Abstract
Cellulosomes are synthesized by anaerobic bacteria and fungi to degrade lignocellulose via synergistic action of multiple enzymes fused to a protein scaffold. Through templating key protein domains (cohesin and dockerin), designer cellulosomes have been engineered from bacterial motifs to alter the activity, stability, and degradation efficiency of enzyme complexes. Recently a parts list for fungal cellulosomes from the anaerobic fungi (Neocallimastigomycota) was determined, which revealed sequence divergent fungal cohesin, dockerin, and scaffoldin domains that could be used to expand the available toolbox to synthesize designer cellulosomes. In this work, multi-domain carbohydrate active enzymes (CAZymes) from 3 cellulosome-producing fungi were analyzed to inform the design of chimeric proteins for synthetic cellulosomes inspired by anaerobic fungi. In particular, Piromyces finnis was used as a structural template for chimeric carbohydrate active enzymes. Recombinant enzymes with retained properties were engineered by combining thermophilic glycosyl hydrolase domains from Thermotoga maritima with dockerin domains from Piromyces finnis. By preserving the protein domain order from P. finnis, chimeric enzymes retained catalytic activity at temperatures over 80 °C and were able to associate with cellulosomes purified from anaerobic fungi. Fungal cellulosomes harbor a wide diversity of glycoside hydrolases, each representing templates for the design of chimeric enzymes. By conserving dockerin domain position within the primary structure of each protein, the activity of both the catalytic domain and dockerin domain was retained in enzyme chimeras. Taken further, the domain positioning inferred from native fungal cellulosome proteins can be used to engineer multi-domain proteins with non-native favorable properties, such as thermostability.
Keywords: Cellulosome, Dockerin, Scaffoldin, Anaerobic fungi, Thermophile, Enzyme
Abbreviations: DDP, Dockerin domain protein; GH, Glycoside hydrolase; CMC, Carboxymethyl cellulose; IPTG, Isopropyl-β- d-thiogalactopyranoside; DNS, Dinitrosalicylate; pNPG, p-Nitrophenyl β-d-glucopyranoside; BSA, Bovine serum albumin
1. Introduction
To lower costs associated with lignocellulose bioprocessing, biomass-degrading enzymes must be engineered to hydrolyze variable feedstocks and be stable and active across a wide range of conditions, including elevated temperatures and acidic environments [1]. Recent efforts to develop more effective lignocellulolytic enzymes have been inspired by nature. For example, many anaerobic bacteria and fungi increase enzyme efficiency by tethering together an assortment of cellulases and related accessory enzymes into complexes called cellulosomes [[2], [3], [4], [5], [6]].
Cellulosome-mediated biomass degradation relies on large, non-catalytic scaffoldins composed of many interspersed cohesin domains that anchor together dockerin-bearing enzymes to synergistically degrade cellulose and hemicellulose. By using truncated scaffolds, as well as sequence variants of cohesin and dockerin domains, synthetic “designer” cellulosomes have been engineered that are more active compared to freely diffusing enzymes [[2], [3], [4], [5], [6]]. Many recent reports have demonstrated that synthetic cellulosomes inspired by bacterial cellulosomes are 2- to 3-fold better than free enzymes at degrading low-accessibility, highly crystalline, insoluble substrates when produced in recombinant systems [[2], [3], [4], [5], [6]]. Additionally, the flexibility offered by modular enzyme domains enables designer cellulosomes to incorporate “new” catalytic activities that are not found in native cellulosomes [7].
While designer cellulosomes have been synthesized from the cohesin, dockerin, and scaffoldin domains found across anaerobic bacteria, analogous domains from anaerobic fungi have not yet been explored as synthetic biology tools. Like cellulosome-producing bacteria, anaerobic fungi are typically found in the rumen and hindgut of large herbivores, where up to 30% of their CAZymes are cellulosome-associated [8,9]. Recently, comparative genomics coupled with proteomic validation revealed a family of repeat-rich, non-catalytic scaffoldins in the genomes of five anaerobic fungi that form a specific interaction with dockerin-fused fungal enzymes [8]. Since the gene sequences that encode fungal cellulosome components are highly divergent from those found in bacteria [8,10], the corresponding structure of these domains is also distinct from bacterial cellulosomes. Limited insight exists into the high-resolution structure of fungal cellulosomes, but an NMR structure of a fungal dockerin verifies how different these structures are from bacteria [11,12]. As fungal dockerins exist at either the N- or C-terminus of proteins and in tandem repeats, elucidating the type and placement of fungal dockerins is necessary for optimal design of synthetic enzymes and synthetic fungal cellulosomes. Furthermore, native fungal cellulosomes offer a much wider repertoire of cellulases with dockerin domains compared to bacterial cellulosomes, including GH3, GH6, and GH45 [8]. Therefore, employing native fungal cellulosomes as templates to engineer chimeric enzymes could enable the inclusion of new classes of enzymes into these complexes, including those that expand the activity and thermal stability of cellulosomes.
Towards the goal of engineering thermostable enzyme complexes for biomass utilization, we fused domains natively found in fungal cellulosomes to cellulases from Thermotoga maritima to engineer thermostable chimeric enzymes that can be recruited into native fungal cellulosomes. Thermostable enzymes are catalytically active at high temperatures (70–90 °C), yet are also stable at moderate temperatures (50–70 °C), resulting in decreased required enzyme titers for biomass hydrolysis [1,13]. T. maritima is a hyperthermophilic bacterium capable of growing at temperatures up to 90 °C in deep sea vents [14], and whose enzymes are likely amenable to high temperature bioprocessing. However, while T. maritima secretes many cellulases and hemicellulases, they are freely diffusive and not contained in cellulosome complexes [15]. Linking thermostable enzymes into cellulosome-inspired complexes could improve the efficiency of cellulose hydrolysis even further through synergistic targeting of the enzymes and alignment of the active sites. With a previously established fungal cellulosome “parts list” (8) for inspiration, we engineered thermostable multi-modular CAZymes for inclusion in synthetic enzyme complexes. Since fungal cellulosomes contain a wide array of catalytic domains, more even than those found in bacterial cellulosomes, this method can assist in the design of a broad range of multi-domain CAZymes with favorable properties such as thermostability.
2. Materials and methods
2.1. Strains, plasmids, and yeast culture conditions
All plasmids and strains used in this study are listed in Table 1. E. coli Tuner (DE3) and E. coli BL21 (DE3) cells were used to produce heterologous proteins. DNA sequences encoding fungal dockerins were PCR-amplified from fungal cDNA libraries using previously published techniques [8]. DNA sequences encoding cellulase GH5 and hemicellulase XylA from T. maritima were PCR-amplified from genomic DNA provided by Prof. Robert Kelly at NCSU, with gene sequences shown in Table 2. For E. coli production, genes were sub-cloned into the pET32a expression system (Addgene), which adds N-terminal TrxA genetic fusions to enhance protein solubility. Protein synthesis was induced when the cells reached an absorbance at 600 nm (OD600) of ~0.6 by adding 0.1 mM isopropyl-β- d-thiogalactopyranoside (IPTG) to the media. E. coli strains were routinely grown aerobically at 37 °C in Luria−Bertani (LB) medium, and antibiotics were supplemented at the following concentrations: ampicillin (Amp, 100 μg/mL) and kanamycin (Kan, 50 μg/mL).
