Abstract
Host-microbiota interactions involving inflammatory and metabolic pathways have been linked to the pathogenesis of multiple immune-mediated diseases and metabolic conditions like diabetes and obesity. Accumulating evidence suggests that alterations in the gut microbiome could play a role in cardiovascular disease. This review focuses on recent advances in our understanding of the interplay between diet, gut microbiota and cardiovascular disease, with emphasis on heart failure and coronary artery disease. Whereas much of the literature has focused on the circulating levels of the diet- and microbiota-dependent metabolite trimethylamine-N-oxide (TMAO), several recent sequencing-based studies have demonstrated compositional and functional alterations in the gut microbiomes in both diseases. Some microbiota characteristics are consistent across several study cohorts, such as a decreased abundance of microbes with capacity for producing butyrate. However, the published gut microbiota studies generally lack essential covariates like diet and clinical data, are too small to capture the substantial variation in the gut microbiome, and lack parallel plasma samples, limiting the ability to translate the functional capacity of the gut microbiomes to actual function reflected by circulating microbiota-related metabolites. This review attempts to give directions for future studies in order to demonstrate clinical utility of the gut-heart axis.
Keywords: Coronary artery disease, Gut microbiota, Microbiome, Heart failure, Atherosclerosis, Butyrate, TMAO, Diet, Fiber, Metabolites
1. Introduction
The gut microbiota, comprising the trillions of bacteria in the gastrointestinal tract, is a complex community whose metabolic activities and interactions with the immune system extend beyond the gut itself [1]. Host-microbiota interactions involving inflammatory and metabolic pathways have been proposed to contribute to the pathogenesis of multiple immune-mediated diseases and metabolic conditions like diabetes and obesity [1].
Cardiovascular disease is the leading cause of death worldwide [2]. Atherosclerosis, the most common cause of cardiovascular disease, is the result of a complex series of events occurring within the arterial wall involving rheology, lipid metabolism, and inflammation [3]. The resulting stenoses in coronary, renal, precerebral and peripheral arteries precipitate end-organ ischemia, thromboembolic infarction, and necrosis.
Heart failure (HF) is a syndrome caused by the impaired ability of the heart to fill or eject blood [4]. Any disorder affecting the structural and/or functional integrity of the heart, such as valvular, coronary or myocardial disease, can commence HF. Hemodynamic stress [5], neurocrine activation [6], and inflammation [7] all contribute to the structural changes observed in advanced HF.
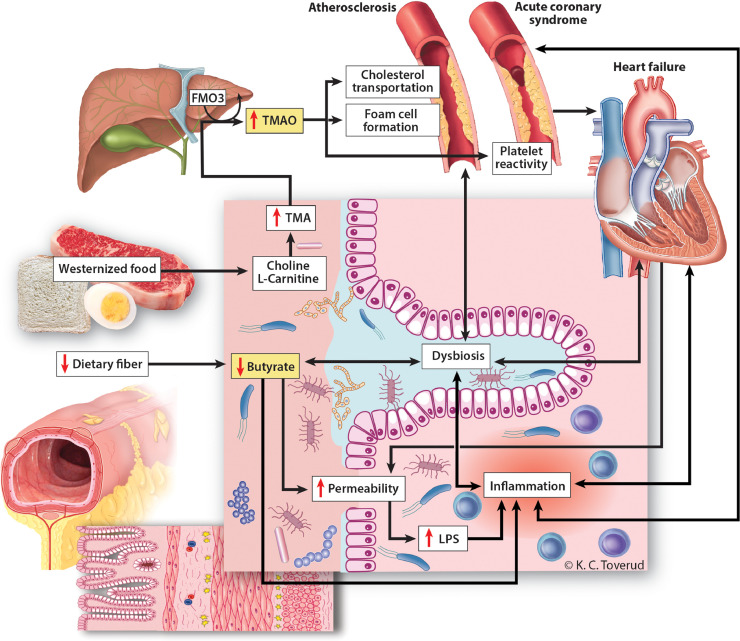
Accumulating evidence suggests that alterations in the gut microbial community could play a role in cardiovascular disease. This review focuses on recent advances in our understanding of the interplay between the gut microbiota and cardiovascular disease, with emphasis on HF and coronary artery disease (CAD). Some microbiota characteristics have consistently been identified in both diseases, such as a decreased abundance of microbes with capacity for producing butyrate and increased circulating levels of the diet- and microbiota-dependent metabolites trimethylamine-N-oxide (TMAO) (Fig. 1). However, most published studies lack essential covariates like diet, and are too small to capture the substantial variation in the gut microbiome. This review attempts to give directions for future studies in order to demonstrate a clinically useful gut-heart axis.
Fig. 1.
Diet-gut-heart interactions: proposed mechanisms. Interactions between diet and the gut microbiome could contribute to atherosclerosis, acute coronary syndromes and heart failure through common and separate mechanisms. Westernized food rich in red meat promotes bacterial production of TMA, which is oxidized in the liver to the pro-atherogenic metabolite TMAO. TMAO may contribute to atherosclerosis by interference with cholesterol transportation, foam cell formation and platelet aggregation, the latter playing a potential role in acute coronary syndromes. Reduced dietary fiber is associated with reduced bacterial production of the short chain fatty acid butyrate, which has immune modulatory effects in the gut mucosa, and also serves as the main energy substrate for colonocytes. Reduction of butyrate levels in the gut could promote local inflammation, aggravate dysbiosis and contribute to impaired gut barrier function, the latter resulting in leakage of bacterial toxins such as LPS, further fueling local and systemic inflammation. FMO3; flavin-containing monooxygenase 3, LPS; lipopolysaccharide, TMA; trimetylamine, TMAO; trimethylamine-N-oxide. Printed with permission from Kari Toverud ©.
2. A brief introduction to microbiota-related laboratory methods
The gut microbiome can be considered an endocrine organ, and each microbe has the capacity to produce hundreds of different known and unknown metabolites which act beyond gut itself [1]. The composition of the microbiota is typically analyzed by high-throughput sequencing and bioinformatic analyses of extracted microbial deoxyribonucleic acid (DNA). The methodologies are extensively reviewed in the literature [8,9], but as basis for this review, we provide a brief overview of the most relevant methods.
This review addresses the bacterial component of the gut microbiome, for which segments of the 16S ribosomal ribonucleic acid (rRNA) marker gene are amplified and submitted to next-generation sequencing [10], [11], [12]. A given bacterial species (e.g. Faecalibacterium prausnitzii) can be described on different taxonomic levels; as part of a phylum (e.g. Firmicutes), class (Clostridia), order (Clostridiales), family (Ruminococcaceae) or genus (Faecalibacterium). With 16S sequencing, bacteria can typically be identified at genus level resolution, more rarely at species level.
When using the full metagenomic (shotgun) sequencing, all the DNA in the sample is sequenced, providing resolution on species level [9]. This method is more expensive and computationally demanding, but provides an overview of potential microbial functions. Similar data may be generated (imperfectly) by predicting the functional gene content from 16S rRNA-based data [13]. Methods for characterizing microbial functions include metatranscriptomics, metaproteomics and metabolomics [14]. Typically, metabolomic methods may be applied to stool or peripheral bloods samples and provide direct measures of microbial activity.
