Abstract
DNA is susceptible to a range of chemical modifications, with one of the most frequent lesions being apurinic/apyrimidinic (AP) sites. AP sites arise due to damage-induced (e.g. alkylation) or spontaneous hydrolysis of the N-glycosidic bond that links the base to the sugar moiety of the phosphodiester backbone, or through the enzymatic activity of DNA glycosylases, which release inappropriate bases as part of the base excision repair (BER) response. Unrepaired AP sites, which lack instructional information, have the potential to cause mutagenesis or to arrest progressing DNA or RNA polymerases, potentially causing outcomes such as cellular transformation, senescence or death. The predominant enzyme in humans responsible for repairing AP lesions is AP endonuclease 1 (APE1). Besides being a powerful AP endonuclease, APE1 possesses additional DNA repair activities, such as 3′–5′ exonuclease, 3′-phophodiesterase and nucleotide incision repair. In addition, APE1 has been shown to stimulate the DNA-binding activity of a number of transcription factors through its ‘REF1’ function, thereby regulating gene expression. In this article, we review the structural and biochemical features of this multifunctional protein, while reporting on new structures of the APE1 variants Cys65Ala and Lys98Ala. Using a functional complementation approach, we also describe the importance of the repair and REF1 activities in promoting cell survival, including the proposed passing-the-baton coordination in BER. Finally, results are presented indicating a critical role for APE1 nuclease activities in resistance to the genotoxins methyl methanesulphonate and bleomycin, supporting biologically important functions as an AP endonuclease and 3′-phosphodiesterase, respectively.
Genotoxic stress can arise from altered cellular metabolism or following exposure to many external agents, including sunlight, ionising radiation, polycyclic aromatic hydrocarbons (e.g. benzo[a]pyrene), aromatic amines (e.g. 2-acetylaminofluorene), aflatoxins, various heavy metals (e.g. cadmium) and numerous therapeutic agents (e.g. X-rays, platins, alkylators, crosslinkers, topoisomerase poisons and antimetabolites). One of the more common forms of intracellular stress originates from an imbalance in reactive oxygen species (ROS) formation, typically stemming from defects in mitochondrial respiration or metabolism, and their sequestration by the protective scavenging systems. In conditions of so-called oxidative stress, the elevated ROS can attack nearby intracellular macromolecules, including proteins, lipids and nucleic acids, potentially giving rise to cellular dysfunction. As damage to one’s genetic material can lead to permanent sequence changes or to alterations in DNA and/or RNA metabolism that can promote disease and ageing, DNA repair processes have evolved as a major front-line defense against the harmful consequences of genotoxic stress. In this article, we review the biochemical and structural features of a key DNA repair enzyme, apurinic/apyrimidinic endonuclease 1 (APE1) and present new data that provides important insights into the biological functions of this multifunctional protein.
DNA Damage and Its Consequences
DNA damage can exist in multiple chemical forms. These include simple base modifications, such as uracil or 8-oxoguanine, bulky base modifications like ultraviolet light-induced photoproducts or platinum adducts, DNA strand break interruptions and linkages that covalently attach the two strands of the helix (so-called interstrand crosslinks). Depending on their chemical composition and structural impact, the various DNA lesions can: (i) alter the coding sequence of the genome, directing mutations during chromosome replication and/or (ii) adversely affect the progression of replicative DNA or RNA polymerases, leading to fork collapse or transcription arrest. Mutational events are known to underlie cancer aetiology, and likely contribute more broadly to disease and ageing, while replication fork collapse can drive chromosome instability commonly associated with carcinogenesis, and persistently stalled transcription apparatuses can contribute to premature ageing phenotypes, as seen in Cockayne syndrome (1).
Apurinic/Apyrimidinic (AP) Sites
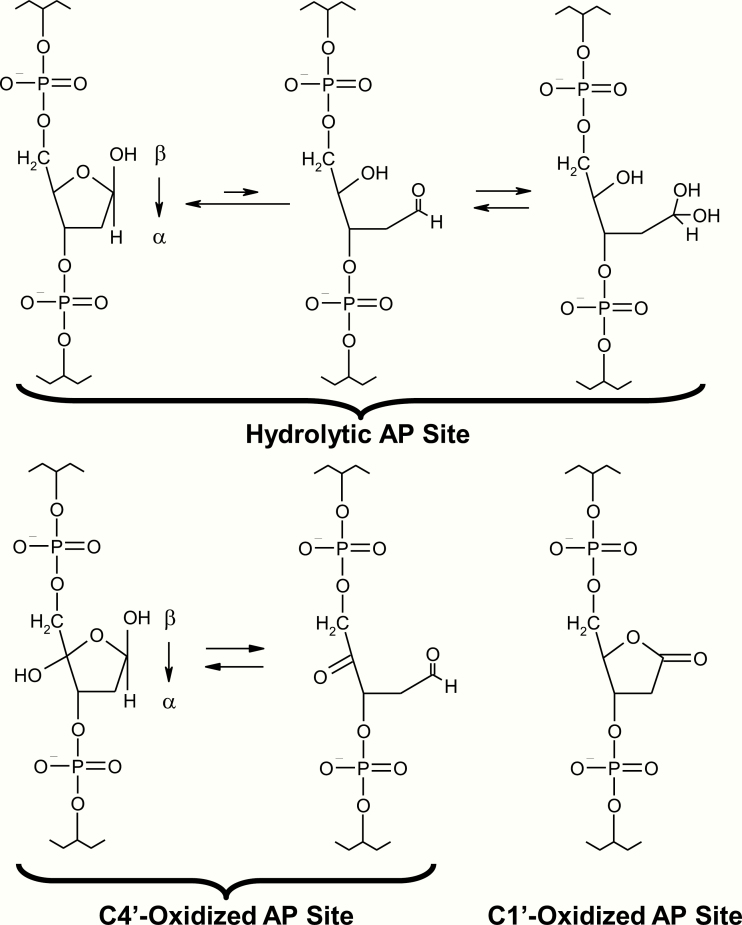
One of the more common modifications to occur in DNA is the apurinic/apyrimidinic (AP) site (Figure 1). Most AP sites arise due to damage-induced or spontaneous hydrolysis of the N-glycosidic bond that attaches the base moiety to the ribose ring of the phosphodiester backbone. Based on chemical extrapolations, Lindahl and colleagues estimated that >10 000 AP sites will be created in a single mammalian genome per day under normal physiological conditions (2). The hydrolytic AP site exists in two major forms: ring-opened and ring-closed (Figure 1, top). The latter is the predominant form, while the former is susceptible to reactions with amines (such as those present in lysine residues) that can establish a covalent intermediate (i.e. an imine or Schiff-base) and bulky adduct, as seen with many DNA binding proteins (see for instance, 3–5). Recent work has shown that AP sites can also generate links between the two strands of DNA by reacting with adenine or guanine in the complementary strand (6,7). Such interstrand crosslinks pose strong impediments to progressing DNA or RNA polymerases, as they prevent unwinding of the DNA duplex. More information on the processing of lethal DNA interstrand crosslinks can be found elsewhere (8,9).
Figure 1.
Chemical form(s) of the hydrolytic and oxidised abasic sites. Hydrolytic AP sites (top) are in an equilibrium mixture of ring-closed [racemeric β- and α-hemiacetals (2-deoxy-D-erythro-pentofuranoses), left] and ring-open (aldehyde, centre; hydrated aldehyde, right) forms. In vitro experiments indicate that the hemiacetal forms predominate, in roughly equal number, with ~1% aldehyde forms being present. The C4′- and C1′-oxidised AP sites are shown at the bottom, along with their alternative forms where relevant.
Besides the hydrolytic AP site, abasic sites can exist in oxidised forms, e.g. C1′-oxidised (a.k.a. 2-deoxyribonolactone) or C4′-oxidised (Figure 1, bottom), produced by free radical-mediated hydrogen abstraction at the C1′ or C4′ position of the ribose sugar ring (10). As with the hydrolytic AP site, oxidised abasic lesions can form covalent crosslinks with several proteins, most notably the major gap-filling DNA polymerase, POLβ (11). The mechanisms for coping with large protein–DNA adducts, which clearly would hinder DNA or RNA polymerase progression, are just now becoming better understood (12,13). Lastly, AP sites are generated as enzymatic intermediates during the process of base excision repair (BER), after substrate base release by a DNA glycosylase (see more later).
AP sites have been reported to have very little effect on the overall helical architecture of DNA, although they appear to allow for a more flexible phosphodiester linkage at the site of the lesion, a feature that is likely important to recognition and incision (14). However, it is noteworthy to point out that the structural effects of AP sites can be influenced by the base opposite and the neighbouring bases ((15) and references therein). For example, when the AP site is flanked by a purine on either side, the duplex is more likely to collapse (due to base-stacking), rotating the abasic lesion out of the helix and often extruding the base opposite. A purine opposite, however, is more likely to favour the normal B-DNA form. Notably, this subtle sequence-context variability can have significant, albeit moderate, effects on cleavage rates, a feature that might influence repair efficiency and the molecular consequences (16,17). Moreover, X-ray crystal structures of APE1 (the major mammalian AP endonuclease; see later) in complex with DNA substrates containing various mismatched base pairings flanking an AP site revealed unique DNA conformations that corresponded to a wide range of incision efficiencies (18).
