Abstract
Glioblastoma is the most common and aggressive primary tumor in the central nervous system, accounting for 12%–15% of all brain tumors. 3-O-Acetyl-11-keto-β-boswellic acid (AKBA), one of the most active ingredients of gum resin from Boswellia carteri Birdw., was reported to inhibit the growth of glioblastoma cells and subcutaneous glioblastoma. However, whether AKBA has antitumor effects on orthotopic glioblastoma and the underlying mechanisms are still unclear. An orthotopic mouse model was used to evaluate the anti-glioblastoma effects of AKBA. The effects of AKBA on tumor growth were evaluated using MRI. The effects on the alteration of metabolic landscape were detected by MALDI-MSI. The underlying mechanisms of autophagy reducing by AKBA treatment were determined by immunoblotting and immunofluorescence, respectively. Transmission electron microscope was used to check morphology of cells treated by AKBA. Our results showed that AKBA (100 mg/kg) significantly inhibited the growth of orthotopic U87-MG gliomas. Results from MALDI-MSI showed that AKBA improved the metabolic profile of mice with glioblastoma, while immunoblot assays revealed that AKBA suppressed the expression of ATG5, p62, LC3B, p-ERK/ERK, and P53, and increased the ratio of p-mTOR/mTOR. Taken together, these results suggested that the antitumor effects of AKBA were related to the normalization of aberrant metabolism in the glioblastoma and the inhibition of autophagy. AKBA could be a promising chemotherapy drug for glioblastoma.
KEY WORDS: Glioblastoma, AKBA, MALDI-MSI, Phospholipids, Autophagy
Abbreviations: AKBA, 3-O-acetyl-11-keto-β-boswellic acid; DAPI, 4′,-6-diamidino-2-phenylindole; GBM, glioblastomas; G3P, glycerol-3-phosphate; GL/FFA, glycerolipid/free fatty acid; G6P, glucose-6-phosphate; IDH1/2, isocitrate dehydrogenases 1/2; ITO, indium tin oxide; LA, linoleic acid; MALDI-MSI, matrix-assisted laser desorption ionization-mass spectrometry imaging; NEDC, N-(1-naphthyl) ethylenediamine dihydrochloride; NAA, N-acetyl-l-aspartic acid; OA, oleic acid; PA, phosphatidic acid; PE, phosphatidylethanolamine; PG, phosphatidylglycerols; PI, phosphatidylinositol; PS, phosphatidylserine; TIC, total ion current; TMZ, temozolomide
Graphical abstract
A natural product, 3-O-acetyl-11-keto-β-boswellic acid (AKBA), exerted antitumor effects via the normalization of aberrant metabolism in the glioblastoma and the inhibition of autophagy.
1. Introduction
Glioblastoma (GBM) is the most common and aggressive primary tumor in the central nervous system, accounting for 12%–15% of all brain tumors1. The current standard treatment for GBM includes maximal surgical resection followed by radiotherapy and chemotherapy with temozolomide (TMZ)2, 3, 4. However, the prognosis of GBM remains poor with the median survival of 15–20 months and the 5-year survival rate of only 3%–5%5,6. Therefore, more studies elucidating the pathogenesis of GBM and developing new chemotherapy drugs are essential.
Abnormal metabolism is a common occurrence in cancers, which exhibits changes in metabolism related to non-neoplastic cells7. Tumor metabolism is mainly dependent on aerobic glycolysis (Warburg effect), resulting in a continuous uptake of glucose which is considered a hallmark of cancer8,9. The metabolisms of other biomolecules essential for cell proliferation10 are also altered in cancer cells, such as nucleotides, amino acids and lipids. Isocitrate dehydrogenases 1 (IDH1) and 2 (IDH2) are the key rate-limiting enzymes for the tricarboxylic acid cycle, and have recently been recognized as major determinants in the molecular differentiation of diffuse gliomas11,12. For that reason, they have been considered as new targets for the treatment of gliomas and studies on the mechanisms behind the aberrant metabolism of glioma cells need to be aggressively pursued to find chemotherapy drugs that can restore normal metabolism in these tumors.
Matrix-assisted laser desorption ionization-mass spectrometry imaging (MALDI-MSI) is a label-free technique that can be used to map the spatial distribution of various molecules in thin tissue sections. This technique has been widely used for in situ imaging of endogenous or exogenous molecules including small molecules13, lipids14, peptides15, proteins16, and drugs17. It is a useful tool for diagnosis and prognosis of diseases18, biomarker discovery19, and drug development20. For example, Wang et al.21 successfully identified the distributions of amino acids, glucose and glycerophospholipids in liver tissues of metastatic colorectal cancer using MALDI-MSI coupled with matrix N-(1-naphthyl) ethylenediamine dihydrochloride (NEDC). These findings could provide a better understanding of the complex biological processes in gliomas, which allow the researchers to monitor the changes of metabolites in gliomas after drug treatment.
Autophagy is an intracellular recycling program that responds to nutrient, energy, oxygen, and hormonal demands under physiologic circumstances to maintain metabolic homeostasis. The occurrence of autophagy was associated with abnormal metabolism, such as starvation. Dysregulated autophagy is emerging as a hallmark of malignancy. Autophagy plays an important role in the survival of cancer22,23. Abnormal metabolism is one of characters of cancers. Therefore, autophagy was closely associated with metabolism. It is important to explore relation between autophagy and metabolism.
Natural products can be widely used to treat various human diseases including cancer24,25, approximately half of which are derived from plants. 3-O-Acetyl-11-keto-β-boswellic acid (AKBA) is isolated from the gum resin of frankincense trees, Boswellia serrata and Boswellia carteri Birdw. This natural product is widely applied in Africa, India, and China26 to treat inflammatory diseases including arthritis27, colitis28, and asthma29, as well as some other illnesses30. In the previous study31, we found that AKBA inhibited the growth of U251 and U87-MG human glioblastoma cell lines by arresting the cell cycle at the G2/M phase via the p21/FOXM1/cyclin B1 pathway, inhibited mitosis by downregulating the aurora B/TOP2A pathway, and induced mitochondrial-dependent apoptosis. However, it is still unknown whether AKBA could inhibit the growth of orthotopic gliomas and the specific mechanisms of its action are still unclear.
In this study, the anti-glioblastoma effects of AKBA were investigated in an in vivo orthotopic model. It was found dramatically suppressing the tumorigenicity, in part by ameliorating the abnormal metabolism of phospholipids, glucose, and other small molecules in the glioma tissue. In addition, our results also showed that AKBA could inhibit autophagy by regulating the ERK/mTOR and P53/mTOR pathways.
2. Materials and methods
2.1. Animals
Female BALB/c-nu nude mice (17–19 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). All animal experiments were conducted according to the principles of the NIH Guide for the Care and Use of Laboratory Animals and were approved by the ethics committee for laboratory animal care and use of the Institute of Materia Medica, Chinese Academy of Medical Science and Peking Union Medical College (Beijing, China).
2.2. U87-MG glioma orthotopic model
Mice were anesthetized by intraperitoneal injection with 0.2 mL 0.6% sodium pentobarbital and immobilized in a stereotactic frame. A hole located 3 mm to the right and 0.5 mm anterior to the bregma was drilled into the skull. U87-MG glioblastoma cells were harvested and resuspended in phosphate buffered saline (PBS) to a concentration of 2 × 105 cells/μL. The needle of a Hamilton micro-syringe was inserted through the borehole to a depth of 3.3 mm where the right striatum is located. An aliquot of 5 μL of cells was slowly injected into this area at a rate of 1 μL/min, and the needle was slowly withdrawn 5 min after cell injection.
