Graphical abstract
Keywords: Functional foods, Gut microbiota, Metabolomics, GCMS, Chemometrics
Abbreviations: GC, Green Coffee; BC, Black Coffee; GT, Green Tea; BT, Black Tea; FI, Opuntia ficus-indica (prickly pear); POM, pomegranate (Punica granatum); SUM, sumac (Rhus coriaria); SCFAs, short chain fatty acids; GI, gastrointestinal; GIT, gastrointestinal tract
Highlights
-
•
Metabolomics was employed to assess 7 functional foods impact on gut microbiota.
-
•
Insights regarding how functional foods alter gut metabolic pathways is presented.
-
•
Increased GABA production was observed in polyphenol rich functional food.
-
•
Purine alkaloids served as direct substrate in microbiota metabolism.
Abstract
Functional food defined as dietary supplements that in addition to their nutritional values, can beneficially modulate body functions becomes more and more popular but the reaction of the intestinal microbiota to it is largely unknown. In order to analyse the impact of functional food on the microbiota itself it is necessary to focus on the physiology of the microbiota, which can be assessed in a whole by untargeted metabolomics. Obtaining a detailed description of the gut microbiota reaction to food ingredients can be a key to understand how these organisms regulate and bioprocess many of these food components. Extracts prepared from seven chief functional foods, namely green tea, black tea, Opuntia ficus-indica (prickly pear, cactus pear), black coffee, green coffee, pomegranate, and sumac were administered to a gut consortium culture encompassing 8 microbes which are resembling, to a large extent, the metabolic activities found in the human gut. Samples were harvested at 0.5 and 24 h post addition of functional food extract and from blank culture in parallel and analysed for its metabolites composition using gas chromatography coupled to mass spectrometry detection (GC-MS). A total of 131 metabolites were identified belonging to organic acids, alcohols, amino acids, fatty acids, inorganic compounds, nitrogenous compounds, nucleic acids, phenolics, steroids and sugars, with amino acids as the most abundant class in cultures. Considering the complexity of such datasets, multivariate data analyses were employed to classify samples and investigate how functional foods influence gut microbiota metabolisms. Results from this study provided a first insights regarding how functional foods alter gut metabolism through either induction or inhibition of certain metabolic pathways, i.e. GABA production in the presence of higher acidity induced by functional food metabolites such as polyphenols. Likewise, functional food metabolites i.e., purine alkaloids acted themselves as direct substrate in microbiota metabolism.
Introduction
In humans, the gastrointestinal (GI) tract harbors approximately 1014 bacterial cells (i.e., 10 times the number of eukaryotic cells in the body), [1]. Bacteria contributing to this intestinal microbiota niche belong to more than 1,000 different species harboring more than three million bacterial genes and amount to a biomass of approximately 2 kg, which can be considered as an internal “organ” and provide many functions that are crucial for the hosts well-being. Moreover, the study of intestinal microbiota cannot be separated from its environmental context. For instance, host genetics, geographical location, nutrition, antibiotics, and other treatments affect the microbiota and its metabolic machinery [2]. The human GI tract hosts a nutrient-rich environment that supports a commensal microbiome providing crucial functions that cannot be carried out alone by the host [3], [4]. These functions are both metabolic (colonic fermentation and production of short chain fatty acids), protective (improving barrier and increasing the resistance to colonization by opportunistic pathogens, secretion of antimicrobial peptides etc.), and structural (maturation of the intestinal epithelium and the immune system) [1]. Microbes present at mucosal sites can also become part of the tumor microenvironment of aerodigestive tract malignancies [5]. In counterpoise, gut microbiota also functions in detoxification of dietary or drug components, can reduce inflammation, and help to maintain a balance in host cell growth and proliferation [6]. Thus, interrogation of gut microbiota metabolism as such and in response to dietary intervention requires a holistic perspective. Assigning microbial communities, their members, and aggregate biomolecular activities into these categories will require a substantial research commitment. Beyond metagenomics, functional approaches, such as metatranscriptomics, metaproteomics, and metabolomics (together referred to as meta-omics), are now also rapidly enhancing our knowledge on the gut microbiome. Meta-omic approaches deliver both qualitative and quantitative data on genetic potential, transcripts, proteins, and metabolites present in specific microbial communities under specific conditions. These approaches also have the potential to highlight the systemic influence of microbial communities beyond the gut, deciphering the intricate crosstalk between humans and their microbial ecosystems [7].
Daily diet has an impact on not only our nutrition but also health and wellness. This has necessitated an increase in the development and characterization of food products with additional effects than just energy, mineral or vitamin supply, the so-called functional foods. Functional foods can be defined as dietary supplements that in addition to their nutritional values, can beneficially modulate body functions towards enhancing physiological responses or reducing a risk of certain disease [8] to the extent that these can be nutraceuticals, i.e. foods with clearly established medicinal properties. Upon food ingestion, several mechanical, chemical and enzymatic processes occur within the GIT to mediate for its digestion into nutrients or active ingredients, which are then absorbed to be suitable for use by the body [9].
Gut microbiota mediated metabolic activity can contribute to the digestion of various dietary compounds as well as transformation of xenobiotics as in functional foods and supply of micronutrients, thus affecting their potential health effects. In contrast, functional food components can themselves also affect the growth and the metabolic activity of gut microbiota and accordingly their composition and or potential functions [10]. For instance, tea phenolics exhibited an inhibitory effect on certain gut microbiota species such as; Bacteroides spp., Clostridium spp. (C. perfringens and C. difficile), E. coli and Salmonella typhimurium with caffeic acid showing the highest inhibitory activity [11]. In parallel, gut microbiota can also affect the pharmacological properties of ingested food products via its biotransformation while in the gut pending if not absorbed earlier in the GIT [12]. For example, tannins are solely metabolized by microbial enzymes leading to the formation of conjugated derivatives, which have a different pharmacological profile and being subject to rapid excretion through urine or bile secretions back into the gastro intestinal tract [13]. Whilst most studies have indeed focused on how gut microbes bio transform functional foods, few reports are known to us, on how gut microbiota metabolism is affected by these food supplements [14]. Including; the wide range alterations in the gut microbiota composition imparted by animal-based vs. plant-based diets [15] as well as, the increased abundance of Bifidobacterium species in breast-fed infants over formula-fed ones [16]. Nevertheless, further studies are needed to investigate the impact of such changes on the normal homeostasis of GI tract.
The main interaction between organisms is of chemical nature. Obtaining a detailed description of how functional foods interact with gut microbiota or do affect its biotransformations can be a key to understand how these organisms regulate our daily food.
Metabolomics is the systematic study of the small-molecule metabolite profiles of living organisms at certain status or phenotype. Such dense chemical information can be acquired through utilizing hyphenated mass spectrometry techniques such as; gas or liquid chromatography-mass spectrometry (GC-MS and LC-MS) [17]. Moreover, untargeted GC/MS-based metabolomics is routinely used to detect and monitor low molecular weight and non-polar primary and secondary metabolites, the latter known to be abundant within a plant matrix [18]. Since the microbiota in humans but also in domestic animals is highly diverse and individual, and thus incomprehensible and irreproducible, we used a simplified but therefore more reliable gut microbiota model system. Hence we established cultivating a selection of eight bacterial species that are representing the core functions of the large intestine microbiota [19]. These consortium is comprised of 8 bacterial species namely; Anaerostipes caccae, Bacteroides thetaiotaomicron, Bifidobacterium longum, Blautia producta, Clostridium butyricum, Clostridium ramosum, Escherichia coli and Lactobacillus plantarum. These species belong to the most abundant phyla of the human gut microbiota representing the extended simplified intestinal human microbiota (SIHUMIx) with functionally important biochemical pathways and interactions that likely occur in the human gut.
In order to cover at least part of the most commonly consumed food products worldwide either as beverages, condiments, food color and or herbal drugs [20], we selected the following plant products: green (GC) and black (BC) coffee (Coffea arabica), green (GT) and black (BT) tea (Camellia sinensis), Opuntia ficus-indica (FI), pomegranate (POM) (Punica granatum) and sumac (SU) (Rhus coriaria). For example, various coffee constituents are reported to exhibit antioxidant properties and to protect against cardiovascular, inflammatory and neurodegenerative diseases [21], [22]. While, many health benefits have been attributed to tea products consumption such as antihypertensive [23], antihyperlipidemic [24], antioxidant [25] and CNS stimulant effects. On the other hand, owing to its rich flavonoid content, Opuntia ficus-indica fruit is reported to possess a powerful anti-inflammatory and antioxidant properties [26]. Numerous studies have reported several health-related benefits from pomegranate, with nearly every part of the plant was tested and pharmacological activities such as antimicrobial, anti-inflammatory and antioxidant effects were reported [27] owing to its rich tannin content. Other functional food enriched in hydrolysable tannins include Rhus coriaria [18] known as sumac. It is suggested to exhibit hypoglycemic, anti-inflammatory, antimicrobial and cytotoxic properties [28].
