Abstract
Overexpression of exogenous lineage-determining factors succeeds in directly reprogramming fibroblasts to various cell types. Several studies have reported reprogramming of fibroblasts into induced cardiac progenitor cells (iCPCs). CRISPR/Cas9-mediated gene activation is a potential approach for cellular reprogramming due to its high precision and multiplexing capacity. Here we show lineage reprogramming to iCPCs through a dead Cas9 (dCas9)-based transcription activation system. Targeted and robust activation of endogenous cardiac factors, including GATA4, HAND2, MEF2C and TBX5 (G, H, M and T; GHMT), can reprogram human fibroblasts toward iCPCs. The iCPCs show potentials to differentiate into cardiomyocytes, smooth muscle cells and endothelial cells in vitro. Addition of MEIS1 to GHMT induces cell cycle arrest in G2/M and facilitates cardiac reprogramming. Lineage reprogramming of human fibroblasts into iCPCs provides a promising cellular resource for disease modeling, drug discovery and individualized cardiac cell therapy.
Key words: Lineage reprogramming, Human foreskin fibroblasts, Induced cardiac progenitor cells, CRISPR/Cas9, SAM, Cardiac transcription factors
Graphical abstract
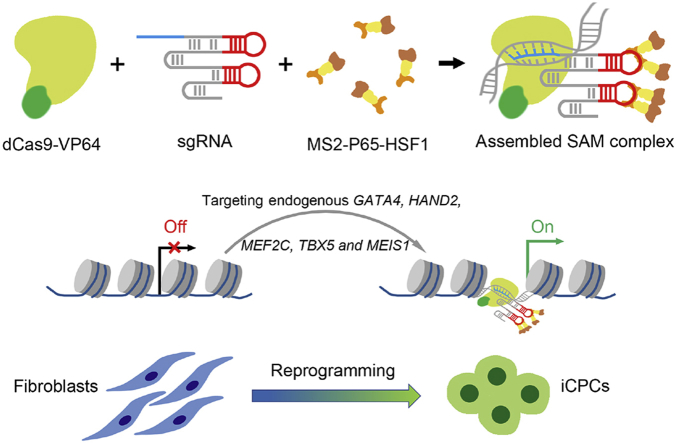
Human foreskin fibroblasts were reprogrammed into induced cardiac progenitor cells via a CRISPR/Cas9-based synergistic activation mediator-mediated gene activation.
1. Introduction
Advances in cellular reprogramming technologies have provided opportunities for facilitating cell fate conversion, with potential applications in disease modeling, drug discovery and cell therapy. Conventional reprogramming has been achieved by forced expression of transgenes encoding lineage-determining factors1, 2, 3. In contrast to direct reprogramming into differentiated cell types such as cardiomyocytes4, 5, hepatocytes6 and neurons7, some groups showed lineage reprogramming into tissue-specific progenitors such as cardiac8, 9, hepatic10, neural11 and hematopoietic12 progenitor cells. Reprogramming into progenitor cells rather than terminally differentiated cell types provides potential advantages for differentiating into necessary cell types to fully reconstitute the diseased or damaged tissue, which lacks the complex paracrine environment and multiple stage-specific signals seen in developing embryos.
Cardiac progenitor cells (CPCs) may provide a potential avenue for treating heart disease and for studying cardiac development. These cells evolve from the mesoderm of developing embryos during cardiogenesis, which can directly differentiate into three cardiac lineage cells including cardiomyocytes (CMs), smooth muscle cells (SMCs) and endothelial cells (ECs). Previous studies have shown several CPC populations from differentiation of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs)13, 14. CPCs have been identified using various markers such as ISL1, NKX2-5 and FLK1. Recently, some groups adopted different strategies to reprogram fibroblasts into iCPCs under defined conditions8, 9, 15. In contrast to ectopic expression of cardiac transcription factors and activation of multiple cardiac signals by small molecules, there are no reports about reprogramming human fibroblasts toward iCPCs through direct activation of endogenous cardiac genes.
Genome editing technologies have been applied in a wide range of scientific and medical fields such as disease modeling and gene therapy. CRISPR/Cas9, discovered in the bacterial adaptive immune system, was repurposed to a remarkably flexible tool for genome manipulation in mammalian cells, as Cas9 nuclease could independently bind to DNA and induce DNA double strand breaks (DSBs)16, 17, 18. Alterations to Cas nuclease broadly expand the applications of the system such as modulating endogenous gene expression and manipulating epigenetic modification of genomic sites without inducing DSBs. This avoids creating unwanted permanent mutations in genome. Specifically, point mutations in Cas9 (D10A and H840A) result in a deactivated form of Cas9, termed as dead Cas9 (dCas9). The dCas9 protein fused with transactivation domains such as VP64 and P300 has been engineered as synthetic transcription activators. This enables the CRISPR/Cas9 system to directly activate the expression of downstream target genes with high precision19, 20. Thus, this system has been thought to be a potential approach for cellular reprogramming through initiating expression of endogenous gene networks.
Several groups have utilized different dCas9-based transcription activation systems for cellular reprogramming21, 22. Black et al.23 demonstrated the direct conversion of mouse fibroblasts to neuronal cells through CRISPR/Cas9-mediated transcriptional activation of endogenous BRN2, ASCL1 and MYTLl genes. Liu et al.24 reported a different strategy for reprogramming mouse embryonic fibroblasts to induced pluripotent stem cells through CRISPR-based chromatin remodeling of the endogenous OCT4 or SOX2 locus. Therefore, CRISPR/Cas9 system holds promise for cardiac reprogramming through direct activation of endogenous cardiac genes of human fibroblasts.
In this study, we tested the hypothesis that targeted activation of endogenous cardiac transcription factors is sufficient for generating iCPCs using synergistic activation mediator (SAM)25, 26, a dCas9-based transcription activation system. Here we show that combinations of cardiac factors GATA4, HAND2, MEF2C and TBX5 (G, H, M and T; GHMT) can reprogram human foreskin fibroblasts into iCPCs under defined conditions. iCPCs are lineage-restricted, capable of differentiating into three cardiovascular cell types including CMs, SMCs and ECs in vitro. Addition of MEIS1 to GHMT facilitates cardiac reprogramming. Lineage reprogramming of human fibroblasts into iCPCs offers a useful cellular resource for disease modeling, drug discovery and individualized cardiac cell therapy. Our data show a promising approach for cardiac reprogramming.
2. Materials and methods
2.1. Cell culture
Human foreskin fibroblasts (HFFs) supplied at a low passage number (p3–p5) were purchased from Merck Millipore (Billerica, MA, USA). HEK293T cells were obtained from Guangdong Institute of Gastroenterology and the Sixth Affiliated Hospital, Sun Yat-sen University (Guangzhou, China). Both cell lines were cultured in Dulbecco's modified Eagle medium (DMEM, Gibco Life Technologies, Carlsbad, CA, USA) supplemented with 10% inactivated fetal bovine serum (Gibco), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco) at 37 °C in a humidified atmosphere with 5% CO2.
2.2. Reagents
Polybrene (Sigma–Aldrich, St. Louis, MO, USA) was dissolved in 0.9% NaCl solution to a concentration of 10 mg/mL and stored at −4 °C. Horse serum, B-27, insulin-selenium-transferrin, blasticidin S HCL and hygromycin B were purchased from Gibco. Matrigel® matrix and PE-Cy™7 mouse anti-human CD90 (THY1) were purchased from BD Biosciences (Bedford, MA, USA). Antibodies against GATA4, MEF2A+MEF2C, TBX5, NKX2-5, ISL1, cTNT, α-actinin, tropomyosin1, CD31, SM-MHC, α-SMA, FSP1 (S100A4), MEIS1, MEIS2, MEIS3, phosphorylated CHK1 at serine296, CHK1, E2F1, STAT3, phosphorylated STAT3 at Y705 and H3K4me3 were purchased from Abcam (Cambridge, MA, USA). Antibodies against HAND2 and tropomyosin were purchased from Sigma–Aldrich. Antibody against CD34 and β-catenin were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Antibody against GAPDH was purchased from GeneTex (Irvine, CA, USA).
