Abstract
The progression of hyperuricemia disease is often accompanied by damage to renal function. However, there are few studies on hyperuricemia nephropathy, especially its association with intestinal flora. This study combines metabolomics and gut microbiota diversity analysis to explore metabolic changes using a rat model as well as the changes in intestinal flora composition. The results showed that amino acid metabolism was disturbed with serine, glutamate and glutamine being downregulated whilst glycine, hydroxyproline and alanine being upregulated. The combined glycine, serine and glutamate could predict hyperuricemia nephropathy with an area under the curve of 1.00. Imbalanced intestinal flora was also observed. Flavobacterium, Myroides, Corynebacterium, Alcaligenaceae, Oligella and other conditional pathogens increased significantly in the model group, while Blautia and Roseburia, the short-chain fatty acid producing bacteria, declined greatly. At phylum, family and genus levels, disordered nitrogen circulation in gut microbiota was detected. In the model group, the uric acid decomposition pathway was enhanced with reinforced urea liver-intestine circulation. The results implied that the intestinal flora play a vital role in the pathogenesis of hyperuricemia nephropathy. Hence, modulation of gut microbiota or targeting at metabolic enzymes, i.e., urease, could assist the treatment and prevention of this disease.
KEY WORDS: Hyperuricemia, Renal function, Gut microbiota, Metabolomics, Urease
Graphical abstract
The combination of metabolomics and 16S rRNA analysis revealed a panel of metabolites and bacterial strains changing with the progression of hyperuricemia nephropathy. In the case of disease, nitrogen circulation in the gut microbiota was dysregulated with enhanced urate decomposition pathway and urea liver-intestinal circulation, causing kidney damage.
1. Introduction
The prevalence of hyperuricemia (HUA) has increased over the past years. HUA is a disease with an excess of urate in the blood, which means the serum urate concentration exceeds the solubility limit of serum urate (6.8 mg/dL at 37 °C)1,2. Uric acid is mainly produced in the liver and is the end-product of dietary and endogenous purine metabolism. Around 4/5 of the total amount of uric acid in the human body is catabolized from nucleic acids in the cells while the remaining 1/5 is produced from the purine-rich foods. The uric acid produced and excreted daily by the human body is 600–700 mg, of which 2/3 is excreted through the kidney and 1/3 is via intestine. When the kidney is impaired, uric acid will be mainly eliminated by intestine as a compensation2.
Renal dysfunction is the most common complication of HUA in addition to gout3. Nephropathy induced by hyperuricemia refers to the deposition of urate crystals in the interstitial of the renal pulp and renal pyramids, with the reaction of surrounding macrophages4. A number of previous studies have investigated the relationship between serum uric acid and chronic kidney disease (CKD), and the results indicated that HUA is a risk factor for kidney disease5. It is worth noting that uric acid is also associated with other risk factors for kidney failure, such as hypertension and metabolic syndromes6,7. Possible mechanisms of uric acid-induced kidney damage include induction of tubular damage, endothelial dysfunction, oxidative stress and intrarenal inflammation8.
The human gut microbiota consist of about 1952 species of bacteria9, the number of which is more than 10 times of the number of cells in the human body. The gut microbiota not only affect the metabolism of the host, but also play a role in maintaining the homeostasis of the intestinal environment, making it a new target for personalized medication10,11. A number of studies have focused on the interactions between gut microbiota and drug interventions12, 13, 14. The small molecule compounds produced by the gut microbiota have also been discovered as an important part of uremic toxins15. In recent years, more and more studies have focused on the relation between intestinal flora and chronic kidney disease. Changes in composition and function of gut microbiota are closely related to the progression of chronic kidney disease as well16, 17, 18. It is known that gut microbiota participate in the metabolism of purine and uric acid19. For example, the gut microbiota could secrete xanthine oxidase which is involved in the oxidative metabolism of purine20. However, there hasn't been any study on the gut microbiota and hyperuricemia nephropathy.
Metabolomics is a study of entire metabolome presenting in cells, tissues, and body fluids under a given set of conditions21. It is the youngest sibling in the family of “-omics” techniques whilst expand rapidly as a science for the understanding of systems biology22,23. It has been successfully applied in drug toxicity prediction, biomarker discovery as well as disease pathology24, 25, 26. Metabolomics has also emerged as an indispensable approach to explore the functional status of host–microbial relationships in a biological specimen27,28. Metabolic profiling of host bio-fluids or tissues for small molecules generated by gut microbiota can provide deep insights on the impact of these gut microbiota on the health/disease status of host. In other word, metabolomics makes it possible to know “who is active” other than “who is there”28,29.
Here, in the present study, we aimed to discover blood metabolites and gut microbiota associated with nephropathy induced by hyperuricemia in a rat model. The abundance of metabolites was measured using gas chromatography–mass spectroscopy (GC–MS) with untargeted metabolomics approach and gut microbiota was analysed with 16S ribosomal RNA (16S rRNA). The selected metabolites and gut microbiota were then compared with kidney function parameters.
2. Materials and methods
2.1. Animals
Male Sprague–Dawley (SD) rats (180–200 g) were provided by the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences (Beijing, China). The animals were placed in the room with a temperature of 22–24 °C and a humidity of 45% with a 12-h day/night cycle (lighting time 8:00–20:00). Rats were fasted for 12 h before the experiment and were free to water. The experiment was carried out in strict accordance with the guidelines and ethical guidelines for experimental animals, and was approved by the Animal Ethics Committee of the Experimental Animal Institute of the Chinese Academy of Medical Sciences and Peking Union Medical College.
2.2. Chemical reagents
High performance liquid chromatography (HPLC) grade acetonitrile and methanol were purchased from Thermo Fisher Scientific Co., Ltd. (Fair Lawn, NJ, USA). Ultra-pure water was obtained from Hangzhou Wahaha Group Co., Ltd. (Hangzhou, China), N,O-bis(trimethylsilyl)trifluoro-acetamide (BSTFA) with 1% trimethylchlorosilane (TMCS), O-methoxyamine-HCl (MOX), succinic-d4 acid were purchased from Sigma–Aldrich (NJ, USA). Adenine, 4% tissue cell fixative, yeast powder, urease activity test kit, hematoxylin-eosin staining (HE) staining kit, Masson staining kit and standards for creatinine, uric acid, hypoxanthine, xanthine, urea, etc. were purchased from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China) and the purity of the above standands are all greater than 98%. The urea nitrogen test kit was purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Other chromatographic reagents were obtained from domestic reagent companies.
2.3. Group information and sample collection
Twenty 6-week-old SD rats were randomly divided into 2 groups, namely control group and model group. The model group was fed with a special folder containing 10% yeast and 0.15% adenine whereas normal group received normal food.
After 6 weeks of modelling, animal faeces were collected and analysed by 16S rRNA gene sequencing technology. Plasma and urine samples were collected for metabolomics analysis. After the rats were anesthetized, the left kidneys were accurately weighed on an electronic balance, and the left kidney weight index was calculated. The right kidney was fixed in 4% tissue cell fixative (4% paraformaldehyde) for 24 h, dehydrated by automatic dehydrator for 16 h, and then routinely embedded in a paraffin embedding machine for preparation of kidney tissue sections. Kidney tissue sections were then subjected to HE staining and Masson staining.
2.3.1. HE staining
The pre-prepared paraffin sections were de-paraffinized with xylene (I) and (II) respectively for 5 min, then treated with a gradient of ethanol (anhydrous ethanol for 5 min, 95% ethanol for 2 min, 80% ethanol for 2 min, 70% ethanol for 2 min), followed by distilled water for 2 min. The de-paraffinized tissue section was dyed with hematoxylin dyeing solution for 20 min, and then washed with tap water. Next, differentiation solution was applied for 30 s and the tissue samples were soaked in water for 15 min. Then samples were stained with eosin dyeing solution for 30 s and rinsed under running water. After the samples were immersed in water for 5 min, ethanol gradient dehydration was performed and followed by xylene transparent and sealed with neutral gum. Finally, an optical microscope was used to observe and photograph.
