Abstract
Probiotics are important bacteria due to their benefit on human health. In this study, four strains of lactic acid bacteria from chicken bile were isolated and the strain with the best antimicrobial activity was selected for further identification and evaluation on its probiotic traits and safety. The strain was identified as Enterococcus faecium by biochemical characterization and 16S rDNA gene sequencing. The strain, named E. faecium MK-SQ-1, was tolerant to acid (pH 3.0), bile salts (up to 0.3%) or trypsin (up to 0.4%) for 3 h and it was able to survive from high temperature (up to 60 °C) for 15 min. This strain inhibited the growth of Salmonella enteritidis and Staphylococcus aureus intermediately. The genes responsible for virulence including asa1, cylA, efaA, esp, gelE and hyl were absent and the mice administrated orally with a very high dose (2 × 109 CFU) of the strain daily for 35 days were not found abnormal. The strain enhanced the serum IgG level and phagocytic index of mice significantly by daily oral administration at a high dose (2 × 108 CFU) for 21 days (p < 0.05). The strain did not have multi-antibiotic resistance and vancomycin resistance. Comprehensive evaluation showed E. faecium MK-SQ-1 could be a candidate as a probiotic strain used in human or animals.
Keywords: Enterococcus faecium, Probiotics, Safety, Probiotic properties
Introduction
Probiotics are a group of living microorganisms which confer a health benefit to the host when administrated in adequate numbers (FAO/WHO 2006). Probiotics can balance the intestinal flora, interfere with the colonization of pathogenic bacteria which prevent from mucosal infection, regulate immunity, and thus improve the health level of the host (Cross 2002; Servin and Coconnier 2003). Probiotics have been reported to be tried in treating such diseases as ulcerative colitis (Floch 2010), acute diarrhea (Canani et al. 2007), antibiotic associated diarrhea (Alam and Mushtaq 2009), colorectal cancer associated diarrhea (Osterlund et al. 2007), irritable bowel syndrome (Krammer et al. 2005), cholesterolemia (Hlivak et al. 2005), hypertension (Lye et al. 2009), and so on.
Enterococci are Gram-positive and facultative anaerobic bacteria and belong to lactic acid bacteria (LAB) for their ability to produce lactic acid during the growth. They are widely distributed in animal intestine, water surface, soil, some plants and fermented food (Gaglio et al. 2016). Although some strains of Enterococcus are pathogenic to human or animals, many strains are nonpathogenic and even some are used as probiotics in medicine, food and animal feed. Compared with bifidobacteria and most lactobacilli, which require strict anaerobic conditions for growth, the facultative anaerobic enterococci are more convenient for cultivation. Enterococcus faecium is the preferred species as probiotic candidate in the genus Enterococcus. Among the various strains of enterococci proven to be probiotic potential by far, E. faecium M-74 (Hlivak et al. 2005) and E. faecium SF68 (Canani et al. 2007) are well known as commercial probiotic strains.
A successful probiotic should be safe and able to resist the digestive effects of various digestive juices of the host, colonize in the host digestive tract and conferring health benefit (Fuller 1989). The candidate bacterial strains should be analyzed comprehensively on their safety and probiotic properties. In addition, isolation and screening from various sources were needed to increase the chance of obtaining competent probiotic strains. In this research, a strain of antibacterial coccus isolated from chicken bile was identified as E. faecium after biochemical and molecular assay, and it was named MK-SQ-1. The growth dynamics, digestive tolerance, antibacterial activity and safety of the isolate were comprehensively studied, which provided the evidence for it to be a successful probiotic for human or animals.
Materials and methods
Strains, reagents and animals
The indicator strain of Salmonella enteritidis CVCC-3377 was purchased from China Veterinary Culture Collection Center and the strain of Staphylococcus aureus CICC-23926 was purchased from China Center of Industrial Culture Collection. The Man, Rogosa and Sharpe (MRS) agar and broth media, the Mueller–Hinton (MH) agar and the biochemical assay tubes were purchased from Hopebiol, Qingdao, China. DNA markers, DNA extract kits and DNA purification kits were purchased from Tiangen, Beijing, China. Ten 5-month-old indigenous chickens were purchased from the local farmers market in Tai’an, China. Kunming mice weighing 18–22 g were purchased from China Biological Products, Inc., Tai’an, China. All animals used in this experiment were handled according to the principles of Shandong Agricultural University Animal Care and Use Committee.
Isolation of bacterial strains and primary screening
The bile was taken out aseptically from the gall bladders of the chickens and plated on MRS agar plates with 0.2 ml per plate. The plates were incubated at 37 °C for 48 h. The colonies with different morphological features were picked randomly and purified by streaking on MRS agar. The pure cultures of the isolates were tested for Gram’s staining and cell morphology under a light microscope. The isolates were grown in MRS broth overnight at 37 °C for further test.
Antibacterial assay was performed on each isolated colony by the routine method of disc agar diffusion (Bauer et al. 1966). Briefly, 0.2 ml suspension of indicator strains, Salmonella enteritidis CVCC-3377 and Staphylococcus aureus CICC-23926, at the concentration of 1.0 × 108 CFU/ml was coated on the surface of MH agar plates, respectively. Discs pre-soaked in the overnight culture of isolated strains were pasted on the surface of MH agar plates coated with the indicator bacteria before incubated at 37 °C for 24 h. Finally, the inhibition zones around the discs were measured and presented as the average inhibition diameter in millimeters. The strain with the best antimicrobial activity was selected for further study.