Table 1.
Number of dockerin-containing genes across representative anaerobic fungal and bacterial genomes.
Table 2.
CAZymes and Dockerin sequences analyzed in this study.
| Gene | Sequence |
|---|---|
| Cel5A (WP_004082283) | atgagcgataaaattattcacctgactgacgacagttttgacacggatgtactcaaagcggacggggcgatcctcgtcgatttctgggcagagtggtgcggtccgtgcaaaatgatcgccccgattctggatgaaatcgctgacgaatatcagggcaaactgaccgttgcaaaactgaacatcgatcaaaaccctggcactgcgccgaaatatggcatccgtggtatcccgactctgctgctgttcaaaaacggtgaagtggcggcaaccaaagtgggtgcactgtctaaaggtcagttgaaagagttcctcgacgctaacctggccggttctggttctggccatatgcaccatcatcatcatcattcttctggtctggtgccacgcggttctggtatgaaagaaaccgctgctgctaaattcgaacgccagcacatggacagcccagatctgggtaccgacgacgacgacaaggccatgggtcaccaccaccatcaccacgagaacttgtactttcaaggtggatcccggccgatgggtgttgatccttttgaaaggaacaaaatattgggaagaggcattaatataggaaatgcgcttgaagcaccaaatgagggagactggggagtggtgataaaagatgagttcttcgacattataaaagaagccggtttctctcatgttcgaattccaataagatggagtacgcacgcttacgcgtttcctccttataaaatcatggatcgcttcttcaaaagagtggatgaagtgataaacggagccctgaaaagaggactggctgttgttataaatattcatcactacgaggagttaatgaatgatccagaagaacacaaggaaagatttcttgctctttggaaacaaattgctgatcgttataaagactatcccgaaactctattttttgaaattctgaatgaacctcacggaaatcttactccggaaaaatggaatgaactgcttgaggaagctctaaaagttataagatcaattgacaaaaagcacactataattataggcacagctgaatgggggggtatatctgcccttgaaaaactgtctgtcccaaaatgggaaaaaaattctatagttacaattcactactacaatcctttcgaatttacccatcaaggagctgagtgggtggaaggatctgagaaatggttgggaagaaagtggggatctccagatgatcagaaacatttgatagaagaattcaattttatagaagaatggtcaaaaaagaacaaaagaccaatttacataggtgagtttggtgcctacagaaaagctgaccttgaatcaagaataaaatggacctcctttgtcgttcgcgaaatggagaaaaggagatggagctgggcatactgggaattttgttccggttttggtgtttatgatactctgagaaaaacctggaataaagatcttttagaagctttaataggaggagatagcattgaagacgtc |
| Cel5A-Dockerin (dockerin fusion shown in bold from ORX48147.1) | atgagcgataaaattattcacctgactgacgacagttttgacacggatgtactcaaagcggacggggcgatcctcgtcgatttctgggcagagtggtgcggtccgtgcaaaatgatcgccccgattctggatgaaatcgctgacgaatatcagggcaaactgaccgttgcaaaactgaacatcgatcaaaaccctggcactgcgccgaaatatggcatccgtggtatcccgactctgctgctgttcaaaaacggtgaagtggcggcaaccaaagtgggtgcactgtctaaaggtcagttgaaagagttcctcgacgctaacctggccggttctggttctggccatatgcaccatcatcatcatcattcttctggtctggtgccacgcggttctggtatgaaagaaaccgctgctgctaaattcgaacgccagcacatggacagcccagatctgggtaccgacgacgacgacaaggccatgggtcaccaccaccatcaccacgagaacttgtactttcaaggtggatcccggccgatgggtgttgatccttttgaaaggaacaaaatattgggaagaggcattaatataggaaatgcgcttgaagcaccaaatgagggagactggggagtggtgataaaagatgagttcttcgacattataaaagaagccggtttctctcatgttcgaattccaataagatggagtacgcacgcttacgcgtttcctccttataaaatcatggatcgcttcttcaaaagagtggatgaagtgataaacggagccctgaaaagaggactggctgttgttataaatattcatcactacgaggagttaatgaatgatccagaagaacacaaggaaagatttcttgctctttggaaacaaattgctgatcgttataaagactatcccgaaactctattttttgaaattctgaatgaacctcacggaaatcttactccggaaaaatggaatgaactgcttgaggaagctctaaaagttataagatcaattgacaaaaagcacactataattataggcacagctgaatgggggggtatatctgcccttgaaaaactgtctgtcccaaaatgggaaaaaaattctatagttacaattcactactacaatcctttcgaatttacccatcaaggagctgagtgggtggaaggatctgagaaatggttgggaagaaagtggggatctccagatgatcagaaacatttgatagaagaattcaattttatagaagaatggtcaaaaaagaacaaaagaccaatttacataggtgagtttggtgcctacagaaaagctgaccttgaatcaagaataaaatggacctcctttgtcgttcgcgaaatggagaaaaggagatggagctgggcatactgggaattttgttccggttttggtgtttatgatactctgagaaaaacctggaataaagatcttttagaagctttaataggaggagatagcattgaagacgtcggtttagaagttaatgtacttcatgctattgaaactgaaccaactgaatgttggtctcttaagtatggttatgaatgctgttctccaaataacactagagttgttgtcaccgatgaaagtggaaagtggggtgttgaaaacagtgattggtgtggtattgtagacagcaaggataaatgctggtctattccattcggttacaaatgctgtgatcactgtaaagtcctcttaactgatgaaagtggtaaatggggtgaattaaacggtgaatggtgtggtattgatactactaaatgtaaaccgcgg |
| XynA (WP_004082550) | atgagcgataaaattattcacctgactgacgacagttttgacacggatgtactcaaagcggacggggcgatcctcgtcgatttctgggcagagtggtgcggtccgtgcaaaatgatcgccccgattctggatgaaatcgctgacgaatatcagggcaaactgaccgttgcaaaactgaacatcgatcaaaaccctggcactgcgccgaaatatggcatccgtggtatcccgactctgctgctgttcaaaaacggtgaagtggcggcaaccaaagtgggtgcactgtctaaaggtcagttgaaagagttcctcgacgctaacctggccggttctggttctggccatatgcaccatcatcatcatcattcttctggtctggtgccacgcggttctggtatgaaagaaaccgctgctgctaaattcgaacgccagcacatggacagcccagatctgggtaccgacgacgacgacaaggccatgggtcaccaccaccatcaccacgagaacttgtactttcaaggtggatcccggccgatgaaagaagtactaaaagactacttcaaagtcggagttgcactgccgtccaaggtcttcctcaacccgaaggacatagaactcatcacgaaacacttcaacagcatcaccgcagaaaacgagatgaaaccagagagcctgctcgcgggcatcgaaaacggtaagctgaagttcaggtttgaaacagcagacaaatacattcagttcgtcgaggaaaacggcatggttataagaggtcacacactggtgtggcacaaccagacacccgactggttcttcaaagacgaaaacggaaacctcctctccaaagaagcgatgacggaaagactcaaagagtacatccacaccgttgtcggacacttcaaaggaaaagtctacgcatgggacgtggtgaacgaagcggtcgatccgaaccagccggatggactgagaagatccacctggtaccagatcatggggcctgactacatagaactcgccttcaagttcgcaagagaagcagatccagatgcaaaactcttctacaacgactacaacacattcgagcccagaaagagagatatcatctacaacctcgtgaaggatctcaaggagaagggactcatcgatgggataggcatgcagtgtcacatcagtcttgcaacagacatcaaacagatcgaagaggccatcaaaaagttcagcaccatacccggtatagaaattcacatcacagaactcgatatgagtgtctacagagattccagttccaactacccagaggcaccgaggacggcactcatcgaacaggctcacaaaatgatgcagctctttgagattttcaagaagtacagcaacgtgatcacgaacgtcacattctggggtctcaaggacgattactcctggagagcaacaagaagaaacgactggccgctcatcttcgacaaagatcaccaggcgaaactcgcttactgggcgatagtggacgtc |
