Abstract
Mutations in non-coding DNA regions are increasingly recognized as cancer drivers. These mutations can modify gene expression in cis or by inducing high-order chormatin structure modifications with long-range effects. Previous analysis reported the detection of recurrent and functional non-coding DNA mutations in the chronic lymphocytic leukemia (CLL) genome, such as those in the 3′ untranslated region of NOTCH1 and in the PAX5 super-enhancer. In this report, we used whole genome sequencing data produced by the International Cancer Genome Consortium in order to analyze regions with previously reported regulatory activity. This approach enabled the identification of numerous recurrently mutated regions that were frequently positioned in the proximity of genes involved in immune and oncogenic pathways. By correlating these mutations with expression of their nearest genes, we detected significant transcriptional changes in genes such as PHF2 and S1PR2. More research is needed to clarify the function of these mutations in CLL, particularly those found in intergenic regions.
Subject terms: Molecular medicine, Chronic lymphocytic leukaemia
Introduction
A major part of mutations in the cancer genome occur in non-coding DNA regions, and their function is still beginning to be understood1. Non-coding DNA comprises approximately 98% of the human genome, but recent research has proven that most of these regions are either part of regulatory motifs or actively transcribed to RNA2,3. These mutations can induce functional genomic changes by altering the binding of transcription factors or by inducing high-order chromatin structural modifications2,4. For example, mutations in 5′ and 3′ untranslated regions (UTRs) may disturb RNA structural conformation, modify microRNA binding sites or disrupt polyadenylation signals2. In a similar fashion, mutations affecting non-protein coding genes such as microRNA and long intergenic RNA genes (lincRNAs) are known cancer driver events2,5. Different studies have evidenced that the expression of genes such as BRCA1, CDH10, CCND1, MALAT1, PAX5, RB1, SDHD, TERT, TOX3, and TAL1 is influenced by non-coding DNA mutations in regulatory regions of the cancer genome1,6,7. The Pancancer Analysis of Whole Genomes (PCAWG) project has revealed the existence of common and tumor-specific recurrently mutated functional elements near known cancer drivers7. Some of these driver mutations can induce long-range changes in genome organization and trigger abnormal expression of distant oncogenes and tumor suppressors8. Furthermore, the sequence distribution of these driver mutations is not random. Hornshøj et al. (2018) identified a significant enrichment in conserved CCCT-binding factor (CTCF) binding sites among recurrently mutated non-coding DNA regions with cancer specificity6. Similarly, Line et al. (2019) identified 21 recurrently altered CTCF-rich insulator regions in the cancer genome, and elegantly demonstrated that some of these mutations drive tumor proliferation9.
Chronic Lymphocytic Leukemia (CLL) is among the most frequent lymphoproliferative disorders, and it is characterized by its remarkable clinical heterogeneity. Recent efforts by Puente et al.10 enabled the discovery of 24 recurrently mutated non-coding genomic regions in the CLL genome, some of which are associated with functional changes such as mutations in the 3′UTR of NOTCH1 and in the PAX5 super-enhancer. Nevertheless, both the sparsity of annotations in non-coding DNA regions and the difficult functional classification of non-coding DNA mutations hinder a better understanding of the non-coding cancer genome, which probably harbors multiple deregulated elements yet to discover. In this analysis, we analyzed whole genome sequencing (WGS) data using a best-practice mutation detection pipeline. Then, we identified signals of positive selection of mutations in regulatory regions. Finally, our last attempt was to analyze if any of these recurrent mutations in non-coding DNA regions were associated with abnormal expression of the nearest gene. Our results point toward the existence of dozens of mutation-enriched regulatory regions near cancer and immune-related genes, some of which influence local gene expression.
Methods
Data origin
Whole genome sequencing files produced by the International Cancer Genome Consortium11 were obtained from the European Genome-Phenome Archive under accession code EGAD00001001466. Gene expression from microarray data of the same set of patients was obtained from EGAD00010000875.
