Abstract
Autism spectrum disorders are associated with some degree of developmental regression in up to 30% of all cases. Rarely, however, is the regression so extreme that a developmentally advanced young child would lose almost all ability to communicate and interact with her surroundings. We applied trio whole exome sequencing to a young woman who experienced extreme developmental regression starting at 2.5 years of age and identified compound heterozygous nonsense mutations in TMPRSS9, which encodes for polyserase-1, a transmembrane serine protease of poorly understood physiological function. Using semiquantitative polymerase chain reaction, we showed that Tmprss9 is expressed in various mouse tissues, including the brain. To study the consequences of TMPRSS9 loss of function on the mammalian brain, we generated a knockout mouse model. Through a battery of behavioral assays, we found that Tmprss9−/− mice showed decreased social interest and social recognition. We observed a borderline recognition memory deficit by novel object recognition in aged Tmprss9−/− female mice, but not in aged Tmprss9−/− male mice or younger adult Tmprss9−/− mice in both sexes. This study provides evidence to suggest that loss of function variants in TMPRSS9 are related to an autism spectrum disorder. However, the identification of more individuals with similar phenotypes and TMPRSS9 loss of function variants is required to establish a robust gene–disease relationship.
Introduction
Autism spectrum disorder (ASD) is a group of neurodevelopmental disorders with a wide range of symptoms. Individuals with ASD manifest difficulties in social interactions, repetitive behaviors, restrictive interests, and impairments in communication. A recent study from the Centers for Disease Control and Prevention estimated the prevalence of ASD in the general population to be around 1/59 (1). With the increased public awareness of ASD as well as the advancement of genome-wide sequencing technology, to date more than 1000 genes implicated in ASD have been curated in the SFARI database (https://gene.sfari.org/), suggesting very high levels of genetic heterogeneity in ASD.
Developmental regression has been found in 15–47% patients diagnosed with ASD among different studies (2). The mean age of the reported developmental regression in the patients typically ranges from 15 to 20 months (3). It is relatively uncommon to see a clear regression after 2 years of age. We identified an individual with profound developmental regression, including speech, social interaction and motor skills, beginning at 2.5 years of age. The regression resulted in ASD and profound intellectual disability. Given the rare nature and time course of the clinical course in this individual, we applied trio whole exome sequencing, expecting that we might learn about the underlying molecular underpinnings.
Animal models are valuable to study the causal role of the disease candidate genes, especially for rare genetic disorders. To understand the consequences of TMPRSS9 loss of function on the mammalian brain, we generated a knockout mouse model. Tmprss9−/− mice display no visible phenotypic difference from wild-type littermates. Through a battery of behavioral assays, we found that Tmprss9−/− mice showed decreased social interest and social recognition using the three-chamber assay and the partition assay. We observed a borderline recognition memory deficit by novel object recognition in 1-year-old Tmprss9−/− female mice, but not in age-matched Tmprss9−/− male mice. This work provides a basis to study rare variants with a personalized medicine approach combining next generation sequencing and animal models.
Results
Identification of compound heterozygous nonsense variants in TMPRSS9
The female patient was born to a then 42-year-old mother and 41-year-old father, both of Italian descent. She was delivered at 24 weeks of gestation via emergency Caesarean section, with a birth weight of 588 g. Her hospital course in the newborn intensive care unit was complicated by bronchopulmonary dysplasia and pneumonia, patent ductus arteriosus (requiring surgical closure) and retinopathy of prematurity (requiring laser surgery). Early childhood development was normal, in fact advanced. At 16–18 months, she was able to speak in 3–4 word sentences with clear diction and count to 100; she could identify and spell the names of most colors; she could draw many shapes, including more complex ones such as a trapezoid and she memorized a number of songs, stories and poems and could fill in the missing phrases from them. However, she never developed normal socialization. She was never able to have an interactive conversation and was unable to interact with other children. Her neurodevelopment peaked at 2.5 years, followed by developmental regression, including the loss of purposeful hand skills, loss of expressive language and the onset of self-stimulating activities. Her brain magnetic resonance imaging showed periventricular leukomalacia, hypoplasia of the right cerebellar hemisphere and an enlarged amygdala. She suffered from multiple neurological problems, including spastic diplegia and decreased truncal tone. At age 5, EEG with video monitoring showed background slowing, along with epileptiform activity, which was much more prominent during sleep. The pattern of EEG changed over time. At age 8, she had an occipital dominant rhythm, with only mild background slowing and rare occurrence of epileptiform activity, which was confined to the left and right centrotemporal regions. At age 9, Mullen Scales of Early Learning were used to assess her language, motor, and perceptual abilities. The results showed that her visual reception skills, receptive language skills, expressive language skills and fine-motor skills were in the 10- to 15-month-old range. Vineland Adaptive Behavior Scales II were used to evaluate her daily living skills, socialization, communication and motor skills. The overall results showed that her performance was below a 3-year-old level, indicative of severe intellectual disability. Other reported concerns included sleep problems, aggressive behavior, self-injurious and repetitive behaviors.
The patient’s father has hypertrophic cardiomyopathy but is otherwise in good health. History of the maternal family included thyroid dysfunction, diabetes and a maternal great aunt with seizures beginning at age 16 years following a febrile illness. There are no members with ASD. Diagnostic records showed negative testing for MECP2 mutations, fragile X syndrome, plasma amino acid analysis, urine organic acid analysis and heavy metal screens. Whole exome sequencing of the patient’s DNA identified compound heterozygous nonsense mutations in TMPRSS9, which were validated by Sanger sequencing (Fig. 1). She inherited a paternal nonsense mutation (c.286C>T; p.R96*) in exon 3 and a maternal nonsense mutation (c.1267C>T; p.R423*) in exon 9 in TMPRSS9. The former nonsense mutation resides between the transmembrane domain and low-density lipoprotein receptor class A (LDLA) domain; the latter resides within the serase-1 domain (Fig. 2).
Figure 1.