| XynA-Dockerin (dockerin fusion shown in bold from ORX55236.1) | atgagcgataaaattattcacctgactgacgacagttttgacacggatgtactcaaagcggacggggcgatcctcgtcgatttctgggcagagtggtgcggtccgtgcaaaatgatcgccccgattctggatgaaatcgctgacgaatatcagggcaaactgaccgttgcaaaactgaacatcgatcaaaaccctggcactgcgccgaaatatggcatccgtggtatcccgactctgctgctgttcaaaaacggtgaagtggcggcaaccaaagtgggtgcactgtctaaaggtcagttgaaagagttcctcgacgctaacctggccggttctggttctggccatatgcaccatcatcatcatcattcttctggtctggtgccacgcggttctggtatgaaagaaaccgctgctgctaaattcgaacgccagcacatggacagcccagatctgggtaccgacgacgacgacaaggccatgggtcaccaccaccatcaccacgagaacttgtactttcaaggtggatcccggccgatgaaagaagtactaaaagactacttcaaagtcggagttgcactgccgtccaaggtcttcctcaacccgaaggacatagaactcatcacgaaacacttcaacagcatcaccgcagaaaacgagatgaaaccagagagcctgctcgcgggcatcgaaaacggtaagctgaagttcaggtttgaaacagcagacaaatacattcagttcgtcgaggaaaacggcatggttataagaggtcacacactggtgtggcacaaccagacacccgactggttcttcaaagacgaaaacggaaacctcctctccaaagaagcgatgacggaaagactcaaagagtacatccacaccgttgtcggacacttcaaaggaaaagtctacgcatgggacgtggtgaacgaagcggtcgatccgaaccagccggatggactgagaagatccacctggtaccagatcatggggcctgactacatagaactcgccttcaagttcgcaagagaagcagatccagatgcaaaactcttctacaacgactacaacacattcgagcccagaaagagagatatcatctacaacctcgtgaaggatctcaaggagaagggactcatcgatgggataggcatgcagtgtcacatcagtcttgcaacagacatcaaacagatcgaagaggccatcaaaaagttcagcaccatacccggtatagaaattcacatcacagaactcgatatgagtgtctacagagattccagttccaactacccagaggcaccgaggacggcactcatcgaacaggctcacaaaatgatgcagctctttgagattttcaagaagtacagcaacgtgatcacgaacgtcacattctggggtctcaaggacgattactcctggagagcaacaagaagaaacgactggccgctcatcttcgacaaagatcaccaggcgaaactcgcttactgggcgatagtggacgtcaatgtcttaaaggaaaccaagccaaaggctactaccactaccaaaacttctcaaccaaccacttctgccggaaagtgttcttctaagattactagccaaggttacaagtgctgtgctagcaattgtgacgtggtttacgttgataatgatggtaactggggtgttgaaaacaatgattggtgtatcattagaaacccgcgg |
2.2. Protein purification and analysis
For purification from E. coli, cells expressing recombinant proteins were fermented in 500 mL cultures at 37 °C agitated at 250 rpm. When the culture reached an OD600 of 0.6, IPTG was added at 10 μM and the temperature was changed to 30 °C. Cultures were harvested 16 h post induction by centrifugation at 3200×g for 15 min in 50 mL conical tubes, and cells were resuspended in 1% of the total culture volume with 20 mM sodium phosphate, 300 mM sodium chloride, 10 mM imidazole (pH 7.4). 0.5 mm Zirconia-Silica (Bio-Spec) beads were added at ~10% of the solution volume and the suspension was vortexed rigorously in 50 mL conical tubes for 10 intervals of 30 s, with a 30 s rest on ice in between each interval. The soluble supernatant was recovered by centrifugation at 10,000×g for 10 min at 4 °C, and then target proteins encoding a 6xHis tag were purified with His-Pur Ni-NTA Resin (Thermo Fisher Scientific) following the manufacturer's instructions. Following elution of target proteins, the buffer was exchanged to PBS (pH 7.4) using Zeba desalting columns, 0.5 mL or 10 mL volumes (Thermo Fisher Scientific). Protein concentration was measured using a BCA Protein Assay Kit (Thermo Fisher Scientific). Purity of proteins were determined to be >95% by SDS-PAGE stained with SYPRO-Ruby (Bio-Rad) following the manufacturer's instructions (Supplemental Fig. 1).
2.3. Isolation of fungal cellulosome fractions
The anaerobic fungus Piromyces finnis was cultured as previously described [16]. Supernatant and native cellulosome preparations were collected between 72 and 96 h post inoculation. Cellulosomes were isolated essentially as described elsewhere via cellulose precipitation [17]. Briefly, the vegetative fungal growth was removed by centrifugation at 4 °C for 10 min at 3220×g. 0.4% (w/v) SigmaCell type 50 (Sigma) was added to the supernatant and incubated with gentle agitation at 4 °C for 2 h. The cellulose and supernatant mixture was centrifuged at 4 °C for 10 min at 3220×g, the supernatant was decanted, and the cellulose pellet was washed once with 100 mM Tris-HCl (pH 7.5) containing 150 mM NaCl and centrifuged again under the same conditions. The wash solution was decanted, and the proteins were eluted from the pelleted cellulose by resuspending it in H2O for 1 h at room temperature.
