Abstract
Background
Glioma has a poor prognosis, and is the most common primary and lethal primary malignant tumor in the central nervous system. Retinoic acid receptor-related orphan receptor A (RORA) is a member of the ROR subfamily of orphan receptors and plays an anti-tumor role in several cancers.
Methods
A cell viability assay, the Edu assay, neurosphere formation assay, and xenograft experiments were used to detect the proliferative abilities of glioma cell line, glioma stem cells (GSCs). Western blotting, ELISAs, and luciferase reporter assays were used to detect the presence of possible microRNAs.
Findings
Our study found for the first time that RORA was expressed at low levels in gliomas, and was associated with a good prognosis. RORA overexpression inhibited the proliferation and tumorigenesis of glioma cell lines and GSCs via inhibiting the TNF-α mediated NF-κB signaling pathway. In addition, microRNA-18a had a promoting effect on gliomas, and was the possible reason for low RORA expression in gliomas.
Interpretation
RORA may be a promising therapeutic target in the treatment of gliomas.
Keywords: Glioma, Glioma stem cells, RORA, Tumorigenesis, microRNA-18a
Research in context.
Evidence before this study
Glioma is the most common and lethal primary malignant tumor of the central nervous system, and the prognosis of glioma patients is not ideal and the median survival time of glioma patients is only 12–15 months. Theories underlying glioma evolution, treatment resistance and recurrence support the existence and key role of glioma stem cells (GSCs). Therefore, our study of disordered genes in GSCs may provide possible treatment target for glioma molecular therapy. RORA can act as a transcription factor and play an important role in the cardiovascular diseases, chronic inflammation, immune insufficiency, cellular stress, tumorigenesis and other pathological processes. Moreover, RORA was reported playing an anti-tumor role in several cancers such as hepatocellular carcinoma, non-small-cell lung carcinoma, breast cancer, melanoma, ovarian cancer. However, there was no study about the possible role and effect of RORA in glioma.
Added value of this study
Our study firstly found that RORA was lower expressed in gliomas and associated with good prognosis. Performing Gene Set Enrichment Analysis (GSEA) based on the TCGA and CGGA databases, we found that lower RORA expression was associated with the tumor necrosis factor mediated signaling pathway. Through both in vivo and in vitro experiments, we found RORA may possibly inhibit glioma proliferation and tumorigenesis via inhibiting TNF-α-mediated NF-κB signaling pathway. Moreover, since microRNAs (miRNAs) can cause degradation of target genes, we found the overexpression of miR-18a in glioma may be the possible reason for lower RORA expression in glioma.
Implications of all the available evidence
We confirmed that RORA expressed lower in higher grade glioma patients with a good prognosis and played tumor suppress effect. RORA can inhibit the proliferation and tumorigenesis of glioma cell lines and GSCs via inhibiting TNF-α-mediated NF-κB signaling pathway. Moreover, we found that lower RORA expression in glioma was caused due to the overexpression of miR-18a, which provided a novel pathway to regulate RORA. RORA might be a promising therapeutic target in glioma treatment.
Alt-text: Unlabelled box
1. Introduction
Glioma is the most common and lethal primary malignant tumor of the central nervous system, accounting for 47.1% of all malignant tumors of this system [1]. Although great progress has been made in recent years in the treatment of glioma using surgery, chemotherapy, and radiotherapy, the therapeutic effects are still not satisfactory [2]. The prognosis of glioma patients are not ideal and the median survival time of these patients is only 12–15 months [3]. However, molecular targeting therapy may be one of the most promising therapies for glioma [4]. Theories underlying glioma evolution, treatment resistance, and recurrence support the existence and key roles of glioma stem cells (GSCs) [5]. Our study of disordered genes in GSCs may therefore provide possible treatment targets for glioma molecular therapy [6].
Retinoic acid receptor-related orphan receptor A (RORA) is the first identified member of the ROR subfamily of orphan receptors. It is widely expressed in multiple tissues, such as the brain, thymus, heart, liver, lung, gastrointestinal tissues, uterus, and skin [7,8]. RORA can act as a transcription factor and bind to the specific DNA response elements in the promoter of target genes, leading to expression and structural changing of target genes [9], [10], [11]. RORA participates in the regulation of human metabolism and plays an important role in cardiovascular diseases, chronic inflammation, immune insufficiency, cellular stress, tumorigenesis, and other pathological processes [12], [13], [14], [15]. Moreover, RORA has been reported to play an anti-tumor role and is expressed in low amounts in several cancers such as hepatocellular carcinoma, non-small-cell lung carcinoma, breast cancer, melanoma, and ovarian cancer [16], [17], [18], [19]. For example, RORA can downregulate β-catenin and function as a tumor suppressor in endometrial cancer [15], and RORA can inhibit tumor cell proliferation via directly binding to the promoter of E2F1 and inhibiting its acetylation [20]. In addition, RORA attenuates Wnt-targeted gene expression in colon cancer cells [21]. Although RORA is expressed in the cerebellum, thalamus, hippocampus, and cerebral cortex, and participates in neurotransmission, neuroplasticity, and circadian rhythms, there has been no report about the possible role and effect of RORA in glioma [22], [23], [24], [25].
In the present study, we first found abnormally low RORA expression in glioma using bioinformatics analyses and detection of clinical specimens. Using gene set enrichment analysis (GSEA) and both in vivo and in vitro experiments, we found that RORA may possibly inhibit glioma proliferation and tumorigenesis via inhibiting the TNF-α-mediated NF-κB signaling pathway. Moreover, because microRNAs (miRNAs) can cause degradation of target genes, we found that overexpression of miR-18a in glioma may be the reason for the low expression of RORA in glioma [26,27].
2. Materials and methods
2.1. Cell culture and cell treatment
Human glioma cell lines, T98G and LN229, were purchased from the American Type Culture Collection (Manassas, VA, USA). Human glioma cell lines, U87, U118, and U251, were purchased from the Chinese Academy of Sciences cell bank (Shanghai, China). Human glioma cell line H4, and U178 cells were purchased from iCell Bioscience (Shanghai, China). All glioma cell lines were maintained in Dulbecco's Modified Eagle's Medium (HyClone, Logan, UT, USA), supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Gibco) at 37 °C with 5% CO2. Normal human astrocytes were purchased from ScienCell Research Laboratories (San Diego, CA, USA) and maintained in astrocyte medium (ScienCell Research Laboratories). Six patient-derived primary GSCs with World Health Organization (WHO) grade II, III, and IV (WHO grade II: GSC2C and GSC2D; III: GSC3C and GSC3D; and IV: GSC4C and GSC4D) were cultured as previously described [28]. The detailed clinicopathological information is presented in Supplementary Table 1. The stemness of GSCs was detected by immunofluorescence staining of CD133 or nestin (Abcam, Cambridge, UK) and the multi-lineage differentiation capacity of GSCs was shown via immunofluorescence staining of GFAP and βIII tubulin (Abcam). Human recombinant TNF-α was reconstituted with distilled H2O to make a final concentration of 1.0 mg/mL, and treated at a concentration of 10 ng/mL.
2.2. Patients and samples
Seventy clinical samples from glioma patients were collected from January 2007 to December 2011 at the First Affiliated Hospital of China Medical University [29]. Another ten acute brain injury samples from patients were collected as the control group during the same period. Clinical information for these samples is outlined in supplementary Table 2. This study was approved by the ethics committee of the First Affiliated Hospital of China Medical University, and written informed consent was obtained from every patient.
2.3. Lentiviral vector construction and transfection
The lentivirus-based vectors for RORA overexpression, RNAi-mediated knockdown of RORA, miR-18a inhibitor, and its negative control were all obtained from Gene-Chem (Shanghai, China). Two siRNA sequences were designed for RORA silencing, the sequences were as follows: RORA-KD1, forward: 5′-GAUGUGUGGUGCUAGACAAGU-3′, reverse: 5′- UUGUCUAGCACCACACAUCAG-3′. RORA-KD2, forward: 5′- GGAGAAGUCAGCAAAGCAAUG-3′, reverse: 5′- UUGCUUUGCUGACUUCUCCUG-3′. MiR-18a inhibitor sequence: 5′-CUAUCUGCACUAGAUGCACCUUA-30. MiR-18a inhibitor negative control sequence: 5′-CAGUACUUUUGUGUAGUACAA-3′. The lentivirus transfection was performed as previously described [27,28].
2.4. Real-time PCR
Real-time PCR was performed as previously described [28]. The total RNA of glioma cells was extracted using the Mini-BEST Universal RNA Extraction kit (TaKaRa, Kyoto, Japan), following first-strand cDNA synthesis using the Prime-Script RT Master Mix (TaKaRa). Finally, the qPCR assays were detected using the SYBR Green Master Mix (TaKaRa) with PCR LightCycler480 (Roche Diagnostics, Basel, Switzerland). The sequences for PCR primer pairs were as follows: RORA, forward: 5′-ACTCCTGTCCTCGTCAGAAGA-3′, and reverse: 5′-CATCCCTACGGCAAGGCATTT-3′; TNF-α, forward: 5′-CCTCTCTCTAATCAGCCCTCTG-3′, and reverse: 5′-GAGGACCTGGGAGTAGATGAG-3′; β-actin, forward: 5′-CATGTACGTTGCTATCCAGGC-3′, and reverse 5′-CTCCTTAATGTCACGCACGAT-3′.
