Abstract
Vascular endothelial growth factors (VEGFs) are hypoxia-inducible secreted proteins to promote angiogenesis, in which VEGF-A is an important molecule that binds and activates VEGF receptor-1 (VEGFR-1) and VEGFR-2. In this study, two DNA aptamers, Apt01 and Apt02, were successfully isolated by alternating consecutive systematic evolution of ligands by exponential enrichment (SELEX) against VEGFR-1 and -2 using deep sequencing analysis in an early selection round. Their binding affinities for VEGFR-2 were lower than that of VEGFR-1, which is similar to that of VEGF-A. Structural analyses with the measurements of circular dichroism spectra and ultraviolet melting curve showed that Apt01 possessed the stem-loop structure in the molecule, whereas Apt02 formed G-quadruplex structures. In addition, Apt02 accelerated a tube formation of human umbilical vein endothelial cells faster than Apt01, which was affected by difference of binding affinity and nuclease resistance due to G-quadruplex structures. These results demonstrated that Apt02 might have a potential to function as an alternative to VEGF-A.
Keywords: aptamer, vascular endothelial growth factor, VEGF, VEGF receptor, angiogenesis, G-quadruplex
Graphical Abstract
Introduction
Vascular endothelial growth factors (VEGFs) are hypoxia-inducible secreted proteins to promote angiogenesis.1 Among them, VEGF-A is the major contributor to physiological and pathological angiogenesis.2 VEGF-A binds and activates vascular endothelial growth factor receptor-1 (VEGFR-1) and VEGFR-2, which are members of the family of receptor tyrosine kinases on endothelial cells. VEGFR-1 and -2 share 43.2% overall sequence similarity.2 The sequences of their extracellular domains are 33.3% similar, and those of their kinase domains are the most similar (70.1%).2 VEGF-A binds to VEGFR-1 with at least 10-fold higher affinity (dissociation constant [KD]: 2–10 pM) than VEGFR-2.3 However, most studies showed that VEGFR-2 is the functional receptor for VEGF-A, which transmits signals that regulate the proliferation and differentiation of endothelial cells. In contrast, there is controversy about the importance of VEGFR-1 in vascular remodeling, macrophage function, inflammatory diseases, and metastasis of cancer.3,4
Recently, VEGF-A has been used as one of the essential molecules for induction of differentiation to endothelial5,6 and cardiac myocyte7 cells from human induced pluripotent stem cells and embryonic stem cells. However, intrinsic drawbacks of recombinant VEGF-A, such as low stability, lot-to-lot variation, and the high cost of production, have been serious problems for use of stem cell-based regenerative therapy and relevant basic research. Therefore, replacing expensive and unstable recombinant protein such as VEGFs with synthetic alternatives is important in the biomedical fields, especially stem cell-based regenerative medicines.8, 9, 10
DNA aptamers are single-stranded oligonucleotides with high affinity to specific targets, such as proteins11 and entire cells.12 DNA aptamers offer several specific advantages owing to their low cost of production and high thermal stability.13 These features are beneficial for applications as therapeutic agents. DNA aptamers are isolated by systematic evolution of ligands by exponential enrichment (SELEX).14,15 The DNA aptamers that bind VEGFR-1 and -2 may serve as the alternatives to VEGF-A. However, compared with SELEX to isolate aptamers that bind to a single target, the consecutive SELEX against multiple target molecules requires numerous selection rounds.16 In addition, in the course of SELEX with multiple rounds, selective pressure shifts the sequence of aptamers from the one with high affinity to the one that is amplified with higher efficiency.16 This PCR amplification bias tends to favor shorter as well as structurally unstable sequences.16 Therefore, rapid acquisition of aptamer candidates within fewer selection rounds is desired for avoiding PCR amplification bias.12,17
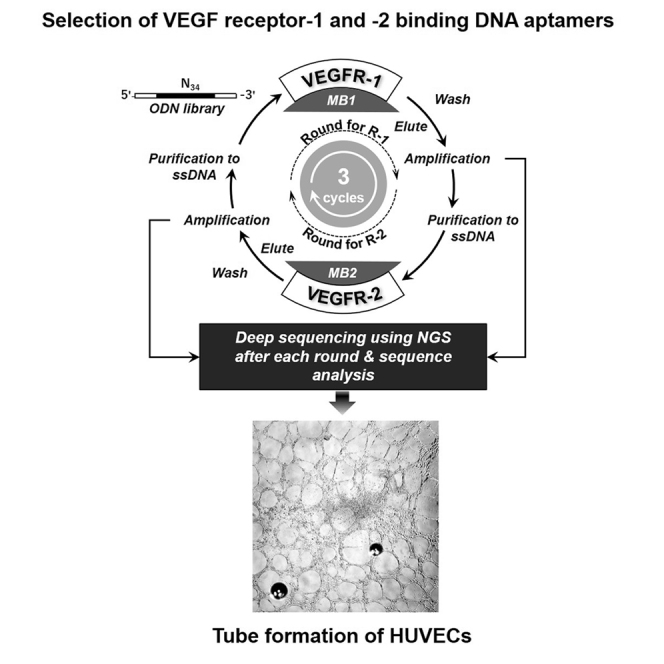
To obtain DNA aptamers with function similar to VEGF-A, in this study, we conducted alternating consecutive SELEX of DNA aptamers against double targets, VEGFR-1 and -2, using deep sequencing analysis with next generation sequencing (NGS).
