Abstract
Acinetobacter baumannii is often highly drug-resistant and causes severe infections in compromised patients. These infections can be life threatening due to limited treatment options. Copper is inherently antimicrobial and increasing evidence indicates that copper containing formulations may serve as non-traditional therapeutics against multidrug-resistant bacteria. We previously reported that A. baumannii is sensitive to high concentrations of copper. To understand A. baumannii copper resistance at the molecular level, herein we identified putative copper resistance components and characterized 21 strains bearing mutations in these genes. Eight of the strains displayed a copper sensitive phenotype (pcoA, pcoB, copB, copA/cueO, copR/cusR, copS/cusS, copC, copD); the putative functions of these proteins include copper transport, oxidation, sequestration, and regulation. Importantly, many of these mutant strains still showed increased sensitivity to copper while in a biofilm. Inductively coupled plasma mass spectrometry revealed that many of these strains had defects in copper mobilization, as the mutant strains accumulated more intracellular copper than the wild-type strain. Given the crucial antimicrobial role of copper-mediated killing employed by the immune system, virulence of these mutant strains was investigated in Galleria mellonella; many of the mutant strains were attenuated. Finally, the cusR and copD strains were also investigated in the murine pneumonia model; both were found to be important for full virulence. Thus, copper possesses antimicrobial activity against multidrug-resistant A. baumannii, and copper sensitivity is further increased when copper homeostasis mechanisms are interrupted. Importantly, these proteins are crucial for full virulence of A. baumannii and may represent novel drug targets.
Keywords: Acinetobacter baumannii, metal, copper, pathogenesis, Galleria
Introduction
Acinetobacter baumannii is responsible for a significant proportion of nosocomial infections worldwide, and an even greater number of ICU-acquired infections; co-morbidities are an important risk factor for A. baumannii infection (Vincent et al., 2009; Lob et al., 2016; Wong et al., 2017; Du et al., 2019). Associated types of infections are diverse, and include pneumonia, urinary tract infections, bacteremia, skin and soft tissue infections, osteomyelitis, and meningitis (Peleg et al., 2008; Wong et al., 2017). Extensive drug-resistance is common among A. baumannii strains, and resistance limits treatment options and leads to higher morbidity and mortality (Antunes et al., 2014; Giammanco et al., 2017; Du et al., 2019). Indeed, multidrug-resistant A. baumannii has been named a “serious” threat by the CDC since 2013, and in 2017 carbapenem-resistant A. baumannii topped the World Health Organization’s Priority Pathogen’s List as a “Level 1: Critical priority” pathogen (Centers for Disease Control and Prevention, 2013; World Health Organization, 2017). Clearly, new therapeutic options are desperately needed to treat multidrug-resistant A. baumannii infections.
Antibiotics target essential functions for bacterial growth and/or survival. Metal homeostasis is a vital process that provides an extensive list of potential new antimicrobial targets. Copper is required for cellular function, e.g., for redox balance and as an enzyme cofactor. However, copper ions become toxic at high concentrations; thus, it is important that intracellular copper levels be tightly controlled. Copper ions cause damage by participating in Fenton-like chemistry to produce hydroxyl radicals that react with and damage essential biomolecules (Liochev and Fridovich, 2002) and also by displacing iron from crucial iron-sulfur cluster proteins (Macomber and Imlay, 2009). Studies in Escherichia coli and Salmonella spp. have shown that when bacteria are placed on copper surfaces, outer membrane integrity is compromised, hydroxyl radicals are produced, respiration is inhibited, and DNA is degraded (Warnes et al., 2012). Fenton-like chemistry-based killing of pathogens also occurs in the host via host-generated reactive oxygen species. Because of the need for new therapeutics to treat antibiotic resistant pathogens, research into the use of copper as an antimicrobial has lately increased. For example, the use of copper-containing surfaces in hospitals has been shown to greatly reduce environmental contamination with nosocomial pathogens and to reduce rates of health care-acquired infections (Salgado et al., 2013; Sifri et al., 2016; von Dessauer et al., 2016). Additionally, copper-containing wound dressings are in development to aid in healing of infected wounds (Borkow et al., 2010; Ahire et al., 2016).
The damaging effects of copper have been harnessed by the host immune system. Indeed, phagocytic immune cells employ a copper burst within the phagosome to kill pathogens (Sheldon and Skaar, 2019). Concentrations of copper upwards of 0.5 mM have been measured in macrophage phagosomes (Wagner et al., 2005). Additionally, host mobilization of copper also occurs in response to infection: increased concentrations of copper have been measured in serum and in wound exudate (Milanino et al., 1993; Jones et al., 2001). Consequently, pathogens with mutations in crucial copper resistance genes have been found to have impaired intracellular survival, colonization, and/or virulence (Djoko et al., 2015). Furthermore, it was recently shown that a copper sensitive mutant strain of A. baumannii demonstrated reduced colonization of the respiratory tract of mice (Alquethamy et al., 2019).
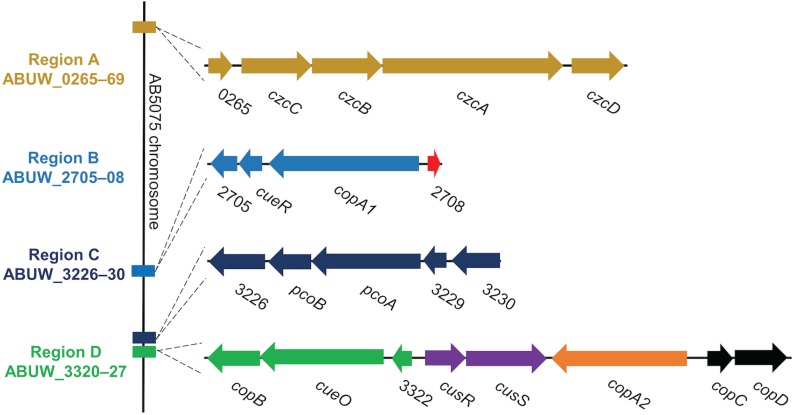
In bacterial species where copper homeostasis has been well-characterized, a variety of proteins are utilized to facilitate copper homeostasis. Though this process remains poorly understood in A. baumannii, recent studies have identified many putative copper resistance gene homologs in A. baumannii clinical isolates (Hernandez-Montes et al., 2012; Williams et al., 2016; Alquethamy et al., 2019). For example, in the model clinical multidrug-resistant isolate, AB5075, copper resistance genes exist in four distinct chromosomal regions named A–D (Figure 1) (Williams et al., 2016). These regions contain genes predicted to encode homologs of the most important copper resistance proteins of bacteria, e.g., copper ATPases, copper oxidases, copper transporters, copper chaperones, and regulatory proteins (Williams et al., 2016).
FIGURE 1.
Organization of regions A–D in the AB5075 chromosome. We previously identified 22 putative copper-related genes in the genome of AB5075 (Williams et al., 2016). These genes are located in four chromosomal regions named A–D. The relative location of each region on the AB5075 chromosome is shown on the left (not to scale), and the genes and regions are depicted to scale relative to one another. Each operon is colored uniquely, and the same colors are maintained in Figure 9. Gene annotations are shown when available, otherwise ORF numbers are used.
Region A includes genes for a large RND family efflux pump, czcCBAD, and a hypothetical protein ABUW_0265, which is located at the start of the operon. A similar type of pump, CusCFBA, has been shown to contribute to copper resistance in E. coli (Franke et al., 2003). While the genes in this operon are annotated as copper-related in AB5075, expression does not increase in response to copper exposure (Williams et al., 2016) and it has been suggested that A. baumannii in fact does not encode cus homologs (Alquethamy et al., 2019), thus the role of this operon in copper resistance is very tentative.
Region B encodes four genes, including homologs of two known copper resistance proteins. CueR, CopA1, and the hypothetical protein ABUW_2705 are encoded as an operon; divergently transcribed from that operon is a putative copper chaperone, ABUW_2708, recently named CopZ (Alquethamy et al., 2019). CueR has been well-studied in E. coli and is a cytoplasmic transcription factor that binds copper ions with exquisite sensitivity (Changela et al., 2003). Upon copper binding, CueR positively regulates transcription of copper resistance genes. In E. coli, CueR is known to regulate the expression of copper ATPase CopA and the copper oxidase CueO (Outten et al., 2000; Yamamoto and Ishihama, 2005). The CueR regulon in A. baumannii is not yet characterized, however, a putative CueR binding site has been identified in the intergenic space of region B, suggesting that all of the genes in region B may be regulated by CueR (Alquethamy et al., 2019). Appropriately, the genes of region B are all upregulated in response to copper in a similar manner (Williams et al., 2016). A CopA1 homolog, also encoded in region B, is a putative copper ATPase, which is another important and well-characterized copper resistance protein. Copper ATPases use energy from ATP to move copper ions into the periplasmic space from the cytoplasm and are known to contribute to copper resistance in a number of bacteria (Rensing et al., 2000). Very recently, the copper ATPase in region B was demonstrated to contribute to copper resistance of A. baumannii AB5075 in vitro and to contribute to colonization in a mouse respiratory infection model (Alquethamy et al., 2019).
Region C encodes a single operon of five genes, ABUW_3226 – 3230. Two of the genes, ABUW_3227 and ABUW_3228, are annotated as pcoB and pcoA, respectively. In E. coli, the pco system is plasmid-based; while the Pco proteins seem to be homologous to known chromosomally encoded copper resistance factors, their functions are less well-studied and understood (Lee et al., 2002). In AB5075, PcoB is homologous to CopB, an outer membrane protein that may function as a copper transporter (Cha and Cooksey, 1993). PcoA is homologous to CueO, a periplasmic copper oxidase that detoxifies by oxidizing copper ions to their less damaging form (Grass and Rensing, 2001). Surprisingly, expression of the genes in region C does not change in response to copper exposure (Williams et al., 2016), however that does not preclude the possibility that these genes are indeed involved in copper resistance.
Region D is the largest of the identified regions in AB5075; it contains a total of eight genes arranged as two pairs of divergently transcribed genes/operons. Among these eight genes are seven homologs of characterized copper resistance proteins: CopB, CueO, CusR, CusS, CopA2, CopC, and CopD. The functions of homologs of some of these copper-related proteins have been previously elucidated in other bacteria. CopB and CopD are membrane proteins and putative copper transporters, though not yet well-characterized (Cha and Cooksey, 1993). CueO is a periplasmic copper oxidase, which detoxifies by oxidizing cuprous copper ions (Cu1+) to their less damaging cupric (Cu2+) form (Grass and Rensing, 2001). CopA2 is a second copper ATPase (the other being CopA1 of region B); previous work did not reveal a role for copA2 in copper resistance in AB5075 (Alquethamy et al., 2019). CopC is a periplasmic copper chaperone protein that likely contributes to copper resistance by binding copper ions as a means to prevent reactivity and/or by shuttling ions between other copper resistance proteins (Puig et al., 2002; Arnesano et al., 2003; Padilla-Benavides et al., 2014). CusR and CusS form a two-component system that senses periplasmic copper ions and activates the CusR regulon in response (Gudipaty et al., 2012). In E. coli, CusR is known to directly regulate expression of the cusRS operon and the divergently transcribed cusCFBA operon (Munson et al., 2000; Yamamoto and Ishihama, 2005; Gudipaty et al., 2012). It is not yet known which genes comprise the CusR regulon in A. baumannii, however, putative CusR binding sites were identified in the intergenic regions upstream of the four operons in region D, suggesting that all of region D may be regulated by CusR (Alquethamy et al., 2019).
Notably, not all clinical isolates of A. baumannii carry all of the copper-related genes found in AB5075. Based on previous analyses of two panels of clinical isolates, as well as the common lab strain ATCC 17978, region A–C genes are encoded by all strains. Conversely, region D genes are found in about 40% of strains (5/12 strains) (Williams et al., 2016; Hassan et al., 2017). Furthermore, the presence of region D genes is linked to increased copper resistance. Indeed, isolates show marked differences in copper sensitivity that directly correlates with the presence of the region D genes; clinical isolates lacking region D demonstrate significantly less copper resistance than isolates that encode the region D genes (Williams et al., 2016). Moreover, expression of the genes in region D dramatically increases in response to copper exposure (Williams et al., 2016). These results strongly suggest that the region D genes are crucial for extensive copper homeostasis capability in A. baumannii.
Given the growing interest in targeting copper homeostasis as a novel antimicrobial strategy and the lack of understanding of which factors contribute to copper resistance in A. baumannii, we sought to identify genes that were important for copper resistance in this important pathogen. Herein, we describe a detailed analysis of 21 mutant strains, each bearing insertions in the genes carried in regions A–D (Figure 1). Marked copper sensitivity of a subset of the mutant strains as well as defects in virulence are described.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains are listed in Table 1. Strains were routinely streaked from freezer stocks and were grown overnight at 37°C on lysogeny broth (LB) 1.5% agar plates (MoBio, Carlsbad, CA, United States). Liquid cultures of A. baumannii were grown at 37°C shaking at 190 rpm in LB or in M9 minimal medium (Sambrook and Russell, 2001) supplemented with 0.1% casamino acids (Difco, Franklin Lakes, NJ, United States); the M9 medium was always supplemented with amino acids, but for simplicity is referred to as ‘M9 medium’ throughout the manuscript. Importantly, overnight cultures were consistently started from a single opaque colony, as phase variation has been shown to affect growth and virulence (Tipton et al., 2015). Overnight growth was restricted to a period of 16–19 h. A. baumannii strains freezer stocks were maintained at −80°C in medium supplemented with 40% glycerol.
TABLE 1.
