Abstract
In order to rapidly identify the phenotypic profile and possible off-target liability effects of novel synthesized thyromimetics for selection of lead compounds for further optimization studies, we performed in vitro screening on a new small library of synthetic thyromimetics. A comprehensive panel of early toxicity assays comprising cytotoxicity on 4 different cell lines (osteosarcoma, U2OS; lung fibroblast, hTERT; human breast adenocarcinoma, MCF7; human embryonic kidney, HEK293), hERG liability, cytochrome P450 inhibition (CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4 isoforms), and off-target liability against selected proteins (Aurora B kinase and phosphodiesterase PDE4C1) and epigenetic enzymes (HDAC4, HDAC6, HDAC8, HDAC9 & SIRT7). All the compounds were screened at 10 μM in at least triplicate using well-established in vitro assays with readouts in luminescence or fluorescence polarization mode. The raw data were processed using Microsoft Excel and the Z′ for each assay was calculated (acceptable Z' >0.40). The processed and normalized data were organized in tables and visualized using spider plots. The results which are reported in the present manuscript can be used in prediction studies of early toxicity and off-target liabilities of other thyromimetics using in silico methods. The data reported herein support our research article entitled “Design, synthesis and biological evaluation of novel TRβ selective agonists sustained by ADME-Toxicity analysis” by Runfola M., Sestito S., et al. [1]
Keywords: Triiodothyronine, Thyronamine, TRβ selective agonist, Fatty-liver disorder, Liver regeneration, ADME-Tox profile, Off-target liability, Screening
Specifications Table
| Subject | DRUG DISCOVERY |
| Specific subject area | Early toxicity and off-target liability |
| Type of data | Tables Figures Spider plots Protocols for early toxicity and off-target liability assays |
| How data were acquired | All in vitro assays were performed after optimization of the manufacturer's kit protocols as reported within the Materials and Methods section. Cytotoxicity effects were evaluated using the CellTiter-Glo (Promega Corp.). Cardiac toxicity was assessed using the Predictor™ hERG Fluorescence Polarization Assay Kit (Thermo). For detection of CYP450 activity, the luminescence-based P450-Glo® (Promega Corp.) assay system was used. HDAC inhibition was determined using the homogeneous, single-addition, bioluminogenic HDAC-Glo® I/II assay (Promega Corp.) for HDAC6 and HDAC8 or the HDAC-Glo® 2A assay (Promega Corp.) for HDAC4 and HDAC9. Inhibition of SIRT7 enzyme was determined using the SIRT-Glo® assay (Promega Corp.). Inhibition of PDE4C1 was determined using the PDE4C1 assay (BPS Bioscience) and inhibition of Aurora B kinase was determined using the Kinase-Glo® Luminescent Kinase Assay Platform (Promega Corp.). Luminescence or fluorescence polarization measurements in the assays were made using an EnVision Multilabel 2103 or 2300 EnSpire Multilabel (PerkinElmer) reader. The raw data were processed and normalized relative to the High and Low controls using Microsoft Excel and visually organized into tables and spider plots, where data for each compound are reported as % inhibition. |
| Data format | Raw data: Microsoft Excel Analyzed data: Microsoft Excel |
| Parameters for data collection | The early toxicity and off-target liability assays were selected using the generally accepted criteria for progressing in the drug discovery value chain to the lead compound stage. |
| Description of data collection | All raw data collected were from luminescence or fluorescence polarization measurements in the early toxicity and off-target liability assays using an EnVision Multilabel 2103 or 2300 EnSpire Multilabel (PerkinElmer) reader. The raw data were processed and normalized relative to the High and Low controls using Microsoft Excel and visually organized into tables and spider plots, where data for each compound are reported as % inhibition. As additional quality control, the Z′ for each were determined and these exceeded the minimum threshold of >0.40. |
| Data source location | Fraunhofer IME ScreeningPort Hamburg Germany |
| Data accessibility | With the article (Supplementary Material: “SM1_raw_data.xlsx” and “SM2_processed_data.xlsx”). |
| Related research article | Massimiliano Runfola, Simona Sestito, Lorenza Bellusci, Valeria La Pietra, Vincenzo Maria D'amore, Marta Anna Kowalik, Grazia Chiellini, Sheraz Gul, Andrea Perra, Amedeo Columbano, Luciana Marinelli, Ettore Novellino, Simona Rapposelli. (2020) Design, synthesis and biological evaluation of novel TRβ selective agonists sustained by ADME-Toxicity analysis. European Journal of Medicinal Chemistry, DOI/In Press. |
Value of the Data
|
1. Data description
Within this study we report data obtained from early toxicity and off-target liability studies together with assay protocols, raw and processed data for newly synthetized thyromimetics [1] as a powerful tool to rapidly identify new potential lead compounds with optimal properties for progression in pre-clinical drug discovery.