Results and Discussion
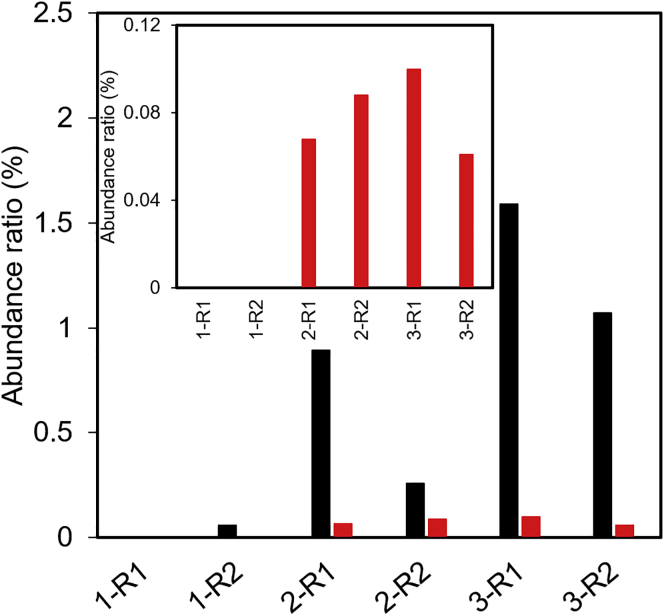
As an initial pool, 70-mer single-stranded oligodeoxynucleotides (ssODNs) containing a random 34-mer sequence with flanking primer regions were used. This ODN library was amplified using alternating consecutive SELEX against VEGFR-1 and -2 immobilized on magnetic beads (MBs), referred to as MB1 and MB2, respectively. The ODN library was first mixed with MB1, and the adsorbed ODNs on the surface of MBs were eluted at 90°C after washing the MB. The small amount of ODNs eluted from the MBs was amplified using PCR, and the enriched double-stranded ODNs (dsODNs) were converted to ssODNs using streptavidin-coated MB, followed by NaOH elution. To achieve alternating consecutive selection against VEGFR-1 and -2, the selection against MB2 was conducted using the amplified ssODN library in the same manner. The process until this selection step is called a “first cycle.” In this study, selection was carried out until third cycles. The sequences of the dsODNs obtained after each round were determined using deep sequencing with NGS. Although, in conventional SELEX, each aptamer sequence can be determined by subcloning and Sanger sequencing, the read number of sequences is limited owing to the colony-picking operation (approximately 50–100 sequences per sample). On the other hand, approximately 107–109 sequences per run are obtained by using deep sequencing with NGS.18 In this study, by using deep sequencing with an Ion PGM with a 314 chip (Life Technologies, USA), readouts of 20,000–50,000 sequences per sample at each round were obtained. Among the obtained readout, we could isolate two aptamers, Apt01 (5′-GTCGTGTTTGTTGTTGTTTTCATTTTTGCGGCCC-3′) and Apt02 (5′-GCTGATAGGATGGGTTGTAGGTCTAGGGGGGGGCC-3′). Their abundance distributions at each round are summarized in Tables S2 and S3, and the changes in the abundance ratios are shown in Figure 1. Although the abundance ratios of both sequences increased with increasing number of rounds, the abundance ratio of Apt01 was higher than that of Apt02. The abundance ratio of Apt01 started to increase from round 2-R1 and reached a maximum of 1.6% after round 3-R1. In contrast, the abundance ratio of Apt02 at each round was ≤0.1% and started to increase from round 2-R1 and reached the maximum (0.1%) after round 3-R1. Probably, the identification of aptamers with these extremely small abundance ratios such as Apt02 must be accomplished by using sequencing analysis with deep sequencing.
Figure 1.
Change in the Abundance Ratio of Apt01 (Black) and Apt02 (Red) at Each Round
In the x axis, first number means the number of cycles, and R1 and R2 indicate selection against VEGFR-1 and -2 at each round, respectively. Inset: enlarged graph of Apt02. The abundance ratio of Apt01 was higher than that of Apt02.
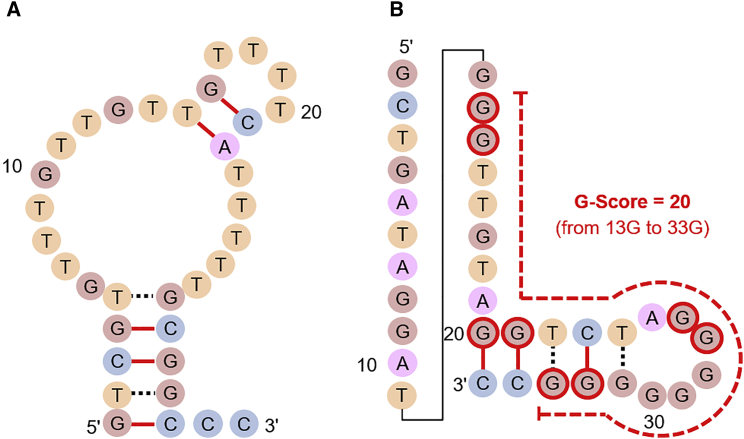
To investigate the binding parameters of the obtained aptamers, we carried out surface plasmon resonance (SPR) analysis (Figure S2). The DNA aptamers, Apt01 and Apt02, had high affinities for VEGFR-1 and -2 as follows: the KD values of Apt01 for VEGFR-1 and -2 were 3.3 and 100 nM, respectively, and the KD values of Apt02 for VEGFR-1 and -2 were 1.5 and 32 nM, respectively (Table 1). The dissociation rate constants (koff) of Apt01 and Apt02 were comparable with VEGF-A, whose koff is 1.32 × 10−3/s, for VEGFR-1 and VEGFR-2.19 This suggested that Apt01 and Apt02 possess the potential to activate the VEGR receptor, similar to VEGF-A. In addition, the binding affinities of sequence-scrambled versions of Apt02 (Apt02-scr: 5′-GGTGTGGCGGAAGGGTGGTCTATTGTGGGGAGCAC-3′) and 35-mer of adenines (A35: AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA) were also measured by SPR (Figure S3). The Apt02-scr and A35 showed lower affinity compared with Apt02. Although there is an overall positive charge in the extracellular domain of VEGFR-1,20 this result showed that the obtained aptamers have specific interaction with VEGFR-1, not electrostatic interactions. In both cases, the binding affinities for VEGFR-2 were lower than that of VEGFR-1. Thus, the binding behavior of these aptamers to VEGFRs is also similar to that of VEGF-A. Although the abundance ratio of Apt02 was significantly lower than that of Apt01, the affinity of Apt02 for VEGFRs was higher than that of Apt01. To define the differences in abundance ratios and binding affinities between Apt01 and Apt02, we conducted structural characterization. First, we used m-fold that is software applications available on the World Wide Web for the prediction of secondary structure of single-strand nucleic acids.21 As shown in Figure 2, it demonstrated that both Apt01 and Apt02 have a potential to form stem-loop structures. Next, we focused on the sequence of Apt02; G-rich sequence was included in Apt02, but not in Apt01. To identify potential G-quadruplex-forming sequences in Apt02, we used the prediction tool of G-quadruplex, QGRS mapper.22 QGRS mapper indicated the likelihood of two G-tetrad layers from guanine residue 13 to guanine residue 33 in Apt02 sequence, a G score of 20 (Figure 2).