As the name implies, an AP site lacks the instructional information provided by the base moiety, and thus, is considered non-coding. Early evidence suggested an A-rule mutational signature upon DNA polymerase bypass of an AP site, particularly in Escherichia coli (19), but there has been unclarity regarding this topic, as much of the mutagenic outcome of DNA damage has been complicated by the identification of numerous translesion synthesis (TLS) DNA polymerases in eukaryotes. These enzymes seemingly have evolved to permit survival via error-prone (often mutagenic) bypass as opposed to persistent replication arrest and cell death, and do not necessarily follow the same insertion strategy to synthesise past a particular blocking lesion (20,21). At present, what is clear is that AP sites can present major blocks to both DNA and RNA polymerases, but in the former situation, can be effectively bypassed by several TLS polymerases, with bypass leading to single nucleotide substitutions or small insertions/deletions (22–26). With the development of new animal models that permit selective deletion of APE1 and consequent cancer formation (see later), it may now be possible to assess disease-associated mutational patterns in the absence of the repair protein.
BER Overview
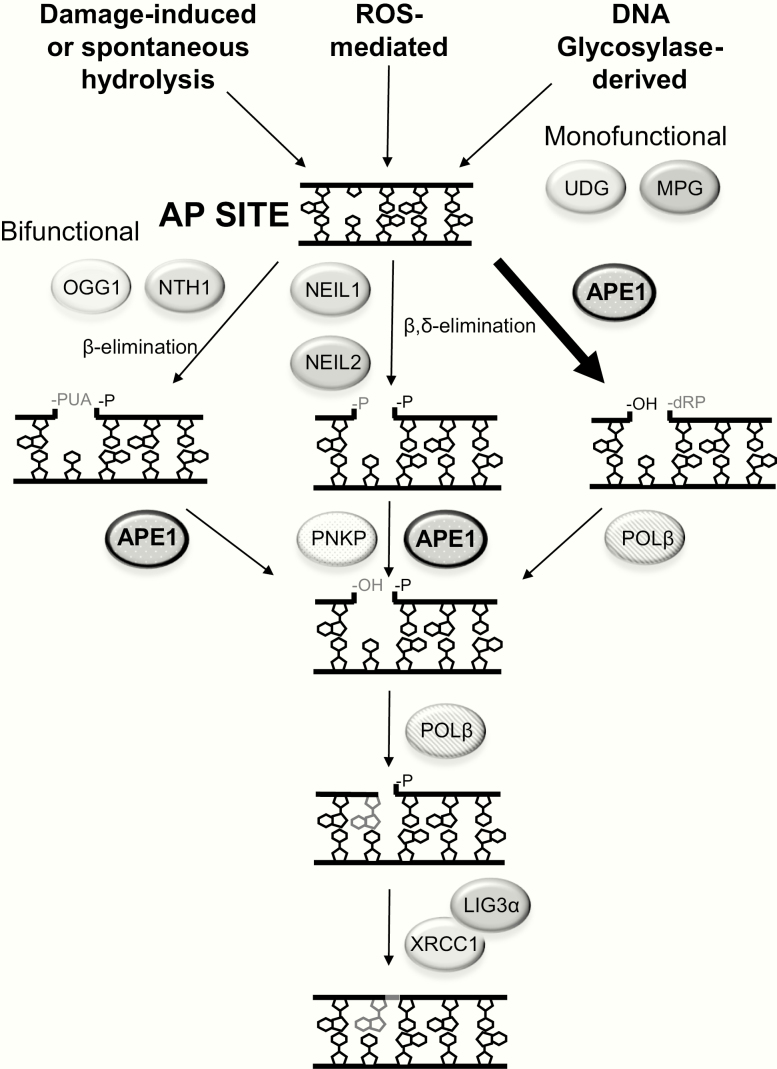
BER is the major pathway for coping with base damage, abasic sites and various single-strand break interruptions (Figure 2). In brief, classic BER can be broken down into five major steps, although the precise order of the steps may vary due to protein concentration variability, efficiency of the different reactions and the nature of the DNA intermediates: (i) base excision by a DNA glycosylase, (ii) incision at the resulting AP site, (iii) clean-up of the 5′ and/or 3′ terminal end(s), (iv) replacement of the damaged and excised nucleotide and (v) nick ligation to complete the process. In mammals, there are 11 DNA glycosylases that remove any number of inappropriate bases from DNA, including uracil (the spontaneous product of cytosine deamination) and 8-oxoguanine (one of the many oxidatively modified base lesions) (27). Besides the monofunctional DNA glycosylases (e.g. UNG, MPG), which only operate to excise base substrates, bifunctional DNA glycosylases can cleave the phosphodiester backbone at the AP site product, creating a strand break with a 3′-α,β-unsaturated aldehyde (β-elimination: OGG1, NTH1) or 3′-phosphate (β,δ-elimination: NEIL1, NEIL2). It is unclear, however, how much the AP lyase activities of the bifunctional enzymes are at play in cells, since they are quite poor enzymatically. Regardless, in situations involving an intact AP site (left behind by either type of DNA glycosylase), the predominant pathway appears to be cleavage by the major AP endonuclease, APE1 (Figure 2, right). Following APE1 incision, the primary gap-filling DNA polymerase, POLβ, inserts the missing nucleotide and excises the remaining 5′-abasic fragment (deoxyribose phosphate). The final nick is sealed by a complex of X-ray cross-complementing protein (XRCC1) and DNA ligase 3α (LIG3α). There exist variations of the BER pathway, including a strand-displacement, long-patch mechanism, but the reader is directed to additional reviews for a more thorough description of the BER system, including its prominent role in both nuclear and mitochondrial DNA repair (28–32).
Figure 2.
AP site formation and repair. The three primary mechanisms for the generation of genomic AP sites are shown at the top, including BER-initiated via the activity of a monofunctional or bifunctional DNA glycosylase. Following AP site formation, the predominant repair process involves APE1 incision, creating a strand break with a 3′-hydroxyl (OH) and 5′-dRP (right). At some low frequency, bifunctional DNA glycosylases can cleave at the AP site via β-elimination (left) or β,δ-elimination (centre) to generate a strand break with a 3′-α,β-unsaturated aldehyde or 3′-phosphate (P), respectively and a 5′-P. The single-strand breaks are then processed by the depicted enzyme(s) to create 3′-OH and 5′-P single-nucleotide gap intermediates. Shown is single-nucleotide gap-filling, executed by DNA POLβ, and nick ligation by an XRCC1/LIG3α complex.
Structure, Mechanism and Biochemical Functions of APE1
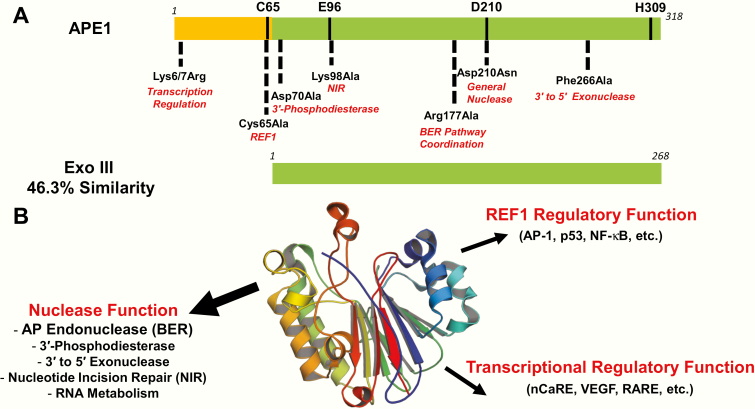
APE1 is a 318 amino acid, multifunctional protein that participates in DNA repair and transcriptional regulation. Indeed, APE1 was independently purified and subsequently cloned based on its ability to: (i) process DNA damage, such as AP sites and 3′-DNA modifications and (ii) stimulate the DNA binding activity of several transcription factors, such as the AP-1 heterodimer (33–36). This latter function led to the nomenclature ‘redox effector factor 1’ (a.k.a., REF1), one of the proteins many aliases. APE1 shares notable sequence homology to E. coli exonuclease III (Xth), an enzyme with AP endonuclease, 3′-phosphodiesterase and 3′–5′ exonuclease activities (37). The conservation occurs mainly in the C-terminal ~260 residues of APE1, which make up the nuclease core of the mammalian protein (Figure 3A). The presumably acquired N-terminal region of APE1 appears to direct intranuclear localisation, refine nucleic acid interactions, mediate protein associations and participate in the aforementioned REF1 function (reviewed in 38,39).
Figure 3.
Key features of APE1. (A) Linear comparison of human APE1 and E. coli Xth (a.k.a., Exo III) proteins. The redox domain of APE1 is shown in orange, and the nuclease domains of APE1 and Exo III are shown in green. Key catalytic and functional residues in APE1 are indicated; see also Table 1. (B) APE1 structural fold and biochemical functions. X-ray structure of the APE1 core displaying its conserved four-layered α/β sandwich structural fold (centre). The major reported biochemical functions of the protein are indicated in red, with either sub-activities or protein/gene targets indicated below or in parentheses, respectively.