2.5. Western blotting
Western blotting was performed as previously described [28]. Briefly, the total proteins of glioma cells or tissues were isolated using a total cell protein extraction kit (KeyGen Biotechnology, Nanjing, China), followed by electrophoresis and transfer to a nitrocellulose membrane, then blocked with 2% bovine serum albumin (KeyGen Biotechnology). The primary antibodies against RORA (1:1000; Abcam), TNF-α (1:1000; Abcam), p-p65 (1:1000; Cell Signaling Technology, Danvers, MA, USA), p65 (1:1000; Cell Signaling Technology), p-IκBα (1:1000; Cell Signaling Technology), IκBα (1:1000; Cell Signaling Technology), p-IKKα/β (1:1000; Cell Signaling Technology), IKKα (1:5,00; Cell Signaling Technology), IKKβ (1:500; Cell Signaling Technology), and β-actin (1:2000; ProteinTech, Chicago, IL, USA) were used to incubate overnight at 4 °C. Following secondary antibodies (ProteinTech) incubation, the bands were detected using a chemiluminescence ECL kit (Beyotime Biotechnology, Beijing, China) and quantified using Image J software (National Institutes of Health, Bethesda, MD, USA).
2.6. Immunohistochemistry (IHC)
IHC was performed as previously described [28]. Briefly, the tumor tissues were embedded in paraffin, sliced into 4 μm sections, and labeled with primary antibody against RORA (1:100; Abcam), Ki-67 (1:100; Abcam), and TNF-α (1:100; Abcam). The slices were then stained with an immunohistochemical labeling kit (MaxVision Biotechnology, Fuzhou, China) and imaged under a light microscope (Olympus, Tokyo, Japan). The German immunohistochemical score(30) was used to evaluate the staining intensity and expression levels.
2.7. Immunofluorescence
Immunofluorescence staining was performed as previously described [28]. Briefly, the glioma cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, blocked with 5% bovine serum albumin, and probed with primary antibodies against CD133, nestin, GFAP, and βIII-tubulin (1:100; Abcam) at 4 °C overnight, followed by treatment with fluorescein isothiocyanate- or rhodamine-conjugated secondary antibodies. The cells were then counterstained with 4′,6-diamidino-2-phenylindole (Sigma-Aldrich, St. Louis, MO, USA). Staining was visualized using a laser scanning confocal microscope (Olympus).
2.8. Cell viability assay
The glioma cells and GSCs treated under different conditions were seeded into 96-well plates at 1 × 103 cells/well and cultured for 5 days. Cell viability was detected using the CellTiter 96® Aqueous Non-Radioactive Cell Proliferation Assay Kit (Promega, Madison, WI, USA) according to the manufacturer's instructions [30].
2.9. Edu assay
The glioma cells and GSCs treated under different conditions were seeded into 24-well plates at 1 × 105 cells/well for 24 h, and an EdU assay kit (Beyotime Biotechnology) was used according to the manufacturer instructions. The percentage of EdU-positive cells was calculated using a laser scanning confocal microscope (Olympus).
2.10. Neurosphere formation assay and in vitro limiting dilution assay
The neurosphere formation assay was performed as previously described [30]. Briefly, GSCs were seeded into 24-well plates at a density of 200 cells/well and cultured in fresh medium for 7 days. The relative neurosphere size and number of neurospheres were counted using a light microscope (Olympus). For in vitro limiting dilution assay, GSCs were seeded into 96-well plates at a gradient of 1, 10, 20, 30, 40 or 50 cells per well, with 10 replicates for each gradient. The number of neurospheres in each well was observed after 7 days incubation, and the sphere formation efficiency were calculated using the Extreme Limiting Dilution Analysis (http://bioinf.wehi.edu.au/software/elda) [31].
2.11. Luciferase reporter assay
Luciferase reporter assays were performed as previously described [29,30]. Briefly, the TNF-α reporter plasmids were constructed by Gene-Chem (Shanghai, China). The glioma cells and GSCs were seeded into 96-well plates at a density of 5 × 103 cells/well and transfected with wild-type TNF-α or mutant type reporter plasmids for 48 h. The luciferase activities were then detected using a Dual-Luciferase Reporter Assay System (Promega).
2.12. Enzyme-linked immunosorbent (ELISA)
An ELISA was performed as previously described [28]. The commercially available ELISA kits (Cusabio, Stratech, UK) were used to detect the concentrations of TNF-α, IL1β, IL6, IL8, IL10, IP-10, MCP1, and RANTES in the media supernatants of the glioma cells and GSCs. All readings were normalized to the protein concentration in the control groups.
2.13. Cell cycle analysis
The glioma cells and GSCs treated under different conditions were harvested, washed with phosphate-buffered saline, and fixed with 70% ice-cold ethanol for 12 h at 4 °C. The fixed cells were then incubated with propidium iodide (Beyotime Biotechnology) and 10 mg/mL RNase I (KeyGEN Biotechnology’) for 30 min, followed by flow cytometry analyses (BD Biosciences, Franklin Lakes, NJ, USA).
2.14. Xenograft experiments
Xenograft experiments were performed in accordance with the Animal Care Committee of China Medical University. Six-week-old female BALB/c nude mice (Beijing Vital River Laboratory Animal Technology, Beijing, China) were bred in the Laboratory Animal Center of China Medical University under specific pathogen free conditions. The GSCs treated under different conditions were orthotopically injected into the mouse brain at 2 mm lateral and 2 mm anterior to the bregma via a stereotaxic apparatus (5 × 104 cells per mouse). Each group with five mice was observed daily for signs of distress or death, and the tumor volume was calculated according to the formula: V = (D × d2)/2, where D represented the longest diameter and d represented the shortest diameter.
2.15. Bioinformatics analysis
The data on RORA mRNA expression, WHO grades, isocitrate dehydrogenase (IDH) status, and survival times of glioma patients were obtained from the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn) using the RNA-seq platform and The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) in HG-U133A platforms. GSEA (http://www.broadinstitute.org/gsea/ index.jsp) was used to find enrichment of signal pathways between high and low RORA expression groups. Four online miRNA databases, Starbase (http://starbase.sysu.edu.cn), TargetScan (www.targetscan.org), microRNA (http://www.microrna.org/microrna/home.do), and miRDB (http://mirdb.org), were used to predict the possible miRNAs that targeted RORA by examining the RORA 3′-UTR with bioinformatics algorithms [32].
2.16. Statistical analysis
All experiments were repeated at least three times and the results are expressed as the mean ± SD. The chi-square test and two-tailed Student's t-test were used for comparisons of two independent groups. One-way analysis of variance was used to evaluate the statistical significance among three or more groups. Pearson's correlation analysis was used to evaluate the correlation between two groups. The log-rank test and Kaplan-Meier analysis were used to analyze the survival rates of each group. Statistical analysis was performed using SPSS 23.0 software (IBM, Armonk, N.Y, USA). Two-tailed P values < 0.05 were considered significant.
