Abstract
Myasthenia gravis (MG) is an autoimmune disorder resulting from antibodies against the proteins at the neuromuscular junction. Emerging evidence indicates that long non-coding RNAs (lncRNAs), acting as competing endogenous RNAs (ceRNAs), are involved in various diseases. However, the regulatory mechanisms of ceRNAs underlying MG remain largely unknown. In this study, we constructed a lncRNA-mediated ceRNA network involved in MG using a multi-step computational strategy. Functional annotation analysis suggests that these lncRNAs may play crucial roles in the immunological mechanism underlying MG. Importantly, through manual literature mining, we found that lncRNA SNHG16 (small nucleolar RNA host gene 16), acting as a ceRNA, plays important roles in the immune processes. Further experiments showed that SNHG16 expression was upregulated in peripheral blood mononuclear cells (PBMCs) from MG patients compared to healthy controls. Luciferase reporter assays confirmed that SNHG16 is a target of the microRNA (miRNA) let-7c-5p. Subsequent experiments indicated that SNHG16 regulates the expression of the key MG gene interleukin (IL)-10 by sponging let-7c-5p in a ceRNA manner. Furthermore, functional assays showed that SNHG16 inhibits Jurkat cell apoptosis and promotes cell proliferation by sponging let-7c-5p. Our study will contribute to a deeper understanding of the regulatory mechanism of MG and will potentially provide new therapeutic targets for MG patients.
Keywords: myasthenia gravis, ceRNA network, SNHG16, IL-10, let-7c-5p
Introduction
Myasthenia gravis (MG) is an autoimmune disorder caused by antibodies that attack proteins of the postsynaptic membrane at the neuromuscular junction, leading to muscle weakness and abnormal fatigability.1 Most MG patients have detectable antibodies against the acetylcholine receptor (AChR), while a small group of patients have antibodies against muscle-specific kinase (MuSK) or lipoprotein receptor-related protein 4 (LRP4).2 The production of antibodies is a T cell-dependent and B cell-mediated process. Cytokines produced by immune cells are crucial regulators of the pathogenesis of MG. For example, cytokines such as interleukin (IL)-2, interferon (IFN)-γ, and tumor necrosis factor (TNF)-α secreted by T helper (Th) cells stimulate the production of pathogenic antibodies, which are mediated by B cells.3 Moreover, both the expression of cytokines, such as IL-2, IFN-γ, and IL-10, and the number of peripheral blood mononuclear cells (PBMCs) secreting these cytokines are higher in patients with MG.4, 5, 6 Increasing evidence shows that noncoding RNAs (ncRNAs) are involved in the regulation of gene expression in the immune system, and thus they provide novel insight into the pathogenesis, diagnosis, and treatment of MG.7
Long ncRNAs (lncRNAs; more than 200 nt) interact with DNA, RNA, or proteins to modulate the expression of protein-coding genes, which play important roles in various biological processes, including the regulation of immune responses.7 It has been reported that aberrant expression of lncRNA IFNG-AS1 in PBMCs regulates CD4+ T cell activation in MG patients partly by influencing human leukocyte antigen (HLA)-DRB1 expression.8 MicroRNAs (miRNAs; ∼22 nt) are small functional ncRNA molecules that repress target gene expression and are involved in a wide range of biological processes.9 Accumulating evidence indicates that miRNAs contribute to the pathogenesis of MG by regulating important genes. For example, downregulation of miR-320a in MG patients induces inflammatory cytokine production by targeting mitogen-activated protein kinase 1 (MAPK1).10 miR-15a is downregulated in MG patients and causes abnormal activation of the immune response by regulating IFN-γ-induced protein 10 (IP-10), which is a highly inducible chemoattractant causing secretion of more IFN-γ by activated Th1 cells.11 Although some miRNAs and lncRNAs have been found to be implicated in MG, few studies have focused on exploring the interactions among miRNAs, lncRNAs, and genes underlying the pathogenesis of MG.
Recently, the competing endogenous RNA (ceRNA) theory was proposed, pointing out that RNA molecules containing miRNA response elements (MREs) could compete with each other by binding to a common miRNA.12 There is increasing evidence that lncRNAs function as ceRNAs, competing with mRNAs by acting as sponges of miRNAs, which relieve miRNA-mediated target repression.13 lncRNAs, acting as ceRNAs, are implicated in various diseases. For example, the lncRNA ZNFX1-AS1 has been shown to play an important role in the progression and metastasis of colorectal cancer by acting as a ceRNA of miR-144, thereby leading to the depression of its endogenous target gene polycomb group protein enhancer of zeste homolog 2 (EZH2).14 Additionally, lncRNAs, acting as ceRNAs, also play crucial roles in the regulation of the immune system and the development of autoimmune diseases. lncRNA MEG3 regulates RORγt expression by sequestering miR-17, which affects the regulatory T (Treg)/Th17 balance in asthma.15 Despite advances in ceRNA regulation, their potential roles in MG remain largely unknown. Thus, there is an urgent need to explore the ceRNA regulatory mechanism of MG and to develop novel biomarkers for the diagnosis and treatment of MG.
In the present study, we first constructed a global lncRNA-mediated ceRNA network involved in MG using a multi-step computational approach. Functional enrichment analysis was performed to reveal the potential roles of these lncRNAs in the network. Through analyzing the network and reviewing reliable publications, we designed biological experiments to further confirm the presence of an SNHG16 (small nucleolar RNA host gene 16)-mediated ceRNA regulatory mechanism through the let-7c-5p/IL-10 axis in MG. Our study provides novel insights into the ceRNA network and reveals potential roles of SNHG16 in MG.