2.3. Grouping and drug administration
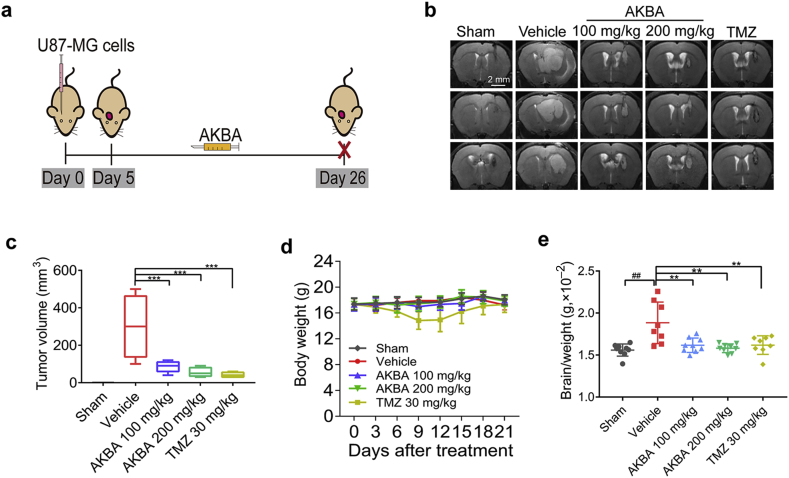
Five days after the surgery, mice bearing intracranial tumors were randomly divided into five groups (10 animals per group) as follows: (1) Sham; (2) Vehicle; (3) Temozolomide (TMZ), 30 mg/kg; (4) AKBA, 100 mg/kg, and (5) AKBA, 200 mg/kg. TMZ and AKBA dissolved in 0.5% carboxymethyl cellulose sodium. 99% AKBA was purchased from National Institutes for Food and Drug Control of China and dissolved in DMSO. U87-MG glioblastoma cells were injected into the right striatum of mice. From the 5th day, AKBA was orally administered to mice once daily. In the 26th day, MRI of the brain was performed and brain tissues were collected. The structural formulas of the drugs and the workflow for the orthotopic tumor experiment are shown in Fig. 1a and Supporting Information Fig. S1a, respectively.
Figure 1.
AKBA suppressed the growth of U87-MG orthotopic tumors. (a) Schematic of in vivo orthotopic tumor experiment for the investigation of anti-tumor effects of AKBA. (b) Representative MRI images of intracranial tumors from various groups of the U87 orthotopic model (Scale bar = 2 mm). (c) Tumor volumes in the U87 orthotopic model. (d) Changes in body weight during the AKBA administration period. (e) Relative brain/weight ratio at the end of the experiment. The data are presented as mean ± SD, n = 10 in each group. ##P < 0.01 vs. Sham group; **P < 0.01, ***P < 0.01 vs. Vehicle group.
2.4. MRI analysis
After administration of AKBA for 14 days, brain tumors were imaged using a small animal MRI scanner (Pharma Scan 70/16 US, Bruker Daltonics, Bremen, Germany). The parameters for MRI were set as: T2_TurboRARE, with TR/TE = 5000/40, six averages, 20 × 20 field-of-view, and 0.5 mm slice thickness. The tumor volume was calculated as: V = L × W × T; where L referred to the maximum length of the tumor, W referred to the maximum width perpendicular to L, and T referred to the thickness of the tumor slice (0.5 mm).
2.5. MALDI-MSI
2.5.1. Tissue preparation
At the end of the experiment, the mice were anesthetized by intraperitoneal injection of sodium pentobarbital (60 mg/kg), and then perfused with 0.9% sodium chloride. Brains were removed, immediately frozen in liquid nitrogen and stored at −80 °C. The frozen brain tissues were sectioned at 10 μm along the coronal plane using a cryostat microtome (Leica CM1950, Leica Microsystems, Wetzlar, Germany). The tissue sections were thaw-mounted on indium tin oxide (ITO) glass slides for MALDI-MSI or onto microscopic glass slides covered with poly-l-lysine for H&E staining. The ITO glass slides were desiccated under vacuum for 1 h at room temperature. The matrix solution, NEDC in methanol/water (50/50, v/v), was prepared and applied using the Bruker ImagePrep device (Bruker Daltonics, Bremen, Germany)21. For ImagePrep deposition, the matrix was typically deposited for 35 cycles, and each cycle included 1 s of spraying at 60% power, 15 s of incubation, and 70 s of drying.
2.5.2. Mass spectrometry imaging of metabolic landscape in the brain tissue sections
The MALDI imaging analysis was performed using the Ultraflextreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) with a 355 nm and 2000 Hz solid-state Nd:YAG/355 nm SmartBeam. The data were collected in the negative ionization mode with a 200 μm spatial resolution (200 laser shots). Spectra were acquired in the mass ranging from m/z 100 to 1000. Imaging data were then recorded and processed by the FlexImaging 3.0 software. Images were normalized to the total ion current (TIC). The metabolites were identified and further confirmed according to a previous study21. The flow chart of MALDI-MSI is shown in Fig. 1c.
2.6. Immunoblotting
Immunoblotting was performed as previously described32. Briefly, normal and glioblastoma tissues of mice were cut into small pieces and the total proteins were extracted in RIPA lysis buffer (Applygen, Beijing, China) with the addition of a protease/phosphatase inhibitor cocktail (Thermo Scientific, Rockford, IL, USA) by sonication for a total of 30 s (3 times, 10 s per time) at 4 °C. The tissue lysate was centrifuged using a centrifuge (Sigma–Aldrich, St. Louis, MO, USA) at 12,500 rpm for 20 min at 4 °C and protein concentrations were measured using a BCA protein quantification kit (CWBIO, Beijing, China). Proteins were separated by electrophoresis on 8% or 10% SDS-PAGE gels and transferred to a 0.45 μm PVDF membrane (Millipore, Billerica, MA, USA). After blocking with 5% fat-free milk in TBST for 1 h, membranes were immunoblotted with primary antibodies to ATG3, ATG5, ATG7, ATG12, ATG16, P62, LC3B, P53, p-ERK1/2, ERK1/2, p-AMPK, AMPK, p-mTOR, m-TOR (Cell Signaling Technology, Danvers, MA, USA), and GAPDH (Proteintech, Rosemont, IL, USA) at the appropriate dilutions with gentle shaking overnight at 4 °C. Blots were incubated with HRP-conjugated goat anti-rabbit or anti-mouse secondary antibody (1:2000; Cell Signaling Technology) and bands were visualized using an enhanced chemiluminescence, ECL Western blot kit (Beyotime Biotechnology, Shanghai, China).
2.7. Immunohistochemistry
Coronal brain tissues were paraffin-embedded, sectioned in 5 μm, and fixed on polylysine-coated slides. The fixed sections were then dried, deparaffinized, rehydrated, immersed in boiled sodium citrate buffer (10 mmol/L, pH 6.0) at a sub-boiling temperature for 25 min, and incubated in a 3% hydrogen peroxide solution for 10 min at room temperature. Then, sections were incubated with primary anti-pERK (1:500, Cell Signaling Technology) and P53 antibodies (1:200, Cell Signaling Technology) in a moist chamber at 4 °C for 14 h. The IHC Detection System (ZSGB-BIO, Beijing, China) was used to detect the brown reaction product of sections incubated in diaminobenzidine (DAB, ZSGB-BIO, Beijing, China). The Nikon Eclipse Ti microscope (Eclipse Ti-E,Nikon, Japan) coupled with NIS elements software (Nikon, Melville, NY, USA) for Windows was used to take photographs of equally exposed slides counterstained with hematoxylin. Quantitative analyses of three randomly selected visual fields were performed by two observers independently.