Data analysis
130 tumor-normal matched CLL whole genomes were processed using the bcbio-nextgen pipeline, which provides best practices for analyzing high throughput sequencing data12. Low complexity regions, areas with abnormally high coverage, sequences with single nucleotide stretches >50 bp and loci with alternative or unplaced contigs in the reference genome were not analyzed. Some polymorphic regions are prone to be classified as highly mutated due to artifacts or biases in the sequencing process, and suspicious elements were manually removed from downstream analysis. Single nucleotide and indel mutation detection was performed with vardict13, varscan14, mutect215 and freebayes16 using default bcbio-nextgen parameters. Only variants with a minimum sequencing depth (DP) of 10 and a genotype quality (GQ) above 20 Phred in both tumor and normal samples were analyzed. A mutation was reported when detected by at least two different mutation callers. Mutations were annotated to the 1000G17, gnomAD18 and ExAc19 databases in order to filter likely germline variants. All mutations with a minimum allele frequency >0.001 in any population were discarded from the analysis.
Region annotation
Annotations corresponding to promoter regions, 5′UTR, 3′UTR and lincRNAs were retrieved from Genecode version 1820. DNAse hypersensitivity (DHS) regions and Transcription Factor Binding Sites (TFBS) tracks from the ENCODE21 project were obtained from Lochovsky et al.22. Similarly, we used enhancer regions from the GeneHancer database23, and analyzed those that were supported by two or more sources of evidence (“elite” enhancers). Regulatory regions within telomeric and centromeric positions were discarded.
Two different methods were used to identify areas with evidence of positive selection of mutations: LARVA22 and OncodriveFML24. LARVA models the mutation counts of each target region as a β-binomial distribution in order to handle overdispersion. Furthermore, LARVA also includes replication timing information in order to estimate local mutation rate, and provides a β-binomial distribution adjusted for replication timing which is used to compute p-values. On the other hand, OncodriveFML is designed to analyze the pattern of somatic mutations across tumors in both coding and non-coding genomic regions. OncodriveFML uses functional predictions in order to identify signals of positive selection. OncodriveFML was run with CADD v1.3 scores and default parameters. TFBS tracks were not analyzed with OncodriveFML due to high computational demands. Regions were labeled as significantly mutated if the q-value was <0.05 with any of the two methods.
Gene expression analysis and association with recurrent non-coding DNA mutations
Background correction, normalization and log2-transformation of microarray gene expression data was performed with the RMA algorithm25. In the case of genes targeted by multiple probes, the median expression was calculated. The Wilcoxon-Rank sum test was used to detect changes in gene expression between mutated and wild-type cases. Non-coding regulatory genomic regions cannot be directly ascribed to any gene, and they can affect the transcription of virtually any part of the genome. However, this study is underpowered to detect long-range interactions due to small sample size and the need of extreme p-values passing multiple-testing correction. Therefore, we centered our efforts on changes in expression of the nearest gene. We annotated the closest gene to each recurrently mutated non-coding genomic region as the nearest transcription start site to the middle position of the corresponding region. In the case of multiple overlapping regulatory regions, we selected the most significant one for downstream analysis. P-values were adjusted for multiple testing using the FDR method, with a significance threshold of 0.05.
Results
Mutation distribution
397,433 non-coding DNA mutations were detected in the genome of this CLL cohort. Most of these were either intergenic (45.46%) or intronic (42.12%). The remaining mutations were located in 5′ flanks (5.83%), 3′ flanks (5.30%), RNA genes (0.64%), 3′UTRs (0.52%) and 5′UTRS (0.13%). Most of the mutations were single nucleotide variants (92.96%), whereas 4.57% and 2.47% were short deletions and insertions, respectively.
