Abstract
One of the most prevalent forms of post-transcriptional RNA modification is the conversion of adenosine-to-inosine (A-to-I), mediated by adenosine deaminase acting on RNA (ADAR) enzymes. The advent of the CRISPR/Cas systems inspires researchers to work actively in the engineering of programmable RNA-guided machines for basic research and biomedical applications. In this regard, CIRTS (CRISPR-Cas-Inspired RNA Targeting System), RESCUE (RNA Editing for Specific C to U Exchange), RESTORE (Recruiting Endogenous ADAR to Specific Transcripts for Oligonucleotide-mediated RNA Editing), and LEAPER (Leveraging Endogenous ADAR for Programmable Editing of RNA) are innovative RNA base-editing platforms that have recently been engineered to perform programmable base conversions on target RNAs mediated by ADAR enzymes in mammalian cells. Thus, these four currently characterized RNA-editing systems constitute novel molecular tools with compelling programmability, specificity, and efficiency that show us some creative ways to take advantage of the engineered deaminases for precise base editing. Moreover, the advanced engineering of these systems permits editing of full-length transcripts containing disease-causing point mutations without the loss of genomic information, providing an attractive alternative for in vivo research and in the therapeutic setting if the challenges encountered in off-target edits and delivery are appropriately addressed. Here, I present an analytical approach of the current status and rapid progress of the novel ADAR-mediated RNA-editing systems when highlighting the qualities of each new RNA-editing platform and how these RNA-targeting strategies could be used to recruit human ADARs on endogenous transcripts, not only for our understanding of RNA-modification-mediated regulation of gene expression but also for editing clinically relevant mutations in a programmable and straightforward manner.
Keywords: ADAR, RNA editing, point mutations, gRNA, editing platforms
Main Text
Role of ADAR in RNA Editing
RNA epitranscriptomic regulatory mechanisms represent a significant part of RNA homeostasis when markedly influencing the transcripts fate and RNA regulatory processes in cells without altering the ribonucleotide sequence.1,2
RNA editing can result from insertion, deletion, or substitution of nucleotides.3 In humans, adenosine-to-inosine (A-to-I) RNA editing is the most common post-transcriptional nucleotide modification.4,5 Based on bioinformatic analyses and deep targeted sequencing, Bazak et al.5 estimated that there are more than 100 million human Alu RNA-editing sites distributed in the human transcriptome.
A-to-I editing is catalyzed by adenosine deaminase acting on RNA (ADAR) enzymes, whose substrates are double-stranded RNAs (dsRNAs).6,7 Three human ADAR genes have been identified (adars 1–3) with ADAR1 (official symbol ADAR) and ADAR2 (ADARB1) proteins having well-characterized adenosine deamination activity.7,8 ADAR3 (ADARB2) is expressed in the human brain, but its function remains unknown because no deaminase activity has been reported for this protein,9 probably because of its inability to homodimerize, and it is thought to act as a competitive inhibitor of ADAR1 and ADAR2 in the brain.10,11 ADARs have a typical modular domain organization that includes at least two copies of a dsRNA binding domain (dsRBD; ADAR1with three dsRBDs; ADAR2 and ADAR3 with two copies) in their N-terminal region followed by a C-terminal deaminase domain.12
Splicing and editing are the two main processes contributing to transcriptome diversity.4 Although infrequently, A-to-I RNA-editing targets canonical splicing acceptor, donor, and branch sites, it was found to affect splicing regulatory elements within exons.4 For instance, Beghini et al.13 showed that RNA editing at the branch site of PTPN6 (protein tyrosine phosphatase, nonreceptor type 6) gene in acute myeloid leukemia patients was found to impair splicing events, with an apparent function in leukemogenesis.
Moreover, in instances where mRNA coding sequence is affected, frameshifts and codon sense changes can have profound effects on protein structure and function. One of the first attempts to correct a mutated RNA by deamination of A-to-I was through the strategy of Woolf et al.14 Initially, a portion of a human dystrophin mutated sequence containing a stop codon was fused in-frame to the luciferase coding region to monitor whether the correction could occur.14 Once authors formed duplexes between the RNA oligonucleotide complementary to the premature stop codon on the target transcript, the oligonucleotide-mRNA hybrids were microinjected into one-cell-stage Xenopus embryos observing a significant increase in luciferase expression as a consequence of the UAG stop codon correction by deamination of A-to-I.14 However, no further mechanism was characterized.