3. Results
3.1. RORA is expressed in low amounts in glioma and is associated with a good prognosis
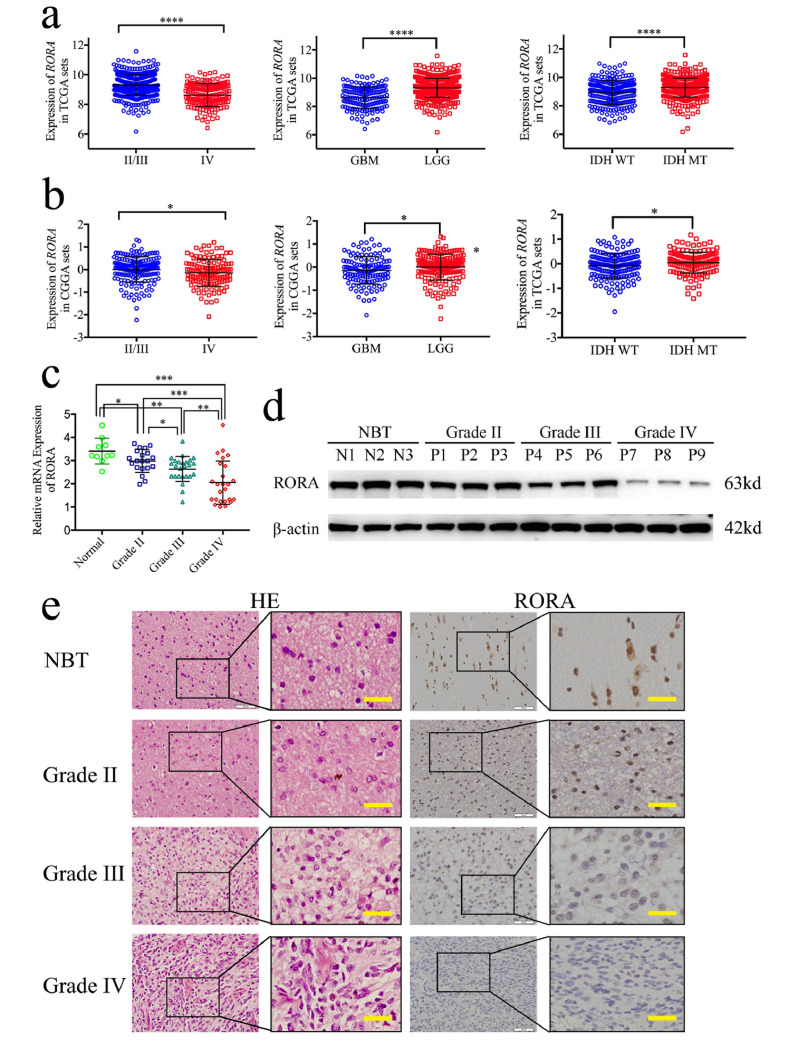
To investigate the possible role of RORA in glioma, we first analyzed the RORA expressions in TCGA and the CGGA databases. Glioblastoma (WHO IV) patients showed low RORA expressions than WHO grade II and grade III glioma in both TCGA and the CGGA databases (Fig. 1(a) and (b); WHO IV vs II/III: TCGA: P < 0.001, CGGA: P= = 0.0306; GBM vs glioma: TCGA: P < 0.001, CGGA: P= = 0.0077; Student's t-test). Then, we characterized the relationship between RORA and IDH status, which showed that RORA was highly enriched in IDH mutant type patients in both the CGGA and TCGA databases (Fig. 1(a); TCGA: P < 0.001; CGGA: P= = 0.016; Student's t-test). Furthermore, RORA overexpression was associated with increased survival rates in both TCGA and the CGGA databases according to Kaplan-Meier survival analyses (Fig. S1(a) and (e)). Moreover, we analyzed the separate prognostic significance of RORA according to glioma WHO grades. Surprisingly, although low RORA expression showed a shorter median survival time than high RORA expression in all grade 2, grade 3, and grade 4 glioma patients, a statistical difference was only found in grade 4 glioma of TCGA datasets (median survival time of low RORA vs high RORA: 12.5 vs 15 months, P= = 0.0318; Fig. S1(b)–(d), (f)–(h)). These results suggested that low RORA expression may be associated with a poor prognosis mainly in GBM or grade 4 gliomas.
Fig. 1.
Retinoic acid receptor-related orphan receptor A (RORA) is expressed at low levels in glioma. (a) and (b): the expressions of RORA are shown according to World Health Organization grades, GBM and LGG, and IDH status in The Cancer Genome Atlas (TCGA) (a) and the Chinese Glioma Genome Atlas (CGGA) (b) glioma datasets. (c), (d) and (e) The qPCR (c), western blots (d), and IHC (e) showed that RORA was expressed at low levels in glioma patients with different grades of disease, compared with normal brain tissue (NBT), (grade II, n = 20; grade III, n = 25; grade IV, n = 25; NBT n = 10; P < 0.001; one-way analysis of variance). Scale bar = 50 μm. All data are shown as the mean ± SD (from three independent experiments). *P < 0.05; ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001.
We next evaluated the expressions of RORA in 70 glioma patients and 10 normal brain tissues. All results (i.e., qPCR, western blotting, and IHC) showed that the expression of RORA in glioma was significantly low than that in normal brain tissues. In addition, the expression was significantly decreased with high glioma WHO grades (Fig. 1(c)–(e), supplementary Table 2). Furthermore, Kaplan–Meier survival analyses showed that the median survival time of all glioma patients with high RORA expression was 70 months compared with 29 months for patients with low RORA expression (P < 0.0001). The survival time of glioma patients with high RORA expression was therefore significantly longer than that of the low expression group (Fig. S1(i); P < 0.0001). The separate prognostic significance of RORA were furtherly analyzed according to glioma WHO grades. Low RORA expression showed a shorter median survival time than high RORA expression in both WHO grade 3 and grade 4 glioma patients (Fig. S1(j)–(l)). Finally, both Cox univariate and multivariate analyses showed that RORA expression, WHO grades and IDH status were independent prognostic factors of glioma patients (Table 1).
Table 1.
Cox univariate and multivariate analysis of glioma patients.
| Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|
| Factors | Categories | Χ2 | P value | HR | P value |
| Gender | Male/female | 0.0585 | 0.8089 | 0.6323 | 0.2735 |
| Age | ≤50/>50 | 0.1426 | 0.7057 | 0.6366 | 0.3324 |
| WHO grade | Grade II | 13.6665 | 0.0011 | 0.2404 | 0.0090 |
| Grade III | |||||
| Grade IV | |||||
| IDH status | Mutant/wild | 11.5877 | 0.0007 | 0.1343 | 0.0006 |
| RORA expression | High/low | 15.6610 | <0.0001 | 0.2559 | 0.0041 |
Moreover, we successfully isolated GSCs from six patient glioma samples representing different pathological diagnoses (e.g., WHO grade II, III, or IV). The six GSC populations were designated GSC2C and GSC2D (WHO grade II), GSC3C and GSC3D (WHO grade III), and GSC4C and GSC4D (WHO grade IV). All GSCs were confirmed via immunofluorescence staining of the stem cell markers, CD133 and nestin (Fig. S2(a)), and differentiation markers, GFAP and β-III tubulin (Fig. S2(b)). Western blotting and qPCR also showed that RORA was expressed at high levels in the WHO grade II GSCs (GSC2C and GSC2D) compared with those in the WHO grade IV GSCs (GSC4C and GSC4D) (Fig. S2(c); d, P < 0.001; one-way analysis of variance). We also compared the expressions of RORA in common glioma cell lines and normal human astrocytes using western blotting and qPCR, and found that the expressions of RORA in common glioma cell lines were all low than those of normal human astrocytes (Fig. S2(c), e; P < 0.001; one-way analysis of variance). Based on these results, there was a significantly low expression of RORA in glioma, with a significant correlation between the expression of RORA and good patient prognoses.
3.2. RORA regulates the growth of glioma cells
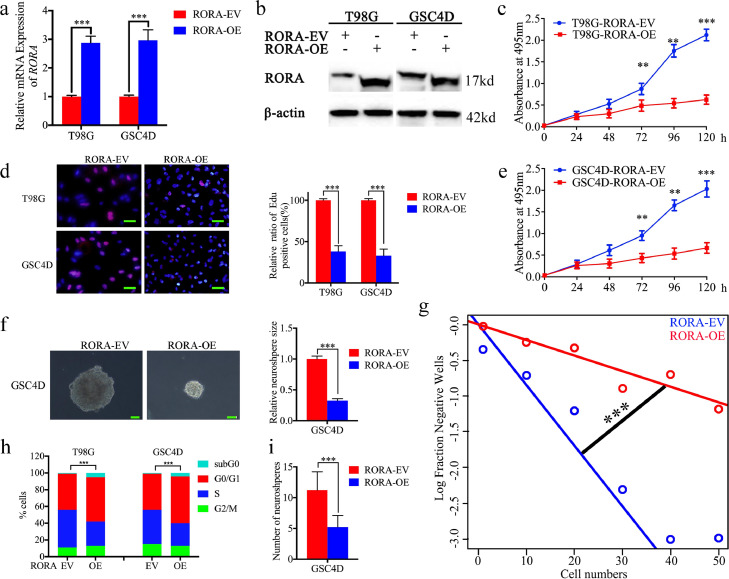
To determine the effect of RORA on the proliferation of glioma tumor cells and GSCs, T98G and GSC4D with the lowest RORA expressions were chosen for overexpression (Fig. S2(c)–(e)). Both western blotting and qPCR confirmed RORA overexpression (Fig. 2(a) and (b)). MTS and EdU assays confirmed that T98G and GSC4D proliferations decreased following RORA overexpression (Fig. 2(c)–(e)). Furthermore, the neurosphere formation rate and relative size of neurospheres formed by GSC4D were significantly smaller than the control group following RORA overexpression (Fig. 2(f)). Limiting dilution assays also showed that the tumor formation incidence decreased after RORA overexpression in GSC4D (Fig. 2(g)). Moreover, cell cycle assays showed that RORA overexpression induced cell cycle arrest and increased both the percentage of cells in subG0 and G0/G1 phases, while the percentage of cells in S phase decreased in both T98G and GSC4D cells (Fig. 2(h)).
Fig. 2.
Retinoic acid receptor-related orphan receptor A (RORA) overexpression inhibited glioma proliferation in vitro. (a) and (b) The lentiviral-based overexpression of RORA was detected by qPCR (a), T98G cells; P < 0.001; GSC4D; P< 0.001; Student's t-test) and western blots (b). (c) and (e) MTS assays showed the cell viability of T98G cells (c) P < 0.001; one-way analysis of variance and GSC4D (e) P < 0.001, one-way analysis of variance was decreased after RORA overexpression. (d) The Edu assay showed that the proliferations of T98G cells and GSC4D were decreased after RORA overexpression. (T98G cells: P < 0.001, GSC4D: P < 0.001; Student's t-test). Scale bar = 50 μm. (f) and (g) The neurosphere formation assay (f) and limiting dilution assays (g) showed the self-renewing capacity of GSC4D decreased after RORA overexpression (neurosphere size: P < 0.001, neurosphere number: P < 0.001; Student's t-test). Scale bar = 20 μm. (h) Cell cycle assay showed the effects of RORA on cell cycle distributions in T98G and GSC4D. (T98G cells: P < 0.001, GSC4D: P < 0.001; Student's t-test). All data are shown as the mean ± SD (from three independent experiments). *P < 0.05; ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001.