2.4. Enzyme activity assays
Enzymatic activity on solubilized CMC (Sigma) or Xylan (from Corn Core, TCI Chemicals) was measured by a microplate activity assay essentially as described elsewhere [18], with the exception that hydrolysis was performed at 80 °C and pH 5 (unless otherwise specified) for 1 h. For thermal stability assays, 0.008 mg of enzyme in 30 μL PBS was incubated with 30 μL 2% (w/v) CMC in 0.1 M sodium acetate buffer (pH 5.5) at increasing temperatures. Enzyme samples that contained Cel5A or Cel5A-Dockerin and fungal cellulosome were pre-incubated (1:1 amounts of protein) for 1 h at 4 °C with gentle shaking. To construct temperature optima curves, endpoint reducing sugar concentrations after 3 h of hydrolysis were taken in triplicate to compute specific activities.
For all activity measurements, reducing sugar concentration was measured by adding 120 μL of 3,5-dinitrosalicylic acid (DNS) reagent to 60 μL of reaction mix and heating the solution at 95 °C for 5 min. DNS reagent was prepared by dissolving 1 g of 3,5-dinitrosalicylic acid in 50 mL of reagent grade water, adding 30 g of sodium potassium tartrate tetrahydrate and 20 mL of 2 N NaOH, and finally diluting to a final volume of 100 mL with reagent grade water. 36 μL of the completed DNS reaction were transferred to 160 μL of water and the absorbance was measured at 540 nm. Rates of activity were calculated by comparing to a standard curve constructed from glucose, and by subtracting a blank measurement where water was added to the substrate. β-Glucosidase activity was determined by adding 30 μL of the enzyme to 970 μL of a reaction mix containing a final concentration of 5 mM (unless otherwise specified) p-Nitrophenyl β-d-glucopyranoside (pNPG) in 50 mM phosphate buffer (pH 5.0) with 2% (w/v) bovine serum albumin. Rates of activity were calculated by tracking the absorbance at 405 nm in a Tecan M220 Infinite Pro spectrophotometer. Unless otherwise stated, samples were performed in triplicate, and all values were normalized by total protein as measured by a BCA protein assay kit (Pierce Biotechnology, Rockford, IL).
2.5. Enzyme linked immunosorbent assay (ELISA)
Fungal cellulosome preparations were diluted 1:2000 in 0.05 M Na2CO3 buffer (pH 9.6) and 100 μL was coated on a 96 well microtiter ELISA plate (Pierce) at 4 °C overnight. Then wells were washed 3 times with 200 μL PBS (pH 7.4) and 100 μL of PBS containing 2% (w/v) bovine serum albumin (BSA) and 0.05% Tween-20 (v/v) was added and the plate was incubated at 4 °C for 1 h. Purified recombinant proteins were serially diluted in the same solvent at the same concentration (3 μM), added to the plate, and incubated at 4 °C for 1 h with gentle agitation. Wells were then washed three times with PBS. StepTactin (Bio-Rad), an HRP-conjugated secondary antibody against the Strep-tag, was diluted 1:5000 in the same solvent, added to the plate, and incubated at 4 °C for 1 h with gentle agitation. Wells were washed four times with PBS, and signals were measured using the TMB chromogen solution (Thermo Fisher Scientific) according to manufacturer instructions. Controls including no recombinant protein, no cellulosome, and no secondary antibody were run in parallel and resulted in no signal above background. All reactions were performed in triplicate.
2.6. Phylogenetic analysis of dockerin domain proteins from Piromyces finnis
The evolutionary history of all GH5 Dockerin Domain Proteins (DDPs) from P. finnis was inferred to determine whether conservation in dockerin placement existed by enzyme subtype. The tree was constructed using the Neighbor-Joining method [19], bootstrapped from 500 replicates [20]. The fractional percent of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. The evolutionary distances were computed using the Poisson correction method (21) and are in the units of the number of amino acid substitutions per site. The analysis involved 12 amino acid sequences. All ambiguous positions were removed for each sequence pair. There was a total of 1410 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [22]. Trees were edited using the Interactive Tree of Life [23].
3. Results
3.1. Sequence analysis of dockerin location on fungal genes informs chimera design
As a first step to systematically design chimeric proteins, the dockerin-containing genes from recently sequenced genomes of the anaerobic fungi Piromyces finnis, Neocallimastix californiae, and Anaeromyces robustus [8] were analyzed. As shown in Table 3, N. californiae contains 422 dockerin domain proteins (DDPs), nearly double the 223 DDPs found in anaerobic bacterium Ruminococcus flavifaciens FD-1, which is widely regarded as the most diverse repertoire in bacterial cellulosomes [24,25]. The other two members of the Neocallimastigomycota that currently have high quality genomes published, P. finnis and A. robustus, also contain more DDPs than R. flavifaciens, with 227 and 276 DDPs, respectively.
Table 3.
Strains and plasmids used in this study.
| Strain or Plasmid | Relevant genotype or features | Source |
|---|---|---|
| Strain | ||
| E. coli BL21 (DE3) |
fhuA2 [lon] ompT gal (λ DE3) [dcm] ΔhsdS λ DE3 = λ sBamHIo ΔEcoRI-B int::(lacI::PlacUV5::T7 gene1) i21 Δnin5 |
Laboratory stock |
| E. coli Tuner (DE3) | F– ompT hsdSB (rB– mB–) gal dcm lacY1 (DE3) | Laboratory stock |
| Piromyces finnis | [16] | |
| Plasmid | ||
| pET32a | E. coli expression vector to create N-terminal genetic fusion to TrxA | Novagen |
| pET32a-Cel5A | DNA encoding T. maritima Cel5A in pET32a | This study |
| pET32a-Cel5A-Dockerin | DNA encoding T. maritima Cel5A fused to C-terminal dockerin from P. finnis in pET32a | This study |
| pET32a-XynA | DNA encoding T. maritima XynA in pET32a | This study |
| pET32a-XynA-Dockerin | DNA encoding T. maritima XynA fused to C-terminal dockerin from P. finnis in pET32a | This study |
One fundamental difference between fungal DDPs and bacterial DDPs is the type and placement of dockerin domains that are fused to the catalytic enzymes. In fungal DDPs, dockerin domains are frequently located on either the N- or C-terminus of catalytic domains and in multiple copies (tandem repeats), whereas bacterial dockerins are almost exclusively found at the C-terminus with only a single domain present [8,26]. Based on this observation, we hypothesized that the dockerin type and placement was specific to each glycoside hydrolase (GH) enzyme type and therefore sought to characterize the GH type and dockerin location in fungal DDPs.