Table 1.
Association/Dissociation Rate Constants and Binding Affinities of Apt01 and Apt02, which Were Analyzed by SPR
| Aptamer Name | Target | kon/M−1s−1 | koff/s−1 | KD (nM) |
|---|---|---|---|---|
| Apt01 | VEGFR-1 | 4.73 × 106 | 1.57 × 10−2 | 3.3 |
| Apt01 | VEGFR-2 | 3.73 × 104 | 3.86 × 10−3 | 100 |
| Apt02 | VEGFR-1 | 9.54 × 105 | 1.39 × 10−3 | 1.5 |
| Apt02 | VEGFR-2 | 1.32 × 105 | 4.16 × 10−3 | 32 |
SPR sensorgrams are shown in Figure S2.
Figure 2.
Secondary Structures of the Aptamers.
(A and B) Secondary structures of (A) Apt01 and (B) Apt02 estimated using m-fold. G-rich sequences were included in Apt02, but not in Apt01. G scores of Apt01 and Apt02 estimated by QGRS mapper were 0 and 20, respectively. G-quadruplex-forming guanine residues in the Apt02 sequence, from 13G to 33G, were estimated by QGRS mapper and indicated as red-edged circles.
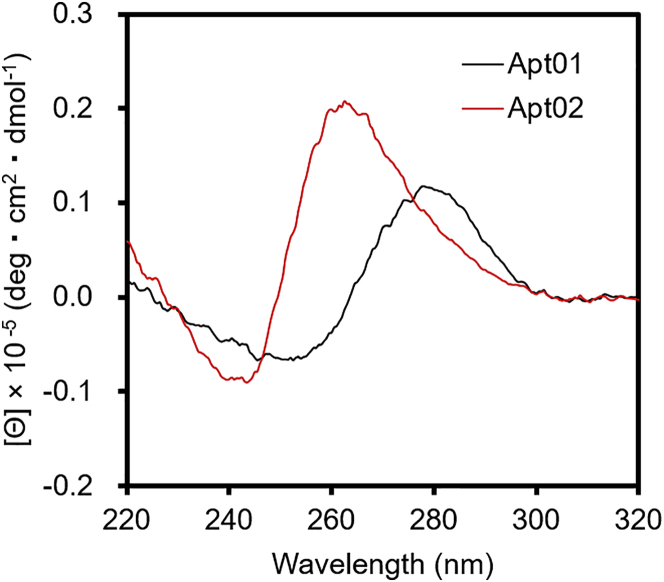
To confirm the formation of G-quadruplexes experimentally, we measured the circular dichroism (CD) spectra of Apt01 and Apt02. CD spectra provide reliable information for identifying DNA structures, and thus are useful for characterizing the G-quadruplex structure.23 Figure 3 shows the CD spectra of Apt01 and Apt02. CD spectrum of Apt02 showed negative and positive peaks at approximately 240 and 260 nm, respectively. On the other hand, Apt01 showed negative and positive peaks at approximately 250 and 280 nm, respectively. Because the CD spectra of parallel-type G-quadruplexes are reported to show negative and positive peaks at approximately 240 and 260 nm, respectively,24 it was suggested that Apt02 formed parallel-type G-quadruplex structures. On the other hand, because typical dsODNs show the negative and positive peaks at approximately 250 and 280 nm, respectively,25 Apt01 might form a stem-loop structure, as shown in Figure 3.
Figure 3.
Circular Dichroism (CD) Spectra of the Aptamers in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.5 mM NaH2PO4 [pH 7.4]).
CD spectrum of Apt02 (red) showed negative and positive peaks at approximately 240 and 260 nm, respectively, which corresponded with the spectrum of parallel-type G-quadruplexes. On the other hand, Apt01 (black) showed negative and positive peaks at approximately 250 and 280 nm, respectively, which corresponded with the spectrum of the typical double-stranded oligodeoxynucleotides.
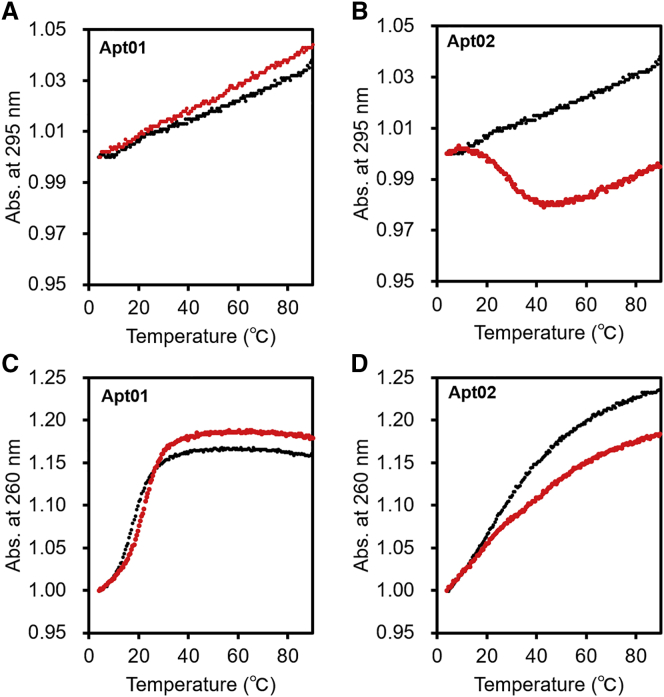
To further define the structures of Apt01 and Apt02, we measured the ultraviolet (UV) melting curves of Apt01 and Apt02. Generally, the melting profile of G-quadruplexes shows a hypochromic sigmoidal transition at 295 nm.26 As shown in the black colored plots of Figures 4A and 4B, not only Apt01 but also Apt02 did not show any transition at 295 nm in 10 mM phosphate buffer containing 100 mM NaCl. As shown in the red colored plots of Figures 4A and 4B, by using 10 mM phosphate buffer containing 100 mM KCl instead of 100 mM NaCl, the hypochromic transition was observed only in the UV melting curve of Apt02 at 295 nm because the structure of G-quadruplexes was stabilized by the presence of potassium ions.26,27 Melting temperature (Tm) value of Apt02 at 295 nm was estimated to be 28.2°C. This result and the CD spectrum of Apt02 shown in Figure 3 strongly suggested that Apt02 possessed a G-quadruplex-forming motif in its structure. On the other hand, the UV melting curves of Watson-Crick duplexes exhibit a hyperchromic transition at 260 nm.26 In the UV melting curves of Apt01 at 260 nm, hyperchromic transitions with a single sigmoidal shape were observed in phosphate buffer containing NaCl and in phosphate buffer containing KCl (Figure 4C), and their Tm values were estimated to be 21.6°C and 17.6°C, respectively. This result indicated that Apt01 possessed the stem-loop structure in the molecule. On the other hand, the melting curves of Apt02 exhibited a gradual increase without the sigmoidal shape at 260 nm; in addition, the variation was reduced in the presence of potassium ions (Figure 4D). Because similar variation of absorbance at 260 nm upon G-quadruplex dissociation has so far been reported,26 Apt02 must form G-quadruplex structures.