Results
Construction of the lncRNA-Mediated MG-Associated ceRNA Network
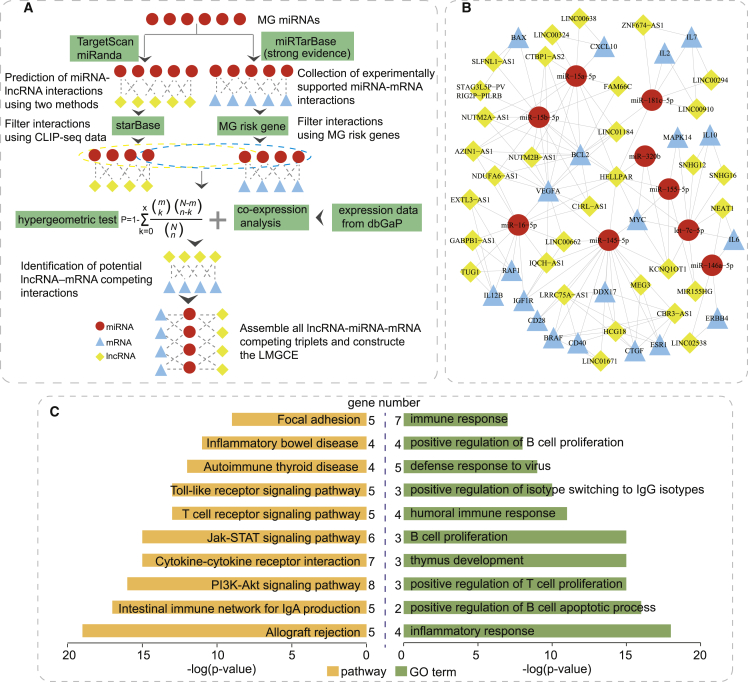
It is well known that lncRNAs can act as ceRNAs by binding miRNAs.13 To evaluate the lncRNA-mediated ceRNA regulation in MG, an lncRNA-mediated MG-associated ceRNA network (LMGCN) was constructed using a multi-step approach (Figure 1A). As a result, the LMGCN contained 9 miRNAs, 20 genes, 32 lncRNAs, and 147 edges (Figure 1B). We referred to the previous study16 to count the number of the primary relationship pairs of lncRNA-miRNA and the secondary relationship pairs of miRNA-mRNA (Table S1). For a given lncRNA(i), the number of the primary relationship pairs of lncRNA-miRNA represents the number of miRNAs linked with lncRNA(i), and the number of the secondary relationship pairs of miRNA-mRNA represents the sum of mRNAs linked with the above miRNAs. HELLPAR had the highest total number of lncRNA-miRNA and miRNA-mRNA pairs, which may play important roles in the network.
Figure 1.
The lncRNA-Mediated MG-Associated ceRNA Network (LMGCN)
(A) Schematic workflow to construct the LMGCN based on the ceRNA hypothesis. (B) LMGCN. Red circles represent miRNAs, blue triangles represent mRNAs, and yellow rhombi represent lncRNAs. Lines represent their regulatory interactions. (C) Pathway enrichment analysis (left) and GO annotation (right) of co-expressed mRNAs with lncRNAs.
To further explore the roles of lncRNAs, we first used RNALocate17 to investigate the subcellular localization of each lncRNA in the LMGCN (Table S2). RNALocate is a comprehensive database that provides experimentally supported high-quality RNA subcellular localizations. Then, we performed functional annotation analysis using the co-expressed mRNA. 58 pathways and 113 Gene Ontology (GO) terms were identified (p < 0.05). Moreover, more than one third of the identified terms and pathways are relevant to immune or inflammatory mechanism (Table S3). We mainly listed several immune-related GO functions and pathways that play important roles in the immunological mechanism of MG (Figure 1C). For example, the significant GO functions included immune response, positive regulation of B cell proliferation, thymus development, positive regulation of T cell proliferation, and inflammatory response. The significant pathways included cytokine-cytokine receptor interaction, T cell receptor signaling pathway, and Toll-like receptor signaling pathway. These findings highlighted the fundamental characteristics of these lncRNAs in the pathogenesis of MG.
Dissection of Potential ceRNA Mechanisms through Manual Literature Mining
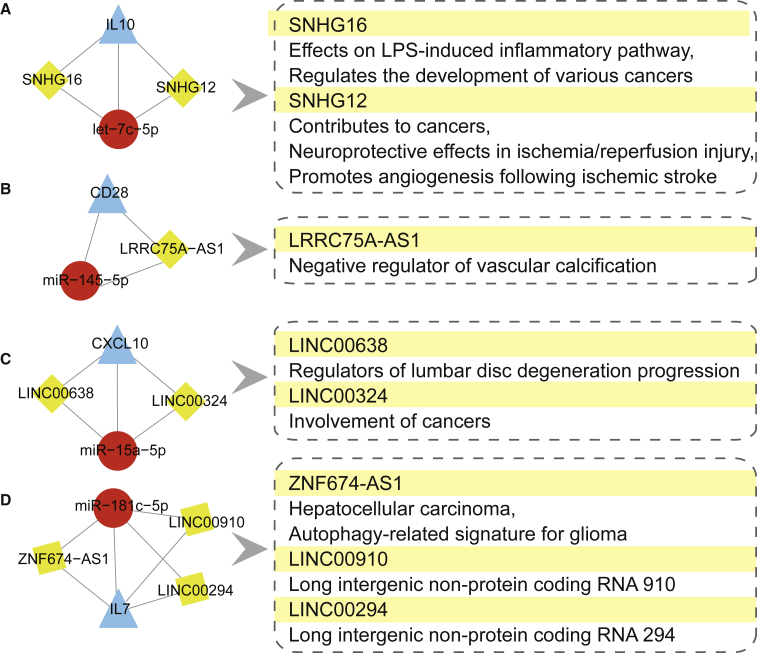
Based on the LMGCN, four miRNA-gene regulation pairs had reportedly been verified in MG: let-7c-5p/IL-10,18 miR-145-5p/CD28,19 miR-15a-5p/CXCL10,11 and miR-181c-5p/IL7.20 We extracted the sub-networks of these four miRNA-gene pairs and their linked lncRNAs in the global triple network. We found eight lncRNAs that may act as ceRNAs to regulate the miRNA-gene pairs mentioned above. Through reviewing reliable publications, we summarized the functions of these lncRNAs (Figure 2; the detailed information is summarized in Table S4). These lncRNAs are mainly involved in the development of various cancers,21, 22, 23 with other functions including immune processes,24 neuroprotective effects in ischemia/reperfusion injury,25 and vascular calcification.26 More importantly, SNHG16 acting as a ceRNA was reported to be involved in inflammatory and immune processes24 that may participate in the pathogenesis of MG. We found that SNHG16 potentially competed with IL-10 in the sub-network. IL-10 is an important growth factor for B cells, augmenting B cells activation into antibody-producing cells, and it plays important roles in the pathogenesis of MG.4 These findings suggest that the IL-10/SNHG16 competing pair might be involved in the immunological pathogenesis of MG. Therefore, we primarily focused on SNHG16/let-7c-5p/IL-10 interaction for further experimental verification.
Figure 2.
The Sub-Network of lncRNA Acting as a ceRNA to Regulate miRNA–Gene Pairs Has Been Verified in MG.