To further evaluate the effects of RORA on the growth of glioma tumor cells and GSCs, we designed two siRNA sequences with a lentivirus envelope, to silence RORA expression. We chose U87 and GSC2C for lentiviral infection due to their highest RORA expressions among all the available cells (Fig. S2(c)–(e)). Both western blotting and qPCR were performed to confirm that the expression of RORA was knocked down in both U87 and GSC2C following transfection of RORA siRNA (Fig. S3(a) and (b)). Furthermore, knockdown of RORA expression significantly increased the growth of U87 cells and GSC2C as determined by the MTS and EdU assays (Fig. S3(c)–(e)). In addition, RORA siRNA infection of GSC2C also increased the rate of neurosphere formation and size of the neurospheres, when compared to the control group (Fig. S3(f), (g), and (h)). Together, these results confirmed that RORA played a role in regulating glioma proliferation, and further confirmed that a high level of RORA expression inhibited the growth of glioma cells.
3.3. RORA negatively affected transcription and downregulated the expression and secretion of TNF-α
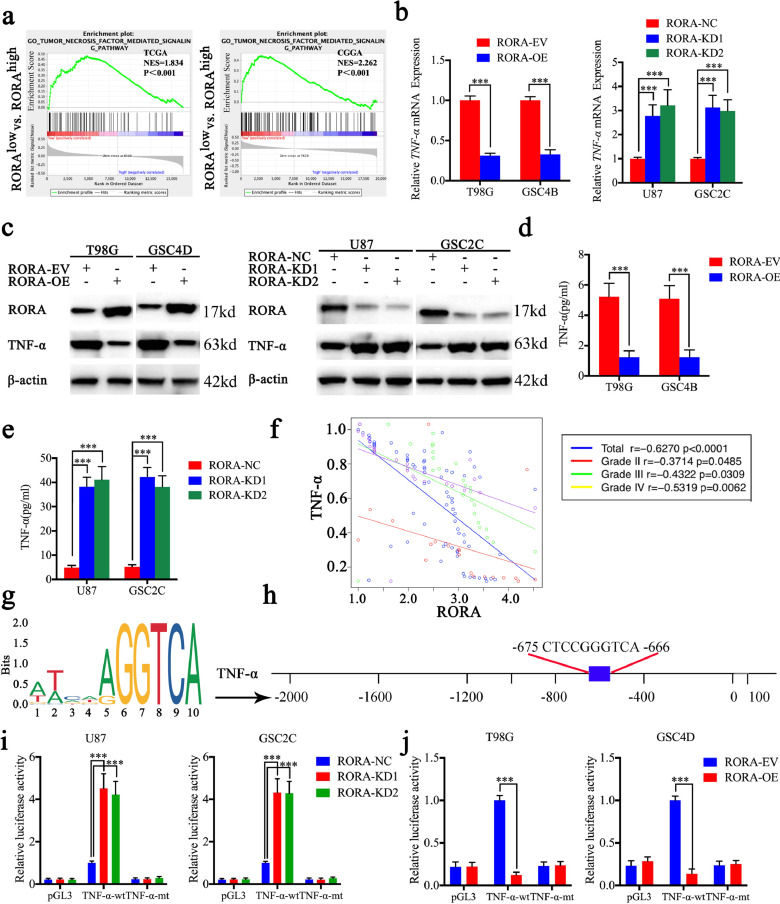
To identify the specific signaling pathways of RORA involved in gliomas, we performed GSEA based on TCGA and the CGGA databases. We found that low RORA expression was associated with the tumor necrosis factor-mediated signaling pathway (Fig. 3(a)). Therefore, we detected the expression changes of TNF-α after RORA overexpression or knockdown. All qPCR, western blotting, and ELISA assays showed that the expression and secretion of TNF-α was downregulated after RORA overexpression, while the opposite results were obtained after RORA knockdown (Fig. 3(b)–(e)). Furthermore, we detected the mRNA expressions of RORA and TNF-α in our clinical glioma specimens, which showed significant negative correlations between RORA and TNF-α expressions in WHO grade II (r = −0.3714, P = 0.0485), grade III (r = −0.4322, P = 0.0309), and grade IV (r = −0.5319, P = 0.0062), separately as well as in total patients (r = −0.6270, P < 0.001; Fig. 3(f)). Moreover, because RORA is a transcription factor, we investigated whether RORA transcriptionally regulated the expression of TNF-α (Fig. 3(g) and (h)). Using luciferase reporter assays, the relative luciferase activity of TNF-α increased after RORA knockdown in U87 cells and GSC2C, while the relative luciferase activity of TNF-α decreased after RORA overexpression in T98G cells and GSC4D (Fig. 3(i) and (j)).
Fig. 3.
Retinoic acid receptor-related orphan receptor A (RORA) inhibits the transcription and expression of TNF-α. (a) Gene set enrichment analysis indicated that high expression of RORA was negatively associated with the tumor necrosis factor-mediated signaling pathway in both TCGA and CGGA databases. (b)–(e) The expression and secretion of TNF-α after RORA overexpression or knockdown were detected by qPCR (b), RORA overexpression: T98G: P < 0.001, GSC4D: P < 0.001, Student's t-test; RORA knockdown: U87 cells: P < 0.001, GSC2C: P < 0.001, one-way analysis of variance, western blots (c) and ELISA (d) and (e). (f) The mRNA expression correlation between RORA and TNF-α in 70 cases of glioma patients were detected by qPCR. (g) Sequence motif representing the consensus RORA binding motif (JASPAR database). (h) Schematic representation of the human TNF-α promoter region. (i) and (j) RORA knockdown ((i), U87 cells: P < 0.001, GSC2C: P < 0.001, one-way analysis of variance) or overexpression ((j), T98G cells: P < 0.001, GSC4D: P < 0.001, Student's t-test) altered the luciferase promoter activities of TNF-α. All data are expressed as the mean ± SD (from three independent experiments). *P < 0.05; ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001.
3.4. RORA inhibits the TNF-α mediated NF-κB signaling pathway
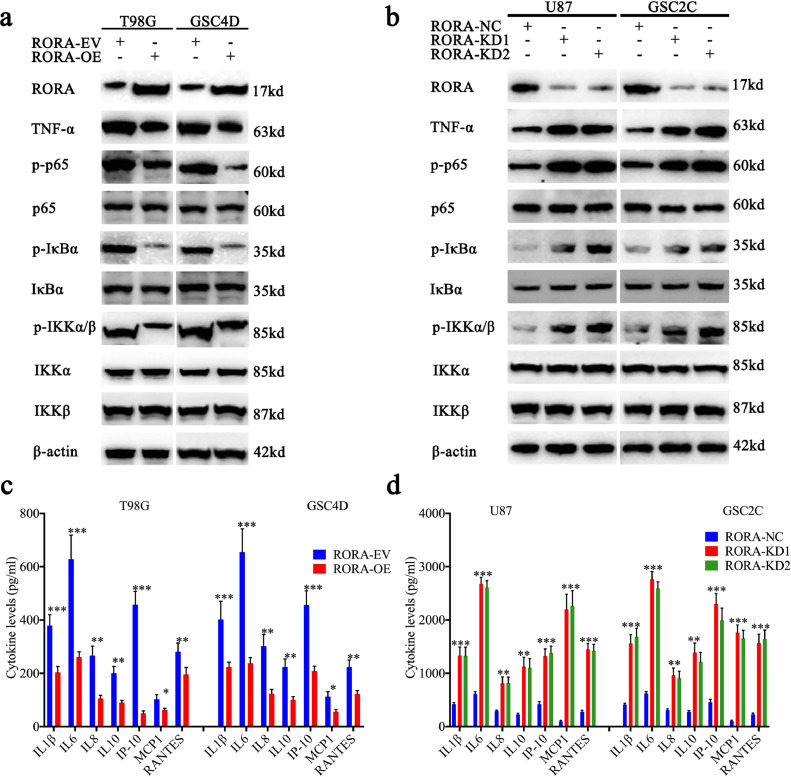
Because TNF-α plays a vital role in promoting tumorigenesis, proliferation, metastasis, and inhibiting apoptosis via the NF- κB signaling pathway, we further detected its downstream molecular expression. Based on the results of western blotting, the expressions of TNF-α, p-P65, p-IκBα, and p-IKKα/β were all significantly downregulated after RORA overexpression in U87 cells and GSC2C (Fig. 4(a)). Conversely, the expressions of all these molecules were significantly upregulated following RORA knockdown in T98G cells and GSC4D (Fig. 4(b)). We further detected the secretion levels of proinflammatory cytokines regulated by the NF-κB signaling pathway using ELISA assays. The results showed that secretion of the NF-κB-regulated cytokines (i.e., IL1β, IL6, IL8, IL10, IP-10, MCP1, and RANTES) were all decreased after RORA overexpression and increased after RORA knockdown (Fig. 4(c) and (d)). Together, these results suggested that RORA inhibited glioma proliferation via negative transcriptional effects and downregulation of the expression and secretion of TNF-α, which further inhibited the NF-κB signaling pathway.