2.3.2. Masson staining
The experimental procedure was as following: first, the paraffin sections were dewaxed using xylene, and then dyed with Weigert iron hematoxylin staining solution for staining 5 min. The acidic ethanol differentiation solution was used for differentiation for 5 s, and washed with water. Masson blue solution was applied to return to blue for 3 min and then the samples were rinsed with distilled water for 1 min. After this, the samples were stained with Lichunhong Magenta staining solution for 5 min, and then washed with a weak acid working solution (mixed solution of distilled water and weak acid solution with a ratio of 2:1) for 1 min, phosphomolybdic acid solution for 2 min and weak acid working solution again for 1 min. Samples were then stained with aniline blue staining solution for 2 min, and washed with weak acid working solution for 1 min. Then samples were dehydrated with 95% ethanol was rapidly dehydrated and then absolute ethanol for 3 times. After xylene transparency for 3 times, the samples were sealed with neutral gum. Finally, an optical microscope was used to observe and photograph.
2.4. Sample preparation for metabolomics analysis
Sample preparation was slightly modified based on our previous protocol30,31. Briefly, 300 μL of methanol containing 10 μg/mL of succinic-d4 acid as internal standard was added to 100 μL plasma/urine for protein precipitation. After centrifuging for 10 min at 10,000×g at 4 °C, 100 μL of supernatant was transferred to a new vial. Then all the samples were dried down under a stream of N2. Subsequently, 50 μL of O-methoxyamine-HCl (MOX) in pyridine (20 mg/mL, Sigma–Aldrich) was added to the residue and kept at 70 °C for 30 min. Samples were then dried down again and reconstituted in a 1:1 (v/v) solution of acetonitrile and the derivatizing agent BSTFA (1% tetramethyl silane, Sigma–Aldrich). The derivatization process was operated at 70 °C for 1 h. Samples were centrifuged again and pipetted to new HPLC vials with inserts.
2.5. GC–MS analysis
Metabolomics analysis was performed on a Shimadzu GC-2010 Plus gas chromatography system coupled to a GCMS-QP2020 SE single quadruple mass spectrometer (Shimadzu, Kyoto, Japan). The conditions were adapted from our previous protocol30,31. One microliter of sample was injected in the split mode with a split ratio of 1:100. The initial oven temperature was maintained at 60 °C for 5 min, and then raised to 300 °C at a rate of 10 °C/min, holding for 5 min. The temperatures for injection, ion source and interface were set at 280, 200 and 300 °C, respectively. Mass data were collected in SCAN mode (m/z 50–600 Da) with an event time 0.2 s. Quality control (QC) samples made from pooled corresponding samples were injected periodically.
2.6. Data pre-processing and analysis
Metabolomics data preprocessing was carried out within “XCMS” package in “R” for peak picking and retention time correction. The pre-processed data were then normalized separately in R which resulted in the best clustering of QC samples in principal component analysis (PCA) score plot and features with relative standard deviation (RSD) < 30% in QC samples were included for following multivariate analysis. Annotation of significantly changed variables was performed by comparing MS fragmentation patterns of detected metabolites with the spectra in National Institute of Standards and Technology (NIST) database32 (Supporting Information Fig. S1).
Different statistical methods were applied to mine features that could distinguish model and control rats. Orthogonal partial least squares-discriminant analysis (OPLS-DA) with corresponding S-plot modelling was accomplished in SIMCA version 14 (MKS Umetrics AB, Sweden). Selection of discriminating features were based on covariance p[1] and correlation p(corr) values in the S-pot (p[1]>0.10, p(corr) > 0.80 and p[1]<–0.15, p(corr)<–0.80 for plasma, p[1]>0.05, p(corr) > 0.80 and p[1]<–0.1, p(corr)<–0.80 for urine). Volcano plot was prepared in R with thresholds of P value < 0.05, fold change >1.2. Interactive cloud plot prepared in “XCMS” online was used as an additional way to display significant features. Overlapping features from these three platforms were chosen for further investigation. Interesting features were measured again in the raw data. Peak areas were normalized to internal standard, and results were expressed as normalised area.
All statistical tests including t-test, Mann–Whitney test and Spearman's correlation analysis were performed in “R”. Receiver operating characteristic (ROC) curve was plotted to compare the prediction ability of selected metabolites and was built in “R” with “pROC” package (with a suitable threshold).
2.7. Targeted analysis of uric acid pathway in faeces
Quantitative determination of faecal hypoxanthine, xanthine, uric acid, allantoin and urea in model group and normal group was performed by high performance liquid chromatography–triple quadrupole mass spectrometry (HPLC–MS/MS 8060, Shimadzu). Chromatographic separation was carried out using an Alltima C18 (150 mm × 4.6 mm, 5 μm) column (Hichrom, Lutterworth, UK). The linear gradient elution flow rate was 0.4 mL/min, with water as mobile phase A, and methanol as mobile phase B: 1 min (95% A and 5% B), 2 min (80% A and 20% B), 4 min (5% A and 95% B), 7.00 min (5% A and 95% B), 7.01 min(5% A and 95% B), 13 min stop. The auto sampler temperature was set to 4 °C and column oven temperature was 40 °C. The mass spectrometer was operated in a multiple reaction monitoring (MRM) mode. The following product ion precursors were monitored: urea (m/z) was 61.10 [M+H]+→44.10, uric acid (m/z) was 167.25 [M–H]–→124.15, hypoxanthine (m/z) was 135.35 [M–H]–→92.10, xanthine (m/z) was 151.20 [M–H]–→108.05, and allantoin (m/z) was 157.00 [M–H]–→97.10. The atomizing gas flow rate, drying gas flow rate and heating gas flow rate were 2.8, 10, and 10 L/min respectively.
2.8. Microbial diversity analysis
At the end of the 6th week of model establishment, the fecal samples of rats within 24 h were collected in a metabolic cage, which can complete separate the faeces and urine. Then the samples were freezed under −80 °C.
The V3–V4 region of the bacteria's 16S rRNA gene was amplified with primers 338F and 806R. AxyPrep DNA GelExtraction Kit (Axygen Biosciences, Union City, CA, USA) was used to purify the amplicons, followed by quantification using QuantiFluor-ST (Promega, Madison, WI, USA). Illumina MiSeq instrument (Illumina, San Diego, CA, USA) was used for sequencing.
The 16S rRNA sequencing data were analysed with Quantitative Insights Into Microbial Ecology platform (V.1.9.1). Operational taxonomic units (OTUs) with similarity over 97% were selected for taxonomy identification with Greengenes database (V.13.8)33.
3. Results
3.1. Animal model evaluation
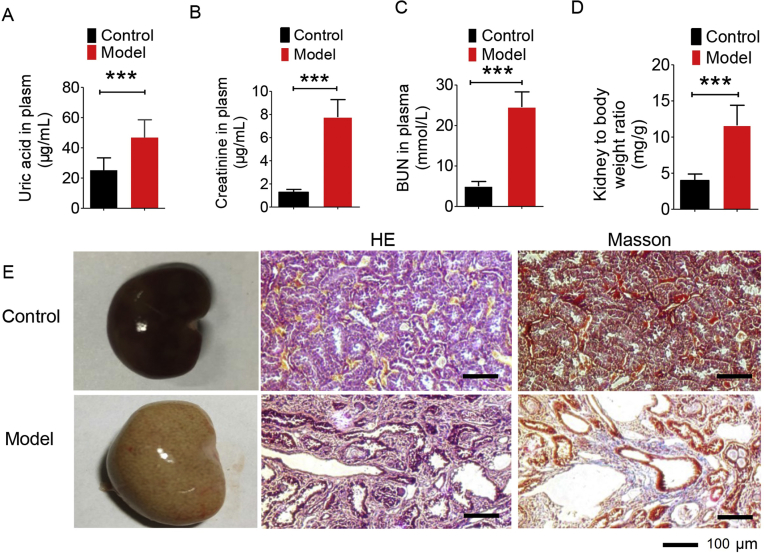
The model of nephropathy induced by hyperuricemia was established by giving the rats a diet containing 10% yeast powder and 0.15% adenine for 6 weeks. In Fig. 1A–C, it can be seen that the levels of plasma uric acid, creatinine and BUN increased significantly. At the end of modelling, the rats were sacrificed to obtain the left kidney and the left kidney weight index was calculated (weight of left kidney/SD rat weight, mg/g). The result is displayed in Fig. 1D, showing the left kidney index in the model group is statistically higher than that in the normal group. Both of them indicate a damaged kidney. In Fig. 1E, the kidney of the normal group was light brown, while the kidney of the model group was pale and significantly increased with white spots on the surface. In Fig. 1E, HE staining results show that the normal rats had normal renal structure and no urate crystallization while the uric acid crystals in the lumen and interstitial of the renal cortex of the model group were observed in a needle-like arrangement. Moreover, the renal tubules were vacuolar degenerated, and the interstitial was significantly broadened. Masson results exhibited that the renal interstitial was broadened and fibrosis was generated in SD rats.
