Abstract
Flavonoids in nature are known to possess various activities such as anti-inflammatory, antimicrobial, anticancer, antioxidant, neuroprotective, anti-HIV activities etc., The molecular docking was performed by 26 naturally occurring flavonoids with selected targets COX-2, hydroxyacyl-ACP dehydratase, tyrosinase from Agaricus bisporus, isomaltase from Saccharomyces cerevisiae, Human IkB kinase beta, Human ABC transporter, topoisomerase II, topoisomerase IV, N-myristoyltransferase from Candida albicans, Peptide deformylase from Pseudomonas aeruginosa, polypeptide deformylase from Streptococcus pneumoniae. The analysis was based on docking score, glide energy, interactions type (bond type and distance) and interaction with amino acids. The top 5 flavonoids with best docking score was reported. The in-silico results provided for 26 naturally occurring flavonoid shows that they reduce the risk of inflammation, cancer and infectious disease if people have taken in diet continuously. The provided docking data of flavonoids may be useful to synthesis novel drug candidate for the mentioned targets.
Keywords: Molecular docking, Flavonoids, Antimicrobial, Anti-cancer, Anti-inflammatory, Glide, Schrodinger maestro
1. Introduction
Specifications Table
| Subject | Pharmaceutical Science |
| Specific subject area | Interdisciplinary field includes organic chemistry, biochemistry, and biology. Drug design and discovery from plant sources. |
| Type of data | Tables Figures |
| How data were acquired | Schrodinger Maestro release 2018-4 |
| Data format | Raw and Analysed |
| Parameters for data collection | The docking score, glide energy and interactions of protein with the ligand. |
| Description of data collection | The Proteins were collected from rcsb wwPDB. The flavonoids structures were obtained from Pubchem online database. The docking was done using glide software. |
| Data source location |
https://www.rcsb.org/ https://pubchem.ncbi.nlm.nih.gov/ |
| Data accessibility | PDB files of the chosen enzyme targets are publically available at https://www.rcsb.org/ Tables and Figures of the docking are accessible in the article. |
Value of the Data
|
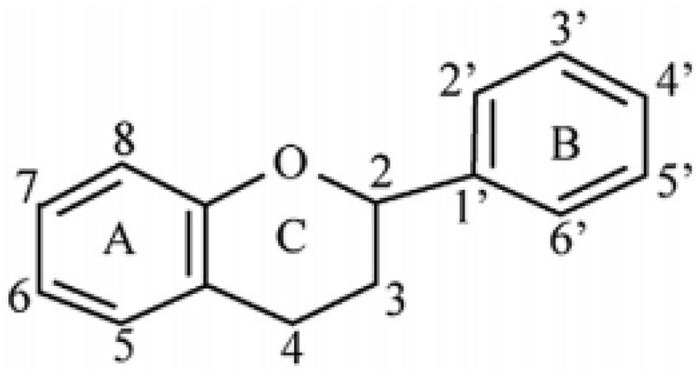
Flavonoids are a group of bioactive compounds which are extensively found in foodstuffs of plant origin. These are plant pigments synthesized from phenylalanine and generally display marvelous colors to the flowering parts of plants. Flavonoids comprise a large group of poly phenolic compounds, characterized by a benzo-4-pyrone structure, which is ubiquitous in vegetables and fruits. More than 9000 flavonoids have been reported in the literature and are present in different types and parts of plants such as vegetables, fruits, grains, legumes, beans, herbs, roots, leaves, seeds etc. The core structure of flavonoids has a three-ring diphenyl-propane (C6–C3–C6) unit, a fifteen-carbon skeleton. The flavonoid contains two benzene rings (A ring and B ring) which are connected by a C3 moiety. The C3 moiety forms a six-membered heterocyclic ring (ring C) attached to ring A. Regular consumption of flavonoids reduces the risk of a number of chronic diseases, including cancer, cardiovascular disease, diabetes, arthrosclerosis, neurodegenerative disorders, anti-ageing, anti-inflammatory, antiallergic, antiviral, and free radical scavenging. Among dietary sources of flavonoids, there are fruits, vegetables, nuts, seeds and spices. So, the provided docking data of flavonoid may be useful to synthesis novel drug candidate for the mentioned targets.