2.3. Single-guide RNA design
Single-guide RNAs (sgRNAs) were designed and assembled as described by Konermann et al25, 26. Briefly, sgRNA sequences were synthesized and annealed in a thermal cycler. Then annealed oligo was cloned into lenti sgRNA(MS2)_zeo backbone (a gift from Feng Zhang, Addgene #61427).
2.4. Lentivirus production
HEK293T cells were cultured in DMEM. One day prior to transfection, cells were seeded in T75 flask at a density of 40%–50% and cultured in growth medium without antibiotics. Cells were transfected with 6.6 μg of pMD2.G (a gift from Didier Trono, Addgene #12259), 8.8 μg of psPAX2 (a gift from Didier Trono, Addgene #12260) and 10.2 μg of plasmid containing the vector of interest for each flask when cells reached 80%–90% confluency on the next day. The transfection was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacture's protocol. Five hours after transfection, the medium was changed. pcDNA3.1-EGFP expression plasmid indicated transfection efficiency. Virus supernatant was harvested 48 h post-transfection, centrifuged at 600×g for 10 min, filtered with a 0.45 μm PVDF filter (Merck Millipore), concentrated using Lenti-X concentrator (Clontech, Mountain View, CA, USA), then aliquoted and stored at −80 °C.
2.5. Lentivirus transduction
Briefly, 5 × 105 HFFs were seeded in T75 flask, cultured in growth medium without antibiotics for 24 h, and then transduced with lentiviral supernatant containing dCas9-VP64 (a gift from Feng Zhang, Addgene #61425) and MS2-P65-HSF1 (a gift from Feng Zhang, Addgene #61426) at a final concentration of 8 μg/mL polybrene. Twenty-four hours after transduction, lentiviral supernatant was replaced by selection medium. The working concentration of selection reagent was determined by a kill curve: 1 μg/mL blasticidin S HCL and 15 μg/mL hygromycin B. Medium was refreshed every 3 days and cells were passaged on day 7. HFFs without virus infection served as a negative control. pLenti CMV GFP Blast (a gift from Eric Campeau & Paul Kaufman, Addgene #17445) transduction control indicated transduction efficiency.
2.6. CPCs induction
For selecting efficient sgRNAs, HFFs stably expressed dCas9-VP64 and MS2-P65-HSF1 were seeded in six-well plates at a density of 30% for 24 h, and then transduced with sgRNA lentiviral supernatant. Cells were cultured for 96 h, harvested for qRT-PCR and Western blot analysis. The efficient sgRNA sequences of GATA4, HAND2, MEF2C, TBX5 and MEIS1 are listed in Table 1. For CPCs induction, HFFs stably expressed dCas9-VP64 and MS2-P65-HSF1 were recovered from selection medium by culturing in normal medium for 2 days. 2.2 × 105 cells were plated in a Matrigel-coated 100 mm dish, cultured in fibroblast culture medium, and then transduced with sgRNA lentivirus expressing GATA4 (GATA4-103), HAND2 (HAND2-136), MEF2C (MEF2C-89), TBX5 (TBX5-178) and MEIS1 (MEIS1-100). Twenty-four hours after transduction, lentiviral supernatant was replaced with induction medium, composed of DMEM/medium 199 (4:1), 10% conditioned medium obtained from neonatal rat cardiomyocyte culture, 10% fetal bovine serum, 5% horse serum, 1% penicillin/streptomycin, 1% B-27, 1% insulin-transferrin-selenium, 1% essential amino acids, 1% non-essential amino acids, 1% vitamin mixture, 1% sodium pyruvate as described by Song et al.27 and 1000 unit/mL leukemia inhibitory factor (Millipore). Medium was changed every 1 day until cells were collected for various experiments.
Table 1.
The efficient sgRNA sequences of GATA4, HAND2, MEF2C, TBX5 and MEIS1.
| Name | Sequence (5ʹ–3ʹ) |
|---|---|
| GATA4-103 Forward | CACCGCGCCCAGCGGAGGTGTAGCC |
| GATA4-103 Reverse | AAACGGCTACACCTCCGCTGGGCGC |
| HAND2-136 Forward | CACCGGAGGTAGCCAATCCTGGAAG |
| HAND2-136 Reverse | AAACCTTCCAGGATTGGCTACCTCC |
| MEF2C-89 Forward | CACCGGAAGACGGAGCACGAATGGT |
| MEF2C-89 Reverse | AAACACCATTCGTGCTCCGTCTTCC |
| TBX5-178 Forward | CACCGGTTCTCCGTAATGTGCCTTG |
| TBX5-178 Reverse | AAACCAAGGCACATTACGGAGAACC |
| MEIS1-100 Forward | CACCGCTTGCAAAGAGGGAGAGAGA |
| MEIS1-100 Reverse | AAACTCTCTCTCCCTCTTTGCAAGC |
2.7. Quantitative real-time PCR (qRT-PCR) and chromatin immunoprecipitation (ChIP) assays
Total RNA was extracted from HFFs-induced CPCs using TRIzol reagent and reverse-transcribed into cDNA using PrimeScript™ RT reagent kit with gDNA eraser (TaKaRa, Dalian, China). qRT-PCR was performed in triplicate for each sample using TB Green™ Premix Ex Taq™ (Tli RNaseH Plus, TaKaRa) on the LightCycler 480 real-time PCR system (Roche Applied Science, Mannheim, Baden-Wuerttemberg, Germany). All expression data were normalized to GAPDH, and relative quantification of gene expression was calculated using ΔΔCt method. The primer sequences are listed in Table 2. ChIP assays were performed with an Enzymatic Chromatin IP Kit (Cell Signaling Technology, Beverly, MA, USA) according to the manufacture's protocol. DNA fragments obtained without antibody served as input controls. DNA fragments obtained with normal rabbit or mouse IgG served as negative controls. Primer sequences used in ChIP-quantitative RT-PCR are listed in Table 3.
Table 2.
Primers used for quantitative RT-PCR.
| Name | Sequence (5ʹ–3ʹ) |
|---|---|
| MESP1 Forward | CGAGTCCTGGATGCTCTCTG |
| MESP1 Reverse | ATGAGTCTGGGGACGAGACG |
| EOMES Forward | CATGCAGGGCAACAAAATGTATG |
| EOMES Reverse | GTGTTGTTGTTATTTGCGCCTTTGT |
| GATA4 Forward | AATGCCTGCGGCCTCTACA |
| GATA4 Reverse | AGATTTATTCAGGTTCTTGGGCTTC |
| HAND2 Forward | CCACCAGCTACATCGCCTACCT |
| HAND2 Reverse | TCGTTGCTGCTCACTGTGCTT |
| MEF2C Forward | GAACGTAACAGACAGGTGACAT |
| MEF2C Reverse | CGGCTCGTTGTACTCCGTG |
| TBX5 Forward | AAATGAAACCCAGCATAGGAGCTGGC |
| TBX5 Reverse | ACACTCAGCCTCACATCTTACCCT |
| MEIS1 Forward | GGCACAAGACACGGGACTCA |
| MEIS1 Reverse | CATGGGCTGTCCATCAGGATTA |
| ISL1 Forward | ATCAGGTTGTACGGGATCAAATG |
| ISL1 Reverse | ATGTGATACACCTTGGAGCG |
| MYOCD Forward | AATTTCAGAGGTAACACAGCCTCCA |
| MYOCD Reverse | CGCTTTCAATAAGCACGTCCAG |
| TNNT2 Forward | GCTGTGGCAGAGCATCTATAACTTG |
| TNNT2 Reverse | GCCCGGTGACTTTAGCCTTC |
| THY1 Forward | ATACCAGCAGTTCACCCATCCAGT |
| THY1 Reverse | ATTTGCTGGTGAAGTTGGTTCGGG |
| FSP1 Forward | TCTTGGTTTGATCCTGACTGCT |
| FSP1 Reverse | ACTTGTCACCCTCTTTGCCC |
| COL1A1 Forward | GATTCCCTGGACCTAAAGGTGC |
| COL1A1 Reverse | AGCCTCTCCATCTTTGCCAGCA |
| COL1A2 Forward | CCTGGTGCTAAAGGAGAAAGAGG |
| COL1A2 Reverse | ATCACCACGACTTCCAGCAGGA |
| GAPDH Forward | GCACCGTCAAGGCTGAGAAC |
| GAPDH Reverse | TGGTGAAGACGCCAGTGGA |
Table 3.
