear HUWE1 (Figure 2E). We also measured the expression of circHUWE1 and linear HUWE1 in a variety of bovine tissues and myoblasts. Consistent with our RNA-seq data, expression was widely expressed and showed a significant decrease during myogenesis (Figures 2F–2I).
Figure 2.
circHUWE1 Structure and Expression Profiles
(A and B) The head-to-tail splicing of circHUWE1 was confirmed by Sanger sequencing (A), and the secondary structure was predicted (B). (C) PCR and agarose gel electrophoresis assay with divergent or convergent primers indicated the existence of circHUWE1, and circHUWE1 was shown to mainly localize to the cytoplasm. (D and E) Resistance to RNase R (D) and actinomycin D (E) was tested using qRT-PCR assay, and the results indicated that circHUWE1 was more stable than linear HUWE1. (F–I) Expression of circHUWE1 and linear HUWE1 in bovine tissues from fetal calf (F) or adult cattle (G), and myoblast-induced proliferation and differentiation (I) was measured, revealing wide expression with a significant decrease during myogenesis (H). *p < 0.05, **p < 0.01.
circHUWE1 Promotes the Proliferation of Myoblasts
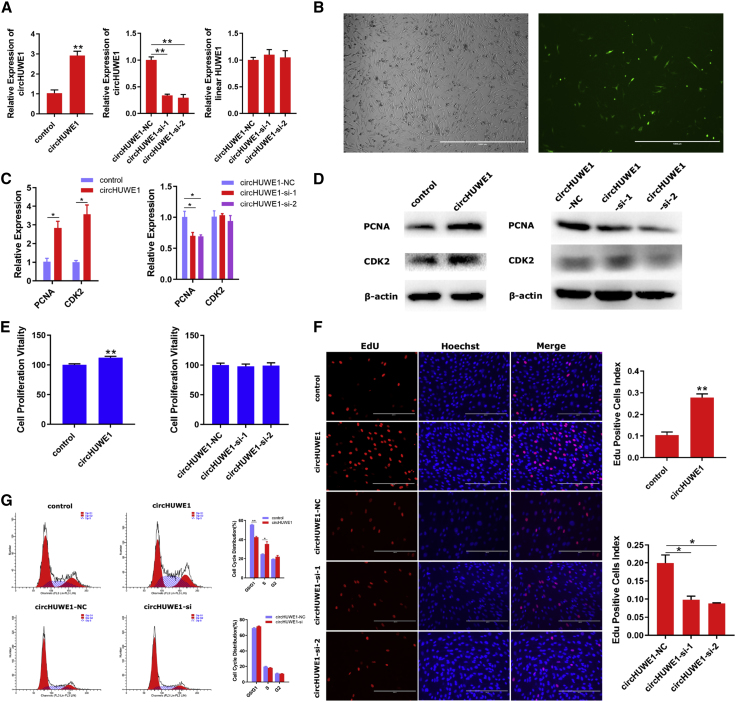
Using a circHUWE1-overexpression vector and backsplice junction-specific small interfering RNA (siRNA), circHUWE1 expression in primary bovine myoblasts was dramatically increased or reduced in cells cultured for 24 h (Figures 3A and 3B). Cells were collected for qRT-PCR and western blot analysis against PCNA and CDK2, markers of proliferation. The overexpression of circHUWE1 markedly increased the mRNA (Figure 3C) and protein (Figure 3D) levels of both PCNA and CDK2. In addition, inhibition of circHUWE1 decreased mRNA (Figure 3C) and protein (Figure 3D) expression of PCNA; however, the knockdown of circHUWE1 did not affect the levels of CDK2. Cell Counting Kit-8 (CCK-8) and 5-ethynyl-2′-deoxyuridine (EdU) proliferation assays revealed that overexpression of circHUWE1 significantly promoted the proliferation of primary bovine myoblasts (Figures 3E and 3F). However, an effect of circHUWE1 knockdown to dramatically restrain cell proliferation was only seen in EdU proliferation assays (Figure 3F). We further evaluated the cell cycle distribution by flow cytometry assay. The results suggested that circHUWE1 overexpression induced cell cycle arrest at the G0/G1 phase and increased the percentage of myoblasts in the S phase, but circHUWE1 knockdown did not affect the cell cycle distribution (Figure 3G), possibly due to its low background expression level. Collectively, these findings support a role of circHUWE1 to promote myoblast proliferation.
Figure 3.
circHUWE1 Promotes the Proliferation of Myoblasts
(A) circHUWE1 was successfully overexpressed and inhibited in primary bovine myoblasts after culturing for 24 h. (B) Microscopic images of myoblasts (green, RFP) after culturing for 24 h. Scale bars, 1,000 μm. (C and D) mRNA (C) and protein (D) expression levels of PCNA and CDK2 24 h after transfection with circHUWE1-overexpression vector and circHUWE1-si. (E and F) CCK-8 (E) and EdU (F) assays were used to measure the ability of proliferation. Scale bars, 200 μm. (G) The cell cycle distribution was evaluated using a flow cytometry assay. Data presented are from three independent experiments. *p < 0.05, **p < 0.01.