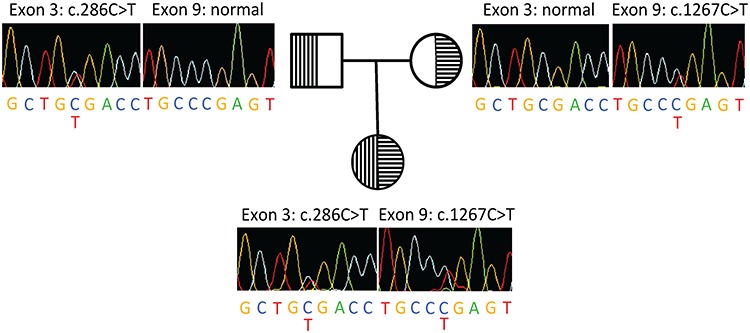
The female proband carries compound heterozygous nonsense mutations in TMPRSS9. Sanger sequencing of the compound heterozygous nonsense mutations in the female proband and her heterozygous parents (carriers).
Figure 2.
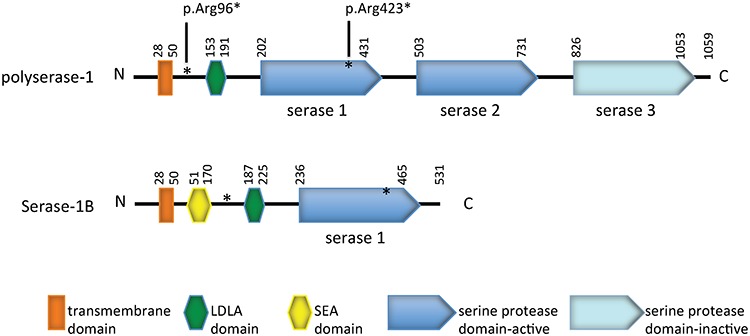
Functional domains of human polyserase-1 and serease-1B. The compound heterozygous nonsense variants (paternal p.Arg96* and maternal p.Arg423*) found in the patient are denoted in asterisks (see context).
Tmprss9 expresses throughout the mouse brain, with the highest expression in cerebellum
Human polyserase-1 shares 79% amino acid sequence identity with its mouse homologue (Supplementary Material, Fig. S1). Mouse Tmprss9 mRNA expression was detected in many tissues, including the brain, kidney, testis and liver, but not in heart, lung and skeletal muscle (Fig. 3A). Considering the clinical features of the patient, we further investigated Tmprss9 expression in mouse brains. Semiquantitative and quantitative polymerase chain reaction (PCR) analysis showed that Tmprss9 is expressed throughout the mouse brain, although the expression is generally low. It has the highest expression in cerebellum, among other brain regions, including cortex, hippocampus, amygdala, brainstem and hypothalamus (Fig. 3B and C).
Figure 3.
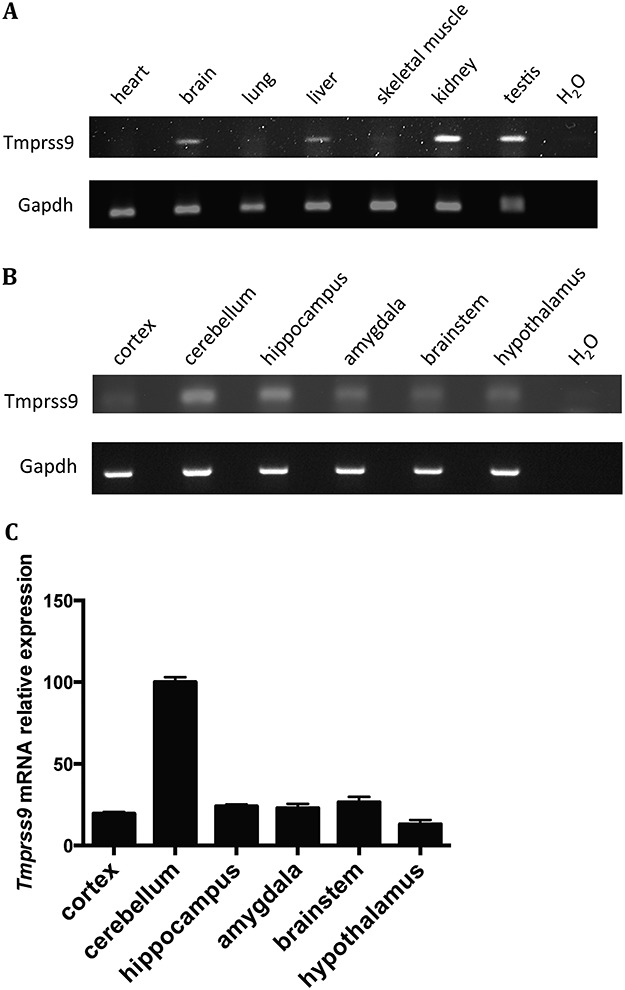
Tmprss9 expression in mouse tissues and specific brain regions. (A) Semiquantitative PCR of Tmprss9 mRNA in multiple tissues in wild-type mice. (B) Semiquantitative PCR of Tmprss9 mRNA in multiple brain regions in wild-type mice. (C) Quantitative RT-PCR of Tmprss9 mRNA expression in various brain regions. Data are normalized to Rps16 mRNA levels. Data are shown as mean ± standard error of mean (N = 2).