Figure 4.
Ultraviolet (UV) Melting Profiles of the Aptamers
(A–D) The UV melting profiles of aptamers in 10 mM phosphate buffer containing 100 mM NaCl (black colored plots) or in 10 mM phosphate buffer containing 100 mM KCl (red colored plots): (A) Apt01 at 295 nm, (B) Apt02 at 295 nm, (C) Apt01 at 260 nm, and (D) Apt02 at 260 nm. The melting profiles normalized by the absorbance at 5°C are shown. The hypochromic transition was observed only in the UV melting curve of Apt02 at 295 nm in 10 mM phosphate buffer containing 100 mM KCl, indicating that Apt02 possessed G-quadruplex. In the UV melting curves of Apt01 at 260 nm, hyperchromic transitions with a single sigmoidal shape, indicating that Apt01 possessed the stem-loop structure in the molecule.
By the results of their structural analyses, the significant lower abundance ratio of Apt02 than Apt01 can be explained. Despite several approaches used to improve PCR, amplification of long, GC-rich, repetitive fragments is challenging and impossible in some cases.17,28 Although the ODNs with G-quadruplex structure are generally known as advantageous architectures for molecular recognition,29,30 their amplification tends to be affected by amplification bias due to high guanine content. However, in the present study, despite alternating consecutive SELEX against double targets, Apt02 with these extremely small abundance ratios could be identified even at earlier rounds by using deep sequencing analyses with NGS.
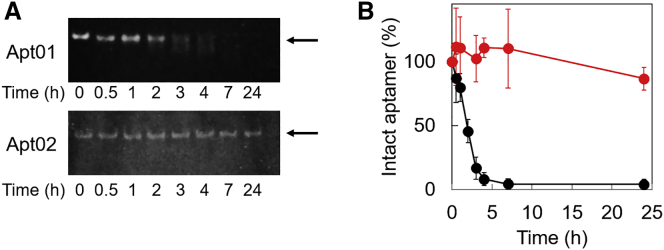
To investigate a nuclease resistance of aptamers, Apt01 and Apt02, we evaluated the stability of the obtained aptamers in cell culture medium. As shown in Figure 5, almost all of Apt01 was degraded in cell culture medium even at 4 h. In contrast, Apt02 was not degraded in cell culture medium at 4 h. Even at 24 h, 86% of Apt02 remained in the culture medium. Because Apt02 formed parallel-type G-quadruplex structures, Apt02 showed higher nuclease resistance than Apt01.
Figure 5.
Stability of the Obtained Aptamer, Apt01 and Apt02, during Incubation in Medium 200PRF with LSGS Kit for 24 h, as Determined by Denaturing Urea Polyacrylamide Gel Electrophoresis (PAGE)
(A) Representative images of denaturing urea PAGE. Bands of intact aptamers are indicated with arrows. (B) Graphical representation of PAGE results. The fraction of intact aptamers (relative to the sample without incubation in Medium 200 with LSGS kit) was plotted as a function of time. Stained DNA was imaged with Digi-Gel Shot (Takara Bio) and quantified using ImageJ software package. Almost Apt01 was degraded in cell culture medium even at 4 h, whereas even at 24 h, 86% of Apt02 remained in the culture medium. The bar graphs represent means ± standard error for three independent experiments.
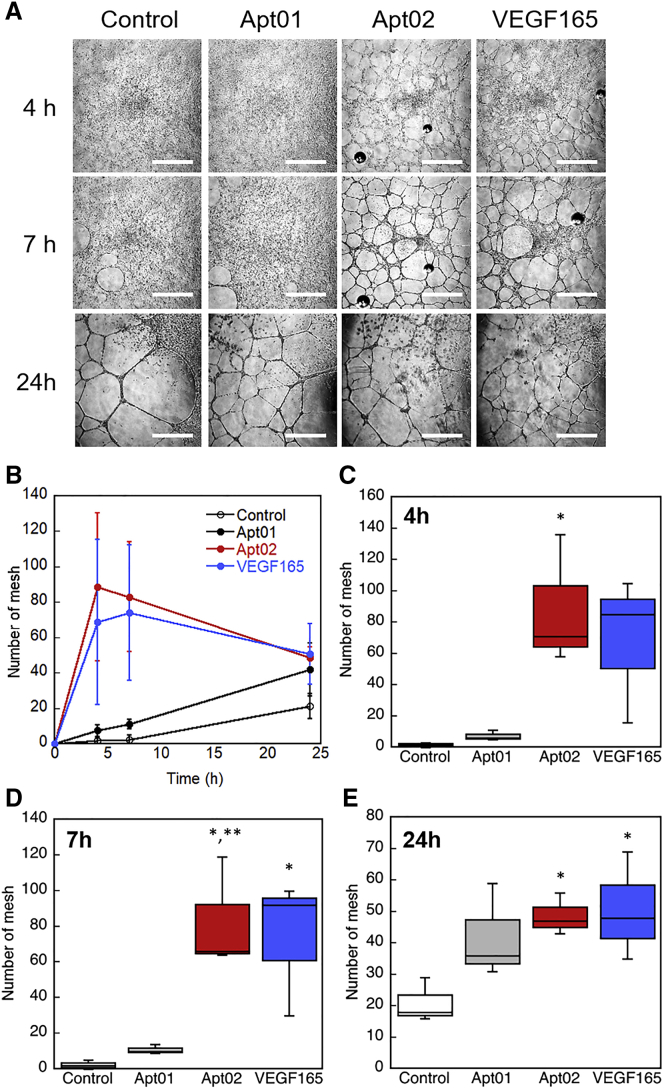
Finally, to evaluate angiogenic activities of the obtained aptamers, Apt01 and Apt02, we performed in vitro tube formation assay using aptamer-treated human umbilical vein endothelial cells (HUVECs) on a three-dimensional gel consisting of Matrigel (Figure 6A). Figure 6B shows the time course of mesh number in the images, and Figures 6C–6E shows the boxplot showing mesh number in the images of the HUVEC networks at 2, 4, 7, and 24 h, respectively. HUVECs formed a capillary-like network about 24 h after plated on a Matrigel (Figure 6A, control). After formation of the tube on the Matrigel, tubes were gradually broken (data not shown). VEGF165-treated HUVECs, which were used as a positive control, started to form capillary-like networks at 4 h; then almost tubes started to be broken at 24 h (Figures 6A and 6B, VEGF165). When HUVECs were plated on a Matrigel in the presence of Apt02 at the concentration of 10 μM, cells started to form a capillary-like network at 4 h, and their tubes started to be broken at 24 h (Figures 6A and 6B, Apt02). On the other hand, in the presence of Apt01 at the concentration of 10 μM, the capillary-like network was not observed at 4 h and formed at 24 h (Figures 6A and 6B, Apt01). The tube formation seemed to be accelerated by Apt02, as well as