Fig. 4.
Retinoic acid receptor-related orphan receptor A (RORA) participates in the TNF-mediated NF-κB signaling pathway. (a) and (b) Western blots showed that the downstream targets of NF-κB signaling pathway were regulated after RORA overexpression (a) or knockdown (b). (c) and (d) RORA overexpression (c) and knockdown (d) regulated the secretion of proinflammatory cytokines in glioma cells and GSCs as measured by an ELISA. All data are shown as the mean ± SD (from three independent experiments). *P < 0.05; ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001.
3.5. Recombinant TNF- α treatment abrogates the inhibiting effects of RORA
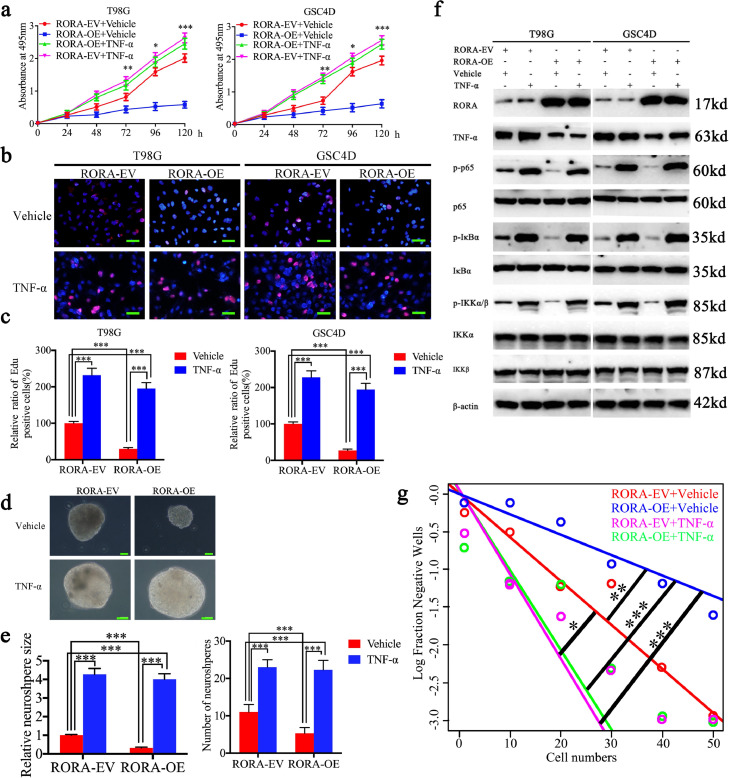
To identify whether RORA inhibited glioma proliferation via the TNF-α-mediated NF-κB signaling pathway, human recombinant TNF-α was treated with RORA-overexpressed T98G cells and GSC4D. Both the MTS and EdU assays confirmed that TNF-α treatment promoted the proliferation of empty vector-transfected T98G cells and GSC4D (Fig. 5(a)–(c)). RORA overexpression-induced proliferation inhibition was also reversed after TNF-α treatment (Fig. 5(a)–(c)). The neurosphere formation rate and relative size of neurospheres formed by GSC4D were also greater after TNF-α treatment, when compared with the control group (Fig. 5(d) and (e)). Also, TNF-α treatment reversed the RORA overexpression-induced low neurosphere formation rate and relative size of neurospheres (Fig. 5(d) and (e)). Limiting dilution assays also showed the similar results (Fig. 5(g)). Moreover, we detected the NF- κB signaling pathway after TNF-α treatment. The expression of p-P65, p-IκBα, and p-IKKα/β were all significantly increased after TNF-α treatment in T98G cells and GSC4D. Similarly, RORA overexpression-induced downregulation of p-P65, p-IκBα, and p-IKKα/β were all reversed following TNF-α treatment in T98G cells and GSC4D (Fig. 5(f)). Based on these results, we suggested that RORA-induced proliferation inhibition of glioma was abrogated following recombinant TNF- α treatment.
Fig. 5.
Recombinant TNF-α treatment abrogates the proliferation inhibiting effects of RORA. (a) The MTS assay showed that the inhibited cell viability of T98G cells and GSC4D after RORA overexpression was reversed after recombinant TNF-α treatment (T98G cells: P < 0.001, GSC4D: P < 0.001, one-way analysis of variance). (b) and ( c) The Edu assay showed that the inhibited proliferation of T98G cells and GSC4D after RORA overexpression was reversed following recombinant TNF-α treatment (T98G cells: P < 0.001, GSC4D: P < 0.001, one-way analysis of variance). Scale bar = 50 μm. (d), (e) and (g) The neurosphere formation assay (d) and (e) and limiting dilution assays (g) showed the inhibited self-renewing capacity of GSC4D after RORA overexpression was reversed after recombinant TNF-α treatment. (neurosphere size: P < 0.001, neurosphere number: P < 0.001, one-way analysis of variance). Scale bar = 20 μm. (f) Western blots showed the inhibited downstream targets of NF-κB signaling pathway after RORA overexpression was activated following recombinant TNF-α treatment. All data are shown as the mean ± SD (three independent experiments). *P < 0.05; ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001.
3.6. miR-18a negatively regulates RORA expression through binding with the 3′-UTR of RORA
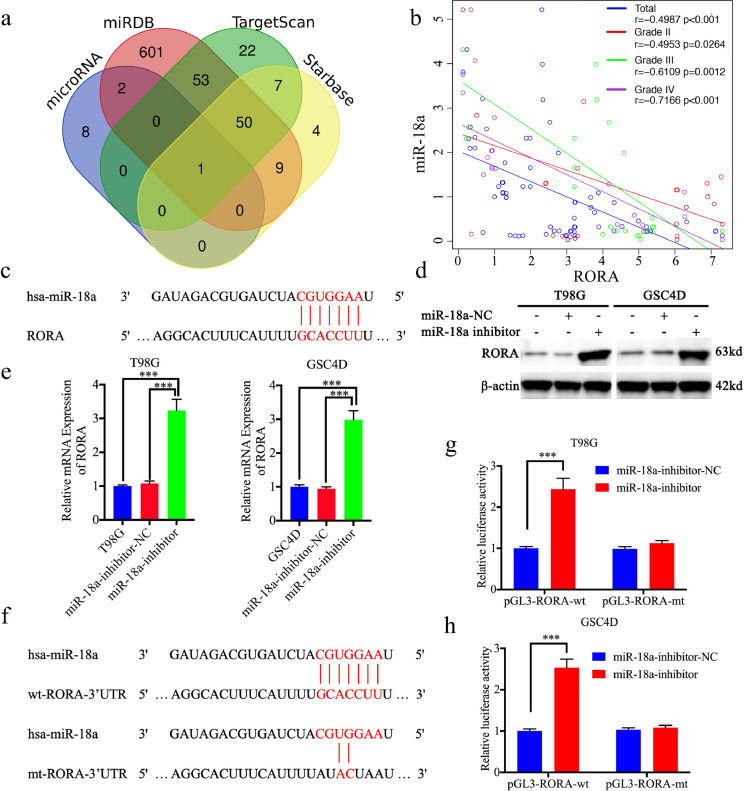
We further studied the possible causes of low RORA expression in gliomas. miRNAs are one of the most common causes of low expression of target genes, which lead to the degradation of downstream target genes via acting on its 3′-UTR [26]. We therefore used four databases, including Starbase, TargetScan, microRNA, and miRDB, to predict if miRNAs regulated RORA. miR-18a was the only intersection among these four databases (Fig. 6(a) and (c); Supplementary Table 3). We first detected mRNA expression of miR-18a and RORA in our clinical glioma specimens, which showed a significant negative correlation between miR-18a and RORA expression (Fig. 6(b)). Moreover, the negative correlation was increased with glioma grades, and was highest in glioma IV (Fig. 6(b)). According to the results of qPCR and western blotting, the expression of RORA was significantly upregulated after miR-18a inhibitor treatment in T98G cells and GSC4D (Fig. 6(d) and (e)), while the opposite results were obtained after miR-18a mimic treatment in U87 cells and GSC2C (Fig. S4(a) and (b)). We further performed luciferase reporter assays (Fig. 6(f)), and found that the relative luciferase activity of RORA increased after miR-18a inhibitor treatment in T98G cells and GSC4D cells (Fig. 6(g) and (h)). The opposite results were also obtained after miR-18a mimic treatment in U87 cells and GSC2C (Fig. S4(c) and (d)). Based on these results, miR-18a was a possible upstream regulatory factor, which negatively regulated RORA expression by binding with the RORA 3′-UTR.