Strains used in this study.
| Transposon mutant strains | ||||||
| Strain name | Shorthand name1 | Locus tag | Strain name in transposon library2 | Tn5 location in ORF2 | Lab designation | Reference |
| AB5075 | Wild-type | DSM1866 | Jacobs et al., 2014a | |||
| AB5075 pcoB140:T26 | pcoB:T26 | ABUW_3227 | tnab1_kr121204p07q140 | 445 (756) | DSM1833 | Gallagher et al., 2015 |
| AB5075 pcoA104:T26 | pcoA:T26 | ABUW_3228 | tnab1_kr121203p02q104 | 607 (1935) | DSM1823 | Gallagher et al., 2015 |
| AB5075 copB156:T26 | copB:T26 | ABUW_3320 | tnab1_kr121203p05q156 | 142 (903) | DSM1837 | Gallagher et al., 2015 |
| AB5075 copA/cueO118:T26 | cueO:T26 | ABUW_3321 | tnab1_kr121128p03q118 | 381 (2118) | DSM1840 | Gallagher et al., 2015 |
| AB5075 copR/cusR106:T26 | cusR:T26 | ABUW_3323 | tnab1_kr121128p07q106 | 195 (684) | DSM1825 | Gallagher et al., 2015 |
| AB5075 copS/cusS128:T26 | cusS:T26 | ABUW_3324 | tnab1_kr121205p01q128 | 328 (1377) | DSM1832 | Gallagher et al., 2015 |
| AB5075 copC160:T26 | copC:T26 | ABUW_3326 | tnab1_kr121119p04q160 | 46 (381) | DSM1844 | Gallagher et al., 2015 |
| AB5075 copD117:T26 | copD:T26 | ABUW_3327 | tnab1_kr121204p06q117 | 280 (882) | DSM1830 | Gallagher et al., 2015 |
| AB5075 ABUW_0265-193:T26 | ABUW_0265 | tnab1_kr130913p10q193 | 177 (399) | DSM1856 | Gallagher et al., 2015 | |
| AB5075 czcC151:T26 | ABUW_0266 | tnab1_kr121127p01q151 | 525 (1311) | DSM1849 | Gallagher et al., 2015 | |
| AB5075 czcB187:T26 | ABUW_0267 | tnab1_kr121205p01q187 | 70 (1218) | DSM1848 | Gallagher et al., 2015 | |
| AB5075 czcA162:T26 | ABUW_0268 | tnab1_kr121204p04q162 | 1523 (3159) | DSM1846 | Gallagher et al., 2015 | |
| AB5075 czcD174:T26 | ABUW_0269 | tnab1_kr121205p05q174 | 486 (957) | DSM1851 | Gallagher et al., 2015 | |
| AB5075 ABUW_2705-139:T26 | ABUW_2705 | tnab1_kr130903p04q139 | 89 (408) | DSM1857 | Gallagher et al., 2015 | |
| AB5075 cueR139:T101 | ABUW_2706 | tnab1_jr130919p01q139 | 238 (402) | DSM1858 | Gallagher et al., 2015 | |
| AB5075 actP2/copA1-181:T26 | ABUW_2707 | tnab1_kr121128p08q181 | 791 (2472) | DSM1822 | Gallagher et al., 2015 | |
| AB5075 ABUW_3226-176:T26 | ABUW_3226 | tnab1_kr121211p04q176 | 193 (996) | DSM1859 | Gallagher et al., 2015 | |
| AB5075 ABUW_3229-182:T26 | ABUW_3229 | tnab1_kr130913p04q182 | 86 (399) | DSM1860 | Gallagher et al., 2015 | |
| AB5075 ABUW_3230-184:T26 | ABUW_3230 | tnab1_kr130904p01q184 | 234 (822) | DSM1861 | Gallagher et al., 2015 | |
| AB5075 ABUW_3322-131:T26 | ABUW_3322 | tnab1_kr130904p04q131 | 25 (366) | DSM1862 | Gallagher et al., 2015 | |
| AB5075 actP1/copA2-159:T26 | ABUW_3325 | tnab1_kr121203p05q159 | 784 (2346) | DSM1828 | Gallagher et al., 2015 | |
| Complemented mutant strains3 | ||||||
| Proper name | Shorthand name1 | Lab designation | Reference | |||
| AB5075 pcoB140:T26 attTn7-hph-pcoB | pcoB:T26C | DSM1904 | This study | |||
| AB5075 pcoA104:T26 attTn7-hph-pcoA | pcoA:T26C | DSM1905 | This study | |||
| AB5075 copB156:T26 attTn7-hph-copB | copB:T26C | DSM1901 | This study | |||
| AB5075 copA/cueO118:T26 attTn7-hph-cueO | cueO:T26C | DSM1906 | This study | |||
| AB5075 copR/cusR106:T26 attTn7-hph-cusR | cusR:T26C | DSM1890 | This study | |||
| AB5075 copS/cusS128:T26 attTn7-hph-cusS | cusS:T26C | DSM1902 | This study | |||
| AB5075 copC160:T26 attTn7-hph-copC | copC:Tn5C | DSM1897 | This study | |||
| AB5075 copD117:T26 attTn7-hph-copD | copD:Tn5C | DSM1898 | This study | |||
| Mating strains | ||||||
| Strain name | Description | Lab designation | Reference | |||
| HB101/pRK2013 | Mating helper strain (oriT helper) | DSM1881 | Kumar et al., 2010 | |||
| DH5αλpir/pTNS2 | Mating helper strain (transposase) | DSM1880 | Kumar et al., 2010 | |||
| DH5α/pUC18T mini-Tn7-hph | Tn7 with hygromycin resistance gene, hph, and MCS | DSM1883 | Jacobs et al., 2014a | |||
| DH5α/pUC18T mini-Tn7-hph-pcoA | Tn7 with hph and pcoA with native promoter | DSM1894 | This study | |||
| DH5α/pUC18T mini-Tn7-hph-pcoB | Tn7 with hph and pcoB with native promoter | DSM1895 | This study | |||
| DH5α/pUC18T mini-Tn7-hph-copB | Tn7 with hph and copB with native promoter | DSM1899 | This study | |||
| DH5α/pUC18T mini-Tn7-hph-cueO | Tn7 with hph and cueO with native promoter | DSM1903 | This study | |||
| DH5α/pUC18T mini-Tn7-hph-cusR | Tn7 with hph and cusR with native promoter | DSM1889 | This study | |||
| DH5α/pUC18T mini-Tn7-hph-cusS | Tn7 with hph and cusS with native promoter | DSM1900 | This study | |||
| DH5α/pUC18T mini-Tn7-hph-copC | Tn7 with hph and copC with native promoter | DSM1893 | This study | |||
| DH5α/pUC18T mini-Tn7-hph-copD | Tn7 with hph and copD with native promoter | DSM1896 | This study | |||
| Additional strains | ||||||
| Strain name | Description | Lab designation | Reference | |||
| AB4857/pAJ100 | AB4857 with empty vector, pAJ100 | DSM1908 | This study | |||
| AB4857/pAJ100 – Region D | AB4857 with pAJ100 carrying Region D | DSM1910 | This study | |||
| AB5711/pAJ100 | AB5711 with empty vector, pAJ100 | DSM1909 | This study | |||
| AB5711/pAJ100 – Region D | AB5711 with pAJ100 carrying Region D | DSM1911 | This study | |||
| E. coli/pAJ100 | DSM1884 | This study | ||||
| E. coli Top 10/pAJ100 – Region D | DSM1907 | This study | ||||
| AB5075 attTn7-hph | AB5075 with a hygromycin resistance gene in the attTn7 site | DSM1926 | This study | |||
| AB5075 copR/cusR106:T26 attTn7-hph | The cusR mutant strain with a hygromycin resistance gene in the attTn7 site | DSM1927 | This study | |||
| AB5075 copC160:T26 attTn7-hph | The copC mutant strain with a hygromycin resistance gene in the attTn7 site | DSM1928 | This study | |||
1For simplicity, mutant strains that were found to have increased copper sensitivity and were included in multiple assays were given a shorthand name. 2The mutant strains in the transposon library were created using a Tn5 transposon carrying a tetracycline resistance cassette (T26) or hygromycin resistance cassette (T101) (Gallagher et al., 2015). Individual strains were purchased from the ordered, annotated library. The transposon’s position in the ORF is indicated as a base pair position out of the total base pairs (denoted in parenthesis) in the ORF; this information was taken directly from the mutant library list (Gallagher et al., 2015). 3Complemented strains were created by tri-parental mating of each transposon mutant strain with E. coli DH5α carrying the appropriate ORF and native promoter on the plasmid pUC18T mini-Tn7-hph, as well as with the two helper strains of E. coli that aid in the mating and transposition processes. The Tn7 from the pUC18T mini-Tn7-hph plasmid, containing a hygromycin resistance gene and the desired complementation ORF and promoter, inserts into the AB5075 chromosome at the attTn7 site located in an intergenic region downstream of the glmS locus. MCS = multiple cloning site.
AB5075 Mutant Strains
Transposon mutant strains were purchased from the comprehensive, ordered transposon library engineered by Dr. Colin Manoil’s group at the University of Washington (Gallagher et al., 2015). In general, we chose strains with transposon insertions near the 5′ end of the gene; most have insertions in the first third of the ORF (Table 1). The isolates were received as stabs, which were streaked onto LB agar plates. The following day, a swab of the growth was used to inoculate an overnight culture in LB medium, which was subsequently frozen as a master stock for each mutant strain. To verify the desired mutations, each master stock was streaked on LB agar containing 10 μg/mL tetracycline to select for cells containing the Tn5 insertion that characterizes the transposon library strains. For each, a single opaque colony was picked and restreaked onto LB agar containing tetracycline. The following day, a single opaque colony was picked and used to inoculate an overnight culture in LB medium containing 10 μg/mL tetracycline; the resulting cultures were frozen as working stocks for each mutant strain and were used for all future experiments. The desired transposon insertion was confirmed by PCR using primers that flank the proper ORF; primers are listed in Table 2. Throughout the manuscript, strains are always organized by ascending gene number, as in Figure 1. A few genes’ annotated names differ from the conventional names for the given protein; these genes’ annotated name and conventional name are both provided here, but the conventional names are used throughout for consistency and clarity: actP2/copA1, copA/cueO, copR/cusR, copS/cusS, actP1/copA2.
TABLE 2.
Oligonucleotides used in this study.
| T26 insertion confirmation primers | |||
| Flanked gene | Annotation1 | Primer name2 | Sequence (5′–3′) |
| ABUW_0265 | ABUW_0265-up ABUW_0265-dw | GTGCAATTTCTAACAGCCATG CAATAAATAAAAGATGCGGTGGG | |
| ABUW_0266 | czcC | czcC-up czcC-dw | TGGCAGCAATTACTCCAACC GCCATCCCGACTTATTGAGCT |
| ABUW_0267 | czcB | czcB-up czcB-dw | CACCAATCCACGCCAGAAC TGGCGAAAGATGCCTTAATGC |
| ABUW_0268 | czcA | czcDA-up czcA-dw | GCAACAGTAGTCGCATTTGTGG GCTTCTTACTGAAGTCCGAATTGG |
| ABUW_0269 | czcD | czcDA-up czcD-dw | GCAACAGTAGTCGCATTTGTGG GGCAACAGTGGTTATTGGTGG |
| ABUW_0611 | ABUW_0611-up ABUW_0611-dw | GGTTTCTAGGAGCGCTGCC GCACCTGCAAAGTATGGCTCTAC | |
| ABUW_2705 | ABUW_2705-up ABUW_2705-dw | CATGACAGCAGGTCTTAAACC CCTGTGGATCGAGGAGATATTAC | |
| ABUW_2706 | cueR | cueR-up-NEW cueR-dw-NEW | GAGGTGTGAGATGAATATCGGTCAG GTACTTGAGTGATGGCCGC |
| ABUW_2707 | copA1 | CopA2_up CopA2_dw | GCTTGACCTTCCCATGATGG CTGACCGATATTCATCTCACACCTC |
| ABUW_3226 | ABUW_3226-up ABUW_3226-dw | GTATTATGGAGCTGGGTTAACAC GTTCCTCCAGTATGTGAAGAC | |
| ABUW_3227 | pcoB | CopB2-up CopB2-dw | CTGAAAATGAGAAAGGAGCTAG CATAAGCTTACTTTTTGTTCGGC |
| ABUW_3228 | pcoA | CopA2-up CopA2-dw | CGTCCACCAAAAGCTTAATCC GAGAAAGCACCGCAACATATC |
| ABUW_3229 | ABUW_3229-up ABUW_3229-dw | CCAAGATAAATAGATTGCTCAAGGC CGAGGCTGCTAATCGTTTTATG | |
| ABUW_3230 | ABUW_3230-up ABUW_3230-dw | GGATAACTGTGTTGATAACCATATGG CCATCTGAGTGGATTGTTGTTAAG | |
| ABUW_3220 | copB | CopB1-up CopB1-dw | CCTAATACTAATGATAAGGGGG CTTTATGAATAGTAGCTTCGGC |
| ABUW_3221 | cueO | ABUW_3322-up CopA1-dw | CCGGTTCCAACCTATAAGTTAG GCAAAAGCTAGACCTGATAATCC |
| ABUW_3322 | ABUW_3322-up ABUW_3322-dw | CCGGTTCCAACCTATAAGTTAG GTTAATTGAGGGCTAAGCAGG | |
| ABUW_3323 | cusR | CusR-up CusR-dw | CTTAATATGCGATACGGGGC GCAATACGGAAACTGATCGC |
| ABUW_3324 | cusS | CusS-up CusS-dw | GGAATGGGGTATGTCTTAGAGG GAAAGGATCATTCTCTGACTACTC |
| ABUW_3325 | copA2 | CopA1_up_NEW CopA1_dw_NEW | GCCGGAGTAGCACTTTCAAG GAGGAAGGCACAGGTTTAGG |
| ABUW_3326 | copC | CopC-up CopC-dw | GGAATTTCACTGGGTCATTAC CTGCTGGCATATAGACTAC |
| ABUW_3327 | copD | CopD-up CopD-dw | GCATATTACTATCGCCGG GAAAGCATTGCTACTACTCC |
| Complementation cloning primers | |||
| Fragment | Primer name | Sequence (5′–3′)3 | |
| pcoABpromoter | pcoAB promoter up | CCCGGTACCcggtagtgtagacgcttaa | |
| pcoAB promoter dw-A | GGCTGCTAATCGTTTTATGAGACATtaatgatctaggtctcttactaaaa | ||
| pcoAB promoter dw-B | AGAAAATAACTTAGTGATGCGCATtaatgatctaggtctcttactaaaa | ||
| pcoA ORF | pcoA complement up | TTTTAGTAAGAGACCTAGATCATTAatgtctcataaaacgattagcagcc | |
| pcoA complement dw | CCCGGATCCgagaaagcaccgcaacatatc | ||
| pcoB ORF | pcoB complement up | TTTTAGTAAGAGACCTAGATCATTAatgcgcatcactaagttattttct | |
| pcoB complement dw | CCCGGATCCcatctttgcaccataactgacac | ||
| copAB promoter | copAB promoter up | CCCGGTACCaagagcttgatgtttacctg | |
| copAB promoter dw | cattgattgctccaaaaataaattttatataactaac | ||
| cueO ORF | NEW copA/cueO complement up | AAATTTATTTTTGGAGCAATCAatgtctagaaaattaagtcatgtcc | |
| copA/cueO complement dw | CCCGGATCCccatgctttgaacatccgtatc | ||
| copB ORF | NEW copB complement up | AAATTTATTTTTGGAGCAATCAatgcgcaccactaaaaaaatatattc | |
| copB complement dw | CCCGGATCCgatttggaacgcttttaagccc | ||
| cusRS promoter and cusR ORF | cusR complement up | CCCGGTACCcccaacacacataaaagcag | |
| cusR complement dw | CCCGGATCCcttctagctgagtacggtc | ||
| cusRS promoter | cusR complement up | CCCGGTACCcccaacacacataaaagcag | |
| NEW cusRS promoter dw – cusS SOE | GATCGCATTAAACAGTTTATGAGTcatttattaagccccgtatcgcatattaag | ||
| cusS ORF | cusS complement up | AATATGCGATACGGGGCTTAATAAatgactcataaactgtttaatgcgatc | |
| cusS complement dw | CCCGGATCCgaaaggatcattctctgactactc | ||
| copCD promoter and copC ORF | copCD promoter up | CCCGGTACCccatgccacagacaggatc | |
| copC complement dw | CCCGGATCCcgtcccagtagaaaatagctaaagg | ||
| copCD promoter | copCD promoter up | CCCGGTACCccatgccacagacaggatc | |
| copCD promoter dw | catttcatcacctttttaaaattagtacttgc | ||
| copD ORF | copD complement up | GTACTAATTTTAAAAAGGTGATGAAatgaatcctgaaacttggatatatgcaac | |
| NEW copD comp dw | CCCGGATCCgacgtggtccttttatgcagc | ||
| Primers used in the creation of pAJ100 | |||
| pWHori NcoI For | GATTACACCATGGgatcgtagaaatatctatgattatcttgaa | ||
| pWHori NcoI Rev | GATTACACCATGGggattttaacattttgcgttgttccaa | ||
1Gene names are given where available. For those whose annotated name is different from the name traditionally given to that protein, the conventional name is listed. 2The primer name may not match the gene name. 3Capital letters signify added primer tail sequences for SOE or restriction site. Underlined sequences are restriction enzyme cut sites.