Figure 1.
Biological index for model establishment. (A) The level of uric acid in normal group and model group; (B) the level of creatinine content in normal group and model group; (C) urea nitrogen content in normal group and model group; (D) specific kidney mass index in normal group and model group; (E) normal kidney morphology, H&E pathological staining and Masson staining. Data are expressed as mean ± SD (n = 8–9) and data were measured at week 6 (***P < 0.001).
3.2. Multivariate model and feature selection from plasma and urine samples
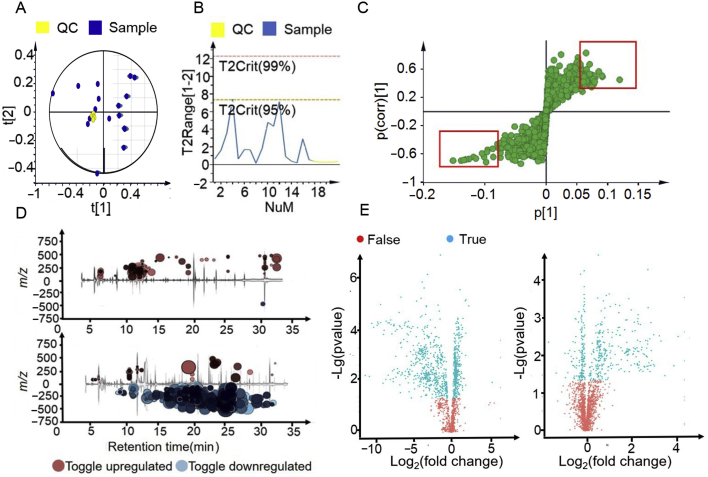
After the model was successfully established, plasma and urine samples were analysed with untargeted metabolomics approach. The pre-processed dataset was first inspected for detection of outlier(s) and the reproducibility of our analysis. In Fig. 2A and B, PCA score plot and Hotelling's T2 plot (observations exceed red dot line are strong outliers) show there is no obvious outliers and all QC samples cluster near the origin in the PCA plot indicating good reproducibility.
Figure 2.
Feature selection based on multivariate analysis. (A) An example of PCA score plot summarising the distribution of QCs and samples; (B) corresponding Hotelling's T2 plot showing strong and moderate outliers; (C) an example of S-plot based on covariant and correlation; (D) interactive cloud plot showing the change of metabolites, with upper being plasma sample and lower being urine sample; (E) volcano plots showing both P value and fold change of metabolites, with left being urine sample and right being plasma sample.
S-plot (Fig. 2C) derived from the OPLS-DA model (Supporting Information Fig. S2, R2Y(cum) = 0.941, Q2(cum) = 0.85 for plasma, R2Y(cum) = 0.988, Q2(cum) = 0.84 for urine), volcano plot and interactive cloud plot (Fig. 2D and E) were employed to generate a list of features of interest that were of importance for group discrimination. Features were selected based on the criteria stated in the method part.
3.3. Univariate analysis of selected features across groups
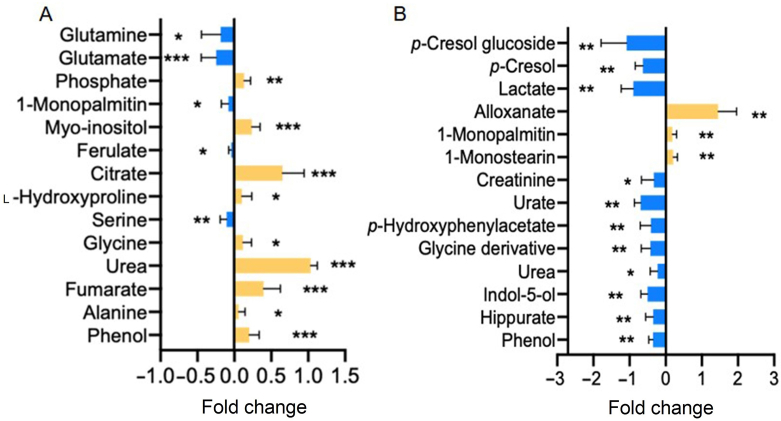
Statistical data analysis was then performed on the selected features to compare how the metabolic features vary across groups. Of these, 12 metabolites in plasma exhibited significant differences between these two phenotypes with 9 metabolites being upregulated in model group whilst 3 being downregulated (Fig. 3A and Supporting Information Table S1). As for urine samples, the levels of 3 metabolites were increased while 11 metabolites went down (Fig. 3B and Table S1).
Figure 3.
An overview of plasma (A) and urine (B) metabolic features showing statistical difference between control and model groups. Data are expressed as mean ± SD with blue representing for downregulation in model group and yellow for upregulation (n = 9 for plasma and n = 6 for urine, *P < 0.05, **P < 0.01, ***P < 0.001).
3.4. Association between plasma amino acids and hyperuricemia
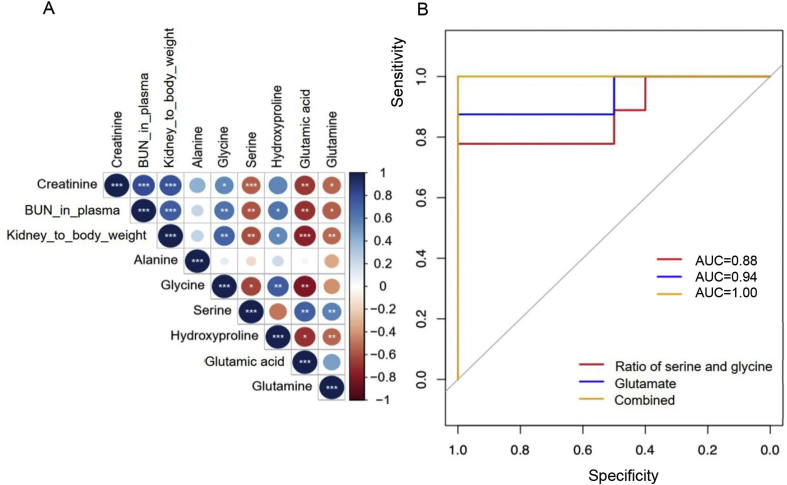
As is illustrated in Fig. 3A, model group had lower plasma concentration of serine glutamate and glutamine with a fold change of −1.22 (P < 0.01), −1.80 (P < 0.001) and −1.61 (P < 0.05) respectively, whilst elevated levels of glycine (FC = 1.38, P < 0.05), hydroxyproline (FC = 1.30, P < 0.05) and alanine (FC = 1.22, P < 0.05). In order to investigate whether these molecules have any association with hyperuricemia, they then underwent a receiver operating characteristic (ROC) analysis and Spearman's correlation analysis. The accuracy, area under the curve (AUC), sensitivity and specificity for individual metabolite in predicting hyperuricemia were listed in Table 1 and correlation parameters were displayed in Fig. 4A. Among the 6 amino acids, glutamate turned out to be the best discriminator with a high prediction ability (AUC = 0.94) and significant correlation with kidney function parameters, followed by serine, glycine and glutamine as potential biomarkers. Since glycine and serine could be converted to each other, we then look into the association between hyperuricemia nephropathy and the ratio of glycine and serine. Data showed that the ratio of serine and glycine improved the prediction ability of hyperuricemia nephropathy with AUC of 0.88. When glutamate and serine to glycine ratio were combined, it returned a ROC curve with all the three parameters (AUC, sensitivity and specificity) equalling to 1, suggesting a perfect biomarker for hyperuricemia induced nephropathy (Fig. 4B).
Table 1.