Generation of a Tmprss9 knockout mouse model
The design and generation of a Tmprss9 constitutive knockout mouse model and a Tmprss9 conditional knockout mouse model were performed by TaconicArtemis GmbH (Cologne, Germany). The targeting vector was designed to generate both a constitutive knockout allele and a conditional knockout allele by a sequential breeding strategy (Fig. 4A). A human growth hormone poly(A) signal sequence (hGH pA) flanked by two FRT sites was inserted between exon 1 and exon 2. The insertion of this gene trap cassette after exon 1 was expected to result in transcriptional termination before exon 2 of 18 in mouse Tmprss9. While homozygous and heterozygous KO mice were viable and fertile, quantitative PCR analysis indicated low knockout efficiency in mouse brains (data not shown). Therefore, we generated a different constitutive knockout allele by a sequential breeding strategy (Fig. 4A). We removed the gene trap cassette by breeding with C57BL/6-Tg(CAG-flpe)2Arte mice, which ubiquitously express flippase, leaving two loxP sites flanking exon 2. Exon 2 was then deleted by breeding with Hprttm1(CAG-cre)Mnn (Fig. 4A). The deletion of exon 2 leads to frameshifting and a premature stop codon in exon 3. Homozygous KO mice were then generated from mating heterozygous KO mice (Fig. 4B). Both homozygous and heterozygous KO mice maintained under C57BL/6J genetic background were viable and fertile, with no visible phenotypic differences from wild-type littermates. They were born at the expected Mendelian ratio (wild-type 26.7%; heterozygous 49.3%; homozygous 23.9%). Overall, the body weight of Tmprss9−/− mice is comparable to wild-type littermates (Supplementary Material, Fig. S2A and B). Based on the quantitative PCR using primers targeting exons 2–3, the knockout efficiency of Tmprss9 was over 95% in the brain and the kidney, and Tmprss9 cDNA was not detectable by semiquantitative RT-PCR (Fig. 4C–E). However, quantitative PCR using primers targeting exons 5–6 and exons 11–12 showed low level of expression in the kidney and similar amount of expression in the brain compared with wild-type littermates, suggesting the escape from nonsense-mediated mRNA decay. Due to a lack of polyserase-1 specific antibodies, we were not able to demonstrate the knockout efficiency at the protein level.
Figure 4.
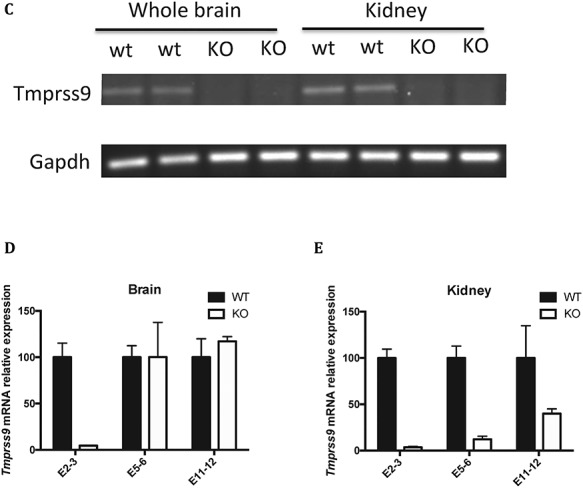
Continued.
Figure 4.
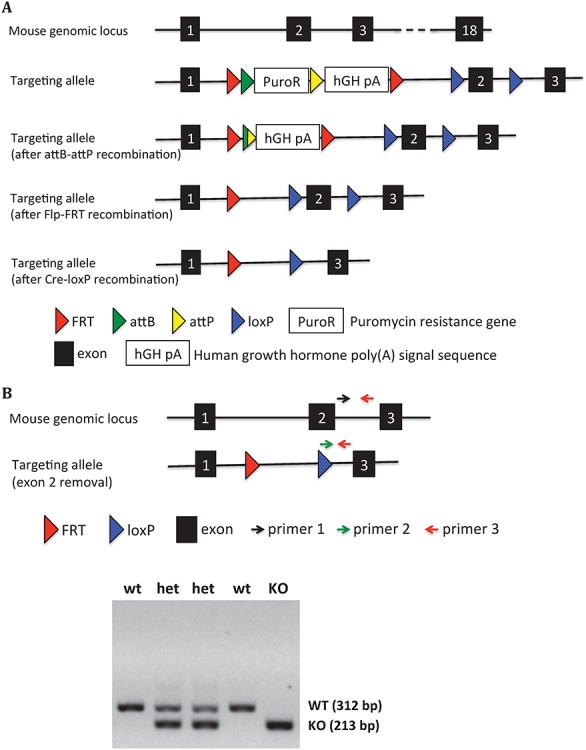
Generation of the Tmprss9 knockout mouse model. (A) The targeting vector was electroporated into ES cells, followed by injection into blastocysts that were implanted into pseudopregnant females. The puromycin resistance gene was removed by breeding with B6-Gt(ROSA)26SorphiC31, generating a constitutive knockout allele due to an insertion of a gene trap cassette between exon 1 and exon 2. The removal of human growth hormone poly(A) signal sequence was performed by breeding with C57BL/6-Tg(CAG-Flpe)2Arte mice, generating a conditional knockout allele. The removal of exon 2 was carried out by breeding with Hprttm1(CAG-cre)Mnn in B6 background, generating a second constitutive knockout allele. (B) Three primers (primer 1, 2 and 3) mixture in PCR reaction were used to genotype constitutive Tmprss9−/− mice (removal of exon 2). The size of WT allele and KO allele PCR products are 312 and 213 bp, respectively. (C) Semiquantitative PCR of Tmprss9 mRNA in Tmprss9−/− mice (removal of exon 2) in brains and kidneys using primers targeting exons 2–3. N = 2. Quantitative PCR of Tmprss9 mRNA in Tmprss9−/− mice (removal of exon 2) in brains and kidneys using primers targeting exons 5–6 (D) and exons 11–12 (E). N = 2.
To investigate whether Tmprss9−/− mice show abnormalities in overall brain cytoarchitecture, we performed immunohistochemical staining in mouse brain sections with anti-NeuN antibody. No remarkable effect on overall brain cytoarchitecture was detected in Tmprss9−/− mice compared with wild-type littermates (Supplementary Material, Fig. S2C and D). Further quantification of NeuN-positive neuronal density in the hippocampus, cerebellum and amygdala showed no significant difference (Supplementary Material, Fig. S2E–G).