Primers used for ChIP-quantitative RT-PCR34.
| Name | Sequence (5ʹ–3ʹ) |
|---|---|
| MESP1 Forward | GAAACAGGCGCAGTCAAGG |
| MESP1 Reverse | GCCGCATCAGCACATCAAAG |
| ACTC1 Forward | CCCTCCCCTTCCTTACATGGT |
| ACTC1 Reverse | GCCGAGGCCATTCATGGA |
| TNNT2 Forward | GGCCCCAGCCCACAT |
| TNNT2 Reverse | GGCGTCTGCTCAGTCTCA |
| THY1 Forward | GGGCTCAGGGAGGAGGATAA |
| THY1 Reverse | ATTGGTGTGAGAGTGGCAGG |
| COL1A1 Forward | TTAGCCCACGCCATTCTGAG |
| COL1A1 Reverse | GGAGAAACTCCCGTCTGCTC |
2.8. Western blot
Briefly, total cells were harvested, washed with PBS and lysed in RIPA lysis buffer (Thermo Fisher Scientific) containing a protease inhibitor cocktail (Roche Diagnostics) for 30 min on ice. Protein concentrations were measured using the Pierce BCA protein assay (Thermo Fisher Scientific). After denaturation, proteins were separated by SDS-PAGE and transferred to PVDF membranes (Merck Millipore). The membranes were blocked with 5% non-fat milk and then probed with primary antibodies against GATA4 (1:1000), HAND2 (1:1000), MEF2A+MEF2C (1:1000), TBX5 (1:1000), FSP1 (1:1000), α-SMA (1:1000), MEIS1 (1:1000), MEIS2 (1:1000), MEIS3 (1:1000), p-STAT3 (1:1000), STAT3 (1:1000), p-CHK1 (1:1000), CHK1 (1:1000), E2F1 (1:1000) and GAPDH (1:1000) overnight at 4 °C. Membranes were washed in TBST buffer on the next day and incubated with HRP-conjugated secondary antibody (Proteintech Group, Chicago, IL, USA) at room temperature for 1 h. After washing three times in TBST buffer, the protein levels were detected using SuperSignal™ West Pico Plus Chemiluminescent Substrate (Thermo Fisher Scientific).
2.9. Immunofluorescence
Cells were washed with PBS three times, fixed with 4% paraformaldehyde for 15 min, and then permeabilized with 0.15% Triton X-100 in PBS for 10 min. After blocking with 3% bovine serum albumin for 1 h, cells were incubated with primary antibodies against GATA4 (1:100), HAND2 (1:150), NKX2-5 (1:250), ISL1 (1:200), cTNT (1:150), α-actinin (1:150), tropomyosin (1:150), FSP1 (1:250), α-SMA (1:200), SM-MHC (1:100), CD31 (1:200), CD34 (1:200) and H3K4me3 (1:250) overnight at 4 °C. Cells were further incubated with goat anti-rabbit Alexa Fluor 488-conjugated antibody or goat anti-mouse Alexa Fluor 647-conjugated antibody (Abcam) for 1 h at room temperature, followed stained with DAPI for 15 min. Images were captured using Zeiss LSM 880 airy scan upright confocal microscope (Carl Zeiss, Jena, Germany) or Leica DMi8 microscope (Leica Microsystems, Wetzlar, Germany).
2.10. Flow cytometry
Before cells were stained with cardiac markers and fibroblast markers, we first detected cell viability through trypan blue staining. Cells were then washed with PBS and dissociated from culture dish with accutase (Gibco). Cells were fixed with 4% paraformaldehyde for 15 min, and then permeabilized with 0.15% Triton X-100 in PBS for 10 min. After blocking with 3% bovine serum albumin for 1 h, cells were incubated with primary antibodies against GATA4 (1:200), HAND2 (1:150), MEF2A+MEF2C (1:50), TBX5 (1:100), cTNT (1:150), tropomyosin1 (1:100), FSP1 (1:250), α-SMA (1:200) and THY1 (1:200) for 30 min at room temperature. Cells were washed with PBS twice, and then incubated with Alexa Fluor 488- or 647-conjugated antibody for 1 h at room temperature. Isotype-matched normal IgG (Abcam) served as negative controls. Cells were collected on the ImageStreamxMark II (Merck Millipore) and analyzed using ImageStream analysis software (Merck Millipore).
For cell cycle analysis, cells were washed with PBS and detached from culture dish with accutase, and then fixed with 70% ethanol overnight at 4 °C. On the next day, cells were washed with PBS twice and labeled with propidium iodide (KeyGen Biotech, Nanjing, China). Cells were then washed with PBS and filtered through a cell strainer. The cell cycle distribution was analyzed by the ImageStreamxMark II.
2.11. Statistical analysis
Statistical analyses were performed using the Student's t-test and one-way analysis of variance by GraphPad Prism software (San Diego, CA, USA). P values of <0.05 were considered statistically significant.
3. Results
3.1. Targeted activation of multiplex endogenous cardiac factors in human foreskin fibroblasts
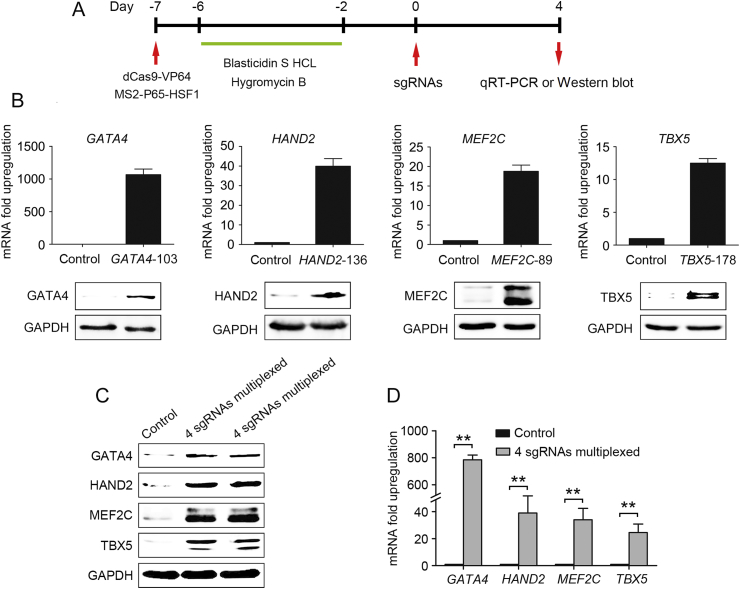
High-efficiency protein transduction of cardiac transcription factors GATA4, HAND2, MEF2C and TBX5 has been shown to effectively convert human dermal fibroblasts to cardiac progenitor cells9. Cardiac early developmental transcription factors are critical for successful reprogramming fibroblasts into iCPCs. Here we hypothesized that SAM is able to activate the expression of endogenous GHMT genes, which provides a new approach for cardiac lineage reprogramming. To select efficient sgRNAs of GATA4, HAND2, MEF2C and TBX5, we first used lentiviral delivery to constitutively express dCas9-VP64 and MS2-P65-HSF1 in HFFs at the same time. The optimal sgRNAs targeted to the proximal promoter region between −200 bp and the +1 transcription start site were selected from a pool of sgRNAs. In separate experiment, a lentivirus-based sgRNA delivery system that targeted each of the endogenous GHMT genes was transduced into HFFs stably expressing dCas9-VP64 and MS2-P65-HSF1 for 4 days (Fig. 1A) and the activation efficiency was measured by qRT-PCR and Western blot analysis. As shown in Fig. 1B, we observed a highly significant increase in both mRNA and protein levels of the corresponding endogenous genes. The original version of the dCas9-VP64 system relies on multiple sgRNAs to synergistically activate target gene expression, diminishing the utility of this epigenetic tool28, 29. The SAM system, a second-generation CRISPR/Cas9 target gene activation system, consists of multiple distinct effector domains. SAM-mediated activation largely depends on the basal expression level of each gene25, 26.