Fig. 6.
miR-18a negatively regulated retinoic acid receptor-related orphan receptor A (RORA) expression by binding with the RORA 3′-UTR. (a) Identification of a miRNA that potentially regulated RORA expression based on Starbase, TargetScan, microRNA, and miRDB. (b) The mRNA expression correlation between RORA and miR-18a in 70 cases of glioma patients were detected by qPCR (r = −0.4987, P < 0.001, Pearson's correlation analysis). (c) and (f) Schematic diagram of the putative miR-18a binding site in the 3′-UTR of RORA in humans. (d) and (e) Western blotting (d) and qPCR (e) showed the expression of RORA increased after miR-18a inhibitor treatment (T98G cells: P < 0.001; GSC4D: P < 0.001, Student's t-test). (g) and (h) MiR-18a inhibitor treatment increased the luciferase promoter activities of RORA ((g), T98G cells: P < 0.001; (h), GSC4D: P < 0.001, Student's t-test). All data are shown as the mean ± SD (from three independent experiments). *P < 0.05; ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001.
3.7. RORA mediates the promoting effects of miR-18a in glioma
miR-18a has been shown to be highly expressed in glioma and can affect proliferation, migration, and invasion of human glioblastoma cells by targeting neogenin. We also detected its expression using the CGGA database. MiR-18a was expressed at the highest level in WHO grade IV glioma and lowest in WHO grade II glioma (Fig. S5(a), P < 0.001; one-way analysis of variance). miR-18a overexpression was associated with shorter survival according to Kaplan-Meier survival analyses (Fig. S5(b)). These results were completely contrary to those of RORA in glioma, so we further determined whether RORA was involved in miR-18a-regulated glioma proliferation.
We detected the proliferation of miR-18a inhibitor-treated T98G and GSC4D cells, and both the MTS and EdU assays showed that miR-18a inhibitor treatment inhibited the proliferation and decreased the percentage of EdU positive cells (Fig. S6(a)–(c)). However, miR-18a mimic treatment promoted the proliferation of U87 cells and GSC2C (Fig. S7(a)–(c)). Furthermore, the inhibitory proliferations of miR-18a inhibitor-treated T98G and GSC4D cells were reversed and even increased after RORA knockdown (Fig. S6(a)–(c)), while the promoted proliferation of the miR-18a mimic was also reversed after RORA overexpression (Fig. S7(a)–(c)). In addition, the neurosphere formation rate and relative size of the neurospheres formed by GSC4D were decreased after miR-18a inhibitor treatment, and were reversed after RORA knockdown (Fig. S6(d) and (e)). Similarly, miR-18a mimic treatment led to the opposite results (Fig. S7(d) and (e)).
3.8. miR-18a activates the TNF-α mediated NF-κB signaling pathway
We also detected whether miR-18a activated the TNF-α-mediated NF-κB signaling pathway. According to the results of western blotting, the expressions of TNF-α, p-P65, p-IκBα, and p-IKKα/β were all significantly upregulated after miR-18a mimic treatment in U87 cells and GSC2C (Fig. S8(a)). Conversely, the expressions of all these molecules were significantly downregulated following miR-18a inhibitor treatment in T98G cells and GSC4D (Fig. S8(b)). Moreover, the secretion of NF-κB-regulated cytokines were also increased after miR-18a mimic treatment in U87 and GSC2C and decreased after miR-18a inhibitor treatment in T98G cells and GSC4D (Fig. S8(c) and (d)).
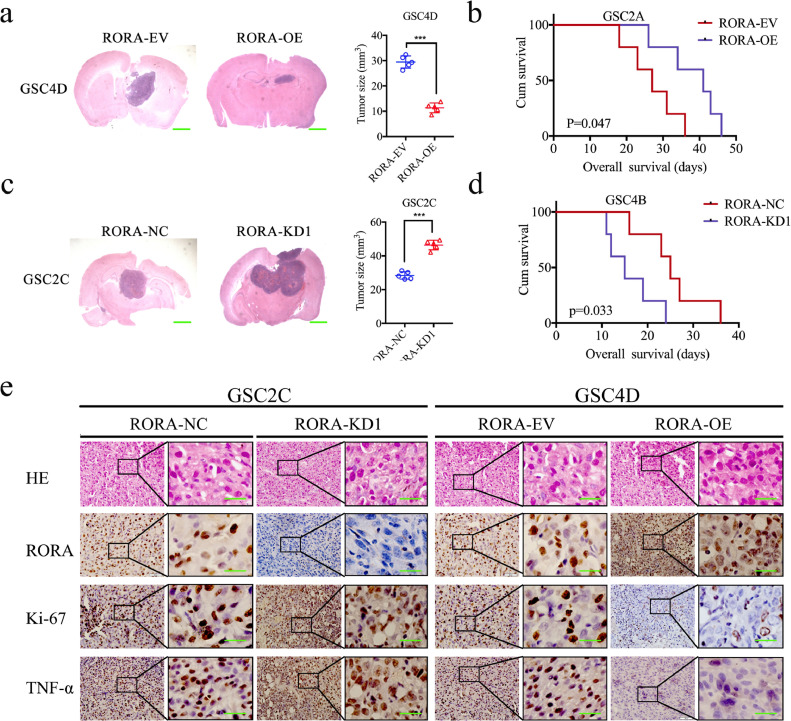
3.9. RORA inhibits glioma tumorigenesis
We then constructed orthotopic xenograft models to evaluate the effects of RORA on glioma tumorigenesis. The tumor volumes were decreased after RORA overexpression (Fig. 7(a), P < 0.0001; Student's t-test), and were accompanied by longer survival times and an increase in the median survival time (MST) from 27 to 41 days (Fig. 7(b); P = 0.0471; log-rank test). However, the tumor volumes were enlarged after RORA silencing (Fig. 7(c); P < 0.0001; Student's t-test), and were accompanied by shorter survival times and decreases in the MST from 25 to 15 days (Fig. 7(d); P = 0.0393; log-rank test). IHC was performed to detect the effects of RORA overexpression or silencing on tumor tissues. In the RORA overexpression group, the staining intensity and expression levels of Ki-67 and TNF-α were all decreased, while the opposite results were obtained in the RORA silenced group (Fig. 7(e)). Taken together, these results suggested that RORA inhibited the tumorigenesis in nude mice.
Fig. 7.
Retinoic acid receptor-related orphan receptor A (RORA) regulates glioma tumorigenesis. (a) and (c) Representative photographs showed that the sizes of intracranial tumors in the coronal position were decreased after RORA overexpression (a), while it increased after RORA was silenced (c). Scale bar = 10 mm. (b) and (d) Kaplan–Meier survival curves showed RORA overexpression shortened the survival times of nude mice (b), while it prolonged the survival times after RORA was silenced (d). For each group, n= = 5. (e) Representative immunohistochemical staining showing the changes in RORA, TNF-α, and Ki-67 after RORA overexpression and knockdown of orthotopic xenograft models. Scale bar = 50 μm. (f) Schematic diagram showing that the downregulation of RORA promoted the proliferation and tumorigenesis of glioma through the TNF-mediated NF-κB signaling pathway. All data are shown as the mean ± SD (three independent experiments). *P < 0.05; ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001.
4. Discussion
The decrease or deletion of the tumor suppressor gene is one of the most important causes of malignant tumors [33]. RORA was reported to be an important tumor suppressor gene in several cancers. RORA is a prognostic marker for hepatocellular carcinoma and suppresses hepatocellular carcinoma tumorigenesis [16,34]. RORA can inhibit adipocyte-conditioned medium-induced colorectal cancer cell proliferation and migration and chick embryo chorioallantoic membrane angiopoiesis [35]. miRNAs cooperatively suppressed RORA and promoted the proliferation of oral squamous cell carcinoma via decreasing p53 protein expression and suppressing p53 phosphorylation activity [26]. RORA can downregulate β-catenin and function as a tumor suppressor in endometrial cancer [15]. However, there have been almost no studies of RORA in glioma. Hypoxia-induced glioma exosomes can lead to immunosuppression by activating the miR-10a/RORA and miR-21/Pten pathways in myeloid-derived suppressor cells [36].
RORA is essential for cerebellar development, and is involved in several central nervous system pathologies [37]. RORA is reduced in the brain and lymphoblastoid cell lines of multiple cohorts of individuals with autism spectrum disorder [25]. RORA is specifically upregulated in the Alzheimer's disease (AD) hippocampus and emerges as a gene with a probable central role in AD pathology/etiology [24]. Microdeletions overlapping RORA on 15q22.2 can also lead to epileptic seizures and mild intellectual disabilities [38]. However, it is not known whether RORA participates in the tumorigenesis and development of gliomas.