In contrast, Stafforst and Schneider15 pioneered in engineering ADAR fusion protein for RNA-guided, site-directed RNA editing. SNAP tag is an engineered mutant of the DNA repair protein O6-alkylguanine-DNA alkyltransferase that forms a covalent bond with O6-benzylguanine (BG).16 The deaminase domain of a human ADAR was fused to SNAP tag that enables the targeting of the fusion with a short (∼20-nt) guide RNA (gRNA).17 The assembly of editase and gRNA is mediated by covalent bond formation (SNAP-ADAR/BG-gRNA).17 As a result, the gRNA directs the engineered fusions to the target RNA to form an RNA duplex, usually containing an A:C mismatch at the target site, to induce site-specific deamination.17 This strategy has been used for the highly specific repair of point mutations in mRNAs by site‐selective editing. In this regard, the potential of SNAP-deaminases for site-directed RNA repair was demonstrated by Vogel et al.17 when repairing, for the first time, the Factor 5 Leiden polymorphism in vitro. This single-point mutation (G1746→A) represents the most abundant genetic risk factor inheritable multifactorial thrombophilia in the Caucasian population.18 More recently, ADAR’s deaminase domain and gRNAs have been covalently linked (SNAP-tagged ADARs approach) for the efficient and simultaneous site-directed editing of transcripts KRAS (two 5′-UAG-3′ sites in KRAS mRNA) and STAT1 (the Tyr701 phosphorylation site [5′-UAU-3′] in STAT1 mRNA), which would be appropriate for the manipulation of signaling proteins.19 Thus, the SNAP tag technology represents a suitable method to assemble gRNA-protein conjugates for transcript-specific RNA editing in vitro, in cell culture, and in vivo.17,19,20
Although A-to-I substitution is a single-nucleotide change, it can have a significant physiological or clinical impact.21 Nevertheless, RNA editing can safely be approached because engineering RNA lowers the risk for permanent genomic changes, and even though off-site RNA editing can occur, it could be reversible.22 Concerning the above subject, efficient programmable RNA-editing tools have previously been designed to achieve targeted RNA editing.19,23,24 For example, human ADAR2’s deaminase domain has been fused (non-covalent interactions) with the RNA binding domain of the λ bacteriophage antiterminator protein (λN peptide-ADAR/boxB RNA hairpin-gRNA) to guide site-specific mRNA editing and correct premature termination in the cystic fibrosis transmembrane conductance regulator anion channel (CFTR) transcript in Xenopus oocytes.23
Another typical example of amino acid substitution is the editing of the glutamate receptor GluR2 transcript at two sites, the R/G and the Q/R site, with the latter one being essential for nervous system function.25 The above led Wettengel et al.26 to develop an elegant strategy to harness wild-type human ADAR2 and stimulate site-selective RNA editing. Thereby, by the ectopic expression of short, structured gRNAs, they mimicked the intronic R/G-motif of the glutamate receptor transcript and recruited human ADAR2 to stimulate A-to-I conversion.26 Also, employing this successful design of gRNAs that enable the re-addressing of human ADAR2 toward specific sites, the authors promoted the recoding of a premature stop codon (UAG) into tryptophan (UIG) to repair a recessive loss-of-function mutation in PINK1 (W437X) in HeLa cells.26 The above showed a functional rescue of PINK1/Parkin-mediated mitophagy26 (process of autophagy by which damaged depolarized mitochondria are eliminated), which is linked to the etiology of Parkinson’s disease (PD).27 Hence this strategy demonstrates the possibility of the approach to repair a neuron-related disease-causing point mutation, and its use could extend to numerous mutations present in other genes associated with inherited forms of PD.
In recent years, several new RNA-targeting platforms based on Cas proteins have been developed, including Cas13.24 For instance, Cox et al.24 fused the ADAR2 deaminase domain (ADAR2DD) to the catalytically inactive Cas13b effector protein (dCas13) to create the RNA Editing for Programmable A-to-I Replacement (REPAIR) system. By using this RNA-editing platform, the authors achieved A-to-I conversion in endogenous transcripts and the correction of two disease-relevant mutations: 878G>A (AVPR2 W293X) in X-linked nephrogenic diabetes insipidus and 1517G>A (FANCC W506X) in Fanconi anemia.24
With the advent of high-throughput detection and coupled with the development of sequencing-based techniques for transcriptome-wide identification (e.g., fourth-generation sequencing technologies such as Oxford Nanopore Technologies [ONT]) and mapping of RNA modifications (e.g., by antibody-based detection methods), new types of RNA chemical modifications are being discovered.1,2 However, the detection of new levels of control in the epitranscriptome emphasizes the need to explore new site-directed RNA-targeting approaches. Interestingly, so far this year, new approaches that add to existing methods have been developed to elicit programmable, ADAR-mediated RNA editing. These elegant strategies enable editing with broad codon range, notable precision, and efficiency, as well as the opportunity for multiplexing, which highlights the scientific progress in site-specific RNA editing (Table 1).
Table 1.