Identification of the isolated strain
Physiological and biochemical feature such as utilization of various sugars, activity of catalase, and growth at 45 °C or in 7% NaCl were tested in terms of routine ways (Dong and Cai, 2001). In these tests, bacterial minimum biochemical reaction tubes (Hopebiol, China) were used according to the manufacturer’s instructions.
The total genomic DNA of the isolated strain was extracted using bacterial genomic DNA extract kit (Tiangen, China.) according to the manufacturer’s instructions and the 16S rRNA gene was amplified by using 16S rRNA universal primers following the procedures described by Feng et al. (2017). The PCR product was purified and sequenced (Songon Biotech, Shanghai, China). Then the sequence was subjected to BLAST in the GenBank database. The strain was identified as E. faecium and named MK-SQ-1.
Bile salts and acid tolerance test
The resistance to bile salts and acid of the strain was assayed following the method of Feng et al. (2017) with minor modifications. Briefly, the inoculum of MK-SQ-1 was individually added to pH 1.0, 2.0, 3.0, 4.0 saline and the pH 7.0 saline as control at 1% (v/v) and incubated for 3 h at 37 °C. The samples were plated on MRS agar plates at a tenfold gradient dilution. After incubated for 36 h at 37 °C, the colony forming units (CFU) and the bacterial survival rate were calculated. Bacterial survival rate (%) = (log CFU N1/log CFU N0) × 100%. N1 represents the treatment groups and N0 represent the control. The bile salts (Songon Biotech, China) was dissolved in pH 7.0 saline as stock solution and prepared for solutions of 0.1%, 0.2%, 0.3%, 1.0% and 2.0% (m/v). The inoculum of MK-SQ-1 was added to the solutions at 1% (v/v) and incubated for 3 h at 37 °C. The survival rate of the strain was determined with the formula described above.
Trypsin and heat tolerance test
The trypsin (Songon Biotech, China) was prepared for solutions of 0.4%, 0.8%, 1.2%, 1.6% and 2.0% (m/v) with 0.9% NaCl as solvent. The inoculum of MK-SQ-1 was added to the solutions at 2% (v/v) and incubated for 3 h at 37 °C. The survival rate of bacteria was determined with the formula described in 2.4. Heat tolerance was evaluated according to Paéz et al. (2012), with some modification. Briefly, the inoculum was diluted with normal saline at 1% (v/v) and incubated in water bath for 15 min at 37 °C, 50 °C, 60 °C and 80 °C, respectively. The bacterial survival rate was measured with the formula described in 2.4.
Dynamics of growth and acid production
The dynamics of growth of MK-SQ-1 was studied in the routine ways (Pellegrino et al. 2019). Briefly, the initial inoculum of MK-SQ-1 was inoculated into MRS broth and cultured at 37 °C and the OD600 value was measured every 2 h with the MRS broth as the blank control. The pH value was also measured at the same time. The growth curve was drawn with the OD600 value as Y-axis and the time as X-axis. The acid-producing curve was drawn with the pH value as Y-axis and the time as X-axis.
Test for surface hydrophobicity
The surface hydrophobicity (H) of the strain MK-SQ-1 was tested according to the methods described by Solieri et al. (2014), after slight modification as follows. The MK-SQ-1 culture was centrifuged at 4000 r/min for 10 min and the bacteria were collected and washed twice with normal saline. The bacteria were resuspended in normal saline and the OD580 value of the suspension was adjusted to about 1.00. Two milliliter suspension was fully mixed with 2 ml xylene on a vortex mixer for 120 s. The water phase was separated from the organic phase after standing for 180 min at room temperature and measured for the OD580 value. The surface hydrophobicity was calculated using the formula below. H (%) = [(A0 − A)/A0] × 100%. A0 and A represent the OD580 value of the bacterial suspension before mixing with xylene and after standing, respectively.
Virulence genes detection and antibiotics susceptibility test
Specific primers (Table 1) were used to detect the presence and absence of virulence genes including asa1, cylA, efaA, esp, gelE and hyl in the genome of E. faecium MK-SQ-1. PCR was performed according to the protocol described in previous report (Lins et al. 2013). The amplified fragments were observed by 1% agarose gel electrophoresis. The strain was tested for the susceptibility to 20 antibiotics by using the method of disc agar diffusion (Bauer et al. 1966).
Table 1.
Primers for virulence genes
| Gene | Primer (5′–3′) | Fragment size (bp) | Annealing temperature (°C) |
|---|---|---|---|
| asa1 | F: GCACGCTATTACGAACTATGA | 375 | 56 |
| R: TAAGAAAGAACATCACCACGA | |||
| cylA | F: ACTCGGGGATTGATAGGC | 688 | 56 |
| R: GCTGCTAAAGCTGCGCTT | |||
| efaA | F: GACAGACCCTCACGAATA | 705 | 52 |
| R: AGTTCATCATGCTGTAGTA | |||
| esp | F: AGATTTCATCTTTGATTCTTGG | 510 | 56 |
| R: AATTGATTCTTTAGCATCTGG | |||
| gelE | F: TATGACAATGCTTTTTGGGAT | 213 | 56 |
| R: AGATGCACCCGAAATAATATA | |||
| hyl | F: ACAGAAGAGCTGCAGGAAATG | 276 | 56 |
| R: GACTGACGTCCAAGTTTCCAA |
Testing for effects of MK-SQ-1 on mice immunity
To evaluate the effects of MK-SQ-1 on mice immunity, 30-day-old Kunming mice were randomly divided into 3 groups, 14 per group. Group 1 were orally administrated 0.2 ml of MK-SQ-1 suspension of 1 × 106 CFU/ml every day for 21 days for each mouse, group 2 were administrated 0.2 ml of the suspension of 1 × 109 CFU/ml every day for 21 days for each, and the controls were administrated 0.2 ml of normal saline. Every administration was carried out after 3 h of fasting. 1 day after the last administration, 6 of each group were detected for serum IgG and IL-2 by using a mouse IgG ELISA kit (Yibang Biotech, China) and a mouse IL-2 ELISA kit (Yibang Biotech, China), respectively, according to the manufacturer’s instructions. 8 of each group were detected for the phagocytic index by using carbon particle clearance test described in previous report (George et al. 2014).