circHUWE1 Inhibited the Apoptosis and Differentiation of Myoblasts
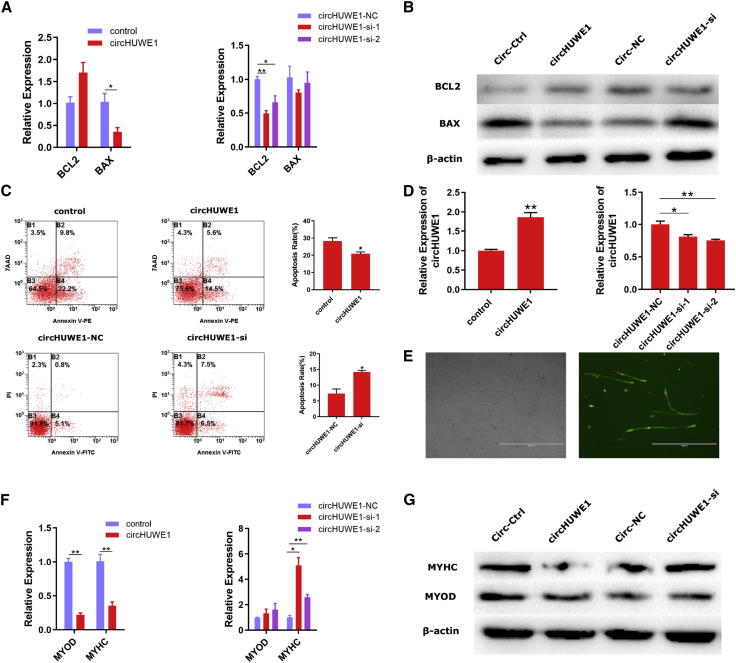
qRT-PCR and western blot analysis were performed against BCL2 and BAX to detect apoptosis (Figures 4A and 4B). Overexpression and knockdown of circHUWE1 markedly affected the protein levels of these markers of apoptosis (Figure 4B), but their mRNA levels showed little change (Figure 4A). Apoptosis assays also revealed that circHUWE1 has an apoptosis-resistance characteristic in primary bovine myoblasts (Figure 4C). For phenotypic validation of differentiation, circHUWE1 was successfully overexpressed and inhibited on day 3 of differentiation (Figures 4D and 4E). At the same time, the mRNA and protein levels of differentiation markers MYOD and MYHC were measured. We observed that circHUWE1 overexpression decreased the mRNA and protein levels of MyoD and MyHC (Figures 4F and 4G), and knockdown of circHUWE1 suggested an increase in the mRNA and protein expression levels of these markers (Figures 4F and 4G). Hence, our results suggest that circHUWE1 inhibited the apoptosis and differentiation of myoblasts.
Figure 4.
circHUWE1 Affects the Apoptosis and Differentiation of Myoblasts
(A and B) The mRNA (A) and protein (B) expression levels of BCL2 and BAX were determined 24 h after transfection with circHUWE1-overexpression vector and circHUWE1-si. (C) Annexin V-APC/7AAD staining revealed that circHUWE1 exhibits an apoptosis-resistance characteristic in primary bovine myoblasts. (D) circHUWE1 was successfully overexpressed and inhibited in primary bovine myoblasts after culturing for 3 days. (E) Microscopic images of myoblasts (green, RFP) cultured for 3 days. Scale bars, 1,000 μm. (F and G) The mRNA (F) and protein (G) expression levels of MYOD and MYHC 3 days after transfection with circHUWE1-overexpression vector and circHUWE1-si were detected by qRT-PCR and western blot, respectively. Values are means ± SEM for three individuals. *p < 0.05, **p < 0.01.
circHUWE1 Functions as a Sponge for miR-29b to Target AKT3 in Myoblasts
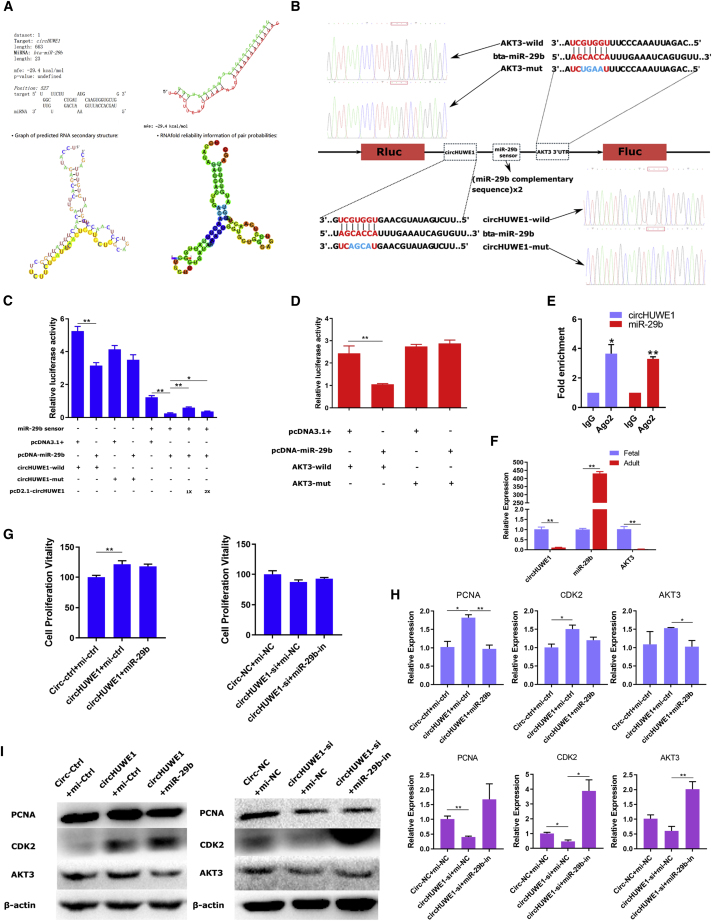
The circHUWE1-miR-29b-AKT3 axis was identified from our constructed circRNA-miRNA-mRNA network. The potential binding site for circHUWE1 to bind miR-29b was visualized using the RNAhybrid and RegRNA 2.0 programs (Figure 5A), and vectors were constructed to confirm this interaction using a luciferase reporter assay (Figure 5B). A notable inhibition was observed upon co-transfection of pcDNA-miR-29b and circHUWE1-wild, but the circHUWE1-mutant (mut) no longer responded to pcDNA-miR-29b inhibition (Figure 5C). To further determine whether circHUWE1 acts as a sponge to sequester miR-29b, a sensor of miR-29b activity was established that contains two copies of the miR-29b binding sites in the 3′ UTR region of a Renilla luciferase gene (Rluc). Co-transfection of pcDNA-miR-29b and this miR-29b sensor resulted in significantly inhibited Rluc expression, and Rluc activity was dramatically restored in a dose-dependent manner by co-transfection with pcD2.1-circHUWE1 (Figure 5C). Analogously, the miR-29b-AKT3 relationship was confirmed by luciferase reporter assay (Figure 5D), as previously established.19 We also