Figure 1.
SAM-mediated gene activation in human foreskin fibroblasts. (A) Schematic of the experimental design for selecting efficient sgRNAs. (B) qRT-PCR and Western blotting analysis of endogenous GHMT genes. (C) and (D) Infection with a mixture of sgRNA lentiviruses expressing GATA4, HAND2, MEF2C and TBX5 for 4 days, cells were collected for Western blotting (C) and qRT-PCR analysis (D). Error bars indicate SEM. ***P < 0.01.
Infection of HFFs stably expressing dCas9-VP64 and MS2-P65-HSF1 with a mixture of sgRNA lentiviruses GATA4 (GATA4-103), HAND2 (HAND2-136), MEF2C (MEF2C-89) and TBX5 (TBX5-178) was sufficient to induce all four endogenous gene up-regulation compared with uninfected HFFs (Fig. 1C and D). These results show that the SAM system is able to stimulate robust target gene activation.
3.2. Reprogramming human foreskin fibroblasts into iCPCs via SAM-mediated gene activation
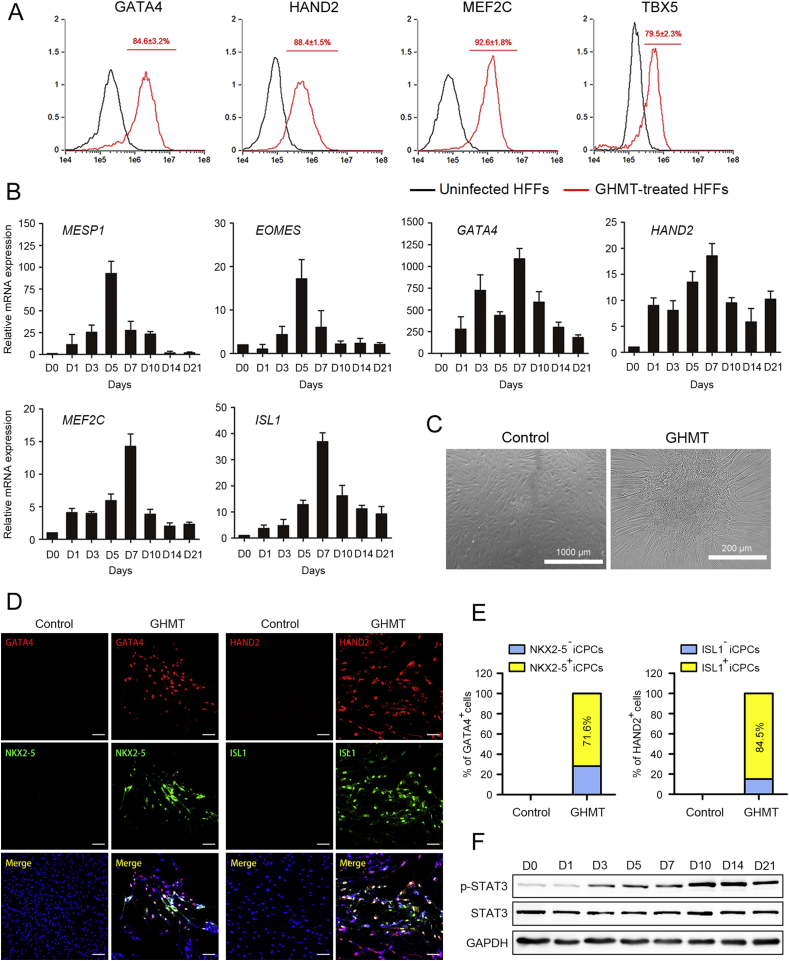
It needs new methods to generate iCPCs for treating heart failure and for studying the molecular basis of cardiac development. Two groups succeed in induction of CPCs from mouse fibroblasts through overexpression of transcription factors8 and activation of multiple cardiac signals by small molecules15. Here we wanted to test whether iCPCs could be reprogrammed from HFFs via SAM-mediated GHMT activation. Initially, we selected the same four cardiac transcription factors, and then a mixture of sgRNA lentiviruses was transduced into HFFs. Twenty-four hours post-transduction, lentiviral supernatant was replaced with induction medium. Flow cytometry analysis demonstrated that the corresponding genes were greatly activated, and more than 77% of cells were positive after 1 week of induction (Fig. 2A).
Figure 2.
Reprogramming human foreskin fibroblasts into iCPCs via SAM-mediated gene activation. (A) Flow cytometry analysis of cardiac factors. (B) qRT-PCR analysis of mesoderm markers and cardiac progenitor markers after infection with GHMT at the indicated time points. (C) Cell colonies were formed around 7–10 days under induction medium. (D) Immunofluorescence labeling of cardiac progenitor markers on day 10 of reprogramming. (E) Quantitative data of (D) showing the percentages of GATA4+ or HAND2+ cells that were positive for NKX2-5 or ISL1. (F) Western blotting analysis of p-STAT3 and STAT3 at the indicated time points. Error bars indicate SEM. Scale bars, 100 μm (D).
Cardiogenesis, a well-orchestrated process in developing embryos, involves induction of pluripotent cells to mesodermal and cardiac precursors prior to terminal differentiation30, 31, 32. We thus analyzed the expression of cardiac-specific genes during SAM-mediated conversion of HFFs into iCPCs by qRT-PCR analysis. After exposure to induction medium, EOMES and MESP1, the key mesoderm markers, were highly expressed during the early cardiac reprogramming stage. Meanwhile, the expression of cardiac progenitor markers, including GATA4, HAND2, MEF2C and ISL1, was significantly up-regulated (Fig. 2B). Infection with GHMT produced 2–3 cell colonies (per 50,000 starting cells) around 7–10 days under induction medium, accompanied with losing their parental fibroblast morphology (Fig. 2C).
Next, we performed immunofluorescence assay to detect several committed cardiac progenitor markers. The cardiac factors, including GATA4, HAND2, NKX2-5 and ISL1, were highly expressed on day 10 of reprogramming. In contrast to uninfected HFFs that were not immunolabelled with GATA4, HAND2, NKX2-5 and ISL1, iCPCs exhibited nuclear localization of these cardiac transcription factors (Fig. 2D and E). It has been shown that JAK/STAT signaling is important for normal cardiogenesis33. Here Western blot analysis exhibited a significant increase of phosphorylated STAT3 during cardiac reprogramming (Fig. 2F). These results suggest that SAM-mediated GHMT activation modulates the expression of mesoderm markers such as EOMES and MESP1 and cardiac progenitor markers such as NKX2-5 and ISL1. This is consistent with the reprogramming process for iCMs34 as well as iPSCs35.
Taken together, these data show a distinct reprogramming strategy. Although the reprogramming efficiency is lower than that in mouse fibroblasts, due to the chromatin state of human fibroblasts becomes more rigid and is hard to access, SAM-induced CPCs are cardiac mesoderm-restricted progenitors.