X-ray crystallographic studies have revealed that APE1 (residues 43–318) belongs to the phosphoesterase superfamily of enzymes that contain a common four-layered α/β sandwich structural core (Figure 3B), showing clear similarity to other nucleases (i.e. Xth, DNase I, RNase H and LINE-1) and also several functionally unrelated proteins (i.e. IP5P, Ccr4p, CdtB, NOCT) (40,41). Key elements of the APE1 core include distinct loop regions and a unique active site that allows APE1 to specifically recognise, bind and slide along a DNA strand in search of an AP site primarily through interactions with the DNA phosphate backbone (42,43).
Crystal structures of APE1 complexed with AP site analogue tetrahydrofuran (THF)-containing duplex DNA have revealed that the protein kinks the DNA by ~35○ and flips the AP site out of the double helix and into the hydrophobic active site binding pocket (comprised of residues Phe266, Trp280 and Leu282) to facilitate complex formation. Once in the active site, the AP site is stabilised in position for cleavage of its 5′-phosphate backbone by residues contained within several loop regions that recognise the AP-DNA (44,45). Specifically, APE1 is proposed to stabilise the flipped-out AP site via a double-loop penetration mechanism, involving interactions of loops α11 and α5 with the minor and the major grooves, respectively. With the AP site positioned into the APE1 active site, an orphan base is left in the opposing strand of the double helix. Side chain Arg177, of loop α5, acts as a surrogate base by intercalating into the major groove and forming a base-stacking interaction (45). Importantly, this interaction is specific to product DNA and thus corroborates biochemical data indicating Arg177 specifically enhances product binding while only moderately affecting substrate binding (46,47). An additional loop region (α8) interacts with the major groove on the 3′ side of the AP site, and along with the hydrophobic pocket, has been shown to dictate substrate specificity as demonstrated by site-specific APE1 variants (48,49).
Prior studies have shown that amino acids Glu96, Asp210 and His309 are critical to the enzymatic nuclease activity of APE1 (50,51). The reported data are consistent with a catalytic mechanism that involves an in-line nucleophilic attack of the AP site phosphorus atom by a nucleophilic water molecule that is highly coordinated through hydrogen bonding to Asn212 and Asp210 (52). This attack is facilitated by charge neutralisation afforded by two hydrogen bonds to the phosphate non-bridging oxygens from Tyr171 and His309. A single Mg2+ is coordinated by Asp70, Glu96 and a water that is in contact with the non-bridging oxygen of the phosphate. Crystal structures imply a pentacovalent intermediate that is stabilised by Mg2+ and key active site contacts (45,53). Moreover, it has been proposed that during catalysis, the metal shifts to coordinate both the phosphate non-bridging oxygen and the newly generated O3′ (45). Importantly, the interactions mentioned here appear to be fundamental for APE1 catalysis, as their independent elimination via site-directed mutagenesis has been shown to significantly diminish catalytic activity (in some cases, by >4-orders of magnitude).
As mentioned above, APE1 participates as a central component of the BER response (Figure 2). In particular, following base excision by a substrate-selective DNA glycosylase, APE1 cleaves at the resulting AP site intermediate to generate a strand break with a 3′-hydroxyl priming group and a 5′-deoxyribose phosphate (5′-dRP). Based on biochemical and structural studies, APE1 has apparently evolved not for enzymatic efficiency, but to optimise pathway efficiency by remaining bound to its incised product to facilitate ‘hand-off’ to the next enzyme in the pathway, POLβ, in a process referred to as ‘passing-the-baton’. This model is supported by the evidence that mutation of the Arg177 residue in APE1 leads to improved turnover of the enzyme (44). However, clear cellular evidence of the significance of this mechanism has not to be reported.
Besides its robust AP endonuclease activity, APE1 has been demonstrated to remove 3′-mismatched or damaged nucleotides through its 3′ to 5′ exonuclease function; excise obstructive 3′-lesions, such as α,β-unsaturated aldehydes, phosphoglycolates and phosphates; initiate repair of bulky oxidative base lesions via a process called nucleotide incision repair (NIR) and degrade damaged RNA molecules as a potential cleansing mechanism (Figure 3B) (reviewed in 31,54). Of note, both biochemical and structural data indicate that unique active site contacts (i.e. Phe266) play a key role in APE1 3′-end processing activities (55,56). In addition to its various nuclease and redox regulatory activities, APE1 has been shown to operate as an integral component of transcription complexes, exhibiting the ability to even control its own gene expression (57).
As mentioned above, APE1 is often referred to by the alias REF1, due to its ability to stimulate the DNA-binding activity of several transcription factors through modulation of their redox status (33). Transcription factors in which APE1 has been shown to activate via this mechanism include AP-1, NFκβ, HIF1α and p53 (58). The APE1 redox activity can be disabled by mutation of residue Cys65 to Ala; however, because APE1 lacks the C-X-X-C motif common to most redox regulatory proteins for the formation, isomerisation and reduction of disulphide bonds, the precise mechanism it employs to activate transcription factor activity remains enigmatic (59–61). There is some evidence indicating Cys93 plays a role in the APE1 redox reaction, but both Cys65 and Cys93 are buried in the protein structure and are positioned too distant from one another to form a disulphide bond. Regardless, a model in which Cys65 acts as the nucleophile for reduction of the disulphide bond in the targeted transcription factor, and Cys93 serves as the resolving reside, has been proposed (62). In this model, APE1 must undergo a large conformational change or become partially unfolded to reveal a third Cys residue in order to facilitate the reaction. Further work is clearly required to determine the precise mechanism of the REF1 function of APE1 (63).
Effect of Amino Acid Substitutions on APE1 Structure–Function
As an extension of the early structure–function work, studies have since identified a set of strategic, site-specific mutations that uniquely affect the different proposed activities of APE1 (Table 1). We hypothesised that these variants would offer a means to evaluate the biological contribution of the different functions of the protein using a previously described complementation approach (see later). However, before determining their cellular complementation effectiveness, we sought to compare the structures of the different APE1 variants to gain insight into their potential biochemical and structural properties.
Table 1.
APE1 mutants
| Mutant | Biochemical consequences | Structural consequences | Function targeted | Predicted complementation outcome | Observed complementation outcome |
|---|---|---|---|---|---|
| Lys6Arg/Lys7Arg | Impaired protein acetylation; impaired transcriptional regulation | Unknown | Transcription regulation | Impaired acetylation will result in defective transcriptional regulation of target genes, cellular dysfunction and possibly inviability | No effect |
| Cys65Ala | Impaired REF1 function | Similar to WT; removal of reducing cysteine residue | REF1 | REF1 defect will result in an impaired transcriptional regulation and reduced cellular viability | Reduced cell viability |
| Asp70Ala | Enhanced 3′-phosphodiesterase activity | Unknown | 3′-Phosphodiesterase | Improved 3′-phosphodiesterase activity will lead to increased resistance to oxidative stress and bleomycin | Enhanced bleomycin resistance |
| Lys98Ala | Reduced NIR activity | Similar to WT; loss of hydrogen bonding between Asp70 | NIR | Reduced NIR activity will lead to increased sensitivity to oxidising agents and base damage accumulation | No effect |
| Arg177Ala | Impaired BER coordination | Unknown | Passing-the-baton | Reduced BER coordination will permit survival, but lead to increased damage accumulation and sensitivity to relevant DNA-damaging agents, such as MMS | Reduced cell viability and MMS resistance |
| Asp210Asn | Inactivation of nuclease activity | Altered coordination of the nucleophilic water and fellow catalytic triad residue Asn68 | All nuclease activities | Absence of nuclease-competent protein will lead to cellular inviability | Reduced cell viability and genotoxin resistance |
| Phe266Ala | Enhanced 3′–5′ exonuclease activity | Larger active site and 120○ rotation of 3′-mismatched base | 3′–5′ Exonuclease | Increased exonuclease function will reduce single nucleotide mutation frequency | Possibly enhanced cell viability |
See text for further details.