In Table 1, % Cytotoxicity or % Inhibition or for each assay are reported as the average (AVG) from triplicate measurements together with the standard deviation (STD). The Z′ is also reported for each assay.
Table 1.
Early toxicity and off-target liability profiles of compounds. For each assay the mean of three replicates (AVG), the standard deviation (STD), and the Z′ for each assay are reported.
|
Compound |
HEK293 % Cytotoxicity 24 h |
HEK293 % Cytotoxicity 48 h |
||
|---|---|---|---|---|
| AVG | STD | AVG | STD | |
| 1 | -8 | 3 | -7 | 2 |
| 2 | -5 | 2 | 2 | 2 |
| 3 | -7 | 3 | -5 | 2 |
| 4 | 7 | 3 | 11 | 2 |
| 5 | 1 | 1 | 1 | 2 |
| 6 | 17 | 5 | 15 | 4 |
| 7 | -3 | 1 | -6 | 0 |
| 8 | 2 | 1 | 4 | 3 |
| 9 | 10 | 3 | 9 | 3 |
| 10 | -4 | 1 | -3 | 0 |
| 11 | -6 | 3 | -5 | 3 |
| Assay Z′ | 0.75 | 0.78 | ||
|
Compound |
hTERT % Cytotoxicity 24 h |
hTERT % Cytotoxicity 48 h |
||
|---|---|---|---|---|
| AVG | STD | AVG | STD | |
| 1 | 1 | 1 | 1 | 2 |
| 2 | -3 | 4 | 3 | 4 |
| 3 | -2 | 7 | -6 | 8 |
| 4 | 14 | 1 | 7 | 5 |
| 5 | 6 | 3 | 5 | 2 |
| 6 | 55 | 7 | 16 | 4 |
| 7 | 3 | 1 | -1 | 5 |
| 8 | 3 | 7 | 0 | 8 |
| 9 | 13 | 3 | 7 | 3 |
| 10 | 4 | 4 | -1 | 1 |
| 11 | 2 | 3 | -2 | 5 |
| Assay Z′ | 0.73 | 0.74 | ||
|