Tmprss9−/− mice display deficits in both social interest and social recognition
We applied two commonly used assays to evaluate the social interest and social recognition in Tmprss9−/− mice, the three-chamber interaction and partition tests. In the three-chamber interaction test, both wild-type and Tmprss9+/− littermates spent significantly more time with a novel mouse partner than an inanimate object. Although Tmprss9−/− mice generally spent more time interacting with a novel mouse partner, comparison to time spent interaction with the inanimate object did not reveal a significant difference (Fig. 5A–D). The results were consistent in both sexes and both age groups (13-week-old and 1-year-old). In the partition test, the amount of time Tmprss9−/− mice spent interacting with familiar mouse partners was comparable to that of wild-type and Tmprss9+/− littermates. However, when the familiar mouse partner was replaced by a novel mouse, Tmprss9−/− mice spent significantly less time interacting with them, both when compared to wild-type littermates and when compared to Tmprss9+/− littermates (Fig. 5E–H). This was true for female Tmprss9−/− mice, both at the 13-week and 1-year time point. Findings were also significant for the 1-year-old male Tmprss9−/− mice. The 15-week-old Tmprss9−/− male mice showed a similar trend without reaching statistical significance (Fig. 5E). Both the three-chamber interaction and partition test suggest a social interaction deficit in Tmprss9−/− mice. The phenotype is similar to the clinical features of the patient, who never developed normal socialization.
Figure 5.
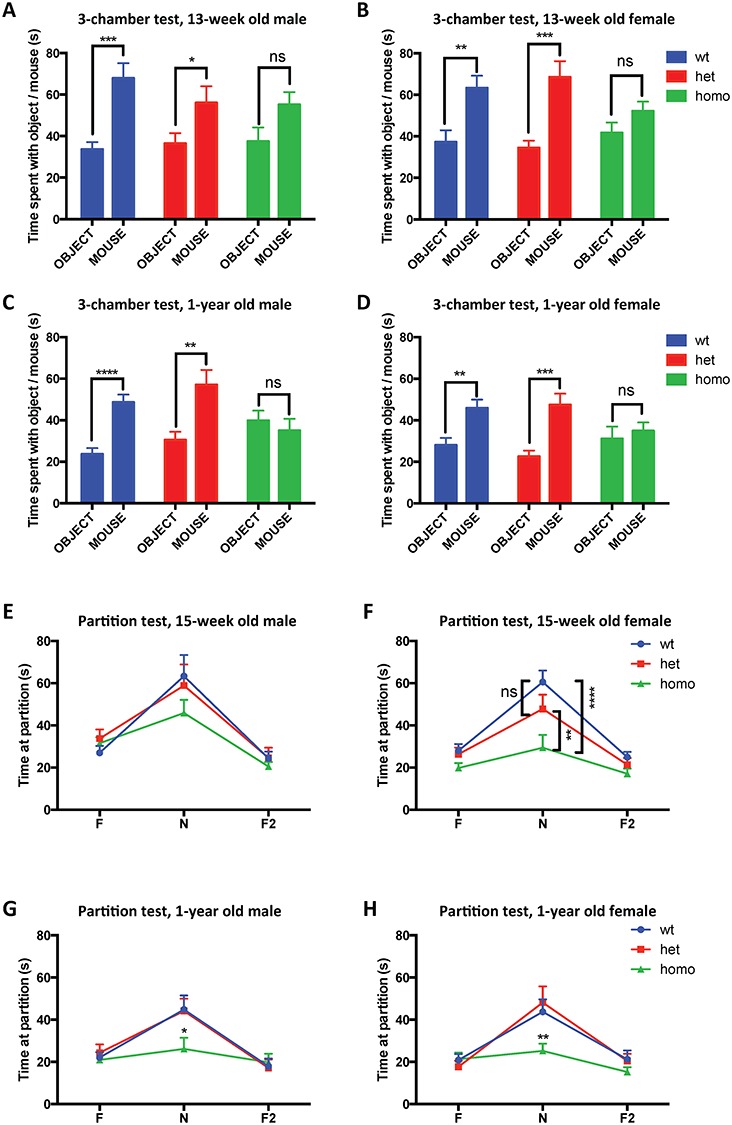
Tmprss9−/− mice display deficits in both social interest and social recognition. (A–D) Tmprss9−/− mice in three-chamber interaction test. (A) 13-week-old males, N = 15–17; (B) 13-week-old females, N = 14–16; (C) 1-year-old males, N = 15–17 and (D) 1-year-old females, N = 14–20. (E–H) Tmprss9−/− mice in partition test. (E) 15-week-old males, N = 15–20; (F) 15-week-old females, N = 16–20; (G) 1-year-old males, N = 15–17 and (H) 1-year-old females, N = 14–20. Data are shown as mean ± standard error of mean. ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Tmprss9−/− mice do not manifest anxiety-like behaviors, motor learning and coordination deficits, nociception defects and sensorimotor gating abnormalities
As part of a broad phenotypic characterization of the mice, three different behavioral assays were performed to assess anxiety-like behavior in Tmprss9−/− mice: elevated plus maze, light dark box and the time spent in the central area in the open field assay. Tmprss9−/− mice did not differ from wild-type mice in all three assays (Supplementary Material, Fig. S3A–L), suggesting no apparent anxiety-like behavior. Given that Tmprss9 expression is relatively high in the cerebellum, we checked whether locomotion or motor coordination was affected in Tmprss9−/− mice. Total distance traveled in the open field assay was used to assess potential locomotion deficits in Tmprss9−/− mice, while the accelerating rotarod and parallel rod footslip tests were performed to evaluate the motor coordination of the mice. We did not notice a striking difference in the three assays (Supplementary Material, Fig. S4A–J), although 11-week-old Tmprss9−/− male mice showed a statistically significant motor coordination or motor learning deficit on the last day of trials (Supplementary Material, Fig. S4E). However, we did not see the same pattern in the age-matched females or 1-year-old Tmprss9−/− mice of both sexes (Supplementary Material, Fig. S4F–H). Additional tests such as the marble burying test, hot plate test and prepulse inhibition (PPI) test were performed to evaluate the repetitive behaviors, nociception and sensorimotor gating, respectively. Tmprss9−/− mice (11–16 weeks) did not show significant deficits in nociception, sensorimotor gating or repetitive behaviors compared to wild-type littermates (Supplementary Material, Fig. S5A–F), although 16-week-old Tmprss9+/− female mice seemed to have delayed pain response (Supplementary Material, Fig. S5D).