(A) The sub-network of let-7c-5p/IL-10 pair. (B) The sub-network of miR-145-5p/CD28 pair. (C) The sub-network of miR-15a-5p/CXCL10 pair. (D)The sub-network of miR-181c-5p/IL-7 pair. The left pipeline are the sub-networks of four experimentally-supported miRNA–gene pairs in MG and their linked lncRNAs. The right pipeline are the summarized functions of lncRNAs.
lncRNA SNHG16 Is Upregulated in MG Patients and Is a Target of let-7c-5p
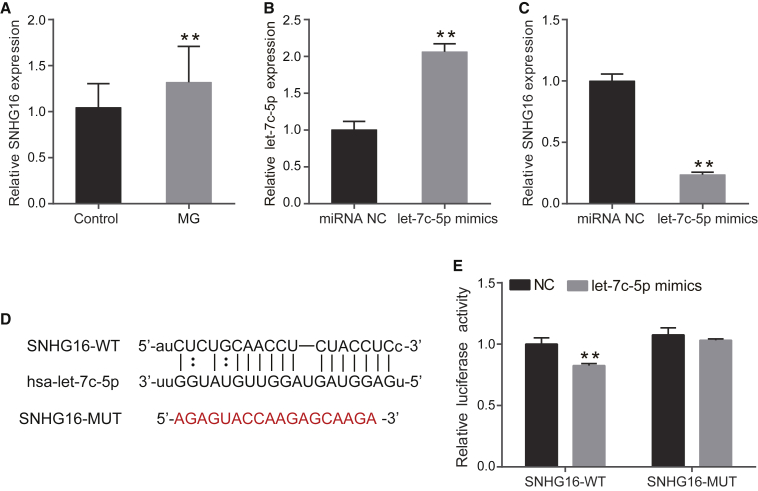
Real-time PCR analysis was performed to examine lncRNA SNHG16 in PBMCs from MG patients and control subjects. The expression of SNHG16 was higher in patients with MG compared with controls (p = 0.004, Figure 3A). Bioinformatics analysis revealed that the SNHG16 sequence contains a putative let-7c-5p binding region. A previous study had indicated that let-7c-5p was downregulated in PBMCs of MG patients.18 To investigate the association between let-7c-5p and SNHG16, let-7c-5p mimics were transfected into Jurkat cells. The transfection efficiency of let-7c-5p mimics was confirmed by real-time PCR (p = 0.0003, Figure 3B). Overexpression of let-7c-5p significantly inhibited the expression of SNHG16 in Jurkat cells (p < 0.01, Figure 3C). To further confirm the direct interaction between SNHG16 and let-7c-5p, we constructed luciferase reporter vectors of SNHG16-wild type (WT) and SNHG16-mutated type (MUT) (Figure 3D). The SNHG16-WT or SNHG16-MUT was then co-transfected with let-7c-5p mimics or negative control into HEK293T cells. The dual-luciferase reporter assay showed that let-7c-5p mimics suppressed the luciferase activity of SNHG16-WT but had no effect on the luciferase activity of SNHG16-MUT (Figure 3E). These results indicated that SNHG16 is the target of let-7c-5p.
Figure 3.
Upregulation of SNHG16 Is a Target of let-7c-5p in MG
(A) SNHG16 expression was examined in 24 MG patients and 29 control subjects by real-time PCR. (B) Transfection efficiency of let-7c-5p mimics was measured by real-time PCR. (C) The relative expression level of SNHG16 in Jurkat cells transfected with miRNA NC or let-7c-5p mimics was measured using real-time PCR. (D) The putative let-7c-5p binding sequence of the wild-type and mutation sequence of SNHG16. (E) The luciferase reporter plasmid containing SNHG16-WT or SNHG16-MUT was co-transfected with let-7c-5p mimics or miRNA NC into HEK293T cells. Luciferase activities were calculated as the ratio of firefly/Renilla activities. The experiment was repeated at least three times, and data are presented as the mean ± SD. **p < 0.01.
SNHG16 Promotes IL-10 Expression by Sponging let-7c-5p
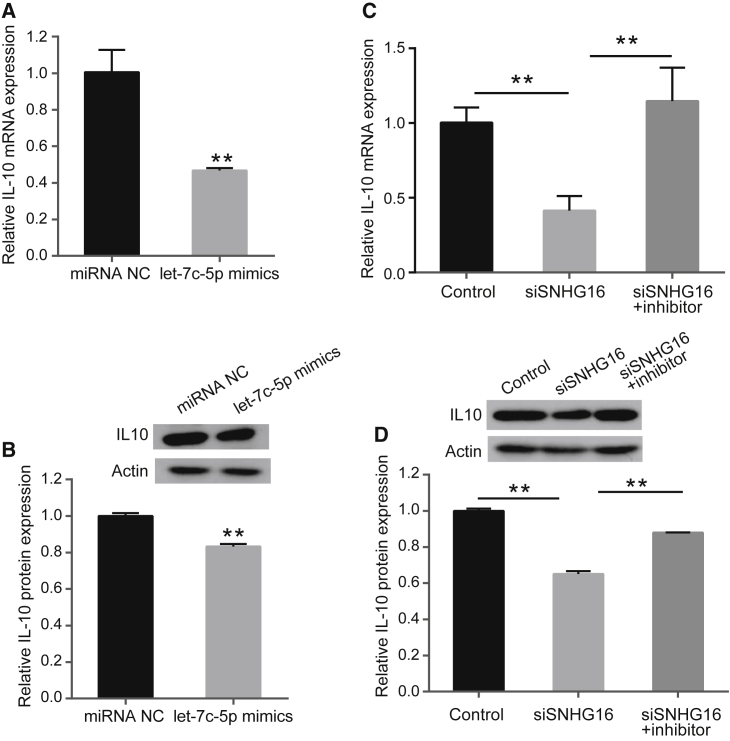
MG is a T cell-dependent and B cell-mediated autoimmune disease. Cytokines play an important role in the regulation of autoantibody production and cell-mediated immunity in MG.2 It has been reported that there is a negative correlation between let-7c-5p and IL-10 mRNA levels in MG patients, and that let-7c-5p mediates regulation of IL-10 by directly targeting IL-10 in Jurkat cells.18 Therefore, we designed experiments to explore whether SNHG16 could regulate IL-10 expression by targeting let-7c-5p. First, we measured the expression of IL-10 mRNA and protein in Jurkat cells after transfection with let-7c-5p mimics or negative control. The results indicated that let-7c-5p overexpression decreased IL-10 expression at both the mRNA and protein level (p < 0.01, Figures 4A and 4B), which was consistent with the results of a previous study.18
Figure 4.
SNHG16 Regulates IL-10 Expression by Binding let-7c-5p in a ceRNA Manner
(A) Relative mRNA levels of IL-10 were determined by real-time PCR after transfection with negative control or let-7c-5p mimics in Jurkat cells. (B) Relative protein expression levels of IL-10 were determined by western blotting after transfection with negative control or let-7c-5p mimics in Jurkat cells. (C) Relative mRNA levels of IL-10 were determined by real-time PCR analysis after transfection with negative control, siSNHG16, and siSNHG16 + let-7c-5p inhibitor in Jurkat cells. (D) Relative protein expression levels of IL-10 were determined by western blotting after transfection with negative control, siSNHG16, and siSNHG16 + let-7c-5p inhibitor in Jurkat cells. The experiment was repeated at least three times, and data are presented as the mean ± SD. **p < 0.01.