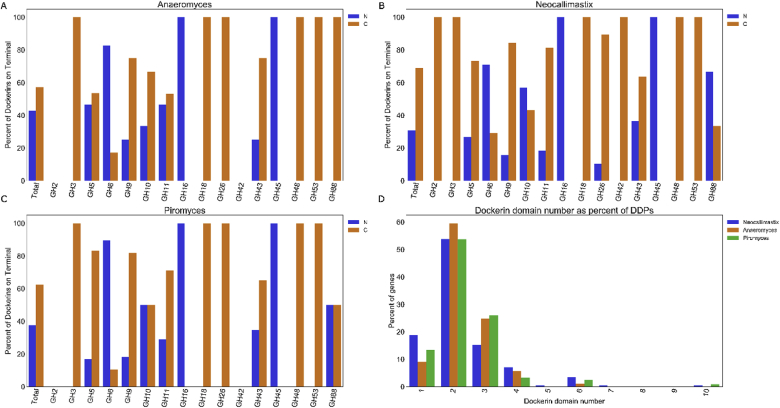
In total from fungal cellulosomes, nearly 60% of dockerin domains are found more proximal to the CAZyme C-terminus compared to the catalytic domain, as shown in Fig. 1. Some CAZymes, including those from the families GH2, GH8, GH11, GH26, GH38, and GH48, contain dockerin domains on the C-terminus exclusively. In these cases, it is possible that the N-terminus is inaccessible or buried within the catalytic domain such that N-terminal dockerins would adversely affect the catalytic activity. Many of the CAZymes thought to originate from bacteria contain dockerin domains at the C-terminus, which mirrors the placement of dockerins on similar CAZymes in bacterial cellulosomes [8]. The GH6 and GH45 are both domains found in fungal cellulosomes, yet are not typically found in bacterial cellulosomes [8], and interestingly, both of these catalytic domains are most likely to fuse fungal dockerins on the N-terminus rather than the C-terminus. Finally, the most common dockerin repeat motif was a double dockerin found in ~60% of DDPs, followed by triple dockerins and then single dockerins as shown in Fig. 1D. While the purpose of tandem repeats among the different fungal dockerins remains unknown, these data suggest that double dockerins are the optimal modular unit to employ for the design of chimeric enzymes.
Fig. 1.
Genomic analysis of dockerin domain proteins (DDPs) in anaerobic fungi. The frequency of dockerin domain placement of DDPs was determined for Anaeromyces robustus (A), Neocallimastix californiae (B), and Piromyces finnis (C). As shown, C-terminal dockerins were more common overall, however several glycoside hydrolase types had dockerins exclusively on the N-terminal. In particular, GH6 and GH45 are both more commonly found with N-terminal dockerins, and they are also families exclusive to fungal cellulosomes. In addition, the number of dockerins per DDP gene was determined for each genome (D). Dockerins are most commonly found in two copies on DPPs occurring just over 50% of all instances.
3.2. Synthesis of thermostable enzyme chimeras to facilitate fungal cellulosome association
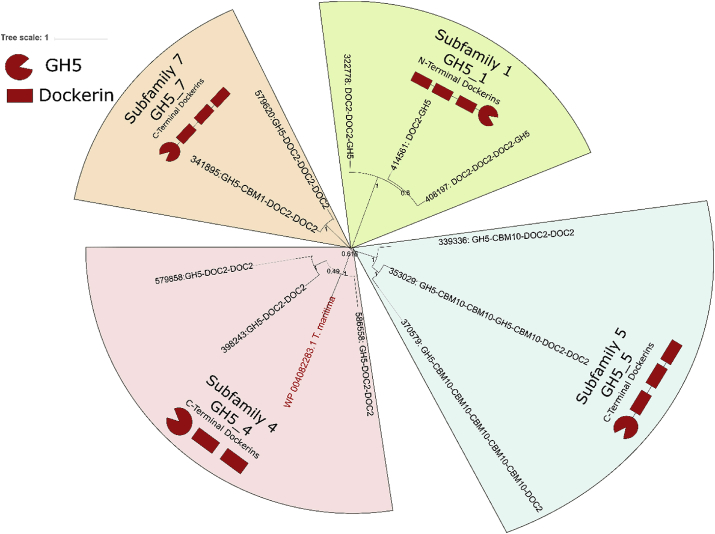
A GH5 from T. maritima was chosen for chimeric enzyme design because GH5s have relatively conserved dockerin placement, as shown in Fig. 1A–C, with the majority existing as double dockerins on the C-terminus in fungi, particularly in P. finnis. The GH5 chosen (Table 2) shared the highest amino acid sequence similarity to several proteins produced by P. finnis each around 26% identity, all of which had two dockerin domains at the C-terminus [8]. The GH5 from T. maritima clustered with Family 5 subfamily 4 (GH5_4) proteins from P. finnis, as shown in Fig. 2. All GH5_4 proteins with dockerin domains contained exactly two repeats at the C-terminus. This level of conservation extended to other enzyme subfamilies as well, where subfamily 1 (GH5_1) proteins all contained N-terminal dockerin domains, subfamily 7 (GH5_7) proteins all contained C-terminal dockerin domains, and subfamily 5 (GH5_5) proteins all contained C-terminal dockerins. These results demonstrate that even within GH families of mixed dockerin type (N- and C-terminal dockerins), there may be some preference at the subfamily level. Accordingly, the catalytic domain was cloned from the T. maritima GH5_4, and the double dockerin was cloned from a P. finnis GH5_4.
Fig. 2.
Phylogenetic analysis of Family 5 Glycoside Hydrolases in the anaerobic fungus P. finnis. The evolutionary history of all GH5 Dockerin Domain Proteins (DDPs) from P. finnis was inferred to determine whether conservation in dockerin placement is a function of enzyme subtype. As shown, the DDPs cluster based on GH5 subfamily [1,4,5,and7]], with dockerin placement conserved based on the subfamily. The GH5 cloned from T. maritima clustered with the DDPs from subfamily 4, and as such a C-terminal double dockerin repeat was used to construct the chimeric protein. The tree was constructed using the Neighbor-Joining method [19], bootstrapped from 500 replicates [20]. The fractional percent of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. The evolutionary distances were computed using the Poisson correction method [21] and are in the units of the number of amino acid substitutions per site.