Aged Tmprss9−/− female mice show borderline learning and memory deficits in novel object recognition test
Novel object recognition was used to evaluate the recognition memory in 1-year-old Tmprss9−/− mice, as the patient manifests intellectual disability. We found that both Tmprss9−/− and Tmprss9+/− female mice showed borderline significant deficits in the discrimination between novel and familiar objects, as indicated by the discrimination index (Fig. 6D). The discrimination index is defined by [(novel object exploration time)—(familiar object exploration time)]/[(novel object exploration time) + (familiar object exploration time)] (4–6). However, we did not see significant differences in their age-matched male counterparts (Fig. 6C) or in younger adults (Fig. 6A and B). Contextual and cued fear conditioning tests were used to assess the fear memory in Tmprss9−/− mice. The results indicated that Tmprss9−/− mice of both sexes and age groups (15-week-old and 1-year-old) do not have fear memory deficits (Supplementary Material, Fig. S6A–H). These memory assays suggested that 1-year-old Tmprss9−/− female mice have minor recognition memory deficits, but normal fear memory.
Figure 6.
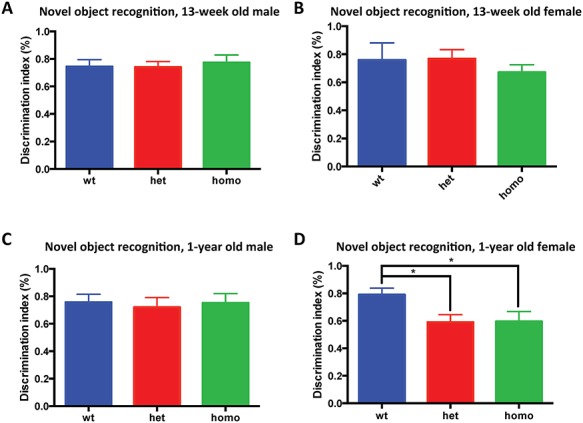
Aged Tmprss9−/− female mice show learning and memory deficits in novel object recognition. (A) 13-week-old males, N = 16–17; (B) 13-week-old females, N = 14–15; (C) 1-year-old males, N = 15–17 and (D) 1-year-old females, N = 14–20. Data are shown as mean ± standard error of mean. *P < 0.05.
Discussion
In this study, we identified TMPRSS9 compound heterozygous nonsense variants in a female patient, who suffered from developmental regression, ASD and intellectual disability. The human genome contains 17 Type II Transmembrane Serine Proteases (TTSP) (7). All of the TTSPs have a short intracellular domain near the amino terminus, followed by a single-pass transmembrane domain and an extracellular serine protease domain at the carboxy terminus. A highly variable ‘stem region’, which may consist of up to 11 protein domains of six different types, is located between the transmembrane domain and the serine protease domain (7). The stem region and the extracellular serine protease domain are considered to dictate the substrate specificity, cellular localization and activation and inhibition of TTSPs. Although some of the TTSPs have been demonstrated to play critical roles in development, homeostasis and pathogenesis of various tumor types, the roles of certain TTSPs remain largely unknown (7–9).
The human TMPRSS9 gene has 17 exons and is located on chromosome 19. TMPRSS9 encodes for polyserase-1, which is considered atypical in the subfamily because it is the only TTSP member with more than one serine protease domain (Fig. 2). Due to its three serine protease domains in the extracellular domain at the carboxy terminus, it was named polyserase-1 (10). Additionally, polyserase-1 has one LDLA domain in its stem region. Human polyserase-1 shares 79 and 78% amino acid identity with mouse and rat orthologs, respectively. The first two serine protease domains (designated serase-1 and serase-2 for the rest of the study) were shown to be catalytically active (10), given that both possess the critical catalytic triad residues (histidine, aspartate and serine) within their typical motif. However, the third serine protease domain (serase-3) was predicted to be inactive due to the replacement of serine (of the catalytic triad) with alanine. A shorter TMPRSS9 isoform, which encodes for serase-1B, possesses only the first serine protease domain (serase-1) with a slightly larger stem region due to an inclusion of a Sea urchin sperm protein, Enteropeptidase, Agrin (SEA) domain that is not seen in the longer isoform (Fig. 2) (11).
The knockout mouse model of Tmprss9 in this study displays no visible phenotypic difference from wild-type littermates. Through a battery of behavioral assays, we found that Tmprss9−/− mice showed decreased social interest and social recognition deficits, using the two gold-standard assays of social interaction in mice, i.e. the three-chamber assay and the partition assay. Although we observed a borderline recognition memory deficit by novel object recognition in 1-year-old Tmprss9−/− female mice, we did not see a similar trend in the age-matched Tmprss9−/− male mice. Fear memory also appeared to be normal in Tmprss9−/− mice, indicating that learning and memory may not be the strongest phenotypes seen in this knockout mouse model. Novel object recognition relies heavily on the perirhinal cortex and hippocampus, although they seem to play different roles in the task. While the former is involved in representing basic information about familiarity or novelty of an object after short retention intervals, the latter is involved in object memorization by encoding the information regarding the experience of object, responsible for long-term object recognition. Lesions of either brain structures are known to impair novel object recognition (12). However, in contextual and cued fear conditioning, the amygdala and hippocampus are believed to play the major roles. Whether the discrepancy shown in these two memory assays suggests a comparatively more important role of Tmprss9 in the perirhinal cortex requires further studies.
The Tmprss9−/− mice generated in this study suggested the escape from nonsense-mediated mRNA decay, based on our quantitative PCR results using additional primers targeting exons other than exon 2, which was removed through Cre/loxP recombination. Variability in nonsense-mediated mRNA decay among different tissue types has been reported both in human patient and rodents (13, 14). However, the deletion of exon 2 is expected to result in frameshifting and a premature stop codon in exon 3 in both Tmprss9 isoforms reported to date (10, 11).
