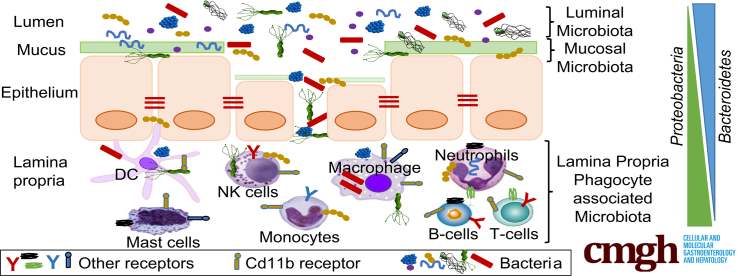
Abstract
Background & Aims
The interaction between intestinal microbiota and the immune system plays a vital role in inflammatory bowel disease (IBD). Although numerous deep-sequencing studies have suggested dysbiosis in IBD, identifying specific bacteria from the stool or mucosa that are responsible for disease susceptibility or severity has remained a challenge. Lamina propria phagocytes ideally are localized to interact with bacteria that are in close proximity to, or have invaded, the tissue. Thus, we examined the microbial populations associated with the lamina propria phagocytes in 20 Crohn’s disease and 12 ulcerative colitis patients. Specifically, we aimed to address whether the phagocyte-associated microbiota differed from the mucosa-associated microbiota and whether this varied based on IBD type or the state of inflammation.
Methods
16S ribosomal RNA gene sequencing and innate immune gene expression profiling was done on CD11b+ lamina propria phagocytes isolated from the biopsies obtained from IBD patients.
Results
Phagocyte-associated microbiota was enriched in bacterial species belonging to phylum Proteobacteria, whereas species belonging to phylum Bacteroidetes were enriched in the mucosal microbiota of IBD patients. Disease type was the most influential factor in driving differences in the microbiota of both the mucosa and the lamina propria phagocytes, irrespective of inflammation state o`r anatomic location. Crohn’s disease and ulcerative colitis specimens showed similar patterns of increased inflammatory gene expression in phagocytes isolated from inflamed areas compared with those isolated from uninflamed regions.
Conclusions
This pilot study shows the feasibility of using lamina propria phagocytes to characterize the microbiota in IBD patients. The approach used in this study can narrow the spectrum of potentially dysbiotic bacterial populations and clinically relevant gene expression signatures in IBD patients.
Keywords: Mucosa, Microbiota, Nanostring, Crohn’s Disease, Ulcerative Colitis
Abbreviations used in this paper: CD, Crohn’s disease; IBD, inflammatory bowel disease; LEfSe, linear discriminant analysis effect size; OSM, oncostatin M; OTU, operational taxonomic unit; PCoA, principle coordinate analysis; rRNA, ribosomal RNA; sPLS-DA, sparse partial least squares–discriminant analysis; UC, ulcerative colitis
Graphical abstract
Summary.
Microbiota associated with intestinal lamina propria phagocytes is distinct from mucosal microbiota and is enriched in Proteobacteria. The lamina propria phagocyte microbiota differs between Crohn’s disease and ulcerative colitis patients whereas inflammatory gene expression does not.
An imbalance between the host and its microbiota can activate immune pathways and play a vital role in the initiation and/or progression of inflammatory bowel diseases (IBDs). In the past decade, alterations in the microbial composition of the gut referred to as microbial dysbiosis have been reported for ulcerative colitis (UC) and Crohn’s disease (CD), the 2 main types of IBD. Decreased bacterial diversity, increased abundance of the phyla Proteobacteria and Actinobacteria, decreases in the phylum Firmicutes, and increases in the abundance of mucosa-associated bacteria have been reported consistently in IBD patients compared with healthy controls and these features are shared between UC and CD.1, 2, 3, 4, 5, 6 However, microbial changes specifically ascribed to either CD or UC have not been clearly addressed. Indeed, these 2 types of IBD share most susceptibility genes despite different phenotypic presentations, suggesting that genes alone do not account for phenotype.7,8 In addition, involvement of certain bacterial taxa such as the phylum Bacteroidetes remains inconclusive in the setting of IBD.
Despite the volume of IBD microbiome studies, identification of specific situational pathogens in IBD has remained a challenge largely owing to high interindividual variation and inconsistencies among published studies. The majority of the previous IBD microbiome studies characterized either stool or biopsy specimens taken from different locations in the gastrointestinal tract.9 Although both stool and mucosa are microbially diverse, the microbiome of stool is fundamentally very different from the mucosal microbiome,2,10 and a recent study with pediatric IBD patients showed that stool samples poorly reflect the dysbiosis associated with IBD.11 On the other hand, characterization of mucosa-associated bacteria from biopsy specimens involves inclusion of bacteria from the mucus layer, epithelium, lamina propria, and any remaining adherent stool; and the depth of the biopsy can influence the ratio of the contribution of each of these factors. Bowel preparation also has been shown to affect microbial composition and diversity,12 and may be a confounding factor in most microbiota studies. To overcome these limitations, we performed this study to examine the microbiota of an important cellular compartment of the intestine with the aim that it would remove several of the aforementioned confounding variables.
IBD patients show impaired intestinal epithelial barrier function, allowing passage of toxins, pathogens, commensals, dietary food components and other small molecules from the lumen into the deeper layers of the intestine. Continuous invasion of the subepithelial lamina propria of the intestine by any of the earlier-described factors results in recruitment and activation of immune cells, leading to inflammatory processes. Neutrophils and mononuclear phagocytes, including macrophages and dendritic cells, located in the lamina propria of the intestine, act as first responders against these intruders. The innate immune cells protect the host through phagocytosis, bacterial killing, and by activating the adaptive immune system. Genome-wide association studies in IBD patients have identified several innate immune genes involved in bacterial recognition and clearance that contribute to IBD pathogenesis, such as NOD2, ATG5, and CARD9.13,14 Lamina propria phagocytes express many of these genes,10,11 highlighting the pivotal role that these cells play in the development of IBD.
In this study, we examined the microbiota associated with lamina propria phagocytes isolated from the biopsy specimens of CD and UC patients. We hypothesized that bacteria translocating into the lamina propria and interacting with phagocytic cells may be a significant driver of tissue inflammation, and that this population may be distinct from that found at the mucosal surface. We were able to sensitively detect the phagocyte-associated microbiota and compare its similarity with the mucosa-associated microbiota.1,9,15 We found that, compared with mucosa-associated microbiota, the phagocyte-associated microbiota of IBD patients represents a distinct microbial population in terms of composition, abundance, and predicted functional profile. We also observed that although the phagocyte-associated microbiota from inflamed and noninflamed sites is quite similar, the gene expression patterns from the 2 sites differ widely. Our findings provide an important proof of principle that lamina propria phagocytes provide a distinct view of the disease-associated microbiome.
Results
Lamina Propria Phagocyte-Associated Microbiota Is Distinct From Matched Mucosal Microbiota in IBD Patients
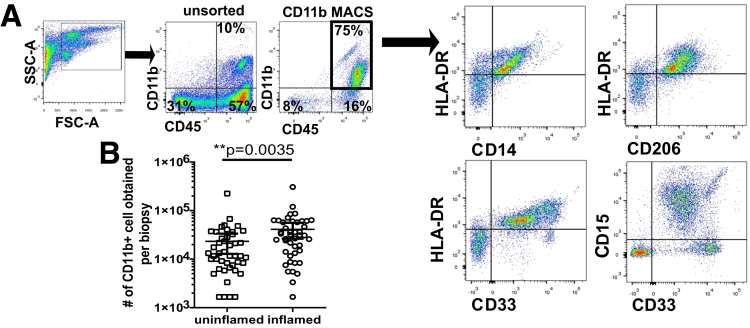
Previous studies have shown differences between stool and mucosa-associated microbiota.16,17 To identify the lamina propria phagocyte-associated microbiota in a thorough and unbiased way, we developed an approach that included isolation of lamina propria cells from mucosal biopsy specimens followed by magnetic bead–based cell sorting of CD11b+ cells. CD11b is a marker of myeloid lineage cells and is expressed on phagocytic cells including neutrophils, macrophages, monocytes, natural killer cells, and dendritic cells.18 By confirmatory flow cytometry, approximately 90% of the sorted cells were CD45+ leukocytes, of which 75% were CD11b+ (Figure 1A). Counting the number of recovered cells showed that inflamed specimens had a significantly higher number of CD11b+ cells compared with biopsy specimens from uninflamed mucosa (Figure 1B). A subset of biopsy specimens was used to estimate the composition of isolated CD11b+ cells. Individual isolations of CD11b+ cells contained varying proportions of cells expressing CD33 (myeloid cells), CD14 (monocytes), HLA-DR and CD206 (dendritic cells and macrophages), and CD15 (granulocytes, including neutrophils and eosinophils).
Figure 1.
Flow cytometric characterization of lamina propria CD11b+ cells isolated from biopsy specimens obtained from IBD patients. (A) The CD11b+ population was examined by determining the expression of CD33 (Siglec-3) (monocytes, macrophages, granulocytes, dendritic cells, and mast cells) and the activation marker CD206 (mannose receptor) (macrophages and dendritic cells). Cells also were assessed for expression of CD14 (part of the lipopolysaccharide-receptor complex) and HLA-DR (antigen presentation) in the 2 subsets. (B) Number (means ± 95% CI) of CD11b+ cells isolated from inflamed/uninflamed patient biopsy specimens. FSC-A, forward scatter area; SSC-A, side scatter area.