2.8. Immunofluorescence staining
Immunofluorescence staining analysis of P62, LC3B expression in brain tissues were done as described previously. Briefly, 5 μm thick tumor biopsy sections were deparaffinized in xylene and subsequently rehydrated with 100%, 95%, and 75% ethanol, and deionized H2O. Sections were then placed in antigen-retrieval solution (10 mmol/L sodium citrate, pH 6.0) and boiled 3 min, and then cooled down to room temperature. Subsequently, sections were stained with 1:200 dilution of secondary antibody at room temperature for 30 min, following incubation with a 1:100 dilution of goat IgG for 1 h at room temperature and stained with dilution of anti-P62 (1:500, Cell Signaling Technology) and anti-LC3B (1:500, Cell Signaling Technology) antibodies overnight at 4 °C. Before analysis, the sections were stained with the fluorescent dye 4′,-6-diamidino-2-phenylindole (DAPI) at room temperature for 5 min. Nikon Eclipse Ti microscope (Eclipse Ti-E, Nikon, Japan) coupled with NIS elements software (Nikon, Melville, NY, USA) for Windows was used to take photographs. Quantitative analyses of three randomly selected visual fields were performed by two observers independently.
2.9. Transmission electron microscope
U87-MG cells were cultured with DMEM without FBS medium overnight. Then, cells were treated by 10 μmol/L AKBA, 20 μmol/L AKBA, and 30 μmol/L AKBA, respectively. After 24 h, cells were harvested and fixed in 0.1 mol/L phosphate buffer containing 3% glutaraldehyde (pH 7.2) at 4 °C overnight. Cells were then washed, post-fixed in 1% OsO4 buffer for 1 h at 4 °C, washed again, dehydrated in a graded series of ethanol, and embedded in spur resin at 56 °C overnight. Sections of 60 nm ultrathin were obtained using the ultramicrotome (Leica, Wetzlar, Germany), stained with uranyl acetate and lead citrate at room temperature for 10 min. The transmission electron microscope (FEI Tecnai Spirit, Hillsboro, OR, USA) was finally used to detect the autophagosomes in U87-MG cells.
2.10. Statistical analysis
Results are presented as mean ± standard deviation (SD). One-way ANOVA was used to calculate differences among groups. P < 0.05 was considered statistically significant.
3. Results
3.1. AKBA suppressed glioblastoma growth in vivo
To investigate the effects of AKBA on the growth of tumor in vivo, U87-MG cells were injected into mouse brains and tumor growth was monitored by MRI. The AKBA (100 and 200 mg/kg) treatment groups showed a significant reduction in tumor volumes (Fig. 1b and c). During the 21 days’ administration of AKBA, no obvious weight loss or abnormal behavior was detected (Fig. 1d). The final brain/weight ratio in the AKBA group was also significantly reduced compared to Vehicle group (Fig. 1e). Taken together, these findings suggested that AKBA inhibited the tumorigenesis of glioma in vivo.
3.2. AKBA ameliorated the aberrant alterations of phospholipids and fatty acid in glioblastoma xenografts
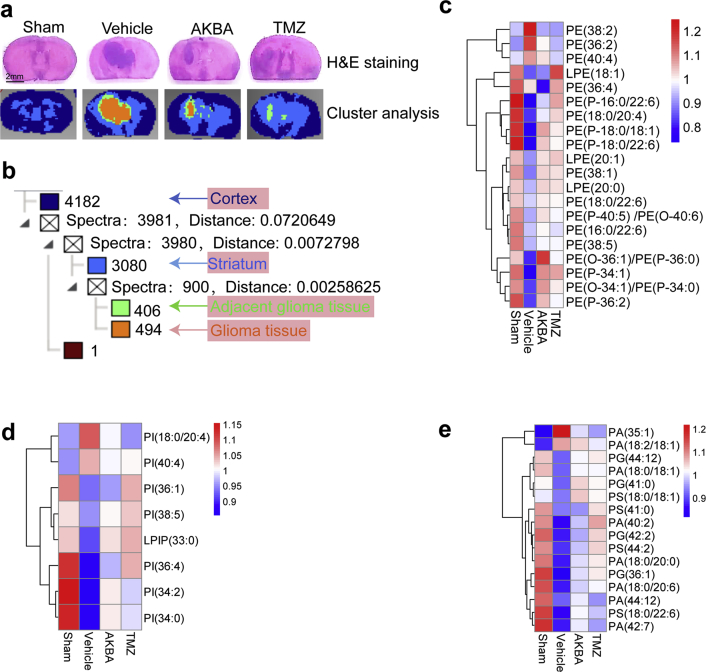
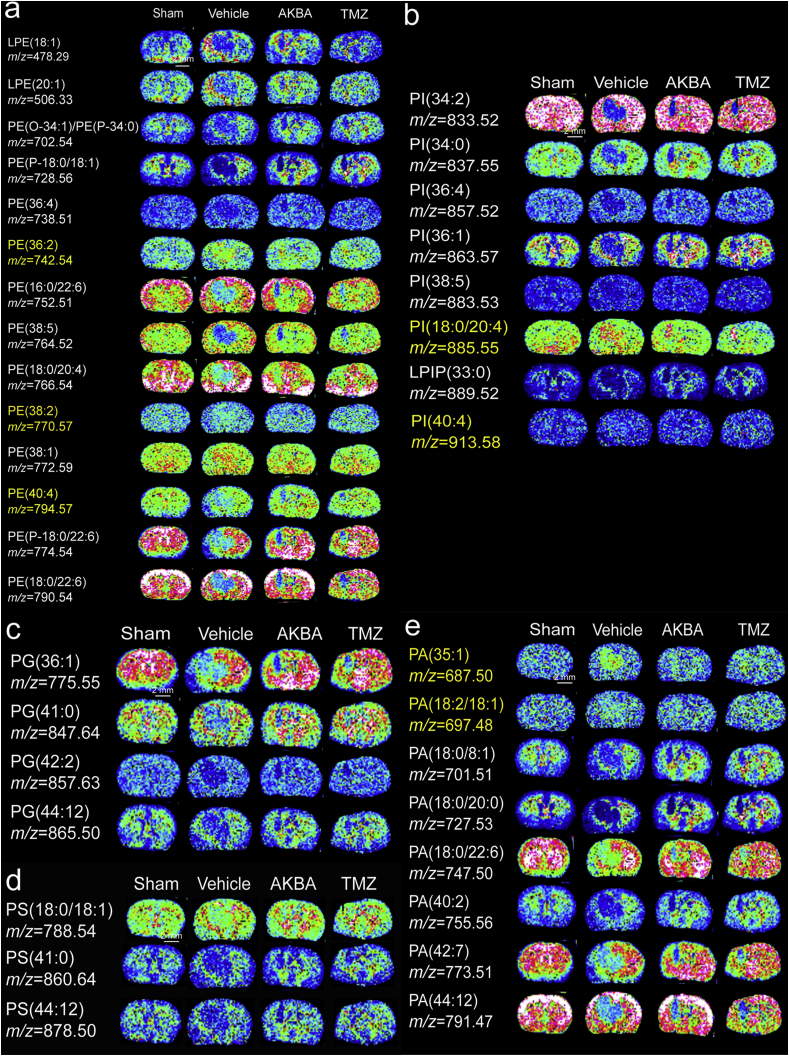
The results of H&E staining and cluster analyses of glioma sections are shown in Fig. 2a. We detected a total of 4182 small molecules in the cortex region, 3080 small molecules in the striatum region, 406 small molecules in adjacent glioma tissue and 494 small molecules in glioma tissue (Fig. 2b). Phospholipid metabolism was different in cancers as compared to normal cells and tissues and this abnormal metabolism can affect numerous cellular processes, including cell growth, proliferation, differentiation and motility33. Using MDALI-MSI, we observed the changes of spatial distribution of phospholipids in the brain. As shown in Fig. 2c–e, 20 changed phosphatidylethanolamines (PEs), 8 changed phosphatidylinositols (PIs), 4 changed phosphatidylglycerols (PGs), 4 changed phosphatidylserines (PSs), and 7 changed phosphatidic acids (PAs) were identified. Among the 20 altered PE species, PE (36:2), PE (38:2), and PE (40:4) were significantly increased in the glioma tissue compared with that in the normal tissue, whereas 17 other PE and LPE were decreased in the glioma tissue compared to normal tissue (Fig. 3a). Among the eight altered PI species, compared with the Sham group, PI (18:0/20:4) and PI (40:4) were significantly increased while six other PIs were decreased in the glioma group (Fig. 3b). Four PG and four PS dramatically decreased in the glioma tissue compared to normal (Fig. 3c and d). PA (35:1) and PA (18:2/18:0) were significantly increased in the glioma tissue compared to normal tissue, whereas five other PAs were decreased in the glioma tissue (Fig. 3e). AKBA, at a dose of 100 mg/kg corrected the abnormal phospholipid metabolism (Fig. 3a–e).