applied the Ago2 RNA immunoprecipitation (RIP) assay to confirm the circHUWE1-miR-29b interaction. As expected, circHUWE1 and miR-29b were efficiently pulled down by anti-Ago2, but not by the non-specific anti-IgG antibody (Figure 5E). Interestingly, the expression trend of circHUWE1, miR-29b, and AKT3 in skeletal muscle from fetal and adult bovines suggested that circHUWE1 sponges miR-29b to reverse degradation or translation repression of AKT3 (Figure 5G). Furthermore, we found that increased miR-29b expression could rescue the effect of circHUWE1 on proliferation in myoblasts; conversely, decreasing miR-29b expression also rescued the effect of circHUWE1-siRNA (circHUWE1-si) at the level of transcription and translation (Figures 5H and 5I). However, this rescue was not evidenced in the CCK-8 assay (Figure 5G).
Figure 5.
circHUWE1 Functions as a miR-29b Sponge to Target AKT3 in Myoblasts
(A) The proposed circHUWE1 binding site on miR-29b was identified using RNAhybrid and RegRNA 2.0 programs. (B) Luciferase reporter vectors. (C) circHUWE1-wild, circHUWE1-mut, and the miR-29b sensor were co-transfected into HEK293T cells with pcDNA-miR-29b, together with 1× or 2× sponge plasmid pcD2.1-circHUWE1. Luciferase activities were measured 24 h after transfection. (D) AKT3 wild and AKT3 mutant were transfected into HEK293T cells together with pcDNA-miR-29b. Luciferase activities were measured 24 h after transfection. (E) A RIP assay was conducted using an antibody against IgG or Ago2, and circHUWE1 and miR-29b were detected by qRT-PCR. (F) Expression of circHUWE1, miR-29b, and AKT3 in skeletal muscle samples from fetal and adult bovines. (G) CCK-8 assay for myoblasts after co-transfection with circHUWE1-overexpression vector and pcDNA-miR-29b, or with the circHUWE1-si and miR-29b inhibitor. (H and I) qRT-PCR (H) and western blot (I) result of proliferation markers in myoblasts co-transfected with the circHUWE1-overexpression vector and pcDNA-miR-29b, or with the circHUWE1-si and miR-29b inhibitor. Values are means ± SEM for three individuals. *p < 0.05, **p < 0.01.
circHUWE1 Facilitated Bovine Myoblast Proliferation and Inhibited Cell Apoptosis and Differentiation through the miR-29b-AKT3 Pathway
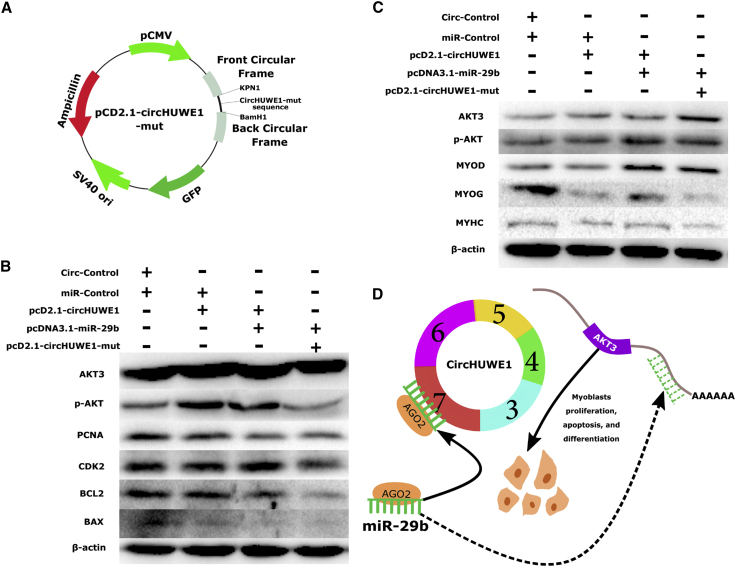
To further characterize the effect of sponging miR-29b in myoblast proliferation, apoptosis, and differentiation, a pcD2.1-circHUWE1-mut lacking the miR-29b binding site was constructed (Figure 6A), and we transfected primary bovine myoblasts with pcD2.1 (circ-control), pcDNA3.1 (miR-control), pcD2.1-circHUWE1, pcDNA3.1-miR-29b, and pcD2.1-circHUWE1-mut to measure the expression levels of AKT3 and marker genes. Transfection with pcDNA3.1-miR-29b partly rescued the effect of pcD2.1-circHUWE1 on the expression of AKT3 and marker genes, and pcD2.1-circHUWE1-mut significantly restored the effect of pcD2.1-circHUWE1 on myoblast proliferation and apoptosis (Figure 6B) and differentiation (Figure 6C), consistent with the results of cell function. Collectively, these observations demonstrate that circHUWE1 promotes proliferation and inhibits the apoptosis and differentiation of myoblasts, at least partly through the miR-29b-AKT3 pathway (Figure 6C).