2. Data
In this article Table 1 provides the details about the targets and their description. Table 2 provide the structure of the naturally occurring flavonoids and plant sources. Table 3 gives docking score, glide energy, interaction type and bond length of the docking. The 3D and 2D interactions of the high scored flavonoids with the target enzymes are shown in Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7.
Table 1.
List of Targets. Shows the PDB ID, resolution and description of the proteins selected for docking with the naturally occurring flavonoids.
| S.No | PDB ID | Resolution (Å) | Description |
| 1 | 3LN0 | 2.20 | Structure of compound 5c-S bound at the active site of COX-2 [1] |
| 2 | 4KIK | 2.83 | Human IkB kinase beta [2] |
| 3 | 2XCS | 2.10 | The crystal structure of Staphylococcus aureus Gyrase complex with GSK299423 and DNA [3] |
| 4 | 4HZ5 | 2.70 | Pyrrolopyrimidine inhibitors of DNA gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity [4] |
| 5 | 4RLJ | 1.75 | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis [5] |
| 6 | 1IYL | 3.20 | Crystal Structure of Candida albicans N-myristoyltransferase with Non-peptidic Inhibitor [6] |
| 7 | 1LRY | 2.60 | Crystal Structure of Pseudomonas aeruginosa Peptide Deformylase Complexed with Antibiotic Actinonin [7] |
| 8 | 2AIE | 1.70 | Streptococcus pneumoniae polypeptide deformylase complexed with inhibitor [7] |
| 9 | 2Y9X | 2.78 | Crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit, with inhibitor tropolone [8] |
| 10 | 3A4A | 1.60 | Crystal structure of isomaltase from Saccharomyces cerevisiae [9] |
| 11 | 6FFC | 3.56 | Structure of an inhibitor-bound human ABC transporter [10] |
Table 2.
List of Flavonoids. This table exemplifies the plant sources of the naturally occurring flavonoids, so that the compounds may be isolated and used for the research purposes.
| S.No | Name of the flavonoid | Major plant source | Structure |
|---|---|---|---|
| 1 | Dabinol | Dalbergia latifolia |  |
| 2 | 5HMF | Citrus X sinensis |  |
| 3 | 6a,12a-Dehydroamorphigenin | Dalbergia sissoo |  |
| 4 | Afromosin | Centrosema pubescens |  |
| 5 | Amorphigenin | Dalbergia cochinchinensis |  |
| 6 | Biochanin- A | fusarium javanicum |  |
| 7 | Catechin | Camelia sinensis | 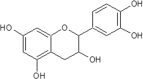 |
| 8 | Chrysin | Scutellaria baicalensis | 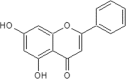 |
| 9 | Demethylnobiletin | Citrus depressa | 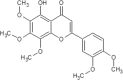 |
| 10 | Epicatechingallate (ECG) | Vicia faba | 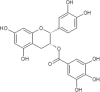 |
| 11 | Epigallocatechin | Camellia sinensis |  |
| 12 | Epigallocatechingallate (EGCG) | Camellia sinensis | 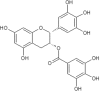 |
| 13 | Floretin (Phloretin) | Manchurian apricot. | 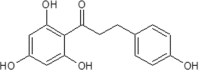 |
| 14 | Formononetin | Trifolium pratense | 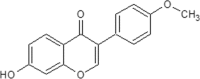 |
| 15 | Hesperidin | Citrus aurantium | 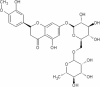 |
| 16 | Morin | Antiaris toxicaria | 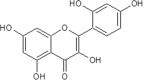 |
| 17 | Naringenin | Citrus paradisi | 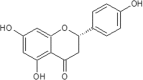 |
| 18 | Naringin | Citrus paradisi | 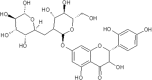 |
| 19 | Neohesperidin | citrus aurantium |  |
| 20 | Quercetin | Malus domestica | 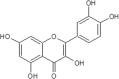 |
| 21 | Robinin | Vinca erecta | 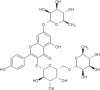 |
| 22 | Rotenone | Pachyrhizus erosus | 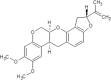 |
| 23 | Sakuranetin | Polymnia fruticosa | 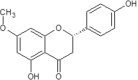 |
| 24 | Silymarin | Silybum marianum |  |
| 25 | Nobiletin | Citrus Unshiu | 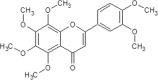 |
| 26 | Kaempferol | Allium cepa | 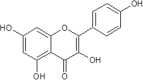 |
Table 3.