3. Coronary artery disease
3.1. Altered gut microbiome with inflammatory properties and reduced capacity for short chain fatty acid (SCFA) production
Three recent studies have investigated the gut microbiota composition in patients with CAD, using different sequencing methodologies (Table 1). Cui et al. reported differences on phylum level, with decreased proportion of Bacteroidetes and increased proportion of Firmicutes in patients with coronary heart disease [15]. In a much more comprehensive study, Jie et al. reported increased levels of several Streptococcus species and genera of the Enterobacteriaceae family, and reduced abundance of Roseburia Intestinalis and Faecalibacterium prausnitzii, known producers of the SCFA butyrate [16]. Zhu and colleagues reported increased abundance of Escherichia-Shigella and Enterococcus and lower abundance of the butyrate producers Faecalibacterium, Roseburia and Eubacterium rectale [17]. The studies by Jie and Zhu are in line with a previous study on symptomatic carotid atherosclerosis by Karlsson et al. reporting decreased relative abundance of Roseburia and Eubacterium, known producers of butyrate [18]. Butyrate and other SCFAs are end products of fermentation of dietary fibers, and the main energy source for colonocytes maintaining the gut mucosal barrier [19]. A reduction in the overall genetic potential for butyrate production could also be observed in the metagenome data in the study by Jie et al. [16]. Furthermore, Karlsson et al. observed a strong negative correlation between genes encoding butyrate production (butyrate-acetoacetate CoA-transferase) and C-reactive protein (CRP) levels in the metagenomes of patients with symptomatic carotid atherosclerosis [18]. Other SCFAs could also be relevant, including acetate, which was shown to attenuate cardiac fibrosis and improve cardiac function in experimental mouse models [20].
Table 1.
Contemporary gut microbiota sequencing studies in patients with coronary artery disease (CAD).
| Study | Cui et al. [15] | Jie et al. [16] | Zhu et al. [17] |
|---|---|---|---|
| Patients | CAD verified by coronary angiography | CAD verified by coronary angiography | CAD verified by coronary angiography |
| Patient age | 68.3 ± 9.5 years | 40–80 years | 63.6 |
| Gender% (f/m) | 85/15 | – | 58/42 |
| Sample size | n = 29 CAD | n = 218 CAD | n = 70 CAD |
| n = 35 controls | n = 187 controls | n = 98 controls | |
| Methods | 16 s rRNA | Metagenomics | 16 s rRNA |
| Parallel plasma/serum | No | No | No |
| Dietary data | No | No | No |
| Increased relative abundance in patients | -Firmicutes phylum | Enterobacteriaceae family and Streptococcus species | Escherichia-Shigella and Enterococcus |
| Decreased relative abundance in patients | -Bacteroidetes phylum | Roseburia Intestinalis and Faecalibacterium prausnitzii | Faecalibacterium, Roseburia, Subdoligranulum and Eubacterium rectale |
| Functional findings | – | -Less fermentative capacity and more inflammatory properties in CAD microbiomes | -Several predicted functions, including lipopolysaccharide biosynthesis and propanoate metabolism enhanced in CAD microbiomes |
3.2. Dysfunctional gut barrier, lipopolysaccharide (LPS) and inflammation
The gut microbial changes affecting butyrate may also influence inflammatory pathways, as butyrate exerts local anti-inflammatory effects in the intestinal mucosa by inducing colonic regulatory T cells [21]. Loss of butyrate producing bacteria may result in a dysfunctional gut mucosal barrier, facilitating passive leakage of microbial toxins such as LPS that binds to Toll-like receptors and other receptors of the innate immune system, thereby triggering inflammation [1,[22], [23], [24]]. Of interest, an increased potential for LPS biosynthesis in the microbiome has been reported among patients with CAD, [17] and previous studies have linked circulating levels of LPS to insulin resistance [22], glycemic control and abdominal obesity [23]. We recently reported that increased plasma levels of LPS-binding protein and soluble CD14 predicted cardiovascular events in a high-risk population [25].
Recent work has demonstrated different bioactivity of LPS, with hexa-acylated, but no penta-acylated LPS triggering inflammation [26,27]. Jie et al. reported that genes required for synthesis of the LPS O-antigen were enriched in CAD, whereas the lipid A module was depleted, most likely due to depletion of Bacteroides, which produce non-inflammatory penta-acylated lipid A [16].
3.3. The microbiota and the diet-dependent metabolite TMAO
The most compelling evidence of a link between the gut microbiome and CAD is related to microbial metabolism of dietary factors like carnitine and choline [28], [29], [30]. In a landmark paper from the Hazen group, the metabolite TMAO was identified as a strong predictor of CAD [28]. More than just a marker of the disease, TMAO is potentially a causative agent in atherosclerosis [29]. The source of TMAO is trimethylamine, which is produced by the gut microbiota from nutrients containing L-carnitine or phosphatidylcholine, and subsequently oxidized by hepatic flavin-containing monooxygenases to TMAO [28,29]. Precursors of TMAO promote foam cell formation and atherosclerosis in animal models, but not when adding antibiotics to the drinking water, suggesting a microbiota dependent mechanism [29]. In several independent cohorts from USA and Europe, plasma levels of TMAO predicted myocardial infarction, stroke and all-cause mortality [28], [29], [30], [31], [32].
In a recent study, TMAO levels increased in healthy individuals after dietary intake of red meat as compared with a non-meat or a white meat enriched diet [33]. The effect of diet on TMAO levels was reversible. Additionally, isotope techniques demonstrated that the increased TMAO production was from carnitine, not choline [33].
Although TMAO is the most studied microbiota-related metabolite in relation to cardiovascular risk, other metabolites along the TMAO pathway are of potential interest. Gammabutyrobetaine (γBB) is a partly microbiota-related metabolite on the pathway from l-carnitine to TMAO, which has also been linked to CAD [34]. Trimethyllysine (TML) is a partly endogenous, partly diet-derived precursor of both γBB and TMAO. It has been linked to increased CAD risk, alone, or in combination with TMAO [35,36]. In recent studies, circulating levels of γBB and TML, but not TMAO, predicted cardiovascular mortality in patients with carotid artery atherosclerosis [37]. Hence, there is a need to include also TMAO precursors in future studies to delineate which pathways are involved in atherosclerosis and CAD.
3.4. From the chronic atherosclerotic process to acute cardiovascular events
The atherosclerotic process starts with the fatty streak and culminates in plaque rupture and acute atherothrombosis, causing acute clinical events such as stroke or myocardial infarction [3,38]. Whereas inflammation is involved in all phases of atherosclerosis, most literature on the microbiome in CAD has not separated clearly between chronic CAD and acute events.
The published studies regarding the role of the microbiome in CAD all investigated patients with mostly stable CAD using cross-sectional designs [16], [17], [18]. Hence, prospective studies powered for clinical events, as well as studies of the microbiome during acute coronary syndromes, should be a priority. Whereas direct analyses of the content of the microbiome could be difficult to perform in patients with acute disease, studies of microbiota-related metabolites could be more feasible in this setting. Interestingly, increased bacterial translocation has been reported in patients with acute myocardial infarction, with LPS and D-lactate blood levels being associated with adverse outcomes [39]. Furthermore, elevated levels of TMAO [32] and TML, alone or in combination with TMAO [36], have been shown to be associated with major adverse cardiac events 30 days after acute coronary syndrome, independent of troponin T levels. TMAO has been shown to interfere with platelet reactivity, which could be relevant for acute thromboembolic events [40]. Hence, although microbiota analyses are not yet ready for clinical use in the emergency room [41], microbiota-dependent biomarkers including LPS and TMA are potential therapeutic targets in patients with CAD.
4. Heart failure
4.1. Altered gut microbiome in HF with reduced capacity for butyrate production
The last two years, several sequencing-based studies have reported that the gut microbiota composition and functions differ between patients with HF and healthy subjects, with some common findings, but also considerable variation between studies (Table 2). Luedde and colleagues observed a reduced abundance of Ruminococcaceae on the family level and reduced abundance of Blautia from the Lachnospiracea family on the genus level in 20 patients with HF [42]. In a similar-sized study, Kamo et al. found a reduced relative abundance of Eubacterium rectale and Dorea longicatena from the Lachnospiracea family, and levels of Faecalibacterium from the Ruminococcaceae family were lower in older patients [43]. Furthermore, Cui et al. reported reduced levels of Faecalibacterium prausnitzii in patients with HF [44]. A common finding in these studies is the relative reduction in taxa from the Lachnospiracea or Ruminococcacea families, known for their capacity for butyrate production.
Table 2.