Figure 2.
MALDI-MSI of glioma tissues. (a) H&E staining and clustering analyses of the glioma brain tissues (Scale bar = 2 mm). (b) Cluster analyses showed the alteration of numerous metabolites in glioma tissues. (c) Heatmap of differentially expressed PEs. (d) Heatmap of differentially expressed PIs. (e) Heatmap of differentially expressed PGs, PSs, and PAs.
Figure 3.
AKBA ameliorated the aberrant alterations of PEs, PIs, PGs, PSs, and PAs in the glioblastoma orthotopic model. In situ MALDI MSI of PEs (a), PIs (b), PGs (c), PSs (d) and Pas (e). The brains were rapidly removed, immediately frozen by immersion in n-hexane at −80 °C, and stored at −80 °C until use. Coronal brain sections were cut at a thickness of 10 μm and NEDC was used as the matrix (n = 3 per group). Mass imaging data were acquired in the negative ionization mode at a 200 μm spatial resolution. Scale bar = 2 mm.
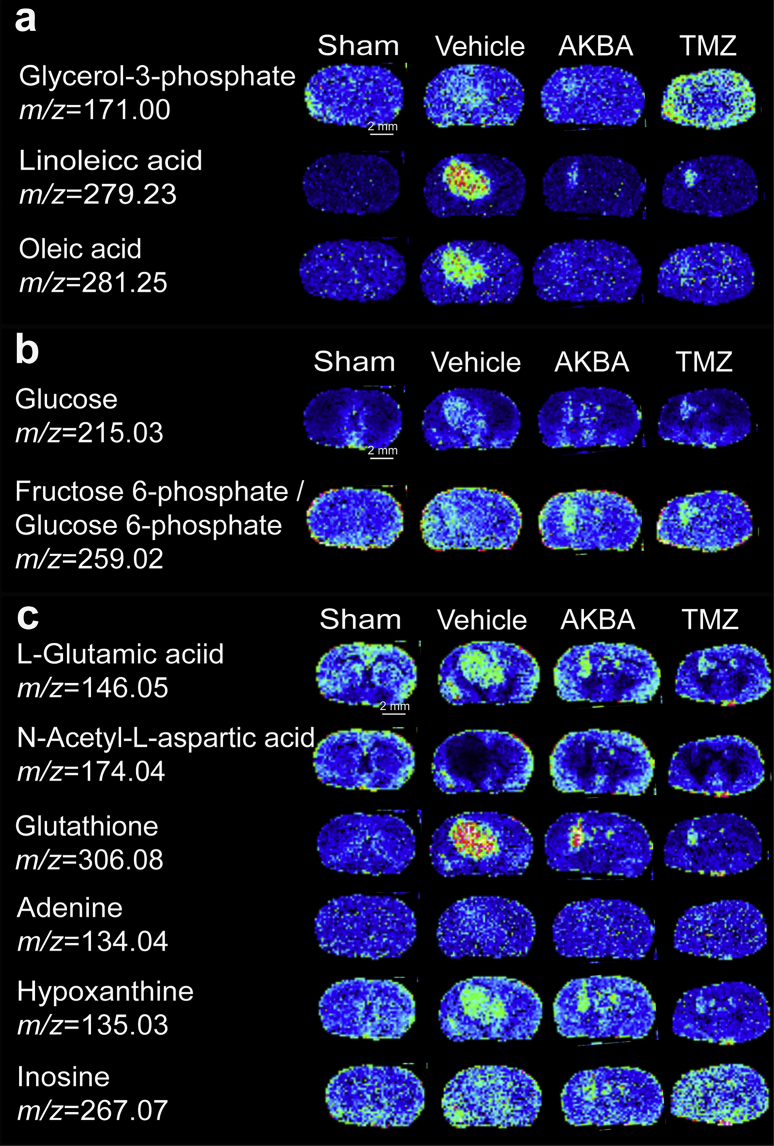
Glycerol-3-phosphate (G3P), which is derived from glucose during glycolysis, is at the crossroads of glucose and lipid metabolism. It is also the starting substrate for the glycerolipid/free fatty acid (GL/FFA) cycle. In this study, compared to the Sham group, the G3P content was increased in Vehicle group and AKBA treatment reduced this aberrant alteration (Fig. 4a).
Figure 4.
AKBA ameliorated the aberrant alterations of fatty acids, glucose and other small molecules in the glioblastoma orthotopic model. (a) In situ MALDI MSI of fatty acids. (b) In situ MALDI MSI of glucose and glucose 6-phosphate. (c) In situ MALDI MSI of amino acids, glutathione and other small molecules. The brains were rapidly removed, immediately frozen by immersion in n-hexane at −80 °C and stored at −80 °C until use. Coronal brain sections were cut at a thickness of 10 μm and NEDC was used as the matrix (n = 3 per group). Mass imaging data were acquired in negative ionization mode at a 200 μm spatial resolution. Scale bar = 2 mm.
FFAs, which are energy source in humans, play important roles in membrane structure, and are able to influence cellular functions and physiological responses34. Linoleic acid (LA) and oleic acid (OA) are two important FFAs. As shown in Fig. 4a, LA and OA were significantly increased in Vehicle group compared with Sham group. AKBA, at a dose of 100 mg/kg, significantly decreased the levels of LA and OA (Fig. 4a) compared to Vehicle group.
3.3. AKBA reduced the abnormal accumulation of glucose in orthotopic gliomas
Glucose is the key source of energy in the brain, and high cellular glucose metabolism has been recognized as one of the hallmarks of cancer35. Glucose-6-phosphate (G6P) is the first and most important metabolic product of glucose metabolism, because G6P is at the convergence point of the glycolytic and the pentose phosphate pathways, the hexosamine pathway and glycogen synthesis35. More and more studies have shown that glucose metabolism is disturbed in cancer cells. Using MDALI-MSI, the changes of spatial glucose and G6P were observed in brain tissue. Compared to Sham group, the concentrations of glucose and G6P increased in the striatum region (Fig. 4b). Compared to Vehicle group, the concentrations of glucose and G6P were decreased in the striatum region in AKBA- and TMZ-treated groups. These results suggested that glucose levels were abnormally high in Vehicle group, and that AKAB reduced the accumulation of glucose.
3.4. AKBA corrected the aberrant changes in the concentrations of amino acids, antioxidants, and other small molecules in glioblastoma xenografts
l-Glutamic acid and N-acetyl-l-aspartic acid (NAA) are the first and second most concentrated amino acids in the brain, respectively36. The alterations of l-glutamic acid and NAA were mapped and quantified by MALDI-MSI (Fig. 4c). Compared to Sham group, the level of l-glutamic acid was increased while the level of NAA was decreased in Vehicle group. After treatment with AKBA (100 mg/kg) for two weeks, the level of l-glutamic acid decreased and NAA increased compared with Vehicle group. The brains of glioma-xenografted mice showed an abnormal cellular redox state, and glutathione has been demonstrated to play a crucial role in the maintenance of the cellular redox state. Results from MALDI-MSI show that the glutathione level much increased in the glioma tissue compared to normal brain, and AKBA reduced this aberrant accumulation (Fig. 4c). Three other identified metabolites, inosine, hypoxanthine and adenine, also increased in glioma tissues of untreated mice. AKBA at a dose of 100 mg/kg significantly decreased the levels of inosine, hypoxanthine, and adenine (Fig. 4c).