Regions significantly enriched in mutations
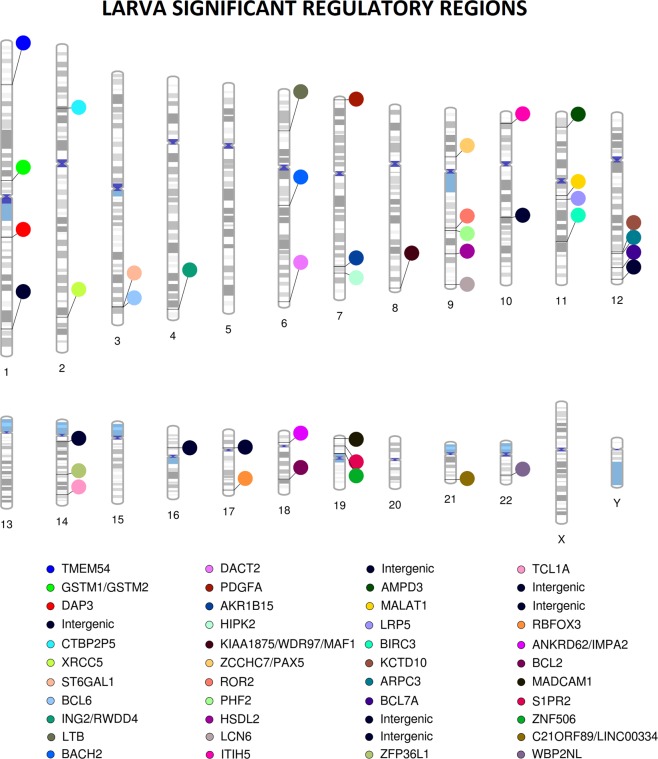
LARVA detected significant mutation enrichments (q-value < 0.05) in 120 TFBS, 16 DHS regions, 10 enhancers, 4 promoters, 2 5′UTRs and 1 lincRNA (Table 1, Supplementary Tables 1–6). No relevant inflation in p-value distribution was observed. (Supplementary Fig. 1). These regions were located in 44 different genomic loci (Fig. 1). The most recurrently mutated promoters were those of TCL1A (q-value 3.32 × 10−4), LCN6 (q-value 4.17 × 10−3), ZFP36L1 (q-value 3.25 × 10−2) and WDR97 (q-value 0.04); and the most significantly mutated enhancers were GH01J229147 (intergenic region chr1:229283343–229284982, q-value 5.79 × 10−6) and GH07J000467 (PDGFA gene, q-value 8.53 × 10−4). The DHS regions chr4:184474905–184475055 (ING2/RWDD4 locus, q-value 1.42 × 10−5), chr21:46673965–46674115 (C21ORF89/LINC00334 locus, q-value 1.38 × 10−4), chr14:96179960–96180110 (TCL1A locus, q-value 3.98 × 10−4) and chr9:115161245–115161395 (HSDL2 locus, q-value 3.98 × 10−4) were the most recurrently mutated among their class (Supplementary Table 2). Furthermore, up to 120 significantly mutated TFBS regions were detected, affecting 19 different genes and 3 intergenic regions. The most recurrently mutated regions were located in chr1:155666495–155666977 (DAP3 gene, q-value 3.14 × 10−10), chr14:96179816–96180607 (TCL1A gene, 1.38 × 10−4), chr3:186782686–186783907 (BCL6 gene, 3.52 × 10−4), chr7:507220–508145 (PDGFA gene, 8.15 × 10−4) and chr18:12086057–12086469 (ANKRD62 gene, 8.30 × 10−4) (Supplementary Table 4).
Table 1.