Contemporary gut microbiota sequencing studies in patients with heart failure (HF).
| Study | Luedde et al. [42] | Kamo et al. [43] | Cui et al. [44] | Kummen et al. [45] |
|---|---|---|---|---|
| Mayerhofer et al. [66] | ||||
| Patients | Chronic HF: 70% exacerbation, 30% stable | Acute HF or exacerbation of chronic HF | Stable chronic HF: Ischaemic or dilated cardiomyopathy | Stable systolic HF |
| Age patients | 65 ± 3.2 years | Two strata: 47.4 ± 2.8 years 73.8 ± 2.8 years | 58.1 ± 13.3 years | 58.9 (39–74) years |
| Gender% (f/m) | 45/55 | 18/82 | 17/83 | 59/41 |
| Sample size | n = 20 HF | n = 12 HF <60years | n = 53 HF | n = 84 HF (discovery- validation) |
| n = 20 controls | n = 10 HF >60years | n = 41 controls | n = 266 controls | |
| n = 12 controls | ||||
| Methods | 16 s rRNA | 16 s rRNA | 16 s rRNA | 16 s rRNA |
| Parallel plasma/serum | No | No | Yes | Yes |
| Dietary data | No | No | No | Yes |
| Increased relative abundance in patients | – | – | Ruminococcus gnavus | Prevotella, Hungatella, Succinclasticum |
| Decreased relative abundance in patients | Coriobacteriaceae, Erysipelotrichaceae, Ruminococcaceae (family level) | -Eubacterium rectale, Dorea longicatena | Faecalibacterium prausnitzii | -Lachnospiracea family: 9 different genera, including Blautia and Eubacterium halli |
| Blautia (genus level) | -Depletion of Faecalibacterium in older patients | - Ruminococcaceae: Faecalibacterium | ||
| -Bifidobacteriaceae: Bifidobacterium | ||||
| Functional findings | – | – | -Increased capacity for lipopolysaccharide biosynthesis and TMA production and reduced capacity for butyrate production in HF microbiomes | -Lower genetic potential for butyrate production in HF microbiomes |
| -Eubacterium halli associated with soluble CD25 and mortality | ||||
| -Dysbiosis related to dietary fiber intake |
In order to define a more robust HF-related gut microbiota signature, we investigated two independent cross-sectional cohorts, finding that patients with HF had reduced biodiversity in the gut microbiome, as well as altered abundance of 15 core taxa. Most of the microbes that were depleted in HF belonged to the Lachnospiracea family, in addition to Faecalibacterium from the Ruminococcacea family [45], again pointing to reduced capacity for butyrate production as a key element, supported by a lower predicted genetic potential for butyrate production (genes encoding butyrate-acetoacetate CoA-transferase). Of relevance, the abundance of several members of the Lachnospiracea family correlated with soluble CD25, a marker of T cell activation, and depletion of the known butyrate producer Eubacterium Halli and increased plasma levels of soluble CD25 were associated with death or heart transplantation [45].
4.2. Gut mucosal biofilm, pathogens and leaky gut
The recent studies mentioned above are partly contrasting older data. In a study by Sandek et al. using fluorescence in situ hybridization, an enrichment of Eubacterium rectale and Faecalibacterium in gut mucosal biofilms were observed in patients with HF [46]. Pasini and colleagues, using traditional culture techniques, found an increased abundance of several pathogenic bacteria in HF, including Campylobacter, Shigella, Salmonella, Yersinia Enterolytica and Candida species [47]. The methodological differences are probably the key to explain these contradictory findings, although the different sampling site is a relevant factor in the former study, as mucosa-adherent microbes might differ from luminal fecal samples [48].
Of interest, the patients in the studies by Sandek and Pasini had increased gut permeability as measured by the lactulose-mannitol test and the cellobiose sugar test [47,49] which fits well with reduced capacity for production of butyrate, the main energy source for colonocytes and critical for maintaining the gut barrier. Finally, the downregulation of a healthy core microbiome in HF patients could facilitate outgrowth of pathogenic microbes as reported by Pasini et al. [47].
4.3. TMAO: of prognostic value in HF?
Inspired by the role of TMAO in CAD, several independent studies have investigated the role of TMAO in chronic HF. It turns out that TMAO is a strong predictor of clinical outcomes in patients with HF, regardless of the underlying etiology [50]. In a study from our hospital, TMAO was elevated in patients with ischemic HF but not with dilated cardiomyopathy, and TMAO levels were associated with increased pulmonary artery pressure and wedge pressure, which are indices of left atrial stress [51]. Hence, the TMAO pathway could be related to decompensated HF and congestion, as TMAO levels were associated with prognosis in patients with acute decompensated HF [52]. An experimental study also supports this concept, since feeding animals with TMAO or its dietary precursors aggravated hemodynamic parameters [53].
Heart transplantation could represent a human “model” to provide additional information on the potential impact of the gut microbiota in HF. Recently, we found increased plasma levels of TMAO and TMAO-precursors in de novo heart transplant recipients, whereas the partly microbiota-dependent metabolite γ-BB increased steadily after transplant. This metabolite was associated with acute rejection and cardiac allograft vasculopathy [54]. Although alloimmunity, treatment with immunosuppressive drugs and other factors could be relevant in these disease processes, there is a major knowledge gap related to the role of the gut microbiota in the post-transplant setting [55].
4.4. Primary and secondary bile acids
We recently analyzed the circulating bile acid pool in patients with HF and healthy controls and found an increased ratio of secondary to primary bile acids in HF, which was associated with reduced overall survival in unadjusted, but not adjusted analyses [56]. Whereas bile acids are traditionally regarded as emulsifiers to facilitate the absorption of dietary fat and fat-soluble vitamins, bile acids are now recognized as signaling molecules that interact with plasma membranes as well as nuclear receptors, exerting regulatory effects on glucose and lipid metabolism [57], energy homeostasis [58] and other physiological processes [59]. In fact, several bile acid receptors are expressed in cardiomyocytes, and it has been proposed that bile acids influence cardiovascular function [60]. In the gut, primary bile acids undergo metabolism to secondary bile acids [61] before reabsorption as a part of the enterohepatic cycle. These microbial bile acid modifications have a major impact on the agonist activity on the bile acid receptors such as the farnesoid X receptor, which has several pleiotropic effects, and could represent a link between the gut microbiome and cardiovascular health [61]. Interestingly, a pilot study targeting the bile acid pool by ursodeoxycholic acid, reported improved peripheral blood flow as well as improved markers of liver function in patients with chronic HF [62].
4.5. Uremic toxins
Although reviewed only briefly here, the role of microbiota-derived uremic toxins could be of particular relevance for targeting cardiovascular risk in patients with chronic kidney disease (CKD) [63], including patients with HF as part of the cardiorenal syndrome.
In CKD, the loss of urinary excretion results in retention of various substances known as uremic retention solutes, many of which have toxic properties, and certain uremic toxins are synthesized by gut microbes [63,64]. Indoles are bacterial metabolites of tryptophan, a semi-essential amino acid found in various food sources such as red meat, egg and fish. Indoles are metabolized into indole derivatives, such as indoxyl sulfate (IS) and indole-3-acetic acid (IAA), which act as endogenous ligands of transcription factors interacting with various regulatory and signaling pathways, thereby mediating cardiotoxicity and vascular inflammation [63].
Another microbiota-generated uremic toxin is P-Cresyl Sulfate (PCS), which is derived from bacterial metabolism of aromatic amino acids that are subsequently sulfonated into PCS in the liver. In several studies, elevated levels of IS, IAA and PCS have been associated with increased mortality and increased risk of cardiovascular events [63]. TMAO is dependent on renal elimination, resulting in elevated plasma levels in CKD. The TMAO pathway has been implicated in the development of renal insufficiency and increased mortality in patients with CKD [31].