Evaluation of the safety of MK-SQ-1 on mice
To evaluate the safety of MK-SQ-1 on mice, 30-day-old Kunming mice were randomly divided into 2 groups, 8 per group. The experimental group was orally administrated 0.2 ml MK-SQ-1 suspension of 1 × 1010 CFU/ml every day for 35 days for each mouse and the controls were administrated 0.2 ml of normal saline in the same way. Each administration was carried out after 3 h of fasting. The mice were observed every day. One day after the last administration, the mice were subjected to blood routine examination and dissected for visceral examination in terms of routine means (Samtiya et al. 2019). Briefly, the concentration of red blood cells (RBC), white blood cells (WBC), hemoglobin (HGB), lymphocytes (LYM), granulocytes (GRA) and platelets (PLT) were detected by an automatic animal blood cell analyzer in the blood routine examination. The visceral organs including the heart, liver, spleen, lung, kidney, stomach, small intestine and large intestine were observed on gross. The tissue sections of the liver, spleen and kidney were made and subjected to observation under a light microscope after stained with hematoxylin and eosin. The hearts, livers, spleens and kidneys were weighed, and the organ indexes were calculated. Organ index equals the percentage of the body weight of the organ.
Statistical analysis
All the data were expressed as mean ± standard deviation (SD). Comparisons of data were performed using T test by SPSS 22.0. Results were considered statistically different at p < 0.05.
Results and discussion
Isolation and identification of MK-SQ-1
Four colonies with different morphological features were picked and purified from the total 18 colonies, and all the four isolates were Gram-positive and like cocci with spherical or ovoid shape by observation under a light microscope. The overnight cultures of the isolates were teste for the antibacterial activity. The inhibition diameter of one isolate was 11 ± 0.82 mm for S. enteritidis CVCC-3377 and 13 ± 1.37 mm for S. aureus CICC-23926, while inhibition diameters of the other three isolates were all below 10 mm for both the indicator bacteria. The strain with be best antibacterial activity was named MK-SQ-1 and selected for further study. The results of biochemical assay (Table 2) showed it was a strain of E. faecium based on the Bergey’s manual of systematic bacteriology, volume 1 (Buchanan and Gibbons 1984). The 16S rRNA gene was amplified by PCR and a gene fragment of 1469 bp was amplified. The sequence of the target gene was analyzed for homology by BLAST in the GenBank and the results showed that the 16S rRNA gene of MK-SQ-1 was 99.8% homologous with that of E. faecium INET2 published in the database. Thus, the isolate was determined to be an E. faecium.
Table 2.
Results of biochemical test of MK-SQ-1
| Items | Results | Items | Results |
|---|---|---|---|
| Aesculin | + | Mannitol | + |
| Cellobiose | + | Salicin | + |
| Maltose | + | Sorbitol | − |
| Saccharose | + | Raffinose | − |
| Synanthrin | + | Lactose | + |
| Hippuric acid | + | Catalase | − |
| Glycerin | + | 45 °C | + |
| Moveability | − | NV7.0 | + |
“+” indicates positive, “−” indicates negative, NV7.0 indicates growing in 7.0% NaCl
While the intestinal mucosa and the contents of man and animals are a recognized source of probiotic candidates (Ambadoyiannis et al. 2004), the trials to isolate probiotic potentials LAB from the bile of animals was lack. In addition, the LAB isolated from bile was expected to be highly bile tolerant. In this study, 10 gall bladders of chicken were used and total 18 colonies were found in the MRS plates, indicating a small number of LAB inhabited in some gall bladders. Because the antimicrobial activity may be one of the key characteristics for exclusion of pathogen survival in the intestine and expression of a probiotic effect for the host (Collado et al. 2005), all 4 selected isolates was tested for their activities against the pathogens S. enteritidis and S. aureus. The tested isolates showed various levels of antibacterial activity. The antibacterial activity of the strains may be attribute to the lactic acid they produced during growth. Other antibacterial substance such as bacteriocin was probable to contribute the activity. Many strains of enterococci have been proved to secrete bacteriocins with strong antibacterial activities (Juturu and Wu 2018). Further studies are needed to determine whether the isolates produce bacteriocin and the types of bacteriocin produced.