Summary of the regions most significantly enriched in mutations according to LARVA.
| Chromosome | Start | Stop | Mutation count | p-value (bbd) | FDR | Gene | SHM target | Type of Regulator |
|---|---|---|---|---|---|---|---|---|
| chr1 | 155666495 | 155666977 | 16 | 2.20E-16 | 3.14E-10 | DAP3 | No | TFBS |
| chr1 | 229283343 | 229284982 | 28 | 1.58E-10 | 5.79E-06 | Intergenic | No | ENHANCER |
| chr3 | 186782686 | 186783907 | 26 | 1.34E-09 | 3.52e-04 | BCL6 | Yes | TFBS |
| chr4 | 184474905 | 184475055 | 13 | 3.92E-11 | 1.42E-05 | ING2/RWDD4 | No | DHS |
| chr7 | 507064 | 509696 | 17 | 4.65E-08 | 8.53e-4 | PDGFA | No | ENHANCER |
| chr7 | 507220 | 508145 | 17 | 3.85E-09 | 8.15E-4 | PDGFA | No | TFBS |
| chr9 | 115161245 | 115161395 | 11 | 2.98E-09 | 3.98e-4 | HSDL2 | No | DHS |
| chr11 | 65265233 | 65273940 | 10 | 2.04E-08 | 4.50e-4 | MALAT1 | Yes | lincRNA |
| chr14 | 96179060 | 96180273 | 25 | 2.34E-08 | 3.32E-4 | TCL1A | Yes | PROMOTER |
| chr14 | 96179721 | 96180690 | 22 | 1.36E-09 | 3.52E-4 | TCL1A | Yes | TFBS |
| chr14 | 96179799 | 96180653 | 21 | 6.70E-10 | 2.25E-4 | TCL1A | Yes | TFBS |
| chr14 | 96179816 | 96180607 | 21 | 2.67E-10 | 1.38E-4 | TCL1A | Yes | TFBS |
| chr14 | 96179960 | 96180110 | 12 | 3.03E-09 | 3.98E-4 | TCL1A | Yes | DHS |
| chr21 | 46673965 | 46674115 | 12 | 7.33E-10 | 1.38E-4 | C21ORF89/LINC00334 | No | DHS |
Figure 1.
Chromosomal ideogram representing the different gene affected by recurrent non-coding mutations according to LARVA.
Other significant enhancer regions were located in the proximity of genes involved in apoptosis (BCL2 and BIRC3), cell cycle control (WBP2NL), cytoskeleton and extracellular matrix formation (ARPC3 and ITIH5), gene expression regulation and chromatin remodelling (BCL7A, PAX5 and PHF2), genome integrity (XRCC5 and ZNF506), gene expression regulation (MALAT1 and RBFOX3), intracellular signalling (DACT2, HIPK2, IMPA2, KCTD10, ROR2 and S1PR2), immune pathways (BACH2, LTB and MADCAM1) and metabolism (AKR1B15, AMPD3, GSTM1/GSTM2, LRP5 and ST6GAL1) (Supplementary Tables 1–6). Recurrent mutations were also found near less well-characterized genes such as TMEM54 and CTBP2P5, as well as within intergenic regions such chr14:26068671–26069217 and chr1:229283491–229285693.
Finally, OncodriveFML identified 4 regions significantly enriched in likely functional mutations (Supplementary Tables 7 and 8). No relevant inflation in p-value distribution was observed (Supplementary Fig. 2). These regions were the enhancer GH14J089855 (q-value 2.54 × 10−3) encoded within an intronic region of EFCAB11, two DHS regions in the proximity of EGR and WBNPL2 (q-values 0.01 and 0.03, respectively), and one intergenic DHS region located in chr8:127155560–127155710 (q-value 1.22 × 10−3).
Mutations associated with changes in gene expression
We studied the association of regions enriched in mutations with changes in the expression of their respective nearest genes. Although this type of analysis is limited by low sample size, we detected significant associations in some cases. We tested if patients with at least one mutation in these regulatory regions were accompanied by changes in expression of the nearest gene. Significant associations were observed in 3 genes, namely PHF2 (q-value 0.02, 95% CI [−0.295, −0.048]), RPL39L (q-value 0.04, 95% CI [0.018, 0.217]) and S1PR2 (q-value 0.03, 95% CI [0.033, 0.38]) (Supplementary Table 9).