Docking score, Glide energy and Protein-Ligand Interactions. The docking score, Glide energy, interacting residues, type of interactions, bond length between residues and ligands are shown for each protein mentioned in Table 1.
| Title | Docking Score |
Glide Energy (Kcal/mol) |
Interactions | Type | Bond length (Å) |
|---|---|---|---|---|---|
| Protein ID: 2XCS Antibacterial by gyrase inhibition | |||||
| Robinin | −8.03 | −84.11 | Asp A:1083 Asp A:1080 Glu A:1088 Arg A:1122 |
H-bond Ar-H-bond 2 H-bond 2 H-bond |
2.03 2.20 1.73, 1.93 2.17, 2.61 |
| Naringin | −7.85 | −62.60 | Lys 581 Asp 1083 |
H-bond Ar-H-bond |
2.43 2.63 |
| Neohesperidin | −7.72 | −70.99 | Asp 1083 | H-bond | 1.90 |
| Silymorin | −7.29 | −44.08 | Glu 1088 Ala 1120 Tyr 1087 |
H-bond H-bond Ar-H-bond |
2.04 2.17 2.70 |
| Hesperidin | −7.25 | −68.40 | Arg 1122 | H-bond | 2.29 |
| Protein ID: 4KIK Anticancer by IkB kinase inhibition | |||||
| Sakuranetin | −9.73 | −44.98 | Asp A:166 Glu A:97 Cys A:99 |
H-bond H-bond H-bond |
1.45 2.40 2.04 |
| Naringenin | −9.53 | −43.50 | Glu A:97 Cys A:99 Asp A:166 |
H-bond H-bond H-bond |
2.42 2.04 1.45 |
| Kaempferol | −9.33 | −45.40 | Lys A:44 Asp A:166 Cys A:99 |
H-bond H-bond H-bond |
2.04 2.16 2.02 |
| Naringin | −9.07 | −59.59 | Asp A:145 Lys A:147 Asp A:166 Glu A:97 Glu A:97 Cys A:99 |
H-bond H-bond H-bond H-bond Ar-H-bond H-bond |
1.78 2.22 1.87 1.87 2.48 2.07 |
| Morin | −9.03 | −46.84 | Asp A:166 Lys A:44 Cys A:99 |
H-bond H-bond H-bond |
2.09 2.02 2.01 |
| Protein ID: 4HZ5 Antibacterial by DNA gyrase b and topoisomerase IV inhibition | |||||
| Phloretin | −7.06 | −50.83 | Asp 26 HOH 407 HOH 408 HOH 401 |
H-bond H-bond H-bond H-bond |
1.87 2.77 1.77 2.95 |
| Catechin | −6.93 | −45.78 | Gly 80 Asp 76 HOH 427 |
H-bond H-bond H-bond |
2.38 1.78 1.64 |
| Morin | −6.92 | −51.77 | Asp 76 Asp 76 Gly 80 |
H-bond Ar-H-bond H-bond |
1.59 2.36 2.45 |
| Kaempferol | −6.75 | −47.93 | Gly 80 Asp 76 |
Ar-H-bond H-bond |
2.40 1.60 |
| Sakuranetin | −6.49 | −41.60 | Asp 76 Gly 80 HOH 401 |
H-bond H-bond H-bond |
1.81 2.69 2.12 |
| Protein ID: 2Y9X Antifungal (Agaricus bisporus) by tyrosinase inhibition | |||||
| Epigallocatechingallate | −7.22 | −51.13 | Arg 268 Asn 260 Phe 264 |
Pi-cation Ar-H-Bond Ar-H-Bond |
6.02 2.66 3.12 |
| Chrysin | −7.20 | −34.87 | His 259 Hid 85 |
Ar-H-Bond Ar-H-Bond |