ROC analysis of amino acids for the prediction of hyperuricemia.
| Metabolite | AUC | Sensitivity | Specificity |
|---|---|---|---|
| Glycine | 0.84 | 0.78 | 0.90 |
| Serine | 0.86 | 0.78 | 0.80 |
| Alanine | 0.77 | 0.67 | 0.78 |
| Glutamine | 0.84 | 0.75 | 0.90 |
| Glutamic acid | 0.94 | 0.88 | 1 |
| Hydroxyproline | 0.63 | 0.58 | 0.60 |
| Ratio of serine and glycine (S/G) | 0.88 | 0.88 | 0.80 |
| Combination of glutamate and S/G | 1.00 | 1.00 | 1.00 |
Figure 4.
(A) Correlation plot between plasma amino acids and biological index; (B) ROC curve prediction of hyperuricemia nephropathy based on ratio of serine and glycine, glutamate, and the combination of them (*P < 0.05, *P < 0.01, ***P < 0.001).
3.5. Gut microbiotal derived metabolic profile in urine
Among the 14 perturbed metabolites in urine, phenol, p-cresol, p-hydroxyphenylacetic acid and indol-5-ol are known to be generated by gut microbiota. From Fig. 3B, it could be observed that all of the four molecules present a declined trend in model group with a fold change of −2.3 (P < 0.01), −4.73 (P < 0.01), −2.28 (P < 0.01), and −3.08 (P < 0.01), respectively. Interestingly, the plasma level of phenol was significantly elevated in mode group (Fig. 3A), which is as expected. Since the kidney doesn't function well, phenols cannot be excreted into the urine, leading to an accumulation in plasma. However, the plasma levels of the other three metabolites were below the limit of detection in this study.
3.6. Microbial diversity analysis
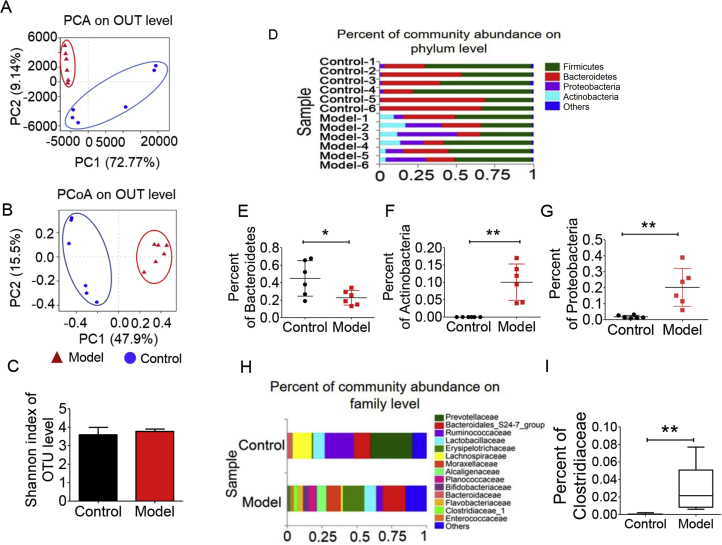
The imbalanced levels of urine metabolites mentioned above leads us to the investigation of the change of gut microbiota. Faecal samples from the normal group and the model group were collected for the analysis of intestinal flora composition. Barcode pyrosequencing analysis of the V3 and V4 regions by 16s rRNA gene sequencing. PCA and PCoA results (Fig. 5A and B) based on 981 OUT demonstrate a great difference between model and control groups in terms of gut microbiota composition. However, Shannon index in Fig. 5C showed there was no observed difference between control and model groups. In Fig. 5D and H, there was also a great variation between model and control groups at both phylum and family levels. The Actinobacteria, Proteobacteria and Clostridiaceae in the model group were significantly increased compared with the normal group (Fig. 5E and F); while the Bacteroidetes in the model group were statistically reduced (Fig. 5G). At family level, Clostridiaceae was dramatically elevated in the model group as is shown in Fig. 5I.
Figure 5.
Gut microbiota diversity analysis between model and control groups (n = 6). (A) PCA score plot; (B) PCoA score plot; (C) Shannon index of each groups on OUT level; (D) community analysis based on phylum level; (E) abundance of Bacteroidetes; (F) abundance of Actinobacteria; (G) abundance of Proteobacteria; (H) community analysis based on family level; (I) abundance of Clostridiaceae. (*P < 0.05, **P < 0.01).
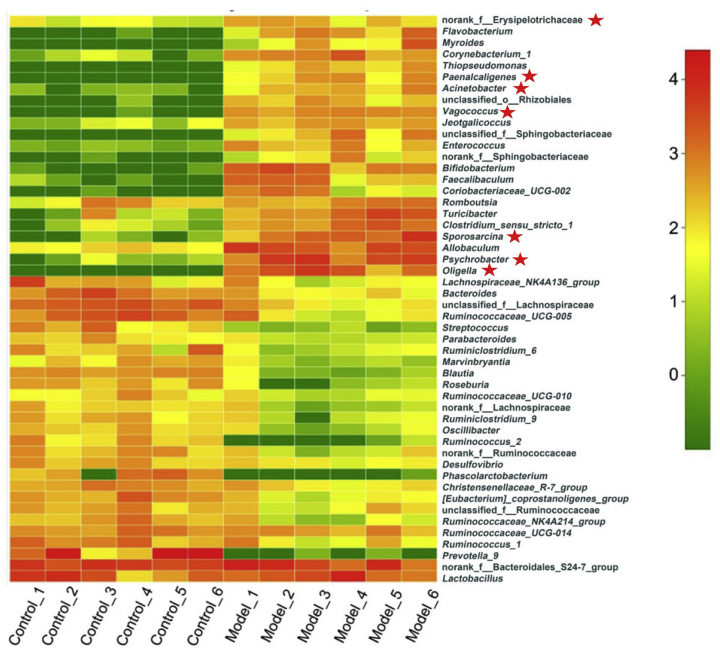
Heat map of the top 50 genus (Fig. 6 and Supporting Information Table S2) clearly illustrates that the gut microbiota composition in model group was significantly changed compared with the normal group. Flavobacterium, Myroides, Corynebacterium, Alcaligenaceae, Oligella and other conditional pathogens increased significantly in the model group, while Blautia, Roseburia and other strains that are beneficial to intestinal microbial ecological restoration were greatly reduced34, 35, 36, 37.
Figure 6.
Heat map showing top 50 genus between model and control groups (n = 6). The bacterial strains with star belong to Actinobacteria or Proteobacteria.
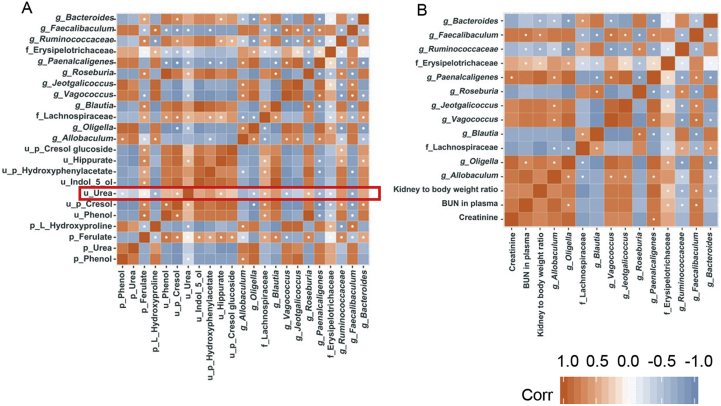
Then we performed t-test among the top 50 genus with false discovery control. Any genus with corrected P value below 0.05 was kept for correlation analysis with biological index as well as intestinal uraemic toxins. Results are shown in Fig. 7. Roseburia, Lachnospiraceae, Blautia and Ruminococcaceae all have a positive correlation with uraemic toxins in urine and negative correlation with uraemic toxins in plasma (Fig. 7A). Erysipelotrichaceae, Vagococcus, Paenalcaligenes and Oligella presented an opposite trend with the four bacteria mentioned previously. The correlation between gut microbiota and biological index is demonstrated in Fig. 7B. Faecalibaculum and Erysipelotrichaceae correlated positively with both BUN in plasma and kidney to body weight ratio while Paenalcaligenes and Oligell only showed positive correlation with plasma creatinine or BUN in plasma. As for Ruminococcacea and Bacteroides, they exhibited negative correlations with biological parameters.
Figure 7.