Comparison of the Main Features of the Novel Programmable RNA-Editing Platforms
| CIRTS30 | RESCUE32 | RESTORE33 | LEAPER36 | |
|---|---|---|---|---|
| Targeted base motifs on the transcripts evaluated | G-to-A mutation in the coding region of firefly luciferase gene | simultaneous targeting of an A and a C in the transcripts | 5′-UAG-3′ triplet in the 3′ UTRs; 5′-UAU-3′ and CAA motifs in the ORF regions | 5′-UAG-3′, 5′-UAC-3′, 5′-AAG-3′, 5′-CAG-3′ motifs |
| Guide RNA (gRNA) | 20–40 nt | 30 nt | short single-stranded sequence (63–95 nt) | ∼111–151 nt long for high editing efficiency |
| Deaminase | ADAR2 | ADAR2 | endogenous ADARs | endogenous ADARs |
| RNA-editing strategy | a gRNA with an engineered hairpin interacts with the hairpin RNA binding domain to drive a ribonucleoprotein complex formation | dRanCas13b-ADAR2 fusion | synthetic ASO with 2′-O-methyl, phosphorothioate modifications | short engineered ADAR-recruiting RNAs (arRNAs) |
| Efficiency | higher gRNA-dependent editing efficiency | RNA-editing rates up to 42% | editing efficiency up to 75%–85% (ADAR1 p150)a | editing efficiencies of up to 80% (arRNA151) |
| Delivery of editing system | viral delivery (AAV) | plasmid transfection | ASOs transfection | plasmid or lentiviral vector, or as a synthetic oligonucleotide |
| Clinically relevant mutation | KRAS4b transcript fused to the luciferase reporter | β-catenin transcript (CTNNB1) | phosphotyrosine 701 in STAT1; PiZZ mutation causing α1-antitrypsin deficiency (E342K in SERPINA1) | α-l-iduronidase catalytic activity (Hurler syndrome); TP53, COL3A1, BMPR2, AHI1, FANCC, MYBPC3, and IL2RG |
| Cellular model | HEK293T cells | HEK293FT and human umbilical vein endothelial cells (HUVECs) | human cell lines and different human primary cells | different human and mouse cell types, including various primary cell types and Hurler patient’s primary fibroblasts |
| Multiplexing approach | target multiple effectors to different transcripts | multiplexed C-to-U and A-to-I editing through the use of tailored guide RNAs | co-transfection of two ASOs | multiplex editing by co-expression of two arRNAs |
ASO, antisense oligonucleotide; ORF, open reading frame; UTR, untranslated region.
One of the two distinct isoforms of ADAR1 (p110 and p150).
Expanding the RNA-Editing Arsenal
CIRTS
The epitranscriptomic code involves all functionally relevant chemical modifications to the transcriptome.28 These chemical modifications in nucleotide bases on RNA are the result of RNA-binding proteins (RBPs)29 which are usually classified as “writers” (enzymes responsible for putting in place the modification), “readers” (RBPs that can distinguish and bind to the sequence upon modification of the RNA), and “erasers” (enzymes responsible for the elimination of the modification).1,28
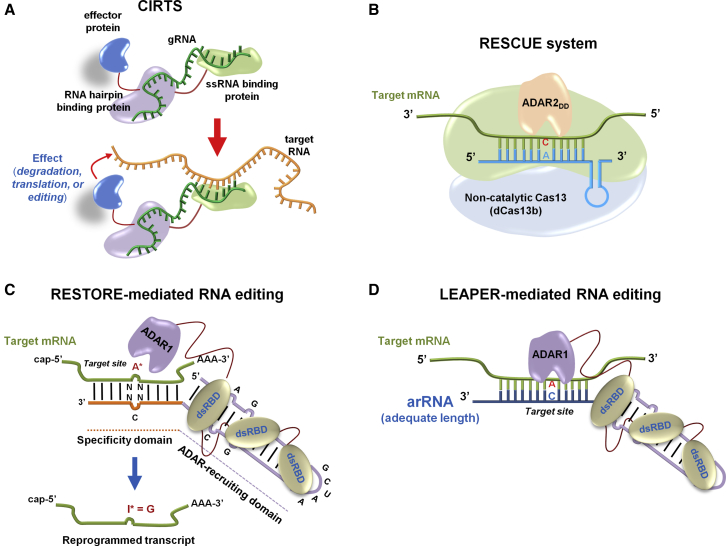
In a recent study in Cell, Rauch et al.30 described the engineering of the CRISPR-Cas-Inspired RNA Targeting System (CIRTS) as a modular protein engineering strategy, developed entirely from human protein parts (Figure 1A). The engineering of this programmable RNA-targeting system consists of: (1) an RNA hairpin-binding protein that provides the core of this system because it is a selective, high-affinity protein binder on an engineered gRNA; (2) a gRNA with a specific RNA structure that interacts with the engineered hairpin binding protein, as well as a complementary sequence to the target RNA of interest; (3) a protein that could bind to the gRNA to stabilize and protect it prior to target interaction; and (4) an epitranscriptomic regulator that acts on the targeted RNA30 (Figure 1A). Hence through Watson-Crick-Franklin base pair interactions between gRNA and the targeted RNA, this ribonucleoprotein complex delivers a range of effector proteins, including ribonucleases, RNA degradation proteins, RNA-editing proteins for A-to-I editing, for example, making use of human ADAR2, and translational activators for enhanced protein production in a gRNA-dependent manner.30 Therefore, CIRTS provides a novel platform easily programmable for the epitranscriptome, because it represents a versatile system that can combine multiple protein domains, where a specific domain might work as “writer,” “reader,” and/or “eraser” to target virtually any transcript of interest. The above could also occur either on the same or different RNA molecules, the latter being possible through rationalized and multiplexed delivery of gRNAs.