In order to cover the process of reaction and metabolization, two different time points at 0.5 and 24 h were choosen. The aim of this study was to evaluate the impact of the seven functional foods of common usage in human diets worldwide on selected bacterial strains, representing the human gut microbiota using a GC-MS approach, immediately after and 24 h post treatment in order to build a hypothesis of how gut microbiota respond to these different treatments and whether a generalized response could be observed.
Materials & methods
Plant material and extraction
Methanol extracts were prepared from finely powdered green GC and black coffee BC seeds, green GT and black tea BT leaf, peeled Opuntia ficus-indica red ‘Rose’ FI fruit powder and sumac SUM lyophilized fruits powders by cold maceration over 2 days using 100% methanol until exhaustion. Extracts were then filtered and subjected to evaporation under vacuum at 40 °C until complete dryness. Extracts were placed in tight glass vials and stored at −20 °C until further analysis. In case of pomegranate POM, seeds were extracted and expressed to obtain a juice, which was then lyophilized until complete dryness and stored as above.
Chemicals and solvents
All solvents and chemicals were of analytical grades and purchased from Sigma-Aldrich, St. Louis, USA.
Gut microbiota culture
The microorganisms consortium used in this study is described as: the extended simplified intestinal human microbiota – SIHUMIx. Microorganisms of the SIHUMIx community, a model for the intestinal microbiota, were selected according to their occurrence in humans, the spectrum of fermentation products formed and the ability to form a stable community [19]. Co-cultured bacterial species included: Anaerostipes caccae (DSMZ 14662), Bacteroides thetaiotaomicron (DSMZ 2079), Bifidobacterium longum (NCC 2705), Blautia producta (DSMZ 2950), Clostridium butyricum (DSMZ 10702), Clostridium ramosum (DSMZ 1402), Escherichia coli K-12 (MG1655) and Lactobacillus plantarum (DSMZ 20174). All bacteria were cultivated in Brain-Heart-Infusion (BHI) medium under anaerobic conditions at 37 °C and 175 rpm shaking for 72 h prior to inoculation. All strains were shown to be able to grow equally in the media. BHI media was prepared by mixing 37 g brain heart infusion, 0.5 g L-cysteine hydrochloride, 0.001 g resazurin, 10 ml Vitamin K hemin solution and 5 g yeast extract in one L of sterile water. Gut bacteria cultured in Brain-Heart-Infusion medium (optical density of 0.1) was left to grow under anaerobic condition at 37 ℃ for 18 h till optical density reached 1.7 prior to functional food extract addition. Details on isolated microbiota consortium strains and its potential metabolic functions is depicted in Suppl. Table S1 and S2, respectively. For the control including only SIHUMIx strains and each functional food extract, cultivation was perfomed in triplicates leading to 48 samples in total.
Gut microbiota functional food incubation assay
A stock solution was prepared of functional food at a concentration of 50 mg/ml in 50:50 methanol: (BHI) growth medium and stored at 4 °C until inoculation. 100 µl and 1 ml of each functional food stock solution was then aliquoted to a final volume of 10 ml BHI media containing the gut microbe culture to achieve a final concentration of 0.5 and 5 mg/ml, respectively. Blank cultures were prepared by adding an equivalent amount of 50 and 500 µl 100% methanol into the culture medium, kept under the same condition, and compared to the culture receiving no solvent treatment.
Metabolites extraction and GCMS analysis
200 µl of aliquoted culture harvested at different time points was spiked with xylitol standard solution dissolved in sterile water to reach a final concentration of 10 µg./ml followed by the addition of 800 µl acetonitrile/methanol mixture with incubation at 4 °C for 30 min till complete protein precipitation. Mixture was then centrifuged at 12,000g using Eppendorf centrifuge for 4 min, with 100 µl of the supernatant then aliquoted and subjected to evaporation under nitrogen stream till complete dryness. For metabolites derivatization, 150 μl of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) was then added to the residue and incubated at 60 ◦C for 45 min. Samples were then analyzed using GC–MS (Shiamdzu, Japan). Silylated derivatives were separated on Rtx-5MS (30 m length, 0.25 mm inner diameter, and 0.25 μm film) column. Injections were made in a (1:15) split mode, conditions: injector 280 °C, column oven 80 °C for 2 min, rate 5 °C/min to 315 °C, kept at 315 °C for 12 min. He carrier gas at 1 mLmin-1. The transfer line and ion–source temperatures were set at 280 and 180 °C, respectively.
GC-MS multivariate data analyses
MS peak abundance of primary silylated metabolites were extracted using MET-IDEA software with default parameter settings for GC–MS [29]. The aligned peak abundance data table was further exported to principal component analysis (PCA) and orthogonal projection least squares discriminant analysis (OPLS-DA) using SIMCA-P version 14.1 software package (Umetrics, Umeå, Sweden). All variables were mean-centered and scaled to Pareto variance (Par).
Results & discussion
Two different time aliquots were obtained from functional foods amended cultures, the first one was at 0.5 h a time at which no significant biotransformation is expected to occur, and at 24 h at which most of the biochemical and enzymatic changes would have occurred. Results revealed that while addition of functional foods alter microbiota metabolism through either stimulation or inhibition of its metabolic pathways, not surprisingly functional food metabolites themselves can also act as substrate for microbiota metabolism.
GC-MS metabolite profiling of gut microbiota treated functional foods
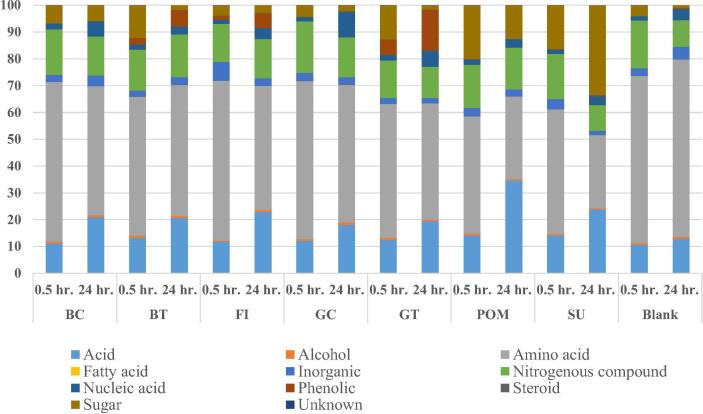
GC/MS was employed to characterize microbiota culture metabolism and monitor changes occurring 30 m and 24 h post exposure to the different functional food extracts. Metabolites detected (as such or as volatile per-trimethylsilylated derivatives) comprised mostly microbial low molecular weight primary metabolites viz. organic acids, alcohols, amino acids, fatty acids, inorganic compounds, nitrogenous compounds, nucleic acids, phenolics, steroids and sugars in addition to few secondary metabolites representative of certain functional foods, e.g. catechins in case of tea [17]. A total of 131 metabolite peaks (Suppl. Table S3) was detected from all untreated blank and functional food treated cultures. The relative percentile levels of all metabolite classes detected for cultures harvested at 0.5 and 24 h from functional foods and blank cultures is presented in (Fig. 1), and a representative chromatogram showing main metabolite classes with their elution regions can be found in (Suppl. Fig. S1). GC-MS Metabolite profiling revealed that amino acids form the major class in cultures harvested at 0.5 h ranging from 44% to 60% in those amended with food extracts compared to 62% in blank culture at the same time point (Suppl. Table S3), suggesting that amino acids are mostly derived from the microbial culture itself. Following amino acids, nitrogenous compounds represented the second most abundant class, ranging from 14% to 19% in blank gut microbial culture and those fortified with food extracts at 0.5 h. Compared to amino acids and nitrogenous compounds that showed comparable levels at 0.5 h incubation, sugars (4–20%) showed a larger variation among cultures fortified with different food extracts compared to blank culture with 4.0%, supporting the conclusion that such difference is attributed to functional foods individual compositions. Sugars are a major primary metabolite class in most plant foods as analysed using GC-MS [30]. Other minor classes identified in microbial cultures included inorganic metabolites (2–7%), phenolics (0.02–5.6%) and nucleic acids (1.6–2%).
Fig. 1.
Relative percentile levels of metabolite classes detected using GC-MS for cultures harvested at 0.5 and 24 h from functional foods amended culture: BC, BT, FI, GC, GT, POM & SU versus blank.