3.5. AKBA inhibited autophagy by regulating ERK and p53 signaling pathways
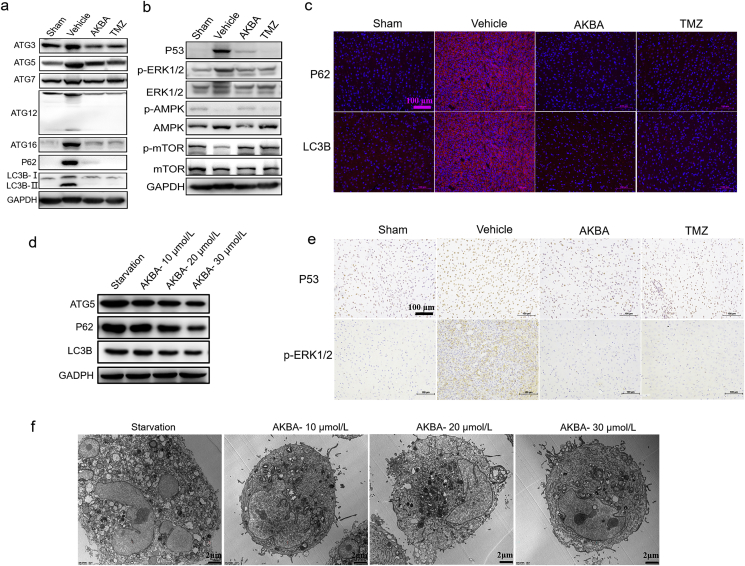
According to the above results, PE is one of rescued phospholipids by the use of AKBA, which is reported to participate in the regulation of cell autophagy37,38. Therefore, we explored if AKBA treatment could affect the cell autophagy. To investigate whether the effect of AKBA on lipid metabolism ultimately affected autophagy, Western blot was used to check the effects of AKAB on key proteins of the autophagy signaling pathway. The protein levels of ATG5, P62, and LC3-II increased in Vehicle group, indicating that the glioma induced an autophagic response (Fig. 5a). The protein levels of ATG3, ATG7, ATG12, and ATG16 were also increased in Vehicle group, further confirming the increased autophagy in glioma tissues. In contrast, AKBA reduced the protein levels of ATG5, P62, LC3-II, ATG3, ATG7, ATG12, and ATG16 (Fig. 5a and Supporting Information Figs. S2 and S3), indicating AKBA inhibited the autophagy and finally suppressed the growth of glioma.
Figure 5.
AKBA inhibited autophagy in vitro and in vivo. (a) Protein levels of key proteins in the autophagy pathway were checked by Western blotting. The protein levels of ATG3, ATG5, ATG7, ATG12, ATG16, P62, and LC3-II were increased in Vehicle group. (b) The protein expression of p-ERK1/2 and P53 was reduced whereas expression of p-mTOR and p-AMPK was increased in AKBA group compared to Vehicle group. (c) Expression of P62 and LC3B was reduced in AKBA group compared with Vehicle group by immunofluorescence. Scale bar = 100 μm. (d) Expressions of ATG 5, P62 and LC3B reduced in U87 cells treated by AKBA in dose-dependent manner. (e) Expression of P53 and p-ERK1/2 was decreased in AKBA group compared Vehicle group by immunohistochemistry. Scale bar = 100 μm. (f) AKBA significantly inhibited autophagy of U87 cells by transmission electron microscope. n = 3 per group. Scale bar = 2 μm.
Furthermore, to explore the specific pathways through which AKBA inhibited the autophagy, the key proteins in AMPK/mTOR, ERK, and P53 pathways were examined by Western blot. The protein expression of p-ERK1/2 and P53 was reduced whereas expression of p-mTOR and p-AMPK was increased in AKBA group compared to Vehicle group (Fig. 5b and Supporting Information Figs. S4 and S5). Results from immunofluorescence showed that expressions of P62 and LC3B reduced in AKBA group compared with Vehicle group (Fig. 5c). Results from immunostaining showed expression of P53 and p-ERK1/2 also was decreased in AKBA group compared Vehicle group (Fig. 5e). In addition, the expression of ATG5, P62 and LC3B was reduced in U87 cells treated by AKBA in dose-dependent manner (Fig. 5d and Supporting Information Fig. S6). Results from transmission electron microscope suggested that AKBA significantly inhibited autophagy of U87 cells (Fig. 5f). Taken together, AKBA inhibited autophagy by regulating ERK and p53 signaling pathways in vitro and in vivo.
4. Discussion
Although there are many studies focusing on the anti-tumor effects of AKBA, no relevant studies about the anti-tumor effects of AKBA in a glioma orthotopic model have been reported. In this study, using an orthotopic xenograft model of glioma in nude mice, we firstly observed that AKBA suppressed tumor growth and reduced the brain/body ratio, suggesting that AKBA could inhibit glioma tumorigenesis in vivo. In addition, using MALDI-MSI, we mapped the alteration of the metabolic landscape in glioma-xenografted mice treated with AKBA, and observed that AKBA corrected the abnormal metabolism of phospholipids, glucose, amino acids, antioxidants, and other endogenous small molecules in the glioma tissue. Furthermore, AKBA was found to inhibit the autophagy formation by regulating ERK and P53 pathways.
Abnormal metabolism is one of hallmarks of cancers. The famous metabolic abnormality in cancers was the Warburg effect, which was awarded the Nobel Prize in Physiology in 1931. Abnormal metabolism promoted tumorigenesis and development of cancers. Metabolism included glucose metabolism, protein metabolism and lipid metabolism39. Lipids not only play an important role in maintaining cellular membrane structure, but also participate in signal transduction as second messengers. Increasing evidence shows that specific alterations in lipid metabolism are associated with the development of cancers. These alterations can affect numerous biological processes including cell growth, proliferation, differentiation, and energy homeostasis through lipid signal transduction pathways. Aberrant lipid metabolism is an early event in tumorigenesis and has been observed in prostate cancers40, breast cancers41, and lung cancers42. Therefore, it is a promising approach to develop new drugs43 that target membrane lipids. MALDI-MS is an emerging method for lipid analysis which can perform rapid and sensitive analyses of numerous lipids in one experiment. Coupled with an imaging method, these techniques can simultaneously visualize the changes of various lipids in situ and provide 2D visualization of the metabolic profiles. For example, Using MALDI-MSI, Fack et al.44 precisely mapped the metabolic landscape in IDH-mutant gliomas. Though the mechanism underlying the aberrant lipid production in glioma tissues is not fully understood, lipidomic analyses provide a new insight into the progression of glioma.
Phospholipids are major components of cell membranes and participate in various biological functions. Their levels are altered in various human cancers45. In this study, we conducted lipidomic analyses in glioma brain tissues using MALDI-MSI. We found that PE (36:2, 38:2, 40:4), PI (18:0/20:4, 40:4), PG (44:2), and PA (35:1) were significantly up-regulated, while all other PE, PS, PI, PG, and PA were significantly down-regulated in glioma tissues. This was the first systematic investigation of changes in phospholipids in glioma tissues and the results could facilitate the discovery of new biomarkers or drug targets for glioma. As the second most abundant phospholipid in mammalian cells, PE participates in the regulation of cell proliferation, metabolism, autophagy, stress responses, apoptosis, and aging. It is also a potent anticancer target for natural products46. In our study, the levels of PE (36:2), PE (38:2) and PE (40:4) significantly were increased while the other PEs were decreased in glioma tissues compared with normal tissues. These findings were consistent with previous studies of aberrant PE metabolism detected in other cancers, such as hepatocellular carcinoma47, colorectal cancer48, breast cancer49, and non-small cell lung cancer50. In conclusion, our results showed that the natural product, AKBA, could alleviate the aberrant changes in PE levels, suggesting that PE might be a potential target for AKBA.