Compound |
MCF7 % Cytotoxicity 24 h |
MCF7 % Cytotoxicity 48 h |
||
|---|---|---|---|---|
| AVG | STD | AVG | STD | |
| 1 | 6 | 35 | 0 | 7 |
| 2 | 10 | 8 | 11 | 1 |
| 3 | 9 | 11 | -1 | 5 |
| 4 | 39 | 10 | 32 | 6 |
| 5 | 17 | 3 | 13 | 7 |
| 6 | 65 | 19 | 53 | 8 |
| 7 | 38 | 19 | 4 | 11 |
| 8 | 36 | 2 | 15 | 2 |
| 9 | 22 | 16 | 28 | 7 |
| 10 | 37 | 15 | 4 | 8 |
| 11 | 24 | 6 | 3 | 12 |
| Assay Z′ | 0.45 | 0.65 | ||
|
Compound |
U2OS % Cytotoxicity 24 h |
U2OS % Cytotoxicity 48 h |
||
|---|---|---|---|---|
| AVG | STD | AVG | STD | |
| 1 | 2 | 10 | -6 | 3 |
| 2 | -8 | 21 | 0 | 10 |
| 3 | 16 | 28 | -4 | 3 |
| 4 | 0 | 12 | 14 | 2 |
| 5 | 33 | 8 | 3 | 3 |
| 6 | 34 | 14 | 13 | 2 |
| 7 | 8 | 15 | 3 | 4 |
| 8 | 9 | 32 | 8 | 2 |
| 9 | 7 | 8 | 4 | 4 |
| 10 | -2 | 6 | 4 | 1 |
| 11 | 2 | 7 | 3 | 4 |
| Assay Z′ | 0.59 | 0.79 | ||
| Compound | CYP1A2 % Inhibition |
CYP2C9 % Inhibition |
CYP2C19 % Inhibition |
CYP2D6 % Inhibition |
CYP3A4 % Inhibition |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| AVG | STD | AVG | STD | AVG | STD | AVG | STD | AVG | STD | |
| 1 | 48 | 48 | 13 | 5 | 7 | 4 | -15 | 5 | 24 | 20 |
| 2 | 1 | 6 | 33 | 5 | 35 | 4 | 7 | 9 | 28 | 4 |
| 3 | 19 | 4 | 14 | 11 | 32 | 5 | 6 | 7 | 86 | 1 |