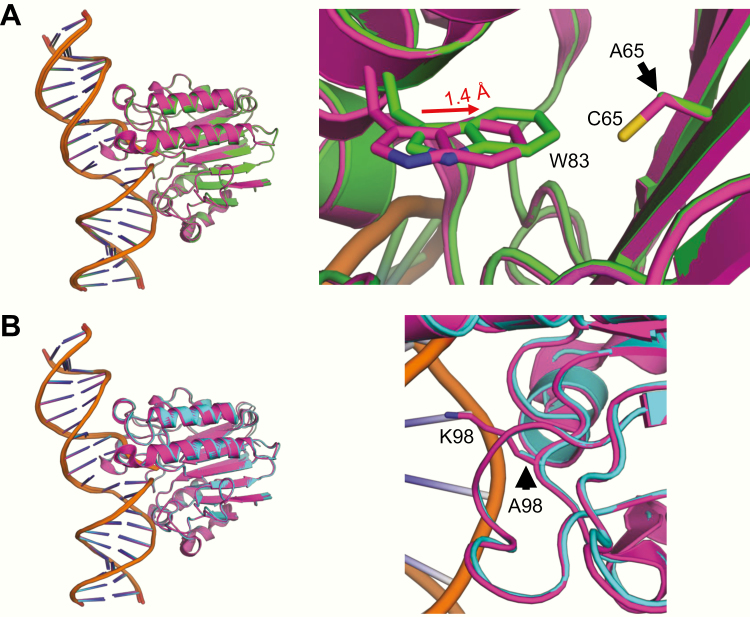
To establish structures for a few of the proteins, we employed a previously described expression system (56) and X-ray crystallographic method (45). Structures of untagged Cys65Ala (the REF1 mutant) and Lys98Ala (the NIR mutant) engaged with an AP endonuclease product (i.e. the cleaved AP site) showed high similarity to wild-type (WT) APE1, with very little change in the orientation of the DNA, protein backbone or sidechains in response to either mutation (Table 2; Figure 4). In the case of Cys65Ala (Figure 4A), the neighbouring residue Trp83 is slightly shifted (1.4 Å); however, it is likely that the absence of the key reducing cysteine residue, and not this modest shift, results in the abolition of redox activity in response to the mutation (see (62) and references therein). While the Lys98Ala structure does not show any structural perturbation in response to the mutation (Figure 4B), the hydrogen bonding interaction between Lys98 and the carboxyl group of Asp70 observed in the WT structure is lost. Given the generally normal structural features of the Lys98Ala mutant, it is not completely clear how the substitution would selectively interfere with the NIR activity as reported (64).
Table 2.
Data collection and refinement statistics of mutant APE1:DNA product complexes with abasic (THF) containing DNA
| C65A | K98A | |
|---|---|---|
| Data collection | ||
| Space group | P1 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 43.59,60.46,71.92 | 44.35,60.49,72.62 |
| α, β, γ (°) | 83.69,79.47,88.74 | 83.72,79.66,88.46 |
| Resolution (Å) | 25–2.15 | 25–2.1 |
| Rmeas (%) | 0.152 (0.798) | 0.084 (0.551) |
| CC1/2 | 0.528 | 0.667 |
| I/σ I | 7.81 (1.08) | 17.2 (2) |
| Completeness (%) | 99.2 (96.1) | 99.8 (98.2) |
| Redundancy | 3.6 (2.1) | 3.9 (2.1) |
| Refinement | ||
| Resolution (Å) | 2.09 | 2.09 |
| No. reflections | 36 463 | 58 444 |
| Rwork/Rfree | 0.1862/0.2445 | 0.1713/0.2272 |
| No. atoms | ||
| Protein | 4321 | 4323 |
| DNA | 836 | 836 |
| Water | 290 | 292 |
| B-factors (Å 2) | ||
| Protein | 33.28 | 30.64 |
| DNA | 43.12 | 41.04 |
| Water | 36.06 | 30.52 |
| R.m.s deviations | ||
| Bond length (Å) | 0.009 | 0.008 |
| Bond angles (º) | 1.091 | 1.025 |
| PDB ID | 6P94 | 6P93 |
Highest resolution shell is shown in parentheses.
Figure 4.
X-ray structures of mutant APE1 proteins bound to incised product DNA. (A) Overlay of Cys65Ala (green) with the WT APE1 protein (magenta, PDBID: 5DFF). (B) Overlay of Lys98Ala (cyan) with the WT APE1 (magenta, PDBID: 5DFF). Panels on the right show a zoomed-in view of the mutation.
Structures of APE1 containing the mutation Asp210Asn or Phe266Ala have been previously reported in the literature. A catalytically dead double mutant enzyme (Asp210Asn Glu96Gln) demonstrated the structural consequence of Asp210Asn to be in its altered coordination of the nucleophilic water and fellow catalytic triad reside Asn68 (45). Consistent with its critical role in the APE1 nuclease reaction, substitution of Asp210 renders the protein essentially inactive (51). In a separate study, a structure of Phe266Ala APE1 bound to DNA containing a C/T mismatch at the 3′ end of a nick (a 3′ exonuclease substrate) revealed an enlarged active site pocket, allowing the mismatched C to occupy an alternate conformation in which it has rotated ~120° relative to WT (56). These data support the observation that a Phe266Ala substitution enhances the enzyme’s 3′–5′ exonuclease function (48).
To our knowledge, there are no reported structures of either Arg177Ala or Asp70Ala. However, based on structures of WT APE1 and complementary biochemical studies, the functional roles of these two residues have been described in detail (see also Table 1). In particular, Arg177 contributes to BER coordination (see earlier), while Asp70 has been implicated in binding the divalent metal ion (45,65), and substitution of this residue with Ala has been reported to enhance the 3′-phosphodiesterase activity of the protein (66). Lastly, any structural consequence of the Lys6Arg/Lys7Arg mutations remains enigmatic, as the N-terminal region of APE1 has yet to be crystallised due to its inherent disorder.
In addition to the many mechanism-based mutations that have been introduced into APE1 (see above), evidence indicates that several missense substitutions, both germline and disease-associated, exist in the protein within the general population. While it is unclear for many of the amino acid variants whether they exhibit altered function, the polymorphic variants Gln51His (~3% frequency), Ile64Val (~0.5%) and Asp148Glu (~45%) have been demonstrated to display WT properties in a number of functional and structural tests (67). Recent work, however, has found that one tumour-associated variant (Arg237Cys) identified in a single endometrial cancer patient may lead to impaired biochemical and cellular activity (68,69). This observation suggests that defects in the function of APE1, either germline or somatic, could contribute to disease susceptibility, although profound deficiencies in APE1 at conception are likely to be incompatible with life (see next).
Backdrop on APE1 Biology
Early work demonstrated that germline deletion of both alleles of APE1 in mice leads to embryonic lethality, indicating a requirement for the protein in multicellular organismal development (70,71). More recent efforts have also found that sufficient depletion of APE1 in cells (around 60% or more) results in apoptotic death (72). The essential nature of APE1 has made investigations into the biological role of the protein and its various biochemical activities complicated. And while useful cell models have been reported using knock-down strategies (see for instance, 73), this approach is complicated by the residual, endogenous WT APE1 protein that remains. Nonetheless, beyond the cell growth complications noted above, evidence indicates that defects in APE1 function give rise to profound sensitivity to alkylating agents, such as methylmethane sulphonate (MMS) and the chemotherapeutic drug temozolomide, and to a lesser extent, increased sensitivity to oxidising agents (such as hydrogen peroxide), ionising radiation, the radiomimetic bleomycin and the chain-terminating nucleoside analogue, β-L-dioxolane-cytidine (clinically known as troxacitabine). These findings are consistent with a major role for the AP endonuclease function of APE1, and a biologically important role for its 3′-repair activities (reviewed in 74). We note that there are reports describing functions of APE1 in cellular processes such as antibody diversification, RNA metabolism and the granzyme A-activated cell death response, which are described in greater detail elsewhere (75–77).
The recent creation of floxed APE1 mouse models has permitted the first glimpse into the consequences of APE1 absence on organismal physiology and health. In the first report describing an APE1 tamoxifen-inducible, Cre-recombinase, conditional knockout mouse, the protein was found to be vital for protecting both gray and white matter from the oxidative stress induced by transient focal cerebral ischemia, and for functional recovery of the central nervous system after mild stroke injury (78). Subsequent analysis examining the consequences of whole-body, conditional-deletion of APE1 revealed that gene inactivation before weaning (i.e. around post-natal day 7–12) resulted in profound growth impairment and animal death (79). APE1 gene deletion later in life (around week 6 post-weaning) caused a more subtle phenotype, which became more evident around 8-months of age and included several premature ageing characteristics, such as loss of hair, reduced wound healing and increased senescence. The most recent paper, where APE1 was selectively deleted in the brain by crossing APE1-floxed animals with mice that express the Cre recombinase via the brain-specific promoter Nestin, found that mutant animals appeared normal at birth (despite embryonic gene inactivation), yet experienced rapid and profound brain-wide degenerative changes that coincided with the change to respiratory oxygenation and led to death within a few months (80). Moreover, loss of APE1, even in a heterozygous state, along with inactivation of the tumour suppressor p53 resulted in a dramatic increase in cancer susceptibility, specifically for glioblastoma and medulloblastoma. These more targeted studies are generally consistent with past work that has found that happloinsufficient animals, relative to controls, exhibit an increase in spontaneous mutagenesis and cancer development (81,82). What is still unclear from the current analysis, however, is the precise contribution of the different functions of APE1 to the pathology observed.