3.63 3.25 |
| Kaempferol | −7.02 | −35.15 | Hie 244 Arg 268 |
Ar-H-Bond Pi-cation |
3.02 5.66 |
| Quercetin | −6.75 | −34.47 | Asn 260 Arg 268 |
Ar-H-Bond Pi-cation |
2.77 2.89 |
| Catechin | −6.67 | −32.30 | Arg 268 | Pi-cation | 6.19 |
| Protein ID: 3LN0 Anti-inflammatory by cyclo-oxygenase inhibition | |||||
| Sakuranetin | −7.31 | −31.36 | Ser 516 | H-Bond | 2.67 |
| Quercetin | −7.00 | −34.33 | Try 371 Try 371 Gly 178 |
Ar-H-Bond H-Bond H-Bond |
3.08 2.72 2.71 |
| Naringenin | −6.99 | −30.27 | Ser 516 | H-Bond | 2.62 |
| Morin | −6.98 | −35.14 | Try 371 His 75 Tyr 341 Arg 106 |
H-Bond Ar-H-Bond Ar-H-Bond Pi-cation |
2.38 3.00 2.51 7.60 |
| Kaempferol | −6.91 | −33.28 | Tyr 371 Tyr 371 |
Ar-H-Bond H-Bond |
3.50 2.24 |
| Protein ID: 4RLJ Antituberculosis (Mycobacterium tuberculosis) by hydroxyacyl-ACP dehydratase inhibition | |||||
| Catechin | −7.47 | −39.00 | HOH 322 HOH 312 |
H-Bond H-Bond |
1.84 2.14 |
| Biochanin A | −6.95 | −32.44 | Gln 86 HOH 322 |
Ar-H-Bond H-Bond |
2.63 1.73 |
| Quercetin | −6.76 | −39.63 | HOH 322 | H-Bond | 1.88 |
| Chrysin | −6.60 | −32.57 | Gln 86 Gln 86 HOH 387 |
H-Bond Ar-H-Bond Ar-H-Bond |
2.65 2.48 2.27 |
| Formononetin | −6.47 | −31.39 | Gln 86 HOH 322 |
Ar-H-Bond Ar-H-Bond |
2.84 2.61 |
| Protein ID: 6FFC Inhibition capacity of human multidrug transporter ABCG2 | |||||
| Robinin | −7.14 | −53.99 | Phe 439 Asn 436 Thr 402 Thr 435 Gln 368 |
H-Bond Ar-H-Bond Ar-H-Bond H-Bond H-Bond |
1.85 2.22 2.57 2.11 2.40 |
| Morin | −6.64 | −34.48 | Thr 402 Phe 489 Asn 436 |
H-Bond Ar-H-Bond H-Bond |
1.68 3.44 1.78 |
| Phloretin | −6.39 | −34.84 | Asn 436 Phe 489 Thr 402 |
H-Bond Ar-H-Bond Ar-H-Bond |
1.86 3.64 2.56 |
| Catechin | −6.36 | −32.35 | Thr 402 Phe 489 Phe 489 Asn 436 Asn 436 |
H-Bond Ar-H-Bond H-Bond Ar-H-Bond H-Bond |
1.85 3.23 1.38 2.22 2.20 |
| Phloretin | −6.36 | −35.65 | Thr 402 Phe 489 Asn 436 |
Ar-H-Bond Ar-H-Bond H-Bond |
2.69 3.63 1.83 |
| Protein ID: 3A4A Antifungal (Saccharomyces cerevisiae) by isomaltase inhibition | |||||
| Epigallocatechin | −6.98 | −44.31 | Glu 411 Tyr 158 Tyr 158 Phe 314 Phe 314 Asp 307 Asp 307 |
H-Bond Ar-H-Bond H-Bond Ar-H-Bond H-Bond H-Bond Ar-H-Bond |
1.76 2.95 2.92 3.12 3.51 1.46 2.60 |
| Morin | −6.50 | −40.65 | Asp 307 Gln 353 HOH 1207 |
H-Bond HBond Ar-H-Bond |
1.69 1.79 2.46 |
