Short abstract
Hepatic fibrogenesis is a pathophysiological outcome of chronic liver injury hallmarked by excessive accumulation of extracellular matrix proteins. Fibrosis is a dynamic process that involves cross-talk between parenchymal cells (hepatocytes), hepatic stellate cells, sinusoidal endothelial cells and both resident and infiltrating immune cells. In this review, we focus on key cell-types that contribute to liver fibrosis, cytokines, and chemokines influencing this process and what it takes for fibrosis to regress. We discuss how mitochondria and metabolic changes in hepatic stellate cells modulate the fibrogenic process. We also briefly review how the presence of fibrosis affects development of hepatocellular carcinoma.
Impact statement
Advanced liver fibrosis results in cirrhosis, portal hypertension, and liver failure and often requires liver transplantation. Advanced liver fibrosis and cirrhosis are also major risk factors for hepatocellular carcinoma (HCC). Hepatic stellate cells (HSCs) play a pivotal role in the pathogenesis of liver fibrosis. In this review, we summarize the basic mechanisms that influence liver fibrosis development and how oxidative stress, mitochondrial dysfunction, and metabolic remodeling modulate HSC activation and indicate areas of potential therapeutic intervention.
Keywords: Liver fibrosis, fibrosis regression, liver cancer, hepatic stellate cells, mitochondria, metabolic pathways, PNPLA3
Introduction
Liver fibrosis is a consequence of sustained wound-healing response to chronic liver injury.1 The main causes include chronic HCV and HBV infection, exposure to toxins (e.g. alcohol liver disease), non-alcoholic steatohepatitis (NASH), and autoimmune diseases such as primary biliary cirrhosis, primary sclerosing cholangitis, and autoimmune hepatitis. Morphologically, liver fibrosis is characterized by accumulation of extracellular matrix (ECM), followed by formation of fibrous scar and subsequent cirrhosis which is defined by the presence of nodules of regenerating hepatocytes with decreased blood supply to the liver.2,3 Hepatic stellate cells (HSCs) are the main ECM-producing cells in the injured liver.4 In healthy livers, HSCs localize in the space of Disse, where they are in a quiescent state and store vitamin A (Figures 1 and 2). However, following continuous liver injury, HSCs activate into myofibroblasts, start expressing alpha-smooth muscle actin (α-SMA), migrate at the site of tissue repair, and secrete large amount of ECM (Figures 1 and 2). Interestingly, when the liver injury is removed, myofibroblasts may undergo apoptosis and inactivation,5 and therefore clearance of causative etiologies underlying liver fibrosis can slow down the process and lead to fibrosis regression. Despite extensive knowledge on liver fibrosis mechanisms, antifibrotic therapies effective in human have not been developed so far.6 In this review, we highlight recent data of molecular mechanisms of liver fibrosis and summarize the current knowledge of targeting each pathway of the pathogenesis process to relieve liver fibrosis.
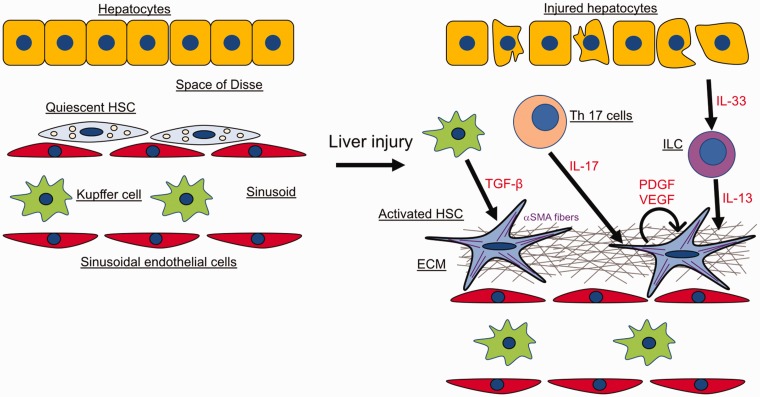
Figure 1.
Cell types in liver fibrosis. Quiescent HSCs in healthy livers are localized in the space of Disse. Following chronic liver damage, injured hepatocytes, immune cells and activated Kupffer cells release pro-fibrogenic molecules which activate HSCs. Activated HSCs upregulate ⍺-SMA expression, secrete growth factors, and produce large amounts of ECM. The figure depicts the different cell types and fibrogenic mediators involved in the fibrotic process and the crosstalk between them (see text for details).