3.3. Fibroblast genes are gradually suppressed along the course of cardiac reprogramming
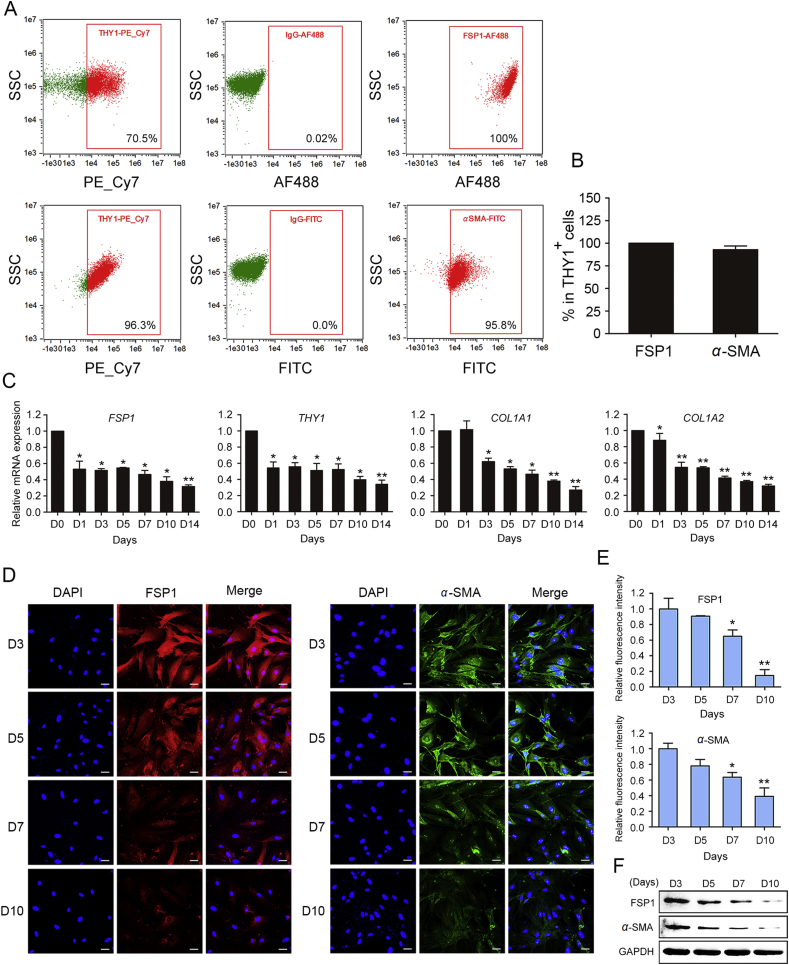
Several reports show that fibroblast genes are gradually suppressed in the reprogramming route for iCPCs8, 9 as well as iCMs3, 34. miR-133a directly targets SNAI1 3ʹUTR and inhibits the expression of fibroblast signatures such as COL1A1, COL1A2 and POSTN, which promotes the transition of fibroblasts toward a cardiomyocyte fate36. In this study we first analyzed the expression of the fibroblast marker FSP1 and the myofibroblast marker α-SMA of human foreskin fibroblasts. THY1 has been identified as a fibroblast marker. Flow cytometry analysis showed a high percentage of cells expressing FSP1 and α-SMA in THY1+ cells (Fig. 3A and B). We then performed qRT-PCR, Western blot and immunofluorescence assay to detect the expression of fibroblast genes during cardiac reprogramming. As shown in Fig. 3C–F, as reprogramming proceeded, the expression of COL1A1, COL1A2, THY1, FSP1 and α-SMA was decreased over time. These results suggest that simultaneous activation of endogenous GHMT genes initiates reprogramming toward iCPCs at the expense of the fibroblast program. However, residual fibroblast signatures may be the major roadblock for cardiac reprogramming. It needs further studies to elucidate the molecular mechanisms underlying cardiac reprogramming.
Figure 3.
Gradual down-regulation of fibroblast genes along the course of reprogramming. (A) and (B) Representative flow cytometry plots (A) and quantification (B) of FSP1+ and α-SMA+ cells in THY1+ cells, showing a high percentage of cells expressing FSP1 and α-SMA. Isotype-matched normal IgG served as negative controls. (C) qRT-PCR analysis of fibroblast genes after infection with GHMT at the indicated time points. (D) and (E) Representative staining images (D) and quantification (E) of FSP1 and α-SMA at the indicated time points. (F) Western blotting analysis of FSP1 and α-SMA at the indicated time points. Error bars indicate SEM. *P < 0.05, **P < 0.01. Scale bars, 50 μm.
3.4. iCPCs differentiate into cardiomyocytes, smooth muscle cells and endothelial cells
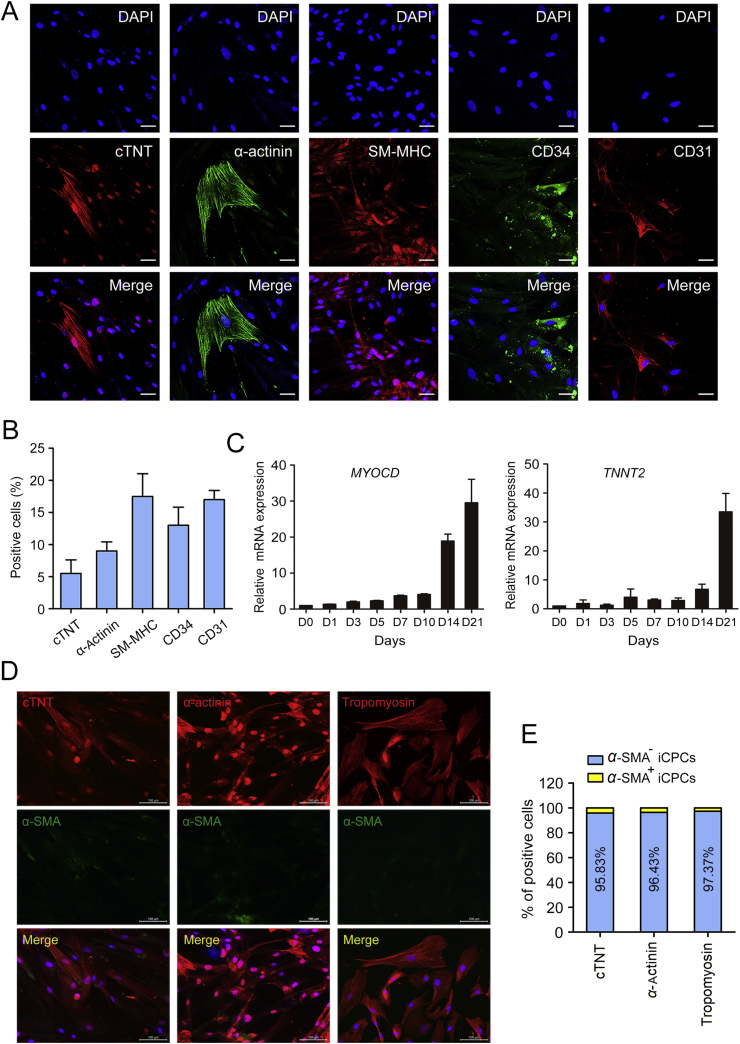
Several CPC populations have been reported from stem cell differentiation13, 14, 32 and cardiac reprogramming8, 9, 15, which can directly differentiate into three types of cardiovascular lineage cells including CMs, SMCs and ECs. Human fibroblasts have been reprogrammed previously into cardiac-like myocytes after maintenance in induction medium for 4–11 weeks37. Therefore, we tried to determine whether SAM-induced CPCs were capable of differentiating into three cardiovascular lineage cells in our study. iCPCs were cultured in induction medium for 4 weeks. We observed that differentiated cells express CM markers such as cTNT (∼6%) and α-actinin (∼9%) with resembling sarcomere striations, SMC marker SM-MHC (∼18%) and EC markers such as CD34 (∼13%) and CD31 (∼17%) (Fig. 4A and B). The expression of CM markers, including MYOCD and TNNT2, was significantly up-regulated during later stage of cardiac reprogramming by qRT-PCR analysis (Fig. 4C). We then tracked the protein expression of cTNT, α-actinin and tropomyosin by co-staining with α-SMA on day 28 of reprogramming. As shown in Fig. 4D and E, cTNT, α-actinin and tropomyosin were expressed, accompanied with down-regulation of α-SMA. Although iCPCs showed differentiation potentials in vitro, iCPC-derived CMs were functionally immature. Given that transdifferentiation is thought to be a long and stochastic process and human fibroblasts are more difficult to reprogram, generating completely reprogrammed CMs will be of great interest.
Figure 4.