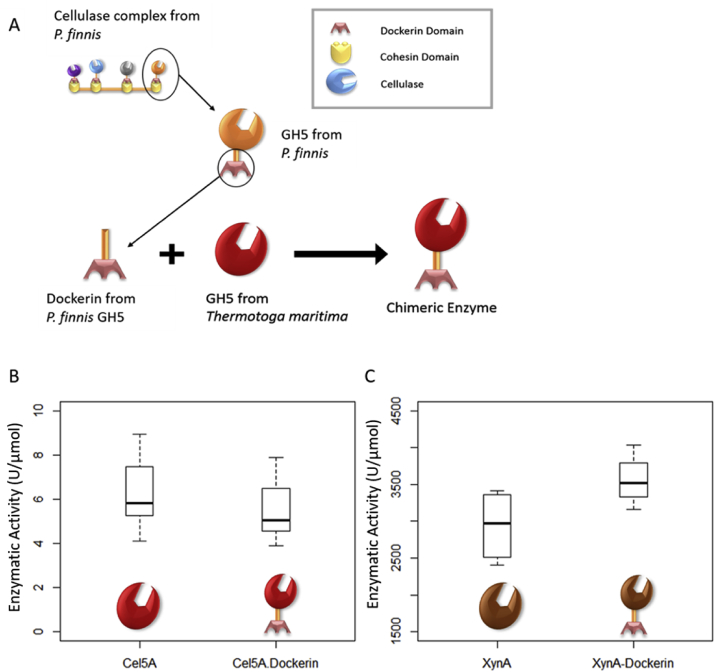
A schematic for the design of the chimeric thermostable enzymes is shown in Fig. 3A. These proteins were termed Cel5A and Cel5A-Dockerin for the T. maritima enzyme with and without the fungal dockerin domain, respectively. The T. maritima GH5 was previously characterized as an endoglucanase active at 80 °C and pH ~5 [27], and therefore carboxymethyl cellulose (CMC) was used as the substrate to characterize its activity at elevated temperature and acidic pH. As seen in Fig. 3B, there was no significant difference between the activities of the wild-type and chimeric enzymes.
Fig. 3.
Design of chimeric CAZymes for synthetic fungal cellulosomes. A) For optimal dockerin placement, the dockerin and linker sequence was taken from the closest-related sequence in P. finnis and grafted in the same location on the on chimeric T. maritima CAZymes. The CMCase or Xylanase activity at 80 °C was comparable for both the native enzyme and the enzyme with the grafted dockerin domain for both a GH5 (Cel5A, panel B) and a GH10 (XynA, panel C). Significance was tested using the Mann-Whitney U test and determined not significant for each set of enzymes. Units (U) are defined as the amount of enzyme required to release 1 μmol of reducing sugar per minute.
The technique for chimeric design was further extended to an endoxylanase (XynA, a GH10) from T. maritima (Table 2). It was identified as similar to dockerin-tagged proteins (C-terminal dockerins) in P. finnis at 36% identity. When the C-terminal double dockerin from the P. finnis GH10 (Table 2) was cloned onto XynA, there was no significant difference between the activities on xylan of XynA and XynA-Dockerin proteins, as shown in Fig. 3C. Collectively, these data suggest that the location-guided grafting approach was successful in engineering active chimeric enzymes with fungal dockerins.
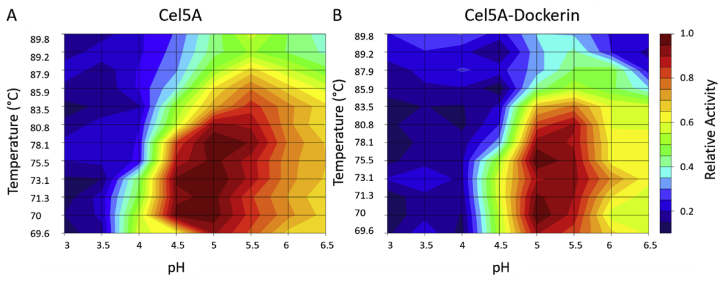
While enzyme activity was preserved in chimeric proteins, it is possible that addition of fungal dockerin domains could perturb other functional properties. Therefore, we measured the impact of the fungal dockerin addition on the optimal pH and temperature of the chimeric enzymes, and we related these changes to the native recombinantly produced enzyme lacking the dockerin. A plate-based assay was used to test a wide range of pH (3–6.5 in 0.5 increments) and temperature (70–90 °C in a gradient thermal cycler), akin to a recently reported method [28]. As shown in Fig. 4, the optimum temperature and pH were similar for both dockerin-fused and dockerin-free proteins, ranging from 70 to 80 °C and centered around pH 5. Cel5A alone spanned a broader range of optimum activity than Cel5A-Dockerin, extending the optimum pH to 4.5–5.5. Anaerobic fungi grow optimally near pH 6 and 39 °C, and therefore the mitigated activity is likely due to instability of the dockerin domain at high temperature and low pH.
Fig. 4.
pH and Temperature effects on the chimeric Cel5A-Dockerin. The optimum pH and temperature of Cel5A and Cel5A-Dockerin was tested to measure the functional impact of grafting a fungal dockerin domain onto Cel5A. While previous experiments showed that the Cel5A-Dockerin was not significantly different in activity than Cel5A at 80 °C and pH 5, there was a slight narrowing of the optimum pH and temperature of the enzyme. Values shown are the average of three blank-subtracted replicates.
3.3. Chimeric enzymes associate with native fungal cellulosomes
Preserving the activity of the catalytic domain is fundamental to demonstrating effectiveness of chimeric enzymes. However, chimeric enzymes must also form an association with complementary fungal cohesin domains to prove useful as synthetic biology tools. In this way, chimeric enzymes could be recruited into native fungal cellulosomes, or they could form the basis for completely synthetic fungal cellulosomes made de novo.
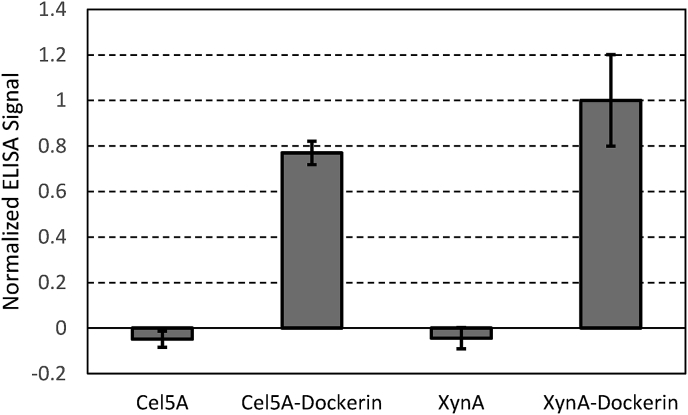
We used an ELISA assay against purified fungal cellulosomes to test whether the grafted dockerin domain could interact with native fungal enzyme complexes. Native cellulosome was bound to a plate and probed for interaction with Cel5A, Cel5A-Dockerin, XynA, or XynA-Dockerin. As shown in Fig. 5, the Cel5A-Dockerin and XynA-Dockerin both showed significant interaction with native cellulosomes isolated from the anaerobic fungus P. finnis, whereas Cel5A and XynA showed negligible binding over background at 3 μM enzyme concentration, ~3 times higher than the previously determined KD. [8] All constructs were cloned into the same vector (pET32) with the same fusion tag (TrxA), and TrxA was previously shown to interact negligibly with fungal cellulosomes [8], and therefore the binding was attributed to the dockerin domains. These results confirm that the dockerin domains grafted onto Cel5A-Dockerin and XynA-Dockerin still retain binding activity, and therefore the two proteins are properly constructed for recruitment into native cellulosomes or inclusion in synthetic enzyme complexes.
Fig. 5.