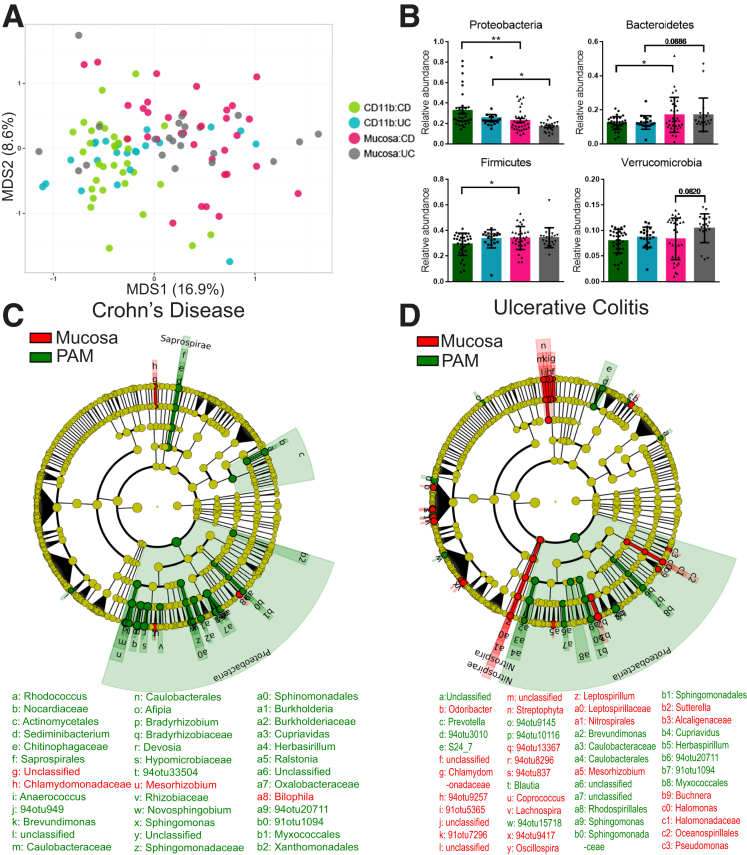
The bacteria associated with isolated CD11b+ cells (referred to as phagocyte-associated microbiota) were identified by 16S ribosomal RNA (rRNA) gene high-throughput sequencing. To compare the microbiota associated with lamina propria phagocytes with the overlying mucosa microbiota, we also analyzed a whole biopsy specimen obtained at the same time from the same location. A total of 22,092,652 paired-end sequences were obtained. After quality filtering, 21,590,854 sequences were retained with an average of 196,280 ± 69,738 sequences per sample. 16S sequencing showed that both CD and UC patients showed considerable interindividual variation in their phagocyte-associated microbiota and mucosal microbiota. Principle coordinate analysis (PCoA) based on weighted Unifrac distances showed that the overall composition and abundance of the microbiota associated with lamina propria phagocytic cells is distinct from the mucosal microbiota (Figure 2A). The PCoA plot also shows the similarity of the phagocyte-associated microbiota in both UC and CD patients by the tight clustering of the CD11b samples.
Figure 2.
Phagocyte-associated microbiota in IBD patients is distinct from the mucosal microbiota. (A) Constrained Analysis of Principal Coordinates (CAP) ordination plot based on Bray–Curtis distance shows separation of mucosal and phagocyte-associated microbiota samples in IBD patients. (B) Relative abundance (means ± SD) of phyla that differed between mucosal and phagocyte-associated microbiota of IBD patients. **P < .01, *P < .05 by 1-way analysis of variance and the Sidak correction for multiple comparisons. (C and D) Cladogram of bacterial taxa after analysis by LEfSe. Yellow represents nonsignificant taxa with linear discriminate analysis (LDA) scores of <2 whereas LDA scores of >2 represent taxa that are significantly enriched in mucosa (red) and phagocyte-associated microbiota (green) of (C) CD and (D) UC patients, respectively. n = 110 obtained from 33 and 22 pairs of matched mucosa and CD11b-positive cells from 20 CD and 12 UC patients, respectively. MDS1 and MDS2, refer to axis 1 and axis 2 of the plot; PAM, phagocyte-associated microbiota.
Strikingly, the phylum Proteobacteria, which is the defining phyla associated with IBD microbiota dysbiosis, was enriched significantly in the phagocyte-associated microbiota of both CD and UC patients when compared with whole mucosa (Figure 2B).1, 2, 3, 4, 5, 6 Furthermore, a decrease in phylum Bacteroidetes was observed in the phagocyte-associated microbiota of both CD and UC patients compared with the mucosal biopsy (Figure 2B). The phylum Firmicutes was observed at a lower abundance only in the phagocyte-associated microbiota of CD patients. Linear discriminant analysis effect size (LEfSe) was used to detect the bacterial taxonomic markers associated with phagocyte-associated microbiota versus mucosa of CD and UC patients. LEfSe analysis resulted in identification of 37 and 67 bacterial features that differed between phagocyte-associated microbiota vs mucosa of CD and UC patients, respectively (Figure 2C and D). In our separate analysis of both CD and UC patients, we focused on microbiota differences between phagocyte-associated microbiota and mucosa only. The biological consistency step of LEfSe ensured that the highly differential bacterial feature in phagocyte-associated microbiota is characteristic of both inflamed and noninflamed sites and is significantly different from that of mucosal samples. In CD patients, operational taxonomic units (OTUs) belonging to genus Bilophila, Bifidobacterium, Paenibacillus, Mezorhizobium, and unclassified Enterobacteraceae were more abundant in the mucosa than phagocytes. Conversely, OTUs belonging to genus Burkholderia, Prevotella, Bifidobacterium, Akkermansia, Stenotrophomonas, Bradyrhizobium, Devosia, Herbaspirillum, Sediminibacterium, Brevundimonas, Ralstonia, Sphingomonas, Cupriavidus, Novosphingobium, Blautia, Afipia, Anaerococcus, and unclassified families of Caulobacteraceae, Oxalobacteraceae, Rikenellaceae, and Clostridiaceae were more abundant in phagocytes than the mucosa in CD patients.
LEfSe analysis of samples from UC patients indicated increased abundance of genera Cupriavidus, Blautia, Akkermansia, Burkholderia, Blautia, Herbaspirillum, Sphingomonas, Eubacterium, Brevundimonas, Prevotella, Paenibacillus, Megasphaera, and unclassified genera from families Rikenellaceae, Enterobacteriaceae, Lachnospiraceae, Clostridiaceae, S24_7, Bifidobacteriaceae, and Ruminococcaceae in the phagocytes. Bacterial OTUs from genus Bacteroides, Paenibacillus, Akkermansia, Prevotella, Mesorhizobium, Bifidobacterium, Streptococcus, Odoribacter, Leptospirillum, Lachnospira, Sutterella, Halomonas, Buchnera, and families Rikenellaceae, Enterobacteriaceae, Lachnospiraceae, Chlamydomonadaceae, and Ruminococcaceae were more abundant in the mucosa of UC patients.
Our data show that it is feasible to perform deep sequencing of an isolated cell subset from the lamina propria. We show that phagocytes have a distinct composition of microbiota compared with the mucosa.
Inferred Metagenomics Reveals Functional Differences Between Lamina Propria Phagocyte and Mucosal Microbiota
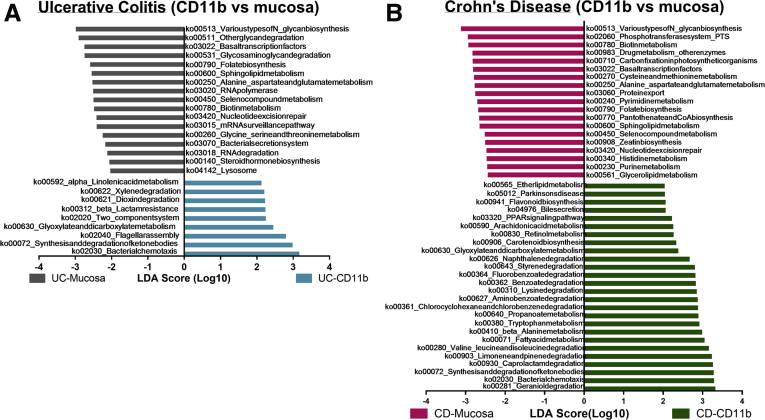
Previous studies have shown that the composition and abundance in microbiota belies similarities in function between bacterial communities across individuals.19 By contrast, a few studies have reported functional metabolic alterations in the microbiota of IBD patients.2,20 To determine if the bacterial abundance differences we observed between the phagocytes and the mucosa resulted in distinct metabolic gene profiles we performed PICRUSt analysis.21 To assess the functional gene profile of the phagocyte-associated microbiota and the matched mucosal biopsy, we used PICRUSt to convert OTU counts from 16S rRNA gene data into functional gene counts. HUMAnN was used to determine the gene pathway abundance in all the samples.22 LEfSe was used to investigate functional differences between mucosa and lamina propria phagocyte-associated microbiota of UC and CD patients (Figure 3A and B). In general, the phagocyte-associated microbiota and mucosa of both CD and UC patients differed in functions associated with metabolism of lipids, xenobiotics, amino acids, carbohydrates, vitamins and cofactors, and genetic information processing. For example, compared with its matched mucosa, the phagocyte-associated microbiota of CD patients was significantly enriched in genes associated with lipid metabolism, metabolism of terpenoids and polyketides (including arachidonic acid metabolism), carbohydrate metabolism, and some amino acid and xenobiotic degradation pathways. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with retinol metabolism, flavonoid biosynthesis, bile secretion, and peroxisome proliferator-activated receptor signaling also were enriched in phagocyte-associated microbiota of CD patients compared with the mucosa. In contrast, we observed a significant decrease in gene pathways associated with genetic information processing (including transcription, translation, replication, and repair) and glycan biosynthesis and metabolism in the phagocyte-associated microbiota of CD patients compared with the mucosa. Genes associated with nucleotide metabolism, amino acid metabolism, metabolism of vitamins and cofactors, and the phosphotransferase system also were decreased in the phagocyte-associated microbiota of CD patients compared with the mucosa.