Induction of CMs, SMCs and ECs during cardiac reprogramming. (A) and (B) Representative staining images (A) and quantification (B) of CM markers such as cTNT and α-actinin, SMC marker such as SM-MHC and EC markers such as CD34 and CD31 after 4 weeks of infection with GHMT. (C) qRT-PCR analysis of cardiac genes MYOCD and TNNT2 at the indicated time points. (D) Immunofluorescence labeling of cTNT, α-actinin and tropomyosin by co-staining with α-SMA after 4 weeks of infection with GHMT. (E) Quantitative data of (D) showing the percentages of cTNT+, α-actinin+ or tropomyosin+ cells that were negative for α-SMA. Error bars indicate SEM. Scale bars, 50 μm (A) and 100 μm (D).
3.5. GHMT-treated fibroblasts have fewer heterochromatin foci
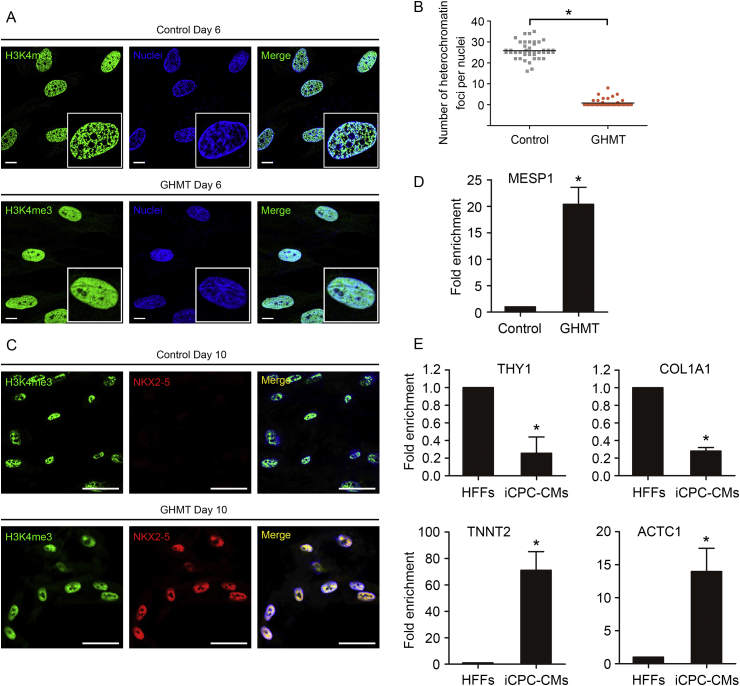
Cardiac reprogramming requires dynamic and coordinated chromatin remodeling for stage-specific gene expression. The chromatin state of human fibroblasts becomes highly compact and less amenable to reprogramming. We hypothesized that transduction of GHMT may promote dynamic transition of chromatin state from compaction to open, which could activate cardiogenic signals. H3K4me3 has been identified as an active chromatin mark that binds on promoters of cardiac genes during cardiac reprogramming34, 38 and cardiac development39. We first performed H3K4me3 immunostaining assay to analyze chromatin structure that was regulated by direct activation of endogenous GHMT genes on day 6 of reprogramming compared with uninfected HFFs. We observed a decrease in the number of heterochromatin foci in GHMT-treated HFFs, accompanied by the expression of NKX2-5 (Fig. 5A–C). SAM-mediated GHMT activation enabled the binding of β-catenin, an effector of cardiogenic WNT signaling, to the promoter region of MESP1 (Fig. 5D). Consistently, the results showed a significantly increased enrichment of H3K4me3 on TNNT2 and ACTC1, and a decreased enrichment of H3K4me3 on THY1 and COL1A1 (Fig. 5E). This indicates that SAM-mediated GHMT activation appears to impact chromatin architecture by decondensing compact chromatin regions, increasing chromatin accessibility at loci that are important for cardiogenesis in HFFs.
Figure 5.
GHMT-treated fibroblasts have fewer heterochromatin foci. (A) Immunofluorescence labeling of H3K4me3 on day 6 of reprogramming. Insets show a single cell nucleus at higher magnification. (B) The graph shows the number of heterochromatin foci per nuclei. (C) Immunofluorescence labeling of H3K4me3 by co-staining with NKX2-5. (D) ChIP analysis of β-catenin on promoter regions of MESP1 on day 6 of reprogramming. (E) ChIP analysis of H3K4me3 on promoter regions of cardiogenic and fibroblast genes on day 21 of reprogramming. Error bars indicate SEM. ∗P < 0.05. Scale bars, 10 μm.
3.6. Addition of MEIS1 to GHMT enhances cardiac reprogramming
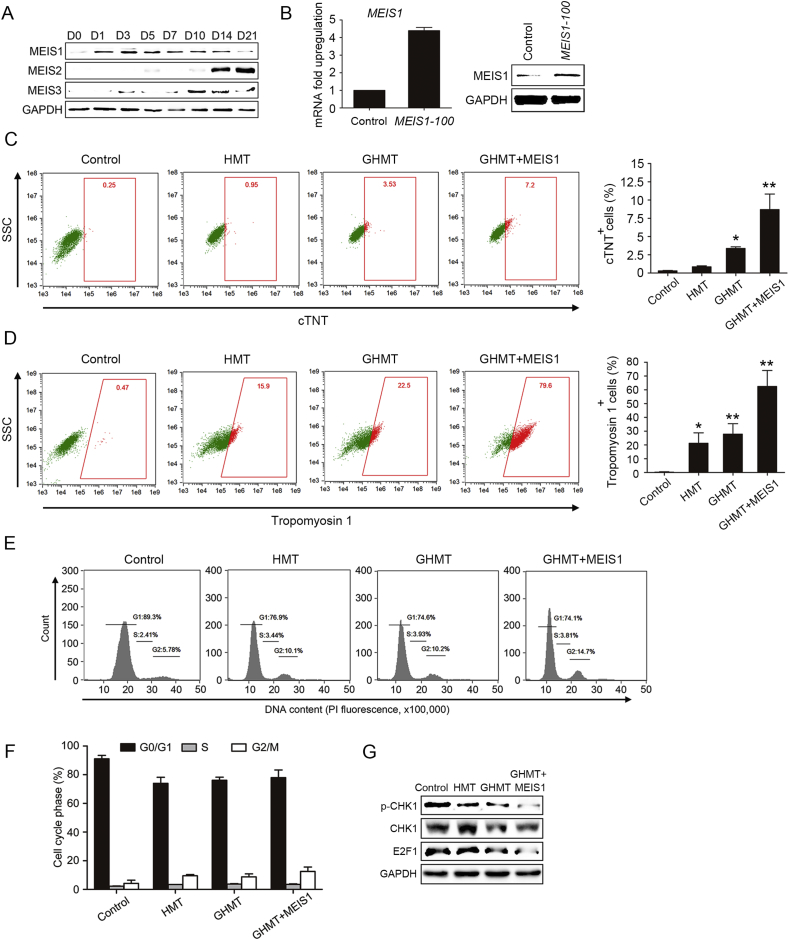
Although a combination of cardiac factors GHMT enables lineage reprogramming of fibroblasts into iCPCs, the reprogramming efficiency is low and wanted further improvement. Previous reports showed that MEIS transcription factors (MEIS1, 2 and 3), belonging to the TALE homeobox family, are critical regulators of cardiac differentiation during embryonic development40, 41. MEIS1 and GATA4 can function together to regulate post-cardiac-mesoderm differentiation31, 40. MEIS2 plays a crucial role in cardiomyocyte differentiation40, 42. These findings reveal an important role of MEIS1 in the early stages of cardiac differentiation. Therefore, MEIS1 may be another important transcription factor that can facilitate cardiac reprogramming.