Biological Significance of the Different APE1 Functions
Several studies have investigated the biological contributions of the different functions of APE1, focusing mainly on their role in preserving cell viability. Three independent efforts, using variations on the functional complementation theme, have found that the repair nuclease activity of APE1 is essential in averting apoptotic cell death of APE1-depleted/deficient cells (72,73,83). Findings regarding the REF1 function are less clear, with two of the studies indicating no obvious role in preserving cell growth, yet the other reporting a critical contribution (73). The work of Izumi et al. (83) also described an important function for the transcriptional regulatory activity of APE1 via its N-terminal lysine residues (Lys6 and Lys7). However, in that work, they did not demonstrate expression of the mutant protein, raising doubt about the conclusions drawn, and Vascotto et al. (73) have since reported near WT rescue of cell viability with the same Lys6/7Arg mutant protein. As mentioned earlier, we developed a complementation strategy to more comprehensively investigate the biological significance of the different functions of this multifunctional protein in an APE1-deficient background, using a set of APE1 mutants that are uniquely altered in a particular activity (Table 1). We acknowledge, however, that although we have taken steps to ensure reproducibility and accuracy of the results, our method of transient overexpression needs to be kept in mind when interpreting results herein.
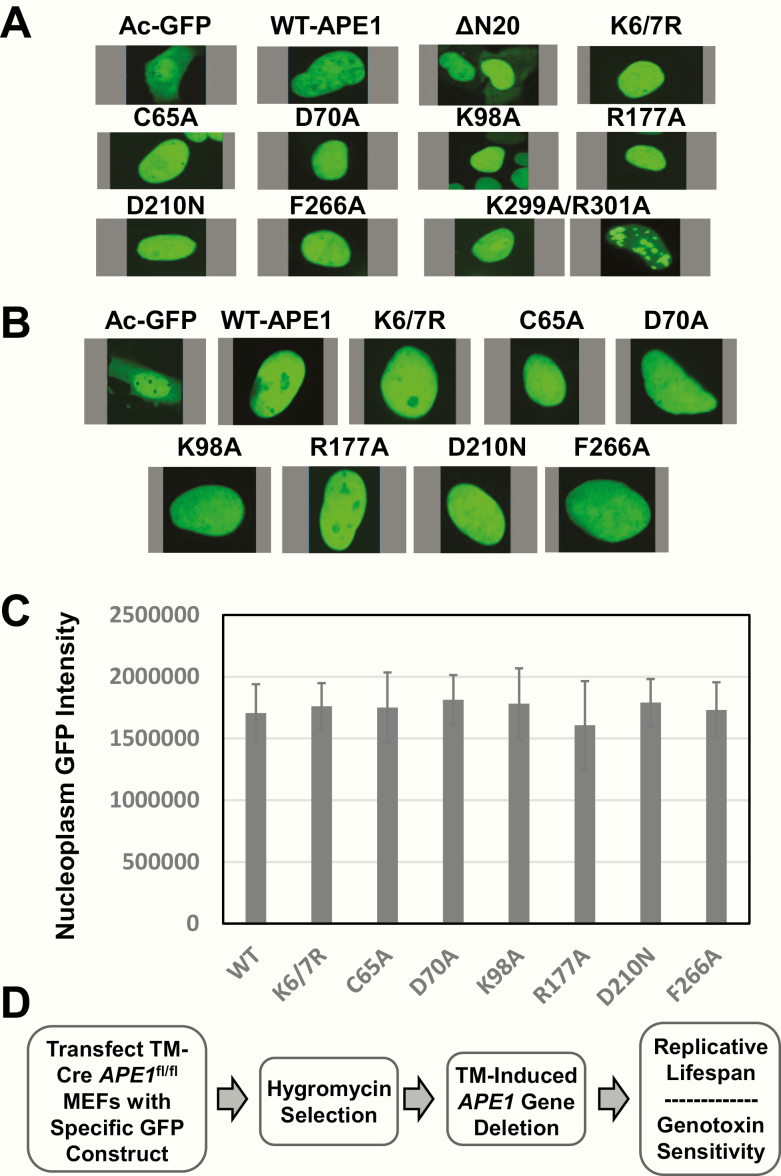
Before carrying out the complementation work, we examined the effect of the different mutations on protein localisation and stability when expressed in mammalian cells. As our complementation studies were to be conducted with GFP-tagged fusion proteins, we were able to easily monitor intracellular distribution using fluorescence confocal microscopy after transfection of the different plasmid DNA constructs. As shown in Figure 5A, when expressed in HeLa cells, GFP alone exhibited the typical pan staining throughout the cell, whereas most APE1 GFP fusion proteins displayed nuclear localisation similar to that of the WT protein. The exceptions were ΔN20 (lacking the 20 N-terminal residues) and Lys299Ala/Arg301Ala, proteins designed to address issues related to nuclear (84) and mitochondrial (85) localisation, respectively. However, given the lack of a defined localisation pattern for these two APE1 variants (Figure 5A), we excluded them from further analysis, as any results would be difficult to interpret with confidence. As seen with the HeLa cells, the remaining APE1 mutants when expressed in the TM-Cre APE1fl/fl mouse embryonic fibroblasts (MEFs) exhibited a nuclear localisation pattern similar to that observed for WT APE1 (Figure 5B). Moreover, examining the fluorescence intensity of each protein as a means of assessing protein expression and stability revealed that each APE1 mutant was present at a steady-state level that was similar to the WT protein (Figure 5C). The findings in total, including the current X-ray structure data (see earlier), support the conclusion that the site-specific mutations have no gross effect on protein conformation and that the different mutants are expressed at comparable levels in the target mouse cells.
Figure 5.
APE1 protein intracellular localisation and expression. HeLa cells (A) or TM-Cre APE1fl/fl MEFs (B) were transfected with the indicated plasmid vector, allowed to grow for 48 h, and then photographed using confocal microscopy for the GFP signal. Representative images of GFP fluorescence are shown. (C) TM-Cre APE1fl/fl MEFs were transfected and photographed as above. Captured images were then analysed by marking a defined spot in the nucleoplasm and quantifying GFP intensity. Shown are the averages and standard deviations of the raw intensity values from at least six individual cells from three independent experiments. (D) Overview of complementation strategy. TM-Cre APE1fl/fl MEFs are transfected with one of the different GFP-containing vectors. Fibroblasts are then placed under hygromycin B selection for 3 days (to enrich for cells carrying the plasmid), before being exposed to TM at an optimised concentration (5 µM) for 96 h to inactive both endogenous APE1 alleles. Cells are subsequently monitored for replicative lifespan or genotoxin sensitivity. A more detailed protocol can be found in (69).
We have described previously the complementation strategy (69), which we have outlined in Figure 5D. In brief, we used MEFs that harbour floxed APE1 alleles (APE1fl/fl) and a tamoxifen-inducible Cre recombinase expression cassette (TM-Cre). In our experimental paradigm, we first transfect TM-Cre APE1fl/fl MEFs with a designated plasmid (i.e. vector or specific APE1 construct), briefly select for the plasmid with hygromycin, and then incubate the cells with TM to induce >99% deletion/inactivation of both endogenous APE1 alleles as assessed by western blot analysis (69). We can then monitor cell viability (replicative lifespan) over time or determine genotoxin sensitivity of the MEFs as a function of the complementing plasmid.
Repair and REF1 function contribute to cell viability
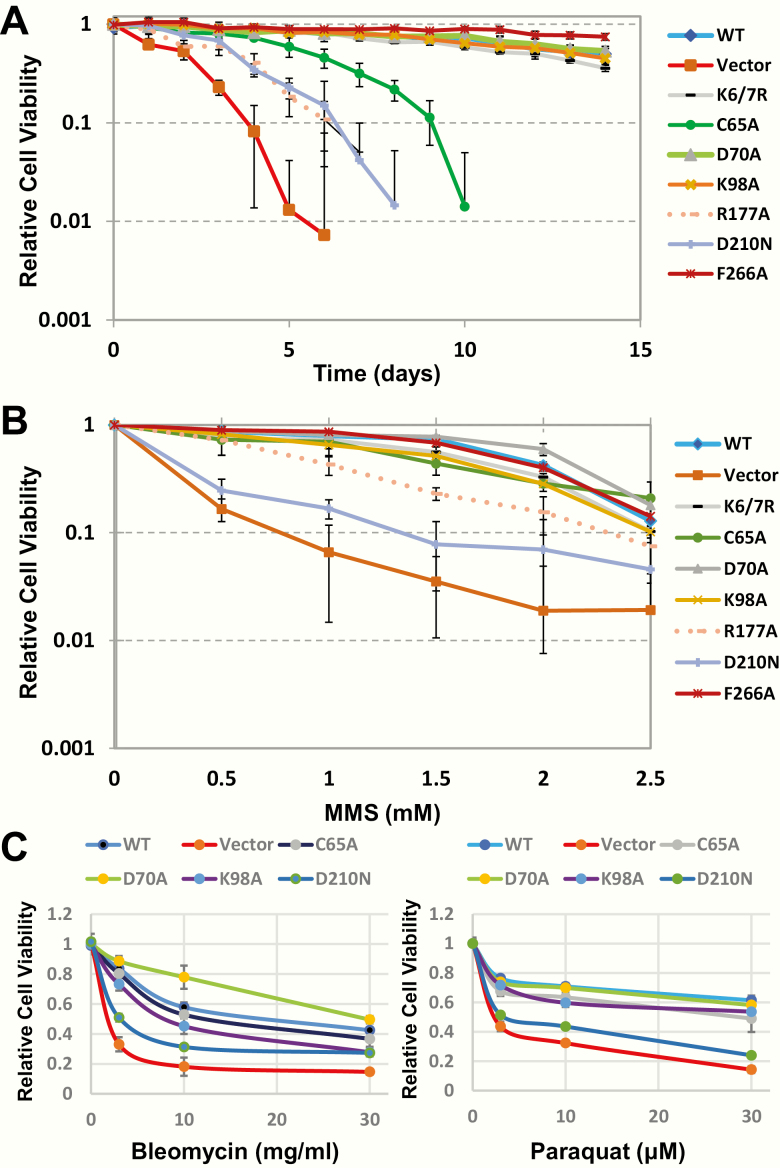
It has been demonstrated that mammalian cells deficient for APE1 have an abbreviated replicative lifespan (see earlier). To address the role of the different APE1 functions in facilitating cell survival, we assessed viability over time following transfection of the indicated plasmid and subsequent inactivation of the endogenous APE1 alleles. As shown in Figure 6A, APE1-deficient TM-Cre APE1fl/fl MEFs harbouring the GFP vector control survived for only ~5 days, whereas the WT-complemented cells grew for >14 days. The acetylation-defective Lys6/7Arg, the 3′-repair-enhanced Asp70Ala and the NIR-defective Lys98Ala mutants all exhibited viability patterns similar to WT-complemented cells; there was a potential slight increase in survival of the Phe266Ala exonuclease-enhanced mutant. Notably, the Asp210Asn nuclease-dead and the Arg177Ala passing-the-baton mutants provided little rescue of cell growth, while the Cys65Ala REF1 mutant displayed intermediate complementation of cell viability. These data clearly support a major role for BER efficiency in supporting cell viability under normal growth conditions, as well as an important role for APE1’s transcriptional regulatory (REF1) function in promoting cell proliferation.