Generation of Complemented Strains
The previously described Tn7-based strategy of complementation was used to complement the transposon mutant strains (Kumar et al., 2010; Jacobs et al., 2014a, b). The wild-type ORF and native promoter were amplified using the primers listed in Table 2. If the ORF was not located directly downstream of the promoter due to operonic structure, the two DNA fragments were fused by SOE PCR. The full construct was amplified with Phusion Hot Start Polymerase (Thermo Fisher Scientific) and then cloned into pUC18T mini-Tn7-hph using the added restriction enzyme sites (Table 2). The resulting plasmids were transformed into electrocompetent DH5α E. coli and transformants were selected for on LB agar with 100 μg/mL ampicillin. Correct plasmid construction was confirmed by digestion and then the resulting E. coli strains were mated with the AB5075 transposon mutant strains using the tri-parental protocol described by Jacobs et al. (2014b). Briefly, 100 μL of four overnight cultures (E. coli carrying the complementation construct in pUC18T-mini Tn7-hph, two helper strains, and the AB5075 mutant strain) grown in LB medium with the appropriate antibiotics were mixed, washed twice, and spotted onto plain LB agar. The mating was allowed to proceed for 1 to 24 h. The cells in the spot were collected with a sterile loop and plated on LB agar with 25 μg/mL chloramphenicol to kill the E. coli strains and 250 μg/mL hygromycin to select for A. baumannii with the Tn7 insertion. Resulting colonies were screened for Tn7 insertion using colony PCR with the previously described attTn7 primers (Jacobs et al., 2014a). Single colonies were restreaked onto LB agar with 10 μg/mL tetracycline and 250 μg/mL hygromycin. A single opaque colony was used to inoculate an overnight culture in LB with tetracycline and hygromycin, which was ultimately used to create a freezer stock of the strain. The correct insertion at the attTn7 site was confirmed by PCR and sequencing. All of the strains carried the Tn7 insertion in the same direction: forward, relative to glmS.
Cloning of Region D From AB5075 Into Other A. baumannii Strain Backgrounds
Creation of pAJ100
An E. coli–A. baumannii shuttle vector with a hygromycin resistance gene was created to move genes into multidrug-resistant A. baumannii. To generate this vector, pMQ300 (Kalivoda et al., 2011) was digested with BspHI to remove the 1.7 kb region that contained yeast replication genes URA3 and CEN6/ARSH4. The A. baumannii origin of replication of pWH1266 (Hunger et al., 1990) was amplified using the pWHori NcoI For and pWHori NcoI Rev primers described in Table 2. This insert was digested with NcoI and then ligated to the remaining 4.7 kb of pMQ300. The resulting 6 kb plasmid was transformed into E. coli and plated on LB agar with 250 μg/mL hygromycin. The resulting vector was named pAJ100. This vector has a lacZ insertion site for genes that contains cut sites for the following enzymes: SmaI, BamHI, HindIII, KpnI, PstI, and PvuI.
Creation of pAJ100 – Region D
Region D was amplified from AB5075 chromosomal DNA with Phusion Hot Start Polymerase (Thermo Fisher Scientific) using the copB complement dw and copD complement dw primers described in Table 2. The fragment was cloned into pAJ100 using the added BamHI restriction enzyme sites. The resulting pAJ100-Region D plasmid was electroporated into E. coli Top 10 cells, which were plated on 250 μg/mL hygromycin, 40 μg/mL X-gal, and 1 mM IPTG. A white colony was restreaked for isolation, and a single colony was used to inoculate an overnight culture of LB medium with 250 μg/mL hygromycin. Plasmid was purified and then electroporated into AB4857 and AB5711, which had been made electrocompetent using the protocol described by Jacobs et al. (2014b). Transformants were selected for on LB with 250 μg/mL hygromycin. Double purified single, opaque colonies were used to inoculate overnight cultures of LB medium with 250 μg/mL hygromycin, which were then used to create freezer stocks of the strains. Correct plasmid construction was confirmed by digestion and PCR.
Determination of A. baumannii Sensitivity to Copper in Liquid Culture
To assess the effect of copper on A. baumannii growth, bacterial strains were grown in the presence of increasing concentrations of copper sulfate as previously described (Williams et al., 2016; Williams and Merrell, 2019). Briefly, bacteria from overnight cultures were subcultured at an optical density (OD600) of 0.05 in 10 mL of M9 medium containing 0.1–1.5 mM CuSO4 (Aldrich, St. Louis, MO, United States). Growth was measured every hour for 6 h by optical density (OD600) and by enumerating CFU every other hour. Three biologically independent experiments were completed for each strain showing noticeable copper sensitivity (Figures 2, 3); if no difference from the wild-type was observed, two biologically independent experiments were completed (Supplementary Figure S1). To ensure that the excess sulfate found in the copper sulfate was not responsible for the growth phenotypes, sodium sulfate was also tested at 1.5 mM using the same growth conditions; CFU were enumerated at 0 and 4 h. Three biologically independent experiments were completed (Supplementary Figure S2).
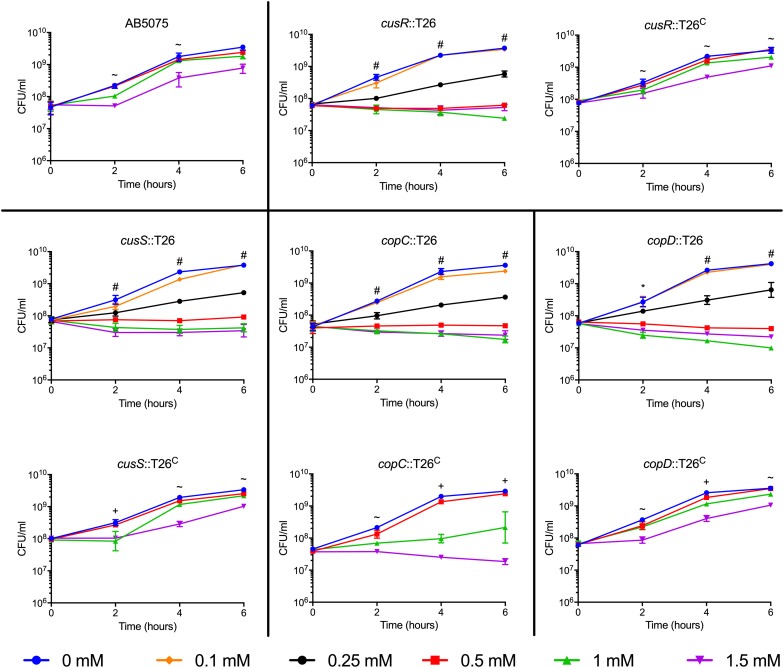
FIGURE 2.
Growth of highly copper sensitive AB5075 mutant and complementation strains. Strains were grown for 6 h at 37°C in M9 medium supplemented with the concentration of CuSO4 indicated in the legend. Growth was measured both by turbidity (OD600, data not shown) and enumeration of viable colonies (CFU/mL). Black lines are used to group the mutant strain with the corresponding complemented derivative. The data are presented as geometric mean and SEM of three biologically independent experiments. Two-way ANOVA with Tukey’s adjustment for multiple comparisons was used to compare growth in each copper concentration at each timepoint. The symbols indicate which copper treatments were statistically different (P < 0.05) from the 0 mM control, as follows: ∼, growth in 1.5 mM CuSO4 was different from the control; +, growth in 1 and 1.5 mM CuSO4 was different from the control; *, growth in 0.5, 1, and 1.5 mM CuSO4 was different from the control; and #, growth in 0.25, 0.5, 1, and 1.5 mM CuSO4 was different from the control.
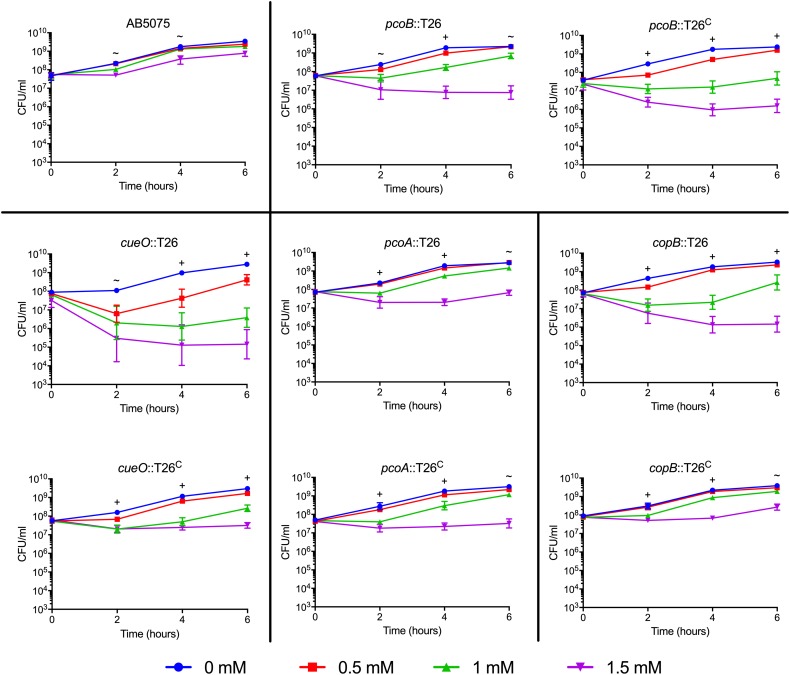
FIGURE 3.
Growth of moderately copper sensitive AB5075 mutant and complementation strains. Strains were grown for 6 h at 37°C in M9 medium supplemented with the concentration of CuSO4 indicated in the legend. Growth was measured both by turbidity (OD600, data not shown) and enumeration of viable colonies (CFU/mL). Black lines are used to group the mutant strain with the corresponding complemented derivative. The data are presented as geometric mean and SEM of three biologically independent experiments. Two-way ANOVA with Tukey’s adjustment for multiple comparisons was used to compare growth in each copper concentration at each timepoint. The symbols indicate which copper treatments were statistically different (P < 0.05) from the 0 mM control, as follows: ∼, growth in 1.5 mM CuSO4 was different from the control; +, growth in 1 and 1.5 mM CuSO4 was different from the control.