Figure 6.
circHUWE1 Promoted Cell Proliferation and Inhibited Cell Apoptosis and Differentiation through the miR-29b-AKT3 Axis in Myoblasts
(A) pcD2.1-circHUWE1-mut vector. (B) pcD2.1-circHUWE1, pcDNA3.1-miR-29b, and pcD2.1-circHUWE1-mut were co-transfected into primary bovine myoblasts to measure AKT3 levels, and proliferation and apoptosis marker expression was measured by western blot. (C) pcD2.1-circHUWE1, pcDNA3.1-miR-29b, and pcD2.1-circHUWE1-mut were co-transfected into primary bovine myoblasts to measure expression of AKT3 and differentiation markers by western blot. (D) Schematic illustration of the circHUWE1-miR-29b-AKT3 axis.
Discussion
Recently, circRNAs have been widely identified in animal cells due to improved methods for high-throughput sequencing and bioinformatics analysis.20 Unlike other ncRNAs, circRNAs have specific properties such as higher stability, RNase R resistance, and longer half-lives due to their unique circular structure.21 In addition, circRNAs act in a tissue- and developmental stage-specific manner, suggesting possible roles in functional regulation.13 Among potential functions of circRNA, ceRNA effects, by which circRNA sponges miRNAs to alleviate degradation or translation repression of target mRNAs, have been increasingly described, especially in processes related to oncology.22 Many studies showed that ceRNA crosstalk could be related to processes of myogenesis, with the emergence of lncRNAs as potent regulators. However, only few ceRNA functions of circRNAs have been characterized to date. For instance, circLMO7 sponges miR-378a-3p to promote proliferation but inhibit apoptosis and differentiation of myoblasts;23 circFUT10 restrains proliferation and facilitates differentiation of myoblasts by sponging miR-133a;24 and circFGFR4 functions as a ceRNA for miR-107 to promote myoblast differentiation.25 We constructed and visualized the potential ceRNA network of circRNAs using our circRNA high-throughput sequencing data of samples at different stages of bovine skeletal myogenesis, with the hypothesis that differentially expressed circRNAs might be associated with myogenesis. In this network, 7-mer sites, the effective canonical site type base pairing to nucleotides 2–8 at the 5′ end of the miRNA defined as the “seed region,” were filtered from the circRNA-miRNA interactions.26 Additionally, miRNA-mRNA pairs were acquired from the experimentally validated miRNA-target interactions database, supporting this circRNA-miRNA-mRNA network.27 Next, the potential functions of circRNAs were inferred by studying the annotated functions of mRNAs, and the results suggested a possible correlation between the circRNA-associated ceRNA crosstalk and the effects of PI3K-AKT signaling on skeletal muscle. Above all, this network allowed screening for potential circRNA-associated ceRNA crosstalk in myogenesis.
The circHUWE1-miR-29b-AKT3 crosstalk was examined more closely, and we observed a decrease in the expression of circHUWE1 and AKT3 developmentally in bovine skeletal muscle from fetal to adult growth, while miR-29b showed the opposite result. Preliminarily, these results are consistent with their possible activities in myogenesis based on the ceRNA mechanism. circHUWE1 is made of five exons (663 bp) from the HUWE1 gene, and it is mainly located in the cytoplasm. Expression results revealed that circHUWE1 expression was lower than that of linear HUWE1, which may arise from the lower efficiency of back-splicing required for the formation of circRNAs.28 Nevertheless, circHUWE1 was differentially expressed in skeletal muscles in samples from the fetal and adult stages, suggesting a potential regulatory role in myogenesis. This is the first report that circHUWE1 can promote bovine myoblast proliferation and inhibit cell apoptosis and differentiation by specifically targeting miR-29b/AKT3. miR-29b has been involved in diverse physiological and pathological processes, including proliferation,29 apoptosis,30 and cell differentiation.31 AKT3 is mainly known for its pathophysiologic role as an AKT subunit in AKT signaling.32 The tumor-suppression effect of miR-29b/AKT3 has been demonstrated extensively.33 Similarly, miR-29b was previously reported to target Akt3 to suppress proliferation and promote differentiation of myoblasts,19 which further supports our conclusion.