VEGF165 (Figures 6C to 6E), although there is no significant difference between control and Apt01 (Figure 6F). In addition, we investigated the effect of oligonucleotide sequence with low affinity in tube formation. The addition of 35-mer of adenines (A35: AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA), which was used as a negative control owing to lower affinity (Figure S3), did not affect tube formation of HUVECs (Figure S4). Furthermore, Apt02-induced HUVEC tubule growth was inhibited by a highly selective small-molecule inhibitor of VEGFR tyrosine kinases, fruquintinib31 (Figure S5). These results suggested that Apt02 possessed the strong affinity with VEGFR-1 and -2 on the cells, resulting in the acceleration of tube formation. The difference between Apt01 and Apt02 must be affected by the differences in binding affinity, nuclease resistance property due to G-quadruplex structures, and their binding sites to VEGFRs. Ramaswamy et al.8 have reported the isolation of an antagonistic aptamer to VEGFR-2 by SELEX and developed its dimer to induce tube formation of HUVECs. In contrast, in this study, monomeric Apt02 itself worked as an agonistic aptamer, probably because of the alternating consecutive SELEX of DNA aptamers against double targets, VEGFR-1 and -2. This is first report of direct selection of agonistic aptamer against VEGFR-1 and -2 to induce tube formation of HUVECs.
Figure 6.
In Vitro Tube Formation Assay Using Human Umbilical Vein Endothelial Cells (HUVECs) on a Three-Dimensional Gel Consisting of Diluted Matrigel
The cells were treated with Apt01 (10 μM), Apt02 (10 μM), or VEGF165 (10 ng/mL, 0.26 nM) for 24 h. (A) Representative images of tube formation of HUVECs on Matrigel, which are treated by aptamers or VEGF165. Scale bars, 1 mm. (B) Time course of mesh number in the images of the HUVEC networks at 4, 7, and 24 h. Error bars represent standard deviation of the mean (n = 3 plots). (C–E) Boxplot showing mesh number in the images of the HUVEC networks at (C) 4, (D) 7, and (E) 24 h (n = 3 plots). *p < 0.05 as compared with control; **p < 0.05 as compared with Apt01.
In conclusion, nuclease-resistant G-quadruplex DNA aptamer with VEGF A-mimic activity, Apt02, was isolated by alternating consecutive SELEX against VEGFR-1 and -2 using deep sequencing analysis, which might have a potential to function as the alternative to VEGF-A.
Materials and Methods
In Vitro Selection of DNA Aptamers against VEGFR-1 and VEGFR-2
Preparation of MBs
VEGFR-1 Fc chimera (Cat. No. 321-FL-055; R&D Systems, USA), VEGFR-2 Fc chimera (Cat. No. 357-KD-050; R&D Systems, USA), and human IgG-Fc fragment (IgG-Fc) (Cat. No. P80-104-35; Bethyl Laboratories, USA) were immobilized onto protein A-coated MBs (Cat. No. 88845; Thermo Fisher Scientific, USA), according to the manufacturer’s instructions. Human albumin (HA; recombinant expressed in plants; Cat. No. 018-21541; Wako Pure Chemical Industries, Japan) was used as a blocking reagent. MBs on which HA and VEGFR-1, HA and VEGFR-2, or HA and IgG-Fc were immobilized were prepared, named MB1, MB2, and Fc-MB, respectively.
Collection and Amplification of Target-Binding ODNs
Seventy-mer ssODNs containing a random 34-mer sequence with flanking primer regions were used. The sequence of ssODNs with random region (N34) is as follows: 5′-GCC TGT TGT GAG CCT CCT N34 CGC TTA TTC TTG TCT CCC-3′ (Nihon Gene Research Laboratories, Japan). The ODN library was first incubated with MB1 in PBS-E buffer (PBS containing 2 mM EDTA and 0.1% w/w HA) for 30 min, and then MB1 was washed with PBS-E three times and collected using a magnet. The ssODNs bound to MB1 were eluted by heating MB1 in TE buffer (10 mM Tris with 1 mM EDTA [pH 8.0]; Cat. No. 316-90025; Nippon Gene) at 95°C for 10 min, and the ODNs in the supernatant were PCR amplified using PrimeSTAR (Cat. No. R010A; Takara Bio, Japan) with the primer set 5′-GCC TGT TGT GAG CCT CCT-3′ and 5′-biotin-GGG AGA CAA GAA TAA GCG-3′. Thermal cycling was performed by incubating the reactions at 94°C for 3 min, followed by 25 cycles at 94°C for 30 s, 66°C for 5 s, and 72°C for 60 s.