Assessing Copper Sensitivity of A. baumannii Biofilm
Biofilms were grown, exposed to copper, and assessed for survival as previously described with minor adjustments to the incubation time and growth media (Williams et al., 2016). Briefly, overnight cultures of A. baumannii strains were grown in CFA medium (Evans et al., 1977), and bacteria were diluted to an OD600 of 0.05 in 24-well tissue-culture treated plates (Corning, Corning, NY, United States) in 1 mL of CFA medium. Plates were incubated statically at 37°C for 24 h. To expose the pre-formed biofilms to copper, the broth was removed and was replaced with 1.5 mL of M9 medium containing 1.5 mM CuSO4; biofilms were incubated for another 6 or 24 h. To determine the number of planktonic cells, at each timepoint (0, 6, and 24 h) the medium was transferred to a 1.5 mL Eppendorf tube and vortexed for approximately 5 s to disrupt any clumped bacteria in the supernatant; samples were visually inspected to confirm the absence of clumped bacteria. To determine the number of biofilm cells, the biofilm was scraped from the sides and bottom of the well with a pipette tip and was resuspended in 1 mL of PBS. Samples of both planktonic cells and biofilm cells were plated to enumerate CFU. Three biologically independent experiments were performed.
Quantification of Intracellular Copper via ICP-MS
Sample Collection
To measure the intracellular copper concentration, dry cell samples were collected and analyzed by inductively coupled plasma mass spectrometry (ICP-MS). Overnight cultures were subcultured to an OD600 of 0.05 in a large 100 mL volume. The subcultures were grown for 2.5 h to reach log phase, and a time zero (T0) sample was collected. The sample collection procedure was as follows: 6 mL of culture was pelleted in three 2-mL tubes using a 4°C table top centrifuge at max speed for 30 s; the supernatant was aspirated. The three pellets were resuspended in 1 mL of cold wash solution (PBS with 0.5 mM EDTA) in a single tube and pelleted again; the supernatant was aspirated. The wash steps were repeated for a total of three washes. After the final aspiration, the tube was left open to begin air drying. A 0.5 mL sample of the culture was separately collected in a cuvette to measure the OD600 of the culture. The cell pellets were fully dried in an 80°C SpeedVac vacuum concentrator for 1 h and stored at −20°C. After collection of the T0 sample, 0.25 mM copper sulfate was added directly to the large culture, swirled immediately, and returned to the shaking incubator. Additional samples were collected along the time course. In experiment two, with the complete set of mutant strains showing a copper sensitive phenotype, the final wash step was conducted in a 15-mL conical tube and spun for 3 min at 4150 rpm/3,700 × g, and the cell pellets were dried overnight in a 37°C incubator. This eliminated the need for transfer of many samples to conical tubes before ICP-MS analysis.
After the copper shock portion of the experiment was completed, the remaining culture was washed and placed into fresh growth medium for recovery. Briefly, the remaining culture was placed in a 50-mL conical tube and spun for 15 min at maximum speed (4150 rpm/3,700 × g) to pellet the cells. The cells were resuspended in 2 mL of wash solution and were transferred to two 2-mL tubes. The cells were pelleted by spinning for 1 min at max speed in a tabletop centrifuge; the supernatants were aspirated. The wash steps were repeated for a total of three washes. The cells were resuspended in 50 mL of pre-warmed growth medium without added copper. Samples were collected in the same way as described above across the recovery time course. Three biologically independent replicates were performed for both experiments.
Copper Measurement
The dry cell pellets were thawed at room temperature and dissolved in 200 μL of concentrated HNO3 (trace metal grade) overnight at 80°C. The following day, the samples were cooled to room temperature and diluted with 2 mL of MilliQ water, for a final HNO3 concentration of 6%. Where necessary, the samples were transferred to metal-free 15-mL conical tubes for ICP-MS analysis. The metal content of the samples was then analyzed by quantitative ICP-MS. Copper concentrations were determined by injecting diluted samples into an Agilent 7700x ICP-MS (Agilent Technologies, Santa Clara, CA, United States). Copper levels were detected using an Octopole Reaction System cell (ORS) in He mode. The ICP-MS parameters used for the analysis were: an RF power of 1550 W, an argon carrier gas flow of 0.99 L/min, helium gas flow of 4.3 mL/min, octopole RF of 200 V, and OctP bias of −18 V. Samples were directly infused using the 7700x peristaltic pump with a speed of 0.1 rps and a micromist nebulizer. Copper concentrations in samples were derived from a calibration curve generated by a series of dilutions of a copper atomic absorption standard (Fluka Analytical, St. Louis, MO, United States) prepared in the same matrix as the samples. Data analysis was performed using Agilent’s Mass Hunter software (4.4 version).
Calculations
The value originally obtained from ICP-MS (ppb, μg/L) was divided by the molecular weight of copper (63.546 g/mol) to determine the concentration of copper in the diluted sample (μM). The sample concentration was multiplied by the total volume (2.2 mL) to determine the quantity of copper in the sample (nmol). The total amount of copper was divided by the sample volume (6 mL) to determine nmol/mL of cells and divided by the OD600 to determine the nmol/ODU of cells. The data was plotted as nmol/ODU vs. time. This protocol was recently described in more detail as a methods chapter (Williams et al., 2019).
Infection of Galleria mellonella Caterpillars
Acinetobacter baumannii strains were grown overnight in LB medium with the appropriate antibiotics: tetracycline for transposon mutant strains, and both tetracycline and hygromycin for complemented strains. Overnight cultures were diluted to an OD600 of 0.05 and were grown 3 h to log phase. The cells were adjusted in PBS to a specific OD600 to achieve the desired CFU/mL, and were then diluted 1:10 in PBS with 0.01% bromophenol blue dye for visibility. For a dose of 5 × 104 CFU, bacteria were adjusted to an OD600 of 0.2–0.3, which corresponded to 107 CFU/mL; for a dose of 5 × 105, bacteria were adjusted to an OD600 of 2–3, or 108 CFU/mL. The exact number of bacterial cells in the inoculum was determined by serial dilution and plating for enumeration of CFU. The actual doses ranged from 3.53–9.63 × 104 to 3.38–9.60 × 105.
Galleria mellonella larvae (Vanderhorst Wholesale, Inc., Saint Marys, OH, United States) were utilized within 4 days of receipt. Larvae weighing 200–300 mg were used. The injections were carried out as described previously (Jacobs et al., 2014a) with minor changes. Briefly, 5 μl of the sample was injected into the last left proleg using a 10-μl glass syringe (Hamilton, Reno, NV, United States) fitted with a 31G needle. Each experiment included control groups of non-injected larvae and PBS-injected larvae. All larvae were incubated at 37°C, and death was assessed at 24 h intervals for 5 days. Larvae were considered dead if they didn’t respond to physical stimulus. All larvae that progressed to the pupation stage of the life cycle were excluded. For consistency, wild-type, mutant strain, and complemented strain infected groups that are plotted together were injected in the same experiment. Experiments were repeated using two or three different orders of larvae with 12 to 18 larvae per experimental group; the total n ranged from 25 to 48 larvae per strain. A single data set was excluded in which the wild-type strain killed only 30% of the caterpillars; no data from this set of infections was used.
Murine Pneumonia Infection Model
Mice were infected with A. baumannii as described previously (Jacobs et al., 2014a). Briefly, 6-week-old female BALB/c mice were first rendered temporarily neutropenic by intraperitoneal injection with cyclophosphamide on days −4 and −1 before infection. On day 0, mice were anesthetized with 2–5% isoflurane gas and inoculated intranasally with approximately 5 × 106 CFU of A. baumannii in a total volume of 25–50 μL PBS. Animals were monitored for morbidity and mortality for 7 days, and humanely euthanized with CO2 inhalation when necessary. All mouse studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 2011), and procedures were approved by the Institutional Animal Care and Use Committee at the Walter Reed Army Institute of Research (protocol 16-BRD-48S). Experiments with the cusR mutant strain and complemented derivative were performed in four biological replicates with five mice per group (n = 20 per strain), spaced two and two over time to ensure reproducibility; two biological replicates were completed with the copD mutant strain and complemented derivative (n = 10 mice per strain).
Data Analysis and Statistics
All graphs and statistical analyses were carried out using GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA, United States). A two-way analysis of variance (ANOVA) with Tukey’s correction for multiple comparisons was used to evaluate differences in growth in various copper treatments. Data are presented as the geometric mean and standard error of the mean. A two-way ANOVA with Sidak’s or Dunnett’s correction was used to compare intracellular copper concentrations. Data are presented as arithmetic mean and range. An ordinary one-way ANOVA with Dunnett’s correction followed by linear contrasts with a Bonferroni adjustment was used to compare biofilm and planktonic cells percent survival at each concentration and time point to the wild-type. Data are presented as arithmetic mean and range. Survival curves were compared using the Mantel–Cox log rank test with Holm’s correction for multiple comparisons. For all tests, a two-sided α level set at 0.05 determined significance.
Results
Mutant Strains of AB5075 Exhibit Copper Sensitivity
Many putative copper-related genes have been identified in A. baumannii; however, their predicted functions are based solely on protein homology and the role of these proteins in copper resistance is largely unknown. To investigate the contribution of individual genes to copper resistance in AB5075, we used the available arrayed transposon mutant strain library (Gallagher et al., 2015) to initiate our studies. To this end, we obtained 21 mutant strains, each of which contained a T26 transposon insertion in a putative copper resistance gene. Specifically we targeted the 22 genes found in regions A–D that we previously identified as putative copper resistance genes in AB5075 (Williams et al., 2016); only one small ORF, ABUW_2708, was not represented in the available mutant strain library. To assess the role of the remaining 21 genes in copper resistance, each of the mutant strains was individually grown in M9 medium containing increasing concentrations of CuSO4. Thirteen of the mutant strains did not differ from the wild-type in their ability to grow in the presence of copper, indicating that the mutated gene was not individually contributing to copper resistance in the tested conditions (Supplementary Figure S1). Conversely, eight of the mutant strains showed significant changes in copper sensitivity relative to the wild-type strain. The phenotypes of these eight strains fell into two categories: moderately and highly sensitive. Strains bearing mutations in cusR, cusS, copC, and copD were all highly sensitive to copper and showed significant growth defects in as little as 0.25 mM CuSO4 (Figure 2). In contrast, strains bearing mutations in pcoB, pcoA, copB, and cueO were moderately copper sensitive and only displayed increased sensitivity relative to the wild-type strain in the presence of at least 1 mM CuSO4 (Figure 3). Of note, nearly all the mutant strains with copper sensitive phenotypes contained mutations in genes carried in region D (Figure 1), and these strains displayed the most substantial defects in copper resistance.
To ensure that excess sulfate found in the copper sulfate was not responsible for the growth phenotypes, we performed a similar growth experiment with 1.5 mM sodium sulfate; sodium sulfate did not inhibit growth of the wildtype or mutant strains (Supplementary Figure S2). To confirm that the copper sensitive phenotype was due to the T26 transposon insertion, we constructed a complemented derivative of each of the eight mutant strains of interest. Complementation was achieved by insertion of a wild-type copy of the ORF and its native promoter in an attTn7 site downstream of the glmS locus (Kumar et al., 2010). Each of the complemented strains was individually tested in the same growth experiment. Complete restoration of copper resistance to wild-type levels was observed in the cusR, cusS, and copD complemented strains (Figure 2). Partial restoration of copper resistance was observed in the copB, cueO, and copC complemented strains (Figures 2, 3). No functional complementation was observed in the pcoA and pcoB complemented strains (Figure 3); the complemented derivative of the pcoB mutant strain actually grew slightly less than its parent strain in 1 mM copper sulfate. The lack of complementation perhaps suggests that the identified phenotypic changes are not purely due to the transposon insertion in pcoA and pcoB, or that this complementation strategy was insufficient for these two particular genes. Taken together, our mutational analyses indicate that many of the identified putative copper resistance genes indeed contribute to copper resistance in A. baumannii. Moreover, the results suggest that the genes found in region D are crucial for the highest levels of copper resistance.
Region D Enhances Copper Resistance in Other A. baumannii Strains
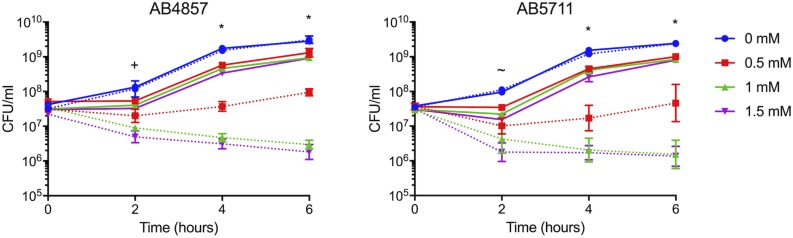
Our previous analysis of multiple clinical isolates of A. baumannii identified two isolates, AB4857 and AB5711, that both showed inherently reduced copper resistance as compared to AB5075. Notably, neither of these strains carried the genes from region D in their genomes (Williams et al., 2016). Given our finding that mutations in the genes located in region D caused the greatest copper sensitivity in AB5075 (Figure 2), we hypothesized that the transfer of region D to AB5711 and AB4857 would increase their overall level of copper resistance. Indeed, when region D from AB5075 was carried on the plasmid pAJ100 in these strains, pAJ100-Region D conferred increased copper resistance comparable to the level seen in AB5075; the empty pAJ100 vector did not affect resistance (Figure 4). Thus, these results also indicate that the genes in region D are important for high levels of copper resistance in A. baumannii.
FIGURE 4.
Growth of AB4857 and AB5711 carrying Region D. Region D from AB5075 was cloned into pAJ100 and transformed into AB4857 and AB5711, which do not naturally encode the region D genes. Strains carrying the empty vector, pAJ100, were used as controls. The strain background is indicated in the graph title (AB4857, left; AB5711, right); strains carrying the control vector pAJ100 are indicated with dotted lines, and strains carrying pAJ100-Region D are indicated with solid lines. Strains were grown for 6 h at 37°C in M9 medium supplemented with the indicated concentration of CuSO4. Growth was measured both by turbidity (OD600, data not shown) and enumeration of viable colonies (CFU/mL). The data are presented as geometric mean and SEM of three biologically independent experiments. Two-way ANOVA with Tukey’s adjustment for multiple comparisons was used to compare growth of the strain carrying the pAJ100 vector to the strain carrying pAJ100-Region D in each copper concentration at each timepoint. The symbols indicate how many comparisons were statistically significant (P < 0.05) at each timepoint, as follows: ∼, growth in 1.5 mM CuSO4 was different between the two strains; +, growth in 1 and 1.5 mM CuSO4 was different between the two strains *, growth in 0.5, 1, and 1.5 mM CuSO4 was different between the two strains.