To verify whether SNHG16 regulates IL-10 expression by targeting let-7c-5p, Jurkat cells were transfected with negative control, siSNHG16, and siSNHG16 in combination with let-7c-5p inhibitor. Then, the IL-10 mRNA and protein levels were determined. The results showed that knockdown of SNHG16 suppressed the IL-10 mRNA and protein expression levels in Jurkat cells, whereas let-7c-5p inhibitor blocked the reduction in IL-10 expression induced by SNHG16 suppression (p < 0.01, Figures 4C and 4D). These findings suggest that SNHG16 regulates the expression of IL-10 by sponging let-7c-5p in a ceRNA manner.
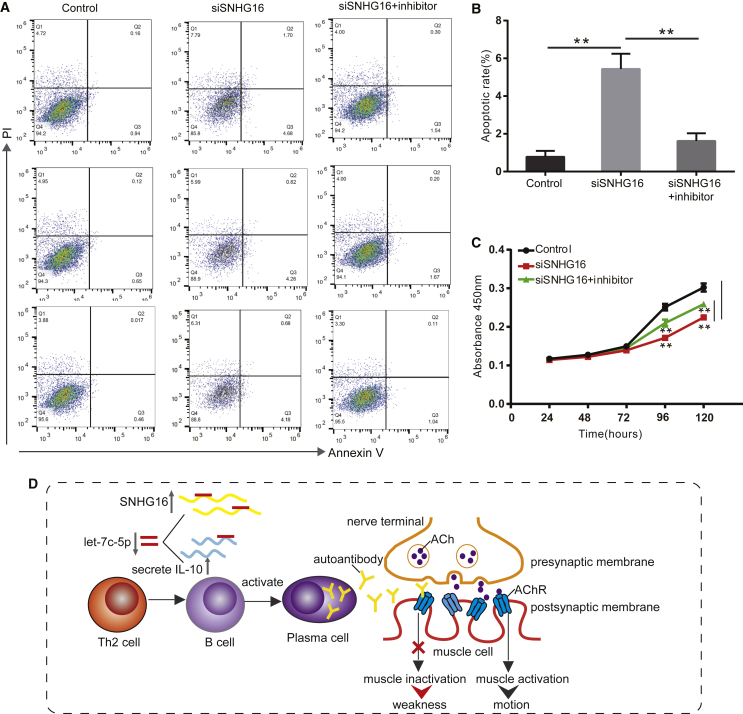
SNHG16 Inhibits Apoptosis and Promotes Proliferation by Sponging let-7c-5p in Jurkat Cells
MG is a T cell-dependent autoimmune disease, and the proliferation and activation of T cells play an important role in the pathogenesis of MG. Therefore, we used Jurkat T cells for functional verification of MG, referring to previous studies.10,18 To determine whether SNHG16 affects apoptosis and proliferation of Jurkat cells by targeting let-7c-5p, Jurkat cells were transfected with negative control, siSNHG16, and siSNHG16 along with let-7c-5p inhibitor. Then, flow cytometry and a Cell Counting Kit-8 (CCK-8) assay were used to assess the rates of apoptosis and proliferation. The rate of apoptosis increased following transfection with siSNHG16, whereas the addition of let-7c-5p inhibitor abrogated the above trend (p < 0.01, Figures 5A and 5B). Moreover, the CCK-8 assay revealed that knockdown of SNHG16 inhibited proliferation of Jurkat cells compared with transfection with negative control, and this effect was reversed by transfecting with let-7c-5p inhibitor (p < 0.01, Figure 5C). These results suggest that SNHG16 inhibits cell apoptosis and promotes proliferation by sponging let-7c-5p in Jurkat cells. Taken together, our results suggest that SNHG16 regulates T cell apoptosis and proliferation by sponging let-7c-5p, which is involved in the immunological pathogenesis of MG (Figure 5D).
Figure 5.
SNHG16 Inhibits Apoptosis and Promotes Cell Proliferation by Sponging let-7c-5p
(A) After transfecting negative control, siSNHG16 or siSNHG16+let-7c-5p inhibitor, Jurkat cells were stained with Annexin-V-FITC/PI, and the apoptosis was detected by flow cytometric analysis. (B) The apoptosis rate of Jurkat cells after transfecting negative control, siSNHG16 or siSNHG16+let-7c-5p inhibitor. (C) Cell proliferation was analyzed using CCK-8 assays by transfecting negative control, siSNHG16, or siSNHG16 + let-7c-5p inhibitor into Jurkat cells. (D) Schematic diagram of SNHG16 involved in the immunological pathogenesis of MG. The experiment was repeated at least three times, and data are presented as the mean ± SD. **p < 0.01.
Discussion
MG is a neurological autoantibody-mediated disease, but the triggering autoimmune processes involved are not clearly defined. Immunomodulatory therapies have been widely used to improve the prognosis for MG patients. Given the complex pathogenesis and heterogeneity of MG, no one treatment therapy is best for all MG patients.27 Extensive evidence indicates that lncRNAs play critical roles in regulation of the immune system and in autoimmune disease.7 The recently uncovered lncRNA-mediated ceRNA regulatory theory and networks improve our understanding of the precise molecular mechanism of MG.
During the past few years, ceRNA regulatory mechanisms have been validated in various diseases. To date, several databases have been developed to curate ceRNA interactions based on experimentally supported evidence or computationally predicted methods, such as LncACTdb,28 starBase,29 and PceRBase30. MNDR v2.031 and RAID v2.032 are also useful resources for analyzing RNA-disease associations. These databases are valuable resources for studying ceRNA regulation underlying complex diseases. However, so far, most studies concerning ceRNA network construction and mechanisms have focused on the cancer field. For example, Wang et al.33 constructed a lncRNA-associated ceRNA network to reveal global patterns and prognostic markers across 12 types of human cancer. However, only a few ceRNA interactions have been reported in autoimmune diseases.16 Accordingly, there is a need to explore the regulatory mechanisms of ceRNAs in MG. In the present study, we first constructed an LMGCN based on the ceRNA theory using a comprehensive approach. The LMGCN was composed of 9 miRNA nodes, 20 mRNA nodes, and 32 lncRNA nodes. The results of functional annotation analysis suggest that these lncRNAs may play crucial roles in the development of MG.