Purification of PCR Amplicons
After analyzing the PCR amplicons using polyacrylamide gel electrophoresis, ssODNs were isolated by incubating the PCR amplicons with streptavidin-coated MBs (Cat. No. 5325; JSR Life Sciences Corporation, Japan) and eluting the non-biotinylated strand with elution buffer (0.1N NaOH and 0.1 M NaCl). Then, the buffer used to elute ssODNs was replaced with storage buffer (TBS; 20 mM Tris, 1 mM EDTA, 200 mM NaCl, 0.1% w/w Tween 20 [pH 7.4]) using an Xpress Micro Dialyzer (Cat. No. 40071-X280; Scienova, Germany), followed by the next selection round as described above.
Alternating Consecutive Selection against MB1 and MB2
The three cycles of consecutive selection against MB1 and MB2 are demonstrated as depicted in Figure S1 (three alternating rounds against MB1 and MB2, six rounds total). The details of the experimental conditions for each selection round are summarized in Table S1.
NGS and Frequency Analysis
Preparation of Tagged ODN Samples
To obtain tagged ODN samples for emulsion PCR, we amplified the PCR amplicons obtained at each round using tagged primer sets with the same sequence of the 18-mer primers used in the previous section (underlined) with barcode region (X10) (forward primer: 5′-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG XXX XXX XXX XGA TGC CTG TTG TGA GCC TCC T-3′, reverse primer: 5′-CCT CTC TAT GGG CAG TCG GTG ATG GGA GAC AAG AAT AAG CG-3′). Thermal cycling was performed by incubating the reactions at 94°C for 3 min followed by 15 cycles at 94°C for 30 s, 66°C for 5 s, and 72°C for 60 s. The sequences of the barcode regions (X10) were as follows: 5′-CAG AAG GAA C-3′ (ID 005; for cycle 1-R1), 5′-CTG CAA GTT C-3′ (ID 006; for cycle 1-R2), 5′-TTC GTG ATT C-3′ (ID 007; for cycle 2-R1), 5′-TTC CGA TAA C-3′ (ID 008; for cycle 2-R2), 5′-TGA GCG GAA C-3′ (ID 009; for cycle 3-R1), and 5′-CTG ACC GAA C-3′ (ID 010; for cycle 3-R2).
Purification of Tagged ODN Samples
PCR products generated using the tagged primer sets described above were PCR amplified using short primer sets (forward primer 5′-CCA TCT CAT CCC TGC GTG TC-3′; reverse primer 5′-CCT CTC TAT GGG CAG TCG GT-3′) as follows: 94°C for 3 min followed by five cycles at 94°C for 30 s, 50°C for 30 s, and 68°C for 30 s. After removing the short primer ODNs and other PCR reagents using the Fast Gene Gel/PCR Extraction Kit (Cat. No. FG-91202; Nippon Genetics, Japan), purified PCR products were obtained (5′-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG XXX XXX XXX XGA TGC CTG TTG TGA GCC TCC T-N34-CGC TTA TTC TTG TCT CCC ATC ACC GAC TGC CCA TAG AGA GG-3′). Polyacrylamide gel electrophoresis was performed to determine the yields and purities of the amplicons.
Emulsion PCR and NGS
Emulsion PCR was performed to prepare monoclonal amplicon-modified beads using Ion OneTouch 2 system (Life Technologies, CA, USA) with a mixture containing equal amounts of the six tagged ODN samples as a template. After bead preparation, the Ion PGM System (Life Technologies, CA, USA) was employed to sequence the beads using an Ion 314 Chip. Emulsion PCR, bead preparation, and sequencing were performed according to the Ion PGM user guides (Publication Number MAN0007220, Rev. 5.0 and MAN0007273, Rev. 3.0, respectively) with the Ion PGM Template OT2 200 Kit, Ion PGM Sequencing 200 Kit v.2, and Ion 314 Chip Kit v.2 (Life Technologies, CA, USA).
Deep Sequence Analysis
All sequencing data for each round were exported as FASTAQ files from the Ion PGM System. Changes in the abundance ratio of each sequence were calculated and compared by Sequence Analyzer (Lifematics, Japan) that contains the CD-HIT-EST32 algorithm for sequence clustering. As shown in Figure S1, using Sequence Analyzer v.1.0.9, sequence acquisition of each round was carried out through the following parameters: quality criteria: 15, maximum low-quality element: 5, length of variable: 32–37. After sequence acquisition, homology clustering was carried out (similarity criteria: 90) and the two enriched sequences, which increase their avoidance ratios with increase in round number, were successfully picked up by “compare window” of Sequence Analyzer. In these processes, sequences with their primers were used.
Measurement of SPR
To determine the binding affinities of aptamers for VEGFR-1 and VEGFR-2, we used a BIAcore X (GE Healthcare, USA) to perform binding analyses at 25°C. Because the sensor chip surface was not regenerated with a regeneration buffer, single-cycle kinetics analyses, which run a series of analyte concentrations in one cycle with no regeneration between sample injections, were used in this study.
In the case of VEGFR-2, DNA aptamer-immobilized sensor surface was constructed, and a concentration series of VEGFR-2 was injected. To construct a DNA aptamer-immobilized sensor surface, a biotinylated ODN fragment (capture-ODN, 5′-dT44-biotin-3′) was first modified on a Sensor Chip SA (GE Healthcare, USA) through the avidin-biotin interaction by injecting the capture-ODN solution for 6 min at a flow rate of 5 μL/min in DPBST containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.5 mM KH2PO4, and 0.05% Tween 20 (pH 7.4). Aptamer candidates (5′-dA30-dT5-Apt01-3′ and 5′-dA30-dT5-Apt02-3′) that hybridized to the capture-ODN were immobilized by hybridization between dA30 and dT30 that was accomplished by injecting the DNA solution for 3.5 min, at a flow rate of 20 μL/min in DPBST. A concentration series of VEGFR-2 in DPBST was injected at a flow rate of 30 μL/min for 1 min, and dissociation was monitored for about 1.5 min using flowing running buffer (DPBST) (30 μL/min). As a reference, 5′-dA30-dT5-3′ was hybridized to the capture-ODN immobilized surface, and its sensorgram was subtracted from those of measuring flow cells.