Biofilms of Mutant Strains Retain Increased Copper Sensitivity
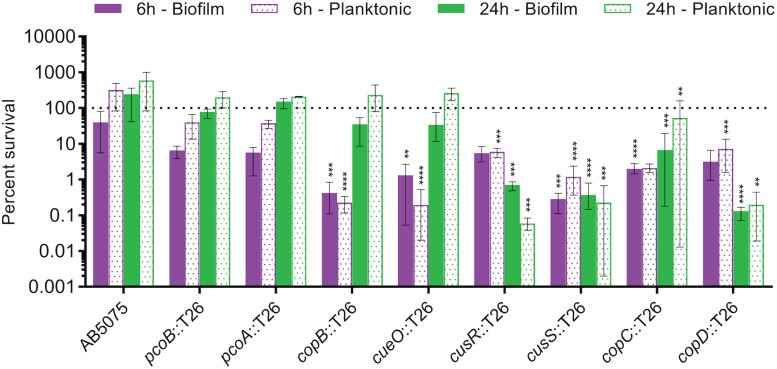
Biofilm formation is an important component of the A. baumannii lifecycle; the bacterium is known to create biofilms on both biotic and abiotic surfaces, which aides in fomite-based transmission and in virulence (Longo et al., 2014; Zurawski et al., 2019). Moreover, biofilms are notorious for their ability to decrease the effectiveness of antibiotic treatment (Hoiby et al., 2010). Because any future copper-based therapeutics would likely need to be effective against A. baumannii found within a biofilm structure, we next tested the copper sensitivity of biofilms formed by the copper sensitive mutant strains as compared to wild-type AB5075. Biofilms were established in complex media for 24 h before switching the medium to M9 medium supplemented with 1.5 mM CuSO4. CFU were enumerated from both the biofilm and supernatant at 0, 6, and 24 h post copper shock, and data were expressed as percent survival relative to T0. For the wild-type AB5075, a slight decrease in biofilm CFU was observed at 6 h, but no obvious sensitivity to copper was observed in the planktonic population at this timepoint or in the biofilm or planktonic populations at 24 h (Figure 5). While the pcoA and pcoB mutant strains of region B did not demonstrate survival differing from the wild-type strain in these biofilm conditions, the strains bearing mutations in region D (copB, cueO, cusR, cusS, copC, copD) each displayed statistically significant decreases in survival of both biofilm and planktonic cells relative to the wild-type strain; a 1–3 log loss in recoverable CFU was observed. The copB and cueO mutant strains displayed an early defect but recovered to wild-type levels by 24 h. Conversely, the cusR, cusS, copC, and copD mutant strains did not recover. These data demonstrate that most mutant strains retain copper sensitivity relative to the wild-type strain even within a biofilm structure, suggesting that future copper resistance targeting therapeutics would likely not be hindered by any inherent copper resistance of a biofilm.
FIGURE 5.
Copper-dependent killing of AB5075 mutant strain biofilms and planktonic cells. Cultures were statically incubated in a 24-well tissue culture-treated plate at 37°C to establish biofilms. After 24 h, the broth was replaced with M9 medium containing 1.5 mM CuSO4. After an additional 0, 6, or 24 h of incubation, bacteria from both the supernatant (planktonic) and the biofilm were plated for enumeration of CFU. Percent survival was calculated as the percentage of bacteria present at time zero under each condition. Data are presented as mean and range for three biologically independent experiments. One-way ANOVA with Dunnett’s correction was used to compare each mutant strain to the wild-type at each indicated condition. Statistically significant comparisons are indicated with the asterisks, as follows: **P < 0.01; ***P < 0.001; ****P < 0.0001.
Copper-Sensitive Mutant Strains Accumulate More Intracellular Copper
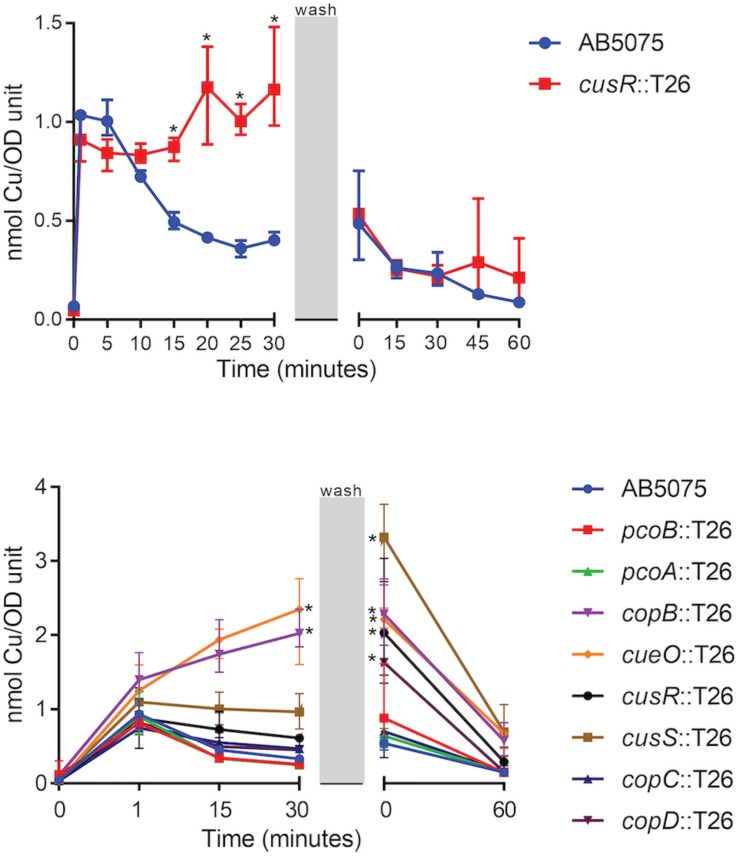
To begin to understand the ability of the wild-type strain to adapt to copper stress as well as the nature of the copper homeostasis defects in the copper sensitive mutant strains, we utilized ICP-MS to temporally measure copper accumulation in strains exposed to a copper shock (0.25 mM). Previous studies in other bacteria have demonstrated that intracellular copper accumulates in mutant strains bearing mutations in copper-related proteins, including pcoB, pcoA, copA, cusR, and cusS (Outten et al., 2001; Lee et al., 2002; Gonzalez-Guerrero et al., 2010; Gudipaty et al., 2012). To optimize the assay, we focused our initial efforts on wild-type AB5075 and the cusR mutant strain. In wild-type AB5075, the intracellular copper concentration increased ∼10x one minute following copper exposure. However, the level reduced to ∼5x in approximately 15 min and fully recovered to baseline levels 60 min following removal of excess copper from the media (Figure 6). Thus, in the wild-type strain, the cells appear to respond to copper stress via the deployment of efflux mechanisms that reduce the level of intracellular copper. In comparison, for the copper sensitive cusR mutant strain, intracellular copper levels immediately increased ∼10x, but showed no reduction while copper was present. However, after copper removal, the level similarly recovered to baseline within 60 min (Figure 6). These data suggest that in the presence of excess copper, the cusR mutant cells are unable to control the intracellular copper concentration and cannot efflux copper ions as well as the wild-type cells.
FIGURE 6.
Accumulation of intracellular copper. Log phase cultures were treated with 0.25 mM CuSO4 and samples were collected at the indicated timepoints. During the wash phase, the remaining culture volume was washed and resuspended in fresh M9 medium without supplemented copper. Cell samples that were collected at each timepoint were washed and dried, and copper content was measured by ICP-MS. Data are presented as the mean and range for three biologically independent experiments. Two-way ANOVA with Sidak’s correction was used to compare the cusR mutant strain to the wild-type at all timepoints (top); two-way ANOVA with Dunnett’s correction was used to compare all the mutant strains to wild-type at all timepoints (bottom). Statistically significant comparisons are indicated with an asterisk (P < 0.01). At 0 min post-wash, five strains were significantly different from wild-type: copB, cueO, cusR, cusS, and copD (bottom).
Based on the above results, we modified the sampled timepoints and collection strategy in order to simultaneously test the wild-type and all eight of the mutant strains that previously demonstrated increased copper sensitivity (Figures 2, 3). Of note, the revised collection strategy resulted in a longer period of copper exposure as the washing steps took ∼20 min longer to collect the cells. Using this procedure, the copB and cueO mutant strains each showed significantly higher accumulation of copper relative to the wild-type strain during the 30 min copper shock. In addition, the cusR, cusS, and copD mutant strain accumulated a higher concentration of intracellular copper within the additional time of copper exposure found with the modified washing steps. All strains demonstrated recovery following the removal of the copper stress (Figure 6). Taken together, these results indicate that many of the mutant strains have defects in copper homeostasis and copper efflux, which likely contribute to their copper sensitivity.
Copper Resistance Genes Are Important for Virulence in Galleria mellonella Larvae
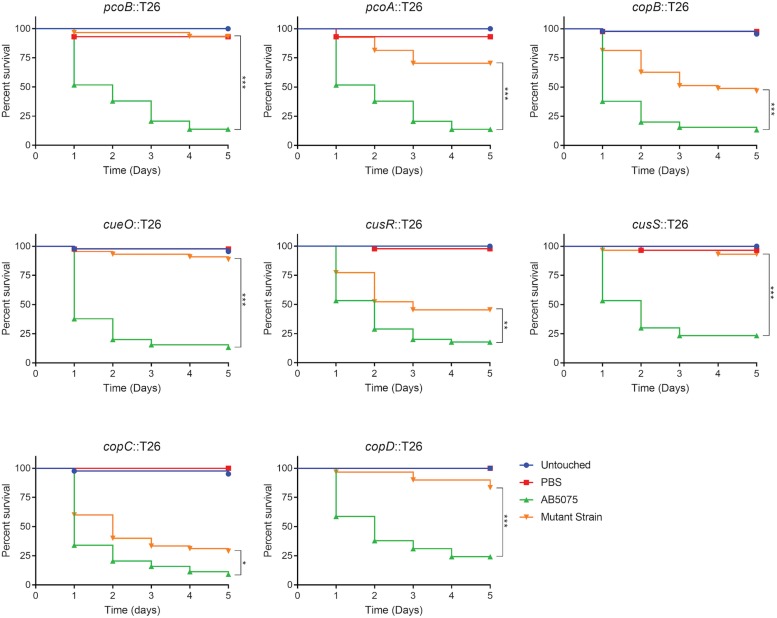
Given that copper is an important component of immune defense, we hypothesized that copper resistance is an important trait that affects virulence of A. baumannii. To begin to test this possibility, we assessed the ability of the copper sensitive mutant strains to kill G. mellonella caterpillars. This model was chosen because: (1) G. mellonella have been established as an inexpensive and simple infection model for many pathogenic bacteria, including A. baumannii, (2) these caterpillars can be maintained at 37°C, (3) these caterpillars have both humoral and cellular immune response pathways, and (4) this invertebrate model has been used for identification of bacterial virulence factors and virulence results often correlate with those obtained in mammalian models (Peleg et al., 2009; Jacobs et al., 2014a; Tsai et al., 2016). Groups of G. mellonella were individually infected with each strain of A. baumannii, incubated at 37°C, and observed for 5 days. Consistent with previous experiments with AB5075 (Jacobs et al., 2014a), at a dose of approximately 5.0 × 104 CFU, wild-type AB5075 killed ∼85% of the G. mellonella, with most death occurring in the first 2 days post-infection (Figure 7). In contrast, the copper sensitive mutant strains were all attenuated in this model and killed significantly less G. mellonella than the wild-type strain (Figure 7). The pcoB, pcoA, cueO, cusS, and copD mutant strains were the most attenuated and killed less than 30% of the G. mellonella. The copB and cusR mutant strains were moderately attenuated and killed ∼55% of G. mellonella. The copC mutant strain was very mildly attenuated and killed ∼70% of G. mellonella. To ensure that the strains from the AB5075 transposon library did not show a general defect in the G. mellonella model, we additionally selected and tested five mutant strains that did not display a copper sensitive phenotype in vitro; none of these strains were attenuated for virulence in the G. mellonella model (Supplementary Figure S5).
FIGURE 7.