Figure 6.
Complementation effectiveness of WT and mutant APE1 proteins. (A) Replicative lifespan. TM-Cre APE1LoxP/LoxP MEFs were transfected with the indicated plasmid construct, put under selection and treated with TM (see Figure 5D), and then monitored for viability over time (every day) in culture using the Cell Counting-8 assay (Dojindo, Rockville, MD). Averages and standard deviations shown represent four biological experiments, each done in duplicate. (B) Cell survival after MMS exposure. Following TM treatment (see above), TM-Cre APE1fl/fl MEFs harbouring the designated plasmid were treated with the indicated dose of MMS for 1 h. Survival (relative to the untreated control) was determined 48 h after MMS exposure as noted in panel A. Averages and standard deviations shown represent four biological experiments, each done in duplicate. (C) Cell survival after bleomycin or paraquat exposure. Relative survival was determined as in panel B, except exposure to bleomycin was for 1 h and paraquat was for 4 h. For these survival assays, averages and standard deviations represent at least four data points from four biological replicates.
Contribution of different APE1 nuclease activities to genotoxin resistance
We next examined the contributions of the different functions in the context of exogenously induced DNA damage. As seen previously for other cell types (reviewed in 74), the vector-complemented APE1-deficient MEFs were far more sensitivity to the alkylator MMS than the WT-complemented counterparts (Figure 6B). Moreover, only the nuclease-dead (Asp210Asn) and, less so, the pathway coordination (Arg177Ala) mutant showed impaired complementation of MMS sensitivity. Since MMS is known to generate a high level of AP sites, these data specifically point to a critical role for the AP endonuclease function of APE1 in coordinating repair and preventing damage-induced cell death. The other activities of APE1, i.e. its transcriptional regulatory, NIR or 3′-processing functions, play a minor role at best in the alkylation survival response.
To more directly interrogate the 3′-repair functions of APE1, we explored sensitivities to oxidative stress-inducing agents, which are more inclined to generate oxidative DNA strand breaks harbouring 3′-blocking lesions, such as phosphates or phosphoglycolates, as well as oxidative base lesions, including those potentially processed by NIR (86). A clear increased sensitivity was observed for the vector-complemented APE1-deficient TM-Cre APE1fl/fl MEFs in comparison to the WT controls for bleomycin and paraquat, but much less so for menadione (Figure 6C and unpublished observation). Focusing on the first two agents, we expectantly observed that the nuclease-dead mutant (Asp210Asn) was unable to rescue sensitivity to both oxidising agents (Figure 6C). The Cys65Ala (REF1) and Lys98Ala (NIR) mutants were generally similar to WT in both paradigms, whereas the Asp70Ala appeared similar to WT in the case of paraquat, but more effective than WT at promoting cell survival in the case of bleomycin. These data indicate APE1 plays an important role in oxidative stress resistance, with its 3′-repair functions operating prominently, and support a particularly significant function in 3′-phosphoglycolate (a prominent product of bleomycin attack) excision (87,88).
Closing Thoughts
AP sites are frequent, non-coding DNA lesions that can block RNA or DNA polymerases or lead to mutagenic by-pass. Such molecular events can ultimately alter cellular behaviour, and in extreme cases, lead to apoptosis, senescence or transformation. The major protein in mammals assigned to cope with AP sites in both the nuclear and mitochondrial genomes is APE1. Besides being the predominant AP endonuclease, APE1 also maintains 3′-repair activities, an NIR function and the ability to regulate transcription factor binding activity via its REF1 function. While many of these activities have been well described in vitro (although in some cases remain inadequately defined mechanistically), given the inviability of cells or animals lacking APE1, the contribution of the different functions of the protein to biology has been more difficult to ascertain. Using a combination of structural and functional complementation approaches, we have added new insights to the existing molecular picture of the multifunctional protein APE1.
There is broad consensus within the current data that the conserved nuclease function of APE1, most likely its AP endonuclease activity, is critical to cell viability, and likely plays a central role in the embryonic lethality observed upon germline deletion of the gene in mice. Notably, when comparing the Arg177Ala mutant to the Asp210Asn mutant (Figure 6A), it is not only the nuclease activity that is critical for efficient cell growth under normal conditions, but also coordination within the BER pathway. The fact that the Arg177Ala mutant played a lesser role in protecting against MMS-induced cell death (Figure 6B), in comparison to its complementation efficiency under normal growth conditions, may suggest that active coordination is most important when DNA damage levels are low and effective communication is more necessary due to the low levels of the proteins downstream of APE1 (e.g. POLβ). Our results also indicate that accumulating DNA strand-breaks can be toxic (in addition to unrepaired AP sites) and that there are not effective backup repair mechanisms for abasic sites in mammals, despite the presence of another Xth ortholog (APE2) or the evidence that pathways such as nucleotide excision repair can handle such damage (89,90). Nevertheless, there are reports of mammalian cells surviving severe depletion or inactivation of APE1, presumably relying upon compensatory mechanisms that are presently unknown (17,91).
Collectively, the current results support an important biological role for the acquired REF1 function of APE1. Our studies, in particular, indicate a specific contribution to normal cell survival, yet no role in genotoxin stress resistance. This finding is consistent with a function for the REF1 activity in maintaining essential gene regulatory networks, particularly of immediately early genes (80), without directly carrying out DNA damage repair. Numerous studies have indeed supported significant roles for the REF1 function in various biological responses, with REF1 proving to be a promising target in various treatment efforts, such as those involving the killing of cancer cells (reviewed in 58).
The collective data are also consistent with an important biological role for the 3′-phosphodiesterase activity of APE1, certainly in the context of oxidative stress or a relevant genotoxin exposure, most notably to the radiomimetic bleomycin that creates 3′-phosphoglycolates (see for instance, 92). The significance of the 3′–5′ exonuclease is less clear, although there is a hint that an enhanced function in this capacity might improve cell growth, possibly by reducing polymerase errors (93). Conversely, we found no clear indication for a biological contribution of the transcriptional regulatory (Lys6/7Arg mutant) or NIR functions of APE1. While our analysis is by no means exhaustive, our data imply either highly specialised roles for these activities or limited significance overall. Further investigations using the developed system within will go a long way towards addressing the remaining issues, as will the creation of animal models designed to alter one function or another.
Funding
This research was supported by the Intramural Research Program of the NIH, National institute on Aging; and by grants [National Institutes of Health R01-ES029203, R35-GM128562] to B.D.F, W.J.S. and A.M.W. and [American Cancer Society PF-1815401-DMC] to A.M.W.
Conflict of interest statement: None declared.
Acknowledgements
We thank Dr Yie Liu and Dr Robert Brosh (NIA) for critical reading of the manuscript.