5. Is dysbiosis of the gut microbiota linked to plasma metabolites?
Circulating microbial metabolites are potential disease-modifying mediators of bacterial functions, provided that the disease-associated dysbiosis correlates with metabolite concentrations. The majority of microbiota analyses in CAD and HF (Tables 1 and 2) have not been accompanied by parallel plasma samples. Even though TMAO is elevated in CAD, Karlsson et al. found no upregulation of the metabolic pathway from phosphatidylcholine to TMA in the gut microbiomes of patients with CAD [18]. In contrast, the larger study by Jie et al. reported enrichment of gut microbial enzymes involved in trimethylamine formation in microbiomes from patients with CAD [16], although none of these studies reported circulating TMAO levels. In HF, Cui et al. found increased genetic potential for TMA production in the gut microbiome, but no association with circulating TMAO [44]. In a study comprising 22 patients with HF and 11 matched controls, elevated TMAO levels in HF correlated with the abundance of Escherichia and Shigella, although the abundance of these genera did not differ between HF patients and controls [65]. In an animal study of TMAO formation and platelet reactivity, taxa of the Lachnospiracea family were negatively associated with circulating TMAO levels [40]. In contrast, in our own HF cohort, in which several members of the Lachnospiracea family were depleted, we found no association between the dysbiosis in HF and circulating TMAO. TMAO generation is determined by a complex interplay between dietary factors, microbiota-dependent activity and hepatic oxidation, and expecting the gut microbiota alterations observed in CAD and HF to correlate directly with circulating TMAO may be too simplistic.
We recently measured circulating butyrate in plasma from patients with HF, but found no association with gut dysbiosis, possibly due to low circulating levels of butyrate [66]. Butyrate and other SCFAs can be measured in fecal samples, providing a more direct measure of microbial activity, but this requires snap frozen samples without preservatives.
6. Search for novel microbiota-related pathways in CAD and HF
With the combined genes of the microbiome outnumbering the human genome by two orders of magnitude, and each microbe having the potential to turn on and off the production of hundreds of metabolites, several undiscovered microbiota-related metabolites are likely to be relevant in human disease. The seminal TMAO-report by Tang et al. reported on several metabolites predictive of CAD, the nature of which are presumably still unknown [29].
There is a need to apply more extensively full metagenomic sequencing to better define changes in the functional genetic alterations in the gut microbiome in patients with HF. Such methods are expensive and resource demanding, but provide species level resolution, as well as the functional potential of the microbes and the host in the gut compartment, as assessed in coronary heart disease in the studies by Karlsson et al. and Jie et al. [16,18]. Ultimately, combined analyses of the actual byproducts of microbial activity (unbiased metabolomics and/or proteomics analyses of parallel plasma samples) and metagenomics analyses must be performed in multi-level bioinformatics controlling for relevant confounders, in order to identify functional alterations influencing the clinical phenotype of interest.
For translation to a clinical setting, biomarkers that are easily measurable in a reproducible way in plasma or urine will probably be of more value. Of relevance, Feng et al. measured metabolites in parallel plasma and urine samples in CAD patients and controls. The results were integrated with metagenomic analyses, identifying several metabolites, including GlcNAc-6-P, mannitol and 15 plasma cholines, as novel candidate biomarkers potentially derived from the gut microbes [67]. In order to make such a biomarker useful, it should preferably provide prognostic information beyond that of established biomarkers or point to novel therapeutic principles. Furthermore, whereas the above-mentioned studied are based upon known proteins and metabolites, a recent study identified thousands of uncharacterized microbiota-generated small molecules, probably small proteins coded from open reading frames [68]. This approach could open up other avenues in microbiota-related studies.
7. Controlling for diet, drugs and comorbidities
Host genetic factors have a small but significant impact on the composition of the gut microbiota [69,70]. However, environmental factors probably play a greater role [71]. One limitation of the studies published so far is the lack of dietary data. In an attempt to address this, we gathered food frequency questionnaires from patients with HF, finding that several characteristics of the dysbiosis observed in HF, including the low diversity and the reduction in butyrate-producing microbes, correlated with a low dietary fiber intake [66]. Importantly, diet has a major impact on the gut microbiota and related metabolites (Fig. 1), and dietary data should preferably be registered among other relevant co-variates in microbiota studies.
Other important confounders include concurrent medication and comorbidities. Recent large-scale studies have identified several commonly used drugs and their metabolism to be associated with microbiota alterations [72]. An interesting in vitro study examined nearly 1000 different drugs, finding that an estimated 24% of the drugs, most of which were not antibiotics, had the ability to suppress the growth of at least one commensal microbe in cultures [73]. Some drugs are even believed to mediate part of their therapeutic effects trough their influence on the gut microbiome, as shown for metformin [74]
The studies summarized in Tables 1 and 2 all have limitations, regarding sample size, methodology, lack of parallel fecal and plasma samples or relevant covariates. All these factors should be considered when planning future microbiota-related studies.
8. Targeting the gut microbiome
Almost 20 years after Ross defined atherosclerosis as an inflammatory disease [38], Paul Ridker published the CANTOS trial [75], showing that inhibition of IL-1β by the monoclonal antibody canakinumab, can reduce cardiovascular events. Clinical translation will hopefully take shorter time in microbiota medicine, but there are several obstacles, including safety issues and substantial inter-individual variation in gut microbiota composition and function.
8.1. Probiotics and prebiotics
Probiotics are live microorganisms that, when administered in adequate amounts, confer a health benefit on the host [76]. Probiotics in clinical use comprise bacterial and fungal microorganisms, including the genera Lactobacillus and Bifidobacterium and the fungus Saccharomyces boulardii [77]. Results from animal models suggest that certain strains of Lactobacilli could have cardioprotective effects. Rats treated with a supplement containing Lactobacillus plantarum 299v before coronary artery ligation had reduced infarct size and improved left ventricular function [78]. Another study showed similar cardioprotective results in a rat model of myocardial ischemia after supplement with Lactobacillus rhamnosus GR-1 [79].
In humans, a pilot study reported not only reduced systemic inflammation, but also improved left ventricular ejection fraction after an intervention with the probiotic yeast Saccharomyces Boulardii in patients with chronic HF [80]. The number of participants was low (n = 20), and the results should be interpreted with caution. We are currently performing a randomized controlled trial (RCT) including 150 patients with systolic HF, powered to detect an increase in left ventricular ejection fraction of 5% [81]. Results are expected during 2020. Given the potential clinical impact of microbiota modulation, as well as the high morbidity and mortality in HF, microbiota modulation is not without risk [82], and close clinical monitoring and pre-defined safety measures should follow the same standards as in other clinical trials [83]. Notably, genomic and epidemiological evidence of bacterial transmission from probiotic capsules to blood was recently reported in patients in intensive care units [84].
Prebiotics are substrates that are selectively utilized by host microorganisms and confer a potential health benefit, e.g. non-digestible dietary fibers and oligosaccharides [85]. Most contemporary deep sequencing studies of patients with cardiovascular disease report depletion of microbes with the capacity of producing SCFAs such as butyrate (Tables 1 and 2). Prebiotics promoting microbial fermentation of dietary fibers to SCFAs may therefore be of potential benefit in the gut, as well as in splanchnic and peripheral tissues, which in total may result in improved metabolic regulation [86]. Some prebiotics, such as inulin, have the potential to counteract harmful effects of antibiotics by promoting the diversity and functional capacity of the gut microbiota [87]. A recent RCT showed that dietary supplementation with inulin or inulin-proprionate ester improved insulin sensitivity and reduced markers of systemic inflammation by raising colonic delivery of the SCFA proprionate [88]. Hence, targeting microbial SCFAs production by supplement of inulin or other prebiotics are attractive strategies for future trials in cardiovascular disease, although current scientific evidence does not support the use of probiotics or prebiotics as supplemental therapy in patients with HF or CAD.
8.2. Antibiotics for gut decontamination
Early studies targeting the gut in patients with HF have focused on gut decontamination with broad-spectrum antibiotics to reduce bacterial translocation and inflammation. Although this approach has been successful in reducing markers of systemic inflammation, a clinical effect has not been demonstrated [89,90].