Figure 1.
Programmable Systems for ADAR-Mediated RNA Editing
(A) CIRTS is composed of a single-stranded RNA (ssRNA) binding protein, an RNA hairpin binding protein, an effector protein, as well as a guide RNA (gRNA). Especially for RNA-guided editing, CIRTS delivers the catalytic domain of human adenosine deaminase acting on RNA 2 (hADAR2) in transcripts containing a G-to-A mutation in the coding region. (B) The design of RESCUE comprises the catalytically inactive Cas13b from Riemerella anatipestifer (dRanCas13b), which is fused to the deaminase domain of human ADAR2 (ADAR2DD). Mismatched adenosine (A) in the crRNA opposite the target cytidine (C) promotes the cytidine deamination to uridine (C-to-U RNA-editing reaction). (C) Site-directed RNA editing by RESTORE is exerted by chemically modified antisense oligonucleotides (ASOs), which can be engineered with a programmable specificity domain that determines target mRNA recognition and an invariant ADAR recruiting domain to guide endogenous ADAR1 or ADAR2 to the ASO:mRNA duplex, although only the recruitment of ADAR1 is depicted. It results in a specific adenosine-to-inosine (A-to-I) conversion at the target site. (D) ADAR-mediated editing by LEAPER is performed through long ADAR-recruiting RNAs (arRNAs) that could anneal with the target transcripts (with an A-C mismatch as specified) and form double-stranded RNA (dsRNA) substrates that in turn recruit endogenous ADAR1 protein (which possess three dsRBDs) for targeted editing. dsRBD, double-stranded RNA binding domain.
Interestingly, CIRTS-1 has an amino acid sequence size of 432, which is even smaller than the smallest DNA-targeting Cas14a protein found to date (529 amino acids [aa]),31 which denotes that this modular system is efficient even in a minimum configuration. In contrast, given the human-derived nature of CIRTS, this would avoid immune concern if this is intended for epitranscriptome-modulating purposes in vivo.30 Regarding this, Rauch et al.30 computationally predicted the likelihood of risk for inducing immune responses of the humanized peptides expressed by the engineered constructs. Specifically, through an analysis of major histocompatibility complex class I (MHC class I) binding peptides, they found a low propensity to cause immune reactions,30 which is particularly attractive for therapeutic interventions.
RESCUE
Previous work established that the deactivated Cas13b moiety recognizes a target sequence of RNA, whereas the ADAR2 moiety promotes site-directed A-to-I RNA editing in full-length transcripts containing pathogenic mutations in mammalian cells without cutting the transcripts.24 Now, a new programmable base-editing tool referred to as RNA Editing for Specific C-to-U Exchange (RESCUE), which is capable of precise cytidine-to-uridine conversion in RNA with increased cytidine deamination activity, was revealed in a recent paper published in Science by the same group of researchers32 (Figure 1B). Via rational mutagenesis approach, with mutations distributed throughout the structure of ADAR2DD, the authors demonstrated that adenosine deaminases could accept other bases, resulting in a novel cytidine deamination mechanism that can edit dsRNA.32 The optimized ADAR2DD was fused to the catalytically inactive Cas13b ortholog32 (Figure 1B), which allows RESCUE to be functional for adenosine and cytidine deamination and, in turn, for the modulation of phosphorylation at specific phosphorylation residues.32 For instance, when the authors applied RESCUE employing a panel of gRNAs targeting the β-catenin transcript at known phosphorylation residues, they observed editing levels between 5% and 28%, resulting in up to 5-fold activation of Wnt/β-catenin signaling, as well as an increasing cell growth in HEK293FT and human umbilical vein endothelial cells (HUVECs).32
Therefore, RESCUE expands the RNA-targeting arsenal with new base-editing functionality. Because this system can be minimized for adeno-associated virus (AAV) packaging for viral delivery,32 it makes it an attractive tool for RNA editing in basic research in life sciences. However, because RESCUE harbors A-to-I and C-to-U deamination activity, this is a problematic issue due to the unintended transcriptomic modifications that limit the potential therapeutic use of this RNA-editing tool. Thus, it suggests the need to more fully define and characterize the RNA off-target effects of deaminase enzymes in this base editor platform before its potential applicability in therapeutics.