Next, we summarized the functions of lncRNA as a ceRNA to regulate miRNA-gene interactions that have been verified in MG. Of note, a recent study showed that SNHG16 affected the LPS-induced inflammatory and immune processes. Subsequently, mechanistic investigations revealed that SHNG16 acts as a ceRNA to positively regulate Toll-like receptor 4 (TLR4) by competitively binding miR-15a/16.24 Indeed, growing evidence shows that SNHG16 serving as a ceRNA is involved in several diseases.34,35 However, the underlying mechanism of the involvement of SNHG16 in MG remains unclear. The present study found that SNHG16 expression is increased in PBMCs from MG patients. Functionally, we found that knockdown of SNHG16 suppresses cell proliferation and promotes apoptosis in Jurkat cells. These results suggest that SNHG16 might be involved in the immunological pathogenesis of MG.
Bioinformatics analysis predicted that SNHG16 was a direct target of let-7c-5p. A previous study has reported that let-7c-5p is downregulated in PBMCs from MG patients, and that IL-10 is a target for let-7c-5p. Meanwhile, let-7c regulates IL-10 secretion in Jurkat cells.18 We found that let-7c-5p overexpression decreased IL-10 mRNA and protein expression levels in Jurkat cells, findings that were consistent with the results of previous study. Hence, we hypothesized that SHNG16 acted as a ceRNA to regulate the let-7c-5p/IL-10 axis in MG (Figure 5D). We found that transfection of let-7c-5p significantly decreased SNHG16 expression in Jurkat cells. Then, we confirmed that SNHG16 is a direct target of let-7c-5p by using luciferase reporter assays. Furthermore, we found that SNHG16 knockdown suppressed the IL-10 mRNA and protein levels, but these effects were reversed by co-transfecting siSHNG16 and let-7c-5p inhibitor into Jurkat cells. Moreover, cell proliferation and apoptosis assays showed that knockdown of SNHG16 inhibited cell proliferation and promoted cell apoptosis, but these were reversed by co-transfecting siSHNG16 and let-7c-5p inhibitor into Jurkat cells. Taken together, our findings suggest that lncRNA SNHG16 functions as a ceRNA to regulate IL-10 by competitively binding let-7c-5p.
Recently, cytokines that drive autoantibody secretion have received widespread attention as specific immunotherapy for MG. Various cytokines have been reported to play a critical role in the immunopathogenic mechanisms of MG.36 Some anti-cytokine agents relevant to MG immunopathogenesis have provided new targeted immunotherapies.37 For example, etanercept (an antagonist of TNF-α) has been proposed to treat patients with MG.38 IL-10 plays a crucial role in B cell activation and autoantibody production.39 It has been found that MG patients had increased numbers of AChR-reactive IL-10 mRNA-expressing PBMCs.4,40 High IL-10 levels were associated with AChR antibody production and treatment response in juvenile MG.5 These findings support the hypothesis that IL-10 plays important roles in the pathogenesis of MG. The symptoms of MG are caused by the autoantibodies that attack the receptors in the postsynaptic muscle membrane, leading to muscle fatigue and weakness. The production of autoantibodies is a T cell-dependent and B cell-mediated process. It has been reported that IL-10 secreted by Th2 cells promotes B cell activation into antibody-producing cells4 (Figure 5D). Thus, the present study showed that SNHG16 regulates the expression of IL-10 by sponging let-7c-5p, and thus improved our understanding of the precise molecular mechanism of MG.
In summary, in the present study, we for the first time constructed a lncRNA-mediated ceRNA network involved in MG. Further experiments revealed a novel regulatory mechanism of lncRNA SNHG16 involved in MG, which regulates IL-10 expression by sponging let-7c-5p. Our findings provide a global view of the mechanisms of ceRNA regulation and candidate lncRNAs as biomarkers for diagnosis and therapies in MG.
Materials and Methods
Human MG Risk miRNA Collection
We mainly applied the following two steps in the data collection process: searching for relevant articles, and extracting useful information from the selected articles. To ensure a high quality, human MG risk miRNA collection referred to our previous studies published online.41 We first obtained all publications from the PubMed database using the keyword combinations “myasthenia gravis” and “microRNA” or “miRNA” or “miR” to collect all experimentally supported MG-associated miRNAs. Then, we manually curated the MG-associated miRNAs that met the following criteria: (1) the species was human; (2) MG patients and healthy controls were included in the study, and at least 10 samples were included in each group; (3) the expression levels of miRNA were analyzed in PBMCs and showed significant differences between the MG patients and controls; (4) selected miRNAs were experimentally confirmed by reliable low-throughput experiments, such as real-time PCR and northern blot; and (5) the collected miRNAs were agreed to by at least two researchers. Finally, detailed information of MG risk miRNAs that we collected is summarized in Table S5.
Identification of miRNA-lncRNA and miRNA-mRNA Interactions
First, miRNA-lncRNA interactions were predicted using TargetScan (release 7.2)42 and miRanda (2010 version)43 computational methods. The miRNA and lncRNA sequences were obtained from miRBase (release 21)44 and GENCODE (v26)45, respectively. We predicted miRNA target binding sites on the whole lncRNA sequences using the intersection of the TargetScan and miRanda methods. Then, predicted miRNA-lncRNA interactions were further filtered by starBase (v3.0)29, which is a database containing the Argonaute (AGO) crosslinking immunoprecipitation sequencing (CLIP-seq) experiment-supported miRNA-lncRNA interactions. The intersections with starBase (v3.0) were selected as candidate miRNA-lncRNA interactions. Then, miRNA-mRNA interactions were obtained from miRTarBase (release 7.0),46 which is a manually curated database that provides experimentally supported miRNA-gene interactions. We only retained high-confidence functional miRNA-gene interactions supported by reporter assay and/or western blot data. The miRNA target genes were further filtered by human MG risk genes, which were manually curated using strict criteria from our previous study.47 We ultimately retained the intersections of experimentally supported genes of miRNA and human MG risk genes as the targets of miRNAs.
Identification of Potential mRNA-lncRNA Competing Interactions
To identify competing mRNA-lncRNA interactions, hypergeometric tests and the Pearson correlation coefficient (PCC) were employed to identify the competing pairs.48 First, the hypergeometric test was used to evaluate the significance of the shared miRNAs between each mRNA and lncRNA. The formula used was as follows:
For each mRNA-lncRNA pair, N denoted the total number of miRNAs in the genome, n represented the number of miRNAs that were associated with one mRNA, m represented the number of miRNAs that were associated with one lncRNA, and x represented the number of common miRNAs that shared the mRNA and lncRNA. The mRNA-lncRNA pairs with a p value <0.05 were considered significant interactions.
Next, we evaluated co-expression correlation of mRNA-lncRNA pairs identified above using the PCC. The lncRNA and mRNA expression data were downloaded from the dbGaP database (the Genotype-Tissue Expression Project, released in 2016; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v7.p2), which contains 32 healthy tissues in 7,862 samples from 552 donors.49 The p values of co-expression analysis were adjusted according to the false discovery rate (FDR). The mRNA-lncRNA pairs with a PCC of ≥0.2 and a FDR of <0.01 were identified as co-expressed pairs. The above analyses were performed by R software.