In a recent study, it was shown that a broad-spectrum cocktail of oral antibiotics markedly increased post- infarction rupture and death in a murine model of ligation of the left anterior descending artery [91], suggesting that an intact microbial community is required around the time of myocardial injury for proper myocardial repair [92]. This study contrasts with previous animal models, reporting that oral vancomycin decreased infarct size and improved post-infarct cardiac function in rats [78], as well as a subsequent study, reporting that a mixture of streptomycin, neomycin, polymyxin B and bacitracin reduced infarct size along with alterations in microbiota-related metabolites [93]. Furthermore, antibiotics reduced bacterial translocation, inflammation and myocardial injury in an experimental mouse model [39]. Taken together, these diverging animal studies strongly suggest a role of the gut microbiota composition in acute myocardial infarction, but the direction of microbiota alterations and the potential metabolic or inflammatory pathways are poorly understood.
Targeting cardiovascular disease with antibiotic therapy is not a new idea. Between 1995 and 2005 > 19 000 patients were included in RCTs aimed to target chlamydia pneumonia in patients with CAD [94]. Positive experimental studies and small RCTs were followed by large trials with adequate sample size, which demonstrated no clinical benefit of antibiotic therapy.
In addition to the obvious risk of antimicrobial resistance, other safety concerns of antibiotics have recently emerged with potential relevance for future trials. Recently, 10 years of follow-up data from the CLARICOR trial demonstrated increased cardiovascular death in clarithromycin-treated patients with stable CAD, [95] leading to an FDA alert in 2018 on the use of clarithromycin in patients with CAD (https://www.fda.gov/Drugs/DrugSafety/ucm597289.htm). In December 2018, another FDA alert on the use of fluoroquinolones warned about increased risk of aortic ruptures and aortic dissection in patients at increased risk, such as elderly patients with hypertension or peripheral atherosclerotic vascular disease (https://www.fda.gov/Drugs/DrugSafety/ucm628753.htm). Finally, a recent study reported increased risk of cardiovascular events in elderly women with increased cumulative exposure to antibiotics in adulthood [96]. The explanation for this increased risk in unknown, but could involve prolongation of the QT-interval and Torsade de Pointes arrhythmia, pro-inflammatory activities mediated by translocation of commensal gut microbes [97], or other effects mediated by the gut microbiota. Given these safety concerns and the lack of clinical effect in numerous trials, antibiotics should be used with caution in future studies targeting the gut microbiota in cardiovascular disease.
8.3. Fecal microbiota transplant (FMT)
FMT is the most radical current intervention for targeting the gut microbiome and an established treatment for recurrent Clostridioides difficile infection. FMT from lean donors was previously shown to normalize insulin sensitivity in obese subjects with the metabolic syndrome [98,99], although the effect was only temporary. In a subsequent study on 20 subjects with the metabolic syndrome, FMT from vegan donors changed the gut microbiota composition toward a vegan profile in some patients; however, without altering TMAO production capacity or parameters related to vascular inflammation [100]. FMT is now tested in several clinical settings, but with the current mode of endoscopic delivery, its use in acute and high-risk settings such as acute coronary syndrome and decompensated HF is limited. FMT is not without risk and was recently shown to transmit drug-resistant E. Coli resulting in bacteremia in two patients, one of whom died [101]. It is critical to standardize and optimize procedures for FMT, including screening for suitable donors, development of non-invasive delivery modes such as capsules, as well as determination of the active components, in order to develop personalized treatment strategies [102].
8.4. Precision medicine targeting enzymatic pathways
Whereas probiotics, antibiotics and FMT are all potential candidate interventions in proof of principle studies, next generation probiotics need to be more goal-directed, targeting specific enzymatic pathways. Interestingly, inhibitors of TMA production that target distinct microbial TMA lyases have been developed. These drugs reduce TMAO levels and reverse atherosclerosis in animal models [103]. TMA lyase has become a current potential therapeutic target of TMAO modulation. An appealing side of this “drug the bug” approach is that microbial pathways may be targeted, apparently without having bactericidal effects.
For now, dietary interventions, including adherence to a Mediterranean diet [104] or discontinuation of dietary red meat [33], are more accessible ways of reducing TMAO levels and possibly, cardiovascular risk. In light of our recent findings of dietary fiber intake being negatively associated with dysbiosis in HF, dietary intervention with a high-fiber diet, alone or in combination with a Mediterranean diet, could be a logical next step for targeting the gut heart axis.
8.5. Personalized approach
One aspect with particular relevance to microbiota research is the tremendous inter-individual variation in the gut microbiota composition [105]. In an elegant study, Zeevi et al. all showed that integration of microbiota profiles and metadata in a machine learning model made it possible to precisely predict individual glycemic responses in order to personalize nutritional advice [106]. These findings have been independently validated [107]. Hence, although certain microbiota-metabolite traits, such as increased TMAO or reduced butyrate production, may be identified in groups of patients, different microbiota-related pathways may be relevant in different individuals. In addition to the requirement of demonstrated effect in RCTs, a personalized approach is probably necessary if gut microbiota interventions should be of clinical significance.
9. Conclusion
Recent studies of the gut microbiome have identified some common traits in CAD and HF, in particular a decreased abundance of gut microbes with capacity for producing butyrate and elevated circulating levels of TMAO. However, a link between gut microbiota alterations and circulating metabolites in cardiovascular disease is yet to be defined, and most published studies lack information about essential covariates like drugs, diet and comorbidities and are too small to capture the substantial variation in the gut microbiome. Given the complexity and magnitude of the gut microbiota and its metabolites, as well as the potential benefits and risks of targeting the gut microbiota in high-risk clinical settings, a multidisciplinary approach is necessary. Close collaboration between dedicated clinicians and microbiota-focused groups with capacity for metagenomics and multi-level bioinformatics will be necessary to demonstrate a clinically relevant gut-heart axis. (Table 3).
Table 3.
Strengths, limitations and future possibilities of potential microbiota-related biomarkers in cardiovascular disease.
| Biomarker | Relevance and main findings | Limitations | Future directions |
|---|---|---|---|
| TMAO | -Predicts clinical end points in numerous studies on HF, stable CAD and acute CAD | -Circulating TMAO weakly linked to disease-specific dysbiosis. | -Potential therapeutic target in dietary interventions and pharmacological products interfering with TMA production |
| -Reproducible measurements with mass spectrometry | -TMAO levels influenced by diet, renal and liver function | -Microbiota-derived precursors such as TML should be studied further | |
| Butyrate | -Low microbial butyrate producing potential linked to dysbiosis in several cohorts of HF and CAD. | -Low circulating levels, not suitable as soluble biomarker | -Potential therapeutic target in high fiber dietary interventions |
| -Measurable in snap frozen fecal samples without preservatives, but rapidly degraded. | |||
| -Confounded by fiber intake | |||
| Gut leakage markers | -Increase in LPS-producing microbes linked to dysbiosis in several cohorts of HF and CAD. | -Direct measurement of gut permeability is so far not feasible in the clinic | -Need of better standardization of LPS measurements |
| -Increased plasma LPS in HF | -Large variability in LPS LAL-assay. | -Other markers of bacterial translocation such as LBP, I-FABP, zonulin, as well as functional measurements of gut leakage should be further studied | |
| -Increased gut permeability measured by lactulose-mannitol test and cellobiose sugar test in HF | -LAL assay does not separate between hexa- and penta-acylated LPS variants. | ||
| Bile acids | -Increased conversion from primary to secondary bile acids in HF | -Large variability and technically difficult to measure | -Circulating bile acid pool should be investigated in relation to disease-specific dysbiosis |
| -Pleiotropic effects of bile acid receptor FXR should be further studied in CVD | |||
| Uremic toxins | -Microbiota-generated toxins such as PCS and IS accumulate as a result of reduced urinary excretion and predict clinical end points in CKD patients | -Mostly relevant for CKD populations | -Interventions targeting uremic toxins, such as oral absorbants and synbiotics, should inspire research also in non-CKD populations |
CAD: coronaray artery disease; HF: heart failure; CKD: chronic kidney disease; LPS: lipopolysaccharide; LBP: LPS-binding protein; LAL-assay: limulus amebocyte lysate assay; I-FABP: intestinal fatty acid binding protein; FXR: farnesoid X receptor; PCS: P-Cresyl Sulfate; IS: indoxyl sulfate TMA: Trimethylamine; TMAO: Trimethylamine-N-Oxide; TML: Trimethyllysine.