RESTORE
Recently, Merkle et al.33 described the RESTORE (Recruiting Endogenous ADAR to Specific Transcripts for Oligonucleotide-mediated RNA Editing) system as another useful alternative for site-directed RNA editing. RESTORE is composed of an antisense oligonucleotide (ASO) chemically modified and engineered in two segments: (1) a programmable specificity domain that determines target mRNA binding, and (2) an invariant ADAR recruiting domain to guide endogenous human ADARs to the ASO:mRNA hybrid to edit transcripts33 (Figure 1C).
This programmable approach has been applied in a panel of immortalized human cell lines and primary human cells with editing yields ranging from 75% to 85%, and these yields were cell line dependent.33 By means of RESTORE, Merkle et al.33 repaired a clinically relevant mutation that causes α1-antitrypsin deficiency, as well as edited the phosphotyrosine 701 site (5′-UAU-3′ codon) of STAT1 in HeLa and primary cells. Furthermore, the PiZZ mutation (E342K) in SERPINA1 (serpin family A member 1), known as the most common cause of α1-antitrypsin deficiency, which, in turn, causes severe damage to the lungs and the liver,34 was another relevant disease-causing mutation edited by RESTORE at the transcript level.33 These examples demonstrate the potential of this RNA-editing technology for therapeutic correction of point mutations in disease-relevant transcripts. In this regard, RNA editing mediated by RESTORE is achieved only through the administration of the ADAR-recruiting ASOs without ectopic expression of ADARs, which gives this approach an advantage by minimizing the off-target editing effect caused by overexpression of these RNA-editing enzymes. The above would considerably simplify their clinical application employing several delivery strategies for oligonucleotides that are being successful in clinical trials, such as liposomal nanoparticle formulations or N-acetylgalactosamine (GalNAc) conjugation.35
LEAPER
Very recently, Qu et al.36 described another RNA-editing approach referred to as Leveraging Endogenous ADAR for Programmable Editing of RNA (LEAPER), which employs short engineered ADAR-recruiting RNAs (arRNAs) to engage endogenous ADAR1 enzymes to change a specific A-to-I (Figure 1D) and achieves editing efficiencies of up to 80%, with minimal global off-target effects and limited editing of non-target adenosines in the target site.36
Interestingly, this study demonstrated that the targeted RNA editing by ADAR proteins could be carried out in the presence or absence of the catalytically inactive Cas13 protein effector (dCas13a-ADAR1 fusion).36 The latter may occur because long gRNAs (∼111–151 nt in length) could anneal with the target transcripts to form dsRNA substrates that, in turn, recruit endogenous ADAR proteins for targeted editing and because of this, the gRNAs were called arRNAs.36 By employing arRNAs of different nucleotides in length, LEAPER can enable effective editing on endogenous transcripts, including PPIB, KRAS, SMAD4, and FANCC, with a tendency to obtain high editing rates.36 Besides, by coexpression of two arRNAs, LEAPER can simultaneously target different sites observing editing events with a synergistic effect compared with those with individual arRNAs,36 which opens up the possibility of multiplex editing through several arRNAs.
In the first instance, to exemplify the therapeutic potential of LEAPER, the authors targeted the tumor suppressor gene TP53, the single most frequently altered gene in human cancers,37 demonstrating that LEAPER is capable of repairing the cancer-relevant premature stop codon of TP53 and restoring its function,36 which represents a promising strategy for precision oncology.
Moreover, the authors restored the α-l-iduronidase (IDUA) catalytic activity, a lysosomal metabolic enzyme responsible for the degradation of mucopolysaccharides,38 in Hurler syndrome patient-derived primary fibroblasts (IDUA-deficient GM06214 cells). Employing this single-molecule system, Qu et al.36 ensure high-efficiency RNA editing without causing an apparent innate immune response. It is noteworthy that LEAPER was delivered either by a plasmid or viral vector, or as a synthetic oligonucleotide into different cell types, including multiple human primary cell types,36 denoting the versatility of this strategy in terms of delivery.