Here we tested whether addition of MEIS1 to GHMT enhanced reprogramming efficiency. First of all, we analyzed the protein level of MEIS transcription factors (MEIS1, 2 and 3) that were regulated by direct activation of endogenous GHMT genes during cardiac reprogramming. MEIS1 was significantly up-regulated during the early cardiac reprogramming stage. However, MEIS2 and MEIS3 showed up-regulation during later stage of cardiac reprogramming (Fig. 6A). Next, we selected an efficient sgRNA for MEIS1. As shown in Fig. 6B, a highly significant increase in both mRNA and protein levels of MEIS1 was confirmed by qRT-PCR and western blot analysis. We then transduced HFFs with HMT and either GATA4 or GATA4 and MEIS1. The expression of endogenous cardiac markers, including cTNT and tropomyosin 1, was quantified by flow cytometry analysis after 2 weeks. HMT and GHMT could activate cTNT expression in ∼0.86% and ∼3.375% of cells. When MEIS1 was added to GHMT, the number of cTNT+ cells was significantly increased to ∼8.7%, indicating the important role of MEIS1 in cardiac reprogramming (Fig. 6C). The percentage of tropomyosin1+ cells induced by HMT and GHMT was ∼21.25% and ∼27.85%. As expected, addition of MEIS1 to GHMT significantly enhanced the percentage of tropomyosin1+ cells (∼62.95%) (Fig. 6D). These results show dynamic changes in the expression of MEIS transcription factors during cardiac reprogramming. MEIS1 in combination with GHMT could activate the cardiac gene program, which facilitates cardiac reprogramming and enhances differentiation of iCPCs into CMs.
Figure 6.
Addition of MEIS1 to GHMT enhances cardiac gene expression. (A) Western blotting analysis of MEIS1, MEIS2 and MEIS3. (B) On day 4 after infection with MEIS1, cells were collected for Western blotting and qRT-PCR analysis. (C) and (D) Representative flow cytometry plots of cTNT+ (C) and tropomyosin1+ cells (D). (E) Cells were stained with propidium iodide for DNA contents, followed by flow cytometric analysis on day 6. (F) Percentages of cells at G0/G1, S and G2/M phases were calculated based on (E). (G) Western blotting analysis of p-CHK1, CHK1 and E2F1. Error bars indicate SEM. *P < 0.05, **P < 0.01.
Recently, it has been shown that MEIS1 is a critical regulator of cardiomyocyte cell cycle43. Few studies involve in cell cycle regulation during cardiac reprogramming. Here we showed that HMT and GHMT increased the proportion of cells at the G2/M phase to ∼9.55% and ∼8.77%. Addition of MEIS1 to GHMT significantly enhanced the percentage of cells to ∼12.55% in G2/M on day 6 of reprogramming (Fig. 6E and F). To determine whether cells were arrested specifically in G2/M, we investigated the expression of CHK-1, a downstream cell cycle regulator of MEIS1, and E2F1 by Western blot analysis. Compared with uninfected HFFs, phosphorylated CHK-1 and E2F1 were significantly down-regulated on day 6. Addition of MEIS1 to GHMT showed a further decrease in the expression of p-CHK1 and E2F1 (Fig. 6G). These results suggest that cell cycle arrest in G2/M may promote cardiac reprogramming.
4. Discussion
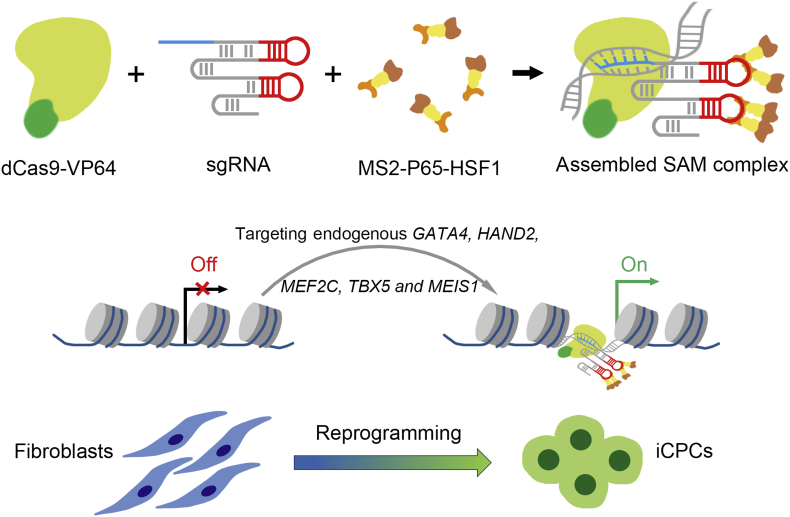
In this study, we show lineage reprogramming to induced cardiac progenitor cells through a dCas9-based transcription activation system (Fig. 7). In contrast to the forced expression of the corresponding transgenes, targeted and robust activation of endogenous cardiac factors can be achieved by precisely accessing the genome loci and rapidly remodeling epigenetic signatures. It is like a natural biological process and acts as an alternate method for lineage reprogramming. Here our data demonstrate that SAM-mediated GHMT activation can reprogram human fibroblasts toward iCPCs under induction medium. iCPCs are capable of differentiating into three cardiac lineage cells including CMs, SMs and ECs.
Figure 7.
Graphic illustration of reprogramming human foreskin fibroblasts into iCPCs via SAM-mediated gene activation. The combination of dCas9-VP64, sgRNA and MS2-P65-HSF1 is assembled SAM complex. Targeted and robust activation of endogenous genes, including GATA4, HAND2, MEF2C, TBX5 and MEIS1, can reprogram human fibroblasts toward iCPCs.
Although mouse embryonic or neonatal fibroblasts are easier to reprogram, these cells are clinically less relevant compared with human cell types. We thus utilized human foreskin fibroblasts as a starting cell source for reprogramming in this study. It is well documented that lineage reprogramming of human fibroblasts is much slower and less efficient than that in mouse fibroblasts37. Human fibroblasts, a kind of terminally differentiated cells, have highly compact chromatin state and show low chromatin accessibility, which are more challenging to reprogram. Compared with efficiencies reported for iCPC reprogramming from adult mouse fibroblasts8, the reprogramming efficiency of human fibroblasts is lower.
In this study, SAM-mediated GHMT activation was able to initiate cardiac-specific gene expression such as ISL1, NKX2-5, cTNT and α-actinin, accompanied with gradual down-regulation of fibroblast genes. Colonies appeared around 7–10 days with showing morphological changes. Likewise, SAM-induced CPCs show potentials to differentiate into CMs, SMCs and ECs. As with induced CMs generated from human fibroblasts37, iCPC-derived CMs are functionally immature, as indicated by their low organized sarcomere. These results suggest that iCPC-CMs remain in a partially reprogrammed state. Cardiac reprogramming probably requires an optimal stoichiometry and certain expression levels of reprogramming factors, which is achieved only in a small subset of fibroblasts. SAM-induced CPCs exhibit some heterogeneity and seem less differentiated. Each cardiac lineage is likely generated from diverse iCPC subpopulations.
Chromatin accessibility is an important precursor for gene expression changes during reprogramming44. Epigenetic changes at cardiac genes have been observed during direct reprogramming of fibroblasts into cardiomyocytes34, 38. In this study, H3K4me3 immunostaining assay exhibited a decrease in the number of heterochromatin foci in GHMT-treated HFFs. SAM-mediated GHMT activation reopens the closed chromatin structure of human fibroblasts for cardiogenic gene expression.