(A) Correlation analysis between gut microbiota and uraemic toxins; (B) correlation analysis between gut microbiota and biological index.
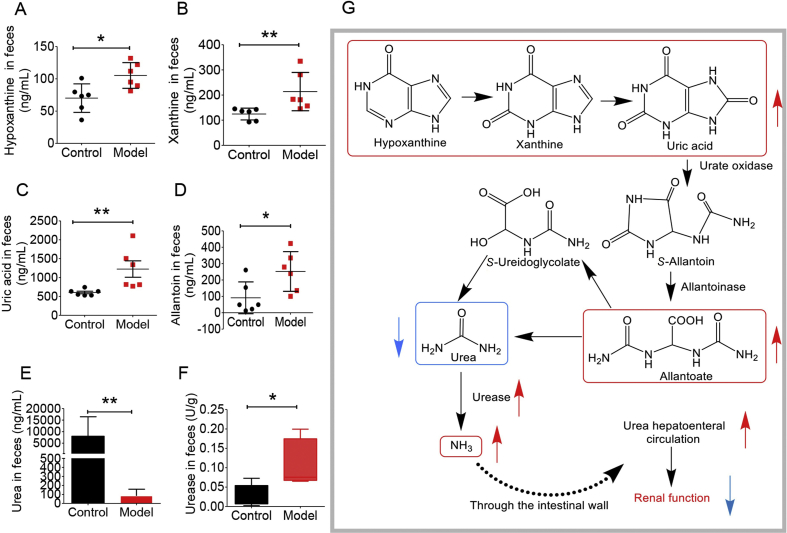
3.7. Targeted analysis of uric acid pathway in faeces
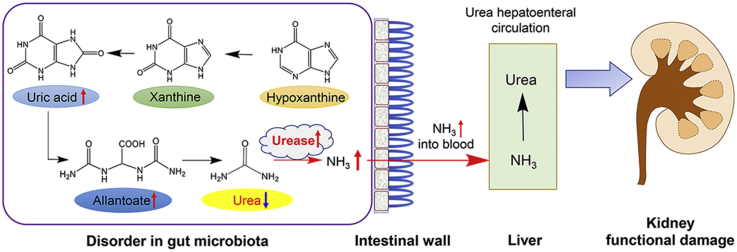
Since plasma urea presented an elevated value in the model group (Fig. 3A) and the 16S rRNA result of the intestinal flora showed that the abundance of uric acid decomposing strains increased with an abnormal metabolism of nitrogen (Fig. 5I), we then performed targeted analysis on urea–urate pathway in stool samples, including uric acid, hypoxanthine, xanthine, allantoin and urea. Hypoxanthine and xanthine are the main precursors of uric acid, and allantoin and urea are uric acid catabolic products. As is shown in Fig. 8A–E, model group presented significantly higher levels of hypoxanthine, xanthine, allantoin, and uric acid than the normal group whilst a decreased level of urea, suggesting a perturbation in the intestinal flora of SD rats with uric acid nephropathy. Next, we used the kit to detect the amount of urease in the stool sample. As is shown in Fig. 8F, the urease content in the stool sample was significantly higher than that in the normal group. The real-time quantitative polymerase chain reaction (qPCR) results of the ureC gene in the model group and the normal group showed that the ureC gene expression was significantly increased in the model group compared with the normal group, which was consistent with the result of enzyme, and the bacteria containing the ureC gene were significantly enriched in the model group (Supporting Information Fig. S3 and Table S3). Fig. 8G summarizes the potential mechanism of uric acid degradation by gut microbiota.
Figure 8.
Targeted analysis of uric acid synthesis pathway (n = 6). (A) hypoxanthine content in SD rats faeces; (B) xanthine content in SD rats faeces; (C) uric acid content in SD rats faeces; (D) allantoin content in SD rats faeces; (E) urea content in SD rats faeces; (F) urease in SD rats faeces G potential molecular mechanism (*P < 0.05, **P < 0.01).
4. Discussion
Nephropathy induced by hyperuricemia is a common complication of hyperuricemia. The main pathological basis is the deposition of urate crystals in the kidney, accompanied by inflammatory reactions to impair the renal function. In this study, the experimental model was established based on the administration of a large amount of urate precursors (including yeast powder and adenine) to SD rats with the diet to make urate accumulate in the kidney and then damage the nephron. This model which induced by yeast powder and adenine has been reported in the literature38,39, and was optimized in our laboratory. At present, this model is simple to operate and repeat.
Imbalanced metabolism of amino acids has been reported to be associated with chronic kidney disease. For example, glutamate, isoleucine and valine presented altered levels in unilateral ureteral obstruction (UUO) rat. Tryptophan was related to tubulointerstitial fibrosis in UUO40. Here, in the present study, we also discovered a panel of amino acids, among which, glutamate, glycine and serine presented strong relation to nephropathy induced by hyperuricemia and showed good prediction ability of this disease with high sensitivity and specificity.
Glutamine is the precursor of glutamate, which is of vital physiological importance to many cells41. Both of them play an essential role in the regulation of acid-base homeostasis with glutamine being the key donor of NH3 in the kidney. Besides, glutamine also acts as an essential nutrient for the maintenance of kidney functions while glutamate is a key intermediate involved in the pathways that are related to energy production42, 43, 44. The uptake of glutamine from the circulating blood of kidney is largely affected by the state of the acid-base45. Research has shown that during acidosis, kidney extraction of glutamine would be enhanced in order to generate ammonium ions which could propel the excretion of acids46, 47, 48. In our present study, the decreased levels of glutamine and glutamate in model group might be associated with the abnormal function of kidney. It is very likely that under the hyperuricemia condition, in which there is excess of blood uric acid, kidney takes up more glutamine and glutamate in order to compensate for the excess acid, leading to the declined plasma levels of glutamine and glutamate. Besides, glutamine was also shown to be able to alter the gut microbiota. Oral supplement of glutamine for a short period could reduce the ratio of Firmicutes to Bacteroidetes40. Hence, the decreased level of glutamine might also play a role in the disturbed microbiota composition.
Serine is generally regarded as a nonessential amino acid in humans, but metabolically, it is of vital importance in several cellular processes and has received increasing attention in the last few years49,50. Kidney is well known as the main site of serine production and one major pathway is the conversion from glycine, making glycine the predominant precursor for serine synthesis45,51. Normally, kidney can take up glycine and release serine52. From Fig. 3A, it could be noted that glycine was more abundant in model group while serine displayed a reduced level in model group. Thus, it could be speculated that hyperuricemia nephropathy might induce the alteration in the conversion of glycine to serine in the kidney. There are two enzymes involved in this pathway, namely glycine cleavage enzyme and serine hydroxymethyltransferase (SHMT). Pyridoxal-5ʹ-phosphate (PLP), which is the active form of vitamin, acts as a coenzyme for SHMT. It has been suggested that vitamin B6 deficiency would result in a reduced activity of hepatic SHMT53. Interestingly, vitamin deficiency is common in chronic renal dysfunction, and a number of studies have observed a reduced plasma level of pyridoxal 5ʹ-phosphate in kidney related disease, such as uremia54,55. Hence, it is very possible that under the hyperuricemia nephropathy status, the activity SHMT is inhibited, thus leading to an increased plasma level of glycine and decreased level of serine.
Additionally, renal interstitial fibrosis is considered as an important pathological feature in chronic kidney disease, which is the result of imbalanced process of collagen synthesis and collagen degradation56,57. Collagen is the most abundant structural protein in mammals, composed of repeated units of triple helix. More than half of total amino acids in collagen is consist of glycine, hydroxyproline and proline with glycine alone contribute to 30%58, 59, 60. Perturbations in serine, glycine and hydroxyproline metabolism are reported to be related with renal and liver fibrosis49,61. From Fig. 1E, an obvious renal fibrosis in the model group was noticed. Therefore, the increased level of glycine and hydroxyproline might partially be the result of an abnormal metabolism of collagen under the diseased condition.
Moreover, research demonstrated that gut microbiota could also cause abnormal metabolism of amino acids. Glycine and tryptophan degradation could be modulated by altered microbial genera in type 2 diabetes gut microbiome62, indicating that the disturbed microbiota observed from 16S rRNA analysis might also contribute to the changed levels of amino acids.