After incubation for a period of 24 h, samples were aliquoted and analyzed using the same protocol to reveal for metabolite changes occurring in culture (Fig. 2). A general decrease in amino acid levels by 16% (0.8 fold) in functional food treated samples, with the largest decrease observed in sumac treated cultures (0.6 fold) indicate that amino acids may serve as nutrient substrate for gut microbial growth. Amino acids are utilized by bacteria as building blocks for microbial protein assembly essential for bacterial growth, or to be fermented as an energy source [31]. This amino acid decrease trend in functional food treated samples 24 h post incubation is contrasting the untreated blank samples, showing even a slight 1.1-fold increase in amino acid content. For nitrogenous compounds a decrease was revealed in all cultures treated and blank. Most pronounced decreases in functional food treated samples were observed with sumac, green tea and green coffee with 0.6, 0.8 and 0.8 fold reductions, respectively. Bacteria can utilize nitrogenous compounds such as amino propanoate as a single carbon source or energy source [32]. Interestingly, a significant decrease in sugar levels is observed in functional food treated samples by 0.7 fold compared to 0.25 fold decrease in blank samples. The most significant decrease of sugars and amino acids in functional food treated samples was observed in GT and BT (0.1 fold), and GC (0.5 fold), i.e. polyphenol rich plant products. Metabolite classes that showed an opposite pattern, i.e. an increase over time (0.5 versus 24 h) included organic acids and low molecular weight phenolics, most evident in case of POM and GT amended cultures compared to blank. Increase is likely attributed for microbiota degradation of high molecular weight plant phenolics to its simpler organic and phenolic acids. Detection of high molecular weight phenolics cannot be achieved using GC-MS and has yet to be performed using liquid chromatography mass spectrometry (LC-MS) [33]. Several bacterial species are reported to possess hydrolytic enzymes necessary for plant polyphenols degradation, e.g. through dehydroxylation, decarboxylation, ring cleavage or oxidation, ultimately generating simpler phenolic and organic compounds [34]. Likewise, nucleic acids increase upon incubation, especially in case of a GC amended culture at 6-fold increase after 24 h compared to 0.5 h treatment versus a 3-fold increase in case of the blank culture. A pie chart showing relative percentile levels of the different metabolite classes analyzed in microbiota culture fortified with the different treatments at 0.5 and 24 h is given in Suppl. Fig. S2A–G. Provided below is an overview of the major changes observed for metabolite classes in blank culture compared to those amended with food extracts.
Fig. 2.
Schematic workflow to assess functional food on gut microbiota metabolism used in this study: I) gut microbiota culture exposure to food extracts i.e., black coffee (BC), green coffee (GC), black tea (BT), green tea (GT), Ficus (FI) pomegranate (POM) and sumac (SU) by addition to growth medium versus untreated blank, II) metabolites extraction & analysis using GC–MS, III) multivariate data analysis i.e. PCA and OPLS.
Canonical amino acids
Ornithine, alanine and isoleucine were the most abundant amongst the detected amino acids, and showed a decline of 0.8, 0.14 and 0.32 fold, respectively, upon incubation in all cultures. An exception to this pattern was observed in case of phenylalanine, glutamic and pyroglutamic acid in both treated and blank cultures. Phenylalanine showed ca. 1.8-fold increase in all functional foods compared to 2.5-fold increase in blank culture, likewise pyroglutamic acid showed an average of 1.7-fold increase in all functional foods compared to 4-fold increase in blank at 24 h treatment, i.e. the increase in treated cultures also was reduced vs. blank. Despite, its reduction or even complete depletion upon GC, FI and POM treatment, in case of the other functional foods, glutamic acid showed a ca. 2.3-fold increase which is slightly reduced vs. the untreated blank with a 4.5-fold increase. This might be attributed to specific producers of these amino acids i.e. E. coli induced production of phenylalanine [35] and L. plantarum mediated production of glutamic and pyroglutamic acid [36], both present in the consortium culture. Amino acids play multiple roles in gut microbiota either via protein fermentation or internally to synthesize essential amino acids to be further utilized as building blocks for cellular composition [37], to serve as signaling molecules between microbial cells [38] or within the host [39], or to be used as an energy source [40]. Nevertheless, such increase failed to lead to an increase in the overall amino acid percentile levels as the aforementioned reduction of other amino acids was much more significant with larger negative fold changes.
Other nitrogenous compounds
The most pronounced decrease in nitrogenous compounds was detected for 3-amino propanoate at comparable levels in functional foods amended and blank cultures (ca. 0.16-fold). Human gut microbiota produces various short chain fatty acids (SCFAs) with propanoate as major metabolic product of anaerobic fermentation. Along with butanoate, propanoate is a major component in microbiota metabolic pathways for the synthesis of other SCFAs [41]. It does play a significant role during irritable bowel syndrome if reduced by medication [42]. Bacterial genera such as Bacteroides, Blautia, Eubacterium, Escherichia and Clostridium can ferment nondigestible dietary carbohydrates and amino acids to propionate through either 1,2-propanediol or succinate pathways (Suppl. Fig. S3) [43], [44].
In contrast, other nitrogenous compounds viz. gamma amino butyric acid (GABA) increased significantly in all food extract amended cultures over time compared to a reduction in case of blank, likely attributed to bacterial fermentation of plant derived amino acids i.e. l-glutamic acid to GABA [45]. The highest increase in GABA levels was detected in FI and POM with ca. 3 and 2 fold increases, respectively. Whether an increase in GABA levels occurs similarly in the gut upon fermentation of functional foods is unclear, but if so it will contribute to a biological effect yet to be determined in its extent. GABA is an inhibitory neurotransmitter that has many reported pharmacological effects. It also is involved in various neurological disorders including epilepsy, seizures, convulsions, Huntington’s disease, and Parkinsonism [46]. The gut microbiota related brain axis effect (Brain-Gut-Microbiome Axis) is a hot topic regarding all CNS disorders, and alterations in brain-gut-microbiome communication is found to be involved in the pathogenesis of several disorders [47].
Other detected nitrogenous compounds include cadaverine derived from lysine via its decarboxylation [48]. Gut microbiota is reported to synthesize cadaverine under high protein diet [49]. This is in general related to biogenic amino compounds including putrescine and spermidine produced via decarboxylation of other amino acids. Polyamines have been reported to stimulate cell division of gut microbiota, e.g., of E. coli [50] and also to impart health benefits to the host such as regulation of growth and aging, and prevention of metabolic and neurodegenerative disorders [51]. Microbiota cell uptake of polyamines may explain the lower cadaverine levels at 24 h. However, the relatively high levels of putrescine may indicate biosynthesis overweighing microbiota cell uptake and can be explained by the fact that putrescine is synthesized from ornithine [51], [52], detected at higher levels at 0.5 h and showing a decrease with incubation time (Suppl. Fig. S4). Polyamine generation reaction is found to favor of SCFA synthesis, driven by certain bacterial strains such as E. coli [53] also present in this microbial consortium (Suppl. Table S3). In this study, a reduction in putrescine was observed to be concurrent with higher lactic acid and succinic acid levels in certain cultures, which support the competition between polyamine and SCFA synthesis pathways on amino acid substrates.
Sugars
Sugars identified included monosaccharides, disaccharides and sugar alcohols. Generally, sugars showed lower abundance 24 h post incubation as expected due to their utilization as energy source and as carbon source for the generation of SCFA [54] as evidenced in strains of Bifidobacterium [55]. For all treatments there was a decrease, with the exception of SUM (Rhus coriaria) that showed higher sugar levels at 24 h compared to 0.5 h (Suppl. Table S3; Suppl. Fig. S5). A neuroprotectant sugar that could exert its effect at the gut level is trehalose which showed one of the most pronounced decreases, being completely depleted upon incubation likely degraded by trehalase enzyme secreted by the microbiota [56]. Only oral intake of trehalose subsequently influenced by gut microbiota but not i.v. injection was found to exert the reportedneuroprotection. An increasing body of evidence for gut microbiota effects on CNS suggests that trehalose exerts its neuroprotective role through microbiota-gut-brain signaling [57]. BT, GT and BC cultures showed the highest levels of trehalose at 4–5% followed by POM and GC (3–4%). Sugar abundance appeared to be dependent on the functional food type i.e., fructose was found most abundant in POM and SUM cultures (Rhus coriaria), Suppl. Table S3, whereas sugars more specific to a certain culture included sucrose and 3-deoxy-D-arabino-hexonic acid found exclusively in GT/BT and GC/BC, respectively. The type of carbohydrates in functional foods was reported to influence gut microbiota composition [14].
Organic acids
Microbiota is known to metabolize carbohydrate into short chain fatty acids (SCFA) i.e. lactic acid to be used as an energy source [58]. Also proteins and free fatty acids act also as precursors for SCFA production by microbiota [38]. For example, glycine, glutamate, ornithine and threonine act as precursor of acetate, whereas threonine, lysine and glutamate yield butyrate, and threonine can be used as substrate for propionate production [38]. A ca. 2-fold increase in SCFA levels was observed upon incubation in all functional food amended cultures. It was not observed in blank untreated culture (Suppl. Table S3, Suppl. Fig. S6). A major acid that showed such a pattern is succinic acid, likely produced as a sugar fermentation product [59], [60], [61] as evident by a decrease in e.g. fructose (Suppl. Table S3). Elevated succinate levels within the gut lumen have been associated with microbiome disturbances (dysbiosis), as well as in patients with inflammatory bowel disease (IBD) [62]. Other sources of succinic acid in the body involve the tricarboxylic acid (TCA) cycle within host cells, though unlikely to function here under hypoxic conditions. Lactate, another carbohydrate fermentation product [63] mediated via lactic acid bacteria (LAB) i.e. Lactobacillus plantarum, [64] also increased upon incubation at 24 h, especially in POM, SUM, and FI treated samples at 18.7 (0.2 fold), 15.3% (0.15 fold) and 6.3% (0.05 fold), respectively, compared to trace levels in blank. A similar (secondary metabolite) profile of changes was observed for hydrolysable tannins, showing enrichment in POM and SUM. This might account for the similar metabolic response observed in their respective microbiota cultures.