| Chrysin | −5.61 | −34.69 | Glu 411 Tyr 158 Gln 353 Asp 307 |
Ar-H-Bond H-Bond Ar-H-Bond Ar-H-Bond |
2.79 1.92 2.35 2.25 |
| Morin | −5.31 | −38.38 | Tyr 158 Tyr 158 Asp 307 Gln 353 Gln 353 |
Ar-H-Bond H-Bond Ar-H-Bond Ar-H-Bond H-Bond |
2.78 1.73 2.50 2.56 1.80 |
| Phloretin | −4.64 | −38.76 | Glu 411 Asn 415 Gln 353 |
H-Bond H-Bond H-Bond |
2.43 2.19 2.10 |
| Protein ID: 1IYL Antifungal (Candida albicans) by N-myristoyltransferase inhibition | |||||
| Epigallocatechingallate | −8.59 | −58.16 | Glu 109 Phe 339 Phe 115 Asn 392 Tyr 225 Tyr 356 Leu 451 Tyr 107 |
Ar-H-bond Ar-H-bond Pi-Pi H-bond Pi-Pi Ar- H-bond 2 H-bond H-bond |
3.09 3.18 4.39 2.07 4.16 3.69 2.07, 1.88 2.60 |
| Neohesperidin | −8.55 | −54.56 | Asn 392 Tyr 354 Tyr 335 Leu 45 |
H-bond 2 Ar- H-bond H-bond H-bond |
1.96 3.44, 3.64 2.25 1.85 |
| Biochanin A | −8.24 | −41.78 | Asn 392 Hie 227 Tyr 354 Tyr 225 |
Ar-H-bond H-bond 2 Pi-Pi 2 Pi-Pi |
2.32 2.19 4.10, 4.13 4.11,4.86 |
| Formononetin | −8.13 | −38.64 | Tyr 354 Tyr 225 Hie 227 Asn 392 |
2 Pi-Pi 2 Pi-Pi H-bond Ar-H-bond |
4.10, 4.13 4.11,4.86 2.24 2.33 |
| Quercetin | −7.91 | −44.10 | Tyr 354 Tyr 225 Hie 227 Hie 227 Glu 109 Leu 451 Leu 450 |
2 Pi-Pi 2 Pi-Pi 2 H-bond Ar-H-bond H-bond H-bond H-bond |
4.10, 4.13 4.11,4.86 2.59, 2.17 3.19 2.33 1.87 1.87 |
| Protein ID: 1LRY Antibacterial (Pseudomonas aeruginosa) by Peptide Deformylase inhibition | |||||
| Silymarin | −8.82 | −63.36 | Glu 134 Ile 144 Pro 42 |
Ar- H-bond H-bond H-bond |
2.53 1.84 1.85 |
| Epigallocatechin | −7.90 | −53.17 | Glu 134 Leu 92 Ile 44 Gln 88 |
2 H-bond H-bond H-bond H-bond |
1.55, 1.67 2.15 2.36 1.78 |
| Naringenin | −7.38 | −42.98 | Glu 134 Glu 134 Gly 44 Gly 44 Tyr 87 |
H-bond Ar-H-bond H-bond Ar-H-bond Ar-H-bond |
1.56 2.35 1.69 2.69 2.58 |
| Quercetin | −7.29 | −48.76 | Glu 134 Glu 134 Gln 88 |
H-bond Ar-H-bond H-bond |
1.67 2.48 1.76 |
| Neohesperidin | −7.28 | −60.23 | Glu 134 | H-bond | 1.56 |
| Protein ID: 2AIE Antibacterial (Streptococcus pneumoniae) by polypeptide deformylase inhibition | |||||
| Epigallocatechin | −6.40 | −46.08 | Gly 68 Leu 131 Glu 174 |
H-bond H-bond 2 H-bond |
1.70 2.21 1.64, 2.03 |
| Catechin | −6.21 | −44.36 | Gly 68 Leu 131 Glu 174 Glu 174 |
Ar- H-bond H-bond H-bond Ar-H-bond |
2.42 2.54 1.77 2.35 |
| Robinin | −6.12 | −64.85 | Glu 125 Arg 67 Glu 174 |
H-bond H-bond 2 H-bond |