Although four isoforms of RORA (RORA1–RORA4) can be produced according to their different N-termini [39] in normal human tissues, RORA1 and RORA4 are the main two isoforms transcribed [14]. RORA1, rather than RORA4, is mainly expressed in the central nervous system and is correlated with unfavorable clinicopathological features of colorectal cancer patients [39,40]. Therefore, isoform RORA1 was chosen in this study. We first detected the expression of RORA in both TCGA and the CGGA databases and found that RORA was expressed in low amounts in high WHO grades and IDH wild-type glioma patients. Similar results were all validated in our 70 cases of glioma patients. Although high RORA was associated with increased survival rates in all TCGA, CGGA databases and our 70 cases of glioma patients according to Kaplan-Meier survival analyses, statistical differences were only found in grade 3 and 4 glioma of our 70 cases of glioma patients and grade 4 of TCGA datasets. These differences may suggest that low RORA expression is associated with a poor prognosis mainly in GBM or grade 4 gliomas. We further conducted in vitro experiments to demonstrate the growth inhibiting effects of RORA in glioma cell lines and GSCs. All results from the MTS, EdU, and neurosphere formation assays showed that RORA overexpression inhibited the proliferation of T98G cells and GSC4D, while RORA knockdown led to the opposite results. Besides the orthotopic xenograft models, RORA overexpression inhibited glioma tumorigenesis and increased the survival times in nude mice. All these results indicated that RORA expression was decreased, which played an anti-tumor role and was associated with a good prognosis in gliomas.
To identify the possible mechanism of RORA's inhibitory proliferative effects in glioma, we conducted GSEA analyses based on TCGA and the CGGA databases. The results suggested that high RORA expression was involved in the negative regulation of the tumor necrosis factor signaling pathway. Inflammation plays a crucial role in growth of glioma tumors. Recent studies reported that inflammation plays an important role in the occurrence, development, and prognosis of glioma [41], [42], [43], [44]. TNF-α is a major regulator of inflammation, and is overexpressed and secreted in the tumor microenvironment [45]. TNF-α promotes the proliferation, migration, and treatment resistance of glioma via activating NF-κB signaling [46,47]. Moreover, it has been reported that RORA plays an anti-inflammatory function in human macrophages, and RORA deletion increases the expression and secretion of a subset of NF-κB regulated genes, such as TNF-α, IL-1β, and IL-6 [48]. RORA suppresses TNF-α-induced VCAM-1 and ICAM-1 expressions, as well as translocation of p50 and p65 to the nucleus in human endothelial cells [49].
We determined whether RORA inhibited the TNF-α-mediated NF-κB signaling pathway. All qPCR, western blotting, and ELISA assays showed that RORA overexpression inhibited the expression and secretion of TNF-α. In addition, luciferase reporter assays showed RORA overexpression inhibited the transcription of TNF-α. Moreover, RORA overexpression in orthotopic xenograft models also showed downregulation of TNF-α expression. Detection of the NF-κB signaling pathway showed that RORA overexpression downregulated the expression of p-P65, p-IκBα, and p-IKKα/β, and secretion of the NF-κB-regulated cytokines (i.e., IL1β, IL6, IL8, IL10, IP-10, MCP1, and RANTES). It is known that IL1β promotes the proliferation of glioma cells via the ERK and JNK signaling pathways [50]. IL6 promotes the proliferation of glioma cells via the JAK2/STAT3 signaling pathway [28]. The inhibition of these NF-κB-regulated cytokines caused by RORA overexpression may be a possible mechanism for its inhibitory effects in glioma. Moreover, human recombinant TNF-α was treated in RORA-overexpressed glioma cells and GSCs, showing that the inhibiting effects of RORA were all abrogated.
We also discovered possible reasons for low RORA expression in glioma. MiRNAs are approximately 22 nucleotide long endogenous non-coding RNAs, which mainly lead to mRNA cleavage or translational repression via binding to the 3′-UTR of mRNAs [26]. It has been reported and confirmed that miR-10a, miR-27a-3p, miR-181a-5p, miR-183-5p miR-450, and miR-503 can all directly target RORA [15,26,36]. In our study, we predicted the miRNAs that might regulate RORA, by using Starbase, TargetScan, microRNA, and miRDB. Furthermore, miR-18a was the only intersection among these four databases. The qPCR, western blotting, and luciferase reporter assays all showed that miR-18a negatively targeted RORA expression. Moreover, miR-18a has already been reported to have a promoting effect in glioma. miR-18a was more highly expressed, and could promote the proliferation, migration, and invasion of glioma cells by targeting neogenin [27]. The opposite effects between RORA and miR-18a also supported the regulation of RORA in glioma by miR-18a. Furthermore, the MTS, Edu, and neurosphere formation assays also showed that RORA knockdown abrogated the inhibiting effects of the miR-18a inhibition in glioma.
5. Conclusion
In summary, RORA played an anti-tumor role in glioma and was expressed in low amounts in high grade glioma patients with a good prognosis. RORA inhibited the proliferation and tumorigenesis of glioma cell lines and GSCs via inhibiting the TNF-α-mediated NF-κB signaling pathway. Moreover, low RORA expression in glioma was possibly caused by the overexpression of miR-18a (Fig. 7(F)). Therefore, RORA may be an important and promising therapeutic target in glioma.
Declarations of Competing Interest
The authors have declared that no conflict of interest exists.
Acknowledgments
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81101917, 81270036, and 30901736), the Plan to Focus on Research and Development from Science and Technology project of Liaoning Province (No. 2017225029), the Natural Science Foundation of Liaoning Province (No. 20170541022), the Liaoning BaiQianWan Talents Program (No. 2019-B45), the Science and Technology Plan Project of Shenyang City (No. 18-014-4-11), the Fund for Scientific Research of the First Hospital of China Medical University (No. FHCMU-FSR), and the Shanghai Sailing Program (No. 19YF1439000). The funding agencies did not participate in the study design, data collection, data analysis, interpretation, or writing of the report.
Acknowledgments
We would like to acknowledge our lab colleagues for their support in the development of this article.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102651.
Appendix. Supplementary materials
References
- 1.Ostrom Q.T., Gittleman H., Liao P. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol. 2017;19(suppl_5):v1–v88. doi: 10.1093/neuonc/nox158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sotoudeh H., Shafaat O., Bernstock J.D. Artificial intelligence in the management of glioma: era of personalized medicine. Front Oncol. 2019;9:768. doi: 10.3389/fonc.2019.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiocca E.A., Yu J.S., Lukas R.V. Regulatable interleukin-12 gene therapy in patients with recurrent high-grade glioma: results of a phase 1 trial. Sci Transl Med. 2019;11(505) doi: 10.1126/scitranslmed.aaw5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajesh Y., Pal I., Banik P. Insights into molecular therapy of glioma: current challenges and next generation blueprint. Acta Pharmacol Sin. 2017;38(5):591–613. doi: 10.1038/aps.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou D., Alver B.M., Li S. Distinctive epigenomes characterize glioma stem cells and their response to differentiation cues. Genome Biol. 2018;19(1):43. doi: 10.1186/s13059-018-1420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marin-Ramos N.I., Thein T.Z., Cho H.Y. NEO212 inhibits migration and invasion of glioma stem cells. Mol Cancer Ther. 2018;17(3):625–637. doi: 10.1158/1535-7163.MCT-17-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kojetin D.J., Burris T.P. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov. 2014;13(3):197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boukhtouche F., Mariani J., Tedgui A. The “CholesteROR” protective pathway in the vascular system. Arterioscler Thromb Vasc Biol. 2004;24(4):637–643. doi: 10.1161/01.ATV.0000119355.56036.de. [DOI] [PubMed] [Google Scholar]
- 9.Giguere V., Tini M., Flock G., Ong E., Evans R.M., Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8(5):538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 10.Jetten A.M. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moraitis A.N., Giguere V. Transition from monomeric to homodimeric DNA binding by nuclear receptors: identification of RevErbAalpha determinants required for RORalpha homodimer complex formation. Mol Endocrinol. 1999;13(3):431–439. doi: 10.1210/mend.13.3.0250. [DOI] [PubMed] [Google Scholar]
- 12.Lopez de Las Hazas M.C., Martin-Hernandez R., Crespo M.C. Identification and validation of common molecular targets of hydroxytyrosol. Food Funct. 2019;10(8):4897–4910. doi: 10.1039/c9fo01159e. [DOI] [PubMed] [Google Scholar]