Chimeric enzymes bind to native fungal cellulosomes. Cel5A, Cel5A-Dockerin, XynA, and XynA-Dockerin were tested for interaction with cellulosome isolated from P. finnis. Only proteins with fungal dockerin domains (Cel5A-Dockerin and XynA-Dockerin) showed appreciable binding over background. Concentrations used were 3 μM for each protein, or 3 times the measured KD previously reported [8], indicative of maximal binding signal for cellulosome-binding proteins. Values displayed are the average of 3 replicates ± standard deviation as shown in error bars.
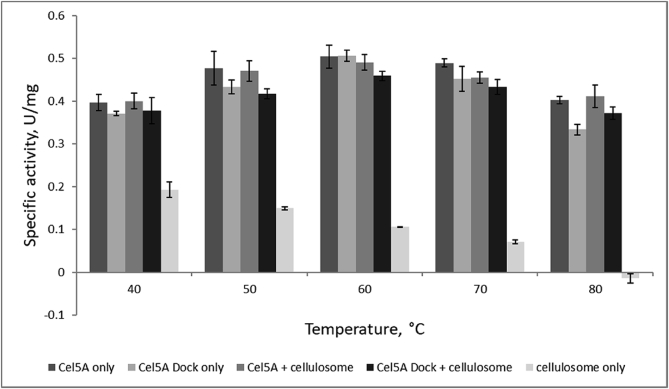
We further hypothesized that the incorporation of thermostable catalytic domains into fungal cellulosomes would enhance their thermal stability beyond that of a free enzyme plus cellulosome system. To test this, the activity of cellulosomes preincubated with Cel5A or Cel5A-Dockerin was measured at increasing temperatures to construct temperature optima curves for both systems. Fig. 6 shows no observable shift in specific activity vs temperature when comparing Cel5A-Dockerin plus cellulosome to Cel5A plus cellulosome. As the activity benefits of enzyme co-localization are well-described [6,29], a plausible explanation for these results is that Cel5A-Dockerin incorporation diminishes at higher temperatures with the unfolding of the dockerin or companion cohesin domains, resulting in loss in binding activity. Additionally, the benefits of co-localization are greatest against insoluble substrates, and therefore a soluble substrate like CMC is not expected to degrade significantly faster by tethered enzymes compared to those that are freely-diffusive.
Fig. 6.
Effect of chimeric enzyme incorporation on fungal cellulosome thermostability. Specific activity of mixtures of Cel5A and Cel5A-Dockerin with fungal cellulosome were compared at increasing temperatures. Values displayed are the average of 3 replicates ± standard deviation as shown in error bars.
4. Discussion
In this work, we demonstrated that the DDPs of anaerobic fungal cellulosomes are templates for designing chimeric proteins with favorable properties such as thermostability. The number of DDPs (>227 in each anaerobic fungal genome) is greater than commonly found in even the most extensive bacterial cellulosome producers (223 in R. flavifaciens), and therefore anaerobic fungi are a rich source of templates for creating fusion proteins. A freely diffusive GH5 and GH10 from T. maritima were successfully adapted to the cellulosome system by placing the dockerin domains in the same place as they are located on the native fungal DDP. The chimeric CAZymes still showed high activity and thermostability, equivalent to the enzymes lacking dockerin domains. Furthermore, the dockerin domains retained activity and allowed the chimeric enzymes to interact with native fungal cellulosome from P. finnis. Unfortunately, that interaction did not persist at higher temperatures, likely due to the thermal instability of the fungal dockerin-cohesin interaction. This remains a challenge for creating thermostable synthetic enzyme complexes based on the fungal cellulosome model, but could be overcome with future protein engineering.
The results shown here are an important first step towards designing chimeric enzymes, however, they represent only a subset of GH families found. Furthermore, the initial attempts at introducing non-cellulosomal GHs were unsuccessful, inferring that design rules for other catalytic domains might be different and still need to be determined. While thermostable enzymes were used as a test case here, the same process could be applied to other types of proteins with favorable properties, such as acid- or alkali-tolerant enzymes. Chimera design could be coupled to other methods of protein engineering, such as mutagenesis to further improve desired properties or in this study, to widen the pH and temperature optima of the Cel5A-Dockerin chimera. Finally, additional future work should seek to optimize the production of a minimal scaffoldin protein alongside the chimeric enzymes to establish a synthetic cellulosome system from anaerobic fungal cellulosomes.
5. Conclusions
In this study, we have shown that chimeric enzymes designed from the “bottom-up” (i.e. sequence modules) can be engineered with favorable properties such as thermostability and thermotolerance. Using native DDPs as templates, chimeric enzymes inspired by anaerobic fungal DDPs were engineered that retained activity at temperatures greater than 80 °C and were capable of interacting with purified native fungal cellulosomes. Applying the method presented here for synthetic DDPs to a wide group of cellulases and hemicellulases may result in a suite of enzymes capable of improving the economic efficiency of biomass degradation processes.
Availability of data and material
All strains and plasmids constructed in this study are freely available by request to the corresponding author.