Discussion
Mutations in the non-coding part of the genome constitute the “dark-matter” of cancer genomics2. Growing evidence indicates that many of these mutations occur in conserved motifs and loci under epigenetic control, and some of these play fundamental roles in cancer biology and disease prognosis1–3,6–9. Using WGS data produced by the ICGC, we identified dozens of recurrently mutated regulatory regions in the CLL genome. Among these, 10 were previously reported by the original analysis performed by Puente et al.10, namely those near BACH2, BLC2, BCL6, BCL7A, BIRC3, S1PR2, PCDH15, ZCCHC7/PAX5 and ZFP36L1. Numerous novel regions were also enriched in non-coding DNA mutations, including transcription factor binding sites, DNAse hypersensitivity regions, 5′UTR regions, promoters, enhancers and non-coding RNAs. These events were frequently found in the vicinity of genes previously vinculated with oncogenic pathways. Indeed, the most significantly mutated regions were a SETB1 binding site within the first intron of DAP3, a GTP-binding protein that participates in the apoptosis pathway26; and a DNAse hypersensitivity region downstream to ING2, a well-characterized tumor suppressor27. Other highly mutated regulatory regions affected cancer-related genes such as DACT228, ERG29,, HIPK230, ITIH531, LRP532, MAF133, MALAT134, PHF235, PDGFA36, RBFOX337, ROR238, ST6GAL139 and XRCC540; and others were detected near genes involved in immunity, such as LTB41 and MADCAM142. Overall, only three of the novel genes (LTB, MALAT1 and ST6GAL1) were previously defined as targets of somatic hypermutation in B cell lymphomas43. Finally, it is worthwhile to mention that recurrent and even highly significant enrichments were detected around barely characterized genes (e.g. C21ORF89/LINC0334) and intergenic regions.
The reported mutations can either be bystander or have functional implications related to their potential to modify gene expression or to induce high-order chromatin structural changes. Although limited by low sample size, we devised significant changes in the expression of PHF2, S1PR2 and RPL39L. These three genes are involved in the regulation of important oncogenic processes. PHF2 encodes a histone demethylase with tumor suppressor activity35. S1PR2 participates in the TGF-β pathway and acts as a tumor suppressor of B cell lymphomas44. Finally, RPL39L45 is involved in cancer stem cell self-renewal and hypoxia response. These results are concordant with other reports of non-coding regulatory mutations driving gene expression changes in B-cell lymphomas46–48.
The combination of an optimized mutation detection pipeline with statistical tests specifically designed to handle non-coding DNA mutations has enabled the detection of novel putative regulatory driver regions in the CLL genome. These regions were mostly located in the vicinity of genes implicated in oncogenic and immune pathways, although several recurrently mutated intergenic regions were detected too. Furthermore, we could confirm the association of some of these events with altered expression of their respective genes. We expect that our results, along with those published by other groups, will promote an improved characterization of the non-coding mutational drivers of CLL.
Supplementary information
Acknowledgements
We would like to thank the International Cancer Genome Consortium for facilitating the data, and to the Supercomputing Center of Galicia (CESGA) for providing informatics support for the analysis. The content of this paper is part of the doctoral thesis of Adrián Mosquera Orgueira to obtain a PhD at the Department of Medicine, University of Santiago de Compostela.
Author contributions
A.M.O. designed the research and performed the analysis. A.M.O., B.A.R. and J.L.B.L. analyzed the results and wrote the paper. J.A.D.A., N.D.V., N.A.V. and M.S.G.P. critically evaluated the paper.
Data availability
There is not data to deposit.
Competing interests
The article processing fee of this paper has been partially funded by Roche Pharmaceuticals. Notwithstandingly, this company did not have any influence on the study design, data analysis, results interpretation or article writing.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-59243-5.