10. Outstanding questions
Adequately powered studies; including well-designed randomized trials with parallel microbiota and plasma samples, controlled for essential covariates like diet; are needed to move the field from associative studies to possible causation.
11. Search and selection criteria
This review is based on a systematic search in PubMed using the term (microbiota OR microbiome) AND (heart failure OR coronary artery disease OR atherosclerosis OR cardiovascular) as of December 10th 2019. We limited our search to articles on adult patients, written in English and published over the last three years.
Declaration of Competing Interest
Dr. Trøseid, Dr Broch and Dr Andersen declare no conflict of interest. Dr. Hov has received funding from Biogen, personal fees from Novartis, and personal fees from Orkla Health.
Funding statement
No external funding source.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102649.
Appendix. Supplementary materials
References
- 1.Sonnenburg J.L., Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth G.A., Johnson C., Abajobir A., Abd-Allah F., Abera S.F., Abyu G. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol. 2017;70(1):1–25. doi: 10.1016/j.jacc.2017.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansson G.K. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 4.Yancy C.W., Jessup M., Bozkurt B., Butler J., Casey D.E., Jr., Drazner M.H. 2013 ACCF/AHA guideline for the management of heart failure: a report of the american college of cardiology foundation/American heart association task force on practice guidelines. Circulation. 2013;128(16):e240–e327. doi: 10.1161/CIR.0b013e31829e8776. [DOI] [PubMed] [Google Scholar]
- 5.Modesti P.A., Vanni S., Bertolozzi I., Cecioni I., Polidori G., Paniccia R. Early sequence of cardiac adaptations and growth factor formation in pressure- and volume-overload hypertrophy. Am J Physiol Heart Circ Physiol. 2000;279(3):H976–H985. doi: 10.1152/ajpheart.2000.279.3.H976. [DOI] [PubMed] [Google Scholar]
- 6.Gajarsa J.J., Kloner R.A. Left ventricular remodeling in the post-infarction heart: a review of cellular, molecular mechanisms, and therapeutic modalities. Heart Fail Rev. 2011;16(1):13–21. doi: 10.1007/s10741-010-9181-7. [DOI] [PubMed] [Google Scholar]
- 7.Yndestad A., Damas J.K., Oie E., Ueland T., Gullestad L., Aukrust P. Systemic inflammation in heart failure–the whys and wherefores. Heart Fail Rev. 2006;11(1):83–92. doi: 10.1007/s10741-006-9196-2. [DOI] [PubMed] [Google Scholar]
- 8.Allaband C., McDonald D., Vazquez-Baeza Y., Minich J.J., Tripathi A., Brenner D.A. Microbiome 101: studying, analyzing, and interpreting gut microbiome data for clinicians. Clin Gastroenterol Hepatol. 2019;17(2):218–230. doi: 10.1016/j.cgh.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan X.C., Huttenhower C. Meta'omic analytic techniques for studying the intestinal microbiome. Gastroenterology. 2014;146(6):1437–1448. doi: 10.1053/j.gastro.2014.01.049. [DOI] [PubMed] [Google Scholar]
- 10.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N. Ultra-high-throughput microbial community analysis on the illumina hiseq and miseq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fadrosh D.W., Ma B., Gajer P., Sengamalay N., Ott S., Brotman R.M. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the illumina miseq platform. Microbiome. 2014;2(1):6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallick H., Ma S., Franzosa E.A., Vatanen T., Morgan X.C., Huttenhower C. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 2017;18(1):228. doi: 10.1186/s13059-017-1359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui L., Zhao T., Hu H., Zhang W., Hua X. Association study of gut flora in coronary heart disease through high-throughput sequencing. Biomed Res Int. 2017;2017 doi: 10.1155/2017/3796359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jie Z., Xia H., Zhong S.L., Feng Q., Li S., Liang S. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8(1):845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Q., Gao R., Zhang Y., Pan D., Zhu Y., Zhang X. Dysbiosis signatures of gut microbiota in coronary artery disease. Physiol Genomics. 2018;50(10):893–903. doi: 10.1152/physiolgenomics.00070.2018. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson F.H., Fak F., Nookaew I., Tremaroli V., Fagerberg B., Petranovic D. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roediger W.E. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology. 1982;83(2):424–429. [PubMed] [Google Scholar]
- 20.Marques F.Z., Nelson E., Chu P.Y., Horlock D., Fiedler A., Ziemann M. High-Fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation. 2017;135(10):964–977. doi: 10.1161/CIRCULATIONAHA.116.024545. [DOI] [PubMed] [Google Scholar]
- 21.Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory t cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 22.Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 23.Troseid M., Nestvold T.K., Rudi K., Thoresen H., Nielsen E.W., Lappegard K.T. Plasma lipopolysaccharide is closely associated with glycemic control and abdominal obesity: evidence from bariatric surgery. Diabetes Care. 2013;36(11):3627–3632. doi: 10.2337/dc13-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandsma E., Kloosterhuis N.J., Koster M., Dekker D.C., Gijbels M.J.J., van d V. A proinflammatory gut microbiota increases systemic inflammation and accelerates atherosclerosis. Circ Res. 2019;124(1):94–100. doi: 10.1161/CIRCRESAHA.118.313234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Awoyemi A., Troseid M., Arnesen H., Solheim S., Seljeflot I. Effects of dietary intervention and n-3 pufa supplementation on markers of gut-related inflammation and their association with cardiovascular events in a high-risk population. Atherosclerosis. 2019;286:53–59. doi: 10.1016/j.atherosclerosis.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Brix S., Eriksen C., Larsen J.M., Bisgaard H. Metagenomic heterogeneity explains dual immune effects of endotoxins. J Allergy Clin Immunol. 2015;135(1):277–280. doi: 10.1016/j.jaci.2014.09.036. [DOI] [PubMed] [Google Scholar]
- 27.Vatanen T., Kostic A.D., d'Hennezel E., Siljander H., Franzosa E.A., Yassour M. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(4):842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang W.H., Wang Z., Levison B.S., Koeth R.A., Britt E.B., Fu X. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., Dugar B. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koeth R.A., Wang Z., Levison B.S., Buffa J.A., Org E., Sheehy B.T. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang W.H., Wang Z., Kennedy D.J., Wu Y., Buffa J.A., Agatisa-Boyle B. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116(3):448–455. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X.S., Obeid S., Klingenberg R., Gencer B., Mach F., Raber L. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes: a prognostic marker for incident cardiovascular events beyond traditional risk factors. Eur Heart J. 2017;38(11):814–824. doi: 10.1093/eurheartj/ehw582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z., Bergeron N., Levison B.S., Li X.S., Chiu S., Jia X. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur Heart J. 2019;40(7):583–594. doi: 10.1093/eurheartj/ehy799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koeth R.A., Levison B.S., Culley M.K., Buffa J.A., Wang Z., Gregory J.C. gamma-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of l-carnitine to tmao. Cell Metab. 2014;20(5):799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X.S., Wang Z., Cajka T., Buffa J.A., Nemet I., Hurd A.G. Untargeted metabolomics identifies trimethyllysine, a TMAO-producing nutrient precursor, as a predictor of incident cardiovascular disease risk. JCI Insight. 2018;3(6) doi: 10.1172/jci.insight.99096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X.S., Obeid S., Wang Z., Hazen B.J., Li L., Wu Y. Trimethyllysine, a trimethylamine N-oxide precursor, provides near- and long-term prognostic value in patients presenting with acute coronary syndromes. Eur Heart J. 2019;40(32):2700–2709. doi: 10.1093/eurheartj/ehz259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skagen K., Troseid M., Ueland T., Holm S., Abbas A., Gregersen I. The carnitine-butyrobetaine-trimethylamine-n-oxide pathway and its association with cardiovascular mortality in patients with carotid atherosclerosis. Atherosclerosis. 