Figure 3.
LEfSe comparison of predicted metabolic pathways from phagocyte-associated microbiota and mucosa-associated microbiota of IBD patients. Metabolic pathways significantly enriched in phagocyte-associated microbiota are shown in green for CD and in blue for UC. The pathways that are significantly enriched in mucosal microbiota are shown in pink for CD and in grey for UC. Pathways were considered significant with P < .05 and linear discriminate analysis (LDA) score >2. (A) Significantly altered pathways in UC. Data were inferred from 44 samples (22 pairs of matched mucosa and CD11b+ cells) from 12 subjects. (B) Significantly altered pathways in CD. Data were inferred from 66 samples (33 pairs of matched mucosa and CD11b+ cells) from 20 subjects.
When examining UC samples, the mucosal samples were more abundant in bacterial secretion systems, whereas genes for cell motility, including flagellar assembly and bacterial chemotaxis, were more abundant in the phagocyte-associated microbiota. Genes for 2-component system signaling and β-lactamase resistance were significantly enriched in the phagocyte-associated microbiota of UC patients compared with their mucosa. Similar to the phagocyte-associated microbiota of CD patients, the UC-associated phagocyte-associated microbiota also was enriched in genes involved in certain xenobiotic degradation pathways. Similar to the phagocyte-associated microbiota in CD patients, the phagocyte-associated microbiota of UC patients was deficient in genes associated with genetic information processing and glycan biosynthesis and metabolism.
The earlier-described results suggest that the observed microbial differences between phagocyte-associated microbiota and mucosa also contribute to functional and metabolic differences between phagocyte-associated microbiota vs mucosa. These differences may be owing to specialized functions needed to facilitate epithelial translocation to the lamina propria and/or survival in the altered environment of the inflamed tissue.
Phagocyte-Associated Microbiota Is Similar Between Matched Inflamed and Uninflamed Samples From IBD Patients
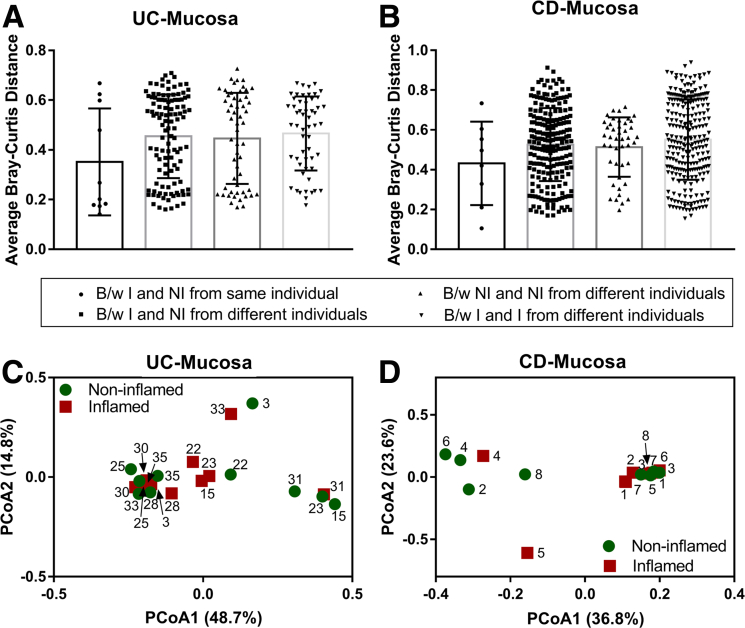
To date, only a few studies have compared the mucosal microbiota differences that occur within a single patient between inflamed and noninflamed mucosal sites,1,4,5,15 and these studies have had inconsistent findings. Here, we compared mucosal microbiota from inflamed mucosa with noninflamed mucosa of UC and CD patients, respectively. Similar to previously published studies,1,9 we found that the mucosal microbiota of inflamed regions is similar to matched noninflamed regions in both CD and UC patients (Figure 4).
Figure 4.
Mucosal microbiota is similar at inflamed and noninflamed sites of IBD patients. Bray–Curtis distance (means ± SD) for each inflamed and noninflamed sample from the same and different individuals are plotted for (A) UC (10 pairs of inflamed and noninflamed samples from 10 subjects) and (B) CD patients (8 pairs of inflamed and noninflamed samples from 8 subjects). PCoA based on Bray–Curtis distances between OTUs detected from matched inflamed and noninflamed mucosa of (C) UC and (D) CD patients as used to generate ordination plots for viewing the relative positioning of inflamed (red squares) and noninflamed (green circles) samples in 2 dimensions. Numbers in parentheses indicate the percentage variation explained by the axis. B/w, between; I, inflamed; NI, non-inflamed.
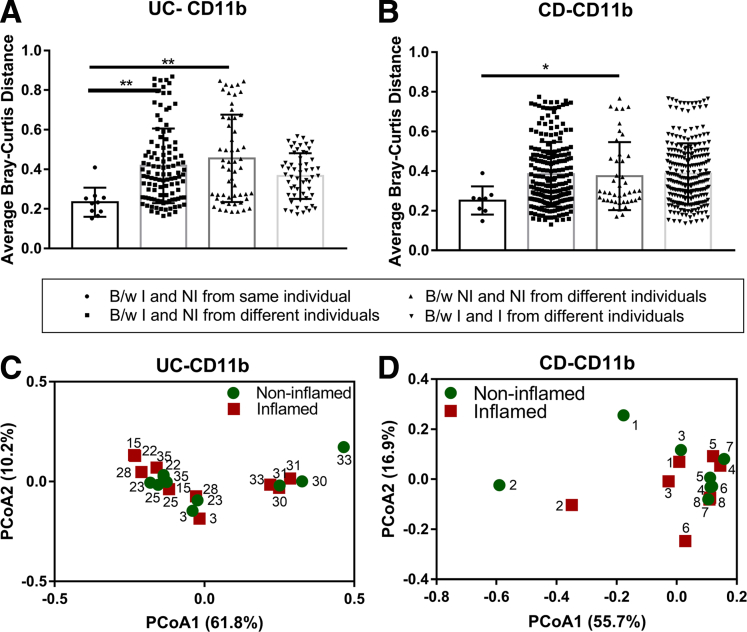
We next examined if inflammation changed the composition of the phagocyte-associated microbiota compared with uninflamed samples. Active inflammation may result in increased epithelial permeability and opportunistic invasion of the lamina propria by pathobionts. In this assessment, UC and CD patients were considered separately, and the phagocyte-associated microbiota from inflamed regions was compared with its matched noninflamed regions. We defined noninflamed samples as those in which no inflammation was present by either endoscopy or histology. For both CD and UC patients, we observed that, within patients, Unifrac distances between phagocyte-associated microbiota from inflamed and noninflamed sites was less than the equivalent distance between the phagocyte-associated microbiota from inflamed or noninflamed sites from different patients (Figure 5A and B). These results, although not reaching statistical significance, indicate that the microbiota from the noninflamed region of a patient is more similar to its own inflamed microbiota than to the noninflamed microbiota of any other person. We also show that the PCoA plot of inflamed and noninflamed phagocyte-associated microbiota overlap, indicating that the phagocyte-associated microbiota from noninflamed regions is similar to inflamed regions (Figure 5C and D). An out-based univariate differential abundance analysis using DESeq2 package, followed by adjustment for false-discovery rate, indicated that with only a few exceptions, the relative abundance of bacterial OTUs were comparable between phagocyte-associated microbiota from paired inflamed and noninflamed samples. Overall, these analyses indicate that the phagocyte-associated microbiota represents an individual’s microbiota regardless of inflammatory state or location of the biopsy.
Figure 5.
Dysbiosis in phagocyte-associated microbiota of IBD patients is not associated with inflammation state. Bray–Curtis distance (means ± SD) for each inflamed and noninflamed sample from the same and different individuals are plotted for (A) UC and (B) CD patients. *P < .05 and **P < .01 by 1-way analysis of variance and the Dunnett post hoc test. PCoA based on Bray–Curtis distances between OTUs detected from matched inflamed and noninflamed lamina propria phagocytic fractions of (C) UC (10 pairs of inflamed and noninflamed samples from 10 subjects) and (D) CD patients (8 pairs of inflamed and noninflamed samples from 8 subjects) as used to generate ordination plots for viewing the relative positioning of inflamed (red squares) and noninflamed (green circles) samples in 2 dimensions. The number close to each symbol inside the plot represents individual patients, with each patient depicted by a unique number. Numbers in parentheses indicate the percentage variation explained by the axis. B/w, between; I, inflamed; NI, non-inflamed.