Phenolics
Phenolics comprise a metabolite class that is almost solely derived from functional food extracts and was close to being absent in the blank culture. Our interest in reporting these metabolites herein is that they are rather important plant natural products with many proven or claimed health benefits including influence on microbes [14]. Thus a potential influence of its composition or its levels on gut microbiota metabolism is likely to impact functional food ultimate health effects. Catechin was found exclusively in GT and BT cultures at 0.5 h and to increase at 24 h ca. 2–3 fold. This can be attributed to its polymers or higher mw conjugates cleaved. e.g. (−)-epicatechin gallate (ECG) is expectedly found enriched in tea samples, and this is supported by a ca. 4-fold increase in gallic acid levels after 24 h. Although catechins were detected almost exclusively in GT and BT, gallic acid was present in most cultures though at lower levels especially for FI, SUM and POM, but it was absent from blank, suggestive of being derived from hydrolysis of plant phenolic conjugates or polymers. e.g. by bacteria such as herein L. plantarum known to act on hydrolysable tannins [24], [26]. L. plantarum is the only bacterial species reported to have esterase (tannase) and gallate decarboxylase enzymes necessary for such a degradation. Gallic acid is an important dietary supplement that has several health benefits including anti-oxidant, anti-inflammatory, antibacterial, anti-allergic, anti-mutagenic and anti-carcinogenic effects [65]. Among the reported significant antibacterial activity of gallic acid was its activity against bacterial strains such as; E. coli [66] and Bifidobacterium [67] available within the used consortium.
Nucleic acids
Nucleic acids i.e. purines were detected in most samples especially GT and GC. Increase in its levels is attributed to gut microbiota metabolism effected by or derived from caffeine catabolism, with caffeine found to be enriched in these two functional foods. Nucleic acids showed a general pattern of an increase upon incubation in most food fortified cultures and blank as observed in case of uracil and to a lesser extent in adenine (Suppl. Table S3). Hypoxanthine was also detected in all amended cultures as well as in blank as a byproduct of microbial oxidation of nucleic acids through a salvage pathway to act as a nitrogen source as well as energy sources and promoting protective functions to the colonic epithelium [60], [68]. Positive correlations of hypoxanthine and uracil with Bacteroidetes species Alloprevotella [69] was reported. In contrast, xanthine was detected exclusively in GC and GT amended cultures as a metabolic microbiota product of methylxanthines i.e. caffeine, theobromine and theophylline enriched in green coffee and tea (Suppl. Fig. S7) [22], [23].
Unsupervised and supervised multivariate data analyses of GC/MS dataset
Considering the complexity of acquired data in terms of the large number of specimens and monitored metabolites (96 × 131), Fig. 1, multivariate data analyses were employed to further determine the impact of treatment on microbiota metabolism in an untargeted manner. Untargeted multivariate analysis tools such as principle component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) can help reveal for differences between samples and postulate hypothesis related to the effect of applied functional food on microbiota metabolism. Principle component analysis (PCA) is an untargeted multivariate analysis tool that is used to bring multidimensional datasets into a two-dimensional plane that can be graphically represented as scatter plots. In the two-dimensional plane, data point coordinates on principle component 1 (PC1) and PC2 vectors are assigned to account for the maximum variability of multidimensional data [70], [71].
PCA analysis of the whole dataset
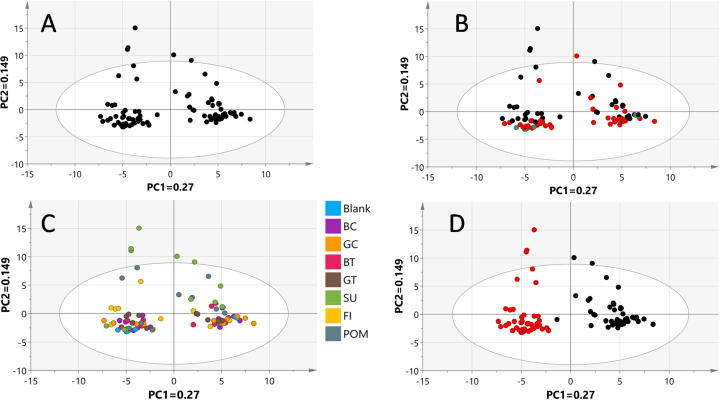
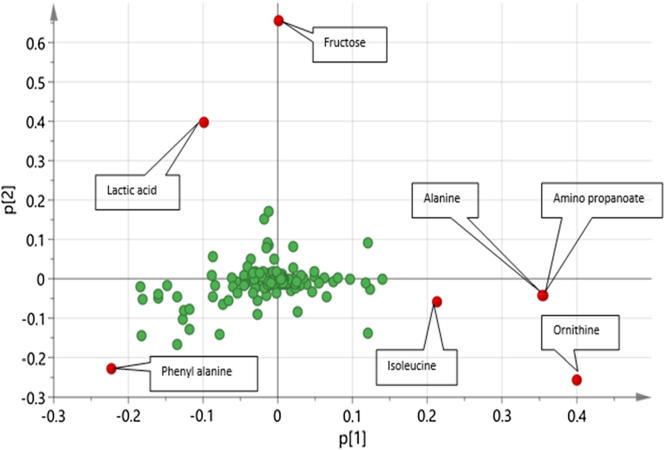
PCA was applied to the whole sample dataset to determine metabolome heterogeneity among all samples examined with no sort of samples classification. The principle component PC1/PC2 score plot derived from the whole GC–MS data accounted for 42% of the total variance (R2). This is not a high value and shows that variance between samples is high and needs more dimensions to cover all aspects. The whole pool of samples in the score plot, however, could be segregated into two big clusters distributed along PC 1 (Fig. 3A). These two major clusters appeared to be based upon incubation time that is (0.5 h versus 24 h incubation time as revealed in (Fig. 3D), suggesting this to be the most-variable parameter overcoming others. Labeling of samples based on the administered functional food extract dose levels (5 mg/mL, 0.5 mg/mL) and blank samples showed no clear segregation of sample in the score plot (Fig. 3B). The same result was also obtained when samples were colored based upon the amended functional food extract type (Fig. 3C), and in all cases they show overlap among specimens. All findings pinpointed that monitored metabolites are mostly derived from microbiota culture and not much represented by metabolites from amended functional foods since either of the identified big clusters (Fig. 3D) contained functional food extract treated samples along with their corresponding blank samples. Hierarchical cluster analysis (HCA) is another multivariate data analysis tool that provides a mean of intuitive graphical abstract of samples clustering pattern [72]. HCA analysis confirmed PCA results (Suppl. Fig. S8) by showing clear samples segregation aliquoted at 0.5 h (highlighted in a green box) compared to samples harvested at 24 h (blue box) disregarding treatment type (either blank or functional food treated samples) or dose level (0.5 mg/mL & 5 mg/mL). Results of whole sample data set modelling either from PCA or HCA fall in agreement with another report showing the impact of transient time on gut microbiota metabolites composition [40]. To pinpoint for metabolites mediating for such segregation and contributing to samples segregation with time course, the corresponding loading plot was inspected. Among detected metabolites, amino acids i.e. phenylalanine, ornithine and to a lesser extent organic acids, especially lactic acid, accounted for most of the variability observed along PC1 (Fig. 4).
Fig. 3.
PCA analysis of GC-MS metabolites dataset at 0.5 and 24 h for all treatments and blank. (A) PCA analysis of whole sample dataset unclassified. (B)PCA analysis of whole sample dataset classified based upon dose level (blank samples [Green], 0.5 mg/mL functional food extract treated sample [Red], and 5.0 mg/mL functional food extract treated sample [Black]). (C) PCA analysis of whole sample dataset classified based on functional food type (Black coffee, green coffee, black tea, green tea, ficus, sumac, pomegranate, and blank). (D) PCA analysis of whole sample dataset classified based on incubation time (0.5 h [Black], and 24 h [Red]).
Fig 4.
PCA Loading plot of whole sample dataset presented in Fig. 3 and showing metabolites prevalent at the 0.5 h incubation with positive p value (to the right) metabolites prevalent at the 24 h with negative p value (to the left). Sugar fructose contributed the some functional food treated sample discrimination (such as pomegranate sample) along with PC2.