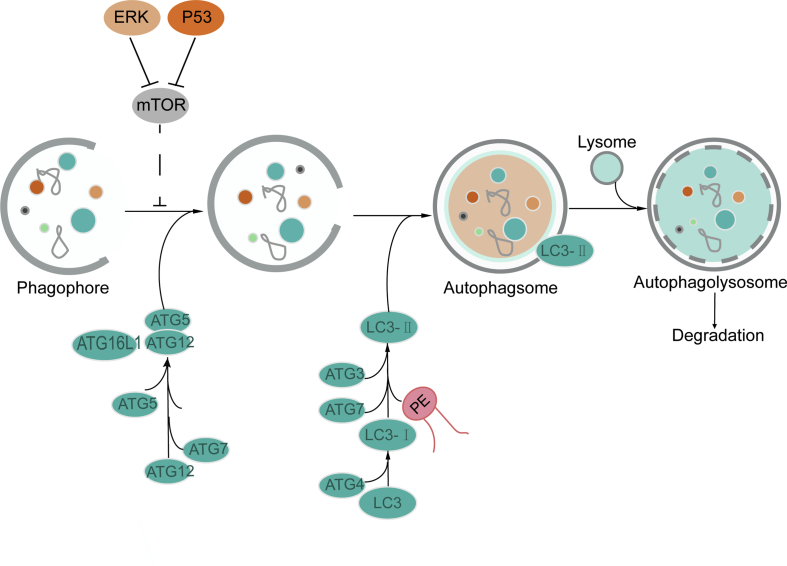
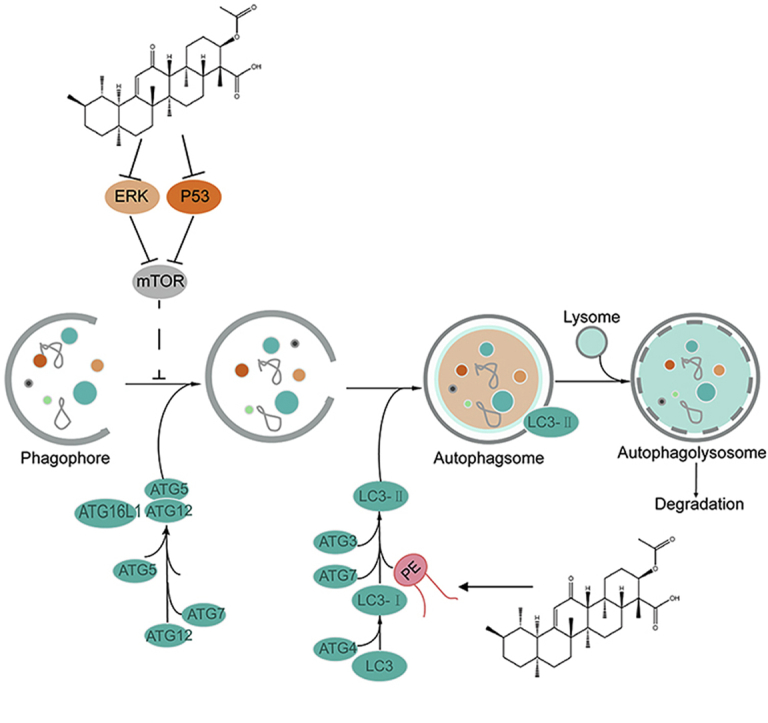
Lipid metabolism is also involved in autophagy, which is a self-degradation process induced by nutrient limitation51. For example, the binding of PE to the autophagy-related protein ATG8 (LC3) is the initiating step in autophagy. During autophagy, ATG7 activated the conjugation of LC3 (ATG8) with PE, ATG12 and ATG5. ATG8/LC3 is essential for autophagosome biogenesis and it functions as an adaptor protein for selective autophagy52. The members of the LC3/ATG8 family of proteins are covalently attached to phagophore and autophagosomal membranes. The ATG8 lipidation, which could turn LC3B-I into LC3B-II, is crucial for autophagosome biogenesis53. In this study, we found that the expression levels of key proteins including ATG5, P62, and LC3B-II in the autophagy pathway were increased in glioma tissues. In contrast, AKBA reduced the expressions of these proteins, suggesting that autophagy was enhanced in glioma tissues and that AKBA-treatment could inhibit the autophagy process. The antitumor effects of AKBA in the previous study were mainly focused on oxidative stress, cell cycle and apoptosis, and this is the first study demonstrating that AKBA could exert its antitumor effect by inhibiting autophagy. Furthermore, we found that AKBA increased p-mTOR/mTOR and decreased the P53 level and ERK1/2 expression compared to Vehicle group, suggesting AKBA inhibited autophagy through the ERK/mTOR and P53/mTOR signaling pathways, but not the AMPK/mTOR pathway. Dysregulated autophagy is emerging as a hallmark of malignancy. Autophagy promotes development of cancers in some situation. AKBA inhibited autophagy to repress growth of glioblastoma by reducing ERK pathway (Fig. 6).
Figure 6.
Proposed mechanistic scheme: AKBA suppressed autophagy in glioblastoma by regulating ERK/mTOR and P53/mTOR pathways. AKBA inhibited growth of glioblastoma in mice by ameliorating their abnormal metabolism to inhibit autophagy by regulating ERK/mTOR and P53/mTOR pathways.
PI, PS, PA and PG are also important lipids which have important physiological functions. PIs constitute a group of phospholipids that commonly act as intracellular second messengers and function in intercellular signaling54. PS, located on the inner leaflet of the cell membrane, is a useful biomarker in the diagnosis of cancer55. Using MALDI-MIS, we found eight PI species, four PS species, nine PA species, and six PG species whose levels were changed in glioma tissues and this could be reversed by AKBA. It has been shown that the decreased abundance of these phospholipids in metastatic tumors may be related with the indirect results of changes in metabolic pathways involving the incorporation/release of oleic acid as a free fatty acid. This is controversial, however, as other scientist found that oleic acid fed highly metastatic carcinoma cells and boosted malignancy56, whereas it actually inhibited cell growth and survival in low metastatic carcinoma cells57. Oleic acid has also been reported to promote migration of breast cancer cells58 while also being linked to cell apoptosis and autophagy in tongue squamous cell carcinomas59. In this study, we found that linoleic acid and oleic acid were increased in glioma tissues, which supported the hypothesis that oleic acid may promote migration of cancer cells in glioma.
The relationship between autophagy and cell-cycle was widely studied, showing that activation of the autophagy flux is observed in mitosis phase. Cell cycle is associated with60 and involved in autophagy61, whereas autophagy regulates cell cycle62. Like the arrest of cell cycle, autophagy is induced in response to various stresses to preserve cellular viability. In addition, P53, a shared key molecule, has been widely reported to orchestrate both cell cycle and activate autophagy63. In our study, we found that P53 and autophagy were inhibited by AKBA, which was consisted with our previous findings that the cell cycle was arrested by AKBA.