- 13.Srividya G., Angayarkanni N., Iyer G., Srinivasan B., Agarwal S. Altered retinoid metabolism gene expression in chronic Stevens–Johnson syndrome. Br J Ophthalmol. 2019;103(8):1015–1023. doi: 10.1136/bjophthalmol-2018-312849. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y., McAvoy S., Kuhn R., Smith D.I. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene. 2006;25(20):2901–2908. doi: 10.1038/sj.onc.1209314. [DOI] [PubMed] [Google Scholar]
- 15.Sun X., Dongol S., Qiu C. miR-652 promotes tumor proliferation and metastasis by targeting RORA in endometrial cancer. Mol Cancer Res. 2018;16(12):1927–1939. doi: 10.1158/1541-7786.MCR-18-0267. [DOI] [PubMed] [Google Scholar]
- 16.Han L., Huang C., Zhang S. The RNA-binding protein SORBS2 suppresses hepatocellular carcinoma tumorigenesis and metastasis by stabilizing RORA mRNA. Liver Int. 2019 doi: 10.1111/liv.14202. [DOI] [PubMed] [Google Scholar]
- 17.Li B., Wang Y., Xu Y. Genetic variants in RORA and DNMT1 associated with cutaneous melanoma survival. Int J Cancer. 2018;142(11):2303–2312. doi: 10.1002/ijc.31243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W., Ma S., Bai X., Pan W., Ai L., Tan W. Long noncoding RNA WDFY3-AS2 suppresses tumor progression by acting as a competing endogenous RNA of microRNA-18a in ovarian cancer. J Cell Physiol. 2019 doi: 10.1002/jcp.29028. [DOI] [PubMed] [Google Scholar]
- 19.Qiu M., Chen Y.B., Jin S. Research on circadian clock genes in non-small-cell lung carcinoma. Chronobiol Int. 2019;36(6):739–750. doi: 10.1080/07420528.2018.1509080. [DOI] [PubMed] [Google Scholar]
- 20.Xiong G., Xu R. RORalpha binds to E2F1 to inhibit cell proliferation and regulate mammary gland branching morphogenesis. Mol Cell Biol. 2014;34(16):3066–3075. doi: 10.1128/MCB.00279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin D., Kim I.S., Lee J.M. The hidden switches underlying RORalpha-mediated circuits that critically regulate uncontrolled cell proliferation. J Mol Cell Biol. 2014;6(4):338–348. doi: 10.1093/jmcb/mju023. [DOI] [PubMed] [Google Scholar]
- 22.Calabro M., Mandelli L., Crisafulli C. Neuroplasticity, neurotransmission and brain-related genes in major depression and bipolar disorder: focus on treatment outcomes in an asiatic sample. Adv Ther. 2018;35(10):1656–1670. doi: 10.1007/s12325-018-0781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayad A., Noroozi R., Omrani M.D., Taheri M., Ghafouri-Fard S. Retinoic acid-related orphan receptor alpha (RORA) variants are associated with autism spectrum disorder. Metab Brain Dis. 2017;32(5):1595–1601. doi: 10.1007/s11011-017-0049-6. [DOI] [PubMed] [Google Scholar]
- 24.Acquaah-Mensah G.K., Agu N., Khan T., Gardner A. A regulatory role for the insulin- and BDNF-linked RORA in the hippocampus: implications for Alzheimer's disease. J Alzheimers Dis. 2015;44(3):827–838. doi: 10.3233/JAD-141731. [DOI] [PubMed] [Google Scholar]
- 25.Hu V.W., Sarachana T., Sherrard R.M., Kocher K.M. Investigation of sex differences in the expression of RORA and its transcriptional targets in the brain as a potential contributor to the sex bias in autism. Mol Autism. 2015;6:7. doi: 10.1186/2040-2392-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng X., Wu K., Liao S. MicroRNA-transcription factor network analysis reveals miRNAs cooperatively suppress RORA in oral squamous cell carcinoma. Oncogenesis. 2018;7(10):79. doi: 10.1038/s41389-018-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Y., Wang P., Zhao W. MiR-18a regulates the proliferation, migration and invasion of human glioblastoma cell by targeting neogenin. Exp Cell Res. 2014;324(1):54–64. doi: 10.1016/j.yexcr.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y., Han S., Cheng W., Wang Z., Wu A. NFAT1-regulated IL6 signalling contributes to aggressive phenotypes of glioma. Cell Commun Signal. 2017;15(1):54. doi: 10.1186/s12964-017-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Y., Zhou J., Luo P. Prosaposin promotes the proliferation and tumorigenesis of glioma through toll-like receptor 4 (TLR4)-mediated NF-κB signaling pathway. EBioMedicine. 2018;37(1):78–90. doi: 10.1016/j.ebiom.2018.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y., Song Y., Wang R. NFAT1-Mediated regulation of NDEL1 promotes growth and invasion of glioma stem-like cells. Cancer Res. 2019;79(10):2593–2603. doi: 10.1158/0008-5472.CAN-18-3297. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J., Jiang Y., Zhang H. Clinicopathological implications of TIM3(+) tumor-infiltrating lymphocytes and the miR-455-5p/Galectin-9 axis in skull base chordoma patients. Cancer Immunol Immunother. 2019;68(7):1157–1169. doi: 10.1007/s00262-019-02349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brand F., Forster A., Christians A. FOCAD loss impacts microtubule assembly, G2/M progression and patient survival in astrocytic gliomas. Acta Neuropathol. 2019 doi: 10.1007/s00401-019-02067-z. [DOI] [PubMed] [Google Scholar]
- 34.Fu R.D., Qiu C.H., Chen H.A., Zhang Z.G., Lu M.Q. Retinoic acid receptor-related receptor alpha (RORalpha) is a prognostic marker for hepatocellular carcinoma. Tumour Biol. 2014;35(8):7603–7610. doi: 10.1007/s13277-014-2007-9. [DOI] [PubMed] [Google Scholar]
- 35.Xiao L., Wang J., Li J. RORalpha inhibits adipocyte-conditioned medium-induced colorectal cancer cell proliferation and migration and chick embryo chorioallantoic membrane angiopoiesis. Am J Physiol Cell Physiol. 2015;308(5):C385–C396. doi: 10.1152/ajpcell.00091.2014. [DOI] [PubMed] [Google Scholar]
- 36.Guo X., Qiu W., Liu Q. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten pathways. Oncogene. 2018;37(31):4239–4259. doi: 10.1038/s41388-018-0261-9. [DOI] [PubMed] [Google Scholar]
- 37.Guissart C., Latypova X., Rollier P. Dual molecular effects of dominant rora mutations cause two variants of syndromic intellectual disability with either autism or cerebellar ataxia. Am J Hum Genet. 2018;102(5):744–759. doi: 10.1016/j.ajhg.2018.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto T., Mencarelli M.A., Di Marco C. Overlapping microdeletions involving 15q22.2 narrow the critical region for intellectual disability to NARG2 and RORA. Eur J Med Genet. 2014;57(4):163–168. doi: 10.1016/j.ejmg.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Matysiak-Scholze U., Nehls M. The structural integrity of ROR alpha isoforms is mutated in staggerer mice: cerebellar coexpression of ROR alpha1 and ROR alpha4. Genomics. 1997;43(1):78–84. doi: 10.1006/geno.1997.4757. [DOI] [PubMed] [Google Scholar]
- 40.Kano H., Takayama T., Midorikawa Y., Nagase H. Promoter hypomethylation of RAR-related orphan receptor alpha 1 is correlated with unfavorable clinicopathological features in patients with colorectal cancer. Biosci Trends. 2016;10(3):202–209. doi: 10.5582/bst.2016.01097. [DOI] [PubMed] [Google Scholar]
- 41.Yeung Y.T., McDonald K.L., Grewal T., Munoz L. Interleukins in glioblastoma pathophysiology: implications for therapy. Br J Pharmacol. 2013;168(3):591–606. doi: 10.1111/bph.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li G., Wang Z., Ye J. Uncontrolled inflammation induced by AEG-1 promotes gastric cancer and poor prognosis. Cancer Res. 2014;74(19):5541–5552. doi: 10.1158/0008-5472.CAN-14-0968. [DOI] [PubMed] [Google Scholar]
- 43.Reynes G., Vila V., Martin M. Circulating markers of angiogenesis, inflammation, and coagulation in patients with glioblastoma. J Neurooncol. 2011;102(1):35–41. doi: 10.1007/s11060-010-0290-x. [DOI] [PubMed] [Google Scholar]
- 44.Michelson N., Rincon-Torroella J., Quinones-Hinojosa A., Greenfield J.P. Exploring the role of inflammation in the malignant transformation of low-grade gliomas. J Neuroimmunol. 2016;297:132–140. doi: 10.1016/j.jneuroim.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 45.Ramaswamy P., Goswami K., Dalavaikodihalli Nanjaiah N., Srinivas D., Prasad C. TNF-alpha mediated MEK-ERK signaling in invasion with putative network involving NF-kappAB and STAT-6: a new perspective in glioma. Cell Biol Int. 2019 doi: 10.1002/cbin.11125. [DOI] [PubMed] [Google Scholar]
- 46.Guo G., Gong K., Ali S. A TNF-JNK-AXL-ERK signaling axis mediates primary resistance to EGFR inhibition in glioblastoma. Nat Neurosci. 2017;20(8):1074–1084. doi: 10.1038/nn.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geeviman K., Babu D., Prakash Babu P. Pantoprazole induces mitochondrial apoptosis and attenuates NF-kappAB signaling in glioma cells. Cell Mol Neurobiol. 2018;38(8):1491–1504. doi: 10.1007/s10571-018-0623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nejati Moharrami N., Bjorkoy Tande E., Ryan L., Espevik T., Boyartchuk V. RORalpha controls inflammatory state of human macrophages. PLoS ONE. 2018;13(11) doi: 10.1371/journal.pone.0207374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Migita H., Satozawa N., Lin J.H., Morser J., Kawai K. RORalpha1 and RORalpha4 suppress TNF-alpha-induced VCAM-1 and ICAM-1 expression in human endothelial cells. FEBS Lett. 2004;557(1–3):269–274. doi: 10.1016/s0014-5793(03)01502-3. [DOI] [PubMed] [Google Scholar]
- 50.Paugh B.S., Bryan L., Paugh S.W. Interleukin-1 regulates the expression of sphingosine kinase 1 in glioblastoma cells. J Biol Chem. 2009;284(6):3408–3417. doi: 10.1074/jbc.M807170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.