References
- 1. Tiwari V. and Wilson D. M. III (2019) DNA damage and associated DNA repair defects in disease and premature aging. Am. J. Hum. Genet., 105, 237–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lindahl T. and Nyberg B (1972) Rate of depurination of native deoxyribonucleic acid. Biochemistry, 11, 3610–3618. [DOI] [PubMed] [Google Scholar]
- 3. Ilina E. S., Lavrik O. I. and Khodyreva S. N (2008) Ku antigen interacts with abasic sites. Biochim. Biophys. Acta, 1784, 1777–1785. [DOI] [PubMed] [Google Scholar]
- 4. Khodyreva S. N., Prasad R., Ilina E. S., Sukhanova M. V., Kutuzov M. M., Liu Y., Hou E. W., Wilson S. H. and Lavrik O. I (2010) Apurinic/apyrimidinic (AP) site recognition by the 5′ -dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1). Proc. Natl. Acad. Sci. U. S. A., 107, 22090–22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kosova A. A., Khodyreva S. N. and Lavrik O. I (2015) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) interacts with apurinic/apyrimidinic sites in DNA. Mutat. Res., 779, 46–57. [DOI] [PubMed] [Google Scholar]
- 6. Dutta S., Chowdhury G. and Gates K. S (2007) Interstrand cross-links generated by abasic sites in duplex DNA. J. Am. Chem. Soc., 129, 1852–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Price N. E., Johnson K. M., Wang J., Fekry M. I., Wang Y. and Gates K. S (2014) Interstrand DNA-DNA cross-link formation between adenine residues and abasic sites in duplex DNA. J. Am. Chem. Soc., 136, 3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lopez-Martinez D., Liang C. C. and Cohn M. A (2016) Cellular response to DNA interstrand crosslinks: the Fanconi anemia pathway. Cell. Mol. Life Sci., 73, 3097–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hashimoto S., Anai H. and Hanada K (2016) Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ., 38, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greenberg M. M. (2014) Looking beneath the surface to determine what makes DNA damage deleterious. Curr. Opin. Chem. Biol., 21, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Quiñones J. L., Thapar U., Yu K., Fang Q., Sobol R. W. and Demple B (2015) Enzyme mechanism-based, oxidative DNA-protein cross-links formed with DNA polymerase β in vivo. Proc. Natl. Acad. Sci. U. S. A., 112, 8602–8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vaz B., Popovic M. and Ramadan K (2017) DNA-protein crosslink proteolysis repair. Trends Biochem. Sci., 42, 483–495. [DOI] [PubMed] [Google Scholar]
- 13. Stingele J., Bellelli R. and Boulton S. J (2017) Mechanisms of DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol., 18, 563–573. [DOI] [PubMed] [Google Scholar]
- 14. Barsky D., Foloppe N., Ahmadia S., Wilson D. M. III and MacKerell A. D. Jr (2000) New insights into the structure of abasic DNA from molecular dynamics simulations. Nucleic Acids Res., 28, 2613–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J., Dupradeau F. Y., Case D. A., Turner C. J. and Stubbe J (2008) DNA oligonucleotides with A, T, G or C opposite an abasic site: structure and dynamics. Nucleic Acids Res., 36, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berquist B. R., McNeill D. R. and Wilson D. M. III (2008) Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J. Mol. Biol., 379, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J., Svilar D., McClellan S., Kim J. H., Ahn E. E., Vens C., Wilson D. M. III and Sobol R. W (2018) DNA repair molecular beacon assay: a platform for real-time functional analysis of cellular DNA repair capacity. Oncotarget, 9, 31719–31743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fairlamb M. S., Whitaker A. M. and Freudenthal B. D (2018) Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5′ mismatches. Acta Crystallogr. D. Struct. Biol., 74, 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strauss B. S. (1991) The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays, 13, 79–84. [DOI] [PubMed] [Google Scholar]
- 20. Powers K. T. and Washington M. T (2018) Eukaryotic translesion synthesis: choosing the right tool for the job. DNA Repair, 71, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vaisman A. and Woodgate R (2017) Translesion DNA polymerases in eukaryotes: what makes them tick? Crit. Rev. Biochem. Mol. Biol., 52, 274–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ling H., Boudsocq F., Woodgate R. and Yang W (2004) Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol. Cell, 13, 751–762. [DOI] [PubMed] [Google Scholar]
- 23. Hogg M., Wallace S. S. and Doublié S (2004) Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J., 23, 1483–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weerasooriya S., Jasti V. P. and Basu A. K (2014) Replicative bypass of abasic site in Escherichia coli and human cells: similarities and differences. PLoS One, 9, e107915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patra A., Zhang Q., Lei L., Su Y., Egli M. and Guengerich F. P (2015) Structural and kinetic analysis of nucleoside triphosphate incorporation opposite an abasic site by human translesion DNA polymerase η. J. Biol. Chem., 290, 8028–8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang W., Walmacq C., Chong J., Kashlev M. and Wang D (2018) Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A., 115, E2538–E2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brooks S. C., Adhikary S., Rubinson E. H. and Eichman B. F (2013) Recent advances in the structural mechanisms of DNA glycosylases. Biochim. Biophys. Acta, 1834, 247–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brenerman B. M., Illuzzi J. L. and Wilson D. M. III (2014) Base excision repair capacity in informing healthspan. Carcinogenesis, 35, 2643–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bauer N. C., Corbett A. H. and Doetsch P. W (2015) The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res., 43, 10083–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carter R. J. and Parsons J. L (2016) Base excision repair, a pathway regulated by posttranslational modifications. Mol. Cell. Biol., 36, 1426–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whitaker A. M., Schaich M. A., Smith M. R., Flynn T. S. and Freudenthal B. D (2017) Base excision repair of oxidative DNA damage: from mechanism to disease. Front. Biosci. 22, 1493–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moor N. A. and Lavrik O. I (2018) Protein-protein interactions in DNA base excision repair. Biochemistry, 83, 411–422. [DOI] [PubMed] [Google Scholar]
- 33. Xanthoudakis S. and Curran T (1992) Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J., 11, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seki S., Hatsushika M., Watanabe S., Akiyama K., Nagao K. and Tsutsui K (1992) cDNA cloning, sequencing, expression and possible domain structure of human APEX nuclease homologous to Escherichia coli exonuclease III. Biochim. Biophys. Acta, 1131, 287–299. [DOI] [PubMed] [Google Scholar]
- 35. Robson C. N. and Hickson I. D (1991) Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res., 19, 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Demple B., Herman T. and Chen D. S (1991) Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl. Acad. Sci. U. S. A., 88, 11450–11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Demple B. and Harrison L (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem., 63, 915–948. [DOI] [PubMed] [Google Scholar]
- 38. Antoniali G., Lirussi L., Poletto M. and Tell G (2014) Emerging roles of the nucleolus in regulating the DNA damage response: the noncanonical DNA repair enzyme APE1/Ref-1 as a paradigmatical example. Antioxid. Redox Signal., 20, 621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thakur S., Dhiman M., Tell G. and Mantha A. K (2015) A review on protein-protein interaction network of APE1/Ref-1 and its associated biological functions. Cell Biochem. Funct., 33, 101–112. [DOI] [PubMed] [Google Scholar]
- 40. Dlakić M. (2000) Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci., 25, 272–273. [DOI] [PubMed] [Google Scholar]
- 41. Abshire E. T., Chasseur J., Bohn J. A., Del Rizzo P. A., Freddolino P. L., Goldstrohm A. C. and Trievel R. C (2018) The structure of human Nocturnin reveals a conserved ribonuclease domain that represses target transcript translation and abundance in cells. Nucleic Acids Res., 46, 6257–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strauss P. R., Beard W. A., Patterson T. A. and Wilson S. H (1997) Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Biol. Chem., 272, 1302–1307. [DOI] [PubMed] [Google Scholar]
- 43. Beloglazova N. G., Kirpota O. O., Starostin K. V., Ishchenko A. A., Yamkovoy V. I., Zharkov D. O., Douglas K. T. and Nevinsky G. A (2004) Thermodynamic, kinetic and structural basis for recognition and repair of abasic sites in DNA by apurinic/apyrimidinic endonuclease from human placenta. Nucleic Acids Res., 32, 5134–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mol C. D., Izumi T., Mitra S. and Tainer J. A (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature, 403, 451–456. [DOI] [PubMed] [Google Scholar]
- 45. Freudenthal B. D., Beard W. A., Cuneo M. J., Dyrkheeva N. S. and Wilson S. H (2015) Capturing snapshots of APE1 processing DNA damage. Nat. Struct. Mol. Biol., 22, 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Izumi T., Schein C. H., Oezguen N., Feng Y. and Braun W (2004) Effects of backbone contacts 3′ to the abasic site on the cleavage and the product binding by human apurinic/apyrimidinic endonuclease (APE1). Biochemistry, 43, 684–689. [DOI] [PubMed] [Google Scholar]
- 47. Peddi S. R., Chattopadhyay R., Naidu C. V. and Izumi T (2006) The human apurinic/apyrimidinic endonuclease-1 suppresses activation of poly(adp-ribose) polymerase-1 induced by DNA single strand breaks. Toxicology, 224, 44–55. [DOI] [PubMed] [Google Scholar]
- 48. Hadi M. Z., Ginalski K., Nguyen L. H. and Wilson D. M. III (2002) Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J. Mol. Biol., 316, 853–866. [DOI] [PubMed] [Google Scholar]