Although Tmprss9−/− mice seem to be a good mouse model for autism-related behaviors, especially social interaction deficits, they do not fully recapitulate the clinical features of the patient, such as severe intellectual disability, self-stimulating activities, spastic diplegia and decreased truncal tone. Notably, although the patient has severe developmental regression, Tmprss9−/− mice did not manifest developmental regression, even in the aged 1-year-old mice. Given that the patient was born prematurely at 24-week gestational age, there is a possibility of neurological effects related to extreme prematurity, leading to a blended clinical phenotype caused by two independent etiologies. To date, we are not aware of additional patients carrying explicitly pathogenic homozygous or compound heterozygous loss of function mutations in TMPRSS9. Based on the Exome Aggregation Consortium (ExAC) database, stop-gain or frameshifting mutations of TMPRSS9 are seen in 1 in 714 individuals. Rough estimation suggested that the prevalence of the individuals carrying both loss-of-function alleles in the general population is about 1 in 2 million people. At this stage, it is also critical to identify more patients with TMPRSS9 loss-of-function variants to provide credence to this study in the context of ASD and related neurological phenotypes. In the absence of additional patients being identified, we cannot rule out the possibility that the patient’s phenotype may be the result of another genetic or environmental cause not yet identified.
Although we argued that preterm birth of the patient might contribute to certain clinical features not recapitulated in our mouse model, it is also possible that the species difference between human and mice underlies the disparity. Future studies focusing on the roles of TMPRSS9 in human neural culture might help address the question, for example comparing the morphology and the electrophysiological properties of the neurons derived from patient induced pluripotent stem cells and their isogenic cell lines, which correct TMPRSS9 mutations through clustered regularly interspaced short palindromic repeats (CRISPR). These results will inform us about whether mouse models could potentially serve as a good disease model or whether we should focus more on the human-based studies in the future.
Materials and Methods
Whole exome sequencing
For each DNA sample, the entire exome was captured using the custom NimbleGen VCRome 2.1 capture reagent that targets coding exons from the consensus coding sequence, NCBI RNA reference sequences and Vega human genome annotations. Capture enrichment was followed by sequencing on the Illumina platform using previously described standard protocols (15). Illumina sequence analysis was performed using the Human Genome Sequencing Center’s integrated Mercury pipeline (16). Briefly, the sequencing reads were mapped to the GRCh37 human reference sequence using Burrows-Wheeler Aligner. The Atlas suite was used to call single-nucleotide and short indel variants. Lastly, variant annotation was accomplished through the Cassandra annotation suite, utilizing different databases to predict functional consequences of genomic variants and place the variants in a biological framework.
Quantitative real-time PCR
Total RNA was extracted from mouse tissues with an miRNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. RNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher). RNA was reverse-transcribed to complementary DNA using a reverse transcription kit (Qiagen). Complementary DNA samples then underwent quantitative real-time PCR (RT-PCR) on the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) with SYBR Green FastMix (Quanta Biosciences). Relative quantities of Tmprss9 messenger RNA were measured with the ΔΔCT method and normalized against the housekeeping gene GAPDH. The quantitative RT-PCR primer sequences were as follows: mouse Tmprss9 forward, 5′-TTGCAGCAGGAGAACTCTGA-3′; mouse Tmprss9 reverse, 5′-GCGCAGCTCGACTCTAACTC-3′; mouse Rps16 forward, 5′-AGGAGCGATTTGCTGGTGTGG-3′ and mouse Rps16 reverse, 5′-GCTACCAGGGCCTTTGAGATG-3′. The semiquantitative reverse transcriptase PCR primer sequences were as follows: mouse Tmprss9 forward, 5′-TTGCAGCAGGAGAACTCTGA-3′; mouse Tmprss9 reverse, 5′- GCGCAGCTCGACTCTAACTC-3′; mouse Gapdh forward, 5′-CGACCACTTTGTCAAGCTCA-3′ and mouse Gapdh reverse, 5′-TTACTCCTTGGAGGCCATGT-3′.
Three-chamber interaction test
Three-chamber interaction was used to assess the sociability in mice as published previously with few modifications (17). During the habituation phase, the mice were placed in the middle chamber and allowed to explore all three chambers for 10 min freely. The left and right chambers each contain an empty wire cage with a weighted bottle on top of it. During the sociability phase, the age- and sex-matched novel partner mouse was placed in the wire cage in either the right or left chamber randomly. A LEGO object, similar in size to a mouse body, was placed in the wire cage in the other chamber. The test mice were then allowed to explore freely for 10 min during the sociability phase. The total amount of time that the test mice spent sniffing, pawing or rearing at each wire cage during the two phases were manually scored. Data are shown as mean ± standard error of mean. Analysis was done by paired Student’s t test with Bonferroni’s correction comparing ‘novel partner mouse’ and ‘inanimate object’ as a measure of sociability.
Partition test
The partition test was used to assess social recognition in mice as previously described (18). Test mice were housed individually for 1 day in standard housing cages separated by a clear perforated Plexiglas barrier into two compartments. Age- and sex-matched partner mice were placed in the other compartment on Day 2, allowing for sight and smell interaction with test mice through the barrier. The total amount of time that the test mice spent on sniffing, pawing or rearing at the barrier was manually scored during the three phases (5 in each): test mice versus familiar partner (F), test mice versus novel partner (N) and repeated test mice versus familiar partner (F2). Data are shown as mean ± standard error of mean and are analyzed by two-way analysis of variance (ANOVA) with Bonferroni’s post hoc analysis.
Elevated plus maze test
The elevated plus maze test was used to evaluate the anxiety in mice, as previously described (19), with few modifications. Mice were put at the cross area of the maze in white, facing the open arm. The time spent in the open arm was recorded over a 10-min period using the Fusion software (AccuScan Instrument, Columbus, OH, USA). Data are shown as mean ± standard error of mean and analyzed by one-way ANOVA with Bonferroni’s post hoc analysis.
Light-dark box assay
The light-dark box assay was used to assess the anxiety in mice as published with few modifications (20). The mice were placed into the illuminated side and allowed to explore freely. The time spent in the illuminated side was recorded over a 10-min period using the Fusion software (AccuScan Instrument). Data are shown as mean ± standard error of mean and analyzed by one-way ANOVA with Bonferroni’s post hoc analysis.
Open field assay
The open field assay was used to evaluate the locomotor activity and the anxiety-like behavior in mice as published previously (18). The mice were placed into an open chamber and allowed to explore freely over a 30-min period using the Fusion software (AccuScan Instrument). Data are shown as mean ± standard error of mean and analyzed by one-way ANOVA with Bonferroni’s post hoc analysis.