2016;247:64–69. doi: 10.1016/j.atherosclerosis.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 38.Ross R. Atherosclerosis–an inflammatory disease. N Engl J Med. 1999;340(2):115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 39.Zhou X., Li J., Guo J., Geng B., Ji W., Zhao Q. Gut-dependent microbial translocation induces inflammation and cardiovascular events after ST-elevation myocardial infarction. Microbiome. 2018;6(1):66. doi: 10.1186/s40168-018-0441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu W., Gregory J.C., Org E., Buffa J.A., Gupta N., Wang Z. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165(1):111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Troseid M. Gut microbiota and acute coronary syndromes: ready for use in the emergency room? Eur Heart J. 2017;38(11):825–827. doi: 10.1093/eurheartj/ehx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luedde M., Winkler T., Heinsen F.A., Ruhlemann M.C., Spehlmann M.E., Bajrovic A. Heart failure is associated with depletion of core intestinal microbiota. ESC Heart Fail. 2017;4(3):282–290. doi: 10.1002/ehf2.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamo T., Akazawa H., Suda W., Saga-Kamo A., Shimizu Y., Yagi H. Dysbiosis and compositional alterations with aging in the gut microbiota of patients with heart failure. PLoS ONE. 2017;12(3) doi: 10.1371/journal.pone.0174099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui X., Ye L., Li J., Jin L., Wang W., Li S. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Sci Rep. 2018;8(1):635. doi: 10.1038/s41598-017-18756-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kummen M., Mayerhofer C.C.K., Vestad B., Broch K., Awoyemi A., Storm-Larsen C. Gut microbiota signature in heart failure defined from profiling of 2 independent cohorts. J Am Coll Cardiol. 2018;71(10):1184–1186. doi: 10.1016/j.jacc.2017.12.057. [DOI] [PubMed] [Google Scholar]
- 46.Sandek A., Anker S.D., von H.S. The gut and intestinal bacteria in chronic heart failure. Curr Drug Metab. 2009;10(1):22–28. doi: 10.2174/138920009787048374. [DOI] [PubMed] [Google Scholar]
- 47.Pasini E., Aquilani R., Testa C., Baiardi P., Angioletti S., Boschi F. Pathogenic gut flora in patients with chronic heart failure. JACC Heart Fail. 2016;4(3):220–227. doi: 10.1016/j.jchf.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Jones R.B., Zhu X., Moan E., Murff H.J., Ness R.M., Seidner D.L. Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci Rep. 2018;8(1):4139. doi: 10.1038/s41598-018-22408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandek A., Bauditz J., Swidsinski A., Buhner S., Weber-Eibel J. Altered intestinal function in patients with chronic heart failure. J Am Coll Cardiol. 2007;50(16):1561–1569. doi: 10.1016/j.jacc.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 50.Tang W.H., Wang Z., Fan Y., Levison B., Hazen J.E., Donahue L.M. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: refining the gut hypothesis. J Am Coll Cardiol. 2014;64(18):1908–1914. doi: 10.1016/j.jacc.2014.02.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Troseid M., Ueland T., Hov J.R., Svardal A., Gregersen I., Dahl C.P. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J Intern Med. 2014 doi: 10.1111/joim.12328. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki T., Heaney L.M., Bhandari S.S., Jones D.J., Ng L.L. Trimethylamine N-oxide and prognosis in acute heart failure. Heart. 2016;102(11):841–848. doi: 10.1136/heartjnl-2015-308826. [DOI] [PubMed] [Google Scholar]
- 53.Organ C.L., Otsuka H., Bhushan S., Wang Z., Bradley J., Trivedi R. Choline diet and its gut microbe-derived metabolite, trimethylamine N-Oxide, exacerbate pressure overload-induced heart failure. Circ Heart Fail. 2016;9(1) doi: 10.1161/CIRCHEARTFAILURE.115.002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Troseid M., Mayerhofer C.C.K., Broch K., Arora S., Svardal A., Hov J.R. The carnitine-butyrobetaine-TMAO pathway after cardiac transplant: impact on cardiac allograft vasculopathy and acute rejection. J Heart Lung Transplant. 2019 doi: 10.1016/j.healun.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 55.Madan S., Mehra M.R. The gut microbiome and transplantation: an amazon jungle. J Heart Lung Transplant. 2018;37(9):1043–1044. doi: 10.1016/j.healun.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 56.Mayerhofer C.C.K., Ueland T., Broch K., Vincent R.P., Cross G.F., Dahl C.P. Increased secondary/primary bile acid ratio in chronic heart failure. J Card Fail. 2017;23(9):666–671. doi: 10.1016/j.cardfail.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 57.Staels B., Fonseca V.A. Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration. Diabetes Care. 2009;32(Suppl 2):S237–S245. doi: 10.2337/dc09-S355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe M., Houten S.M., Mataki C., Christoffolete M.A., Kim B.W., Sato H. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 59.Vallim T.Q., Edwards P.A. Bile acids have the gall to function as hormones. Cell Metab. 2009;10(3):162–164. doi: 10.1016/j.cmet.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 60.Khurana S., Raufman J.P., Pallone T.L. Bile acids regulate cardiovascular function. Clin Transl Sci. 2011;4(3):210–218. doi: 10.1111/j.1752-8062.2011.00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang J.Y. Bile acids: regulation of synthesis. J Lipid Res. 2009;50(10):1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von H.S., Schefold J.C., Jankowska E.A., Springer J., Vazir A., Kalra P.R. Ursodeoxycholic acid in patients with chronic heart failure: a double-blind, randomized, placebo-controlled, crossover trial. J Am Coll Cardiol. 2012;59(6):585–592. doi: 10.1016/j.jacc.2011.10.880. [DOI] [PubMed] [Google Scholar]
- 63.Velasquez M.T., Centron P., Barrows I., Dwivedi R., Raj D.S. Gut microbiota and cardiovascular uremic toxicities. Toxins (Basel) 2018;10(7) doi: 10.3390/toxins10070287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Onal E.M., Afsar B., Covic A., Vaziri N.D., Kanbay M. Gut microbiota and inflammation in chronic kidney disease and their roles in the development of cardiovascular disease. Hypertens Res. 2019;42(2):123–140. doi: 10.1038/s41440-018-0144-z. [DOI] [PubMed] [Google Scholar]
- 65.Hayashi T., Yamashita T., Watanabe H., Kami K., Yoshida N., Tabata T. Gut microbiome and plasma microbiome-related metabolites in patients with decompensated and compensated heart failure. Circ J. 2018;83(1):182–192. doi: 10.1253/circj.CJ-18-0468. [DOI] [PubMed] [Google Scholar]
- 66.Mayerhofer C.C.K., Kummen M., Holm K., Broch K., Awoyemi A., Vestad B. Low fibre intake is associated with gut microbiota alterations in chronic heart failure. ESC Heart Fail. 2020 doi: 10.1002/ehf2.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng Q., Liu Z., Zhong S., Li R., Xia H., Jie Z. Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease. Sci Rep. 2016;6:22525. doi: 10.1038/srep22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sberro H., Fremin B.J., Zlitni S., Edfors F., Greenfield N., Snyder M.P. Large-Scale analyses of human microbiomes reveal thousands of small. Novel Genes. Cell. 2019;178(5):1245–1259. doi: 10.1016/j.cell.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J., Thingholm L.B., Skieceviciene J., Rausch P., Kummen M., Hov J.R. Genome-wide association analysis identifies variation in vitamin d receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48(11):1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 72.Zhernakova A., Kurilshikov A., Bonder M.J., Tigchelaar E.F., Schirmer M., Vatanen T. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maier L., Pruteanu M., Kuhn M., Zeller G., Telzerow A., Anderson E.E. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555(7698):623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu H., Esteve E., Tremaroli V., Khan M.T., Caesar R., Manneras-Holm L. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23(7):850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 75.Ridker P.M., Everett B.M., Thuren T., MacFadyen J.G., Chang W.H., Ballantyne C. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 76.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B. Expert consensus document. the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 77.Patel R., DuPont H.L. New approaches for bacteriotherapy: prebiotics, new-generation probiotics, and synbiotics. Clin Infect Dis. 2015;60(Suppl 2):S108–S121. doi: 10.1093/cid/civ177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lam V., Su J., Koprowski S., Hsu A., Tweddell J.S., Rafiee P. Intestinal microbiota determine severity of myocardial infarction in rats. FASEB J. 2012;26(4):1727–1735. doi: 10.1096/fj.11-197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gan X.T., Ettinger G., Huang C.X., Burton J.P., Haist J.V., Rajapurohitam V. Probiotic administration attenuates myocardial hypertrophy and heart failure after myocardial infarction in the rat. Circ Heart Fail. 2014;7(3):491–499. doi: 10.1161/CIRCHEARTFAILURE.113.000978. [DOI] [PubMed] [Google Scholar]