Tolerance to acid, bile salts, trypsin and heat
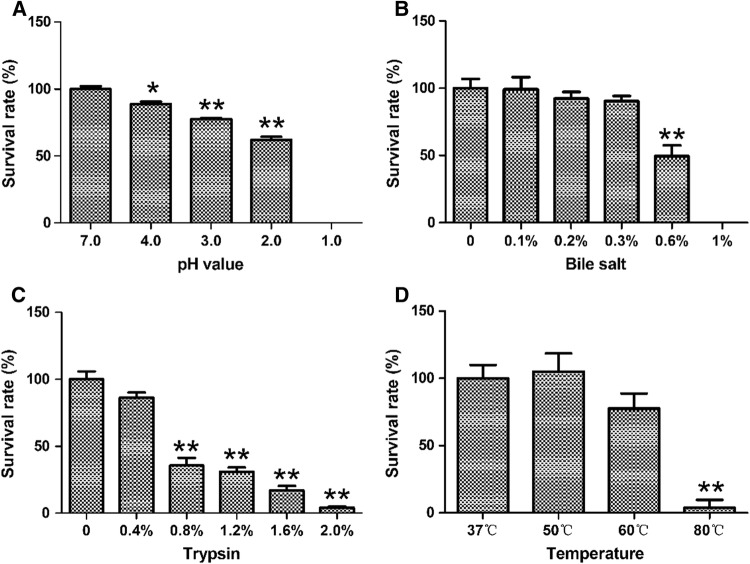
The tolerance of E. faecium MK-SQ-1 was tested by acid, bile salts, trypsin and high temperature. The survival rate of the strain was nearly 90% after staying in the solution with pH 4.0 for 3 h and decreased to about 80% by staying in the solution with pH 3.0 for 3 h (Fig. 1a). The strain showed strong tolerance to bile salts with 90% of survival rate in 0.3% bile salts for 3 h and kept nearly 50% vitality in 0.6% bile salts for 3 h (Fig. 1b). The strain was able to tolerate the hydrolysis of 0.4% trypsin for 3 h and kept nearly 90% of vitality (Fig. 1c). The heat tolerance tests showed that the strain kept nearly total vitality in 50 °C and about 80% of vitality in 60 °C for 15 min (Fig. 1d).
Fig. 1.
The survival rates of E. faecium MK-SQ-1 after treatments of acid (a), bile salt (b) or trypsin (c) for 3 h, or high temperature (d) for 15 min. *Indicate significant difference from control (p < 0.05) and **indicate very significant difference from control (p < 0.01)
Potential probiotic strains must tolerate the acidic environments, secretions of bile, pancreatic juice and intestinal juice in order to successfully pass through the stomach and small intestine. The pH of the pure gastric juice is around 2.0–3.0 and the pH of the gastric contents post food-intake goes up to more than 3.0, which causes most ingested microorganisms to die (Singh et al. 2012). The results of this experiment showed that the strain MK-SQ-1 could tolerate pH 3.0 for 3 h, indicating that it could pass through the stomach successfully. By the way, it was recommended that the strain should be taken after meal. The results match with the reports on different strains of E. faecium with acid-resistance. (Yoon et al. 2008; Rehaiem et al. 2014; Ayyash et al. 2018). The H+ in the acid changes cell membrane charge and deactivates various proteins by entering microbial cell. Common mechanisms for acid resistance in bacteria are pumping out protons through F1-F0-ATPase, generation of protective ammonia from arginine, glutamine and urea, repair or protection of macromolecules, and biofilm formation (Liu et al. 2015).
In this experiment, the strain MK-SQ-1 was able to tolerate 0.3% of bile salts for 3 h, indicating that it was able to survive through the small intestine which is believed to be with no more than 0.3% of bile salts (Prasad et al. 1998). This strain presented a higher or similar resistance level to bile salts compared to other reported bile-resistant strains of Enterococcus faecium (Yoon et al. 2008; Rehaiem et al. 2014; Ayyash et al. 2018). The high bile-salts-tolerance of this strain might relate to the fact that it was isolated from chicken bile. Bile salts can inhibit the growth of microorganisms in many ways. The main is that as an emulsifier of lipids, bile salts damage the phospholipid structure of bacterial cell membranes and organelles (Coleman et al. 1980). In addition to basic barriers constructed by the bacterial cell wall and cell membrane, one of the mechanisms of bacterial tolerance to bile salts depends on bacterial bile salt hydrolase (BSH). BSH hydrolyzes bile salts to form bile acids, which combines cholesterol to generate products with low solubility, which are difficult of entering bacteria. Also, the bacteria with high BSE have the potential to reduce the host’s absorption of cholesterol (Bustos et al. 2018; Li 2012). The BSE activity of this strain could be considered tested in the further studies.
Oral probiotics undergo digestion by powerful digestive enzymes in the small intestine before reaching the large intestine. In this study, a series concentration of trypsin was used to test the resistance level of MK-SQ-1 to trypsin. The strain was able to resist 0.4% trypsin digestion for 3 h, indicating that it can resist the proteolysis by pancreatic enzymes in the contents of intestine. Since 0.1% trypsin was used to simulate intestinal juice by Fernandez et al. (2003) in the previous research, 0.4% trypsin could be considered an overcritical condition for testing resistance strains. This research demonstrated the low pH in the stomach has more influences on bacterial survival than the presence of digestive enzymes in the small intestine, in accordance with the previous reports (Feng et al. 2017).
Heat-tolerance of bacteria is another property needed to be tested for probiotic potential evaluation. Not only the probiotic bacteria must be active under human body temperature, they must also be stable under higher temperature condition when being processed or packed for commercial purpose. Therefore, the survival rates at various temperatures of the strain were studied. The results showed during the 15 min of incubation time, nearly 80% of MK-SQ-1 could survive at 60 °C, indicating that it could tolerate the moderately-elevated-temperature when being processed or packed. The heat-tolerance of this strain was close to the enterococcal strain reported by Tinrat et al. (2018).