Phenol, p-cresol, p-hydroxyphenylacetic acid and indol-5-ol are produced metabolically from precursors originating from intestinal microbial fermentation (in casu tyrosine and tryptophan)63,64. The increased blood levels of phenols and p-cresol have been associated with end-stage renal disease63,65. In addition, phenols, p-cresol and indols are protein-bound solutes and belong to bioactive uraemic toxins with the first two being regarded as the biomarkers of a disturbed gut environment and abnormal level of p-hydroxyphenylacetic acid has been linked with irregular growth of Clostridia species66,67. The imbalanced levels of these metabolites might indicate a disrupted intestinal flora under the condition of nephropathy induced by hyperuricemia.
The 16S rRNA gene sequencing results revealed that there was a significant difference between model and control groups in terms of intestinal flora. The pathogens and opportunistic pathogens in the model group were more abundant than that in the normal group. Meanwhile, the probiotics in the model group were significantly eliminated. It is worth noting that the intestinal flora of the model group was able to respond to the abnormal nitrogen circulation. More specifically, in the model group, nitrogen-fixing strains were elevated significantly, suggesting that it may have a stronger nitrogen-fixing capacity. Proteobacteria is a major phylum of Gram-negative bacteria, including a wide variety of pathogens and opportunistic pathogens. It is reported that the increase in the abundance of Proteobacteria phylum is linked with the metabolism of nitrogen in the intestinal flora as these microorganisms contain genes that could promote nitrogen use68. Moreover, Proteobacteria strains are also able to increase the nitrogen fixation capacity of the intestinal flora of the model group68. A number of studies have proved that many strains in the family of Clostridiaceae have the ability to degrade uric acid69,70. However, at genus level, except for pathogens and opportunistic pathogens, the four SCFA-generating bacteria declined, such as Roseburia, Lachnospiraceae, Blautia and Ruminococcaceae. Supplementation with short-chain fatty acids can significantly improve renal function71. Hence, the reduction of the above genus might be associated with dysfunction of kidney.
For the correlation analysis, Roseburia, Lachnospiraceae, Blautia, and Ruminococcaceae all exhibited strong correlations with uraemic toxins. These four bacteria are short-chain fatty acid producing bacteria, which were depleted in the model group. This result is in accordance with literature, demonstrating that short chain fatty acids might be beneficial to kidney function. For example, Blautia genus is associated with the increased survival rate in patients undergoing allogeneic transplantation35 Lachnospiraceae, Ruminococcaceae and Roseburia could module the immune system by generating butyric acid. Meanwhile, Roseburia was shown to be related with the improvement of renal function17. On the other hand, Vagococcus, Paenalcaligenes and Oligella, showing negative correlation with uraemic toxins, were reported in relation with urinary tract infection, wound infection and increased levels of inflammation72, 73, 74, 75. These strains might contribute to the progression of kidney degeneration. In the meantime, the strong correlation between gut microbiota and urea and ferulate levels revealed that gut microbiota and these uraemic toxins could affect each other under disease condition68. Faecalibaculum, Erysipelotrichaceae, Paenalcaligenes and Oligella showed possible correlation with biological index whilst Ruminococcaceae and Bacteroides had negative correlation, which implies that the first four strains might not be good for kidney function improvement, whereas the other two strains might enhance renal function.
Targeted analysis of uric acid pathway pointed out that the uric acid synthesis pathway of intestinal flora in the model group was remarkably enhanced together with intensified urease activity. With increased uric acid synthesis, more uric acid would enter the gut microbiota to produce urea. Urease could mediate the decomposition of uric acid in the gut microbiota which would produce ammonia entering the body or being reused by the gut microbiota. Thus, the level of urease is closely related with reconstruction of gut microbiota under diseased condition. Besides, the enhanced urease activity was directly related to the decrease of urea level in gut microbiota, thereby promoting urea liver and intestinal circulation76. This would lead to elevated plasma level of urea, which in turn could aggravate renal function damage. In addition, the urease has non-enzymatic activity, which is associated with pro-inflammatory mechanism.
5. Conclusions
To sum up, nephropathy induced by hyperuricemia is a common complication of hyperuricemia and gout, which causes renal dysfunction in patients. This study combines metabolomics and 16S rRNA sequencing approach to reveal the association between amino acid metabolism disorder, abnormal gut microbiota composition, and the progression of hyperuricemia nephropathy. Our results indicated that glycine, serine, glutamate and glutamine in plasma were highly correlated to hyperuricemia nephropathy with an AUC of 0.84, 0.86, 0.94 and 0.84 respectively. The combined glycine, serine and glutamate even returned an AUC of 1, representing a good prediction ability. Hence the levels of these amino acids could possibly aid the diagnosis of hyperuricemia nephropathy. 16S rRNA revealed a number of bacterial strains that were diminished or increased in the model group. Targeted analysis of uric acid pathway demonstrated a dysregulated nitrogen circulation in gut microbiota under diseased condition, such as enhanced uric acid decomposition pathway, and urea liver-intestinal circulation. Therefore, modifying the composition of gut microbiota or targeting at the characteristic metabolic enzymes of the gut microbiota such as urease could be a useful way to treat or prevent this disease.
Acknowledgments
The project was supported by the National Natural Science Foundation of China (Nos. 81573493 and 81973290), CAMS Innovation Fund for Medical Sciences (CIFMS, No. 2016-I2M-3-011, China), Beijing Key Laboratory of Non-Clinical Drug Metabolism and PK/PD study (Z141102004414062, China), the National Megaproject for Innovative Drugs (Nos. 2018ZX09711001-002-002 and 2018ZX09302015, China), and Beijing Natural Sciences Fund Key Projects (NO. 7181007). We also thank Shimadzu (China) Co., Ltd. for the technological supports in this study.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.10.007.
Contributor Information
Yan Wang, Email: wangyan@imm.ac.cn.
Jiandong Jiang, Email: jiang.jdong@163.com.
Author contribution list
Yan Wang and Jiandong Jiang designed and conceptualized this study. Yan Wang, Pei Han and Li-Bin Pan drafted the manuscript. Li-Bin Pan, Pei Han, Shu-Rong Ma, Wei-Jia Kong, Zheng-Wei Zhang, Hang Yu, and Can Wang conducted the animal experiments and data analysis. Ran Peng, Li-Bin Pan, Jie Fu and Lin Cong performed the molecular biology study.