Survival of Galleria mellonella infected with AB5075 mutant strains. G. mellonella were injected with approximately 5.0 × 104 CFU of the indicated strain and survival was monitored for 5 days. The gene interrupted in each mutant strain is indicated in the graph title. Experiments were repeated using two or three different orders of larvae with 12–15 larvae per experimental group; the total n ranged from 25 to 45 larvae per strain. Kaplan–Meier survival curves were compared (excluding Untouched and PBS controls) using the Mantel–Cox test with Holm’s correction for multiple comparisons. Statistically significant comparisons of the mutant strain to the wild-type strain are indicated by bars with asterisks signifying the P-value as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
As with the in vitro phenotypes, we also sought to functionally complement the virulence defects seen in the G. mellonella model. However, infection with the complemented strains resulted in similar rates of G. mellonella death as those infected with the corresponding mutant strain; in some cases, the complemented strains appeared slightly more attenuated (Supplementary Figures S3, S4). Based on these data, we speculated that the insertion at the attTn7 site was causing unintended attenuation in this model; to our knowledge, no published studies have shown functional complementation using the Tn7-based strategy in assays of G. mellonella survival. To test this possibility, a strain carrying an attTn7 insertion of only the hygromycin resistance gene was created and then compared in virulence to the wild-type background strain. Attenuation of the AB5075:Tn7 strain relative to AB5075 was not consistently observed (Supplementary Figure S5). We reasoned that perhaps the effect was only evident in strains already carrying the T26 transposon insertion and that the size of the attTn7 insertion might be important. Thus, derivatives of the cusR and copC mutant strains were engineered to carry an attTn7 insertion of only the hygromycin resistance gene and were then compared to the parental mutant strain as well as the complemented derivatives, which each carried much larger attTn7 insertions. These hygromycin only insertion strains were both slightly more attenuated than the parental strains (Supplementary Figure S5). Furthermore, the complemented strains carrying the larger attTn7 insertion were even further attenuated (Supplementary Figure S5). These results suggest that the inability to functionally complement the mutant virulence phenotypes is due to the fact that insertion at the attTn7 site is attenuating in the G. mellonella survival model. The mechanistic reason for this effect is unknown, but may be due to disruption of an important genetic element near the attTn7 site, or perhaps polar effects from expression of the hygromycin resistance gene; all of our complemented strains contain insertions oriented in the same forward direction, therefore the hygromycin resistance gene and its promoter face out from Tn7 and downstream of glmS. While the reason for this attenuation currently remains unclear, overall, the data strongly suggest that copper resistance, and many individual proteins in regions B and D, are specifically needed for full virulence of A. baumannii in the G. mellonella model. These conclusions are strengthened by a lack of attenuation of other transposon mutant strains that did not demonstrate increased copper sensitivity (Supplementary Figure S5), however, these conclusions would clearly be further strengthened by functional complementation in this model.
CusR and CopD Are Important for Virulence in a Murine Pneumonia Infection Model
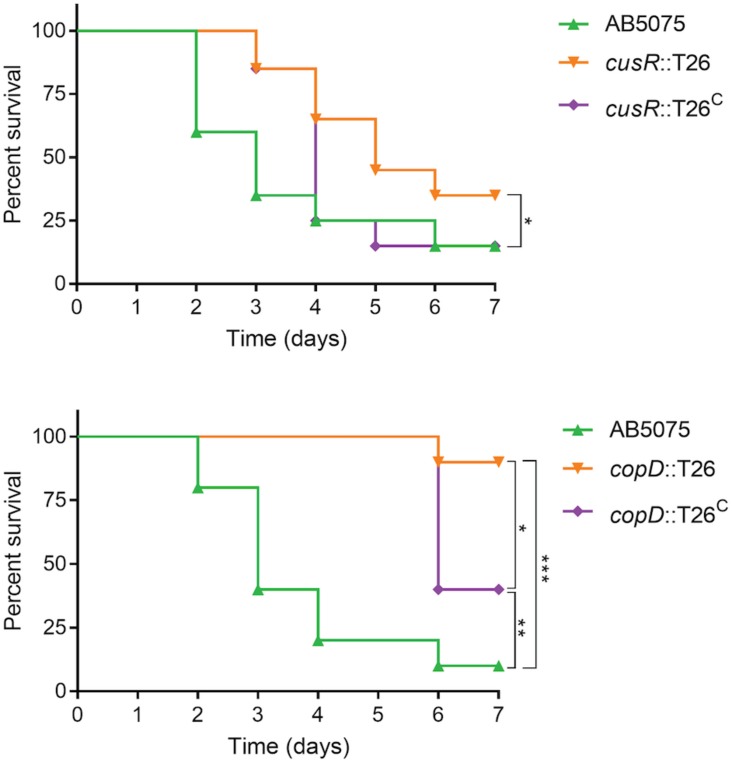
Given the attenuated virulence observed in the G. mellonella model, we next wished to determine if A. baumannii copper resistance is also an important virulence determinant during infection of a mammalian host. To this end, we selected two of the mutant strains and their complemented derivatives to test in a murine pneumonia model; the cusR and copD mutant strains were selected due to their high level of copper sensitivity and successful in vitro complementation (Figure 2), as well as their phenotypic differences from the wild-type strain in all other assays, including virulence in G. mellonella (Figures 5–7). The murine pneumonia model was chosen because lung infections are among the most common A. baumannii infections, and AB5075 virulence has been characterized in this model (Jacobs et al., 2014a). As shown in Figure 8, the wild-type strain killed 85% of mice by 7 days post-infection. However, the cusR mutant strain was attenuated in this model; death was delayed relative to wild-type and only 65% of the mice died. Virulence of the cusR mutant strain was restored by complementation of the cusR gene in trans; 85% of the mice were killed by the complemented strain by 7 days post-infection. The copD mutant strain showed an even more dramatic virulence defect and only killed 10% of mice as compared to 90 and 60% by the wild-type and complemented strains, respectively. The ability to functionally complement both the copD and cusR mutant strain phenotypes in the pneumonia model further supports our conclusions concerning failure to achieve complementation in the G. mellonella model. In summary, both CusR and CopD individually contribute to virulence of A. baumannii in the murine pneumonia model.
FIGURE 8.
Assessment of virulence of AB5075 mutant and complemented strains in a murine pneumonia model. Mice were infected intranasally with approximately 5 × 106 CFU of the indicated strain and survival was monitored for 5 days. Experiments with the cusR mutant strain and complemented derivative were performed in four biological replicates with five mice per group (n = 20 per strain), spaced two and two over time to ensure reproducibility; two biological replicates were completed with the copD mutant strain and complemented derivative (n = 10 mice per strain). Kaplan–Meier survival curves were compared using the Mantel–Cox test with Holm’s correction for multiple comparisons. Statistically significant comparisons are indicated by bars with asterisks signifying the P-value as follows: *P < 0.05; **P < 0.01; ***P < 0.001. The only significant comparison in the top panel is wild-type AB5075 vs. the cusR mutant strain.
Discussion
Novel therapeutics are desperately needed to treat infections with drug-resistant A. baumannii and other superbugs. Because copper homeostasis is an essential process and copper ions themselves have potent toxicity, we sought to identify important copper resistance determinants in A. baumannii; we theorize that these could potentially be targeted with future therapeutics. Using a focused approach, we identified genes in A. baumannii with putative roles in copper resistance, and we demonstrated that 8 of 21 tested genes individually contributed to in vitro copper resistance of AB5075; mutant strains displayed significant copper sensitive phenotypes during planktonic growth (Figures 2, 3). Mutation of genes carried in region D caused the largest increase in copper sensitivity, indicating that this region is crucial for high levels of resistance. Furthermore, when a plasmid-borne copy of region D was introduced into A. baumannii strains that naturally lacked region D, copper resistance was dramatically increased (Figure 4). Moreover, strains bearing mutations of genes in region D showed enhanced copper-dependent killing even when found in a biofilm structure (Figure 5). The copper sensitivity is likely due to the fact that many of the mutant strains (copB, cueO, cusR, cusS, copD) demonstrated markedly increased accumulation of intracellular copper as compared to wild-type AB5075, which was able to efficiently efflux copper ions (Figure 6). Finally, when tested in vivo in G. mellonella and a murine pneumonia model, the copper sensitive mutant strains showed distinct attenuation in virulence. In the G. mellonella model, the mutant strains killed less G. mellonella than the wild-type strain, with the strongest attenuation observed with the pcoB, cueO, cusS, and copD mutant strains (Figure 7). Similarly, in the murine mouse model, the cusR and copD mutant strains were also attenuated and the attenuation phenotype was able to be complemented (Figure 8). En masse, our data indicate that copper resistance is mediated by many genes in AB5075 and show that copper resistance contributes to virulence.
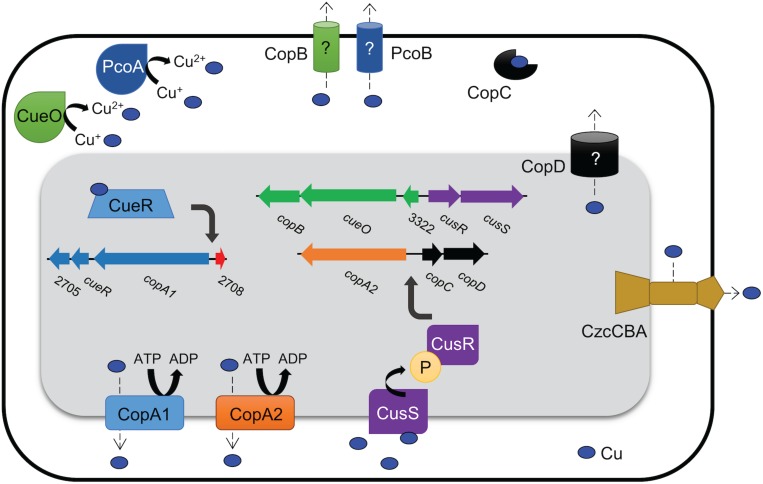
Though the absolute functions of the proteins encoded by each of the genes showing homology to copper related systems remains to be determined, based on predicted functions, a model for the mechanisms of copper resistance in AB5075 was created (Figure 9). When thinking about this model, it is important to keep in mind that based on the mutational analysis, proteins putatively involved in transport (PcoB, CopB, CopD), oxidation (PcoA, CueO), chaperoning (CopC), and regulation (CusR, CusS) individually contributed to copper resistance in AB5075 in our tested in vitro conditions. However, this does not negate a possible role for the other predicted copper-related factors within other environments. Also, it should be noted that the model proposed by another group for copper resistance proteins in AB5075 differs slightly from ours (Alquethamy et al., 2019). Furthermore, our model does not include any potential interactions with other biologically relevant heavy metals. In copper-related systems that have been studied in other bacteria, it has been shown that there is sometimes interaction with other heavy metals, e.g., silver (Rensing et al., 2000; Gudipaty et al., 2012); therefore, a limitation of our study is that we only investigated the role of these genes in copper resistance. Thus, there is potential that these genes could also contribute to resistance to other metals; such possible roles warrant further study.
FIGURE 9.
Model of putative copper resistance mechanisms in AB5075. Predicted protein identifications were determined by amino acid sequence homology to known proteins from other bacteria, and proteins with putative copper resistance functions are indicated. In the mutational analysis described herein, proteins putatively involved in transport (PcoB, CopB, CopD), oxidation (PcoA, CueO), chaperoning (CopC), and regulation (CusR, CusS) individually contributed to copper resistance in AB5075. Copper ATPases are highly conserved inner-membrane proteins that efflux copper ions out of the cytoplasm; AB5075 appears to have two copper ATPases: CopA1 and CopA2. The RND family efflux pump CzcCBA may efflux copper from the periplasm to the extracellular milieu. Three other putative membrane proteins in AB5075 may also have roles in copper transport, though the function of these proteins is not yet well-defined in any bacteria (CopB, PcoB, and CopD). Periplasmic copper oxidases reduce damage by converting Cu+ to the less toxic Cu2+; CueO and PcoA are both predicted copper oxidases. CopC is a highly conserved copper chaperone that is found in the periplasm; it prevents damage by binding and sequestering copper ions and may facilitate movement of copper ions between other copper-related proteins, i.e., delivering copper ions to an efflux pump. Two well-conserved putative regulatory systems are also present in AB5075: CueR senses cytoplasmic copper levels and then upregulates copper resistance genes in its copper-bound form, and CusRS is a two-component system that is thought to be activated by periplasmic copper. The regulons of CueR and CusR have not yet been defined in AB5075, though putative binding sites have been identified in regions B and D, respectively, and these putative target genes are indicated in the figure.
We utilized multiple assays to analyze in vitro copper resistance and to assess virulence. In many cases, we observed corroborating results across multiple assays. For instance, we observed similar patterns of sensitivity in our planktonic and biofilm growth assays. The four strains with the greatest increase in copper sensitivity during planktonic growth (cusR, cusS, copC, copD) also demonstrated sustained, increased copper sensitivity in the context of biofilm growth. The four additional strains that showed more moderate increases in copper sensitivity in the planktonic growth assay, displayed temporarily increased (copB, cueO) or no change in sensitivity (pcoA, pcoB) relative to the wild-type background in the context of biofilm growth. Interestingly, relative to the most copper sensitive mutant strains, the copB and cueO mutant strains seemed to be more copper sensitive in the biofilm assay than in the planktonic growth assay; the reason for this change in the pattern of sensitivity among the strains is unknown, however, different culture conditions (rich vs. minimal media) could play a role. Additionally, we did not observe a direct correlation between in vitro copper sensitivity and attenuation of virulence in the G. mellonella infection model. For example, the two most attenuated mutant strains in the G. mellonella model (pcoB and cueO) had only moderate copper sensitivity in the planktonic growth and biofilm survival assays, while the cusR and copC mutant strains demonstrated strong phenotypes in in vitro assays but were only mildly attenuated for virulence in G. mellonella. These differences among assays highlight the importance of the methods chosen to assess genes of interest; the roles of individual genes may vary depending on environmental conditions, and this will affect the experimental outcomes and the conclusions drawn. Indeed, experimental differences may account for contrasting results in this work and those published recently by another group (Alquethamy et al., 2019). Under the conditions we tested, we did not observe increased copper sensitivity of either of the copper ATPase mutant strains (Supplementary Figure S1) or attenuation in the G. mellonella infection model (Supplementary Figure S5). We originally hypothesized that perhaps CopA1 and CopA2 were functionally redundant, resulting in an inability to observe a copper-sensitive phenotype. However, a recently published study of the copper ATPases in AB5075 found that the gene referred to as copA1 in our publication (ABUW_2707, region B), but not copA2 (ABUW_3325, region D), contributed to copper resistance (Alquethamy et al., 2019); those results suggest that the CopA1 and CopA2 proteins are not functionally redundant. While the exact reason for the difference in study results is not clear, it is worth noting that the experimental methods used were different, e.g., growth medium (which dramatically affects free copper concentrations) and infection model; additionally, the various utilized mutant strains differed in their site of transposon insertion and transposon orientation, which may cause differential polar effects. Given the disparity in the results of the studies, it is currently unclear what the individual roles of these copper ATPases are in A. baumannii, and additional studies will be required to clearly define the role of these factors in copper resistance.