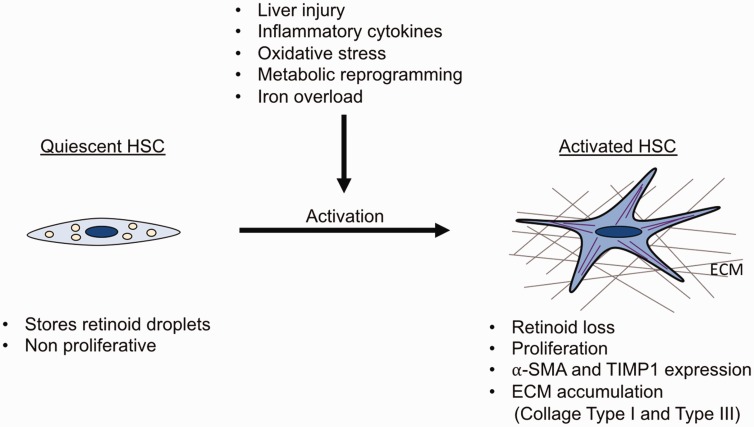
Figure 2.
Features of quiescent and activated HSCs. Quiescent HSCs store retinoid droplets, proliferate slowly, and express high levels of GFAP and LRAT. Upon liver injury, increase of inflammatory cytokines, ROS production, metabolic reprogramming or iron overload, HSCs become activated. Activated HSCs are characterized by loss of retinoid droplets, increased proliferation, and expression of fibrogenic markers (⍺-SMA, TIMP1, Lox and LoxL2). Moreover, activated HSCs produce ECM proteins such as collagen type I and collagen type III which are hallmarks of liver fibrosis.
Origin of fibrogenic myofibroblasts
The majority of the studies aiming at identifying the origin of myofibroblasts have suggested that most likely HSCs are the main source of myofibroblasts in the injured liver.7–9 However, this topic is still controversial and other than HSCs several other cell types have been proposed to give rise to myofibroblasts. For example, epithelial cells which in physiological conditions are located on the surface of blood vessels and organs could potentially lose their polarity, migrate and originate myofibroblasts through a process called epithelial–mesenchymal transition (EMT). Interestingly, some studies have shown that cholangiocytes and hepatocytes under prolonged culturing conditions increase the expression of α-SMA and downregulate epithelial markers.10,11 Nonetheless, lineage-tracing experiments that permanently label cholangiocytes, hepatocytes, and epithelial precursor cells have shown that epithelial cells do not give rise to hepatic myofibroblasts.12–14 These observations therefore suggest that EMT is not crucial for liver fibrogenesis in vivo.15
Bone marrow (BM)-derived cells like mesenchymal stem cells (MSCs) and fibrocytes have also been proposed to originate myofibroblasts. MSCs are multipotent cells that differentiate into osteoblasts, adipocytes, myocytes, and chondrocytes. Surprisingly, recently it has been reported that MSCs may protect the tissue in which they are recruited from developing fibrosis.16
Fibrocytes are cells with a spindle-like shape which were first described in 1994.17 They express fibroblast markers such as fibronectin, vimentin, collagen type I as well as hematopoietic cell markers like CD45, MHCII, CD34, CD11b, Gr1, CD86, CCR2, Ly6c, CD54, CD80, CCR1, CCR7, and CCR5.18,19 Studies have reported that both cholestatic and CCl4-induced liver injury can trigger the recruitment of fibrocytes into the injured liver where they can start expressing α-SMA. However, their contribution to liver myofibroblasts range between 3% and 50%.20–22
Mesothelial cells are similar to epithelial cells, and they can be found in internal organs and serous cavities where they are organized into cell monolayers.23 They originate from the embryonic mesoderm layer and some studies have suggested that both portal fibroblasts (PFs) and HSCs may derive from mesothelial cells.24,25 Interestingly, it has been shown that after CCl4-induced liver injury, both HSCs and myofibroblasts originate from mesothelial cells; however, in cholestatic liver injury, mesothelial cells give rise only to HSCs, but not myofibroblasts.26,27 Therefore, some controversies remain whether mesothelial cells can differentiate into myofibroblasts in fibrotic livers. Nonetheless, a study has suggested that mesothelial cells may be involved in fibrosis of the liver capsule.22
PFs consist of heterogeneous cell populations which are localized underneath the bile duct epithelium. PFs were first described in cholestatic liver disease by immunohistochemistry, histology, and electron miscroscopy.28–30 During biliary fibrosis, PFs start expressing α-SMA and produce ECM.31–33 However, identification or purification of quiescent PFs is challenging due to the lack of markers which can efficiently discriminate fibroblasts from other mesenchymal cells. Nevertheless, markers such as Ntpdas2, elastin, and Thy1 were recently reported to be expressed specifically by PFs, but not by HSCs.34 Intriguingly, by using transgenic mice, two collagen-producing cell populations were identified in the injured liver: Vitamin A-positive HSCs and Vitamin A-negative PFs.35 In CCl4-induced liver fibrosis, myofibroblasts originate mainly from HSCs, whereas PFs are the major source of myofibroblasts in early biliary fibrosis. However, as cholestatic disease progresses, HSCs contribute to the majority of myofibroblasts. More recently, it was reported that activation of PFs during cholestatic liver fibrosis is mediated by TGF-β1 and involves the interaction of mesothelin with a MUC16-Thy1-TGFβRI complex.36
In summary, current studies regarding the origin of hepatic myofibroblasts indicate that, depending on the type of liver injury, they arise mostly from liver-resident HSCs, mesothelial cells, and activated PFs. The contribution of BM-derived cells to hepatic fibrosis is quantitatively small, suggesting that these cells may be important in the modulation of other myofibroblast populations.