References
- 1.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diederichs S, et al. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non‐coding RNA and synonymous mutations. EMBO Mol. Med. 2016;8:442–457. doi: 10.15252/emmm.201506055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat. Rev. Genet. 2010;11:559–571. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- 4.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Sci. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palamarchuk A, et al. 13q14 deletions in CLL involve cooperating tumor suppressors. Blood. 2010;115:3916–3922. doi: 10.1182/blood-2009-10-249367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornshøj H, et al. Pan-cancer screen for mutations in non-coding elements with conservation and cancer specificity reveals correlations with expression and survival. NPJ Genom. Med. 2018;3:1. doi: 10.1038/s41525-017-0040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rheinbay, E. et al. Discovery and characterization of coding and non-coding driver mutations in more than 2,500 whole cancer genomes. bioRxiv 237313, 10.1101/237313.
- 8.Wadi, L. et al. Candidate cancer driver mutations in superenhancers and long-range chromatin interaction networks. bioRxiv 236802, 10.1101/236802
- 9.Liu EM, et al. Identification of Cancer Drivers at CTCF Insulators in 1,962 Whole Genomes. Cell Syst. 2019;8:446–455. doi: 10.1016/j.cels.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nat. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 11.International Cancer GC, et al. International network of cancer genome projects. Nat. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valls-Guimera R. Bcbio-nextgen: Automated, distributed, next-gen sequencing pipeline. EMBnet J. 2012;17:30. doi: 10.14806/ej.17.B.286. [DOI] [Google Scholar]
- 13.Lai Z, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.do Valle IF, et al. Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data. BMC Bioinforma. 2016;17:341. doi: 10.1186/s12859-016-1190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv, 1207.3907 [q-bio.GN] (2012)
- 17.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature526 68–74, 10.1038/nature15393 (2015). [DOI] [PMC free article] [PubMed]
- 18.Karczewski, K.J. et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 531210, 10.1101/531210.
- 19.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nat. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis CA, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:794–801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lochovsky L, Zhang J, Fu Y, Khurana E, Gerstein M. LARVA: an integrative framework for large-scale analysis of recurrent variants in noncoding annotations. Nucleic Acids Res. 2015;43:8123–8134. doi: 10.1093/nar/gkv803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fishilevich, S. et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford), 10.1093/database/bax028 (2017). [DOI] [PMC free article] [PubMed]
- 24.Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016;17:128. doi: 10.1186/s13059-016-0994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2013;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Wazir U, et al. The role of death-associated protein 3 in apoptosis, anoikis and human cancer. Cancer Cell Int. 2015;15:39. doi: 10.1186/s12935-015-0187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guérillon C, Larrieu D, Pedeux R. ING1 and ING2: multifaceted tumor suppressor genes. Cell Mol. Life Sci. 2013;70:3753–3772. doi: 10.1007/s00018-013-1270-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, et al. Methylation of DACT2 promotes breast cancer development by activating Wnt signaling. Sci. Rep. 2017;7:3325. doi: 10.1038/s41598-017-03647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adamo P, Ladomery MR. The oncogene ERG: a key factor in prostate cancer. Oncogene. 2016;35:403–414. doi: 10.1038/onc.2015.109. [DOI] [PubMed] [Google Scholar]