Construction of the LMGCN
We constructed the LMGCN based on the theory that lncRNAs share common miRNA-binding sites with mRNAs and function as miRNA sponges to regulate mRNAs. For a given lncRNA-miRNA-mRNA interaction, both mRNA and lncRNA shared common miRNAs and were co-expressed for merging into a competing triplet. After assembling all lncRNA-miRNA-mRNA competing triplets, we constructed the LMGCN. The network was visualized using Cytoscape software, in which nodes represent miRNAs, genes, and lncRNAs, and edges represent their interactions.
Functional Enrichment Analysis
To further confirm the roles of lncRNAs, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and investigated biological processes in GO annotation of co-expressed mRNAs in the LMGCN using Database for Annotation, Visualization and Integrated Discovery (DAVID),50 which is an online functional annotation tool. Pathways and GO terms with p < 0.05 were considered to be significantly enriched function annotations.
Clinical Samples
A total of 24 MG patients who were followed at The Second Affiliated Hospital of Harbin Medical University were included in this study. All the patients were diagnosed initially and met the diagnostic criteria for MG.51 A total of 29 sex- and age-matched healthy donors with no history of autoimmune disease were included as the control. This study was approved by the Ethics Committee of The Second Affiliated Hospital of Harbin Medical University. Written informed consent was obtained from all subjects. The study was carried out according to the World Medical Association Declaration of Helsinki. Peripheral blood samples were collected from each participant in tubes containing ethylenediaminetetraacetic acid, and PBMCs were isolated using lymphocyte separation medium.
Cell Culture
The T cell leukemia line (Jurkat cells) and human embryonic kidney 293T (HEK293T) cells were purchased from the American Type Culture Collection (Manassas, VA, USA). Jurkat cells, which had been used for functional verification of MG according to previous studies,10,18 were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA). HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco). All media were supplemented with 10% fetal bovine serum (Gibco), together with 100 IU/mL penicillin and 100 μg/mL streptomycin (KeyGen Biotech, Nanjing, China). All cells were incubated at 37°C in a humidified atmosphere of 5% CO2.
Cell Transfection
lncRNA Smart Silencer for human SNHG16, let-7c-5p mimics, let-7c-5p inhibitor, and negative control (NC), designed and synthesized by Ribobio (Guangzhou, China), was transfected into Jurkat cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). SNHG16 Smart Silencer (siSNHG16) was a pool containing three siRNAs and three antisense oligonucleotides, which was applied to knock down the expression of SNHG16. The sequences of Smart Silencer were as follows: siSNHG16-1 target sequence, 5′-CCTTCCAGGAACAGATCTT-3′; siSNHG16-2 target sequence, 5′-TGTTGACTCACCAAGGCAA-3′; siSNHG16-3 target sequence, 5′-GGAACAGATCTTTGCATAG-3′; antisense oligonucleotides target sequence-1, 5′-CCCAGCTATTTTTTCTTTCG-3′; antisense oligonucleotides target sequence-2, 5′-CGTGCCCTAAATTGACCAAC-3′; and antisense oligonucleotides target sequence-3, 5′-CCACTTACAATAAACTTGGG-3′.
Real-Time PCR Analysis
Total RNA was extracted from PBMCs or Jurkat cells using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Reverse transcription of total RNA into cDNA was performed using Primescript RT reagent kit (Takara, Dalian, China) according to the manufacturer’s instructions. The sequences of the primers used are listed in Table S6. Quantitative real-time PCR was performed using the SYBR Premix Ex Taq kit (Takara, Dalian, China) in a Roche LightCycler 480 instrument. The relative expression level was calculated using the 2−ΔΔCt method. RPL13A was used as the reference for SNHG16 and IL-10, and U6 was used as the reference for let-7c-5p.
Western Blot Analysis
Total protein was extracted from Jurkat cells using a radioimmunoprecipitation assay lysis buffer (Beyotime Biotechnology, Shanghai, China). The protein concentration was determined using a bicinchoninic acid protein assay kit (Beyotime). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membrane was incubated with the following antibodies at 4°C overnight: IL-10 (1:1,000, ABclonal, Wuhan, China) and β-actin (1:10,000, Abways, Shanghai, China). The membranes were then washed and incubated with anti-rabbit horseradish peroxidase (HRP)-linked secondary antibodies (Cell Signaling Technology, Danvers, MA, USA) for 2 h at room temperature. The PVDF membranes were washed with Tris-buffered saline with Tween 20 (TBST), and the blots were visualized using enhanced chemiluminescence (ECL) (Carestream, Wuxi, China). β-Actin was used as the internal control. The experiments were independently repeated in triplicate.
Luciferase Reporter Assay
A fragment from SNHG16 containing the predicted let-7c-5p binding site was cloned into a PHY-811 vector (Hanyin Biotechnology, Shanghai, China) to construct the luciferase reporter vector, named SNHG16-WT. To mutate the putative binding site of let-7c-5p in SNHG16, the sequence of the putative binding site was replaced, and the vector was named SNHG16-MUT. The SNHG16-WT or SNHG16-MUT plasmids were co-transfected together with let-7c-5p mimics or negative control into HEK293T cells using Lipofectamine 3000 (Invitrogen). A dual-luciferase reporter assay system (Promega, Madison, WI, USA) was used to assess luciferase activity after 48 h of transfection according to the manufacturer’s protocol. Relative luciferase activity was measured and normalized to Renilla luciferase activity.
Cell Proliferation Assay
Cell proliferation was evaluated using a CCK-8 assay (Dojindo, Tokyo, Japan). Cells from different groups were seeded onto 96-well plates at a density of 1,500 cells/well and incubated at 37°C in a humidified atmosphere of 5% CO2. According to the instructions, 10 μL of CCK-8 reagent was added to the wells at time points of 24, 48, 72, 96, and 120 h. The absorbance at 450 nm was measured using a microplate reader (BioTek Instruments, Winooski, VT, USA) after the plates were incubated at 37°C for 2 h. The experiments were independently repeated in triplicate.
Apoptosis Analysis
Apoptosis was analyzed using an annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s protocol. Jurkat cells were transfected and cultured in a six-well plate. After a 48-h incubation, cells were digested using trypsin (Gibco) and then stained with annexin V-FITC and PI. Then, stained cells were analyzed by flow cytometry using CellQuest software (BD Biosciences, Franklin Lakes, NJ, USA). The experiments were independently repeated in triplicate.
Statistical Analysis
SPSS 17.0 software was used to perform the statistical analyses. Data are expressed as mean ± standard deviation (SD). The differences between the two groups were analyzed using Student’s t test, while the differences among multiple groups were analyzed using ANOVA. p < 0.01 was considered statistically significant.