Looking Ahead to the Future
All of these strategies constitute programmable systems that are harnessing the nature of Watson-Crick-Franklin base pair to interact with specific nucleic acid sequences just by shifting the sequence complementarity of the guide strand. As a result, these programmable RNA-guided tools allow identifying and correcting RNA modifications in coding RNAs, although they could also edit non-coding RNAs for modulating gene-regulatory networks. In this respect, because ADARs have the ability to bind any dsRNA >20 bp in length,39 the number of possible targets of these enzymes also includes small non-coding RNA sequences, such as microRNAs (miRNAs), small non-coding RNA molecules of 20–24 nt in length, whose primary transcripts can form hairpins containing bulges and loops that may act as double-stranded targets.40 Concerning this, A-to-I RNA editing within miRNAs precursor sequences (pre-miRNAs) could alter their processing into mature miRNAs, which would lead to a decrease in the availability of miRNA sequences. To give an example, Kawahara et al.41 reported that A-to-I RNA editing of pri‐miR‐151 at two specific sites within its foldback dsRNA structure completely inhibits its cleavage by the Dicer-TRBP complex, leading to an accumulation of edited pre‐miR‐151. In contrast, by integrating deep sequencing and array approaches with bioinformatics analyses along with molecular studies, Tomaselli et al.42 showed that ADAR2 is critical to edit a small number of mature miRNAs and to significantly modulate the expression of approximately 90 miRNAs in glioblastoma cells. After the mature miRNAs editing events, the transient “gRNA/miRNA duplexes” could be unwinding by the action of the AGO2 proteins to release to single-stranded miRNAs, which would eventually be bound to AGO2 and give, as a result, the formation of active RNA-induced silencing complexes (RISCs) for miRNAs function.43 Hence through the implementation of these editing technologies, the processing of specific miRNAs could be corrected and, as a result, restore critical signaling pathways regulated by these non-coding RNAs.
Although RNA base editing via RNA-guided adenosine deaminases of human origin represents an attractive approach for correction of disease-causing mutations,44 one aspect that should be considered is the immunogenicity issues. This is relevant because recently it was identified that a substantial fraction of people already have circulating antibodies to CRISPR-Cas proteins,45,46 suggesting immunogenic aspects might be a limitation for therapeutic interventions if it is desired to overexpress ADAR proteins or nucleic acid sequences to recruit these editing proteins. Regarding this, computational prediction strategies could be an excellent approach to predict low propensity to cause immunogenicity of the different components that are part of each platform.30 However, although bioinformatics predictions are usually useful tools only until the experimental assessment, we could know the degree of immunogenicity that these systems may confer.
Now, to achieve RNA editing with high efficiency and high accuracy, most of these systems require the co-existence of a deaminase (either by endogenous or ectopic expression) together with one or more gRNAs and the target RNA in suitable stoichiometry. Therefore, it is necessary to direct efforts to improve these strategies in terms of efficiency and specificity, and consequently enhance editing yield for potential applications in clinical and biomedical research, also addressing the challenges encountered in delivery.
For example, the supporters of the RESTORE system could focus efforts on minimizing the number of chemical modifications included in the ASOs, thinking in large-scale and low-cost production for applications in vivo (e.g., pilot in vivo experiments), without losing long shelf-life in an intracellular environment and ensuring and even improving higher editing. In contrast, various bulges and loops could be incorporated in dsRNA regions to increase the ability to recruit endogenous ADAR enzymes in a highly specific manner.40 Those supporters of LEAPER could focus on improving the ADAR-recruiting scaffold fused to arRNA for increasing binding ADAR proteins, or stabilizing the complete sequence of arRNA, or even increasing its endogenous expression in the different cellular environments.36 In allusion to this, an alternative for increasing binding ADAR proteins could be the use of aptamers, small non-coding RNAs capable of recognizing, with high specificity and affinity, a wide variety of molecules in a manner that resembles antibodies,47 which have certain advantages in stability and non-immunogenicity. By in vitro selection or SELEX (systematic evolution of ligands by exponential enrichment), it is possible to iteratively evolve the best combinations of oligonucleotides with chemical modifications, through an oligonucleotide library,48 to improve not only the high-affinity ADAR-binding proteins but also improve their stability by prolonging their residence in biofluids, thinking in a translational perspective.
It is clear that off-target editing is a significant concern when deaminases are ectopically overexpressed, and this is related to the enzyme activity. On this issue, RESTORE and LEAPER produce almost no off-target editing because both approaches rely on the harnessing of the endogenous enzyme, which implies that both RNA-editing tools require only the administration of the ADAR-recruiting ASOs. The above considerably simplify research studies, and clinical application due to unwanted RNA editing can be preferentially reduced. Nevertheless, RESTORE employs chemosynthetic ASOs for recruiting endogenous ADARs with complex chemical modification,33 which could not be functional through expression in cells, whereas LEAPER represents an alternative approach that uses arRNA with a length that allows it to use endogenous ADAR for RNA editing without incorporating dense chemical modifications.36 As a result, with LEAPER it is possible to express arRNAs by plasmids and viral agents, or as a synthetic oligonucleotide, with what is feasible to achieve efficient and precise editing of endogenous RNAs and correction of pathogenic mutations in a broader spectrum of cell types.36
Conclusions
We not only are witnessing the emergence of new CRISPR-Cas immune systems in nature, in its smaller versions,31 but also the rapid evolution of base editors, as well as being able to engineer sophisticated RNA-editing systems in a programmable manner (Table 1). As a result, by these base-editing platforms, it is possible to modulate RNA activities and edit or correct human disease-associated mutation that would be a valuable complement to existing CRISPR/Cas systems.