Dysbiosis in Lamina Propria Phagocytes of IBD Patients Relates to the Disease Phenotype
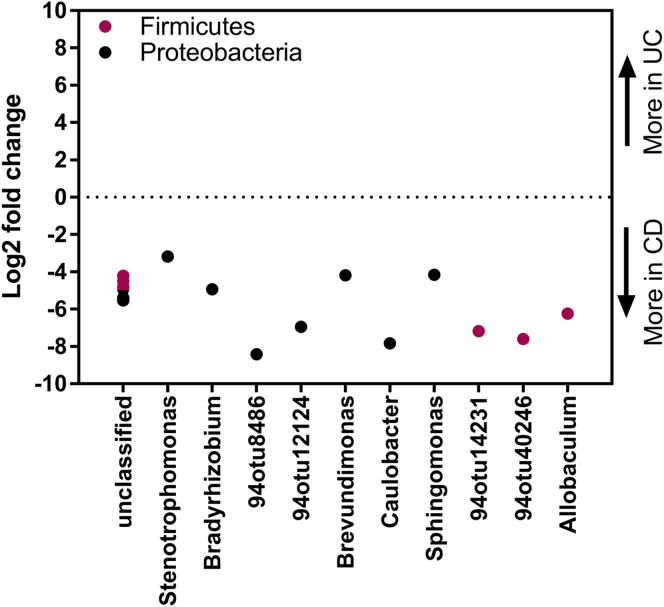
Similar to other studies, we compared the mucosal microbiota of UC patients vs CD patients obtained from the same intestinal anatomic site and inflammation status. Similar to other studies,9 we found more differences between CD and UC patients at noninflamed mucosal sites (Figure 6) than inflamed mucosa (data not shown).
Figure 6.
Mucosa-associated microbiota is distinct in UC and CD. Graphic summary of bacterial features that differed significantly (false-discovery rate corrected P ≤ .05 by the Wald test from the DESeq2 package) between noninflamed ascending colon mucosa of CD (N = 7) and UC patients (N = 9). The y-axis represents the log2 fold change in bacterial relative abundance in UC over CD, with a negative log2 fold change indicating a significant increase in CD mucosa and a positive log2 fold change indicating a significant increase in UC mucosa. Points are OTUs belonging to each genus.
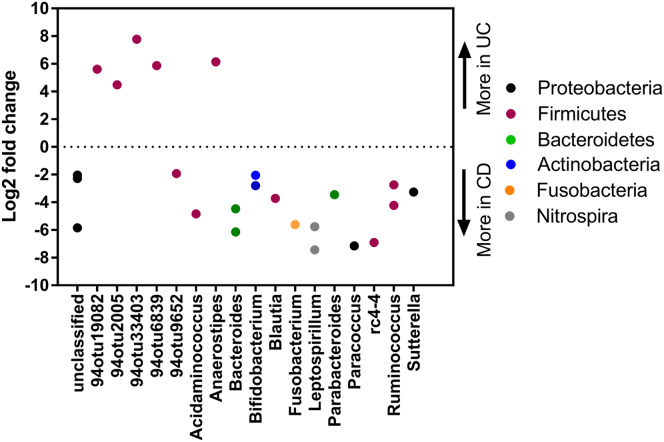
We next examined if the same observations were reflected in the phagocyte-associated microbiota of IBD patients. We compared phagocyte-associated microbiota isolated from inflamed CD tissue (n = 8) with phagocyte-associated microbiota isolated from inflamed UC tissue (n = 10) obtained from the rectosigmoid colon. We also separately compared phagocyte-associated microbiota isolated from noninflamed tissue obtained from the ascending colon of CD patients (n = 7) with noninflamed phagocyte-associated microbiota isolated from ascending colon of UC patients (n = 9). Contrary to our data on the mucosal microbiota, numerous significant differences were observed in the inflamed phagocyte-associated microbiota compared between UC and CD patients. After adjusting for false-discovery rate (q ≤ 0.05), the abundance of 17 OTUs (Figure 7) differed significantly between inflamed phagocyte-associated microbiota of UC patients compared with inflamed phagocyte-associated microbiota of CD patients. Of the 17 OTUs, 11 belonged to phylum Proteobacteria and 6 belonged to phylum Firmicutes, with all the OTUs relatively more abundant in CD patients compared with UC patients. The abundance of only 2 OTUs were significantly different between phagocyte-associated microbiota from noninflamed colon of UC and CD patients, with genera Slackia (phylum Actinobacteria) more abundant in the phagocyte-associated microbiota of UC patients and 94otu22202 (phylum Cyanobacteria) more abundant in phagocyte-associated microbiota of CD patients. These results suggest that the phagocyte-associated microbiota distinguishes CD and UC in the setting of inflammation.
Figure 7.
Dysbiosis in phagocyte-associated microbiota of IBD patients relates to disease phenotype. Graphic summary of bacterial features that differed significantly (false-discovery rate corrected P ≤ .05 by the Wald test from the DESeq2 package) between inflamed sigmoid colon phagocyte fractions of CD (N = 8) and UC (N = 10) patients. The y-axis represents the log2 fold change in bacterial relative abundance in UC over CD, with a negative log2 fold change indicating a significant increase in CD and a positive log2 fold change indicating a significant increase in UC mucosa. Points are OTUs belonging to each genus.
Immune Gene Expression in CD11b+ Cells Relates to the Inflammation State of the Disease
Given that we have shown that lamina propria phagocytes have a microbiota pattern that reflects the inflammation state and type of IBD, we next asked whether IBD-related genes are expressed differentially in the same cell types. We performed gene expression profiling of an aliquot of the CD11b+ cells by nCounter technology (NanoString, Seattle, WA) using a custom-designed panel consisting of IBD susceptibility genes curated for their specific expression in innate immune cells.
The most differentially expressed genes in inflammation included LILRA5, S100A9, S100A12, CXCR2, S100A8, CLEC4E, CXCL1, SLC11A1, OSM, LILRB3, CXCR1, CHI3L1, IL1B, and CXCL8. Interestingly, oncostatin M (OSM), a member of the IL6 cytokine family, was highly up-regulated in inflamed CD11b+ cells in this data set. West et al23 recently showed that a high pretreatment level of OSM was associated with primary nonresponsiveness to anti–tumor necrosis factor therapy. In their study, OSM messenger RNA levels were assessed in published data sets of mixed mucosal biopsy specimens and confirmed in activated CD4+ memory T cells and HLA-DR+ mononuclear cells. Consistent with these data, we show an increase in OSM gene expression in isolated CD11b+ cells from inflamed biopsy specimens in both CD and UC patients compared with uninflamed regions. Likewise, up-regulation of CHI3L1 and CLEC4E as observed in this study, was reported previously in IBD patients and may play a role in facilitating entry of adherent invasive Escherichia coli.24,25 Thus, our choice of examined transcripts reflects a biologically relevant list of genes in the pathogenesis of IBD and links back to local and generalized dysbiosis in IBD patients.
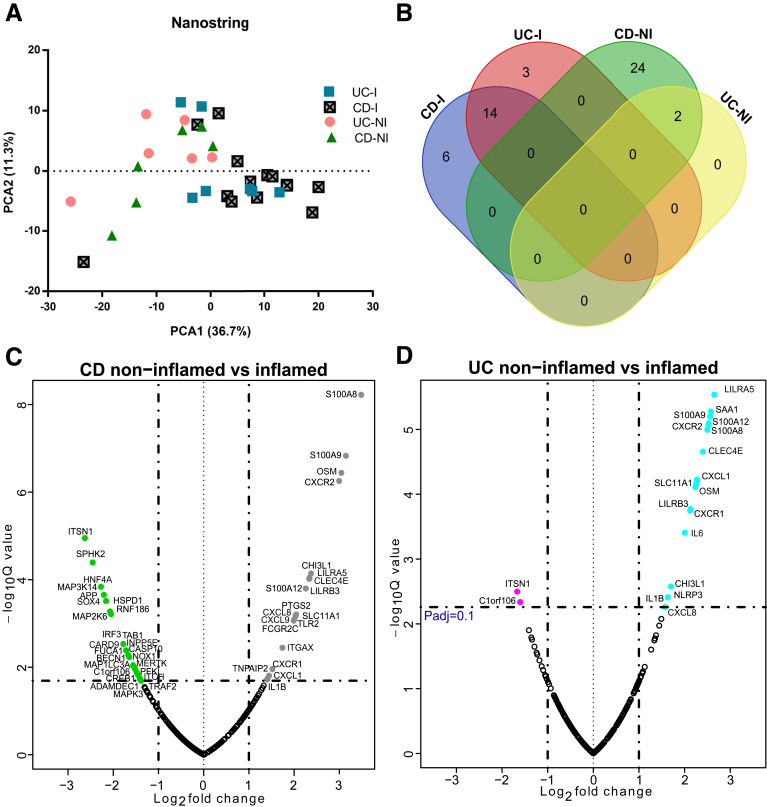
Principle component analysis of gene expression counts showed clustering of inflamed UC and CD patients away from the noninflamed samples (Figure 8A). No significant gene expression differences were observed when comparing the inflamed UC vs CD phagocytes. In UC patients, we observed significant differential expression of 19 genes in lamina propria phagocytes isolated from noninflamed vs inflamed biopsy specimens (Figure 8B). Of the 19 genes that showed differential expression, 17 were up-regulated in phagocytes from inflamed tissues and only ITSN1 (an endocytosis-associated protein) and C1orf106 mRNA were increased in noninflamed CD11b+ phagocytes.
Figure 8.