Author Contributions
H.Z., L.W., and S.N. designed the study. J.W. and Y.C. wrote the manuscript. J.W. and X.L. performed the bioinformatics analysis. X.W., X.K., C.B., and S.L. conducted the experiments. J.W. performed the statistical analyses. M.B., Y.J., H.G., and X.Y. provided valuable opinions in the process of writing the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81820108014, 81771361, 81701190, 81701155, 81801190, and 81671169); the Applied Technique Research and Development Project of Harbin (grant 2016RAXYJ067); the Postdoctoral Foundation of Heilongjiang Province (grants LBH-Z17138 and LBH-Z18204); and the China Postdoctoral Science Foundation (grant 2018M640307).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.005.
Contributor Information
Shangwei Ning, Email: ningsw@ems.hrbmu.edu.cn.
Lihua Wang, Email: wanglh211@163.com.
Huixue Zhang, Email: zhanghuixue2009@163.com.
Supplemental Information
References
- 1.Gilhus N.E. Myasthenia gravis. N. Engl. J. Med. 2016;375:2570–2581. doi: 10.1056/NEJMra1602678. [DOI] [PubMed] [Google Scholar]
- 2.Gilhus N.E., Tzartos S., Evoli A., Palace J., Burns T.M., Verschuuren J.J.G.M. Myasthenia gravis. Nat. Rev. Dis. Primers. 2019;5:30. doi: 10.1038/s41572-019-0079-y. [DOI] [PubMed] [Google Scholar]
- 3.Conti-Fine B.M., Milani M., Kaminski H.J. Myasthenia gravis: past, present, and future. J. Clin. Invest. 2006;116:2843–2854. doi: 10.1172/JCI29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y.M., Kivisäkk P., Ozenci V., Pirskanen R., Link H. Increased levels of circulating acetylcholine receptor (AChR)-reactive IL-10-secreting cells are characteristic for myasthenia gravis (MG) Clin. Exp. Immunol. 1999;118:304–308. doi: 10.1046/j.1365-2249.1999.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yapici Z., Tüzün E., Altunayoğlu V., Erdoğan A., Eraksoy M. High interleukin-10 production is associated with anti-acetylcholine receptor antibody production and treatment response in juvenile myasthenia gravis. Int. J. Neurosci. 2007;117:1505–1512. doi: 10.1080/00207450601125840. [DOI] [PubMed] [Google Scholar]
- 6.Link J., Navikas V., Yu M., Fredrikson S., Osterman P.O., Link H. Augmented interferon-γ, interleukin-4 and transforming growth factor-β mRNA expression in blood mononuclear cells in myasthenia gravis. J. Neuroimmunol. 1994;51:185–192. doi: 10.1016/0165-5728(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y.G., Satpathy A.T., Chang H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017;18:962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo M., Liu X., Meng H., Xu L., Li Y., Li Z., Liu C., Luo Y.B., Hu B., Xue Y. IFNA-AS1 regulates CD4+ T cell activation in myasthenia gravis though HLA-DRB1. Clin. Immunol. 2017;183:121–131. doi: 10.1016/j.clim.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Z., Qiu S., Jiang L., Zhang A., Bao W., Liu P., Liu J. miR-320a is downregulated in patients with myasthenia gravis and modulates inflammatory cytokines production by targeting mitogen-activated protein kinase 1. J. Clin. Immunol. 2013;33:567–576. doi: 10.1007/s10875-012-9834-5. [DOI] [PubMed] [Google Scholar]
- 11.Liu X.F., Wang R.Q., Hu B., Luo M.C., Zeng Q.M., Zhou H., Huang K., Dong X.H., Luo Y.B., Luo Z.H., Yang H. miR-15a contributes abnormal immune response in myasthenia gravis by targeting CXCL10. Clin. Immunol. 2016;164:106–113. doi: 10.1016/j.clim.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P., Zhi H., Zhang Y., Liu Y., Zhang J., Gao Y., Guo M., Ning S., Li X. miRSponge: a manually curated database for experimentally supported miRNA sponges and ceRNAs. Database (Oxford) 2015;2015:bav098. doi: 10.1093/database/bav098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi L., Hong X., Ba L., He X., Xiong Y., Ding Q., Yang S., Peng G. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting as a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis. 2019;10:150. doi: 10.1038/s41419-019-1332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu Y.Y., Wu Y., Lin M.J., Bian T., Xiao Y.L., Qin C. LncRNA-MEG3 functions as a competing endogenous RNA to regulate Treg/Th17 balance in patients with asthma by targeting microRNA-17/RORγt. Biomed. Pharmacother. 2019;111:386–394. doi: 10.1016/j.biopha.2018.12.080. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H., Ma R., Zou S., Wang Y., Li Z., Li W. Reconstruction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in rheumatoid arthritis. Mol. Biosyst. 2017;13:1182–1192. doi: 10.1039/c7mb00094d. [DOI] [PubMed] [Google Scholar]
- 17.Zhang T., Tan P., Wang L., Jin N., Li Y., Zhang L., Yang H., Hu Z., Zhang L., Hu C. RNALocate: a resource for RNA subcellular localizations. Nucleic Acids Res. 2017;45(D1):D135–D138. doi: 10.1093/nar/gkw728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang L., Cheng Z., Qiu S., Que Z., Bao W., Jiang C., Zou F., Liu P., Liu J. Altered let-7 expression in myasthenia gravis and let-7c mediated regulation of IL-10 by directly targeting IL-10 in Jurkat cells. Int. Immunopharmacol. 2012;14:217–223. doi: 10.1016/j.intimp.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Sengupta M., Wang B.D., Lee N.H., Marx A., Kusner L.L., Kaminski H.J. MicroRNA and mRNA expression associated with ectopic germinal centers in thymus of myasthenia gravis. PLoS ONE. 2018;13:e0205464. doi: 10.1371/journal.pone.0205464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Guo M., Xin N., Shao Z., Zhang X., Zhang Y., Chen J., Zheng S., Fu L., Wang Y. Decreased microRNA miR-181c expression in peripheral blood mononuclear cells correlates with elevated serum levels of IL-7 and IL-17 in patients with myasthenia gravis. Clin. Exp. Med. 2016;16:413–421. doi: 10.1007/s10238-015-0358-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z.B., Tang C., Jin X., Liu S.H., Pi W. Increased expression of lncRNA SNHG12 predicts a poor prognosis of nasopharyngeal carcinoma and regulates cell proliferation and metastasis by modulating Notch signal pathway. Cancer Biomark. 2018;23:603–613. doi: 10.3233/CBM-181873. [DOI] [PubMed] [Google Scholar]