Our studies identified many copper resistance proteins that individually contributed to virulence in the chosen models; these proteins have a variety of putative functions including copper efflux, oxidation, chaperoning, and regulation. Copper-related genes with similar functions have been shown to be important for virulence in many bacterial and fungal pathogens including Pseudomonas aeruginosa, Pseudomonas fluorescens, Xanthomonas campestris pv. campestris, Xanthomonas citri subsp. citri, Streptococcus pneumoniae, Streptococcus mutans, Salmonella Typhimurium, Mycobacterium tuberculosis, Neisseria gonorrhoeae, Listeria monocytogenes, E. coli, Staphylococcus aureus, Campylobacter jejuni, Aspergillus fumigatus, and A. baumannii (Francis and Thomas, 1997; Schwan et al., 2005; Zhang and Rainey, 2007; Hu et al., 2009; White et al., 2009; Achard et al., 2010; Gonzalez-Guerrero et al., 2010; Osman et al., 2010; Ward et al., 2010; Corbett et al., 2011; Hsiao et al., 2011; Shafeeq et al., 2011; Wolschendorf et al., 2011; Yan and Wang, 2011; Djoko et al., 2012; Marrero et al., 2012; Chaturvedi et al., 2014; Shi et al., 2014; Subashchandrabose et al., 2014; Johnson et al., 2015; Garcia et al., 2016; Park et al., 2017; Wiemann et al., 2017; Gardner and Olson, 2018; Purves et al., 2018; Zapotoczna et al., 2018; Alquethamy et al., 2019). Of note, three types of copper-related proteins that have been previously shown to contribute to virulence of other bacteria were found to also contribute to virulence of A. baumannii: copper ATPases, copper oxidases, and copper chaperones. In the majority of the previous studies, the mutated copper resistance gene of interest was a copper ATPase, homologous to copA1 and copA2 of AB5075. Of note, a role for CopA1 in virulence of A. baumannii was recently described (Alquethamy et al., 2019). However, we did not observe copper sensitivity or reduced virulence in either of our copper ATPase mutant strains (Supplementary Figures S1, S5). Copper oxidases have been shown to be important for virulence of S. pneumoniae, C. jejuni, and S. aureus (Achard et al., 2010; Gardner and Olson, 2018; Zapotoczna et al., 2018). Here, we demonstrated that both copper oxidases of AB5075, PcoA and CueO, contribute to virulence of A. baumannii as well. Similarly, chaperone proteins have been previously linked to virulence in S. pneumoniae and S. mutans (Johnson et al., 2015; Garcia et al., 2016), and we found that the putative chaperone CopC contributes in A. baumannii. Though their functions may be similar, it is unclear how related CopC of A. baumannii is to the chaperones of other bacteria, as these chaperone proteins are small and have minimal sequence identity or similarity. While these three types of proteins are interesting as putative drug targets, an important consideration in identifying novel drug targets is assessment of homology with host proteins and potential toxicity from off-target activity. Copper ATPase and oxidase proteins have amino acid homology to human proteins, and the copper ATPases are also functionally similar. Thus, designing therapeutics that target these functions may be complicated by homology to the essential human proteins, e.g., human copper ATPases ATP7A and ATP7B. While not insurmountable, homology to host proteins certainly makes design of novel therapeutics more challenging.
Of particular interest, we identified a number of copper-associated proteins that have not previously been shown to contribute to virulence in other bacterial species. To our knowledge, this is the first publication to report attenuated virulence in any bacterial species of strains lacking the putative transporters PcoB, CopB, or CopD (Wolschendorf et al., 2011; Subashchandrabose et al., 2014). We also observed attenuated virulence of the strains bearing mutations in either portion of the two-component system CusRS. To our knowledge, this is the first report that this copper-sensing two-component system contributes to virulence of a bacterial species during mammalian infection. Additionally, none of these proteins share amino acid sequence homology with human proteins, and therefore may be targeted more readily.
While the focus of this work was copper resistance in A. baumannii, we note that copper homeostasis mechanisms are well-conserved across bacterial species; the open reading frames studied here were originally identified in AB5075 due to their conserved predicted protein sequence. Therefore, we expect that any novel therapeutics that are targeted against these proteins may have broad-spectrum antibacterial activity. Despite the fact that individual proteins are well-conserved, the presence of these proteins or their homologs varies across bacterial species and even amongst strains (Hernandez-Montes et al., 2012). Indeed, we observed that not all the same copper resistance proteins are present in all clinical isolates of A. baumannii. Thus, these differences will be an important consideration when choosing which copper resistance genes may be the best therapeutic targets. Given all of these possible targets, copper homeostasis components could provide an attractive starting place for the development of future antimicrobial therapies.
The immune system has harnessed the antimicrobial power of copper for use against pathogens and copper-mediated immune mechanisms are known to play a role in clearance of bacterial pathogens (Sheldon and Skaar, 2019). Because the innate immune response of phagocytic cells is crucial for clearance of A. baumannii infection, we hypothesize that copper resistance contributes to virulence of this pathogen by enhancing immune evasion and survival in vivo. Therefore, we predict that novel therapeutics targeting copper resistance of bacterial pathogens would enhance immune clearance by reducing bacterial survival in the copper-rich phagosomal compartment. The role of copper in the immune system has been demonstrated by prior studies that utilized copper-replete and copper-deficient cell lines and animal models. For example, macrophages that are pre-incubated with copper are able to kill more internalized bacteria than controls (White et al., 2009). Conversely, copper-deficient macrophages or macrophages that are unable to mobilize copper to the phagosomal compartment due to loss of function of ATP7A are less efficient at killing bacterial pathogens (White et al., 2009; Achard et al., 2012; Ladomersky et al., 2017). The importance of copper in immunity has also been demonstrated on an organismal level. Animals with copper-deficient diets are more susceptible to infection, while those with copper-rich diets more efficiently clear infection (Newberne et al., 1968; Wolschendorf et al., 2011; Gardner and Olson, 2018). Furthermore, mice with a myeloid-specific knockout of ATP7A, the copper ATPase that pumps toxic copper ions into the macrophage phagosome, are more susceptible to infection with Salmonella Typhimurium (Ladomersky et al., 2017). Comparable future studies using A. baumannii will help to shed light on whether copper resistance of this organism aids in virulence by enabling evasion of host copper-mediated immune strategies.
In summary, we identified several additional copper resistance proteins that may serve as potential therapeutic targets, including copper transporters PcoB, CopB, and CopD, chaperone CopC, oxidases CueO and PcoA, and regulatory proteins CusR and CusS. Future work using targeted antibodies and small molecules will seek to determine the efficacy of targeting these copper resistance proteins as a successful treatment strategy. Given the relative conservation of these proteins and pathways among bacteria, we believe several are viable therapeutic targets to treat A. baumannii infection as well as other multidrug-resistant bacterial pathogens, for which novel treatments are urgently needed.
Data Availability Statement
The datasets generated for this study are available on request from the corresponding author.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee at the Walter Reed Army Institute of Research.
Author Contributions
CW, SM, DZ, and DM conceived and designed the experiments. CW, HN, YA, RR, AJ, SS, and RA-T performed the experiments. CW and DM wrote the manuscript.
Disclaimer
Material has been reviewed by the Walter Reed Army Institute of Research (WRAIR). There is no objection to its presentation and/or publication. The opinions and assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense, the Uniformed Services University of the Health Sciences, the Department of the Army, or any other agency of the U.S. Government. Research was conducted in an AAALACi-accredited facility in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Cara Olsen for guidance with statistics. We thank Colin Manoil’s group for the construction of the transposon mutant library in AB5075. CW is thankful to members of the Merrell and Zurawski labs for sharing their time and knowledge, especially Faith Blum for her help with the final experiment.
Footnotes
Funding. Research in the Merrell Laboratory was funded by the NIH and DoD. SM was grateful to the NSF (CHE 1708732). DZ lab was funded by the Military Infectious Diseases Research Program and the Defense Health Agency.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00016/full#supplementary-material
References
- Achard M. E., Stafford S. L., Bokil N. J., Chartres J., Bernhardt P. V., Schembri M. A., et al. (2012). Copper redistribution in murine macrophages in response to Salmonella infection. Biochem. J. 444 51–57. 10.1042/BJ20112180 [DOI] [PubMed] [Google Scholar]
- Achard M. E., Tree J. J., Holden J. A., Simpfendorfer K. R., Wijburg O. L., Strugnell R. A., et al. (2010). The multi-copper-ion oxidase CueO of Salmonella enterica Serovar typhimurium is required for systemic virulence. Infect. Immun. 78 2312–2319. 10.1128/IAI.01208-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahire J. J., Hattingh M., Neveling D. P., Dicks L. M. (2016). Copper-containing anti-biofilm nanofiber scaffolds as a wound dressing material. PLoS One 11:e0152755. 10.1371/journal.pone.0152755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alquethamy S. F., Khorvash M., Pederick V. G., Whittall J. J., Paton J. C., Paulsen I. T., et al. (2019). The role of the CopA copper efflux system in Acinetobacter baumannii virulence. Int. J. Mol. Sci. 20:E575. 10.3390/ijms20030575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes L. C., Visca P., Towner K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71 292–301. 10.1111/2049-632X.12125 [DOI] [PubMed] [Google Scholar]
- Arnesano F., Banci L., Bertini I., Mangani S., Thompsett A. R. (2003). A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc. Natl. Acad. Sci. U.S.A. 100 3814–3819. 10.1073/pnas.0636904100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkow G., Gabbay J., Dardik R., Eidelman A. I., Lavie Y., Grunfeld Y., et al. (2010). Molecular mechanisms of enhanced wound healing by copper oxide-impregnated dressings. Wound Repair Regen. 18 266–275. 10.1111/j.1524-475X.2010.00573.x [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2013). Antibiotic Resistance Threats in the United States, 2013. Atlanta, GA: CDC. [Google Scholar]
- Cha J. S., Cooksey D. A. (1993). Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon. Appl. Environ. Microbiol. 59 1671–1674. 10.1128/aem.59.5.1671-1674.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changela A., Chen K., Xue Y., Holschen J., Outten C. E., O’Halloran T. V., et al. (2003). Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301 1383–1387. 10.1126/science.1085950 [DOI] [PubMed] [Google Scholar]
- Chaturvedi K. S., Hung C. S., Giblin D. E., Urushidani S., Austin A. M., Dinauer M. C., et al. (2014). Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic. ACS Chem. Biol. 9 551–561. 10.1021/cb400658k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett D., Schuler S., Glenn S., Andrew P. W., Cavet J. S., Roberts I. S. (2011). The combined actions of the copper-responsive repressor CsoR and copper-metallochaperone CopZ modulate CopA-mediated copper efflux in the intracellular pathogen Listeria monocytogenes. Mol. Microbiol. 81 457–472. 10.1111/j.1365-2958.2011.07705.x [DOI] [PubMed] [Google Scholar]
- Djoko K. Y., Franiek J. A., Edwards J. L., Falsetta M. L., Kidd S. P., Potter A. J., et al. (2012). Phenotypic characterization of a copA mutant of Neisseria gonorrhoeae identifies a link between copper and nitrosative stress. Infect. Immun. 80 1065–1071. 10.1128/IAI.06163-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoko K. Y., Ong C. L., Walker M. J., McEwan A. G. (2015). The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J. Biol. Chem. 290 18954–18961. 10.1074/jbc.R115.647099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Xu X., Yao J., Deng K., Chen S., Shen Z., et al. (2019). Predictors of mortality in patients infected with carbapenem-resistant Acinetobacter baumannii: a systematic review and meta-analysis. Am. J. Infect. Control 47 1140–1145. 10.1016/j.ajic.2019.03.003 [DOI] [PubMed] [Google Scholar]
- Evans D. G., Evans D. J., Jr., Tjoa W. (1977). Hemagglutination of human group a erythrocytes by enterotoxigenic Escherichia coli isolated from adults with diarrhea: correlation with colonization factor. Infect. Immun. 18 330–337. 10.1128/iai.18.2.330-337.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis M. S., Thomas C. J. (1997). Mutants in the CtpA copper transporting P-type ATPase reduce virulence of Listeria monocytogenes. Microb. Pathog. 22 67–78. 10.1006/mpat.1996.0092 [DOI] [PubMed] [Google Scholar]
- Franke S., Grass G., Rensing C., Nies D. H. (2003). Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185 3804–3812. 10.1128/jb.185.13.3804-3812.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., Ramage E., Weiss E. J., Radey M., Hayden H. S., Held K. G., et al. (2015). Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J. Bacteriol. 197 2027–2035. 10.1128/JB.00131-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S. S., Du Q., Wu H. (2016). Streptococcus mutans copper chaperone, CopZ, is critical for biofilm formation and competitiveness. Mol. Oral Microbiol. 31 515–525. 10.1111/omi.12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. P., Olson J. W. (2018). Interaction of copper toxicity and oxidative stress in Campylobacter jejuni. J. Bacteriol. 200:e0208-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammanco A., Cala C., Fasciana T., Dowzicky M. J. (2017). Global assessment of the activity of tigecycline against multidrug-resistant gram-negative pathogens between 2004 and 2014 as part of the tigecycline evaluation and surveillance trial. mSphere 2:e0310-16. 10.1128/mSphere.00310-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M., Raimunda D., Cheng X., Arguello J. M. (2010). Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 78 1246–1258. 10.1111/j.1365-2958.2010.07402.x [DOI] [PubMed] [Google Scholar]