- 30.D’Orazi G, Rinaldo C, Soddu S. Updates on HIPK2: a resourceful oncosuppressor for clearing cancer. J. Exp. Clin. Cancer Res. 2012;31:63. doi: 10.1186/1756-9966-31-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose M, et al. ITIH5 induces a shift in TGF-β superfamily signaling involving Endoglin and reduces risk for breast cancer metastasis and tumor death. Mol. Carcinog. 2018;57:167–181. doi: 10.1002/mc.22742. [DOI] [PubMed] [Google Scholar]
- 32.Ren DN, et al. LRP5/6 directly bind to Frizzled and prevent Frizzled-regulated tumour metastasis. Nat. Commun. 2015;6:6906. doi: 10.1038/ncomms7906. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, et al. MAF1 suppresses AKT-mTOR signaling and liver cancer through activation of PTEN transcription. Hepatology. 2016;63:1928–1942. doi: 10.1002/hep.28507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Peng WX, Mo YY, Luo D. MALAT1-mediated tumorigenesis. Front. Biosci. 2017;22:66–80. doi: 10.2741/4572. [DOI] [PubMed] [Google Scholar]
- 35.Lee KH, et al. PHF2 histone demethylase acts as a tumor suppressor in association with p53 in cancer. Oncogene. 2015;34:2897–2909. doi: 10.1038/onc.2014.219. [DOI] [PubMed] [Google Scholar]
- 36.Palomero J, et al. SOX11 promotes tumor angiogenesis through transcriptional regulation of PDGFA in mantle cell lymphoma. Blood. 2014;124:2235–2247. doi: 10.1182/blood-2014-04-569566. [DOI] [PubMed] [Google Scholar]
- 37.Liu T, et al. RBFOX3 Promotes Tumor Growth and Progression via hTERT Signaling and Predicts a Poor Prognosis in Hepatocellular Carcinoma. Theranostics. 2017;7:3138–3154. doi: 10.7150/thno.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debebe Z, Rathmell WK. Ror2 as a therapeutic target in cancer. Pharmacol. Ther. 2015;150:143–148. doi: 10.1016/j.pharmthera.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antony P, et al. Epigenetic inactivation of ST6GAL1 in human bladder cancer. BMC Cancer. 2014;14:901. doi: 10.1186/1471-2407-14-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, et al. XRCC5 cooperates with p300 to promote cyclooxygenase-2 expression and tumor growth in colon cancers. PLoS One. 2017;12:e0186900. doi: 10.1371/journal.pone.0186900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagy B, et al. Lymphotoxin beta expression is high in chronic lymphocytic leukemia but low in small lymphocytic lymphoma: a quantitative real-time reverse transcriptase polymerase chain reaction analysis. Haematologica. 2003;88:654–658. [PubMed] [Google Scholar]
- 42.Sakai Y, Kobayashi M. Lymphocyte ‘homing’ and chronic inflammation. Pathol. Int. 2015;65:344–354. doi: 10.1111/pin.12294. [DOI] [PubMed] [Google Scholar]
- 43.Khodabakhshi AH, et al. Recurrent targets of aberrant somatic hypermutation in lymphoma. Oncotarget. 2012;3:1308–1319. doi: 10.18632/oncotarget.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stelling A, et al. The tumor suppressive TGF-β/SMAD1/S1PR2 signaling axis is recurrently inactivated in diffuse large B-cell lymphoma. Blood. 2018;131:2235–2246. doi: 10.1182/blood-2017-10-810630. [DOI] [PubMed] [Google Scholar]
- 45.Dave B., Granados-Principal S., Zhu R., Benz S., Rabizadeh S., Soon-Shiong P., Yu K.-D., Shao Z., Li X., Gilcrease M., Lai Z., Chen Y., Huang T. H.- M., Shen H., Liu X., Ferrari M., Zhan M., Wong S. T. C., Kumaraswami M., Mittal V., Chen X., Gross S. S., Chang J. C. Targeting RPL39 and MLF2 reduces tumor initiation and metastasis in breast cancer by inhibiting nitric oxide synthase signaling. Proceedings of the National Academy of Sciences. 2014;111(24):8838–8843. doi: 10.1073/pnas.1320769111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Batmanov K, Wang W, Bjørås M, Delabie J, Wang J. Integrative whole-genome sequence analysis reveals roles of regulatory mutations in BCL6 and BCL2 in follicular lymphoma. Sci. Rep. 2017;7:7040. doi: 10.1038/s41598-017-07226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arthur SE, et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat. Commun. 2018;9:4001. doi: 10.1038/s41467-018-06354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathelier A, et al. Cis-regulatory somatic mutations and gene-expression alteration in B-cell lymphomas. Genome Biol. 2015;23(16):84. doi: 10.1186/s13059-015-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There is not data to deposit.