| 4 | 10 | 1 | 13 | 7 | -2 | 7 | -13 | 10 | 30 | 8 |
| 5 | 62 | 2 | 100 | 0 | 99 | 0 | 83 | 1 | 96 | 1 |
| 6 | 97 | 0 | 100 | 0 | 100 | 0 | 89 | 1 | 97 | 0 |
| 7 | 82 | 2 | 99 | 0 | 100 | 0 | 51 | 12 | 70 | 3 |
| 8 | 91 | 0 | 97 | 0 | 98 | 0 | 56 | 5 | 67 | 2 |
| 9 | 18 | 5 | 56 | 2 | 39 | 1 | 47 | 3 | 47 | 2 |
| 10 | 6 | 2 | -3 | 2 | -28 | 11 | -30 | 14 | -2 | 9 |
| 11 | 14 | 4 | 43 | 2 | 8 | 4 | -15 | 29 | 9 | 7 |
| Assay Z′ | 0.83 | 0.76 | 0.83 | 0.78 | 0.77 | |||||
| Compound | HDAC4 % Inhibition |
HDAC6 % Inhibition |
HDAC8 % Inhibition |
HDAC9 % Inhibition |
SIRT7 % Inhibition |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| AVG | STD | AVG | STD | AVG | STD | AVG | STD | AVG | STD | |
| 1 | -15 | 0 | -13 | 7 | -2 | 2 | -21 | 1 | 7 | 8 |
| 2 | -12 | 1 | -8 | 4 | 11 | 21 | -14 | 1 | 5 | 2 |
| 3 | -9 | 1 | -20 | 18 | 1 | 3 | -9 | 0 | 6 | 5 |
| 4 | 14 | 2 | -19 | 11 | 17 | 1 | 4 | 5 | 5 | 9 |
| 5 | -10 | 1 | -15 | 10 | -2 | 2 | -10 | 1 | 6 | 3 |
| 6 | -11 | 3 | -32 | 3 | 1 | 4 | -15 | 1 | 5 | 2 |
| 7 | -7 | 1 | -21 | 2 | -2 | 2 | -10 | 1 | 5 | 1 |
| 8 | -2 | 3 | -16 | 8 | 2 | 4 | -2 | 3 | -9 | 30 |
| 9 | 15 | 1 | -26 | 8 | 12 | 2 | 11 | 4 | 10 | 7 |
| 10 | -8 | 3 | -17 | 5 | 3 | 2 | -11 | 3 | 6 | 2 |