Accelerating rotarod test
A 4-day trial accelerating rotarod test was used to evaluate motor learning and motor coordination in mice as previously published (21). The mice were placed on an accelerating rotarod (Ugo Basile) for 16 trials (four trials per day on four consecutive days). The rod accelerated linearly from 4 to 40 rpm for the first 5 min, with 40 rpm being the maximum speed. The latency for each mouse to fall from the rod was recorded for each trial. Data are shown as mean ± standard error of mean and analyzed by two-way ANOVA with repeated measurement with Bonferroni’s post hoc analysis.
Parallel rod footslip test
The parallel rod footslip test was used to assess the motor coordination in mice as published previously (22). Mice were placed in a chamber to explore on a floor made of parallel rods. The travel distance and number of footslips were recorded by the ANY-maze system (Stoelting). Data are shown as mean ± standard error of mean and analyzed by one-way ANOVA with Bonferroni’s post hoc analysis.
Marble burying test
The marble burying test was used to evaluate the repetitive digging behavior in mice as published previously (23). The mice were placed in cage covered with bedding, which is about 8 cm high, and were allowed to explore freely for 30 min. The mice were then carefully removed from the cage followed by manually counting the number of marbles buried into the bedding. Data are shown as mean ± standard error of mean and analyzed by one-way ANOVA with Bonferroni’s post hoc analysis.
Hot plate assay
Hot plate assay was used to test the pain recognition in mice as previously described (24). The mice were placed on a 55°C hot plate (Columbus Instruments). The latency to hindlimb response (jumping, shaking, or licking) was manually recorded. Data are shown as mean ± standard error of mean and analyzed by one-way ANOVA with Bonferroni’s post hoc analysis.
PPI of acoustic startle
PPI test was used to assess schizophrenia-associated behavior in mice as previously described with few modifications (25). The mice were placed in a Plexiglas chamber to habituate for 5 min with 70 dB background white noise. Three 20 ms prepulse sounds (72, 78 or 82 dB) presented either alone or 100 ms before a 40 ms 120 dB startle stimulus. The instrument used for this experiment was SR-LAB Startle Response System (San Diego Instruments, San Diego, CA, USA). Percent PPI (% PPI) was calculated as 100—[(startle response on acoustic prepulse + startle stimulus trials/startle response alone trials) × 100]. PPI data are analyzed by two-way ANOVA with Bonferroni’s post hoc analysis.
Novel object recognition assay
Novel object recognition was performed to evaluate the recognition memory in mice with the following protocol. On Day 1, the mice were placed in the test arena without any objects and allowed to explore for 5 min. The mice were then placed in the test arena with two identical LEGO objects, which had been placed with equal distance to the walls of the arena. The mice were allowed to explore and investigate the objects for 5 min (test trial) and then return to their home cage. One trial was performed each day for five consecutive days following the same protocol, except Day 4. On Day 4, one of the familiar objects was replaced with a novel object (a LEGO object of different shape and color) in the test trial. The amount of time that mice spent investigating the objects was manually recorded. The data were analyzed and a discrimination index was calculated, i.e. the difference in exploration time between familiar and novel object, divided by the total amount of exploration time of the familiar and novel object on Day 4 (4–6). Data are shown as mean ± standard error of mean and analyzed by one-way ANOVA with Bonferroni’s post hoc analysis.
Fear conditioning
Contextual fear conditioning was used to evaluate learning and memory in mice as previously described with few modifications (26, 27). The mice were trained on the first day in a chamber with a grid floor that could deliver an electric shock (Med Associates, Inc.). Each mouse was habituated in the chamber for 2 min, after which a tone (30 s, 5 kHz, 85 dB) coincided with a scrambled foot shock (2 s, 0.72 mA). The tone/foot-shock stimuli were repeated 2 min later. The mice were then returned to the home cage. Contextual fear memory was evaluated the next day by returning the mice to the same chamber for 5 min without tone or foot stimuli. After a rest period of about an hour, cued fear memory was tested by placing the mice in the same chamber with altered visual and olfactory cues, including a plastic cover on the grid floor, a vanilla odor in the chamber and different shape of the chamber. In cued fear memory, mice were left in the altered chamber without any sound for the first 3 min, followed by a 3-min tone (85 dB) without foot stimuli. The freezing of the mice was recorded and scored automatically by ANY-maze (Stoelting). Data are shown as mean ± standard error of mean and are analyzed by two-way ANOVA with Bonferroni’s post hoc analysis.
Immunohistochemistry
Three- and 4-month-old Tmprss9−/− and age-matched wild-type littermates were anaesthetized with isoflurane and transcardially perfused with PBS followed by 4% buffered paraformaldehyde in 0.1 M sodium phosphate buffer, pH 7.4. Brains were subsequently removed and postfixed overnight. Before sectioning, the brains were cryoprotected in a solution containing 30% sucrose in Tris-buffered saline (TBS: 50 mM Tris, pH 7.6). Consecutive 20 μm sagittal sections were cut and then sections were then mounted on glass slides. Sections were washed three times with 1X TBS followed by incubation with 1.8% hydrogen peroxide (H2O2) for 15 min at room temperature (RT) to inhibit internal peroxidase activity. All sections were then washed three times with 1X TBS and then blocked for 1 h at RT with 15% NGS made in TBST (Tween20:TBS 1:1000). Brain sections were then incubated with primary antibody against NeuN (Chemicon, cat# MAB377, 1:200 dilution in blocking buffer) for 48 h in cold room (4°C), followed by washing with TBST for three times. Sections were then stained with anti-mouse (VECTOR, cat# BA9200) secondary antibody, and immunoreactivity detected with Vectastain ABC (avidin-biotin) kit (VECTOR) prior to imaging.
Supplementary Material
Acknowledgements
The work was supported in part by the BCM Intellectual and Developmental Disabilities Research Center and Neurobehavioral Cores (NIH grant U54HD083092).
Conflict of interest statement. The authors declare no competing interests.