Dynamic of growth and acid-producing and surface hydrophobicity
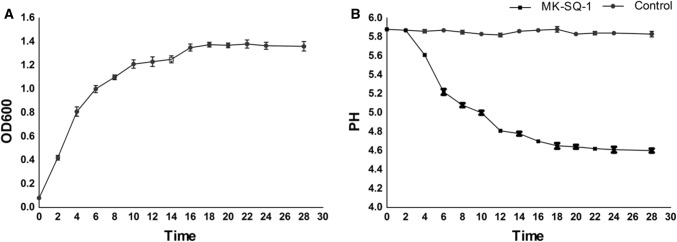
The growth curve of MK-SQ-1 (Fig. 2a) showed that the bacteria grew fast between 2 and 8 h post incubation and kept stable 12 h post incubation. The acid-producing curve (Fig. 2b) showed that pH value dropped steeply between 2 and 8 h post incubation and dropped slowly after that. The results of growth and acid-producing test illustrate that the MK-SQ-1 reaches the maximum amount between 10 and 14 h post incubation and should be collected during this period.
Fig. 2.
Growth curve (a) and acid-producing curve (b) of E. faecium MK-SQ-1
The surface hydrophobicity of E. faecium MK-SQ-1 was 34.53% ± 1.69% by triplicate testing, meaning that the strain was middle hydrophobic. Probiotics need to adhere and colonize on the surface of intestine for their probiotic effects. The bacteria that adhere to the surface of intestine avoid being excreted in feces, and at the same time, they prevent other microbes including pathogens from colonizing on the surface of intestine. The colonized probiotics also inhibit the growth of other microbes by taking over the nutrients and generating inhibitory secretions such as organic acid and bacteriocin (Juturu and Wu 2018). The adhesion capacity of bacteria is positively correlated to their cell surface hydrophobicity, which represents a benefit for bacterial maintenance in the gastrointestinal tract (Kos et al. 2003). Bacteria with high surface hydrophobicity tend to move towards the intestinal wall in order to avoid hydrophilic electrolytes in the intestinal environment. The strain MK-SQ-1 had a medium surface hydrophobicity, suggesting that it has a medium adhesive capacity.
Effects of feeding E. faecium MK-SQ-1 on mouse immunity
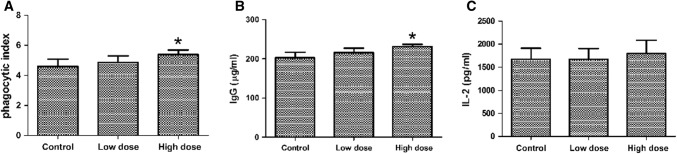
The mice orally given E. faecium MK-SQ-1 for 21 days were detected for the phagocytic index, the IgG and IL-2 levels in the serum. The results showed that the phagocytic index and the IgG level of the high dose group were significantly higher than the control (p < 0.05), while the IL-2 level was not obviously increased (p > 0.05) (Fig. 3). All the three immune indices were not detected significantly different between the low dose group and the control (p > 0.05).
Fig. 3.
Phagocytic index (a), serum IgG level (b) and serum IL-2 level of mice administrated with E. faecium MK-SQ-1. *Indicate significant difference from control (p < 0.05)
The results in this experiment showed that the strain MK-SQ-1 regulate the host’s immunity only when administrated in adequate numbers. It has been reported that the probiotic E. faecium S68 were able to modulate the intestinal mucosa and overall immunity of the host (Broom et al. 2006; Sun et al. 2010). The results in our experiments agree with these reports on the augment of serum IgG due to the probiotic administration. It is deemed that the administrated bacteria stimulate macrophages, dendritic cells and other immune cells in the Peyer’s patches to secrete some types of cytokines for activating Th cells and B lymphocytes which secret some types of cytokines or antibodies based on the general immune knowledge. The activation of macrophages could upregulate the phagocytic receptors for its increased phagocytic capacity. This reasoning could explain the augment of IgG and phagocytic index in our experiments. The non-significant change of IL-2 level in our experiments was also in accord with the report by Sun et al. (2010). Why the IL-2 secretion was not affected positively by the administration of the probiotics need further study.
Detection of virulence genes, antibiotic susceptibility and safety test on mice
The results of gene amplification showed the E. faecium MK-SQ-1 did not contain the genes encoding virulence factors in enterococcus, esp, cylA, Asa1, efaA, gelE and hyl. The susceptibility of the strain to 20 different antibiotics was tested and the results showed that it was sensitive to amoxicillin–clavulanate potassium, ampicillin, furadantin, phosphonomycin, chloromycetin, cefalexin, cefaclor and vancomycin, and resistant to polymyxin B, norfloxacin, doxycycline, gentamycin, tetracycline, cefalotin, cefotaxime and neomycin (Table 3). The mice were orally given E. faecium MK-SQ-1 at very high doses for 35 days were observed every day. During the experiment, the mice were in good spirit, with bright coat color, flexible movement, smooth respiration, normal diet, normal defecation and urine, and no death. There were no obvious ocular lesions in the heart, liver, spleen, lung, kidney, stomach, small intestine and large intestine by gross organ examination. Tissue sections of the liver, spleen and kidney in each group were observed and no significant pathological changes was found in these organs. The organ indexes of hearts, liver, spleen and kidney were measured, and no statistic difference was found between the groups (data was not shown). The blood routine indices including the concentrations of RBC, WBC, LYM, GRA, HGB and PLT were not significantly different in statistic (data was not shown) between the groups.
Table 3.