| 11 | 13 | 7 | -33 | 14 | 5 | 4 | 21 | 2 | 9 | 1 |
| Assay Z′ | 0.72 | 0.83 | 0.85 | 0.71 | 0.89 | |||||
| Compound | Aurora B kinase % Inhibition |
PDE4C1 % Inhibition |
||
|---|---|---|---|---|
| AVG | STD | AVG | STD | |
| 1 | 6 | 1 | 7 | 24 |
| 2 | 3 | 2 | 23 | 20 |
| 3 | 4 | 2 | 31 | 4 |
| 4 | 14 | 3 | 15 | 2 |
| 5 | 7 | 4 | 26 | 14 |
| 6 | 4 | 5 | 23 | 32 |
| 7 | 10 | 6 | 45 | 20 |
| 8 | 8 | 3 | 17 | 16 |
| 9 | 18 | 6 | 17 | 8 |
| 10 | 14 | 3 | 15 | 2 |
| 11 | 18 | 6 | 18 | 8 |
| Assay Z′ | 0.81 | 0.62 | ||
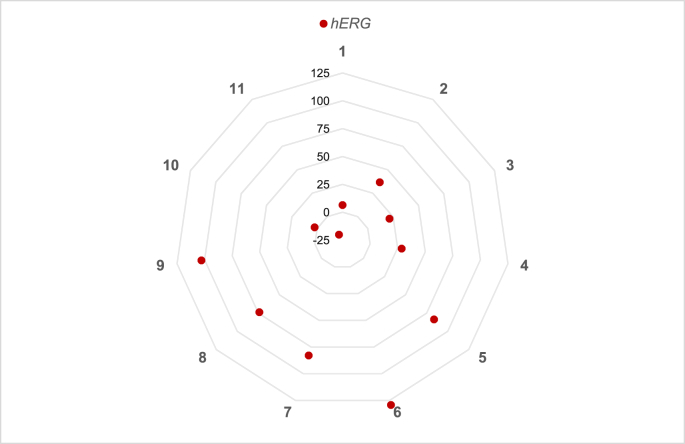
| Compound |
hERG % Inhibition |
|
|---|---|---|
| AVG | STD | |
| 1 | 6 | 16 |
| 2 | 37 | 6 |
| 3 | 21 | 5 |
| 4 | 29 | 8 |
| 5 | 84 | 5 |
| 6 | 129 | 14 |
| 7 | 83 | 10 |
| 8 | 74 | 7 |
| 9 | 103 | 14 |
| 10 | 3 | 16 |
| 11 | -19 | 16 |
| Assay Z′ | 0.43 | |
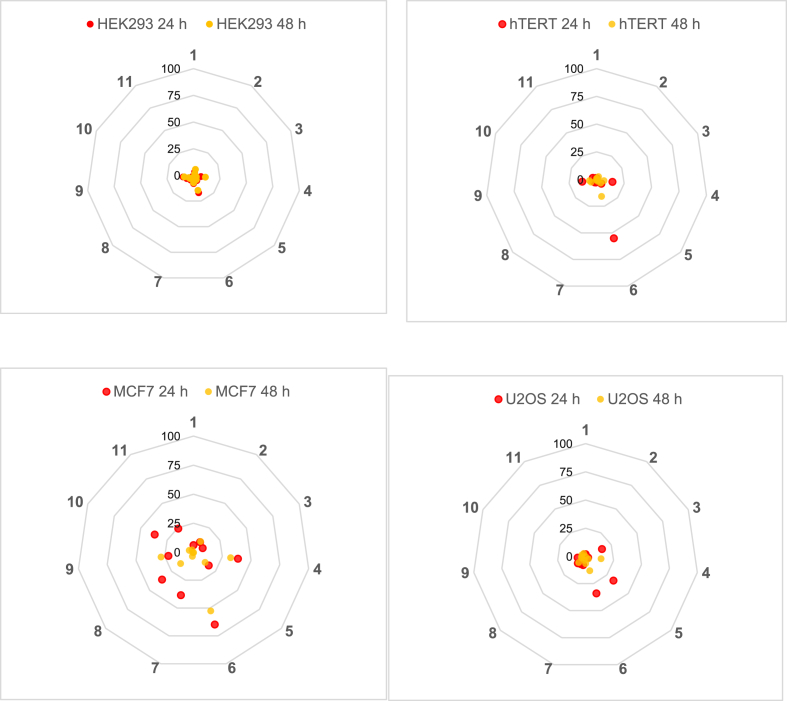
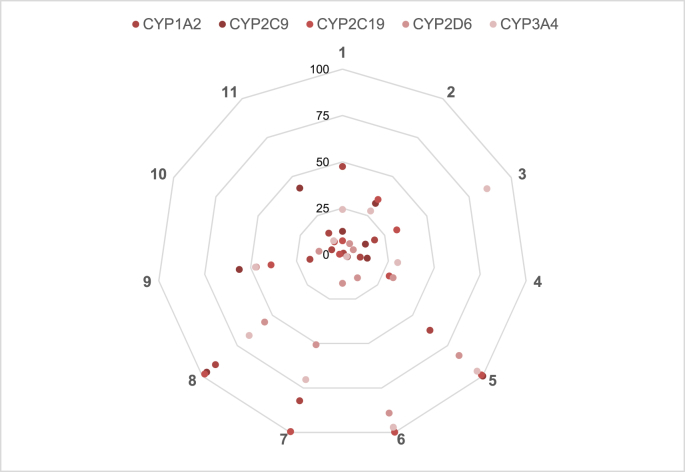
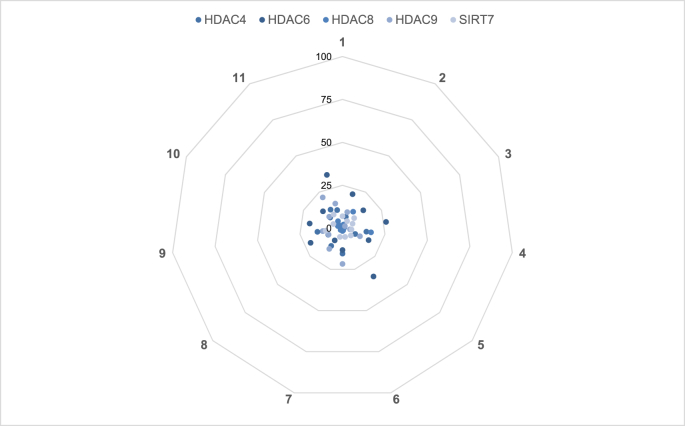
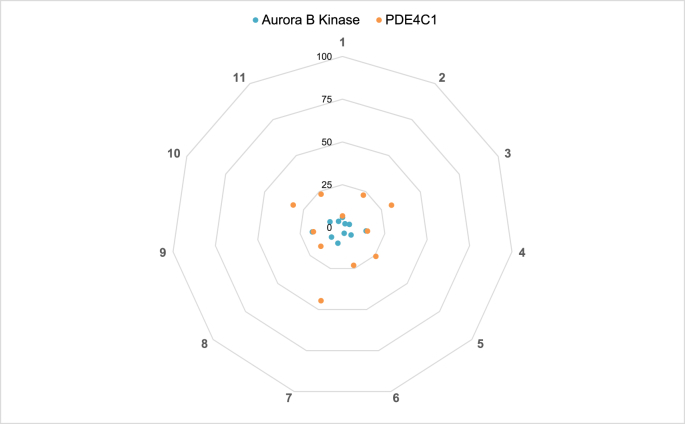
Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5 consist of data collected in Table 1 organized in the form of spider plots in order to optimize the visualization of generated data from each early toxicity and off-target liability assay.