- Grass G., Rensing C. (2001). CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 286 902–908. 10.1006/bbrc.2001.5474 [DOI] [PubMed] [Google Scholar]
- Gudipaty S. A., Larsen A. S., Rensing C., McEvoy M. M. (2012). Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol. Lett. 330 30–37. 10.1111/j.1574-6968.2012.02529.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan K. A., Pederick V. G., Elbourne L. D., Paulsen I. T., Paton J. C., McDevitt C. A., et al. (2017). Zinc stress induces copper depletion in Acinetobacter baumannii. BMC Microbiol. 17:59. 10.1186/s12866-017-0965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Montes G., Arguello J. M., Valderrama B. (2012). Evolution and diversity of periplasmic proteins involved in copper homeostasis in gamma proteobacteria. BMC Microbiol. 12:249. 10.1186/1471-2180-12-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35 322–332. 10.1016/j.ijantimicag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- Hsiao Y. M., Liu Y. F., Lee P. Y., Hsu P. C., Tseng S. Y., Pan Y. C. (2011). Functional characterization of copA gene encoding multicopper oxidase in Xanthomonas campestris pv. campestris. J. Agric. Food Chem. 59 9290–9302. 10.1021/jf2024006 [DOI] [PubMed] [Google Scholar]
- Hu Y. H., Wang H. L., Zhang M., Sun L. (2009). Molecular analysis of the copper-responsive CopRSCD of a pathogenic Pseudomonas fluorescens strain. J. Microbiol. 47 277–286. 10.1007/s12275-008-0278-9 [DOI] [PubMed] [Google Scholar]
- Hunger M., Schmucker R., Kishan V., Hillen W. (1990). Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87 45–51. 10.1016/0378-1119(90)90494-c [DOI] [PubMed] [Google Scholar]
- Jacobs A. C., Thompson M. G., Black C. C., Kessler J. L., Clark L. P., McQueary C. N., et al. (2014a). AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5:e001076-14. 10.1128/mBio.01076-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. C., Thompson M. G., Gebhardt M., Corey B. W., Yildirim S., Shuman H. A., et al. (2014b). Genetic Manipulation of Acinetobacter baumannii. Curr. Protoc. Microbiol. 35 1G–6G. [DOI] [PubMed] [Google Scholar]
- Johnson M. D., Kehl-Fie T. E., Klein R., Kelly J., Burnham C., Mann B., et al. (2015). Role of copper efflux in pneumococcal pathogenesis and resistance to macrophage-mediated immune clearance. Infect. Immun. 83 1684–1694. 10.1128/IAI.03015-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. W., Taylor D. M., Williams D. R., Finney M., Iorwerth A., Webster D., et al. (2001). Using wound fluid analyses to identify trace element requirements for efficient healing. J. Wound Care 10 205–208. 10.12968/jowc.2001.10.6.26084 [DOI] [PubMed] [Google Scholar]
- Kalivoda E. J., Horzempa J., Stella N. A., Sadaf A., Kowalski R. P., Nau G. J., et al. (2011). New vector tools with a hygromycin resistance marker for use with opportunistic pathogens. Mol. Biotechnol. 48 7–14. 10.1007/s12033-010-9342-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Dalton C., Cortez-Cordova J., Schweizer H. P. (2010). Mini-Tn7 vectors as genetic tools for single copy gene cloning in Acinetobacter baumannii. J. Microbiol. Methods 82 296–300. 10.1016/j.mimet.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Ladomersky E., Khan A., Shanbhag V., Cavet J. S., Chan J., Weisman G. A., et al. (2017). Host and pathogen copper-transporting P-Type ATPases function antagonistically during Salmonella infection. Infect. Immun. 85:e0351-17. 10.1128/IAI.00351-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. M., Grass G., Rensing C., Barrett S. R., Yates C. J., Stoyanov J. V., et al. (2002). The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 295 616–620. 10.1016/s0006-291x(02)00726-x [DOI] [PubMed] [Google Scholar]
- Liochev S. I., Fridovich I. (2002). The Haber-weiss cycle – 70 years later: an alternative view. Redox Rep. 7 55–57. 10.1179/135100002125000190 [DOI] [PubMed] [Google Scholar]
- Lob S. H., Hoban D. J., Sahm D. F., Badal R. E. (2016). Regional differences and trends in antimicrobial susceptibility of Acinetobacter baumannii. Int. J. Antimicrob. Agents 47 317–323. 10.1016/j.ijantimicag.2016.01.015 [DOI] [PubMed] [Google Scholar]
- Longo F., Vuotto C., Donelli G. (2014). Biofilm formation in Acinetobacter baumannii. New Microbiol. 37 119–127. [PubMed] [Google Scholar]
- Macomber L., Imlay J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106 8344–8349. 10.1073/pnas.0812808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero K., Sanchez A., Gonzalez L. J., Ledon T., Rodriguez-Ulloa A., Castellanos-Serra L., et al. (2012). Periplasmic proteins encoded by VCA0261-0260 and VC2216 genes together with copA and cueR products are required for copper tolerance but not for virulence in Vibrio cholerae. Microbiology 158 2005–2016. 10.1099/mic.0.059345-0 [DOI] [PubMed] [Google Scholar]
- Milanino R., Marrella M., Gasperini R., Pasqualicchio M., Velo G. (1993). Copper and zinc body levels in inflammation: an overview of the data obtained from animal and human studies. Agents Actions 39 195–209. 10.1007/bf01998974 [DOI] [PubMed] [Google Scholar]
- Munson G. P., Lam D. L., Outten F. W., O’Halloran T. V. (2000). Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182 5864–5871. 10.1128/jb.182.20.5864-5871.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (2011). Guide for the Care and Use of Laboratory Animals. Washington, DC: The National Academies Press. [Google Scholar]
- Newberne P. M., Hunt C. E., Young V. R. (1968). The role of diet and the reticuloendothelial system in the response of rats to Salmonella typhimurium infection. Br. J. Exp. Pathol. 49 448–457. [PMC free article] [PubMed] [Google Scholar]
- Osman D., Waldron K. J., Denton H., Taylor C. M., Grant A. J., Mastroeni P., et al. (2010). Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 285 25259–25268. 10.1074/jbc.M110.145953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten F. W., Huffman D. L., Hale J. A., O’Halloran T. V. (2001). The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276 30670–30677. [DOI] [PubMed] [Google Scholar]
- Outten F. W., Outten C. E., Hale J., O’Halloran T. V. (2000). Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J. Biol. Chem. 275 31024–31029. 10.1074/jbc.m006508200 [DOI] [PubMed] [Google Scholar]
- Padilla-Benavides T., George Thompson A. M., McEvoy M. M., Arguello J. M. (2014). Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. J. Biol. Chem. 289 20492–20501. 10.1074/jbc.M114.577668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. S., Kim T. H., Yun C. W. (2017). Functional characterization of the copper transcription factor AfMac1 from Aspergillus fumigatus. Biochem. J. 474 2365–2378. 10.1042/BCJ20170191 [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Jara S., Monga D., Eliopoulos G. M., Moellering R. C., Jr., Mylonakis E. (2009). Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob. Agents Chemother. 53 2605–2609. 10.1128/AAC.01533-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21 538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S., Rees E. M., Thiele D. J. (2002). The ABCDs of periplasmic copper trafficking. Structure 10 1292–1295. 10.1016/s0969-2126(02)00863-8 [DOI] [PubMed] [Google Scholar]
- Purves J., Thomas J., Riboldi G. P., Zapotoczna M., Tarrant E., Andrew P. W., et al. (2018). A horizontally gene transferred copper resistance locus confers hyper-resistance to antibacterial copper toxicity and enables survival of community acquired methicillin resistant Staphylococcus aureus USA300 in macrophages. Environ. Microbiol. 20 1576–1589. 10.1111/1462-2920.14088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C., Fan B., Sharma R., Mitra B., Rosen B. P. (2000). CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 97 652–656. 10.1073/pnas.97.2.652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado C. D., Sepkowitz K. A., John J. F., Cantey J. R., Attaway H. H., Freeman K. D., et al. (2013). Copper surfaces reduce the rate of healthcare-acquired infections in the intensive care unit. Infect. Control Hosp. Epidemiol. 34 479–486. 10.1086/670207 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schwan W. R., Warrener P., Keunz E., Stover C. K., Folger K. R. (2005). Mutations in the cueA gene encoding a copper homeostasis P-type ATPase reduce the pathogenicity of Pseudomonas aeruginosa in mice. Int. J. Med. Microbiol. 295 237–242. 10.1016/j.ijmm.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Shafeeq S., Yesilkaya H., Kloosterman T. G., Narayanan G., Wandel M., Andrew P. W., et al. (2011). The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81 1255–1270. 10.1111/j.1365-2958.2011.07758.x [DOI] [PubMed] [Google Scholar]
- Sheldon J. R., Skaar E. P. (2019). Metals as phagocyte antimicrobial effectors. Curr. Opin. Immunol. 60 1–9. 10.1016/j.coi.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Festa R. A., Ioerger T. R., Butler-Wu S., Sacchettini J. C., Darwin K. H., et al. (2014). The copper-responsive RicR regulon contributes to Mycobacterium tuberculosis virulence. mBio 5:e0876-13. 10.1128/mBio.00876-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifri C. D., Burke G. H., Enfield K. B. (2016). Reduced health care-associated infections in an acute care community hospital using a combination of self-disinfecting copper-impregnated composite hard surfaces and linens. Am. J. Infect. Control 44 1565–1571. 10.1016/j.ajic.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Subashchandrabose S., Hazen T. H., Brumbaugh A. R., Himpsl S. D., Smith S. N., Ernst R. D., et al. (2014). Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc. Natl. Acad. Sci. U.S.A. 111 18327–18332. 10.1073/pnas.1415959112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton K. A., Dimitrova D., Rather P. N. (2015). Phase-variable control of multiple phenotypes in Acinetobacter baumannii strain AB5075. J. Bacteriol. 197 2593–2599. 10.1128/JB.00188-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. J., Loh J. M., Proft T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7 214–229. 10.1080/21505594.2015.1135289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J. L., Rello J., Marshall J., Silva E., Anzueto A., Martin C. D., et al. (2009). International study of the prevalence and outcomes of infection in intensive care units. JAMA 302 2323–2329. 10.1001/jama.2009.1754 [DOI] [PubMed] [Google Scholar]
- von Dessauer B., Navarrete M. S., Benadof D., Benavente C., Schmidt M. G. (2016). Potential effectiveness of copper surfaces in reducing health care-associated infection rates in a pediatric intensive and intermediate care unit: a nonrandomized controlled trial. Am. J. Infect. Control 44 e133–e139. 10.1016/j.ajic.2016.03.053 [DOI] [PubMed] [Google Scholar]
- Wagner D., Maser J., Lai B., Cai Z., Barry C. E., III, Honer, et al. (2005). Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J. Immunol. 174 1491–1500. 10.4049/jimmunol.174.3.1491 [DOI] [PubMed] [Google Scholar]
- Ward S. K., Abomoelak B., Hoye E. A., Steinberg H., Talaat A. M. (2010). CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 77 1096–1110. 10.1111/j.1365-2958.2010.07273.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes S. L., Caves V., Keevil C. W. (2012). Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ. Microbiol. 14 1730–1743. 10.1111/j.1462-2920.2011.02677.x [DOI] [PubMed] [Google Scholar]
- White C., Lee J., Kambe T., Fritsche K., Petris M. J. (2009). A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 284 33949–33956. 10.1074/jbc.M109.070201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P., Perevitsky A., Lim F. Y., Shadkchan Y., Knox B. P., Landero Figueora J. A., et al. (2017). Aspergillus fumigatus copper export machinery and reactive oxygen intermediate defense counter host copper-mediated oxidative antimicrobial offense. Cell Rep. 19 1008–1021. 10.1016/j.celrep.2017.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. L., Merrell D. S. (2019). Testing metal sensitivity of a baumannii strains: survival in copper-supplemented liquid media and on copper-containing surfaces. Methods Mol. Biol. 1946 51–60. 10.1007/978-1-4939-9118-1_5 [DOI] [PubMed] [Google Scholar]
- Williams C. L., Neu H. M., Gilbreath J. J., Michel S. L., Zurawski D. V., Merrell D. S. (2016). Copper resistance of the emerging pathogen Acinetobacter baumannii. Appl. Environ. Microbiol. 82 6174–6188. 10.1128/aem.01813-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. L., Neu H. M., Michel S. L. J., Merrell D. S. (2019). Measuring intracellular metal concentration via ICP-MS following copper exposure. Methods Mol. Biol. 1946 195–205. 10.1007/978-1-4939-9118-1_19 [DOI] [PubMed] [Google Scholar]
- Wolschendorf F., Ackart D., Shrestha T. B., Hascall-Dove L., Nolan S., Lamichhane G., et al. (2011). Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 108 1621–1626. 10.1073/pnas.1009261108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D., Nielsen T. B., Bonomo R. A., Pantapalangkoor P., Luna B., Spellberg B. (2017). Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 30 409–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: WHO. [Google Scholar]
- Yamamoto K., Ishihama A. (2005). Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56 215–227. 10.1111/j.1365-2958.2005.04532.x [DOI] [PubMed] [Google Scholar]
- Yan Q., Wang N. (2011). The ColR/ColS two-component system plays multiple roles in the pathogenicity of the citrus canker pathogen Xanthomonas citri subsp. citri. J. Bacteriol. 193 1590–1599. 10.1128/JB.01415-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapotoczna M., Riboldi G. P., Moustafa A. M., Dickson E., Narechania A., Morrissey J. A., et al. (2018). Mobile-genetic-element-encoded hypertolerance to copper protects Staphylococcus aureus from killing by host phagocytes. mBio 9:e00550-18. 10.1128/mBio.00550-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. X., Rainey P. B. (2007). The role of a P1-type ATPase from Pseudomonas fluorescens SBW25 in copper homeostasis and plant colonization. Mol. Plant Microb. Interact. 20 581–588. 10.1094/mpmi-20-5-0581 [DOI] [PubMed] [Google Scholar]
- Zurawski D. V., Black C. C., Alamneh Y. A., Biggemann L., Banerjee J., Thompson M. G., et al. (2019). A porcine wound model of Acinetobacter baumannii infection. Adv. Wound Care 8 14–27. 10.1089/wound.2018.0786 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request from the corresponding author.