In response to the AKBA treatment, small molecules other than lipids were also changed, including amino acids and anti-oxidants in glioma tissues. For example, l-glutamic acid and NAA are the first and second most concentrated amino acids in the brain, respectively36. NAA is a type of neurotransmitter that can be hydrolyzed by aspartoacylase to produce aspartate and acetate64, which can serve as major bioenergetic substrates for highly proliferative brain tumors65. In patient-derived glioma xenograft models, the NAA content was reported to be decreased44. Our MSI results showed that l-glutamic acid increased while NAA was decreased in gliomas compared with normal brain tissue, and that AKBA corrected these abnormal changes. Glutathione is an important anti-oxidant in the brain and plays a crucial role in the maintenance of the cellular redox state. MSI showed that the glutathione level was much higher in glioma tissues compared to normal brain, and that AKBA increased this abnormal accumulation.
5. Conclusions
Using an orthotopic glioma xenograft model, AKBA was found significantly inhibiting the glioma growth in vivo. MALDI-MSI results showed that AKBA could affect lipid metabolism and the levels of amino acids, nucleotides, and anti-oxidants in gliomas. AKBA was further shown to inhibit autophagy by regulating the ERK and P53 signaling pathways. Taken together, these findings suggest that AKBA could be a novel natural product for the treatment of gliomas.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81573454 for Jinhua Wang and No. 81703536 for Wan Li) and supported by Beijing Natural Science Foundation (7172142, China). This work was also supported by CAMS Innovation Fund for Medical Sciences (2016-I2M-3-007, China) and Science and Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711001-005-025, China).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.12.012.
Contributor Information
Jinhua Wang, Email: wjh@imm.ac.cn.
Tengfei Ji, Email: jitf@imm.ac.cn.
Guanhua Du, Email: dugh@imm.ac.cn.
Author contributions
Jinhua Wang and Guanhua Du developed the hypothesis, designed the experiments, and revised the manuscript. Tengfei Ji designed and synthesized the compound. Wan Li and Liwen Ren conducted whole experiments and wrote the main manuscript. Xiangjin Zheng and Jinyi Liu performed the statistical analyses. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Iacob G., Dinca E.B. Current data and strategy in glioblastoma multiforme. J Med Life. 2009;2:386–393. [PMC free article] [PubMed] [Google Scholar]
- 2.Franceschi E., Minichillo S., Brandes A.A. Pharmacotherapy of glioblastoma: established treatments and emerging concepts. CNS Drugs. 2017;31:675–684. doi: 10.1007/s40263-017-0454-8. [DOI] [PubMed] [Google Scholar]
- 3.Nishikawa R. Standard therapy for glioblastoma—a review of where we are. Neurol Med Chir. 2010;50:713–719. doi: 10.2176/nmc.50.713. [DOI] [PubMed] [Google Scholar]
- 4.Gao C., Liang J., Zhu Y., Ling C., Cheng Z., Li R. Menthol-modified casein nanoparticles loading 10-hydroxycamptothecin for glioma targeting therapy. Acta Pharm Sin B. 2019;9:843–857. doi: 10.1016/j.apsb.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K., Burger P.C., Jouvet A. The 2007 who classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran B., Rosenthal M.A. Survival comparison between glioblastoma multiforme and other incurable cancers. J Clin Neurosci. 2010;17:417–421. doi: 10.1016/j.jocn.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Koppenol W.H., Bounds P.L., Dang C.V. Otto warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 10.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research N., Brat D.J., Verhaak R.G., Aldape K.D., Yung W.K., Salama S.R. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilsson A., Fehniger T.E., Gustavsson L., Andersson M., Kenne K., Marko-Varga G. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornett D.S., Reyzer M.L., Chaurand P., Caprioli R.M. Maldi imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 15.Clemis E.J., Smith D.S., Camenzind A.G., Danell R.M., Parker C.E., Borchers C.H. Quantitation of spatially-localized proteins in tissue samples using MALDI-MRM imaging. Anal Chem. 2012;84:3514–3522. doi: 10.1021/ac202875d. [DOI] [PubMed] [Google Scholar]
- 16.Chaurand P., Norris J.L., Cornett D.S., Mobley J.A., Caprioli R.M. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J Proteome Res. 2006;5:2889–2900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 17.Oppenheimer S.R., Wehr A.Y. Imaging mass spectrometry in drug discovery and development. Bioanalysis. 2015;7:2609–2610. doi: 10.4155/bio.15.202. [DOI] [PubMed] [Google Scholar]
- 18.Agar N.Y., Malcolm J.G., Mohan V., Yang H.W., Johnson M.D., Tannenbaum A. Imaging of meningioma progression by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Anal Chem. 2010;82:2621–2625. doi: 10.1021/ac100113w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calligaris D., Longuespee R., Debois D., Asakawa D., Turtoi A., Castronovo V. Selected protein monitoring in histological sections by targeted MALDI-FTICR in-source decay imaging. Anal Chem. 2013;85:2117–2126. doi: 10.1021/ac302746t. [DOI] [PubMed] [Google Scholar]
- 20.Fehniger T.E., Vegvari A., Rezeli M., Prikk K., Ross P., Dahlback M. Direct demonstration of tissue uptake of an inhaled drug: proof-of-principle study using matrix-assisted laser desorption ionization mass spectrometry imaging. Anal Chem. 2011;83:8329–8336. doi: 10.1021/ac2014349. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Qiu S., Chen S., Xiong C., Liu H., Wang J. MALDI-TOF MS imaging of metabolites with a n-(1-naphthyl) ethylenediamine dihydrochloride matrix and its application to colorectal cancer liver metastasis. Anal Chem. 2015;87:422–430. doi: 10.1021/ac504294s. [DOI] [PubMed] [Google Scholar]
- 22.Ianniciello A., Rattigan K.M., Helgason G.V. The ins and outs of autophagy and metabolism in hematopoietic and leukemic stem cells: food for thought. Front Cell Dev Biol. 2018;6:120. doi: 10.3389/fcell.2018.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leone R.D., Amaravadi R.K. Autophagy: a targetable linchpin of cancer cell metabolism. Trends Endocrinol Metab. 2013;24:209–217. doi: 10.1016/j.tem.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H., Zhu G., Fan Y., Du Y., Lan M., Xu Y. Natural products research in China from 2015 to 2016. Front Chem. 2018;6:45. doi: 10.3389/fchem.2018.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koehn F.E., Carter G.T. Rediscovering natural products as a source of new drugs. Discov Med. 2005;5:159–164. [PubMed] [Google Scholar]
- 26.Siddiqui M.Z. Boswellia serrata, a potential antiinflammatory agent: an overview. Indian J Pharm Sci. 2011;73:255–261. doi: 10.4103/0250-474X.93507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabina E.P., Indu H., Rasool M. Efficacy of boswellic acid on lysosomal acid hydrolases, lipid peroxidation and anti-oxidant status in gouty arthritic mice. Asian Pac J Trop Biomed. 2012;2:128–133. doi: 10.1016/S2221-1691(11)60206-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarkate A., Dhaneshwar S.S. Investigation of mitigating effect of colon-specific prodrugs of boswellic acid on 2,4,6-trinitrobenzene sulfonic acid-induced colitis in wistar rats: design, kinetics and biological evaluation. World J Gastroenterol. 2017;23:1147–1162. doi: 10.3748/wjg.v23.i7.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X., Cai J.G., Zhu W.W., Zhao H.Y., Wang K., Zhang X.F. Boswellic acid attenuates asthma phenotype by downregulation of GATA3 via inhibition of pSTAT6. Genet Mol Res. 2015;14:7463–7468. doi: 10.4238/2015.July.3.22. [DOI] [PubMed] [Google Scholar]