- 49. Shen J. C. and Loeb L. A (2003) Mutations in the alpha8 loop of human APE1 alter binding and cleavage of DNA containing an abasic site. J. Biol. Chem., 278, 46994–47001. [DOI] [PubMed] [Google Scholar]
- 50. Barzilay G., Mol C. D., Robson C. N., Walker L. J., Cunningham R. P., Tainer J. A. and Hickson I. D (1995) Identification of critical active-site residues in the multifunctional human DNA repair enzyme HAP1. Nat. Struct. Biol., 2, 561–568. [DOI] [PubMed] [Google Scholar]
- 51. Erzberger J. P. and Wilson D. M. III (1999) The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: new insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J. Mol. Biol., 290, 447–457. [DOI] [PubMed] [Google Scholar]
- 52. Tsutakawa S. E., Shin D. S., Mol C. D., et al. (2013) Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J. Biol. Chem., 288, 8445–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aboelnga M. M. and Wetmore S. D (2019) Unveiling a single-metal-mediated phosphodiester bond cleavage mechanism for nucleic acids: a multiscale computational investigation of a human DNA repair enzyme. J. Am. Chem. Soc., 141, 8646–8656. [DOI] [PubMed] [Google Scholar]
- 54. Whitaker A. M. and Freudenthal B. D (2018) APE1: a skilled nucleic acid surgeon. DNA Repair, 71, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gross G. and McCullough E. C (1975) Exposure values around an x-ray scanning transaxial tomograph (EMI scanner). Med. Phys., 2, 282. [DOI] [PubMed] [Google Scholar]
- 56. Whitaker A. M., Flynn T. S. and Freudenthal B. D (2018) Molecular snapshots of APE1 proofreading mismatches and removing DNA damage. Nat. Commun., 9, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Izumi T., Henner W. D. and Mitra S (1996) Negative regulation of the major human AP-endonuclease, a multifunctional protein. Biochemistry, 35, 14679–14683. [DOI] [PubMed] [Google Scholar]
- 58. Shah F., Logsdon D., Messmann R. A., Fehrenbacher J. C., Fishel M. L. and Kelley M. R (2017) Exploiting the Ref-1-APE1 node in cancer signaling and other diseases: from bench to clinic. NPJ Precision Oncology, 1, 19. doi:10.1038/s41698-017-0023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Walker L. J., Robson C. N., Black E., Gillespie D. and Hickson I. D (1993) Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol. Cell. Biol., 13, 5370–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Georgiadis M. M., Luo M., Gaur R. K., Delaplane S., Li X. and Kelley M. R (2008) Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat. Res., 643, 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fomenko D. E. and Gladyshev V. N (2003) Identity and functions of CxxC-derived motifs. Biochemistry, 42, 11214–11225. [DOI] [PubMed] [Google Scholar]
- 62. Luo M., Zhang J., He H., Su D., Chen Q., Gross M. L., Kelley M. R. and Georgiadis M. M (2012) Characterization of the redox activity and disulfide bond formation in apurinic/apyrimidinic endonuclease. Biochemistry, 51, 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bazlekowa-Karaban M., Prorok P., Baconnais S., et al. (2019) Mechanism of stimulation of DNA binding of the transcription factors by human apurinic/apyrimidinic endonuclease 1, APE1. DNA Repair, 82, 102698. [DOI] [PubMed] [Google Scholar]
- 64. Timofeyeva N. A., Koval V. V., Ishchenko A. A., Saparbaev M. K. and Fedorova O. S (2011) Lys98 substitution in human AP endonuclease 1 affects the kinetic mechanism of enzyme action in base excision and nucleotide incision repair pathways. PLoS One, 6, e24063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gorman M. A., Morera S., Rothwell D. G., de La Fortelle E., Mol C. D., Tainer J. A., Hickson I. D. and Freemont P. S (1997) The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J., 16, 6548–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Castillo-Acosta V. M., Ruiz-Pérez L. M., Yang W., González-Pacanowska D. and Vidal A. E (2009) Identification of a residue critical for the excision of 3′ -blocking ends in apurinic/apyrimidinic endonucleases of the Xth family. Nucleic Acids Res., 37, 1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Illuzzi J. L., Harris N. A., Manvilla B. A., Kim D., Li M., Drohat A. C. and Wilson D. M. III (2013) Functional assessment of population and tumor-associated APE1 protein variants. PLoS One, 8, e65922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lirussi L., Antoniali G., D’Ambrosio C., Scaloni A., Nilsen H. and Tell G (2016) APE1 polymorphic variants cause persistent genomic stress and affect cancer cell proliferation. Oncotarget, 7, 26293–26306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Illuzzi J. L., McNeill D. R., Bastian P., et al. (2017) Tumor-associated APE1 variant exhibits reduced complementation efficiency but does not promote cancer cell phenotypes. Environ. Mol. Mutagen., 58, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xanthoudakis S., Smeyne R. J., Wallace J. D. and Curran T (1996) The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. U. S. A., 93, 8919–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ludwig D. L., MacInnes M. A., Takiguchi Y., Purtymun P. E., Henrie M., Flannery M., Meneses J., Pedersen R. A. and Chen D. J (1998) A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat. Res., 409, 17–29. [DOI] [PubMed] [Google Scholar]
- 72. Fung H. and Demple B (2005) A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell, 17, 463–470. [DOI] [PubMed] [Google Scholar]
- 73. Vascotto C., Bisetto E., Li M., et al. (2011) Knock-in reconstitution studies reveal an unexpected role of Cys-65 in regulating APE1/Ref-1 subcellular trafficking and function. Mol. Biol. Cell, 22, 3887–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li M. and Wilson D. M. III (2014) Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox Signal., 20, 678–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schrader C. E., Guikema J. E., Wu X. and Stavnezer J (2009) The roles of APE1, APE2, DNA polymerase beta and mismatch repair in creating S region DNA breaks during antibody class switch. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 364, 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Antoniali G., Malfatti M. C. and Tell G (2017) Unveiling the non-repair face of the base excision repair pathway in RNA processing: a missing link between DNA repair and gene expression? DNA Repair, 56, 65–74. [DOI] [PubMed] [Google Scholar]
- 77. Lieberman J. and Fan Z (2003) Nuclear war: the granzyme A-bomb. Curr. Opin. Immunol., 15, 553–559. [DOI] [PubMed] [Google Scholar]
- 78. Stetler R. A., Gao Y., Leak R. K., et al. (2016) APE1/Ref-1 facilitates recovery of gray and white matter and neurological function after mild stroke injury. Proc. Natl. Acad. Sci. U. S. A., 113, E3558–E3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li M., Yang X., Lu X., et al. (2018) APE1 deficiency promotes cellular senescence and premature aging features. Nucleic Acids Res., 46, 5664–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dumitrache L. C., Shimada M., Downing S. M., Kwak Y. D., Li Y., Illuzzi J. L., Russell H. R., Wilson D. M. III and McKinnon P. J (2018) Apurinic endonuclease-1 preserves neural genome integrity to maintain homeostasis and thermoregulation and prevent brain tumors. Proc. Natl. Acad. Sci. U. S. A., 115, E12285–E12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Meira L. B., Devaraj S., Kisby G. E., Burns D. K., Daniel R. L., Hammer R. E., Grundy S., Jialal I. and Friedberg E. C (2001) Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res., 61, 5552–5557. [PubMed] [Google Scholar]
- 82. Huamani J., McMahan C. A., Herbert D. C., Reddick R., McCarrey J. R., MacInnes M. I., Chen D. J. and Walter C. A (2004) Spontaneous mutagenesis is enhanced in Apex heterozygous mice. Mol. Cell. Biol., 24, 8145–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Izumi T., Brown D. B., Naidu C. V., Bhakat K. K., Macinnes M. A., Saito H., Chen D. J. and Mitra S (2005) Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl. Acad. Sci. U. S. A., 102, 5739–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jackson E. B., Theriot C. A., Chattopadhyay R., Mitra S. and Izumi T (2005) Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (APE1/Ref1). Nucleic Acids Res., 33, 3303–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li M., Zhong Z., Zhu J., et al. (2010) Identification and characterization of mitochondrial targeting sequence of human apurinic/apyrimidinic endonuclease 1. J. Biol. Chem., 285, 14871–14881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gros L., Saparbaev M. K. and Laval J (2002) Enzymology of the repair of free radicals-induced DNA damage. Oncogene, 21, 8905–8925. [DOI] [PubMed] [Google Scholar]
- 87. Suh D., Wilson D. M. III and Povirk L. F (1997) 3′ -phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res., 25, 2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Parsons J. L., Dianova I. I. and Dianov G. L (2004) APE1 is the major 3′ -phosphoglycolate activity in human cell extracts. Nucleic Acids Res., 32, 3531–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hadi M. Z. and Wilson D. M. III (2000) Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ. Mol. Mutagen., 36, 312–324. [PubMed] [Google Scholar]
- 90. Huang J. C., Hsu D. S., Kazantsev A. and Sancar A (1994) Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc. Natl. Acad. Sci. U. S. A., 91, 12213–12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Masani S., Han L. and Yu K (2013) Apurinic/apyrimidinic endonuclease 1 is the essential nuclease during immunoglobulin class switch recombination. Mol. Cell. Biol., 33, 1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fung H. and Demple B (2011) Distinct roles of Ape1 protein in the repair of DNA damage induced by ionizing radiation or bleomycin. J. Biol. Chem., 286, 4968–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chou K. M. and Cheng Y. C (2002) An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature, 415, 655–659. [DOI] [PubMed] [Google Scholar]