In the case of VEGFR-1, strong non-specific binding of VEGFR-1 on the flow-cell and sensor surfaces was observed. Thus, VEGFR-1-immobilized sensor surface was constructed, and a concentration series of DNA aptamer was injected. To construct a VEGFR-1-immobilized sensor surface, we injected 50 mg/mL VEGFR-1 in acetic acid buffer onto Sensor Chip CM5 (GE Healthcare, USA) for 7 min at a flow rate of 5 μL/min in HBS-EP buffer (GE Healthcare, USA) containing 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.005% Surfactant P20 (pH 7.4). A concentration series of VEGFR-1 in HBS-EP was injected at a flow rate of 30 μL/min for 1 min, and dissociation was monitored for about 1.5 min using flowing running buffer (HBS-EP) (30 μL/min). The data were fitted with a 1:1 binding model using the BIAevaluation software version 4.1.1 (GE Healthcare, USA), and the KD values were determined using single-cycle kinetics.
Prediction of the Secondary and G-Quadruplex Structures of DNA Aptamers
Secondary structures of Apt01 and Apt02 were predicted using m-fold (25°C, 1 M [Na+]). Estimation of G-quadruplex formation by these aptamers was performed using QGSR mapper (maximum length, 35; minimum group, 2; loop size, 0–36).
Measurement of CD
CD analysis of the obtained aptamers was performed using 4 μM ODNs in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.5 mM NaH2PO4 [pH 7.4]). The ODN samples were denatured at 95°C before analysis and cooled slowly to room temperature. Their CD spectra were measured using a J-725 CD Spectrometer (JASCO Corporation, Japan) with a 1-cm path-length quartz cuvette using a constant flow of dry nitrogen. Scans were performed twice from 220 to 350 nm at 500 nm/min with a 1-s response time, 0.5-nm pitch, 1-nm bandwidth, and 100-millidegree sensitivity. The CD spectrum of PBS (blank) was measured in the same manner and subtracted from the collected data.
Measurement of UV Melting Curve
UV absorbance versus temperature profiles (melting curves) was measured for each aptamer at two wavelengths, 260 and 295 nm, by TMSPC-8 with a SHIMAZU UV-2450 spectrophotometer. ODNs were prepared at 4 μM in phosphate buffer containing NaCl (10 mM Na2HPO4, 10 mM NaH2PO4, and 100 mM NaCl [pH 7.4]) or phosphate buffer containing KCl (10 mM Na2HPO4, 10 mM NaH2PO4, and 100 mM KCl [pH 7.4]). These samples were then heat annealed to 90°C and allowed to slowly cool to 4°C over a period of several hours. Then 110 μL of a sample was transferred to a 1-cm path-length quartz cuvette and covered with a layer of 30 μL liquid paraffin. It was heated to 95°C and cooled to room temperature at 0.5°C/min, with data collection occurring every 0.5 min on the annealing and melting steps.
HUVEC Culture
HUVECs (HUVEC-2) derived from single donors were obtained from Corning (San Jose, CA, USA). Medium 200 phenol red free (PRF) (Cat No. M200PRF500) and Low Serum Growth Supplement kit (Cat. No. S-003-K; GIBCO) were obtained from Thermo Scientific (MA, USA). HUVEC-2 was cultured in Medium 200PRF supplemented with low serum growth supplement (LSGS). The cells were incubated in a 37°C, 5% CO2/95% air, humidified cell culture incubator. The media were changed every 5 days. When confluent or for seeding on different structures, cells were detached with trypsin/EDTA, and then trypsin neutralization solution was added. Cells were centrifuged at 200 × g for 5 min and dispersed in growth medium.
In Vitro Tube Formation Assay
In brief, after defrosting on ice, BD Matrigel Matrix (∼10 mg/mL; Cat. No. 354234, Corning, NY, USA) was added to the pre-cooled 24-well plates at 298 μL/well followed by incubation for 1 h at 37°C. HUVECs were trypsinized, pelleted by low-speed centrifugation, and suspended in Medium 200PRF supplemented with LSGS. HUVECs (1.1 × 105 cells/well) were placed into 24-well flat-bottomed plates pre-coated with Matrigel and then incubated with aptamers (10 μM) or VEGF165 (Recombinant Human VEGF 165 Protein, Cat. No. 293-VE [R&D Systems, USA]; 10 ng/mL, 0.26 nM) for 24 h. In the experiment of dose dependency, the Apt02 concentration of more than 10 μM showed the significant difference from control group (data not shown). After incubation, cell tube or network formation was observed using a phase-contrast microscope (IX51S8F-3; Olympus, Japan). The number of mesh-like circles was manually counted.
Stability Assay of Aptamer in Cultured Medium
The aptamers (0.4 μM) were incubated in Medium 200PRF supplemented with LSGS for 0.5, 1, 2, 3, 4, 7, or 24 h at 37°C, followed by heating sample for 20 min at 80°C for inactivation of nucleases. Samples were analyzed by denaturing PAGE using a 10% polyacrylamide/urea gel and stained with Lonza GelStar Nucleic Acid Gel Stain (Cat. No. 50535; Lonza). Stained DNA was imaged with Digi-Gel Shot (Takara Bio) and quantified using ImageJ software package. The fraction of intact aptamers was normalized to the DNA sample without incubation at each time point.
Statistical Analysis
All values are expressed as mean ± standard error. Statistical difference was analyzed by Kaleida Graph 4.1 version 4.13 (Synergy Software, USA). All one-way analyses of variance (ANOVAs) were performed with one-way ANOVA tests followed by post hoc Tukey’s honestly significantly different (HSD) test. Differences with a p value <0.05 were considered statistically significant.
Author Contributions
Conception and Design: K.Y.; Acquisition of Data: M.H., T.O., K.K., and F.W.; Analysis and Interpretation of Data: T.Y., M.H., T.O., K.K., F.W., and K.Y.; Writing – Review and/or Revision of the Manuscript: T.Y. and K.Y.; Administrative, Technical, or Material Support: H.F.; Study Supervision: K.Y.; Equal Contribution: M.H. and T.O.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study is part of a collaborative research project between Nissan Chemical Industries, Ltd. (Japan) and the Yoshimoto research group at The University of Tokyo (Japan), and was partly funded by PRESTO-JST (grant number JPMJPR16FB), JST-SCORE (Japan), JSPS KAKENHI (grant number 18H02002), and Iketani Science and Technology Foundation (Japan).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.12.034.