PCA classification based upon functional food type versus blank
Due to the low model variance coverage (R2) and predictability (Q2) as revealed from whole sample datasets (Fig. 3), another PCA attempt was adopted to model each functional food extract amended culture vs. the blank at the two time points at 0.5 and 24 h (Suppl. Fig. S9). Both functional food and blank samples aliquoted at 0.5 h were positioned on the right side of PC1 with positive p values. Whereas its counterpart aliquots harvested at 24 h were aligned on the left side of PC1, with an overall covered variance from PC1 and PC2 from each dataset (Suppl. Fig. S9) showing in general higher variance coverage than of the whole sample dataset (Fig. 3). The score plots of BC (A), GC (B), BT (C) and GT (D) treated cultures showed consistent patterns of samples segregation being mostly based upon harvest time. Both blank and treated samples aliquoted at 0.5 h showed overlapping masses clustering at the positive side of PC1 versus samples aliquoted at 24 h showing two well-separated clusters distributed along PC2, which indicated no metabolome difference detected at the 0.5 h harvest point between blank samples and functional food. The separation of functional food treated samples at the positive side of PC2 versus its blank samples being located at the opposite side suggested for contributions of functional food metabolites or its biotransformed products in such a discrimination. It should be noted, that other the three additives FI (E), POM (F) and SUM (G) showed atypical behavior, in which both functional food and blank sample aliquoted at 24 h showed non-separable masses contrary to the previous models (Suppl. Fig. S9). In contrast, in both POM model score plot (Suppl. Fig. S9F) high scattering along PC2 was observed at either 0.5 h or 24 h incubation time indicating a higher influence of functional food composition on the microbiota metabolism. The same holds for the case of SUM (Suppl. Fig. S9G).
PCA classification based upon functional food type versus blank at 0.5 h
To better reveal for contributions from untransformed functional food metabolites in sample segregations as observed in Suppl. Fig. S9, we attempted to model each functional culture amended along with its corresponding blank sample aliquoted at 0.5 h, a time at which no significant biotransformation by microbiota is expected to occur (Suppl. Fig. S10). For BC (A), GC (B), BT (C), and GT (D) lower variance coverage was revealed compared to that in (Suppl. Fig. S9), and overlap between treatment and blank samples at both dose levels indicate that the functional food metabolite interference at 0.5 h is negligible. Such pattern was nevertheless not observed in case of POM (F) and SUM (G) models showing no overlap between functional food treated samples and blank, suggestive for significant functional food native metabolite interference within 0.5 h, and accounting for specimens segregation. The loading plot of pomegranate model revealed that sugars, especially fructose, short-chain fatty acid (SCFA), here lactic and succinic acid, and the nitrogenous compound and neuroactive amino acid GABA were already upregulated in the treated cultures compared to blank.
OPLS-DA classification based upon incubation time at 0.5 h versus 24 h
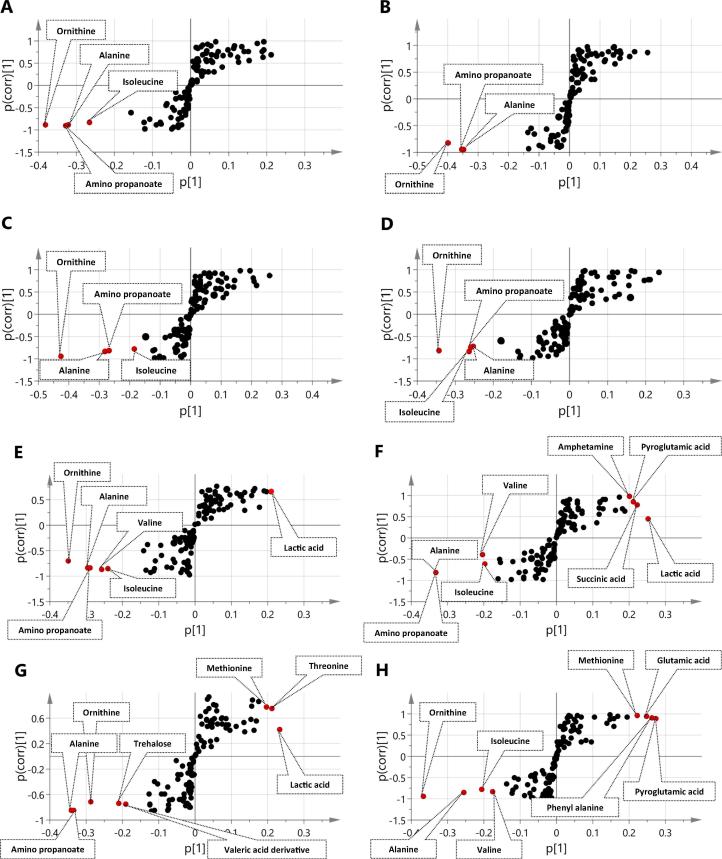
Considering that unsupervised data analysis failed to provide clear segregation in response to treatment for each respective time point, supervised orthogonal partial least squares discriminant analysis (OPLS-DA) was further employed to model each treatment separately viz. BC, GC, BT etc. Samples were classified as such by pooling samples for each treatment for the 2 dose levels 0.5 and 5 mg/ml as one class group at 0.5 h (a) versus 24 h (b) as another class group (Suppl. Fig. S11). Compared to PCA (Suppl. Fig. S9), each OPLS model showed clearer sample segregations (Suppl. Fig. S11). Model validation was based on estimating the total variance (R2), prediction goodness parameter (Q2) and p-value as detailed in (Suppl. Table S4). All of the models showed high repeatability, prediction and significantly low regression p-values suggestive of no overfitting of these models. Investigation of the S-loading plot allows to visualize both the covariance and the correlation structure between the X-variables and the predictive score t[1] [73] for each respective OPLS-DA model (Fig. 5) revealed for multiple findings. Metabolites with a positive p[1] value indicate an increase upon incubation, whereas metabolites with a negative p[1] value indicate higher abundance at 0.5 h and a decrease upon incubation (Fig. 5A–H).
Fig 5.
S-Plot of OPLS model of black coffee (A), Green coffee (B), Black tea (C), Green tea (D), Ficus (E), Pomegranate (F), Sumac (G) and Blank (G) classified based on incubation time. Samples were classified by pooling samples for each treatment at the 2 dose levels 0.5 and 5 mg/ml as one class group at 0.5 h (a) versus 24 h (b) as another class group. Metabolites increasing with time have positive p value while metabolites decreasing with time have negative p value.
Metabolites belonging to amino acids/nitrogenous compounds i.e., alanine, amino propanoate, isoleucine, valine and ornithine exhibited significant model influence at 0.5 h (negative p[1] on S-plot). In contrast, other metabolites related to the same classes such as leucine, methionine, glutamic acid and phenyl alanine showed significant model influence at 24 h (positive p[1] on S-plot). These findings propose mixed microbial metabolic activities that are either to utilize some of these amino acids as a substrate to form other compounds (catabolism) [38] or to be synthesized from other metabolites (anabolism). To provide stronger evidence for metabolites reprograming and to understand metabolic pathways, a Spearman rank correlation was employed to compute all pairwise correlations between metabolites across the entire dataset, depicted as a metabolite-metabolite correlation heat map (Suppl. Fig. S12). This metabolite correlation analysis shows a negative correlation of ornithine with succinic acid (r2 = −0.4) and tartaric acid (r2 = −0.35). Similar to ornithine, alanine shows a negative correlation with succinic acid and tartaric acid, in addition to fumaric acid (r2 = 0.43) and malic acid (r2 = 0.65). Alanine reduced levels are attributed to either serve as building unit in cell wall formation (peptidoglycan) or in its catabolism via aminotransferase into pyruvate [74]. In contrast to this, amino acids such as phenylalanine, threonine and pyroglutamic acid show positive p[1]-values in the S-plot with an increase upon incubation time (Fig. 5F–H). Though threonine is reported to be catabolized into acetate, microbiota can also synthesize it internally [38] and this can account for its increase with elapsed time. Such hypothesis is supported by the correlation analysis showing positive correlation of threonine with acids i.e. maleate (r2 = 0.64), succinate (r2 = 0.5) and tartarate (r2 = 0.5) which all showed upregulation with incubation time. Studies revealed that microbiota consumption and synthesis of amino acids is dependent on strain [75] and incubation time [40]. Sampling times for more than 24 h or utilization of several strains for each microorganism in the future can provide better evidence for such a hypothesis.
SCFA major metabolite markers of gut microbiota, i.e. lactic acid (Fig. 5E–G) and succinic acid, show higher p[1] values in the S-plot of (Fig. 5F), indicating an increase over time at 24 h by up to 17 fold in the pomegranate case. This most probably is due to its role as by-products of microbial metabolism. Other than primary metabolites converting into SCFA, succinic acid showed an increase with time (+p value in S-plot) acting as fermentation product of secondary metabolites. e.g., chlorogenic acid (CA) was reported to be fermented into succinic acid by microbiota [76], and could account for its 2.4-fold increase in BC at 24 h (Suppl. Table S3). CA is the main active constituent in coffee recognized as its slimming factor.