Immune gene expression in CD11b+ cells relates to the inflammation state of the disease. (A) Principal component analysis (PCA) based on log2 transformed gene expression counts obtained from inflamed and noninflamed lamina propria phagocytes of UC and CD patients (n = 6–14 samples per group). Numbers in parentheses indicate the percentage variation explained by the axis. (B) Venn diagram showing the number of differentially expressed genes that were shared between inflamed and noninflamed samples obtained from CD and UC patients. (C and D) Volcano plots showing genes differentially expressed between inflamed and noninflamed samples from CD and UC patients, respectively. Colored symbols in the volcano plots represent genes that were significantly more abundant (log2 fold change greater than 1 and Padjusted ≤ .1) in the respective groups: CD inflamed (grey), CD noninflamed (green), UC inflamed (blue), UC noninflamed (pink). The P values were obtained by t test followed by adjustment for false-discovery rate. I, inflamed; NI, non-inflamed.
In CD patients, we found differential expression of 46 genes between noninflamed and inflamed samples (Figure 8C). Phagocytes isolated from inflamed areas in CD patients showed 20 genes increased and 26 genes decreased compared with their noninflamed counterparts. Similar to UC samples, ITSN1 and C1orf106 were up-regulated in the noninflamed CD11b+ phagocytes of CD patients compared with inflamed phagocytes. Of the 18 genes up-regulated in inflamed UC phagocytic cells, 15 genes were similarly up-regulated in inflamed CD phagocytic cells (Figure 8D). These data suggest there is a CD11b+ signature of inflammation that is largely shared between UC and CD.
Phagocyte-Associated Microbiota Correlates With Innate Immune Gene Expression in IBD Patients
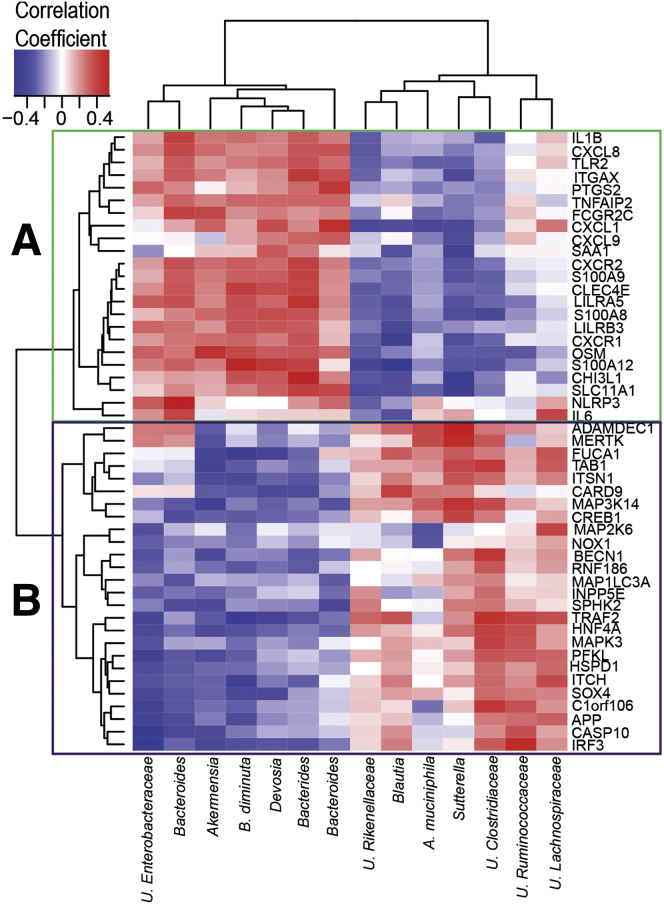
We further investigated potential associations between the phagocyte-associated microbiota and gene expression in the same CD11b+ cells. Here, we focused only on bacterial OTUs significantly enriched in lamina propria phagocytes and host gene expression significantly altered between lamina propria inflamed and noninflamed sites. The lamina propria phagocyte gene expression that correlated with more than 2 bacterial OTUs (P ≤ .05) are represented in a heatmap (Figure 9). Although after adjusting for the false-discovery rate we did not observe any significant Spearman correlation between microbes and genes, we observed some notable trends with correlation coefficients ranging from 0.3 to 0.5 and -0.3 to -0.5. The genes that were associated with microbial abundance formed 2 clusters (Figure 9). Cluster A was expressed at high levels in phagocytes isolated from inflamed tissue, whereas cluster B consisted of genes expressed at low levels in these cell types when compared with phagocytes isolated from noninflamed sites. Up-regulated genes in cluster A showed a noticeable positive correlation with genera Bacteroides, Akkermansia, Devosia, and Brevundimonas diminuta. Down-regulated genes in cluster B showed noteworthy positive correlation with genera Sutterella, Akkermansia, and Blautia, and OTUs belonging to family Clostridiaceae. These results suggest that the variation in the abundance of specific groups of microbiota may affect gene expression levels in host lamina propria phagocyte cell types.
Figure 9.
Bacterial correlations to gene expression in lamina propria phagocytes. Heat map showing the Spearman correlation coefficient (ρ) values of genes expressed in lamina propria phagocytes and phagocyte-associated microbiota. The data represent correlation values obtained from 33 samples collected from inflamed and noninflamed regions of UC (N = 7) and CD (N = 12) patients. Columns correspond to OTUs and rows correspond to genes that had more than 2 correlations with P < .05 without adjusting for false-discovery rate. Scale bar: (left to right) represents a negative correlation in blue to a positive correlation in red. The genes and bacterial OTUs were clustered by Euclidean distance. Cluster A (green) and B (purple) labeled at the nodes of the dendrogram represent group of genes that were expressed significantly more (cluster A) or less (cluster B) in CD11b+ cells from inflamed areas compared with noninflamed areas, respectively. Within the OTU labels, U refers to unclassified, B refers to genus Brevundimonas and A refers to genus Akermensia.
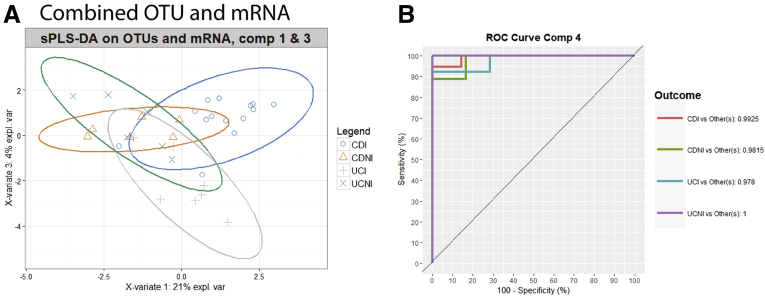
We have shown that the phagocyte-associated microbiota is different between inflamed biopsy specimens of UC and CD patients (Figure 7) and that gene expression of several genes is different between inflamed and noninflamed CD11b+ cells (Figure 8B). We next aimed to determine if the combination of these 2 measures in CD11b+ cells could be used to classify the samples based on both their disease phenotype and their inflammation status. Using the earlier-described edited lists of significantly altered OTUs and gene expression data, we assessed the ability of the combined list to predict the inflammation state and disease type by using sparse partial least squares–discriminant analysis (sPLS-DA) (Figure 10). Although both the OTU and gene expression data independently were able to cluster the different groups of samples to some extent, the concatenated list of both data sets was better at differentiating the 4 groups (Figure 10). Using the sPLS-DA model to build receiver operating characteristic curves and assessing area under the curves showed that the concatenated list using 4 components could discriminate the groups from each other with area under the curve values ranging from 0.978 to 1 (Figure 10B). This is a promising suggestion that the microbiota and gene expression of lamina propria phagocytes may be used to predict disease type and inflammation. Because this was a pilot study we were not able to confirm the model with an independent cohort.
Figure 10.
Combining gene expression data with bacterial OTUs discriminates the type of IBD and inflammatory state. sPLS-DA analysis was performed on a curated concatenated list of gene expression data and bacterial OTU abundance and grouped by class (CDI, CD inflamed; CDNI, CD noninflamed; UCI, UC inflamed; UCNI, UC noninflamed). (A) Graph of sPLS-DA visualized on components (comp) 1 and 3. (B) Receiver operating characteristics (ROC) curves were generated based on the sPLS-DA model using the area under the ROC curve function. Area under the curve calculations are shown. mRNA, messenger RNA.
Discussion
In the past decade, there have been extensive studies examining the intestinal microbiome in patients with IBD. Cataloging the microbiota has led to some broad observations. Most studies have compared stool or mucosal biopsy specimens from IBD patients with healthy controls or symptom controls without IBD.1, 2, 3, 4, 5, 6 Generally, these studies have shown a decrease in diversity of the microbiota and an increase in Proteobacteria and Actinobacteria. Studies also have suggested that few differences exist between inflamed and noninflamed tissue. However, the methods used in the different studies varied widely. Other trends that recently have emerged are that effective treatment, whether with biologics or diet, generally results in increased diversity and normalization of the microbiota.26 Unfortunately, none of these studies has resulted in actionable strategies to inform the treatment of IBD. Moreover, none of these studies has endeavored to link gene expression of isolated cells with the microbiota.
In this study, we aimed to determine if the lamina propria microbiota was a better indicator of disease pathogenesis than the mucosa-associated microbiota and whether it could be correlated with inflammatory gene expression. These approaches may provide insights into immune responses driving inflammation and provide targets for intervention. Our study provides an important proof of principle that there are differences between the mucosa-associated microbiota and the microbiota that is found in lamina propria immune cells. We describe an approach that allowed us to amplify and detect bacteria that are found at very low abundance in the gastrointestinal tract. The presence of these bacteria in the phagocyte-associated microbiota may participate in initiating or promoting IBD.