Supplemental Information
References
- 1.Olsson A.K., Dimberg A., Kreuger J., Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat. Rev. Mol. Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 2.Rahimi N. VEGFR-1 and VEGFR-2: two non-identical twins with a unique physiognomy. Front. Biosci. 2006;11:818–829. doi: 10.2741/1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibuya M. Vascular endothelial growth factor receptor-1 (VEGFR-1/Flt-1): a dual regulator for angiogenesis. Angiogenesis. 2006;9 doi: 10.1007/s10456-006-9055-8. 225–230, discussion 231. [DOI] [PubMed] [Google Scholar]
- 4.Dias S., Hattori K., Zhu Z., Heissig B., Choy M., Lane W., Wu Y., Chadburn A., Hyjek E., Gill M. Autocrine stimulation of VEGFR-2 activates human leukemic cell growth and migration. J. Clin. Invest. 2000;106:511–521. doi: 10.1172/JCI8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nourse M.B., Halpin D.E., Scatena M., Mortisen D.J., Tulloch N.L., Hauch K.D., Torok-Storb B., Ratner B.D., Pabon L., Murry C.E. VEGF induces differentiation of functional endothelium from human embryonic stem cells: implications for tissue engineering. Arterioscler. Thromb. Vasc. Biol. 2010;30:80–89. doi: 10.1161/ATVBAHA.109.194233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikuno T., Masumoto H., Yamamizu K., Yoshioka M., Minakata K., Ikeda T. Efficient and robust differentiation of endothelial cells from human induced pluripotent stem cells via lineage control with VEGF and cyclic AMP. PLoS ONE. 2017;12:e0173271. doi: 10.1371/journal.pone.0173271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye L., Zhang S., Greder L., Dutton J., Keirstead S.A., Lepley M., Zhang L., Kaufman D., Zhang J. Effective cardiac myocyte differentiation of human induced pluripotent stem cells requires VEGF. PLoS ONE. 2013;8:e53764. doi: 10.1371/journal.pone.0053764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramaswamy V., Monsalve A., Sautina L., Segal M.S., Dobson J., Allen J.B. DNA Aptamer Assembly as a Vascular Endothelial Growth Factor Receptor Agonist. Nucleic Acid Ther. 2015;25:227–234. doi: 10.1089/nat.2014.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueki R., Ueki A., Kanda N., Sando S. Oligonucleotide-Based Mimetics of Hepatocyte Growth Factor. Angew. Chem. Int. Ed. Engl. 2016;55:579–582. doi: 10.1002/anie.201508572. [DOI] [PubMed] [Google Scholar]
- 10.Ueki R., Atsuta S., Ueki A., Hoshiyama J., Li J., Hayashi Y., Sando S. DNA aptamer assemblies as fibroblast growth factor mimics and their application in stem cell culture. Chem. Commun. (Camb.) 2019;55:2672–2675. doi: 10.1039/c8cc08080a. [DOI] [PubMed] [Google Scholar]
- 11.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 12.Saito S., Hirose K., Tsuchida M., Wakui K., Yoshimoto K., Nishiyama Y., Shibukawa M. Rapid acquisition of high-affinity DNA aptamer motifs recognizing microbial cell surfaces using polymer-enhanced capillary transient isotachophoresis. Chem. Commun. (Camb.) 2016;52:461–464. doi: 10.1039/c5cc07268a. [DOI] [PubMed] [Google Scholar]
- 13.Kristian S.A., Hwang J.H., Hall B., Leire E., Iacomini J., Old R., Galili U., Roberts C., Mullis K.B., Westby M., Nizet V. Retargeting pre-existing human antibodies to a bacterial pathogen with an alpha-Gal conjugated aptamer. J. Mol. Med. (Berl.) 2015;93:619–631. doi: 10.1007/s00109-015-1280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 15.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 16.Blind M., Blank M. Aptamer Selection Technology and Recent Advances. Mol. Ther. Nucleic Acids. 2015;4:e223. doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acinas S.G., Sarma-Rupavtarm R., Klepac-Ceraj V., Polz M.F. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzker M.L. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 19.Mac Gabhann F., Popel A.S. Dimerization of VEGF receptors and implications for signal transduction: a computational study. Biophys. Chem. 2007;128:125–139. doi: 10.1016/j.bpc.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markovic-Mueller S., Stuttfeld E., Asthana M., Weinert T., Bliven S., Goldie K.N., Kisko K., Capitani G., Ballmer-Hofer K. Structure of the Full-length VEGFR-1 Extracellular Domain in Complex with VEGF-A. Structure. 2017;25:341–352. doi: 10.1016/j.str.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikin O., D’Antonio L., Bagga P.S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tóthová P., Krafčíková P., Víglaský V. Formation of highly ordered multimers in G-quadruplexes. Biochemistry. 2014;53:7013–7027. doi: 10.1021/bi500773c. [DOI] [PubMed] [Google Scholar]
- 24.Masiero S., Trotta R., Pieraccini S., De Tito S., Perone R., Randazzo A., Spada G.P. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 2010;8:2683–2692. doi: 10.1039/c003428b. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov V.I., Minchenkova L.E., Schyolkina A.K., Poletayev A.I. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 1973;12:89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- 26.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 27.Yang X., Liu D., Lu P., Zhang Y., Yu C. Nucleic acid G-quadruplex based label-free fluorescence turn-on potassium selective sensing. Analyst (Lond.) 2010;135:2074–2078. doi: 10.1039/c0an00106f. [DOI] [PubMed] [Google Scholar]
- 28.Orpana A.K., Ho T.H., Stenman J. Multiple heat pulses during PCR extension enabling amplification of GC-rich sequences and reducing amplification bias. Anal. Chem. 2012;84:2081–2087. doi: 10.1021/ac300040j. [DOI] [PubMed] [Google Scholar]
- 29.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tucker W.O., Shum K.T., Tanner J.A. G-quadruplex DNA aptamers and their ligands: structure, function and application. Curr. Pharm. Des. 2012;18:2014–2026. doi: 10.2174/138161212799958477. [DOI] [PubMed] [Google Scholar]
- 31.Sun Q., Zhou J., Zhang Z., Guo M., Liang J., Zhou F., Long J., Zhang W., Yin F., Cai H. Discovery of fruquintinib, a potent and highly selective small molecule inhibitor of VEGFR 1, 2, 3 tyrosine kinases for cancer therapy. Cancer Biol. Ther. 2014;15:1635–1645. doi: 10.4161/15384047.2014.964087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.