- 80.Costanza A.C., Moscavitch S.D., Faria Neto H.C., Mesquita E.T. Probiotic therapy with saccharomyces boulardii for heart failure patients: a randomized, double-blind, placebo-controlled pilot trial. Int J Cardiol. 2015;179:348–350. doi: 10.1016/j.ijcard.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 81.Mayerhofer C.C.K., Awoyemi A.O., Moscavitch S.D., Lappegard K.T., Hov J.R., Aukrust P. Design of the gutheart-targeting gut microbiota to treat heart failure-trial: a phase II, randomized clinical trial. ESC Heart Fail. 2018 doi: 10.1002/ehf2.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He Z., Wang J., Chen Y., Cong X., Li N., Ding R. Potential risk associated with direct modulation of the gut flora in patients with heart failure. ESC Heart Fail. 2019;6(3):555–556. doi: 10.1002/ehf2.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mayerhofer C.C.K., Awoyemi A., Hov J.R., Troseid M., Broch K. Reply: potential risk associated with direct modulation of the gut flora in patients with heart failure. ESC Heart Fail. 2019;6(3):557–558. doi: 10.1002/ehf2.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yelin I., Flett K.B., Merakou C., Mehrotra P., Stam J., Snesrud E. Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in ICU patients. Nat Med. 2019;25(11):1728–1732. doi: 10.1038/s41591-019-0626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gibson G.R., Hutkins R., Sanders M.E., Prescott S.L., Reimer R.A., Salminen S.J. Expert consensus document: the international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14(8):491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 86.Chambers E.S., Preston T., Frost G., Morrison D.J. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutr Rep. 2018;7(4):198–206. doi: 10.1007/s13668-018-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson L.P., Walton G.E., Psichas A., Frost G.S., Gibson G.R., Barraclough T.G. Prebiotics modulate the effects of antibiotics on gut microbial diversity and functioning in vitro. Nutrients. 2015;7(6):4480–4497. doi: 10.3390/nu7064480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chambers E.S., Byrne C.S., Morrison D.J., Murphy K.G., Preston T., Tedford C. Dietary supplementation with inulin-propionate ester or inulin improves insulin sensitivity in adults with overweight and obesity with distinct effects on the gut microbiota, plasma metabolome and systemic inflammatory responses: a randomised cross-over trial. Gut. 2019;68(8):1430–1438. doi: 10.1136/gutjnl-2019-318424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Conraads V.M., Jorens P.G., De Clerck L.S., van Saene H.K., Ieven M.M., Bosmans J.M. Selective intestinal decontamination in advanced chronic heart failure: a pilot trial. Eur J Heart Fail. 2004;6(4):483–491. doi: 10.1016/j.ejheart.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 90.Fox M.A., Peterson S., Fabri B.M., van Saene H.K. Selective decontamination of the digestive tract in cardiac surgical patients. Crit Care Med. 1991;19(12):1486–1490. doi: 10.1097/00003246-199112000-00008. [DOI] [PubMed] [Google Scholar]
- 91.Tang T.W.H., Chen H.C., Chen C.Y., Yen C.Y.T., Lin C.J., Prajnamitra R.P. Loss of gut microbiota alters immune system composition and cripples postinfarction cardiac repair. Circulation. 2019;139(5):647–659. doi: 10.1161/CIRCULATIONAHA.118.035235. [DOI] [PubMed] [Google Scholar]
- 92.McMillan A., Hazen S.L. Gut microbiota involvement in ventricular remodeling post-myocardial infarction. Circulation. 2019;139(5):660–662. doi: 10.1161/CIRCULATIONAHA.118.037384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lam V., Su J., Hsu A., Gross G.J., Salzman N.H., Baker J.E. Intestinal microbial metabolites are linked to severity of myocardial infarction in rats. PLoS ONE. 2016;11(8) doi: 10.1371/journal.pone.0160840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andraws R., Berger J.S., Brown D.L. Effects of antibiotic therapy on outcomes of patients with coronary artery disease: a meta-analysis of randomized controlled trials. JAMA. 2005;293(21):2641–2647. doi: 10.1001/jama.293.21.2641. [DOI] [PubMed] [Google Scholar]
- 95.Winkel P., Hilden J., Hansen J.F., Kastrup J., Kolmos H.J., Kjoller E. Clarithromycin for stable coronary heart disease increases all-cause and cardiovascular mortality and cerebrovascular morbidity over 10years in the claricor randomised, blinded clinical trial. Int J Cardiol. 2015;182:459–465. doi: 10.1016/j.ijcard.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 96.Heianza Y., Zheng Y., Ma W., Rimm E.B., Albert C.M., Hu F.B. Duration and life-stage of antibiotic use and risk of cardiovascular events in women. Eur Heart J. 2019 doi: 10.1093/eurheartj/ehz231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knoop K.A., McDonald K.G., Kulkarni D.H., Newberry R.D. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 2016;65(7):1100–1109. doi: 10.1136/gutjnl-2014-309059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vrieze A., van N.E., Holleman F., Salojarvi J., Kootte R.S., Bartelsman J.F. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–916. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 99.Kootte R.S., Levin E., Salojarvi J., Smits L.P., Hartstra A.V., Udayappan S.D. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26(4):611–619. doi: 10.1016/j.cmet.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 100.Smits L.P., Kootte R.S., Levin E., Prodan A., Fuentes S., Zoetendal E.G. Effect of vegan fecal microbiota transplantation on Carnitine- and Choline-Derived trimethylamine-n-oxide production and vascular inflammation in patients with metabolic syndrome. J Am Heart Assoc. 2018;7(7) doi: 10.1161/JAHA.117.008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DeFilipp Z., Bloom P.P., Torres S.M., Mansour M.K., Sater M.R.A., Huntley M.H. Drug-Resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med. 2019;381(21):2043–2050. doi: 10.1056/NEJMoa1910437. [DOI] [PubMed] [Google Scholar]
- 102.Wortelboer K., Nieuwdorp M., Herrema H. Fecal microbiota transplantation beyond clostridioides difficile infections. E Bio Med. 2019;44:716–729. doi: 10.1016/j.ebiom.2019.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Z., Roberts A.B., Buffa J.A., Levison B.S., Zhu W., Org E. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163(7):1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.De F.F., Pellegrini N., Vannini L., Jeffery I.B., La S.A., Laghi L. High-level adherence to a mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65(11):1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 105.Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 107.Mendes-Soares H., Raveh-Sadka T., Azulay S., Edens K., Ben-Shlomo Y., Cohen Y. Assessment of a personalized approach to predicting postprandial glycemic responses to food among individuals without diabetes. JAMA Netw Open. 2019;2(2) doi: 10.1001/jamanetworkopen.2018.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.