Because this is a proof of principle, we chose to look broadly at lamina propria phagocytes. Isolating CD11b-positive cells allowed us to survey the majority of different subsets of phagocytes that may be present in both inflamed and noninflamed biopsy specimens. We did this for several reasons. We reasoned that lamina propria phagocytes would be the first line of defense for microbiota that breached the epithelial barrier and therefore could act as sentinels for microbiota that had more invasive properties. In addition, genetic data in IBD implicate macrophages and defects in innate immunity in pathogenesis of disease. We also wanted to be able to isolate sufficient cells per site to allow us to perform both microbiota 16S sequencing as well as gene expression from the same cells. Future studies should examine specific subsets of phagocytic cells to determine if there is even more specificity of microbiota based on myeloid cell subtype.
The majority of the microorganisms identified in this study are normal inhabitants of the gut. Some of these bacteria including Akkermensia, Bifidobacterium, Streptococcus, Sutterella, Prevotella, and Blautia have been associated previously with inflamed mucosa. Other bacteria, such as Herbaspirillum and Cupriavidus (mainly from phyla Proteobacteria) are not encountered frequently in the gastrointestinal tract. On the other hand, some culture and deep sequencing studies have reported their presence in human beings, albeit at different body sites, under diseased conditions or as pathogens in immune-compromised individuals or nosocomial infections.27, 28, 29, 30 Under inflammatory conditions, as observed in IBD, these bacteria can become opportunistic survivors, inhabiting the deeper layers of inflamed mucosa and ultimately finding their way to lamina propria phagocytes.
Our results showed significant differences between the mucosal microbiota and that associated with lamina propria phagocytes. IBD is associated with increased nitric oxide and methane production in the gut.31 The higher abundance of Proteobacteria in phagocyte-associated microbiota indicates a switch from the anaerobic gut environment to auxotrophic, which promotes the growth of pathobionts. Sphingomonas species previously have been reported to modulate the function of invariant natural killer T cells.32 Similarly, Prevotella species have been shown to augment inflammation by promoting T helper 17 cell–mediated mucosal inflammation.33 Increased abundance of both of these bacteria in phagocytes indicates a selective invasion of bacteria capable of modulating immune responses toward inflammation. Intriguingly, we observed that some bacterial families and genera (such as Prevotella, Akkermensia, Bifidobacterium, Rikkenellaceae, and Enterobacteraceae) were abundant in both mucosa and phagocyte-associated microbiota. These bacteria have high species diversity, and even a single species of these bacteria can have different strains. Therefore, the distribution of these bacteria from the same species can be strain-specific, with some strains possessing more invasive characteristics and thus being more abundant in phagocyte-associated microbiota, whereas others are more abundant in mucosa.
Although meta-transcriptomic analysis is better for assessing functional pathways in microbiota, our starting cells are of mammalian origin and therefore most of the RNA sequences are from the host, not bacteria. Thus, PICRUSt represents the best way to perform this analysis. The results from PICRUSt analysis indicated that xenobiotic degradation pathways are more abundant in the phagocyte-associated microbiota, whose metabolites are known to promote oxidative stress and bacterial virulence. Adult IBD patients have lower folate and biotin concentrations.34 Abnormal folate levels can play a role in IBD pathogenesis by modifying DNA methylation. In our study, we found that the folate biosynthesis pathway was less abundant in the phagocyte-associated microbiota of both CD and UC patients. Likewise, there was an underrepresentation of core genetic processing pathways in the phagocyte-associated microbiota compared with mucosa. These results indicate that compared with the mucosa, the microbiota associated with lamina propria phagocytes more closely represents compositional and functional characteristics representative of the inflamed state described previously by various studies.1, 2, 3, 4, 5, 6 Collectively, these results also indicate that the lamina propria of IBD patients consists of an environment that favors bacteria with properties of invasion, virulence, auxotrophy, and possibly immune system modulation.
Whole-genome gene expression analyses of IBD patients have shown important candidate genes in IBD. However, these studies were not able to identify any major differences between CD and UC gene expression patterns.35 These studies were based on heterogeneous samples of the mucosa, which is a complex tissue composed of different cell types. The numbers of these cells differ from patient to patient. A strength of our study was the use of a subset of these cell types, namely CD11b+ lamina propria phagocytes. In this study, we observed that the total number of genes that were expressed differentially between CD11b+ lamina propria phagocytes of inflamed and noninflamed sites were far higher in CD patients as opposed to UC patients. This observation may be owing to deeper penetrating inflammation in Crohn's disease vs more superficial involvement of mucosa in ulcerative colitis. Consistent with Granlund et al,35 our results showed no major differences in innate immune gene expression of inflamed lamina propria phagocytes between CD and UC patients. This indicates that host inflammatory gene expression does not differentiate CD vs UC. Interestingly, OSM was highly expressed in CD11b+ cells in inflamed tissue regardless of UC or CD. Many of the patients enrolled in this study were receiving, or had been receiving, anti–tumor necrosis factor therapy and had ongoing inflammation, suggesting that OSM is associated with failure of response to therapy.23 In contrast, we did observe significant differences in the phagocyte-associated microbiota found in inflamed areas between UC and CD patients. Further studies should determine the role of these bacteria in driving the different disease phenotypes.
Despite offering a novel approach for studying the role of microbiota in intestinal diseases, this study was not without limitations. For example, absence of healthy controls in this study limited the information about the bacteria that may be present in lamina propria of the healthy gut. In addition, we were not able to distinguish if the bacteria amplified from lamina propria fractions were attached or engulfed by the phagocytic cells. Regardless of these limitations, we believe that the ability to use nonconventional lamina propria phagocyte fractions as a microbiota sample source provides a unique perspective for future studies aimed at studying the role of microbiota in the gastrointestinal tract.
Ultimately, our goal was to translate our findings to the care of IBD patients. Antibiotics have had very modest efficacy in the treatment of IBD,36 and fecal microbial transplant has shown efficacy only in UC.37,38 It is possible that certain bacteria are impervious or less accessible to systemic antibiotic treatment because they linger in lamina propria phagocytes. Quite possibly, the genetic perturbations underlying IBD contribute to their persistence in phagocytic cells. The finding that the phagocyte-associated microbiota varied little between inflamed and uninflamed regions of the same patient allowed us to speculate that biopsy specimens taken in the distal colon may inform us of what is happening proximally and may someday aid in therapeutic decisions. To validate these observations, large-scale studies would need to be performed to identify whether genetic risk or aberrant innate immune signaling in IBD contributes to the microbiota community in the lamina propria of IBD and whether the lamina propria microbiota contributes to IBD. It is our hope that approaches such as these could be used in personalized strategies for the treatment of IBD patients.
Methods
Subjects, Ethics Statement, and Biopsy Specimen Acquisition
Six to 8 pinch biopsy specimens were collected from a total of 32 patients (20 CD and 12 UC) at the time of colonoscopy from the ileum, ascending colon, and/or sigmoid colon for a total of 55 samples. An additional 4 biopsy specimens were collected from each site for pathology. One biopsy for each sample was transferred to RNAlater solution (cat #AM7020; Thermo Fisher Scientific, Waltham, MA) and stored at -80°C for studying mucosal microbiota. The remaining biopsy specimens were transferred to 1 mL of HypoThermosol FRS preservation solution (cat #H4416-100ML; Sigma-Aldrich, St. Louis, MO) to be used immediately for isolating lamina propria CD11+ phagocyte cell populations. Of the 32 patients, paired biopsy specimens of inflamed and noninflamed tissue were collected from 8 CD and 10 UC patients, whereas from the 14 remaining patients (12 CD and 2 UC), biopsy specimens were collected from either inflamed or noninflamed mucosa only. The use of human samples for this study was approved by the University of Miami Institutional Review Board. The clinical and demographic characteristics of the patients included in the study are shown in Table 1.
Table 1.
Characteristics of the Patients and Biopsy Specimens Included in This Study
| CD (N = 20) | UC (N = 12) | |
|---|---|---|
| Male/female | 7/13 | 6/6 |
| Treatment | ||
| None | 3 | 1 |
| Anti-TNFs | 5 | 4 |
| Anti-p40 (ustekinumab) | 2 | 0 |
| Vedolizumab | 3 | 0 |
| Others | 7 | 7 |
| Biopsy location | Inflamed/noninflamed | Inflamed/noninflamed |
| Ileum | 10/2 | 0/1 |
| Rectosigmoid colon | 8/1 | 11/1 |
| Ascending colon | 4/7 | 0/9 |
| Transverse colon | 1/0 | 0/0 |
TNF, tumor necrosis factor.
Lamina Propria Phagocytic Cell Isolation and Magnetic-activated Cell Sorting Sorting
CD11b+ cells from intestinal lamina propria were isolated from 6–7 mucosal biopsy specimens. Lamina propria cells were isolated as previously described.39 Briefly, biopsy specimens were rinsed with phosphate-buffered saline and incubated in 10 mmol/L dithiothreitol (cat #646563; Sigma-Aldrich) in Dulbecco’s modified Eagle medium on an orbital shaker at room temperature for 20 minutes. The supernatant was discarded and the biopsy specimens were rinsed and treated with 1 mmol/L EDTA (cat #15575020; Thermo Fisher Scientific) with shaking at room temperature for 20 minutes. Again, the supernatants were discarded and the biopsy specimens were digested to a single-cell suspension in 2.5 mg/mL Liberase (cat #5401127001; Roche, Indianapolis, IN) with 1 U/μL DNase (cat #D9905K; Epicentre, Madison, WI) in rotation at 37°C for 30 minutes. After 2 washes, cells were labeled with CD11b+ microbeads (cat #130-049-601; Miltenyi-Biotec, Auburn, CA) as per the manufacturer’s instructions and positively selected after passage through an LS MACS column (cat #130-042-401; Miltenyi-Biotec). The purity of selected isolations was tested by flow cytometry. Isolated cells were stained with antibodies against CD11b-APCcy7 (cat #557657, M1/70; BD Biosciences, San Jose, CA), CD15-eF450 (cat #557657, HI98; Thermo Fisher Scientific), CD206-APC (cat #550889, 19.2; BD Biosciences), HLA-DR–fluorescein isothiocyanate (cat #555559, TU36; BD Biosciences), and CD14-BV786 (cat #563698, M5E2; BD Biosciences), and were analyzed on a BD Fortessa instrument (BD Biosciences, San Jose, CA). To minimize contamination and remove external bacteria, isolated lamina propria CD11b+ cells were incubated with 50 ng/mL gentamicin (cat #G1397-10ML; Sigma-Aldrich) for 20 minutes followed by 3 washes with phosphate-buffered saline.