OPLS-DA classification based upon functional food type versus blank at 24 h
Considering that all of the above metabolic changes (Fig. 5) were related to the time course metabolism of microbiota and to help identify other metabolic changes specifically influenced by certain functional food treatment, OPLS-DA was performed by modelling each treatment at 24 h versus its blank sample at the same point for the 2 different dose levels. Derived S-plots for each case are presented in (Suppl. Fig. S4) and with metabolites that showed the strongest variation influence as reflect by their p-values listed in Table 1.
Table 1.
Major metabolites differentiating functional food treated samples against blank at 24 h post incubation as revealed from OPLS analysis.
| Metabolite | Category | BC | BT | Fi | GC | GT | POM | SU |
|---|---|---|---|---|---|---|---|---|
| Lactic acid | Acid | (+) | (+) | (+++) | (++) | (+) | (+++) | (+++) |
| Succinic acid | (++) | (++) | (+) | (++) | (+) | (++) | (+) | |
| 3-Deoxytetronic acid | (+) | |||||||
| Fumaric acid | (+) | |||||||
| Glycine | Amino acid | (+) | (+) | (+) | ||||
| Aspartic acid | (+) | (+) | ||||||
| Isoleucine | (+) | (++) | (++) | (+) | ||||
| Threonine | (+) | (+) | (+) | (+) | ||||
| Ornithine | (+) | |||||||
| Valine | (++) | (+++) | (++) | (+) | (+) | |||
| GABA | Nitrogenous compound | (++) | (++) | (++) | (+) | (+) | (++) | (+) |
| Amphetamine | (++) | (++) | (++) | (+) | (++) | (+) | ||
| Norvaline ester derivative | (+) | (+) | (+) | |||||
| Purine | Nucleic acid | (++) | (++) | |||||
| Uracil | (+) | |||||||
| 2-Hydroxy-3-methylvaleric acid | Phenolic | (++) | (++) | (++) | (++) | (++) | (+) | |
| Catechin | (+) | (+++) | ||||||
| Gallic acid | (++) | (+++) | (++) | |||||
| D-arabino-3-deoxy Hexonic acid | Sugar | (++) | (+) | (++) | (+) | (+) | ||
| Ribose-O-methyloxime | (+) | |||||||
| Fructofuranose | (+) | |||||||
| Fructose | (++) | (++++) | ||||||
| Sorbose | (+) | |||||||
| Unknown sugar | (+) |
Symbols (++++) indicate very high influence of S-plot, (+++) high influence, (++) intermediate, and (+) low influence.
Among the treatment models, SUM and POM amended cultures exhibited the highest discrimination from blank samples at 24 h incubation time owing to their high content in fructose followed by lactic acid and to a lesser extent in case of FI (Opuntia) treatment (Suppl. Fig. S4G & F). With regard to major markers indicative of secondary metabolite biotransformations by gut microbiota as revealed from S-plots, gallic acid was enriched in black tea treated samples (Suppl. Fig. S4C) owing to gut microbiota effect on theaflavin-3-gallate, a major polyphenol in black tea [77]. Other phenolics revealed as markers for a treatment effect (at 24 h) include catechin, gallic acid, and valeric acid derivatives especially in GT and BT samples (Suppl. Fig. S4C & D). Cleavage of ring C in catechin by microbiota is reported to yield valeric acid derivatives [77].
In contrast to these treatment specific markers revealed exclusively for GT and BT, metabolites such as sugars, organic acids, i.e. lactic and succinic acid, and GABA were detected as significant contributors to variation in almost all functional food treated cultures compared with blank samples at 24 h (Suppl. Fig. S4A–G). Gut microbiota such as Lactobacillus species and E. coli have been reported to secrete GABA as an acid resistance mechanism at low pH which may explain co-appearance of GABA with organic acid increase and other low acidic metabolites such as polyphenols in OPLS –derived S plot of several functional food treatments (Suppl. Fig. S4A, C, E & F). GABA can also be utilized by bacteria as carbon source through conversion to succinate which enters the tricarboxylic acid cycle [78] and this may explain the coexistence of GABA and succinic acid (r2 = 0.40) as strong influencers in S-Plots of functional food treated samples (Suppl. Fig. S4C & F). Such metabolite correlations could not be readily revealed from visual inspection of results and highlight the value of modelling data in results interpretation.
Accumulation of a norvaline derivative (Table 1), a branched chain non-proteinogenic amino acid has been observed in BT and FI treatments (Suppl. Fig. S4C & E). Previous research showed that this compound could only accumulate in high glucose based mineral salt media at limited oxygen supply as modified metabolic route [79]. However, norvaline ester showed higher abundance with elapsed time in functional food treated samples except in case of POM and SUM (Rhus coriaria). Pyruvate, a sugar metabolism intermediate, may convert to norvaline via α-isopropyl malate synthase instead of being converted to SCFA, i.e. lactate, which may explain its absence in POM and SUM both showing upregulation of lactate and succinate at 24 h (Suppl. Fig. S4F & G).
Conclusion
Our results provide insights into gut microbiota altered metabolisms in response to different functional food extracts at the metabolite level and define general and specific biomarkers for each functional food type. In general, functional food amendment to gut microbiota appeared to alter its metabolism in two different scenarios. First, functional food components can serve as a substrate to microbiota metabolism as in case of purine alkaloids such as caffeine acting as precursors of purine by microbiota demethylation. An alternative mechanism is that functional food components modify the extent, existing metabolic pathways are activated within microbiota, showing e.g. GABA production in presence of higher acidity induced by metabolites such as polyphenols and organic acids. Nevertheless, it should be noted that current work does not look at microbial strains growth in response to different functional food treatments, further analysis using 16S ribosomal RNA sequencing and as well viability of each individual strain in the presence of each tested functional food should be pursued.
This study demonstrated the metabolic pathways adopted by microbiota in the generation of SCFA through either nitrogen metabolism of amino acids or carbon metabolism of sugars lead to an ultimate increase of SCFA under consumption of (most members) of these metabolic precursors (Fig. 6A and B). However, most of these changes are related to the change the microbiota environment encounters with time and is not specific for certain functional foods with a few exceptions.
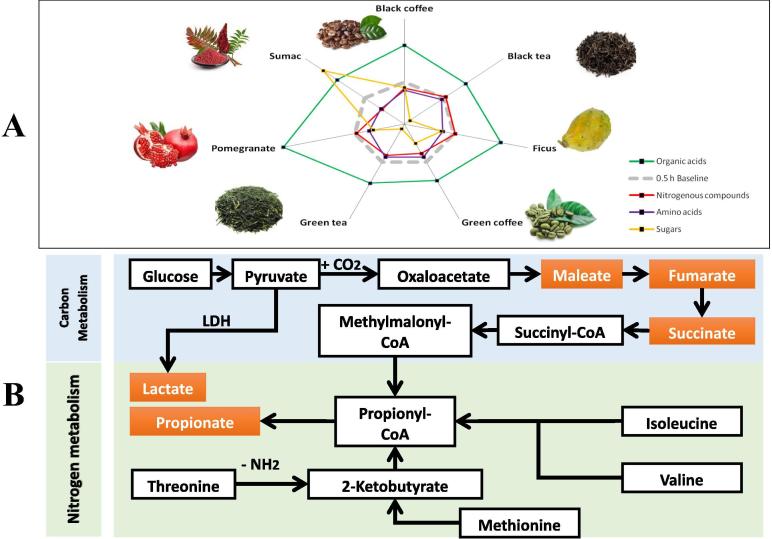
Fig. 6.
Diagram sketch outlining the major alterations in microbiota metabolic pathways. (A) Radar Chart summarizing the relative change in abundance of primary metabolites amino acids, nitrogenous compounds, sugars, and organic acids after incubation for 24 h compared to their relative abundance at 0.5 h (identified in dashed gray line). Measurement points indicate abundance in functional food treated samples. All points outside the gray dashed frame indicate increased abundance with time whereas points within the gray frame indicate lower abundance with time. While organic acid increase in all functional food treated samples, all of amino acids, nitrogenous compounds and sugars are reduced with time (except for sumac treated sample with regard to sugar abundance 24 h post incubation). (B) Schematic diagram outlining the metabolic pathways adopted by microbiota to convert amino acids and sugars into SCFA through either Carbon metabolism or Nitrogen metabolism and explaining the part A findings of the Radar chart.