Susceptibility of E. faecium MK-SQ-1 to antibiotics
| Antibiotics | Dosage per disc (μg) | Criterion for R (mm) | Criterion for I (mm) | Criterion for S (mm) | Inhibition zone (mm)* | Result |
|---|---|---|---|---|---|---|
| Amoxicillin–clavulanate potassium | 20/10 | ≤ 13 | 13–18 | ≥ 18 | 25 ± 0.15 | S |
| Ampicillin | 10 | ≤ 16 | – | ≥ 17 | 23 ± 0.15 | S |
| Polymyxin B | 300 | ≤ 8 | 8–12 | ≥ 12 | 7 ± 0.10 | R |
| Furadantin | 300 | ≤ 14 | 14–17 | ≥ 17 | 24 ± 0.20 | S |
| Lincomycin | 2 | ≤ 14 | 14–21 | ≥ 21 | 20 ± 0.05 | I |
| Phosphonomycin | 200 | ≤ 12 | 13–18 | ≥ 19 | 24 ± 0.12 | S |
| Chloromycetin | 30 | ≤ 12 | 12–18 | ≥ 18 | 19 ± 0.10 | S |
| Norfloxacin | 10 | ≤ 12 | 12–16 | ≥ 16 | 12 ± 0.03 | R |
| Doxycycline | 30 | ≤ 18 | 18–22 | ≥ 22 | 8 ± 0.05 | R |
| Gentamycin | 10 | ≤ 12 | 12–15 | ≥ 15 | 10 ± 0.06 | R |
| Tetracycline | 30 | ≤ 14 | 14–19 | ≥ 19 | 9 ± 0.10 | R |
| Cefalexin | 30 | ≤ 14 | 14–17 | ≥ 17 | 18 ± 0.15 | S |
| Cefaclor | 30 | ≤ 14 | 14–18 | ≥ 18 | 18 ± 0.11 | S |
| Cefradine | 30 | ≤ 14 | 14–19 | ≥ 19 | 16 ± 0.05 | I |
| Cefoperazone | 75 | ≤ 15 | 15–21 | ≥ 21 | 16 ± 0.12 | I |
| Cefalotin | 30 | ≤ 14 | 14–18 | ≥ 18 | 9 ± 0.11 | R |
| Cefotaxime | 30 | ≤ 16 | 16–22 | ≥ 22 | 8 ± 0.03 | R |
| Vancomycin | 30 | ≤ 14 | 14–18 | ≥ 18 | 19 ± 0.13 | S |
| Neomycin | 30 | ≤ 17 | 17–23 | ≥ 23 | 11 ± 0.08 | R |
| Levofloxacin | 5 | ≤ 13 | 13–17 | ≥ 17 | 14 ± 0.10 | I |
S, sensitive; I, intermediate sensitive; R, resistant. *Values are given as mean ± standard deviation (SD) from triplicate experiments
Probiotics should have a health-promoting effect rather than disease-causing on the host. Genetic tests showed that the strain did not contain the 6 main virulence genes of enterococci. The absence of the virulence genes was in accordance to the results of safety test on mice. Taking the strain orally for 35 days at a very high dose, the mice were not found abnormal for the appearance, gross observation and tissue sections of the main organs, indicating that MK-SQ-1 was safe to mice. Although some strains of E. faecium are pathogenic to host, lots of strains were reported safe and probiotic potential to the hosts by in vivo evaluations (Pollmann et al. 2005; Scharek et al. 2005; Broom et al. 2006; Martin et al. 2012; Zhang et al. 2016), in vitro evaluations (Yoon et al. 2008; Rehaiem et al. 2014; Ayyash et al. 2018) or by both (Khalkhali and Mojganic 2018).
The safety of probiotics also involves their antimicrobial resistance. If probiotics are resistant to most of frequently-used antibiotics, they can avoid be killed when the host take the antibiotics. On the other hand, the drug resistant probiotics could transmit the resistance genes into pathogens to generate resistant pathogens, therefore a safe probiotic should not contain multi-antibiotic resistance genes. In particular, the probiotic enterococci should not contain the resistance gene to vancomycin, which is the last line of defense against enterococcal infections. The cases of infection of vancomycin-resistant enterococci (VRE) occurred occasionally around the world (Reik et al. 2008; Reyes et al. 2016) and VRE were reported to be isolated from meat and some environmental samples (Messi et al. 2006). The isolate E. faecium MK-SQ-1 was sensitive to 8 antibiotics including vancomycin and ampicillin and resistant to other 8 antibiotics such as tetracycline of the total 20 tested antibiotics, indicating it was relatively safe in terms of antimicrobial resistance. Palmer et al. (2012) divided E. faecium into two distinct clades by based on whole genome phylogeny. Clade A consists mainly of the hospital-associated strains and most of them are ampicillin-resistant. Clade B consists predominantly of isolates from the feces of healthy individuals and is characterized by susceptibility to ampicillin. According to this classification, the strain MK-SQ-1 should belongs to clade B. In order to ensure the reliable safety, more genes responsible for antibiotic resistance and virulence should be detected in the further studied. Nevertheless, the strain MK-SQ-1 was considered safe based on the acquired data.
Conclusion
A strain of LAB with anti-pathogenic-bacterial activity was isolated from chicken bile and identified as an E. faecium using biochemical and molecular methods in this study. The characteristics of the strain MK-SQ-1 such as acid-resistance, bile-salts-resistance, trypsin-resistance, heat-resistance and surface hydrophobicity suggested its application in food. The further investigation on safety showed the strain did not contain the main virulence genes of enterococcus and did not have pathogenicity to mice. In addition, the strain was not vancomycin-resistant and multi-antibiotic-resistant. Some immune indices of the mice including serum IgG and phagocytic index were detected elevated upon adequate administration of the strain. Comprehensive evaluation showed that the strain has potential as a probiotic used in human or animals.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Youfei Shi, Email: shiyoufei@163.com.
Mengkai Zhai, Email: zhaimengkai@foxmail.com.
Jinlian Li, Email: 815023462@qq.com.
Baoquan Li, Email: libq72@163.com.