Consistently, our results showed that circHUWE1 overexpression significantly upregulates the protein level of phosphorylated (p-)Akt (S473), a marker of AKT activation, whose level was significantly downregulated by circHUWE1-mut treatment. Therefore, circHUWE1 may serve as an important activator of Akt signaling in myoblasts. Skeletal muscle development in vertebrates is a complex process that is orchestrated by a combination of genetic and environmental factors, in which hormones and growth factors act synergistically to impact muscle phenotype at the cellular level.34 As a serine (S)/threonine (T) kinase, Akt can phosphorylate and change the activity of many critical metabolic targets in myoblasts. For example, Akt stimulates protein synthesis and decreases protein catabolism by phosphorylating substrates TSC2 and transcription factor FoxO,35,36 respectively. Akt also phosphorylates the pro-apoptotic protein Bad to inhibit apoptosis in skeletal muscle.37 AKT has also been shown to phosphorylate IKKα and facilitate its activation of nuclear factor κB (NF-kB) signaling,38 which inhibits myogenic differentiation by stimulating cell cycle progression,39 indirectly repressing the synthesis of late-stage differentiation genes via recruiting YinYang1 (YY1),40 and even reducing the MyoD protein level by post-transcriptional mechanisms.41 Accordingly, Akt is situated at a critical juncture in muscle signaling, and the regulation effect of circHUWE1 in our study is probably mediated by the Akt signaling pathway.
Notably, miR-29b and AKT3 may be involved in muscle diseases, such as atrophy, hypertrophy, and primary muscle disorders. A previous study showed that knockdown of miR-29b in vivo induced cardiac fibrosis in mice, suggesting that miR-29b regulates cardiac fibrosis and represents a potential therapeutic target for tissue fibrosis.42 Cardiac hypertrophy induced by AKT3 overexpression was found in cardiac-specific AKT3 transgenic mice, suggesting that inhibition of AKT3 might represent a novel therapeutic approach for cardiac hypertrophy.43 In addition, there is significantly increased expression of miR-29b in multiple in vivo and in vitro models of muscle atrophy. Mechanistically, miR-29b can inhibit the IGF-1-PI3K-AKT pathway by targeting both IGF-1 and PI3K(p85a), and inhibition of AKT leads to the activation of FOXO3, which will induce muscle atrophy by inducing increased expression of atrophy-linked ubiquitin ligases, including Atrogin-1 (MAFBX) and muscle-specific RING-finger 1 (MURF-1).44 To date, there have been few studies of regulation of circRNAs in muscle disorders, and based on our previous analysis, the circHUWE1-miR-29b-AKT3 axis may include potential targets for treatment of muscular diseases.
One limitation of our study is that the circHUWE1 functional research was performed only at the cellular level (in vitro), due to convenience. However, the complex modulation of the extracellular matrix during skeletal muscle development is overlooked by in vitro assays.45 Most in vivo studies of ncRNA in vivo have focused on the pathological state of mature muscles and changes due to atrophy and muscular dystrophies, but the in vivo requirement of circHUWE1 in mammal myogenesis through targeted deletion remains unknown. Furthermore, existing results suggest that the activity of ceRNA is affected by a series of factors, including the relative abundances of ceRNAs and shared miRNAs,46,47 binding affinity,15,48 RNA binding proteins (RBPs),49 RNA editing,50 and the subcellular localization of ceRNAs51 in pathophysiologic environments. Accordingly, it is unclear whether ceRNA crosstalk is possible within a physiological range of transcript abundance in vivo. Intriguingly, in contrast to the positive effect of muscle-specific miRNAs such as miR-1,52 miR-133,53 and miR-20654 in cultured skeletal myotubes, their genomic ablation in mouse models had no apparent effect on normal muscle development.55 Thus, little is known of the roles of the circRNA ceRNA network in skeletal myogenesis, and comprehensive elucidation of ceRNA functions of circRNAs is significantly needed.
To our knowledge, this is the first study that constructs a circRNA-associated ceRNA network for Chinese Qinchuan bovine skeletal muscle development. Functionally and mechanistically, circHUWE1 was verified as a sponge of miR-29b, which eliminates the suppressive effect on its target gene AKT3 and leads to the development of myoblasts by activating AKT signaling. This is the first study to elucidate the role of circHUWE1 in skeletal myogenesis, and the circHUWE1-miR-29b-AKT3 axis may, therefore, be a potential therapeutic target for muscle-wasting-related disorders.
Materials and Methods
Ethics Statement
This study was approved by the Ethics Committee of Northwest A&F University and carried out according to the Administration of Affairs Concerning Experimental Animals published by the Ministry of Science and Technology. Heart, liver, spleen, lung, kidney, and longissimus muscle samples were collected from Qinchuan cattle at the embryonic stage (90 days) and the adult stage (24 months old) at Shannxi Kingbull Animal Husbandry (Baoji, China). Adult cattle were not fed the night before they were humanely slaughtered.
circRNA-miRNA-mRNA Network Construction and Analysis
The circRNA-miRNA-mRNA network was constructed and visualized using Cytoscape software (http://www.cytoscape.org/). The network was based on our circRNA sequencing data, which were obtained from Chinese Qinchuan bovine skeletal muscles in different stages (embryonic, 90 days old; adult, 24 months old).23 In data processing, differentially expressed circRNAs between the two developmental stages were screened as the |log2 fold change (FC)| >1 and p values <0.05 standard, and co-expression of RNA pairs was assessed by bioinformatics analysis using different data analysis tools such as RNAhybrid, RegRNA 2.0, and miRTarBase programs. Functional annotations such as GO analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis were performed for the exploration of circRNA ceRNA in myogenesis.