Conflicts of interest
All authors state that there are no conflicts of interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Qin Z., Wang S., Lin Y., Zhao Y., Yang S., Song J. Antihyperuricemic effect of mangiferin aglycon derivative J99745 by inhibiting xanthine oxidase activity and urate transporter 1 expression in mice. Acta Pharm Sin B. 2018;8:306–315. doi: 10.1016/j.apsb.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalbeth N., Merriman T.R., Stamp L.K. Gout. Lancet. 2016;388:2039–2052. doi: 10.1016/S0140-6736(16)00346-9. [DOI] [PubMed] [Google Scholar]
- 3.Johnson R.J., Nakagawa T., Jalal D., Sánchez-Lozada L.G., Kang D.H., Ritz E. Uric acid and chronic kidney disease: which is chasing which?. Nephrol Dial Transplant. 2013;28:2221–2228. doi: 10.1093/ndt/gft029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Towiwat P., Chhana A., Dalbeth N. The anatomical pathology of gout: a systematic literature review. BMC Muscoskelet Disord. 2019;20:140. doi: 10.1186/s12891-019-2519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou Y.C., Kuan J.C., Yang T., Chou W.Y., Hsieh P.C., Bai C.H. Elevated uric acid level as a significant predictor of chronic kidney disease: a cohort study with repeated measurements. J Nephrol. 2015;28:457–462. doi: 10.1007/s40620-014-0158-9. [DOI] [PubMed] [Google Scholar]
- 6.Yan D., Tu Y., Jiang F., Wang J., Zhang R., Sun X. Uric acid is independently associated with diabetic kidney disease: a cross-sectional study in a Chinese population. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129797. e0129797-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartáková V., Kuricová K., Pácal L., Nová Z., Dvořáková V., Švrčková M. Hyperuricemia contributes to the faster progression of diabetic kidney disease in type 2 diabetes mellitus. J Diabet Complicat. 2016;30:1300–1307. doi: 10.1016/j.jdiacomp.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Sharaf El Din U.A.A., Salem M.M., Abdulazim D.O. Uric acid in the pathogenesis of metabolic, renal, and cardiovascular diseases: a review. J Adv Res. 2017;8:537–548. doi: 10.1016/j.jare.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Tong Q., Shou J.W., Zhao Z.X., Li X.Y., Zhang X.F. Gut microbiota-mediated personalized treatment of hyperlipidemia using berberine. Theranostics. 2017;7:2443–2451. doi: 10.7150/thno.18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Jiang J. A new research mode of drug PK–PD mediated by the gut microbiota: insights into the pharmacokinetics of berberine. Yao Xue Xue Bao. 2018;53:659–666. [Google Scholar]
- 12.Feng R., Shou J.W., Zhao Z.X., He C.Y., Ma C., Huang M. Transforming berberine into its intestine-absorbable form by the gut microbiota. Sci Rep. 2015;5:12155. doi: 10.1038/srep12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Shou J.W., Li X.Y., Zhao Z.X., Fu J., He C.Y. Berberine-induced bioactive metabolites of the gut microbiota improve energy metabolism. Metabolism. 2017;70:72–84. doi: 10.1016/j.metabol.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Z.X., Fu J., Ma S.R., Peng R., Yu J.B., Cong L. Gut–brain axis metabolic pathway regulates antidepressant efficacy of albiflorin. Theranostics. 2018;8:5945–5959. doi: 10.7150/thno.28068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Y.L., Cao G., Chen D.Q., Vaziri N.D., Chen L., Zhang J. Microbiome–metabolomics reveals gut microbiota associated with glycine-conjugated metabolites and polyamine metabolism in chronic kidney disease. Cell Mol Life Sci. 2019 doi: 10.1007/s00018-019-03155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung S., Barnes J.L., Astroth K.S. Gastrointestinal microbiota in patients with chronic kidney disease: a systematic review. Adv Nutr. 2019;10:888–901. doi: 10.1093/advances/nmz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aron-Wisnewsky J., Clément K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol. 2015;12:169–181. doi: 10.1038/nrneph.2015.191. [DOI] [PubMed] [Google Scholar]
- 19.Guo Z., Zhang J., Wang Z., Ang K.Y., Huang S., Hou Q. Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep. 2016;6:20602. doi: 10.1038/srep20602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crane J.K., Naeher T.M., Broome J.E., Boedeker E.C. Role of host xanthine oxidase in infection due to enteropathogenic and Shiga-toxigenic Escherichia coli. Infect Immun. 2013;81:1129–1139. doi: 10.1128/IAI.01124-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beger R.D., Dunn W., Schmidt M.A., Gross S.S., Kirwan J.A., Cascante M. Metabolomics enables precision medicine: "A white paper, community perspective". Metabolomics. 2016;12:149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rochfort S. Metabolomics reviewed: a new "omics" platform technology for systems biology and implications for natural products research. J Nat Prod. 2005;68:1813–1820. doi: 10.1021/np050255w. [DOI] [PubMed] [Google Scholar]
- 23.Tachibana C. What's next in omics: the metabolome. Science. 2014;345:1519–1521. [Google Scholar]
- 24.Carter R.A., Pan K., Harville E.W., McRitchie S., Sumner S. Metabolomics to reveal biomarkers and pathways of preterm birth: a systematic review and epidemiologic perspective. Metabolomics. 2019;15:124. doi: 10.1007/s11306-019-1587-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Y., Li W., Chen J., Wang X., Lv Y., Huang Y. Pharmacometabolomic prediction of individual differences of gastrointestinal toxicity complicating myelosuppression in rats induced by irinotecan. Acta Pharm Sin B. 2019;9:157–166. doi: 10.1016/j.apsb.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tso V.K., Sydora B.C., Foshaug R.R., Churchill T.A., Doyle J., Slupsky C.M. Metabolomic profiles are gender, disease and time specific in the interleukin-10 gene-deficient mouse model of inflammatory bowel disease. PLoS One. 2013;8:e67654. doi: 10.1371/journal.pone.0067654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M.X., Wang S.Y., Kuo C.H., Tsai I.L. Metabolome analysis for investigating host–gut microbiota interactions. J Formos Med Assoc. 2019;118:S10–S22. doi: 10.1016/j.jfma.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Holmes E., Li Jia V., Marchesi Julian R., Nicholson Jeremy K. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16:559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Vernocchi P., Del Chierico F., Putignani L. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front Microbiol. 2016;7:1144. doi: 10.3389/fmicb.2016.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han P., Huang Y., Xie Y., Yang W., Xiang W., Hylands P.J. Metabolomics reveals immunomodulation as a possible mechanism for the antibiotic effect of Persicaria capitata (Buch.-Ham. ex D. Don) H. Gross. Metabolomics. 2018;14:91. doi: 10.1007/s11306-018-1388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snowden S.G., Ebshiana A.A., Hye A., An Y., Pletnikova O., O'Brien R. Association between fatty acid metabolism in the brain and Alzheimer disease neuropathology and cognitive performance: a nontargeted metabolomic study. PLoS Med. 2017;14 doi: 10.1371/journal.pmed.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D.Q., Cao G., Chen H., Argyopoulos C.P., Yu H., Su W. Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan. Nat Commun. 2019;10:1476. doi: 10.1038/s41467-019-09329-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang R., Wei Y., Li Y., Chen W., Chen H., Wang Q. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. 2018;67:534–541. doi: 10.1136/gutjnl-2016-313332. [DOI] [PubMed] [Google Scholar]
- 34.Zhernakova D.V., Le T.H., Kurilshikov A., Atanasovska B., Bonder M.J., Sanna S. Individual variations in cardiovascular-disease-related protein levels are driven by genetics and gut microbiome. Nat Genet. 2018;50:1524–1532. doi: 10.1038/s41588-018-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenq R.R., Taur Y., Devlin S.M., Ponce D.M., Goldberg J.D., Ahr K.F. Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. 2015;21:1373–1383. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamanai-Shacoori Z., Smida I., Bousarghin L., Loreal O., Meuric V., Fong S.B. Roseburia spp.: a marker of health?. Future Microbiol. 2017;12:157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 37.Sanders M.E., Merenstein D.J., Reid G., Gibson G.R., Rastall R.A. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16:605–616. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- 38.Yang L., Chang B., Guo Y., Wu X., Liu L. The role of oxidative stress-mediated apoptosis in the pathogenesis of uric acid nephropathy. Ren Fail. 2019;41:616–622. doi: 10.1080/0886022X.2019.1633350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye Y., Zhang Y., Wang B., Walana W., Wei J., Gordon J.R. CXCR1/CXCR2 antagonist G31P inhibits nephritis in a mouse model of uric acid nephropathy. Biomed Pharmacother. 2018;107:1142–1150. doi: 10.1016/j.biopha.2018.07.077. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y.N., Ma S.X., Chen Y.Y., Chen L., Liu B.L., Liu Q.Q. Chronic kidney disease: biomarker diagnosis to therapeutic targets. Clin Chim Acta. 2019;499:54–63. doi: 10.1016/j.cca.2019.08.030. [DOI] [PubMed] [Google Scholar]