1.68 2.07 1.50, 1.78 |
| Hesperidin | −5.93 | −52.09 | Glu 125 Arg 67 |
H-bond Pi-cation |
1.60 5.98 |
| Epigallocatechingallate | −5.88 | −56.62 | Gly 68 Glu 174 |
H-bond 2 H-bond |
1.77 1.92, 1.65 |
Fig. 1.
3D and 2D interactions of COX-2 (PDB ID: 3LN0) with flavonoid Sakuranetin. Figure shows Sakuranetin binding and interactions with human cyclo-oxygenase-2 with docking score of −7.31.
Fig. 2.
3D and 2D interactions of Human Ikb Kinase Beta (PDB ID: 4KIK) with flavonoid Sakuranetin. Figure shows Sakuranetin binding and interactions with Human Ikb Kinase Beta with docking score of −9.73.
Fig. 3.
3D and 2D interactions of DNA gyrase (PDB ID: 2XCS) with Flavonoid Robinin. Figure shows Robinin binding and interactions with DNA gyrase Staphylococcus aureus with docking score of −8.03.
Fig. 4.
3D and 2D interactions of tyrosinase (PDB ID: 2Y9X) with flavonoid Epigallocatechingallate. Figure shows Epigallocatechingallate binding and interactions with Agaricus bisporus tyrosinase with docking score of −7.22.
Fig. 5.
3D and 2D interactions of DNA gyrase (PDB ID: 4HZ5) with flavonoid Phloretin. Figure shows Phloretin binding and interactions with DNA gyrase with docking score of −7.06.
Fig. 6.
3D and 2D interactions of Isomaltase (PDB ID: 34A4) with flavonoid Epigallocatechin. Figure shows Epigallocatechin binding and interactions with Isomaltase of Saccharomyces cerevisiae with docking score of −6.98.
Fig. 7.
3D and 2D interactions of ABC transporter (PDB ID: 6FFC) with flavonoid Robinin. Figure shows Robinin binding and interactions with Human ABC transporter with docking score of −7.14.
3. Experimental design, materials, and methods
3.1. Protein selection and preparation
The crystal structures of the selected proteins were retrieved from protein data bank. (PDB database, www.rcsb.org). The downloaded protein structure was prepared prior to docking using Schrodinger Maestro release 2018-4. Protein preparation was done by preprocessing the structures by assignment of bonds and bond orders, addition of hydrogens, filling in missing loops or side chains, capping uncapped C and N termini, adjusting bonds and formal charges for metals, and correcting mislabeled elements, removing water molecules, removing unwanted chains and optimization of hydrogen bonded structures followed by minimization.