circRNA Screening and Specificity Confirmation
The circHUWE1-miR-29b-AKT3 axis was selected from the constructed network, and the circHUWE1 was immediately confirmed by Sanger sequencing, divergent primer PCR, RNase R, and actinomycin D treatment. Divergent and convergent primers of circHUWE1 were designed to amplify circHUWE1 or linear HUWE1 mRNA using cDNA and gDNA templates, respectively, from bovine skeletal muscles. The circHUWE1 was only amplified by divergent primers in cDNA, and the PCR products were then identified by Sanger sequencing. For RNase R treatment, total RNA (1 mg) from bovine skeletal muscles was treated with 2 U/mg RNase R for 20 min at 37°C, and then the myoblasts were incubated with actinomycin D (1 mg/mL) and total cellular RNA was isolated according to the manufacturer’s instructions. These treated RNAs were then reverse transcribed into cDNA to further assess the stability of circHUWE1 and linear HUWE1 mRNA using qRT-PCR.
Cell Culture
Primary bovine myoblasts were isolated from bovine longissimus muscle as previously described, and were then cultured in DMEM/high-glucose medium with 20% fetal bovine serum (Gibco, USA) and 1% penicillin-streptomycin (HyClone, USA). HEK293T cell lines were cultured in 10% fetal bovine serum and 1% penicillin-streptomycin, which were provided by our laboratory. In order to induce differentiation, primary bovine myoblasts were incubated with differentiation medium (DMEM supplemented with 2% horse serum and 1% penicillin-streptomycin). All cell lines were cultured at 37°C with 5% CO2.
Vector Construction, siRNAs, and Transfection
To overexpress circHUWE1 and miR-29b, the full-length cDNA of circHUWE1 or the primary (pri)-miR-29b fragment were separately cloned into the pcD2.1 vector (Geneseed, China) and the pcDNA3.1(+) vector (Invitrogen, USA), respectively. The siRNAs to target circHUWE1 and miR-29b inhibitors were synthesized by RiboBio (Guangzhou; China) to knock down circHUWE1 and miR-29b. The circHUWE1 full-length sequence with mutational sites pairing to the miR-29b seed region was also amplified using overlap PCR, and was then inserted into the pcD2.1 vector to construct the pcD2.1-circHUWE1-mut vector. Here, all vectors were verified by sequencing, and transfections were conducted with R0531 (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
RNA Extraction and Real-Time qRT-PCR Analysis
Total RNA from tissues and cell lysates was generally extracted using TRIzol reagent (Takara, China). However, RNA from nucleus and cytoplasm was isolated with a nucleoprotein extraction kit (Sangon Biotech, China) according to the kit instructions. For circRNA, miRNA, and mRNA, cDNA was synthesized using random, stem-loop, and oligo(dT)18 primers, respectively, using a RT reagent kit (Takara, China). qRT-PCR was performed in triplicate using a SYBR Premix Ex Taq II kit (Takara, China) on the CFX connect real-time system (Bio-Rad, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as the internal control for circRNA and mRNA, while U6 was used for miRNA, and relative gene quantitation was calculated by the 2−ΔΔCT method. All primers are listed in Table S4.
Western Blotting Assay
Total protein from primary bovine myoblasts was extracted using radioimmunoprecipitation assay (RIPA) buffer supplemented with 1 mM PMSF (Solarbio, China), and harvested protein was quantified by bicinchoninic acid (BCA) analysis (Solarbio, China) and an equal amount was boiled in SDS. Proteins in the supernatant were then separated by SDS-PAGE gels at 120 V for 2 h and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Germany) at 200 mA for 2.5 h. Afterward, the membranes were sealed with 5% skim milk and incubated overnight at 4°C with primary antibodies as follows: AKT3, p-AKT, PCNA, CDK2, BCL2, BAX, MyoD, MyoG, MyHC, and β-actin. The next day, the PVDF membranes were further incubated with horseradish peroxidase (HRP)-conjugated secondary antibody; after washes, signals were finally visualized using a chemiluminescence system (Bio-Rad, USA).
Cell Proliferation, Cell Cycle, and Apoptosis Assays
For cell proliferation assays, primary bovine myoblasts were seeded into 96-well plates at a density of 2,000 cells/well and then cultured for 24 h after transfection. Each treatment group in this study had eight independent replicates. 10 μL of CCK-8 (Solarbio, China) solution was then added to each well, and after 2 h of incubation at 37°C in the dark, the absorbance of each experimental well was measured with a microplate reader (BioTek, USA) at 450 nm. Cell viability was also measured with an EdU DNA cell proliferation kit (RiboBio, China) according to the manufacturer’s instructions, and images were finally obtained by fluorescence microscopy (DM5000B; Leica Microsystems, Germany). To assess the cell cycle and apoptosis, myoblasts were seeded in 60-mm plates and cultured for 24 h after transfection. Transfected cells for cell cycle analysis were harvested to stain with propidium iodide (PI) and then measured by flow cytometry (BD Biosciences, USA). For apoptosis detection, cells were double stained with allophycocyanin (APC)-conjugated annexin V and 7-aminoactinomycin D (7AAD) using an annexin V-APC/7AAD apoptosis kit (eBiosciences, USA) and analyzed on a flow cytometer. Experiments were independently performed in triplicate.