DNA extraction and 16S rRNA gene sequencing
Genomic DNA was extracted from the whole biopsy and from CD11b+ lamina propria phagocytic cell pellets using the QIAmp Tissue DNA extraction kit (catalog #51304) and the QIAamp DNA micro kit (catalog #56304) (Qiagen, Germantown, MD), respectively, and according to the manufacturer’s instructions with few modifications. Although the 2 kits were similar in the DNA extraction methodology as well as chemicals used, we found that the micro kit (cat #56304) was more suitable for small numbers of CD11b+ cells with very low total DNA concentration owing to its requirement of a small elution volume. DNA quantification was performed with the Qubit Quant-iT dsDNA High Sensitivity Kit (Invitrogen, Grand Island, NY). Bacterial 16S V4 ribosomal DNA was amplified using 2 differently barcoded V4 fusion primers. Pooled polymerase chain reaction samples were purified and paired-end sequenced on MiSeq instrument for 250 cycles (Illumina, San Diego, CA). The steps from DNA quantification to sequencing were conducted at Second Genome, Inc. (San Francisco, CA)
Microbiota data analysis
The OTU count table, α diversity metrics, and β diversity metrics were generated by Second Genome, Inc, using the secondgenomeR package: 0.2.4. Briefly, paired-end reads were aligned, quality-filtered, and dereplicated with USEARCH.40 After removing chimeric sequences, representative OTUs were picked by de novo OTU clustering at 97% similarity by UPARSE.41 Bacterial taxonomic classification was assigned to the representative sequences using the Mothur Bayesian classifier trained against the Greengenes reference database of 16S rRNA gene sequences clustered at 99%. Weighted sample-to-sample distance matrices were calculated using Bray–Curtis dissimilarity.42 Unweighted distance matrices were calculated with the Jaccard43 index. PCoA plots were used to position samples relative to each other based on their dissimilarity values. LEfSe was performed to detect bacterial biomarkers that differed significantly between assigned groups.44 Bacterial taxon significance testing was performed using the DESeq2 package,45 a negative binomial noise model for overdispersion, and the Poisson process as described for microbiome applications.46 False-discovery rates were calculated with the Benjamini–Hochberg procedure to obtain adjusted P values. The sequences obtained in this study have been submitted to the European Genome-phenome Archive database under accession number EGAS00001003105.
Nanostring gene expression data analysis
CD11b-positive cell lysates were obtained by disrupting the cells with RLT buffer (cat #79216; Qiagen) at a concentration of 1 uL per 10,000 cells. The cell lysates were hybridized with a custom-designed code set of 347 genes related to innate immunity according to the manufacturer’s instructions (NanoString, Seattle, WA). The genes were selected based on published IBD-associated gene signatures as well as genes associated with common bacterial recognition pathways and autophagy. The raw data were analyzed using nSolver Analysis software (NanoString). Only 33 of 48 samples passed the quality control. The data were normalized to average gene expression of the housekeeping genes. During the data normalization process, 110 genes were below the detection limits in 80% of the samples. These genes were removed from further analyses, leaving a total of 237 genes for downstream analysis. Further statistical analysis was performed on the log2 transformed normalized data. The Student t test was used to compare 2 groups and the P values obtained were adjusted for false-discovery rate by the Benjamini–Hochberg procedure. Results were considered significant when q ≤ 0.10 (adjusted P value). The Spearman correlation coefficient and significance was determined in R using the psych package. A volcano plot and heat map were constructed in R.
sPLS-DA
A subset of significantly altered messenger RNAs (49) and OTUs (84) were either assessed separately, or concatenated and subjected to sPLS-DA using the mix0mics package available in R.47,48 ROC curves based on the sPLS-DA model were plotted using the area under the receiver operator characteristic function.
Acknowledgment
The authors thank Dr Sion Williams and Yoslayma Cardentey at the University of Miami Sylvester Comprehensive Cancer Center Oncogenomics Core Facility for performing nanostring experiments. The authors also thank Micky and Madeleine Arison as well as Joanne Trempala for their philanthropic support of the Crohn’s and Colitis Research Laboratory.
Current affiliations of R.D.: Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan; of J.M.D.: Mater Research Institute, The University of Queensland, Brisbane, Queensland, Australia.
Footnotes
Author contributions Rishu Dheer, Julie M. Davies, and Maria T. Abreu designed the study; Maria A. Quintero, Oriana M. Damas, Amar R. Deshpande, David H. Kerman, and Maria T. Abreu collected samples and clinical data; Rishu Dheer, Julie M. Davies, Juan F. Burgueno, Matthew C. Phillips, and Irina Fernandez performed the experiments; Rishu Dheer, Julie M. Davies, Yuguang Ban, and W. Peter Sawyer interpreted the data; and Rishu Dheer, Julie M. Davies, Judith Pignac-Kobinger, and Maria T. Abreu wrote the manuscript. All authors critically reviewed and approved the manuscript.
Conflicts of interest This author discloses the following: Maria T. Abreu reports grants from Prometheus laboratories, grants from Takeda Pharmaceuticals, grants from Pfizer Inc, has served as a consultant to AbbVie Laboratories, Janssen Pharmaceuticals, Focus Medical Communications, Celgene Corporation, Eli Lilly Pharmaceuticals, Shire Pharmaceuticals, Roche Pharmaceuticals, Boehringer Ingelheim Pharmaceuticals, Amgen, Allergan and serves on the scientific advisory board of Gilead. The remaining authors disclose no conflicts.
Funding This work was supported by grant 5R01CA137869-05 from the National Institutes of Health, National Cancer Institute; grant R01DK099076 from the National Institute of Diabetes and Digestive and Kidney Diseases; and a Senior Investigator Award (3786-SRA) from the Crohn's and Colitis Foundation of America (M.T.A.). Additional funding was provided by The Micky & Madeleine Arison Family Foundation Crohn's & Colitis Discovery Laboratory and the Martin Kalser Chair in Gastroenterology at the University of Miami.