References
- Alam S, Mushtaq M. Antibiotic associated diarrhea in children. Indian Pediatr. 2009;46:491–496. [PubMed] [Google Scholar]
- Ambadoyiannis G, Hatzikamari M, Litopoulou-Tzanetaki E, Tzanetakis N. Probiotic and technological properties of enterococci isolates from infants and cheese. Food Biotechnol. 2004;18:307–325. [Google Scholar]
- Ayyash M, Abushelaibi A, Al-Mahadin S, et al. In-vitro investigation into probiotic characterization of Streptococcus and Enterococcus isolated from camel milk. LWT Food Sci Technol. 2018;87:478–487. [Google Scholar]
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- Broom LJ, Miller HM, Kerr KG, Knapp JS. Effects of zinc oxide and Enterococcus faecium SF68 dietary supplementation on the performance, intestinal microbiota and immune status of weaned piglets. Res Vet Sci. 2006;80:45–54. doi: 10.1016/j.rvsc.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Buchanan RE, Gibbons NE. Bergey’s manual of systematic bacteriology. Beijing: Science Press; 1984. pp. 347–395. [Google Scholar]
- Bustos AY, de Valdez GF, Fadda S, Taranto MP. New insights into bacterial bile resistance mechanisms: the role of bile salt hydrolase and its impact on human health. Food Res Int. 2018;112:250–262. doi: 10.1016/j.foodres.2018.06.035. [DOI] [PubMed] [Google Scholar]
- Canani RB, Cirillo P, Terrin G, et al. Probiotics for treatment of acute diarrhoea in children: randomised clinical trial of five different preparations. BMJ. 2007;335(7615):340. doi: 10.1136/bmj.39272.581736.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R, Lowe PJ, Billington D. Membrane lipid composition and susceptibility to bile salt damage. BBA Biomembranes. 1980;599:294–300. doi: 10.1016/0005-2736(80)90075-9. [DOI] [PubMed] [Google Scholar]
- Collado MC, Gueimonde M, Herna’ndez M, Sanz Y, Salminen S. Adhesion of selected bifidobacterium strains to human intestinal mucus and the role of adhesion in enteropathogen exclusion. J Food Prot. 2005;68:2672–2678. doi: 10.4315/0362-028x-68.12.2672. [DOI] [PubMed] [Google Scholar]
- Cross ML. Microbes versus microbes: immune signals generated by probiotic lactobacilli and their role in protection against microbial pathogens. FEMS Immunol Med Microbiol. 2002;34:245–253. doi: 10.1111/j.1574-695X.2002.tb00632.x. [DOI] [PubMed] [Google Scholar]
- Dong X, Cai M. Manual of system determinative common bacteriology. Beijing: Science Press; 2001. pp. 370–398. [Google Scholar]
- FAO/WHO (2006) Probiotics in food. Health and nutritional properties and guidelines for evaluation. FAO Food and Nutritional paper No. 85 (ISBN 92-5-105513-0)
- Feng Y, Qiao L, Liu R, et al. Potential probiotic properties of lactic acid bacteria isolated from the intestinal mucosa of healthy piglets. Ann Microbiol. 2017;67:239–253. [Google Scholar]
- Fernandez MF, Boris S, Barbes C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J Appl Microbiol. 2003;94:449–455. doi: 10.1046/j.1365-2672.2003.01850.x. [DOI] [PubMed] [Google Scholar]
- Floch MH. Probiotic therapy for ulcerative colitis. J Clin Gastroenterol. 2010;44:237–238. doi: 10.1097/MCG.0b013e3181cf837f. [DOI] [PubMed] [Google Scholar]
- Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–378. [PubMed] [Google Scholar]
- Gaglio R, Couto N, Marques C, et al. Evaluation of antimicrobial resistance and virulence of enterococci from equipment surfaces, raw materials, and traditional cheeses. Int J Food Microbiol. 2016;236:107–114. doi: 10.1016/j.ijfoodmicro.2016.07.020. [DOI] [PubMed] [Google Scholar]
- George A, Chinnappan S, Choudhary Y, Bommu P, Sridhar M. Immunomodulatory activity of an aqueous extract of Polygonum minus Huds on Swiss albino mice using carbon clearance assay. Asian Pac J Trop Dis. 2014;4:398–400. [Google Scholar]
- Hlivak P, Odraska J, Ferencik M, et al. One-year application of probiotic strain Enterococcus faecium M-74 decreases serum cholesterol levels. Bratisl Lek Listy. 2005;106:67–72. [PubMed] [Google Scholar]
- Juturu V, Wu J. Microbial production of bacteriocins: latest research development and applications. Biotechnol Adv. 2018;36:2187–2200. doi: 10.1016/j.biotechadv.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Khalkhali S, Mojganic N. In vitro and in vivo safety analysis of Enterococcus faecium 2C isolated from human breast milk. Microb Pathog. 2018;116:73–77. doi: 10.1016/j.micpath.2018.01.012. [DOI] [PubMed] [Google Scholar]
- Kos B, Suskovic J, Vukovic S, et al. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. 2003;94:981–987. doi: 10.1046/j.1365-2672.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- Krammer HJ, Schlieger F, Harder H, et al. Probiotics as therapeutic agents in irritable bowel syndrome. Z Gastroenterol. 2005;43:467–471. doi: 10.1055/s-2004-813934. [DOI] [PubMed] [Google Scholar]
- Li G. Intestinal probiotics: interactions with bile salts and reduction of cholesterol. Procedia Environ Sci. 2012;12(Part B):1180–1186. [Google Scholar]