References
- 1. Baio J., Wiggins L., Christensen D.L., Maenner M.J., Daniels J., Warren Z., Kurzius-Spencer M., Zahorodny W., Robinson Rosenberg C., White T. et al. (2018) Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2014. MMWR. Surveill. Summ., 67, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Szabo R., Wu Q., Dickson R.B., Netzel-Arnett S., Antalis T.M. and Bugge T.H. (2003) Type II transmembrane serine proteases. Thromb. Haemost., 90, 185–193. [DOI] [PubMed] [Google Scholar]
- 3. Ozonoff S., Heung K., Byrd R., Hansen R. and Hertz-Picciotto I. (2008) The onset of autism: patterns of symptom emergence in the first years of life. Autism. Res., 1, 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aggleton J.P., Albasser M.M., Aggleton D.J., Poirier G.L. and Pearce J.M. (2010) Lesions of the rat perirhinal cortex spare the acquisition of a complex configural visual discrimination yet impair object recognition. Behav. Neurosci., 124, 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burke S.N., Wallace J.L., Nematollahi S., Uprety A.R. and Barnes C.A. (2010) Pattern separation deficits may contribute to age-associated recognition impairments. Behav. Neurosci., 124, 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silvers J.M., Harrod S.B., Mactutus C.F. and Booze R.M. (2007) Automation of the novel object recognition task for use in adolescent rats. J. Neurosci. Methods., 166, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanabe L.M. and List K. (2017) The role of type II transmembrane serine protease-mediated signaling in cancer. FEBS. J., 284, 1421–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finberg K.E., Heeney M.M., Campagna D.R., Aydinok Y., Pearson H.A., Hartman K.R., Mayo M.M., Samuel S.M., Strouse J.J., Markianos K. et al. (2008) Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat. Genet., 40, 569–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scott H.S., Kudoh J., Wattenhofer M., Shibuya K., Berry A., Chrast R., Guipponi M., Wang J., Kawasaki K., Asakawa S. et al. (2001) Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet., 27, 59–63. [DOI] [PubMed] [Google Scholar]
- 10. Cal S., Quesada V., Garabaya C. and Lopez-Otin C. (2003) Polyserase-I, a human polyprotease with the ability to generate independent serine protease domains from a single translation product. Proc. Natl. Acad. Sci. USA, 100, 9185–9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Okumura Y., Hayama M., Takahashi E., Fujiuchi M., Shimabukuro A., Yano M. and Kido H. (2006) Serase-1B, a new splice variant of polyserase-1/TMPRSS9, activates urokinase-type plasminogen activator and the proteolytic activation is negatively regulated by glycosaminoglycans. Biochem. J., 400, 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Antunes M. and Biala G. (2012) The novel object recognition memory: neurobiology, test procedure and its modifications. Cogn. Process, 13, 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bateman J.F., Freddi S., Nattrass G. and Savarirayan R. (2003) Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Hum. Mol. Genet., 12, 217–225. [DOI] [PubMed] [Google Scholar]
- 14. Zetoune A.B., Fontaniere S., Magnin D., Anczukow O., Buisson M., Zhang C.X. and Mazoyer S. (2008) Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC. Genet., 9, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bainbridge M.N., Wang M., Wu Y., Newsham I., Muzny D.M., Jefferies J.L., Albert T.J., Burgess D.L. and Gibbs R.A. (2011) Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol., 12, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C. et al. (2014) Launching genomics into the cloud: deployment of mercury, a next generation sequence analysis pipeline. BMC. Bioinformatics, 15, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crawley J.N. (2004) Designing mouse behavioral tasks relevant to autistic-like behaviors. Ment. Retard. Dev. Disabil. Res Rev., 10, 248–258. [DOI] [PubMed] [Google Scholar]
- 18. Spencer C.M., Alekseyenko O., Serysheva E., Yuva-Paylor L.A. and Paylor R. (2005) Altered anxiety-related and social behaviors in the Fmr1 knockout mouse model of fragile X syndrome. Genes Brain Behav., 4, 420–430. [DOI] [PubMed] [Google Scholar]
- 19. Holmes A. (2001) Targeted gene mutation approaches to the study of anxiety-like behavior in mice. Neurosci. Biobehav. Rev., 25, 261–273. [DOI] [PubMed] [Google Scholar]
- 20. Bouwknecht J.A. and Paylor R. (2002) Behavioral and physiological mouse assays for anxiety: a survey in nine mouse strains. Behav. Brain Res., 136, 489–501. [DOI] [PubMed] [Google Scholar]
- 21. Park J., Al-Ramahi I., Tan Q., Mollema N., Diaz-Garcia J.R., Gallego-Flores T., Lu H.C., Lagalwar S., Duvick L., Kang H. et al. (2013) RAS-MAPK-MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature, 498, 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chao H.T., Chen H., Samaco R.C., Xue M., Chahrour M., Yoo J., Neul J.L., Gong S., Lu H.C., Heintz N. et al. (2010) Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature, 468, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas A., Burant A., Bui N., Graham D., Yuva-Paylor L.A. and Paylor R. (2009) Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology, 204, 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Samaco R.C., Fryer J.D., Ren J., Fyffe S., Chao H.T., Sun Y., Greer J.J., Zoghbi H.Y. and Neul J.L. (2008) A partial loss of function allele of methyl-CpG-binding protein 2 predicts a human neurodevelopmental syndrome. Hum. Mol. Genet., 17, 1718–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paylor R., Yuva-Paylor L.A., Nelson D.L. and Spencer C.M. (2008) Reversal of sensorimotor gating abnormalities in Fmr1 knockout mice carrying a human Fmr1 transgene. Behav. Neurosci., 122, 1371–1377. [DOI] [PubMed] [Google Scholar]
- 26. Hao S., Tang B., Wu Z., Ure K., Sun Y., Tao H., Gao Y., Patel A.J., Curry D.J., Samaco R.C. et al. (2015) Forniceal deep brain stimulation rescues hippocampal memory in Rett syndrome mice. Nature, 526, 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zeitlin R., Patel S., Solomon R., Tran J., Weeber E.J. and Echeverria V. (2012) Cotinine enhances the extinction of contextual fear memory and reduces anxiety after fear conditioning. Behav. Brain Res., 228, 284–293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