Fig. 1.
Cytotoxicity of new thyromimetics. The % Cytotoxicity determinations were made in four different cell-lines (HEK293, hTERT, MCF7, and U2OS) at 24 h (red) and 48 h (yellow). See Table 1 for further details.
Fig. 2.
CYP450 liability of new thyromimetics. % Inhibition of five CYP450 enzymes (CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP3A4). See Table 1 for further details.
Fig. 3.
Epigenetic off-target enzyme liability of new thyromimetics. % Inhibition of epigenetic off-targets (HDAC4, HDAC6, HDAC8, HDAC9 and SIRT7). See Table 1 for further details.
Fig. 4.
Aurora B kinase and PDE4C1 off-target enzyme liability of new thyromimetics. % Inhibition of Aurora B kinase and PDE4C1. See Table 1 for further details.
Fig. 5.
hERG liability of new thyromimetics. % Inhibition of hERG. See Table 1 for further details.
Fig. 1 depicts the cytotoxicity effects assessed for each compound at 24 and 48 hours of treatment in different cell cultures.
In Fig. 2 data on metabolic interference of thyromimetics towards CYP450 isoforms are reported.
Fig. 3 depicts data collected on possible interaction between tested compounds and HDACs and SIRT7 activity, while Fig. 4 is a representation of inhibitory effect of tested compounds on Aurora B kinase and PDE4C1.
Finally, data collected on potential inhibitory effect of test compounds on hERG ion-channel are reported in Fig. 5.d
2. Experimental design, materials, and methods
2.1. Protocols for in vitro early toxicity and off-target liability assays
2.1.1. Cytotoxicity assays
Test compounds (10 μM final concentration, 0.1% of DMSO), negative control (Valinomycin, 10 μM final concentration, 0.1% of DMSO), or DMSO as positive control were dispensed using an Echo 550 Liquid Handler (20 nL/well) into white 384-well microtiter plates. Cell-lines (osteosarcoma, U2OS; lung fibroblast, hTERT; human breast adenocarcinoma, MCF7; human embryonic kidney, HEK293) were from ATCC and grown on surface-modified T175 cell culture flasks in Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS), 5% of l-glutamic acid, 5% of antibiotics (streptomycin and penicillin G). At about 80% confluency, cells were washed, trypsinized, and resuspended. Then, cells were counted in DMEM and seeded at 4000 cells/well (in triplicate) in plate containing test compounds and controls (20 μL/well). Plates were incubated at 37 °C in presence of 5% CO2 for 24 h or 48 h. Then again, 20 μL/well of CTG detection mix from CellTiter-Glo Assay Kit (Promega Corp.) were added, and plates were gently mixed, incubated for 10-min in the dark and read using an EnVision Multilabel 2103 reader (PerkinElmer). Raw data were normalized to percentage of cell growth by using the corresponding NC containing only 0.1% v/v DMSO. The luminescence signal of each sample (S) was converted into percentage of cell growth compared with the average signal of NC. The following formula was used: % effect = (S – PC)/NC × 100.
2.1.2. hERG cardiotoxicity assay
Compounds were tested for potential cardiotoxicity (in triplicate) using The Predictor hERG fluorescence polarization assay (Thermo). 100 nL of the test/control compound was added to each well of an assay plate, followed by addition of 5 μL homogenized membrane solution (undiluted) and 5 μL of tracer (1 nM final concentration in assay). Then, plates were incubated for 2 h at 25 °C in a humidity-controlled incubator, and the fluorescence polarization was measured using an EnVision Multilabel 2103 Reader (PerkinElmer). The NCs (0% inhibition) and positive controls with E−4031, a blocker of hERG-type potassium channels (yielding 100% inhibition), were used to normalize the raw data.