Luciferase Reporter Assay
For the luciferase reporter assay, the full-length cDNA of circHUWE1 and AKT3-3′ UTR and their corresponding mutant versions without miR-29b binding sites were cloned into a psiCHECK-2 vector (Promega, USA) and confirmed by sequencing, termed circHUWE1-wild, circHUWE1-mut, AKT3-wild, and AKT3-mut, respectively. Furthermore, the miR-29b biosensor (miR-29b sensor) was created by inserting two copies of miR-29b consecutive sites into the psiCHECK-2 vector. These plasmids and miR-29b overexpression vector were then co-transfected into 293T cells separately; after 24 h, cells were lysed and luciferase activities were examined by a dual-luciferase reporter assay system (Promega, USA).
RIP Assay
A RIP assay was performed using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, USA). Specifically, primary bovine myoblasts were harvested and lysed in RIP lysis buffer after 24 h of culture. The cells were then divided into two equal parts and incubated at 4°C overnight with magnetic beads that were conjugated with an antibody against non-specific immunoglobulin G (IgG) (Millipore, USA) or Ago2 (Abcam, USA). After washing with wash buffer, the RNA that was bound to the magnetic beads was purified using an RNA extraction reagent (phenol/chloroform/isoamylol at 25:24:1), and the abundance of circHUWE1 and miR-29b was detected by qRT-PCR.
Statistical Analysis
GraphPad Prism 8.0 (GraphPad, USA) and SPSS 22.0 (SPSS, USA) were used for data analyses. All data presented herein are expressed as the mean ± standard error of the mean (SEM). An independent-sample t test was used to compare two groups, and one-way analysis of variance (ANOVA) was used to compare three or more groups. The statistical significance is indicated as *p < 0.05 and **p < 0.01.
Author Contributions
B.Y. and H.C. conceived the ideas and designed the work. B.Y. performed the experiments and drafted the manuscript. J. Wang, W.R., J. Wu, X.C., and H.Y. helped perform the experiments and analyzed the data. Y.H. and X.L. provided suggestions about the experiments. B.H. provided some samples. C.L. revised the manuscript critically. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (no. 31772574), the Program of National Beef Cattle and Yak Industrial Technology System (CARS-37), and the Program of Yunling Scholar.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.12.039.
Supplemental Information
References
- 1.Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S., Montarras D., Rocancourt D., Relaix F. The formation of skeletal muscle: from somite to limb. J. Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo B., Greenwood P.L., Cafe L.M., Zhou G., Zhang W., Dalrymple B.P. Transcriptome analysis of cattle muscle identifies potential markers for skeletal muscle growth rate and major cell types. BMC Genomics. 2015;16:177. doi: 10.1186/s12864-015-1403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin H., Price F., Rudnicki M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hindi S.M., Shin J., Gallot Y.S., Straughn A.R., Simionescu-Bankston A., Hindi L., Xiong G., Friedland R.P., Kumar A. MyD88 promotes myoblast fusion in a cell-autonomous manner. Nat. Commun. 2017;8:1624. doi: 10.1038/s41467-017-01866-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kassar-Duchossoy L., Gayraud-Morel B., Gomès D., Rocancourt D., Buckingham M., Shinin V., Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 6.Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H.K., Lee Y.S., Sivaprasad U., Malhotra A., Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong C., Li Z., Ramanujan K., Clay I., Zhang Y., Lemire-Brachat S., Glass D.J. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell. 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Kallen A.N., Zhou X.B., Xu J., Qiao C., Ma J., Yan L., Lu L., Liu C., Yi J.S., Zhang H. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu M., Liu J., Xiao J., Yang L., Cai M., Shen H., Chen X., Ma Y., Hu S., Wang Z. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 2017;8:14718. doi: 10.1038/ncomms14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 14.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbagallo D., Caponnetto A., Brex D., Mirabella F., Barbagallo C., Lauretta G., Morrone A., Certo F., Broggi G., Caltabiano R. circSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers (Basel) 2019;11:E194. doi: 10.3390/cancers11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 17.Wesselhoeft R.A., Kowalski P.S., Parker-Hale F.C., Huang Y., Bisaria N., Anderson D.G. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell. 2019;74:508–520.e4. doi: 10.1016/j.molcel.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Wei W., He H.B., Zhang W.Y., Zhang H.X., Bai J.B., Liu H.Z., Cao J.H., Chang K.C., Li X.Y., Zhao S.H. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013;4:e668. doi: 10.1038/cddis.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo L., Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patop I.L., Wüst S., Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S., Huang V., Xu X., Livingstone J., Soares F., Jeon J., Zeng Y., Hua J.T., Petricca J., Guo H. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176:831–843.e22. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 23.Wei X., Li H., Yang J., Hao D., Dong D., Huang Y., Lan X., Plath M., Lei C., Lin F. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017;8:e3153. doi: 10.1038/cddis.2017.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Yang J., Wei X., Song C., Dong D., Huang Y., Lan X., Plath M., Lei C., Ma Y. circFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J. Cell. Physiol. 2018;233:4643–4651. doi: 10.1002/jcp.26230. [DOI] [PubMed] [Google Scholar]