- 30.Roy N.K., Deka A., Bordoloi D., Mishra S., Kumar A.P., Sethi G. The potential role of boswellic acids in cancer prevention and treatment. Cancer Lett. 2016;377:74–86. doi: 10.1016/j.canlet.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 31.Li W., Liu J., Fu W., Zheng X., Ren L., Liu S. 3-O-Acetyl-11-keto-beta-boswellic acid exerts anti-tumor effects in glioblastoma by arresting cell cycle at G2/M phase. J Exp Clin Cancer Res. 2018;37:132. doi: 10.1186/s13046-018-0805-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin Z., Wang S., Lin Y., Zhao Y., Yang S., Song J. Antihyperuricemic effect of mangiferin aglycon derivative J99745 by inhibiting xanthine oxidase activity and urate transporter 1 expression in mice. Acta Pharm Sin B. 2018;8:306–315. doi: 10.1016/j.apsb.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santos C.R., Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 34.Burdge G.C., Calder P.C. Introduction to fatty acids and lipids. World Rev Nutr Diet. 2015;112:1–16. doi: 10.1159/000365423. [DOI] [PubMed] [Google Scholar]
- 35.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy?. Nat Rev Cancer. 2016;16:635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sreekumar A., Poisson L.M., Rajendiran T.M., Khan A.P., Cao Q., Yu J. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 37.Oh-oka K., Nakatogawa H., Ohsumi Y. Physiological pH and acidic phospholipids contribute to substrate specificity in lipidation of ATG8. J Biol Chem. 2008;283:21847–21852. doi: 10.1074/jbc.M801836200. [DOI] [PubMed] [Google Scholar]
- 38.Mitroi D.N., Karunakaran I., Graler M., Saba J.D., Ehninger D., Ledesma M.D. SGPL1 (sphingosine phosphate lyase 1) modulates neuronal autophagy via phosphatidylethanolamine production. Autophagy. 2017;13:885–899. doi: 10.1080/15548627.2017.1291471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu W., Zhao S. Metabolic changes in cancer: beyond the warburg effect. Acta Biochim Biophys Sin. 2013;45:18–26. doi: 10.1093/abbs/gms104. [DOI] [PubMed] [Google Scholar]
- 40.Swinnen J.V., Roskams T., Joniau S., Van Poppel H., Oyen R., Baert L. Overexpression of fatty acid synthase is an early and common event in the development of prostate cancer. Int J Cancer. 2002;98:19–22. doi: 10.1002/ijc.10127. [DOI] [PubMed] [Google Scholar]
- 41.Chajes V., Lanson M., Fetissof F., Lhuillery C., Bougnoux P. Membrane fatty acids of breast carcinoma: contribution of host fatty acids and tumor properties. Int J Cancer. 1995;63:169–175. doi: 10.1002/ijc.2910630204. [DOI] [PubMed] [Google Scholar]
- 42.Marien E., Meister M., Muley T., Fieuws S., Bordel S., Derua R. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int J Cancer. 2015;137:1539–1548. doi: 10.1002/ijc.29517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan L.T., Chan K.G., Pusparajah P., Lee W.L., Chuah L.H., Khan T.M. Targeting membrane lipid a potential cancer cure?. Front Pharmacol. 2017;8:12. doi: 10.3389/fphar.2017.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fack F., Tardito S., Hochart G., Oudin A., Zheng L., Fritah S. Altered metabolic landscape in IDH-mutant gliomas affects phospholipid, energy, and oxidative stress pathways. EMBO Mol Med. 2017;9:1681–1695. doi: 10.15252/emmm.201707729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagace T.A., Ridgway N.D. The role of phospholipids in the biological activity and structure of the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833:2499–2510. doi: 10.1016/j.bbamcr.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 46.Patel D., Witt S.N. Ethanolamine and phosphatidylethanolamine: partners in health and disease. Oxid Med Cell Longev. 2017;2017:4829180. doi: 10.1155/2017/4829180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Q., Tan Y., Yin P., Ye G., Gao P., Lu X. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res. 2013;73:4992–5002. doi: 10.1158/0008-5472.CAN-13-0308. [DOI] [PubMed] [Google Scholar]
- 48.Dobrzynska I., Szachowicz-Petelska B., Sulkowski S., Figaszewski Z. Changes in electric charge and phospholipids composition in human colorectal cancer cells. Mol Cell Biochem. 2005;276:113–119. doi: 10.1007/s11010-005-3557-3. [DOI] [PubMed] [Google Scholar]
- 49.Vance J.E. Molecular and cell biology of phosphatidylserine and phosphatidylethanolamine metabolism. Prog Nucleic Acid Res Mol Biol. 2003;75:69–111. doi: 10.1016/s0079-6603(03)75003-x. [DOI] [PubMed] [Google Scholar]
- 50.Zinrajh D., Horl G., Jurgens G., Marc J., Sok M., Cerne D. Increased phosphatidylethanolamine n-methyltransferase gene expression in non-small-cell lung cancer tissue predicts shorter patient survival. Oncol Lett. 2014;7:2175–2179. doi: 10.3892/ol.2014.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dall'Armi C., Devereaux K.A., Di Paolo G. The role of lipids in the control of autophagy. Curr Biol. 2013;23:R33–R45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krick R., Thumm M. Atg8 lipidation is coordinated in a Ptdins3p-dependent manner by the proppin Atg21. Autophagy. 2016;12:2260–2261. doi: 10.1080/15548627.2016.1221564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juris L., Montino M., Rube P., Schlotterhose P., Thumm M., Krick R. Pi3p binding by atg21 organises atg8 lipidation. EMBO J. 2015;34:955–973. doi: 10.15252/embj.201488957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waugh M.G. PIPs in neurological diseases. Biochim Biophys Acta. 2015;1851:1066–1082. doi: 10.1016/j.bbalip.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Sharma B., Kanwar S.S. Phosphatidylserine: a cancer cell targeting biomarker. Semin Cancer Biol. 2018;52:17–25. doi: 10.1016/j.semcancer.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Paine M.R.L., Liu J., Huang D., Ellis S.R., Trede D., Kobarg J.H. Three-dimensional mass spectrometry imaging identifies lipid markers of medulloblastoma metastasis. Sci Rep. 2019;9:2205. doi: 10.1038/s41598-018-38257-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S., Zhou T., Li C., Dai Z., Che D., Yao Y. High metastaticgastric and breast cancer cells consume oleic acid in an AMPK dependent manner. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Navarro-Tito N., Soto-Guzman A., Castro-Sanchez L., Martinez-Orozco R., Salazar E.P. Oleic acid promotes migration on MDA-MB-231 breast cancer cells through an arachidonic acid-dependent pathway. Int J Biochem Cell Biol. 2010;42:306–317. doi: 10.1016/j.biocel.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 59.Jiang L., Wang W., He Q., Wu Y., Lu Z., Sun J. Oleic acid induces apoptosis and autophagy in the treatment of tongue squamous cell carcinomas. Sci Rep. 2017;7:11277. doi: 10.1038/s41598-017-11842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H., Peng X., Wang Y., Cao S., Xiong L., Fan J. Atg5-mediated autophagy deficiency in proximal tubules promotes cell cycle G2/M arrest and renal fibrosis. Autophagy. 2016;12:1472–1486. doi: 10.1080/15548627.2016.1190071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Azzopardi M., Farrugia G., Balzan R. Cell-cycle involvement in autophagy and apoptosis in yeast. Mech Ageing Dev. 2017;161:211–224. doi: 10.1016/j.mad.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 62.Laggner M., Pollreisz A., Schmidinger G., Schmidt-Erfurth U., Chen Y.T. Autophagy mediates cell cycle response by regulating nucleocytoplasmic transport of PAX6 in limbal stem cells under ultraviolet—a stress. PLoS One. 2017;12 doi: 10.1371/journal.pone.0180868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White E. Autophagy and p53. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsen A.R., Long P.M., Driscoll H.E., Davies M.T., Teasdale B.A., Penar P.L. Triacetin-based acetate supplementation as a chemotherapeutic adjuvant therapy in glioma. Int J Cancer. 2014;134:1300–1310. doi: 10.1002/ijc.28465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mashimo T., Pichumani K., Vemireddy V., Hatanpaa K.J., Singh D.K., Sirasanagandla S. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.