2.1.3. Cytochrome (CYP) P450 assays
These assays (at least in duplicate) make use of microsomal preparations of CYP450 (1A2, 2C9, 2C19, 2D6, and 3A4 isoforms) from baculovirus-infected insect cells (Corning Inc.) and cytochrome c reductase (and cytochrome b5 for CYP450 3A4). For detection of CYP450 activity, the luminescence-based P450-Glo (Promega Corp.) assay system was used that contained a luminogenic CYP450 substrate, lyophilized luciferin detection reagent, and reconstitution buffer. The substrates were luciferin derivatives of CYP450-specific substrates that produce (4S)-4,5-dihydro-2-(6-hydroxybenzothiazolyl)-4-thiazolecarboxylic acid (D-luciferin) after cleavage by CYP450 (CYP450 1A2 luciferin-ME; CYP450 3A4, luciferin-IPA; CYP450 2C19, luciferin-H EGE; CYP450 2C9, luciferin-H; CYP450 2D6, luciferin-ME EGE). CYP450 reactions were initiated by addition of the NADPH regeneration system to the enzyme-substrate mixture with the luciferin detection reagent stopping the reaction and the D-luciferin being converted to oxyluciferin under production of light being proportional to the CYP450 activity. Compounds were added into an empty 384-well plate (10 nL/well in 0.1% v/v DMSO) using the Echo 550 Liquid Handler followed by addition of 5 μL/well of CYP450/substrate mixture and incubation for 30 min at 37 C, after which the reaction was initiated by addition of 5 μL/well NADPH regeneration system. After a further 30 min incubation at 37 °C, the CYP450 reaction was stopped, and the luciferase reaction was simultaneously initiated by addition of 10 μL/well of luciferin detection reagent, followed by an additional 30 min incubation at 37 °C. The luminescence signal was detected using an EnVision Multilabel 2103 reader (PerkinElmer).
2.1.4. HDAC4, HDAC6, HDAC8 and HDAC9 assays
Inhibition of HDAC enzymes (in triplicate) was determined using the homogeneous, single-addition, bioluminogenic HDAC-Glo I/II assay (Promega Corp.) for HDAC6 and HDAC8 or the HDAC-Glo 2A assay (Promega Corp.) for HDAC 4 and HDAC9. Human recombinant HDAC enzymes (BPS Bioscience), and standard inhibitor trichostatin A (Sigma-Aldrich) was dissolved to a yield stock solution in 100% v/v DMSO and stored at −20 °C. Plate handling was performed using an Echo 550 Liquid Handler and luminescence measurements taken using a 2300 EnSpire Multilabel reader (PerkinElmer). Test compounds and positive control (Trichostatin A with final concentration of 10 μM and 0.1% v/v DMSO) and high control (final 0.1% v/v DMSO) were added into the 384-well plates (10 nL/well; 0.1% v/v DMSO) using the Echo 550 Liquid Handler. Enzyme solutions containing optimized concentration of substrate and enzyme were prepared according to each kit. 5 μL of each solution were added to each well (in assay concentrations were 0.002 nM for HDAC4, 2.5 nM for HDAC6, 0.2 nM for HDAC8, and 0.10.025 nM for HDAC9). The HDAC-Glo I/II assay reagent and HDAC-Glo 2A assay reagent were prepared according to each kit manual and 5 μL of them were added to each well. The microtiter plates were mixed briefly by orbital shaking (500–700 rpm), and luminescence was measured at steady-state signal: background, which was achieved after 20 min.