Fig. 6A outlines the major changes in microbiota primary metabolite classes viz. sugars, nitrogenous compounds, acids and amino acids in response to functional food treatments and the underlying mechanism (Fig. 6B). Here three (partially overlapping) groups can be separated: caffeine containing additives, changing expectedly nitrogeneous compound metabolism, polyphenolics rich additives like tea, changing a number of properties outlined in detail above, and pomegranate and sumac, which inflict the largest differentiations through their sugar (fructose) and phenolics content.
Further studies that apply high resolution visualization of metabolite pools, such as nanoscale secondary ion mass spectrometry, coupled with stable isotope tracers will serve to provide further insight into the potential roles of free metabolites and their exact metabolic origin considering the complexity of microbial metabolisms. It also remains to clarify, if additional information on non-volatile metabolites, which might be captured by LC-MS, and a correlation to the strain composition changes with time, will be useful or not. These data, coupled with the ongoing outputs from rapidly developing additional “omics” studies (i.e., genomics, transcriptomics, and proteomics) will aid in further elucidating the metabolic cross-talk both within and between partners, which is essential for maintaining a functional healthy gut environment.
Obviously neither our functional food selection nor that of the microorganisms does cover all types, and no setup can do that. But we see that the platform presented here can be employed to assess other systems, containing e.g. other metabolite classes like saponins, sulfur organics like in garlic or onion, or even of complex mixture as typical in traditional Chinese medicine TCM.
The above mentioned limitation of this study to metabolites amenable to GC–MS detection, i.e. those of primary origin except for a few simple phenolics, may underpin the impact of secondary metabolites not detected by this analytical platform such as plant polyphenols or not represented in this study at all as with plant terpenoids in leading to a variation in microbiota composition and metabolism. Still, clear evidence for functional food comparable metabolites composition impact on microbiota culture metabolism was observed, exemplified in xanthine rich extracts i.e. GC, GT, BC, BT versus hydrolysable tannins in POM and SUM. Further, clinical studies ought to be considered in the future by analyzing stool metabolism in humans exposed to selected functional foods for defined periods, which would provide a true translation of this in vitro yet fundamental assay results to future research in the field of functional food – gut microbiota interaction.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Dr. Mohamed A. Farag acknowledges the funding received by Jesour grant number 30 from the Academy of Scientific Research & Technology (ASRT) and Cairo University research grant number 16-57. MvB is thankful for partial funding by DFG in the framework of the SFB 1052 “Mechanisms of obesity”. We are thankful for technical assistance from Dorothee Düsterhöft.
Functional foods gut microbiota interaction
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.01.001.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Qin J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasan N., Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7 doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z, et al. Gut microbiota-mediated biotransformation of food components: the key for their biological functions. Abstracts of Papers of the American Chemical Society; 2018. 256.
- 4.Tengeler A.C., Kozicz T., Kiliaan A.J. Relationship between diet, the gut microbiota, and brain function. Nutr Rev. 2018;76(8):603–617. doi: 10.1093/nutrit/nuy016. [DOI] [PubMed] [Google Scholar]
- 5.Pushalkar S. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012;12(1):1. doi: 10.1186/1471-2180-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowland I.R. The role of the gastrointestinal microbiota in colorectal cancer. Curr Pharm Des. 2009;15(13):1524–1527. doi: 10.2174/138161209788168191. [DOI] [PubMed] [Google Scholar]
- 7.Mayer C., Brachhold K. Molecular nutrition-from gut microbiota to metabolomics and inter-individual nutrition. Mol Nutr Food Res. 2019;63(2) doi: 10.1002/mnfr.201970005. [DOI] [PubMed] [Google Scholar]
- 8.Nicoletti M. Nutraceuticals and botanicals: overview and perspectives. Int J Food Sci Nutr. 2012;63(Suppl 1):2–6. doi: 10.3109/09637486.2011.628012. [DOI] [PubMed] [Google Scholar]
- 9.Lentle R.G., Janssen P.W. Manipulating digestion with foods designed to change the physical characteristics of digesta. Crit Rev Food Sci Nutr. 2010;50(2):130–145. doi: 10.1080/10408390802248726. [DOI] [PubMed] [Google Scholar]
- 10.Laparra J.M., Sanz Y. Interactions of gut microbiota with functional food components and nutraceuticals. Pharmacol Res. 2010;61(3):219–225. doi: 10.1016/j.phrs.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Lee H.C. Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res Microbiol. 2006;157(9):876–884. doi: 10.1016/j.resmic.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Guthrie L., Wolfson S., Kelly L. The human gut chemical landscape predicts microbe-mediated biotransformation of foods and drugs. Elife. 2019;8 doi: 10.7554/eLife.42866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozdal T. The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients. 2016;8(2):78. doi: 10.3390/nu8020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thursby E., Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David L.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Z.T. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology. 2013;23(2):169–177. doi: 10.1093/glycob/cws138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farag M.A. Variation in Ceratonia siliqua pod metabolome in context of its different geographical origin, ripening stage and roasting process. Food Chem. 2019;283:675–687. doi: 10.1016/j.foodchem.2018.12.118. [DOI] [PubMed] [Google Scholar]
- 18.Farag M.A. Gas chromatography/mass spectrometry-based metabolite profiling of nutrients and antinutrients in eight lens and lupinus seeds (Fabaceae) J Agric Food Chem. 2018;66(16):4267–4280. doi: 10.1021/acs.jafc.8b00369. [DOI] [PubMed] [Google Scholar]
- 19.Becker N. Human intestinal microbiota: characterization of a simplified and stable gnotobiotic rat model. Gut Microbes. 2011;2(1):25–33. doi: 10.4161/gmic.2.1.14651. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Amaya D.B. Natural food pigments and colorants. Curr Opin Food Sci. 2016;7:20–26. [Google Scholar]
- 21.Caprioli G. Quantification of isoflavones in coffee by using solid phase extraction (SPE) and high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) J Mass Spectrom. 2016;51(9):698–703. doi: 10.1002/jms.3802. [DOI] [PubMed] [Google Scholar]
- 22.Svilaas A. Intakes of antioxidants in coffee, wine, and vegetables are correlated with plasma carotenoids in humans. J Nutr. 2004;134(3):562–567. doi: 10.1093/jn/134.3.562. [DOI] [PubMed] [Google Scholar]
- 23.Peng X. Effect of green tea consumption on blood pressure: a meta-analysis of 13 randomized controlled trials. Sci Rep. 2014;4:6251. doi: 10.1038/srep06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhary N. Anti-hyperlipidemic and fat pad lowering effect of standardized tea seed cake extract in mice fed high-fat and high-carbohydrate diet. Biotechnol Bioprocess Eng. 2015;20(1):157–167. [Google Scholar]
- 25.Forester S.C., Lambert J.D. The role of antioxidant versus pro-oxidant effects of green tea polyphenols in cancer prevention. Mol Nutr Food Res. 2011;55(6):844–854. doi: 10.1002/mnfr.201000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antunes-Ricardo M. Topical anti-inflammatory effects of isorhamnetin glycosides isolated from Opuntia ficus-indica. Biomed Res Int. 2015;2015:847320. doi: 10.1155/2015/847320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassiri-Jahromi S. Punica granatum (Pomegranate) activity in health promotion and cancer prevention. Oncol Rev. 2018;12(1):345. doi: 10.4081/oncol.2018.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shidfar F. The effect of sumac (Rhus coriaria L.)powder on serum glycemic status, ApoB, ApoA-I and total antioxidant capacity in type 2 diabetic patients. Iran J Pharm Res. 2014;13(4):1249–1255. [PMC free article] [PubMed] [Google Scholar]
- 29.Broeckling C.D. MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal Chem. 2006;78(13):4334–4341. doi: 10.1021/ac0521596. [DOI] [PubMed] [Google Scholar]
- 30.Farag M.A. Rats' urinary metabolomes reveal the potential roles of functional foods and exercise in obesity management. Food Funct. 2017;8(3):985–996. doi: 10.1039/c6fo01753c. [DOI] [PubMed] [Google Scholar]
- 31.Diether N.E., Willing B.P. Microbial fermentation of dietary protein: an important factor in diet-microbe-host interaction. Microorganisms. 2019;7(1) doi: 10.3390/microorganisms7010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suvorova I.A., Ravcheev D.A., Gelfand M.S. Regulation and evolution of malonate and propionate catabolism in proteobacteria. J Bacteriol. 2012;194(12):3234–3240. doi: 10.1128/JB.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farag M.A. Comparative mass spectrometry & nuclear magnetic resonance metabolomic approaches for nutraceuticals quality control analysis: a brief review. Recent Pat Biotechnol. 2014;8(1):17–24. doi: 10.2174/1389201014666131218125035. [DOI] [PubMed] [Google Scholar]