- Lins RX, Oliveira-Andrade A, Hirata JR, et al. Antimicrobial resistance and virulence traits of Enterococcus faecalis from primary endodontic infections. J Dent. 2013;41:779–786. doi: 10.1016/j.jdent.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tang H, Lin Z, Xu P. Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnol Adv. 2015;33:1484–1492. doi: 10.1016/j.biotechadv.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Lye HS, Kuan CY, Ewe JA, et al. The improvement of hypertension by probiotics: effects on cholesterol, diabetes, renin, and phytoestrogens. Int J Mol Sci. 2009;109:3755–3775. doi: 10.3390/ijms10093755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Pieper R, Kroger S, et al. Influence of age and Enterococcus faecium NCIMB 10415 on development of small intestinal digestive physiology in piglets. Anim Feed Sci Technol. 2012;175:65–75. [Google Scholar]
- Messi P, Guerrieri E, de Niederhäusern S, Sabia C, Bondi M. Vancomycin-resistant enterococci (VRE) in meat and environmental samples. Int J Food Microbiol. 2006;107:218–222. doi: 10.1016/j.ijfoodmicro.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Osterlund P, Ruotsalainen T, Korpela R, et al. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: a randomised study. Br J Cancer. 2007;97:1028–1034. doi: 10.1038/sj.bjc.6603990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paéz R, Lavari L, Vinderola G, et al. Effect of heat treatment and spray drying on lactobacilli viability and resistance to simulated gastrointestinal digestion. Food Res Int. 2012;48:748–754. [Google Scholar]
- Palmer KL, Godfrey P, Griggs A, et al. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio. 2012;3(1):e00318-11. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MS, Frola ID, Natanael B, et al. In vitro characterization of lactic acid bacteria isolated from bovine milk as potential probiotic strains to prevent bovine mastitis. Probiotics Antimicro Proteins. 2019;11:74–84. doi: 10.1007/s12602-017-9383-6. [DOI] [PubMed] [Google Scholar]
- Pollmann M, Nordhoff M, Pospischil A, Tedin K, Wieler LH. Effects of a probiotic strain of Enterococcus faecium on the rate of natural chlamydia infection in swine. Infect Immun. 2005;73:4346–4353. doi: 10.1128/IAI.73.7.4346-4353.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad J, Gill H, Smart J, Gopal PK. Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int Dairy J. 1998;8:993–1002. [Google Scholar]
- Rehaiem A, Belgacem ZB, Edalatian MR, Martínez B, Guerra NP. Assessment of potential probiotic properties and multiple bacteriocin encoding-genes of the technological performing strain Enterococcus faecium MMRA. Food Control. 2014;37:343–350. [Google Scholar]
- Reik R, Tenover F, Klein E, McDonald C. The burden of vancomycin-resistant enterococcal infections in US hospitals, 2003 to 2004. Diagn Microbiol Infect Dis. 2008;62:81–85. doi: 10.1016/j.diagmicrobio.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Reyes K, Bardossy AC, Zervos M. Vancomycin-resistant enterococci: epidemiology, infection prevention, and control. Infect Dis Clin N Am. 2016;30:953–965. doi: 10.1016/j.idc.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Samtiya M, Bhat MI, Gupta T, et al. Safety assessment of potential probiotic Lactobacillus fermentum MTCC-5898 in murine model after repetitive dose for 28 days (sub-acute exposure) Probiotics Antimicrob Prot. 2019 doi: 10.1007/s12602-019-09529-6. [DOI] [PubMed] [Google Scholar]
- Scharek L, Guth J, Reiter K, et al. Influence of a probiotic Enterococcus faecium strain on development of the immune system of sows and piglets. Vet Immunol Immunopathol. 2005;105:151–161. doi: 10.1016/j.vetimm.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Servin AL, Coconnier MH. Adhesion of probiotic strains to the intestinal mucosa and interaction with pathogens. Best Pract Res Clin Gastroenterol. 2003;17:741–754. doi: 10.1016/s1521-6918(03)00052-0. [DOI] [PubMed] [Google Scholar]
- Singh TP, Kaur G, Malik RK, Schillinger U, Guigas C, Kapila S. Characterization of intestinal Lactobacillus reuteri strains as potential probiotics. Probiotics Antimicrob Prot. 2012;4:47–58. doi: 10.1007/s12602-012-9090-2. [DOI] [PubMed] [Google Scholar]
- Solieri L, Bianchi A, Mottolese G, Lemmetti F, Giudici P. Tailoring the probiotic potential of non-starter Lactobacillus strains from ripened Parmigiano Reggiano cheese by in vitro screening and principal component analysis. Food Microbiol. 2014;38:240–249. doi: 10.1016/j.fm.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Sun P, Wang J, Jiang Y. Effects of Enterococcus faecium (SF68) on immune function in mice. Food Chem. 2010;123:63–68. [Google Scholar]
- Tinrat S, Khuntayaporn P, Thirapanmethee K, Chomnawang MT. In vitro assessment of Enterococcus faecalis MTC 1032 as the potential probiotic in food supplements. J Food Sci Technol. 2018;55:2384–2394. doi: 10.1007/s13197-018-3155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MY, Kim YJ, Hwang H. Properties and safety aspects of Enterococcus faecium strains isolated from Chungkukjang, a fermented soy product. LWT Food Sci Technol. 2008;41:925–933. [Google Scholar]
- Zhang ZF, Rolando AV, Kim IH. Effects of benzoic acid, essential oils and Enterococcus faecium SF68 on growth performance, nutrient digestibility, blood profiles, faecal microbiota and faecal noxious gas emission in weanling pigs. J Appl Anim Res. 2016;44:173–179. [Google Scholar]