We also discover potentially new transcription factors for efficient reprogramming. It has been shown that both MEIS1 and GATA4 activate certain cardiac enhancers such as GATA5, MYOCD, HCN4 and IRX4, which regulates post-cardiac-mesoderm differentiation during embryonic development31, 40. MEIS1 disruption results in congenital heart defects45. Therefore, we hypothesized that MEIS1 may be another key cardiac reprogramming factor. Western blot analysis showed that MEIS1 is transiently upregulated during the early cardiac reprogramming stage, consistent with a role in cardiac progenitors. The percentage of cTNT+ and tropomyosin1+ cells was significantly increased when MEIS1 was added to GHMT. However, a combination of HMT was ineffective in activating cardiac gene expression in human fibroblasts. This suggest that MEIS1 and GATA4 function together to activate gene networks critical for cardiac reprogramming. Although MEIS1 in combination with GHMT showed higher efficiency of generating cTNT+ and tropomyosin1+ cells, iCPC-CMs remain in an immature state. Further studies will be necessary for increasing the maturity of iCPC-CMs. MEIS1 has been identified as an important regulator of cardiomyocyte cell cycle43. Addition of MEIS1 to GHMT could induce cell cycle arrest in G2/M on day 6 of reprogramming. Western blot analysis showed a further decrease in the expression of p-CHK1 and E2F1. Therefore, cell cycle arrest may promote cardiac reprogramming at the early stage. Collectively, our data provides novel insights for MEIS1 as a potential cardiac reprogramming factor.
iCPC reprogramming provides a potentially useful cellular resource for cardiac regenerative therapy and basic cardiovascular research. In this study we utilize the SAM system to reprogram human foreskin fibroblasts into iCPCs. The iCPCs are capable of differentiating into CMs, SMs, and ECs. However, iCPC-CMs are partially reprogrammed. Further studies are urgently needed to determine how to increase the maturity of iCPC-CMs. Additionally, we have identified MEIS1 as a potentially novel cardiac reprogramming factor. Together, our work exhibits a promising approach to reprogram human fibroblasts toward iCPCs and provides a foundation for further optimization of this process.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant numbers 81330007 and U1601227 to Xi-Yong Yu; 81700382 to Lingmin Zhang), and the Science and Technology Programs of Guangdong Province (grant numbers 2015B020225006 to Xi-Yong Yu, China).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Tanabe K., Ang C.E., Chanda S., Olmos V.H., Haag D., Levinson D.F. Transdifferentiation of human adult peripheral blood T cells into neurons. Proc Natl Acad Sci U S A. 2018;115:6470–6475. doi: 10.1073/pnas.1720273115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z., Wang L., Welch J.D., Ma H., Zhou Y., Vaseghi H.R. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature. 2017;551:100–104. doi: 10.1038/nature24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohamed T.M., Stone N.R., Berry E.C., Radzinsky E., Huang Y., Pratt K. Chemical enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation. 2017;135:978–995. doi: 10.1161/CIRCULATIONAHA.116.024692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang P., Zhang L., Gao Y., He Z., Yao D., Wu Z. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell. 2014;14:370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Luo C., Lee Q.Y., Wapinski O., Castanon R., Nery J.R., Mall M. Global DNA methylation remodeling during direct reprogramming of fibroblasts to neurons. Elife. 2019;8 doi: 10.7554/eLife.40197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lalit P.A., Salick M.R., Nelson D.O., Squirrell J.M., Shafer C.M., Patel N.G. Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell. 2016;18:354–367. doi: 10.1016/j.stem.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X.H., Li Q., Jiang L., Deng C., Liu Z., Fu Y. Generation of functional human cardiac progenitor cells by high-efficiency protein transduction. Stem Cells Transl Med. 2015;4:1415–1424. doi: 10.5966/sctm.2015-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu B., He Z.Y., You P., Han Q.W., Xiang D., Chen F. Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell Stem Cell. 2013;13:328–340. doi: 10.1016/j.stem.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Tang Y., Xiong S., Yu P., Liu F., Cheng L. Direct conversion of mouse fibroblasts into neural stem cells by chemical cocktail requires stepwise activation of growth factors and Nup210. Cell Rep. 2018;24:1355–1362. doi: 10.1016/j.celrep.2018.06.116. [DOI] [PubMed] [Google Scholar]
- 12.Batta K., Florkowska M., Kouskoff V., Lacaud G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 2014;9:1871–1884. doi: 10.1016/j.celrep.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veevers J., Farah E.N., Corselli M., Witty A.D., Palomares K., Vidal J.G. Cell-surface marker signature for enrichment of ventricular cardiomyocytes derived from human embryonic stem cells. Stem Cell Rep. 2018;11:828–841. doi: 10.1016/j.stemcr.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J.H., Protze S.I., Laksman Z., Backx P.H., Keller G.M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell. 2017;21:179–194. doi: 10.1016/j.stem.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Cao N., Huang Y., Spencer C.I., Fu J.D., Yu C. Expandable cardiovascular progenitor cells reprogrammed from fibroblasts. Cell Stem Cell. 2016;18:368–381. doi: 10.1016/j.stem.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilton I.B., D'Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E. Epigenome editing by a CRISPR/Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavez A., Scheiman J., Vora S., Pruitt B.W., Tuttle M., Iyer E P.R. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty S., Ji H., Kabadi A.M., Gersbach C.A., Christoforou N., Leong K.W. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Reports. 2014;3:940–947. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weltner J., Balboa D., Katayama S., Bespalov M., Krjutškov K., Jouhilahti E.M. Human pluripotent reprogramming with CRISPR activators. Nat Commun. 2018;9:2643. doi: 10.1038/s41467-018-05067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Black J.B., Adler A.F., Wang H.G., D'Ippolito A.M., Hutchinson H.A., Reddy T.E. Targeted epigenetic remodeling of endogenous loci by CRISPR/Cas9-based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell. 2016;19:406–414. doi: 10.1016/j.stem.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P., Chen M., Liu Y., Qi L.S., Ding S. CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell. 2018;22:252–261. doi: 10.1016/j.stem.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joung J., Konermann S., Gootenberg J.S., Abudayyeh O.O., Platt R.J., Brigham M.D. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans S.M., Yelon D., Conlon F.L., Kirby M.L. Myocardial lineage development. Circ Res. 2010;107:1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wamstad J.A., Alexander J.M., Truty R.M., Shrikumar A., Li F., Eilertson K.E. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skelton R.J.P., Kamp T.J., Elliott D.A., Ardehali R. Biomarkers of human pluripotent stem cell-derived cardiac lineages. Trends Mol Med. 2017;23:651–668. doi: 10.1016/j.molmed.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Foshay K., Rodriguez G., Hoel B., Narayan J., Gallicano G.I. JAK2/STAT3 directs cardiomyogenesis within murine embryonic stem cells in vitro. Stem Cells. 2005;23:530–543. doi: 10.1634/stemcells.2004-0293. [DOI] [PubMed] [Google Scholar]
- 34.Cao N., Huang Y., Zheng J., Spencer C.I., Zhang Y., Fu J.D. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science. 2016;352:1216–1220. doi: 10.1126/science.aaf1502. [DOI] [PubMed] [Google Scholar]
- 35.Brambrink T., Foreman R., Welstead G.G., Lengner C.J., Wernig M., Suh H. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muraoka N., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Isomi M. miR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–1581. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam Y.J., Song K., Luo X., Daniel E., Lambeth K., West K. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z., Chen O., Zheng M., Wang L., Zhou Y., Yin C. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016;16:507–518. doi: 10.1016/j.scr.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q., Jiang C., Xu J., Zhao M.T., Van Bortle K., Cheng X. Genome-wide temporal profiling of transcriptome and open chromatin of early cardiomyocyte differentiation derived from hiPSCs and hESCs. Circ Res. 2017;121:376–391. doi: 10.1161/CIRCRESAHA.116.310456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Górnikiewicz B., Ronowicz A., Krzemiński M., Sachadyn P. Changes in gene methylation patterns in neonatal murine hearts: implications for the regenerative potential. BMC Genomics. 2016;17:231. doi: 10.1186/s12864-016-2545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paige S.L., Thomas S., Stoick-Cooper C.L., Wang H., Maves L., Sandstrom R. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahmoud A.I., Kocabas F., Muralidhar S.A., Kimura W., Koura A.S., Thet S. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D., Liu J., Yang X., Zhou C., Guo J., Wu C. Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell. 2017;21:819–833. doi: 10.1016/j.stem.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Stankunas K., Shang C., Twu K.Y., Kao S.C., Jenkins N.A., Copeland N.G. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ Res. 2008;103:702–709. doi: 10.1161/CIRCRESAHA.108.175489. [DOI] [PMC free article] [PubMed] [Google Scholar]