3.2. Ligand preparation and molecular docking
The structures of the selected 26 flavonoids were downloaded from Pubchem https://pubchem.ncbi.nlm.nih.gov/) and saved in mol format. The energy minimization was done using Ligprep. The minimized structures were docked on the prepared protein. The best flavonoid was identified based on the binding energy and interaction with amino acid residues for each protein.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2020.105243.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wang J.L., Limburg D., Graneto M.J., Springer J., Hamper J.R.B., Liao S., Pawlitz J.L., Kurumbail R.G., Maziasz T., Talley J.J., Kiefer J.R., Carter J. The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: the second clinical candidate having a shorter and favorable human half-life. Bioorg. Med. Chem. Lett. 2010;20(23):7159–7163. doi: 10.1016/j.bmcl.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 2.Thomas N.S., George K., Selvam A.A.A. Anticancer mechanism of troxerutin via targeting Nrf2 and NF-κB signalling pathways in hepatocarcinoma cell line. Toxicol. Vitro. 2019;54:317–329. doi: 10.1016/j.tiv.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Yao L., Wu L.-L., Li Q., Hu Q.-M., Zhang S.-Y., Liu K., Jiang J.-Q. Novel berberine derivatives: design, synthesis, antimicrobial effects, and molecular docking studies. Chin. J. Nat. Med. 2018;16(10):774–781. doi: 10.1016/S1875-5364(18)30117-1. [DOI] [PubMed] [Google Scholar]
- 4.Szulczyk D., Dobrowolski M.A., Roszkowski P., Bielenica A., Stefańska J., Koliński M., Kmiecik S., Jóźwiak M., Wrzosek M., Olejarz W., Struga M. Design and synthesis of novel 1H-tetrazol-5-amine based potent antimicrobial agents: DNA topoisomerase IV and gyrase affinity evaluation supported by molecular docking studies. Eur. J. Med. Chem. 2018;156:631–640. doi: 10.1016/j.ejmech.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 5.Dong Y., Qiu X., Shaw N., Xu Y., Sun Y., Li X., Li J., Rao Z. Molecular basis for the inhibition of β-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors. Protein & Cell. 2015;6(7):504–517. doi: 10.1007/s13238-015-0181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerrero-Perilla C., Bernal F.A., Coy-Barrera E.D. Molecular docking study of naturally occurring compounds as inhibitors of N-myristoyl transferase towards antifungal agents discovery. Rev. Colomb. Ciencias Quím. Farm. 2015;44(2):162–178. [Google Scholar]
- 7.Fieulaine S., Alves de Sousa R., Maigre L., Hamiche K., Alimi M., Bolla J.-M., Taleb A., Denis A., Pagès J.-M., Artaud I., Meinnel T., Giglione C. A unique peptide deformylase platform to rationally design and challenge novel active compounds. Sci. Rep. 2016;6(1):35429. doi: 10.1038/srep35429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H., Zhu Y., Wang T., Qi J., Liu X. Enzyme-site blocking combined with optimization of molecular docking for efficient discovery of potential tyrosinase specific inhibitors from Puerariae lobatae radix. Molecules. 2018;23(10):2612. doi: 10.3390/molecules23102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murugesu S., Ibrahim Z., Ahmed Q.-U., Nik Yusoff N.-I., Uzir B.-F., Perumal V., Abas F., Saari K., El-Seedi H., Khatib A. Characterization of α-glucosidase inhibitors from Clinacanthus nutans Lindau leaves by gas chromatography-mass spectrometry-based metabolomics and molecular docking simulation. Molecules. 2018;23(9):2402. doi: 10.3390/molecules23092402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson S.M., Manolaridis I., Kowal J., Zechner M., Taylor N.M.I., Bause M., Bauer S., Bartholomaeus R., Bernhardt G., Koenig B., Buschauer A., Stahlberg H., Altmann K.-H., Locher K.P. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat. Struct. Mol. Biol. 2018;25(4):333–340. doi: 10.1038/s41594-018-0049-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.