For now, we cannot state that some of these novel approaches are better than the other because all could be complemented depending on the transcripts that need to be edited and the cellular and tissue model where these could be used. Regardless of the RNA-editing strategy chosen, further experimental testing is needed to demonstrate not only the low immunogenic capability of these effector proteins and all its components but also minimal off-target effects. Undoubtedly these challenges have to be overcome before these platforms are directed toward therapeutic intervention.
Conflicts of Interest
The author declares no competing interests.
Acknowledgments
G.A.-J. is supported by grant SEP-CONACyT-CB-286421 from the Mexican Council of Sciences and Technology (CONACyT).
References
- 1.Kadumuri R.V., Janga S.C. Epitranscriptomic Code and Its Alterations in Human Disease. Trends Mol. Med. 2018;24:886–903. doi: 10.1016/j.molmed.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linder B., Jaffrey S.R. Discovering and Mapping the Modified Nucleotides That Comprise the Epitranscriptome of mRNA. Cold Spring Harb. Perspect. Biol. 2019;11:a032201. doi: 10.1101/cshperspect.a032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim H., Kim J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 4.Solomon O., Oren S., Safran M., Deshet-Unger N., Akiva P., Jacob-Hirsch J., Cesarkas K., Kabesa R., Amariglio N., Unger R. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR) RNA. 2013;19:591–604. doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F.J., Rechavi G., Li J.B., Eisenberg E., Levanon E.Y. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X., Wang Y., Liang H. The role of A-to-I RNA editing in cancer development. Curr. Opin. Genet. Dev. 2018;48:51–56. doi: 10.1016/j.gde.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walkley C.R., Li J.B. Rewriting the transcriptome: adenosine-to-inosine RNA editing by ADARs. Genome Biol. 2017;18:205. doi: 10.1186/s13059-017-1347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phelps K.J., Tran K., Eifler T., Erickson A.I., Fisher A.J., Beal P.A. Recognition of duplex RNA by the deaminase domain of the RNA editing enzyme ADAR2. Nucleic Acids Res. 2015;43:1123–1132. doi: 10.1093/nar/gku1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho D.S., Yang W., Lee J.T., Shiekhattar R., Murray J.M., Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 2003;278:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 11.Mingardi J., Musazzi L., De Petro G., Barbon A. miRNA Editing: New Insights into the Fast Control of Gene Expression in Health and Disease. Mol. Neurobiol. 2018;55:7717–7727. doi: 10.1007/s12035-018-0951-x. [DOI] [PubMed] [Google Scholar]
- 12.Barraud P., Allain F.H. ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr. Top. Microbiol. Immunol. 2012;353:35–60. doi: 10.1007/82_2011_145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beghini A., Ripamonti C.B., Peterlongo P., Roversi G., Cairoli R., Morra E., Larizza L. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum. Mol. Genet. 2000;9:2297–2304. doi: 10.1093/oxfordjournals.hmg.a018921. [DOI] [PubMed] [Google Scholar]
- 14.Woolf T.M., Chase J.M., Stinchcomb D.T. Toward the therapeutic editing of mutated RNA sequences. Proc. Natl. Acad. Sci. USA. 1995;92:8298–8302. doi: 10.1073/pnas.92.18.8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stafforst T., Schneider M.F. An RNA-deaminase conjugate selectively repairs point mutations. Angew. Chem. Int. Ed. Engl. 2012;51:11166–11169. doi: 10.1002/anie.201206489. [DOI] [PubMed] [Google Scholar]
- 16.Juillerat A., Gronemeyer T., Keppler A., Gendreizig S., Pick H., Vogel H., Johnsson K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem. Biol. 2003;10:313–317. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 17.Vogel P., Schneider M.F., Wettengel J., Stafforst T. Improving site-directed RNA editing in vitro and in cell culture by chemical modification of the guideRNA. Angew. Chem. Int. Ed. Engl. 2014;53:6267–6271. doi: 10.1002/anie.201402634. [DOI] [PubMed] [Google Scholar]
- 18.Rees D.C., Cox M., Clegg J.B. World distribution of factor V Leiden. Lancet. 1995;346:1133–1134. doi: 10.1016/s0140-6736(95)91803-5. [DOI] [PubMed] [Google Scholar]
- 19.Vogel P., Moschref M., Li Q., Merkle T., Selvasaravanan K.D., Li J.B., Stafforst T. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat. Methods. 2018;15:535–538. doi: 10.1038/s41592-018-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanswillemenke A., Stafforst T. Protocols for the generation of caged guideRNAs for light-triggered RNA-targeting with SNAP-ADARs. Methods Enzymol. 2019;624:47–68. doi: 10.1016/bs.mie.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Azad M.T.A., Bhakta S., Tsukahara T. Site-directed RNA editing by adenosine deaminase acting on RNA for correction of the genetic code in gene therapy. Gene Ther. 2017;24:779–786. doi: 10.1038/gt.2017.90. [DOI] [PubMed] [Google Scholar]