- 34.Rowland I. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57(1):1–24. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teufel R. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci USA. 2010;107(32):14390–14395. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zareian M. A glutamic acid-producing lactic acid bacteria isolated from Malaysian fermented foods. Int J Mol Sci. 2012;13(5):5482–5497. doi: 10.3390/ijms13055482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapot-Chartier M.P., Kulakauskas S. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact. 2014;13(Suppl 1):S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin R. A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids. 2017;49(12):2083–2090. doi: 10.1007/s00726-017-2493-3. [DOI] [PubMed] [Google Scholar]
- 39.Heiss C.N., Olofsson L.E. Gut microbiota-dependent modulation of energy metabolism. J Innate Immun. 2018;10(3):163–171. doi: 10.1159/000481519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tottey W. Colonic transit time is a driven force of the gut microbiota composition and metabolism: in vitro evidence. J Neurogastroenterol Motil. 2017;23(1):124–134. doi: 10.5056/jnm16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gill T. Novel inter-omic analysis reveals relationships between diverse gut microbiota and host immune dysregulation in HLA-B27-induced experimental spondyloarthritis. Arthritis Rheumatol. 2019 doi: 10.1002/art.41018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuang X. Fecal microbiota alterations associated with diarrhea-predominant irritable bowel syndrome. Front Microbiol. 2018;9:1600. doi: 10.3389/fmicb.2018.01600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerdo T. Gut microbial functional maturation and succession during human early life. Environ Microbiol. 2018;20(6):2160–2177. doi: 10.1111/1462-2920.14235. [DOI] [PubMed] [Google Scholar]
- 44.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19(1):29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 45.Hou C.W., Jeng K.C., Chen Y.S. Enhancement of fermentation process in Pu-erh tea by tea-leaf extract. J Food Sci. 2010;75(1):H44–H48. doi: 10.1111/j.1750-3841.2009.01441.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhao M. Determination and comparison of gamma-aminobutyric acid (GABA) content in pu-erh and other types of Chinese tea. J Agric Food Chem. 2011;59(8):3641–3648. doi: 10.1021/jf104601v. [DOI] [PubMed] [Google Scholar]
- 47.Martin C.R. The brain-gut-microbiome axis. Cell Mol Gastroenterol Hepatol. 2018;6(2):133–148. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakanaka A. Distinct signatures of dental plaque metabolic byproducts dictated by periodontal inflammatory status. Sci Rep. 2017;7:42818. doi: 10.1038/srep42818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu C. The colonic microbiome and epithelial transcriptome are altered in rats fed a high-protein diet compared with a normal-protein diet. J Nutr. 2016;146(3):474–483. doi: 10.3945/jn.115.223990. [DOI] [PubMed] [Google Scholar]
- 50.Inouye M., Pardee A.B. Requirement of polyamines for bacterial division. J Bacteriol. 1970;101(3):770–776. doi: 10.1128/jb.101.3.770-776.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tofalo R., Cocchi S., Suzzi G. Polyamines and gut microbiota. Front Nutr. 2019;6(16) doi: 10.3389/fnut.2019.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tassoni A., Awad N., Griffiths G. Effect of ornithine decarboxylase and norspermidine in modulating cell division in the green alga Chlamydomonas reinhardtii. Plant Physiol Biochem. 2018;123:125–131. doi: 10.1016/j.plaphy.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 53.Gao K. Time-course responses of ileal and fecal microbiota and metabolite profiles to antibiotics in cannulated pigs. Appl Microbiol Biotechnol. 2018;102(5):2289–2299. doi: 10.1007/s00253-018-8774-2. [DOI] [PubMed] [Google Scholar]
- 54.Collins J. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature. 2018;553(7688):291–294. doi: 10.1038/nature25178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albert K., Rani A., Sela D.A. The comparative genomics of Bifidobacterium callitrichos reflects dietary carbohydrate utilization within the common marmoset gut. Microb Genom. 2018;4(6) doi: 10.1099/mgen.0.000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee H.J., Yoon Y.S., Lee S.J. Mechanism of neuroprotection by trehalose: controversy surrounding autophagy induction. Cell Death Dis. 2018;9(7):712. doi: 10.1038/s41419-018-0749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Felice V.D. Microbiota-gut-brain signalling in Parkinson's disease: implications for non-motor symptoms. Parkinsonism Relat Disord. 2016;27:1–8. doi: 10.1016/j.parkreldis.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 58.Bernalier-Donadille A. Fermentative metabolism by the human gut microbiota. Gastroentérologie Clin Biol. 2010;34:S16–S22. doi: 10.1016/S0399-8320(10)70016-6. [DOI] [PubMed] [Google Scholar]
- 59.Nghiem N.P., Kleff S., Schwegmann S. Succinic acid: technology development and commercialization. Fermentation. 2017;3(2):26. [Google Scholar]
- 60.Liu Y.-P. Economical succinic acid production from cane molasses by Actinobacillus succinogenes. Bioresour Technol. 2008;99(6):1736–1742. doi: 10.1016/j.biortech.2007.03.044. [DOI] [PubMed] [Google Scholar]
- 61.Guettler M.V., Rumler D., Jain M.K. Actinobacillus succinogenes sp. nov., a novel succinic-acid-producing strain from the bovine rumen. Int J Syst Bacteriol. 1999;49(Pt 1):207–216. doi: 10.1099/00207713-49-1-207. [DOI] [PubMed] [Google Scholar]
- 62.Connors J., Dawe N., Van Limbergen J. The role of succinate in the regulation of intestinal inflammation. Nutrients. 2019;11(1) doi: 10.3390/nu11010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Detman A. Cell factories converting lactate and acetate to butyrate: clostridium butyricum and microbial communities from dark fermentation bioreactors. Microb Cell Fact. 2019;18(1):36. doi: 10.1186/s12934-019-1085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Behera S.S., Ray R.C., Zdolec N. Lactobacillus plantarum with functional properties: an approach to increase safety and shelf-life of fermented foods. Biomed Res Int. 2018;2018:9361614. doi: 10.1155/2018/9361614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samuel K.G. Effects of dietary gallic acid supplementation on performance, antioxidant status, and jejunum intestinal morphology in broiler chicks. Poult Sci. 2017;96(8):2768–2775. doi: 10.3382/ps/pex091. [DOI] [PubMed] [Google Scholar]
- 66.Kang J. The specific effect of gallic acid on Escherichia coli biofilm formation by regulating pgaABCD genes expression. Appl Microbiol Biotechnol. 2018;102(4):1837–1846. doi: 10.1007/s00253-017-8709-3. [DOI] [PubMed] [Google Scholar]
- 67.Gwiazdowska D. The impact of polyphenols on Bifidobacterium growth. Acta Biochim Pol. 2015;62(4):895–901. doi: 10.18388/abp.2015_1154. [DOI] [PubMed] [Google Scholar]
- 68.Lee J.S. Hypoxanthine is a checkpoint stress metabolite in colonic epithelial energy modulation and barrier function. J Biol Chem. 2018;293(16):6039–6051. doi: 10.1074/jbc.RA117.000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng W. Effects of a galacto-oligosaccharide-rich diet on fecal microbiota and metabolite profiles in mice. Food Funct. 2018;9(3):1612–1620. doi: 10.1039/c7fo01720k. [DOI] [PubMed] [Google Scholar]
- 70.Lyu J. Characterization of Chinese white-flesh peach cultivars based on principle component and cluster analysis. J Food Sci Technol. 2017;54(12):3818–3826. doi: 10.1007/s13197-017-2788-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farag M.A. Cytotoxic effects of Sarcophyton sp. soft corals-is there a correlation to their NMR fingerprints? Mar Drugs. 2017;15(7) doi: 10.3390/md15070211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farag M.A. Salicylic acid and its derivatives elicit the production of diterpenes and sterols in corals and their algal symbionts: a metabolomics approach to elicitor SAR. Metabolomics. 2018;14(10):127. doi: 10.1007/s11306-018-1416-y. [DOI] [PubMed] [Google Scholar]
- 73.Wiklund S. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80(1):115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 74.Pena-Soler E. Structural analysis and mutant growth properties reveal distinctive enzymatic and cellular roles for the three major L-alanine transaminases of Escherichia coli. PLoS ONE. 2014;9(7):e102139. doi: 10.1371/journal.pone.0102139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Biggs M.B. Systems-level metabolism of the altered Schaedler flora, a complete gut microbiota. ISME J. 2017;11(2):426–438. doi: 10.1038/ismej.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomas-Barberan F. In vitro transformation of chlorogenic acid by human gut microbiota. Mol Nutr Food Res. 2014;58(5):1122–1131. doi: 10.1002/mnfr.201300441. [DOI] [PubMed] [Google Scholar]
- 77.Chen H. The microbiota is essential for the generation of black tea theaflavins-derived metabolites. PLoS ONE. 2012;7(12):e51001. doi: 10.1371/journal.pone.0051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Strandwitz P. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol. 2019;4(3):396–403. doi: 10.1038/s41564-018-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soini J. Norvaline is accumulated after a down-shift of oxygen in Escherichia coli W3110. Microb Cell Fact. 2008;7:30. doi: 10.1186/1475-2859-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.