2.1.5. SIRT7 assay
Inhibitory effect of test compounds on SIRT7 enzyme (in triplicate) was determined using the SIRT-Glo assay kit (Promega Corp.). Human recombinant SIRT7 enzyme (BPS Bioscience). Plate handling was performed using an Echo 550 Liquid Handler and luminescence measurements taken using a 2300 EnSpire Multilabel reader (PerkinElmer). Test compounds and DMSO as control were added into the 384-well plates (10 nL/well; final concentration of 10 μM; 0.1% v/v DMSO) using the Echo 550 Liquid Handler. SIRT-Glo reagent was prepared as described in the manual kit and added to each well after 20 min of incubation (10 μL/well). The microtiter plates were mixed briefly by orbital shaking (500–700 rpm), and luminescence was measured at steady-state signal: background, which was achieved after 30 min.
2.1.6. PDE4C1 assay
Inhibitory effect of test compounds on PDE4C1, a protein necessary to assure normal cell growth, was determined (in triplicate) using the PDE4C1 assay kit (BPS Bioscience). Test compounds (10 nL, in assay concentration of 10 μM and 0.1% v/v DMSO) and DMSO as control were dispensed into a black 384-well no-binding low-volume plate. Substrate and enzyme solution were prepared and 2.5 μL of each mix were dispensed in each well, and the plate was gently mixed and incubated for 1 h at rt in the dark. Then, binding agent solution were prepared according to kit protocol, and 10 μL of this solution were added to each well. The plate was then sealed, centrifuged and incubated for 20 min at rt with slow shaking. Fluorescence polarization measurement were performed on an EnVision Multilabel 2103 reader (PerkinElmer). Each signal was normalized to positive (100% inhibition, no substrate) and to negative control (0% inhibition, DMSO control).
2.1.7. Aurora B assay
Inhibitory effect of test compounds on Aurora B kinase was determined (in triplicate) using the Kinase-Glo® Luminescent Kinase Assay Platform (Promega Corp.). 5 nL of compound dissolved in 100% DMSO and DMSO as control were dispensed to each well of a 384-well plate (final concentration 10 μM; final concentration of 0.1% v/v DMSO) using an Echo 550 Liquid Handler. An enzyme master mix containing 1× buffer, 50 μM DTT, and 300 nM Aurora B (all reagents provided in the kit) was prepared and 2.5 μL of solution were added to each well. 2.5 μL of reaction mix containing of 1× buffer, 3.33 μM adenosine triphosphate (ATP), and 7.5 ng/μL (18.75 ng/well) myelin basic protein (MBP) as substrate (buffer and MBP were provided in the Aurora B Kinase Enzyme System (Promega Corp.); ultrapure ATP (Sigma Aldrich) were added to each well in order to start reaction. Plate was then sealed using thermowell sealing taper, briefly mixed, and incubated for 30 min at rt. Following this, 5 μL of Kinase-Glo® Reagent were added to each well, the plate was then mixed and incubated at rt for 20 min, and luminescence measurements were read using a 2003 EnSpire Multilabel reader (PerkinElmer). The luminescence signal of each sample (S) was converted into percentage of Aurora B inhibition and compared with the average signal of no substrate (100% Inhibition).
Acknowledgments
We thank the COST action CA15135 (Multitarget Paradigm for Innovative Ligand Identification in the Drug Discovery Process MuTaLig) for support the working experience of MR at the Fraunhofer-IME SP (Hamburg, Germany). We also thank POR FSE 2014–2020 – Regione Toscana and the University of Pisa for financing the fellowship to SS.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2020.105206.
Contributor Information
Sheraz Gul, Email: Sheraz.Gul@ime.fraunhofer.de.
Simona Rapposelli, Email: simona.rapposelli@unipi.it.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Runfola M., Sestito S., Bellusci L., La Pietra V., D’amore V.M., Kowalik M.A., Chiellini G., Gul S., Perra A., Columbano A., Marinelli L., Novellino E., Rapposelli S. Design, synthesis and biological evaluation of novel TRβ selective agonists sustained by ADME-Toxicity analysis. Eur. J. Med. Chem. 2020 Feb 15;188:112006. doi: 10.1016/j.ejmech.2019.112006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.