- 41.Newsholme P., Procopio J., Lima M.M., Pithon-Curi T.C., Curi R. Glutamine and glutamate—their central role in cell metabolism and function. Cell Biochem Funct. 2003;21:1–9. doi: 10.1002/cbf.1003. [DOI] [PubMed] [Google Scholar]
- 42.Tapiero H., Mathe G., Couvreur P., Tew K.D. II. Glutamine and glutamate. Biomed Pharmacother. 2002;56:446–457. doi: 10.1016/s0753-3322(02)00285-8. [DOI] [PubMed] [Google Scholar]
- 43.Spanaki C., Plaitakis A. The role of glutamate dehydrogenase in mammalian ammonia metabolism. Neurotox Res. 2012;21:117–127. doi: 10.1007/s12640-011-9285-4. [DOI] [PubMed] [Google Scholar]
- 44.Dryer S.E. Glutamate receptors in the kidney. Nephrol Dial Transplant. 2015;30:1630–1638. doi: 10.1093/ndt/gfv028. [DOI] [PubMed] [Google Scholar]
- 45.Tizianello A., De Ferrari G., Garibotto G., Gurreri G., Robaudo C. Renal metabolism of amino acids and ammonia in subjects with normal renal function and in patients with chronic renal insufficiency. J Clin Invest. 1980;65:1162–1173. doi: 10.1172/JCI109771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor L., Curthoys N.P. Glutamine metabolism: role in acid-base balance. Biochem Mol Biol Educ. 2004;32:291–304. doi: 10.1002/bmb.2004.494032050388. [DOI] [PubMed] [Google Scholar]
- 47.Kamm D.E., Strope G.L. The effects of acidosis and alkalosis on the metabolism of glutamine and glutamate in renal cortex slices. J Clin Invest. 1972;51:1251–1263. doi: 10.1172/JCI106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinay P., Khoury N., Soowamber M., Gougoux A. Renal extraction of glutamine from plasma and whole blood: studies in dogs and rats. Can J Physiol Pharmacol. 1985;63:886–892. doi: 10.1139/y85-145. [DOI] [PubMed] [Google Scholar]
- 49.Øvrehus M.A., Bruheim P., Ju W., Zelnick L.R., Langlo K.A., Sharma K. Gene expression studies and targeted metabolomics reveal disturbed serine, methionine, and tyrosine metabolism in early hypertensive nephrosclerosis. Kidney Int Rep. 2018;4:321–333. doi: 10.1016/j.ekir.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalhan S.C., Hanson R.W. Resurgence of serine: an often neglected but indispensable amino acid. J Biol Chem. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lowry M., Hall D.E., Hall M.S., Brosnan J.T. Renal metabolism of amino acids in vivo: studies on serine and glycine fluxes. Am J Physiol. 1987;252:F304–F309. doi: 10.1152/ajprenal.1987.252.2.F304. [DOI] [PubMed] [Google Scholar]
- 52.van de Poll M.C., Soeters P.B., Deutz N.E., Fearon K.C., Dejong C.H. Renal metabolism of amino acids: its role in interorgan amino acid exchange. Am J Clin Nutr. 2004;79:185–197. doi: 10.1093/ajcn/79.2.185. [DOI] [PubMed] [Google Scholar]
- 53.Martinez M., Cuskelly G.J., Williamson J., Toth J.P., Gregory J.F., 3rd Vitamin B-6 deficiency in rats reduces hepatic serine hydroxymethyltransferase and cystathionine beta-synthase activities and rates of in vivo protein turnover, homocysteine remethylation and transsulfuration. J Nutr. 2000;130:1115–1123. doi: 10.1093/jn/130.5.1115. [DOI] [PubMed] [Google Scholar]
- 54.Lacour B., Parry C., Drueke T., Touam M., Kreis H., Bailly M. Pyridoxal 5ʹ-phosphate deficiency in uremic undialyzed, hemodialyzed, and non-uremic kidney transplant patients. Clin Chim Acta. 1983;127:205–215. doi: 10.1016/s0009-8981(83)80005-9. [DOI] [PubMed] [Google Scholar]
- 55.Busch M., Gobert A., Franke S., Ott U., Gerth J., Muller A. Vitamin B6 metabolism in chronic kidney disease—relation to transsulfuration, advanced glycation and cardiovascular disease. Nephron Clin Pract. 2010;114:38–46. doi: 10.1159/000245068. [DOI] [PubMed] [Google Scholar]
- 56.Hijmans R.S., Rasmussen D.G.K., Yazdani S., Navis G., van Goor H., Karsdal M.A. Urinary collagen degradation products as early markers of progressive renal fibrosis. J Transl Med. 2017;15:63. doi: 10.1186/s12967-017-1163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKleroy W., Lee T.-H., Atabai K. Always cleave up your mess: targeting collagen degradation to treat tissue fibrosis. Am J Physiol Lung Cell Mol Physiol. 2013;304:L709–L721. doi: 10.1152/ajplung.00418.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arata J., Hatakenaka K., Oono T. Effect of topical application of glycine and proline on recalcitrant leg ulcers of prolidase deficiency. Arch Dermatol. 1986;122:626–627. [PubMed] [Google Scholar]
- 59.Li P., Wu G. Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth. Amino Acids. 2018;50:29–38. doi: 10.1007/s00726-017-2490-6. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y., Fan P.-S., Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54:2271–2279. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- 61.Song Y.N., Dong S., Wei B., Liu P., Zhang Y.Y., Su S.B. Metabolomic mechanisms of gypenoside against liver fibrosis in rats: an integrative analysis of proteomics and metabolomics data. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niwa T. Phenol and p-cresol accumulated in uremic serum measured by HPLC with fluorescence detection. Clin Chem. 1993;39:108–111. [PubMed] [Google Scholar]
- 64.Nowak A., Libudzisz Z. Influence of phenol, p-cresol and indole on growth and survival of intestinal lactic acid bacteria. Anaerobe. 2006;12:80–84. doi: 10.1016/j.anaerobe.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Nakabayashi I., Nakamura M., Kawakami K., Ohta T., Kato I., Uchida K. Effects of synbiotic treatment on serum level of p-cresol in haemodialysis patients: a preliminary study. Nephrol Dial Transplant. 2011;26:1094–1098. doi: 10.1093/ndt/gfq624. [DOI] [PubMed] [Google Scholar]
- 66.Vanholder R., Glorieux G. The intestine and the kidneys: a bad marriage can be hazardous. Clin Kidney J. 2015;8:168–179. doi: 10.1093/ckj/sfv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyazaki K., Masuoka N., Kano M., Iizuka R. Bifidobacterium fermented milk and galacto-oligosaccharides lead to improved skin health by decreasing phenols production by gut microbiota. Benef Microbes. 2014;5:121–128. doi: 10.3920/BM2012.0066. [DOI] [PubMed] [Google Scholar]
- 68.Ni J., Shen T.-C.D., Chen E.Z., Bittinger K., Bailey A., Roggiani M. A role for bacterial urease in gut dysbiosis and Crohn's disease. Sci Transl Med. 2017;9:eaah6888. doi: 10.1126/scitranslmed.aah6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hartwich K., Poehlein A., Daniel R. The purine-utilizing bacterium Clostridium acidurici 9a: a genome-guided metabolic reconsideration. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051662. e51662-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barker H.A., Beck J.V. Clostridium acidi-uridi and Clostridium cylindrosporum, organisms fermenting uric acid and some other purines. J Bacteriol. 1942;43:291–304. doi: 10.1128/jb.43.3.291-304.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marzocco S., Fazeli G., Di Micco L., Autore G., Adesso S., Dal Piaz F. Supplementation of short-chain fatty acid, sodium propionate, in patients on maintenance hemodialysis: beneficial effects on inflammatory parameters and gut-derived uremic toxins, a pilot study (PLAN Study) J Clin Med. 2018;7:315. doi: 10.3390/jcm7100315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shewmaker P.L., Whitney A.M., Gulvik C.A., Humrighouse B.W., Gartin J., Moura H. Vagococcus bubulae sp. nov., isolated from ground beef, and Vagococcus vulneris sp. nov., isolated from a human foot wound. Int J Syst Evol Microbiol. 2019;69:2268–2276. doi: 10.1099/ijsem.0.003459. [DOI] [PubMed] [Google Scholar]
- 73.Mukhopadhyay R., Joaquin J., Hogue R., Kilaru A., Jospin G., Mars K. Complete genome sequence of a Paenalcaligenes hominis strain isolated from a paraplegic patient with neurogenic bladder using single-molecule real-time sequencing technology. Genome Announc. 2017;5 doi: 10.1128/genomeA.00252-17. e00252-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pearce M.M., Hilt E.E., Rosenfeld A.B., Zilliox M.J., Thomas-White K., Fok C. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio. 2014;5 doi: 10.1128/mBio.01283-14. e01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gootenberg D.B., Paer J.M., Luevano J.-M., Kwon D.S. HIV-associated changes in the enteric microbial community: potential role in loss of homeostasis and development of systemic inflammation. Curr Opin Infect Dis. 2017;30:31–43. doi: 10.1097/QCO.0000000000000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lau W.L., Vaziri N.D. Urea, a true uremic toxin: the empire strikes back. Clin Sci (Lond) 2017;131:3–12. doi: 10.1042/CS20160203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.