- 22.Vogel P., Stafforst T. Critical review on engineering deaminases for site-directed RNA editing. Curr. Opin. Biotechnol. 2019;55:74–80. doi: 10.1016/j.copbio.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Montiel-Gonzalez M.F., Vallecillo-Viejo I., Yudowski G.A., Rosenthal J.J. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc. Natl. Acad. Sci. USA. 2013;110:18285–18290. doi: 10.1073/pnas.1306243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rueter S.M., Burns C.M., Coode S.A., Mookherjee P., Emeson R.B. Glutamate receptor RNA editing in vitro by enzymatic conversion of adenosine to inosine. Science. 1995;267:1491–1494. doi: 10.1126/science.7878468. [DOI] [PubMed] [Google Scholar]
- 26.Wettengel J., Reautschnig P., Geisler S., Kahle P.J., Stafforst T. Harnessing human ADAR2 for RNA repair—Recoding a PINK1 mutation rescues mitophagy. Nucleic Acids Res. 2017;45:2797–2808. doi: 10.1093/nar/gkw911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 28.Esteller M., Pandolfi P.P. The Epitranscriptome of Noncoding RNAs in Cancer. Cancer Discov. 2017;7:359–368. doi: 10.1158/2159-8290.CD-16-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neelamraju Y., Hashemikhabir S., Janga S.C. The human RBPome: from genes and proteins to human disease. J. Proteomics. 2015;127:61–70. doi: 10.1016/j.jprot.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 30.Rauch S., He E., Srienc M., Zhou H., Zhang Z., Dickinson B.C. Programmable RNA-Guided RNA Effector Proteins Built from Human Parts. Cell. 2019;178:122–134.e12. doi: 10.1016/j.cell.2019.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abudayyeh O.O., Gootenberg J.S., Franklin B., Koob J., Kellner M.J., Ladha A., Joung J., Kirchgatterer P., Cox D.B.T., Zhang F. A cytosine deaminase for programmable single-base RNA editing. Science. 2019;365:382–386. doi: 10.1126/science.aax7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merkle T., Merz S., Reautschnig P., Blaha A., Li Q., Vogel P., Wettengel J., Li J.B., Stafforst T. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 2019;37:133–138. doi: 10.1038/s41587-019-0013-6. [DOI] [PubMed] [Google Scholar]
- 34.Lomas D.A., Mahadeva R. Alpha1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J. Clin. Invest. 2002;110:1585–1590. doi: 10.1172/JCI16782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsui M., Corey D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu L., Yi Z., Zhu S., Wang C., Cao Z., Zhou Z., Yuan P., Yu Y., Tian F., Liu Z. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 2019;37:1059–1069. doi: 10.1038/s41587-019-0178-z. [DOI] [PubMed] [Google Scholar]
- 37.Duffy M.J., Synnott N.C., Crown J. Mutant p53 as a target for cancer treatment. Eur. J. Cancer. 2017;83:258–265. doi: 10.1016/j.ejca.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 38.Ou L., DeKelver R.C., Rohde M., Tom S., Radeke R., St Martin S.J., Santiago Y., Sproul S., Przybilla M.J., Koniar B.L. ZFN-Mediated In Vivo Genome Editing Corrects Murine Hurler Syndrome. Mol. Ther. 2019;27:178–187. doi: 10.1016/j.ymthe.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawahara Y., Zinshteyn B., Chendrimada T.P., Shiekhattar R., Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomaselli S., Galeano F., Alon S., Raho S., Galardi S., Polito V.A., Presutti C., Vincenti S., Eisenberg E., Locatelli F., Gallo A. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015;16:5. doi: 10.1186/s13059-014-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwak P.B., Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 44.Chen G., Katrekar D., Mali P. RNA-Guided Adenosine Deaminases: Advances and Challenges for Therapeutic RNA Editing. Biochemistry. 2019;58:1947–1957. doi: 10.1021/acs.biochem.9b00046. [DOI] [PubMed] [Google Scholar]
- 45.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., Vakulskas C.A., Collingwood M.A., Zhang L., Bode N.M. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner D.L., Amini L., Wendering D.J., Burkhardt L.M., Akyüz L., Reinke P., Volk H.D., Schmueck-Henneresse M. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat. Med. 2019;25:242–248. doi: 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- 47.Aquino-Jarquin G., Toscano-Garibay J.D. RNA aptamer evolution: two decades of SELEction. Int. J. Mol. Sci. 2011;12:9155–9171. doi: 10.3390/ijms12129155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon A.J., d’Oelsnitz S., Ellington A.D. Synthetic evolution. Nat. Biotechnol. 2019;37:730–743. doi: 10.1038/s41587-019-0157-4. [DOI] [PubMed] [Google Scholar]