- 22.Wen Q., Zhao L., Wang T., Lv N., Cheng X., Zhang G., Bai L. lncRNA SNHG16 drives proliferation and invasion of papillary thyroid cancer through modulation of miR-497. OncoTargets Ther. 2019;12:699–708. doi: 10.2147/OTT.S186923. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Luan F., Chen W., Chen M., Yan J., Chen H., Yu H., Liu T., Mo L. An autophagy-related long non-coding RNA signature for glioma. FEBS Open Bio. 2019;9:653–667. doi: 10.1002/2211-5463.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W., Lou C., Gao J., Zhang X., Du Y. lncRNA SNHG16 reverses the effects of miR-15a/16 on LPS-induced inflammatory pathway. Biomed. Pharmacother. 2018;106:1661–1667. doi: 10.1016/j.biopha.2018.07.105. [DOI] [PubMed] [Google Scholar]
- 25.Yao X., Yao R., Huang F., Yi J. lncRNA SNHG12 as a potent autophagy inducer exerts neuroprotective effects against cerebral ischemia/reperfusion injury. Biochem. Biophys. Res. Commun. 2019;514:490–496. doi: 10.1016/j.bbrc.2019.04.158. [DOI] [PubMed] [Google Scholar]
- 26.Jeong G., Kwon D.H., Shin S., Choe N., Ryu J., Lim Y.H., Kim J., Park W.J., Kook H., Kim Y.K. Long noncoding RNAs in vascular smooth muscle cells regulate vascular calcification. Sci. Rep. 2019;9:5848. doi: 10.1038/s41598-019-42283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders D.B., Wolfe G.I., Benatar M., Evoli A., Gilhus N.E., Illa I., Kuntz N., Massey J.M., Melms A., Murai H. International consensus guidance for management of myasthenia gravis: executive summary. Neurology. 2016;87:419–425. doi: 10.1212/WNL.0000000000002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P., Li X., Gao Y., Guo Q., Wang Y., Fang Y., Ma X., Zhi H., Zhou D., Shen W. LncACTdb 2.0: an updated database of experimentally supported ceRNA interactions curated from low- and high-throughput experiments. Nucleic Acids Res. 2019;47(D1):D121–D127. doi: 10.1093/nar/gky1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan C., Meng X., Li X., Illing N., Ingle R.A., Wang J., Chen M. PceRBase: a database of plant competing endogenous RNA. Nucleic Acids Res. 2017;45(D1):D1009–D1014. doi: 10.1093/nar/gkw916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui T., Zhang L., Huang Y., Yi Y., Tan P., Zhao Y., Hu Y., Xu L., Li E., Wang D. MNDR v2.0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018;46(D1):D371–D374. doi: 10.1093/nar/gkx1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi Y., Zhao Y., Li C., Zhang L., Huang H., Li Y., Liu L., Hou P., Cui T., Tan P. RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 2017;45(D1):D115–D118. doi: 10.1093/nar/gkw1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P., Ning S., Zhang Y., Li R., Ye J., Zhao Z., Zhi H., Wang T., Guo Z., Li X. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43:3478–3489. doi: 10.1093/nar/gkv233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Kan J., Han J., Zhang W., Bai L., Wu H. lncRNA SNHG16 functions as an oncogene by sponging miR-135a and promotes JAK2/STAT3 signal pathway in gastric cancer. J. Cancer. 2019;10:1013–1022. doi: 10.7150/jca.29527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Q., Zheng H., Xu J., Zhang F., Pan H. lncRNA SNHG16 aggravates tumorigenesis and development of hepatocellular carcinoma by sponging miR-4500 and targeting STAT3. J. Cell. Biochem. 2019;120:11604–11615. doi: 10.1002/jcb.28440. [DOI] [PubMed] [Google Scholar]
- 36.Yoshikawa H., Satoh K., Yasukawa Y., Yamada M. Cytokine secretion by peripheral blood mononuclear cells in myasthenia gravis. J. Clin. Neurosci. 2002;9:133–136. doi: 10.1054/jocn.2001.1028. [DOI] [PubMed] [Google Scholar]
- 37.Dalakas M.C. Immunotherapy in myasthenia gravis in the era of biologics. Nat. Rev. Neurol. 2019;15:113–124. doi: 10.1038/s41582-018-0110-z. [DOI] [PubMed] [Google Scholar]
- 38.Fee D.B., Kasarskis E.J. Myasthenia gravis associated with etanercept therapy. Muscle Nerve. 2009;39:866–870. doi: 10.1002/mus.21280. [DOI] [PubMed] [Google Scholar]
- 39.Spellberg B., Edwards J.E., Jr. Type 1/type 2 immunity in infectious diseases. Clin. Infect. Dis. 2001;32:76–102. doi: 10.1086/317537. [DOI] [PubMed] [Google Scholar]
- 40.Matusevicius D., Navikas V., Palasik W., Pirskanen R., Fredrikson S., Link H. Tumor necrosis factor-α, lymphotoxin, interleukin (IL)-6, IL-10, IL-12 and perforin mRNA expression in mononuclear cells in response to acetylcholine receptor is augmented in myasthenia gravis. J. Neuroimmunol. 1996;71:191–198. doi: 10.1016/s0165-5728(96)00152-x. [DOI] [PubMed] [Google Scholar]
- 41.Wang J., Cao Y., Zhang H., Wang T., Tian Q., Lu X., Lu X., Kong X., Liu Z., Wang N. NSDNA: a manually curated database of experimentally supported ncRNAs associated with nervous system diseases. Nucleic Acids Res. 2017;45(D1):D902–D907. doi: 10.1093/nar/gkw1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kersey P.J., Lawson D., Birney E., Derwent P.S., Haimel M., Herrero J., Keenan S., Kerhornou A., Koscielny G., Kähäri A. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S., Cao Y., Li L., Zhang H., Lu X., Bo C., Kong X., Liu Z., Chen L., Liu P., Jiao Y. Building the drug-GO function network to screen significant candidate drugs for myasthenia gravis. PLoS One. 2019;14:e0214857. doi: 10.1371/journal.pone.0214857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao Y., Wang P., Ning S., Xiao W., Xiao B., Li X. Identification of prognostic biomarkers in glioblastoma using a long non-coding RNA-mediated, competitive endogenous RNA network. Oncotarget. 2016;7:41737–41747. doi: 10.18632/oncotarget.9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tryka K.A., Hao L., Sturcke A., Jin Y., Wang Z.Y., Ziyabari L., Lee M., Popova N., Sharopova N., Kimura M., Feolo M. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 51.Jaretzki A., 3rd, Barohn R.J., Ernstoff R.M., Kaminski H.J., Keesey J.C., Penn A.S., Sanders D.B., Task Force of the Medical Scientific Advisory Board of the Myasthenia Gravis Foundation of America Myasthenia gravis: recommendations for clinical research standards. Neurology. 2000;55:16–23. doi: 10.1212/wnl.55.1.16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.