References
- 1.Bibiloni R., Mangold M., Madsen K.L., Fedorak R.N., Tannock G.W. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J Med Microbiol. 2006;55:1141–1149. doi: 10.1099/jmm.0.46498-0. [DOI] [PubMed] [Google Scholar]
- 2.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., Reyes J.A., Shah S.A., LeLeiko N., Snapper S.B., Bousvaros A., Korzenik J., Sands B.E., Xavier R.J., Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagalingam N.A., Lynch S.V. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2012;18:968–984. doi: 10.1002/ibd.21866. [DOI] [PubMed] [Google Scholar]
- 4.Sepehri S., Kotlowski R., Bernstein C.N., Krause D.O. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:675–683. doi: 10.1002/ibd.20101. [DOI] [PubMed] [Google Scholar]
- 5.Walker A.W., Sanderson J.D., Churcher C., Parkes G.C., Hudspith B.N., Rayment N., Brostoff J., Parkhill J., Dougan G., Petrovska L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willing B.P., Dicksved J., Halfvarson J., Andersson A.F., Lucio M., Zheng Z., Jarnerot G., Tysk C., Jansson J.K., Engstrand L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854 e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 7.de Lange K.M., Moutsianas L., Lee J.C., Lamb C.A., Luo Y., Kennedy N.A., Jostins L., Rice D.L., Gutierrez-Achury J., Ji S.G., Heap G., Nimmo E.R., Edwards C., Henderson P., Mowat C., Sanderson J., Satsangi J., Simmons A., Wilson D.C., Tremelling M., Hart A., Mathew C.G., Newman W.G., Parkes M., Lees C.W., Uhlig H., Hawkey C., Prescott N.J., Ahmad T., Mansfield J.C., Anderson C.A., Barrett J.C. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., Abedian S., Cheon J.H., Cho J., Dayani N.E., Franke L., Fuyuno Y., Hart A., Juyal R.C., Juyal G., Kim W.H., Morris A.P., Poustchi H., Newman W.G., Midha V., Orchard T.R., Vahedi H., Sood A., Sung J.Y., Malekzadeh R., Westra H.J., Yamazaki K., Yang S.K., International Multiple Sclerosis Genetics Consortium, International IBD Genetics Consortium. Barrett J.C., Alizadeh B.Z., Parkes M., Bk T., Daly M.J., Kubo M., Anderson C.A., Weersma R.K. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbes J.D., Van Domselaar G., Bernstein C.N. Microbiome survey of the inflamed and noninflamed gut at different compartments within the gastrointestinal tract of inflammatory bowel disease patients. Inflamm Bowel Dis. 2016;22:817–825. doi: 10.1097/MIB.0000000000000684. [DOI] [PubMed] [Google Scholar]
- 10.Momozawa Y., Deffontaine V., Louis E., Medrano J.F. Characterization of bacteria in biopsies of colon and stools by high throughput sequencing of the V2 region of bacterial 16S rRNA gene in human. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gevers D., Kugathasan S., Denson L.A., Vazquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S.J., Yassour M., Morgan X.C., Kostic A.D., Luo C., Gonzalez A., McDonald D., Haberman Y., Walters T., Baker S., Rosh J., Stephens M., Heyman M., Markowitz J., Baldassano R., Griffiths A., Sylvester F., Mack D., Kim S., Crandall W., Hyams J., Huttenhower C., Knight R., Xavier R.J. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shobar R.M., Velineni S., Keshavarzian A., Swanson G., DeMeo M.T., Melson J.E., Losurdo J., Engen P.A., Sun Y., Koenig L., Mutlu E.A. The effects of bowel preparation on microbiota-related metrics differ in health and in inflammatory bowel disease and for the mucosal and luminal microbiota compartments. Clin Transl Gastroenterol. 2016;7:e143. doi: 10.1038/ctg.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ning K., Gettler K., Zhang W., Ng S.M., Bowen B.M., Hyams J., Stephens M.C., Kugathasan S., Denson L.A., Schadt E.E., Hoffman G.E., Cho J.H. Improved integrative framework combining association data with gene expression features to prioritize Crohn's disease genes. Hum Mol Genet. 2015;24:4147–4157. doi: 10.1093/hmg/ddv142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., Essers J., Mitrovic M., Ning K., Cleynen I., Theatre E., Spain S.L., Raychaudhuri S., Goyette P., Wei Z., Abraham C., Achkar J.P., Ahmad T., Amininejad L., Ananthakrishnan A.N., Andersen V., Andrews J.M., Baidoo L., Balschun T., Bampton P.A., Bitton A., Boucher G., Brand S., Buning C., Cohain A., Cichon S., D'Amato M., De Jong D., Devaney K.L., Dubinsky M., Edwards C., Ellinghaus D., Ferguson L.R., Franchimont D., Fransen K., Gearry R., Georges M., Gieger C., Glas J., Haritunians T., Hart A., Hawkey C., Hedl M., Hu X., Karlsen T.H., Kupcinskas L., Kugathasan S., Latiano A., Laukens D., Lawrance I.C., Lees C.W., Louis E., Mahy G., Mansfield J., Morgan A.R., Mowat C., Newman W., Palmieri O., Ponsioen C.Y., Potocnik U., Prescott N.J., Regueiro M., Rotter J.I., Russell R.K., Sanderson J.D., Sans M., Satsangi J., Schreiber S., Simms L.A., Sventoraityte J., Targan S.R., Taylor K.D., Tremelling M., Verspaget H.W., De Vos M., Wijmenga C., Wilson D.C., Winkelmann J., Xavier R.J., Zeissig S., Zhang B., Zhang C.K., Zhao H., Silverberg M.S., Annese V., Hakonarson H., Brant S.R., Radford-Smith G., Mathew C.G., Rioux J.D., Schadt E.E., Daly M.J., Franke A., Parkes M., Vermeire S., Barrett J.C., Cho J.H. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davenport M., Poles J., Leung J.M., Wolff M.J., Abidi W.M., Ullman T., Mayer L., Cho I., Loke P. Metabolic alterations to the mucosal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2014;20:723–731. doi: 10.1097/MIB.0000000000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L., Wang W., Zhou R., Ng S.C., Li J., Huang M., Zhou F., Wang X., Shen B., M A.K., Wu K., Xia B. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine. 2014;93:e51. doi: 10.1097/MD.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malizia G., Calabrese A., Cottone M., Raimondo M., Trejdosiewicz L.K., Smart C.J., Oliva L., Pagliaro L. Expression of leukocyte adhesion molecules by mucosal mononuclear phagocytes in inflammatory bowel disease. Gastroenterology. 1991;100:150–159. doi: 10.1016/0016-5085(91)90595-c. [DOI] [PubMed] [Google Scholar]
- 19.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenblum S., Turnbaugh P.J., Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D. a Reyes J, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology. 2013;8:1–10. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abubucker S., Segata N., Goll J., Schubert A.M., Izard J., Cantarel B.L., Rodriguez-Mueller B., Zucker J., Thiagarajan M., Henrissat B., White O., Kelley S.T., Methe B., Schloss P.D., Gevers D., Mitreva M., Huttenhower C. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West N.R., Hegazy A.N., Owens B.M.J., Bullers S.J., Linggi B., Buonocore S., Coccia M., Gortz D., This S., Stockenhuber K., Pott J., Friedrich M., Ryzhakov G., Baribaud F., Brodmerkel C., Cieluch C., Rahman N., Muller-Newen G., Owens R.J., Kuhl A.A., Maloy K.J., Plevy S.E., Oxford IBD Cohort Investigators. Keshav S., Travis S.P.L., Powrie F. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23:579–589. doi: 10.1038/nm.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buisson A., Vazeille E., Minet-Quinard R., Goutte M., Bouvier D., Goutorbe F., Pereira B., Barnich N., Bommelaer G. Faecal chitinase 3-like 1 is a reliable marker as accurate as faecal calprotectin in detecting endoscopic activity in adult patients with inflammatory bowel diseases. Aliment Pharmacol Ther. 2016;43:1069–1079. doi: 10.1111/apt.13585. [DOI] [PubMed] [Google Scholar]
- 25.Te Velde A.A. The C-type lectin mincle: clues for a role in Crohn's disease adjuvant reaction. Front Immunol. 2017;8:1304. doi: 10.3389/fimmu.2017.01304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis J.D., Chen E.Z., Baldassano R.N., Otley A.R., Griffiths A.M., Lee D., Bittinger K., Bailey A., Friedman E.S., Hoffmann C., Albenberg L., Sinha R., Compher C., Gilroy E., Nessel L., Grant A., Chehoud C., Li H., Wu G.D., Bushman F.D. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spilker T., Uluer A.Z., Marty F.M., Yeh W.W., Levison J.H., Vandamme P., Lipuma J.J. Recovery of Herbaspirillum species from persons with cystic fibrosis. J Clin Microbiol. 2008;46:2774–2777. doi: 10.1128/JCM.00460-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajilic-Stojanovic M., de Vos W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langevin S., Vincelette J., Bekal S., Gaudreau C. First case of invasive human infection caused by Cupriavidus metallidurans. J Clin Microbiol. 2011;49:744–745. doi: 10.1128/JCM.01947-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaakoush N.O., Day A.S., Huinao K.D., Leach S.T., Lemberg D.A., Dowd S.E., Mitchell H.M. Microbial dysbiosis in pediatric patients with Crohn's disease. J Clin Microbiol. 2012;50:3258–3266. doi: 10.1128/JCM.01396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faber F., Baumler A.J. The impact of intestinal inflammation on the nutritional environment of the gut microbiota. Immunol Lett. 2014;162:48–53. doi: 10.1016/j.imlet.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wingender G., Stepniak D., Krebs P., Lin L., McBride S., Wei B., Braun J., Mazmanian S.K., Kronenberg M. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–428. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan Y., Liu Y., Guo H., Jabir M.S., Liu X., Cui W., Li D. Associations between folate and vitamin B12 levels and inflammatory bowel disease: a meta-analysis. Nutrients. 2017;9 doi: 10.3390/nu9040382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granlund A., Flatberg A., Ostvik A.E., Drozdov I., Gustafsson B.I., Kidd M., Beisvag V., Torp S.H., Waldum H.L., Martinsen T.C., Damas J.K., Espevik T., Sandvik A.K. Whole genome gene expression meta-analysis of inflammatory bowel disease colon mucosa demonstrates lack of major differences between Crohn's disease and ulcerative colitis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan K.J., Ullman T.A., Ford A.C., Abreu M.T., Abadir A., Marshall J.K., Talley N.J., Moayyedi P. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011;106:661–673. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- 37.Jacob V., Crawford C., Cohen-Mekelburg S., Viladomiu M., Putzel G.G., Schneider Y., Chabouni F., O'Neil S., Bosworth B., Woo V., Ajami N.J., Petrosino J.F., Gerardin Y., Kassam Z., Smith M., Iliev I.D., Sonnenberg G.F., Artis D., Scherl E., Longman R.S. Single delivery of high-diversity fecal microbiota preparation by colonoscopy is safe and effective in increasing microbial diversity in active ulcerative colitis. Inflamm Bowel Dis. 2017;23:903–911. doi: 10.1097/MIB.0000000000001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paramsothy S., Kamm M.A., Kaakoush N.O., Walsh A.J., van den Bogaerde J., Samuel D., Leong R.W.L., Connor S., Ng W., Paramsothy R., Xuan W., Lin E., Mitchell H.M., Borody T.J. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 39.Ramesh R., Kozhaya L., McKevitt K., Djuretic I.M., Carlson T.J., Quintero M.A., McCauley J.L., Abreu M.T., Unutmaz D., Sundrud M.S. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J Exp Med. 2014;211:89–104. doi: 10.1084/jem.20130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 41.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 42.Bray J.R., Curtis J.T. An ordination of the upland forest communities of southern Wisconsin. Ecologic Monogr. 1957;27:325–349. [Google Scholar]
- 43.Jaccard P. The distribution of the flora in the Alpine zone. New Phytol. 1912;11:37–50. [Google Scholar]
- 44.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Cao K.A., Boitard S., Besse P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics. 2011;12:253. doi: 10.1186/1471-2105-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohart F., Gautier B., Singh A., Le Cao K.A. mixOmics: an R package for 'omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]