- 25.Li H., Wei X., Yang J., Dong D., Hao D., Huang Y., Lan X., Plath M., Lei C., Ma Y. circFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Mol. Ther. Nucleic Acids. 2018;11:272–283. doi: 10.1016/j.omtn.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 29.Mundy-Bosse B.L., Scoville S.D., Chen L., McConnell K., Mao H.C., Ahmed E.H., Zorko N., Harvey S., Cole J., Zhang X. MicroRNA-29b mediates altered innate immune development in acute leukemia. J. Clin. Invest. 2016;126:4404–4416. doi: 10.1172/JCI85413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mott J.L., Kobayashi S., Bronk S.F., Gores G.J. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L., Zhou L., Jiang P., Lu L., Chen X., Lan H., Guttridge D.C., Sun H., Wang H. Loss of miR-29 in myoblasts contributes to dystrophic muscle pathogenesis. Mol. Ther. 2012;20:1222–1233. doi: 10.1038/mt.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Cai B., Shen L., Dong Y., Lu Q., Sun S., Liu S., Ma S., Ma P.X., Chen J. miRNA-29b suppresses tumor growth through simultaneously inhibiting angiogenesis and tumorigenesis by targeting Akt3. Cancer Lett. 2017;397:111–119. doi: 10.1016/j.canlet.2017.03.032. [DOI] [PubMed] [Google Scholar]
- 34.Frost R.A., Lang C.H. Skeletal muscle cytokines: regulation by pathogen-associated molecules and catabolic hormones. Curr. Opin. Clin. Nutr. Metab. Care. 2005;8:255–263. doi: 10.1097/01.mco.0000165003.16578.2d. [DOI] [PubMed] [Google Scholar]
- 35.Potter C.J., Pedraza L.G., Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 36.Sakaguchi M., Cai W., Wang C.H., Cederquist C.T., Damasio M., Homan E.P., Batista T., Ramirez A.K., Gupta M.K., Steger M. FoxK1 and FoxK2 in insulin regulation of cellular and mitochondrial metabolism. Nat. Commun. 2019;10:1582. doi: 10.1038/s41467-019-09418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou B.H., Tan P.P., Jia L.S., Zhao W.P., Wang J.C., Wang H.W. PI3K/AKT signaling pathway involvement in fluoride-induced apoptosis in C2C12 cells. Chemosphere. 2018;199:297–302. doi: 10.1016/j.chemosphere.2018.02.057. [DOI] [PubMed] [Google Scholar]
- 38.Barré B., Perkins N.D. Retraction. EMBO J. 2014;33:1978. doi: 10.15252/embj.201470010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitin N., Kudla A.J., Konieczny S.F., Taparowsky E.J. Differential effects of Ras signaling through NFκB on skeletal myogenesis. Oncogene. 2001;20:1276–1286. doi: 10.1038/sj.onc.1204223. [DOI] [PubMed] [Google Scholar]
- 40.Wang H., Hertlein E., Bakkar N., Sun H., Acharyya S., Wang J., Carathers M., Davuluri R., Guttridge D.C. NF-κB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol. Cell. Biol. 2007;27:4374–4387. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dogra C., Changotra H., Mohan S., Kumar A. Tumor necrosis factor-like weak inducer of apoptosis inhibits skeletal myogenesis through sustained activation of nuclear factor-κB and degradation of MyoD protein. J. Biol. Chem. 2006;281:10327–10336. doi: 10.1074/jbc.M511131200. [DOI] [PubMed] [Google Scholar]
- 42.van Rooij E., Sutherland L.B., Thatcher J.E., DiMaio J.M., Naseem R.H., Marshall W.S., Hill J.A., Olson E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taniyama Y., Ito M., Sato K., Kuester C., Veit K., Tremp G., Liao R., Colucci W.S., Ivashchenko Y., Walsh K., Shiojima I. Akt3 overexpression in the heart results in progression from adaptive to maladaptive hypertrophy. J. Mol. Cell. Cardiol. 2005;38:375–385. doi: 10.1016/j.yjmcc.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Li J., Chan M.C., Yu Y., Bei Y., Chen P., Zhou Q., Cheng L., Chen L., Ziegler O., Rowe G.C. miR-29b contributes to multiple types of muscle atrophy. Nat. Commun. 2017;8:15201. doi: 10.1038/ncomms15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M., FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherji S., Ebert M.S., Zheng G.X., Tsang J.S., Sharp P.A., van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Figliuzzi M., Marinari E., De Martino A. MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophys. J. 2013;104:1203–1213. doi: 10.1016/j.bpj.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M. Impact of microRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol. Cell. 2016;64:565–579. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawahara Y., Megraw M., Kreider E., Iizasa H., Valente L., Hatzigeorgiou A.G., Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stalder L., Heusermann W., Sokol L., Trojer D., Wirz J., Hean J., Fritzsche A., Aeschimann F., Pfanzagl V., Basselet P. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013;32:1115–1127. doi: 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 53.Liu N., Bezprozvannaya S., Williams A.H., Qi X., Richardson J.A., Bassel-Duby R., Olson E.N. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams A.H., Valdez G., Moresi V., Qi X., McAnally J., Elliott J.L., Bassel-Duby R., Sanes J.R., Olson E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sayed D., Abdellatif M. MicroRNAs in development and disease. Physiol. Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.