CRediT authorship contribution statement
Sean P. Gilmore: Conceptualization, Investigation, Writing - original draft. Stephen P. Lillington: Investigation, Writing - original draft. Charles H. Haitjema: Methodology. Randall de Groot: Investigation. Michelle A. O'Malley: Conceptualization, Writing - review & editing, Funding acquisition, Project administration.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgements
The authors are grateful to funding sources from the Office of Science (BER), U.S. Department of Energy (DE-SC0010352), the National Science Foundation (MCB-1553721), the Institute for Collaborative Biotechnologies through grants W911NF-09-0001 and W911NF-19-D-0001 from the U.S. Army Research Office, and the Camille Dreyfus Teacher-Scholar Awards Program. All authors acknowledge support from the California NanoSystems Institute (CNSI) Challenge Grant Program, supported by the University of California, Santa Barbara and the University of California, Office of the President. SPG also acknowledges support from the National Science Foundation Graduate Research Fellowship Program under Grant DGE 1144085. We thank Prof. Robert Kelly for providing us with T. maritima DNA. The authors acknowledge the use of the Biological Nanostructures Laboratory within the California NanoSystems Institute, supported by the University of California, Santa Barbara and the University of California, Office of the President.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2020.01.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Viikari L., Alapuranen M., Puranen T., Vehmaanpera J., Siika-Aho M. Thermostable enzymes in lignocellulose hydrolysis. Adv Biochem Eng Biotechnol. 2007;108:121–145. doi: 10.1007/10_2007_065. Epub 2007/06/26. [DOI] [PubMed] [Google Scholar]
- 2.Fierobe H.P., Bayer E.A., Tardif C., Czjzek M., Mechaly A., Belaich A. Degradation of cellulose substrates by cellulosome chimeras - substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277(51):49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- 3.You C., Zhang X.Z., Zhang Y.H.P. Mini-scaffoldin enhanced mini-cellulosome hydrolysis performance on low-accessibility cellulose (Avicel) more than on high-accessibility amorphous cellulose. Biochem Eng J. 2012;63:57–65. [Google Scholar]
- 4.Morais S., Barak Y., Hadar Y., Wilson D.B., Shoham Y., Lamed R. Assembly of xylanases into designer cellulosomes promotes efficient hydrolysis of the xylan component of a natural recalcitrant cellulosic substrate. mBio. 2011;2(6) doi: 10.1128/mBio.00233-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai S.-L., DaSilva N.A., Chen W. Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth Biol. 2013;2(1):14–21. doi: 10.1021/sb300047u. [DOI] [PubMed] [Google Scholar]
- 6.Gefen G., Anbar M., Morag E., Lamed R., Bayer E.A. Enhanced cellulose degradation by targeted integration of a cohesin-fused beta-glucosidase into the Clostridium thermocellum cellulosome. Proc Natl Acad Sci. 2012;109(26):10298–10303. doi: 10.1073/pnas.1202747109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srikrishnan S., Chen W., Da Silva N.A. Functional assembly and characterization of a modular xylanosome for hemicellulose hydrolysis in yeast. Biotechnol Bioeng. 2013;110(1):275–285. doi: 10.1002/bit.24609. Epub 2012/07/19. [DOI] [PubMed] [Google Scholar]
- 8.Haitjema C.H., Gilmore S.P., Henske J.K., Solomon K.V., de Groot R., Kuo A. A parts list for fungal cellulosomes revealed by comparative genomics. Nat Microbiol. 2017;2:17087. doi: 10.1038/nmicrobiol.2017.87. Epub 2017/05/31. [DOI] [PubMed] [Google Scholar]
- 9.Henske J.K., Gilmore S.P., Knop D., Cunningham F.J., Sexton J.A., Smallwood C.R. Transcriptomic characterization of Caecomyces churrovis: a novel, non-rhizoid-forming lignocellulolytic anaerobic fungus. Biotechnol Biofuels. 2017;10:305. doi: 10.1186/s13068-017-0997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilmore S.P., Henske J.K., O'Malley M.A. Driving biomass breakdown through engineered cellulosomes. Bioengineered. 2015;6(4):204–208. doi: 10.1080/21655979.2015.1060379. Epub 2015/06/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagy T., Tunnicliffe R.B., Higgins L.D., Walters C., Gilbert H.J., Williamson M.P. Characterization of a double dockerin from the cellulosome of the anaerobic fungus Piromyces equi. J Mol Biol. 2007;373(3):612–622. doi: 10.1016/j.jmb.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Raghothama S., Eberhardt R.Y., Simpson P., Wigelsworth D., White P., Hazlewood G.P. Characterization of a cellulosome dockerin domain from the anaerobic fungus Piromyces equi. Nat Struct Biol. 2001;8(9):775–778. doi: 10.1038/nsb0901-775. [DOI] [PubMed] [Google Scholar]
- 13.Frock A.D., Notey J.S., Kelly R.M. The genus Thermotoga: recent developments. Environ Technol. 2010;31(10):1169–1181. doi: 10.1080/09593330.2010.484076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber R., Langworthy T., Köning H., Thomm M., Woese C., Sleytr U. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch Microbiol. 1986;144:324–333. [Google Scholar]
- 15.Nelson K.E., Clayton R.A., Gill S.R., Gwinn M.L., Dodson R.J., Haft D.H. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399(6734):323–329. doi: 10.1038/20601. Epub 1999/06/09. [DOI] [PubMed] [Google Scholar]
- 16.Solomon K.V., Haitjema C.H., Henske J.K., Gilmore S.P., Borges-Rivera D., Lipzen A. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science. 2016;351(6278):1192–1195. doi: 10.1126/science.aad1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali B.R.S., Zhou L.Q., Graves F.M., Freedman R.B., Black G.W., Gilbert H.J. Cellulases and hemicellulases of the anaerobic fungus Piromyces constitute a multiprotein cellulose-binding complex and are encoded by multigene families. FEMS Microbiol Lett. 1995;125(1):15–21. doi: 10.1111/j.1574-6968.1995.tb07329.x. [DOI] [PubMed] [Google Scholar]
- 18.Xiao Z., Storms R., Tsang A. Microplate-based carboxymethylcellulose assay for endoglucanase activity. Anal Biochem. 2005;342(1):176–178. doi: 10.1016/j.ab.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 19.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. Epub 1987/07/01. [DOI] [PubMed] [Google Scholar]
- 20.Felenstein J. Confidence limits on phylogenies: an approach using the bootstrap on JSTOR. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 21.Zuckerkandl E., Pauling L., editors. Evolutionary divergence and convergence in proteins. Academic Press; New York: 1965. [Google Scholar]
- 22.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. Epub 2018/05/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. Epub 2016/04/21. PubMed PMID: 27095192; PubMed Central PMCID: PMCPMC4987883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dassa B., Borovok I., Ruimy-Israeli V., Lamed R., Flint H.J., Duncan S.H. Rumen cellulosomics: divergent fiber-degrading strategies revealed by comparative genome-wide analysis of six ruminococcal strains. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0099221. Epub 2014/07/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg Miller M.E., Antonopoulos D.A., Rincon M.T., Band M., Bari A., Akraiko T. Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1. PLoS One. 2009;4(8) doi: 10.1371/journal.pone.0006650. Epub 2009/08/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayer E.A., Belaich J.P., Shoham Y., Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 27.Pereira J.H., Chen Z., McAndrew R.P., Sapra R., Chhabra S.R., Sale K.L. Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima. J Struct Biol. 2010;172:372–379. doi: 10.1016/j.jsb.2010.06.018. United States: Published by Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- 28.Herlet J., Kornberger P., Roessler B., Glanz J., Schwarz W.H., Liebl W. A new method to evaluate temperature vs. pH activity profiles for biotechnological relevant enzymes. Biotechnol Biofuels. 2017;10(1):234. doi: 10.1186/s13068-017-0923-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fierobe H.P., Mingardon F., Mechaly A., Belaich A., Rincon M.T., Pages S. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J Biol Chem. 2005;280(16):16325–16334. doi: 10.1074/jbc.M414449200. [DOI] [PubMed] [Google Scholar]
- 30.Zhivin O., Dassa B., Morais S., Utturkar S.M., Brown S.D., Henrissat B. Unique organization and unprecedented diversity of the Bacteroides (Pseudobacteroides) cellulosolvens cellulosome system. Biotechnol Biofuels. 2017;10:211. doi: 10.1186/s13068-017-0898-6. Epub 2017/09/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dassa B., Borovok I., Lamed R., Henrissat B., Coutinho P., Hemme C.L. Genome-wide analysis of Acetivibrio cellulolyticus provides a blueprint of an elaborate cellulosome system. BMC Genomics. 2012;13:210. doi: 10.1186/1471-2164-13-210. Epub 2012/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains and plasmids constructed in this study are freely available by request to the corresponding author.