Key cytokines and chemokines involved in liver fibrosis
In liver fibrosis, death of hepatocytes and cholangiocytes causes activation of HSCs directly or through several cytokines which are released by immune cells including innate lymphoid cells (ILC), Kupffer cells, Th17 cells, and bone marrow-derived monocytes (Figure 1). Those inflammatory cytokines have been reported to affect liver fibrosis in vivo and in vitro.37 Interestingly, while it has been shown that monocyte chemotactic protein type I (MCP-1) and CCL5 promote fibrogenesis, IL-10 and IFN-γ have the opposite effect.38,39 IL-17 is secreted by Th17 cells and its levels are elevated in hepatitis B and C, alcoholic liver disease (ALD), and autoimmune hepatitis.40 Neutrophils and mast cells can also be a major producer of IL-17 in fibrotic liver.41 IL-17 is a profibrogenic cytokine which stimulates HSCs to increase levels of collagen type I, α-SMA, and TGF-β by activating NF-κB and STAT3.42 Deletion of IL-17 or IL-17RA protects mice from cholestatic and toxin-induced liver fibrosis.42 High blood levels of IL-22, another cytokine produced by Th17 cells, are observed in cirrhotic patients.43,44 Although IL-22 was shown to promote human hepatocellular carcinoma,45 other studies demonstrated the protective effects of IL-22 in murine models of ALD, acetaminophen-induced liver injury, and T-cell mediated hepatitis.46–48 Accordingly, it has been shown that IL-22 induces HSC senescence and inhibits liver fibrosis in mice.49 However, in contrast to these findings, other studies have suggested that IL-22 is proinflammatory and profibrogenic in hepatitis B patients and hepatitis B mice model.50–52 Additionally, increased IL-17 and IL-22 is a common signature for advanced liver fibrosis and pharmacological inhibition of IL-22 and IL-17 reduced hepatic fibrosis in murine models.41
IL-33 and its receptor ST2 are elevated in cirrhotic patients.53,54 In experimental models of mouse liver fibrosis, IL-33 is released from injured hepatocytes that stimulate ILC to produce IL-13 which in turn stimulates HSCs via the IL-4Rα and STAT6 axis.53 Liver sinusoidal endothelial cells and HSCs can also be a potential source of IL-33 in fibrotic liver.55 Mice lacking IL-33 or ST2 are protected from liver fibrosis.53,54
Chemokines are small chemotactic cytokines that direct chemotaxis and recruitment of leukocytes into the injured liver.56 Within the chemokine family, C-C motif chemokine (CCL2), also known as monocyte chemoattractant protein 1 (MCP-1), is released by activated macrophages, fibroblasts, and parenchymal cells during injury.57 CCL2 stimulates the recruitment of monocyte-derived macrophages in the injured liver causing activation of myofibroblasts.58 Therefore, the CCR2-CCL2 axis can potentially be an excellent target for therapeutic interventions.59–61
Among growth factors, transforming growth factor-β (TGF-β) plays a central role in liver fibrogenesis.62 Liver macrophages including Kupffer cells are the main producer or TGF-β, but it can also be secreted by HSCs. TGF-β binds to the TGF-β receptor type II which phosphorylates TGF-β receptor type-I which follows the activation of both Smad-dependent and Smad-independent pathways.62 In HSCs, TGF-β activates Smad2/3 stimulating the synthesis of ECM proteins such as type I and II collagen and inhibiting their degradation. Overall, TGF-β promotes activation of HSCs into myofibroblast and liver fibrosis through various mechanisms. For example, it has been shown that the Rho GTPase signaling mediates the TGF-β-induced HSCs migration.63 MicroRNAs (miRNA) have also been suggested to be involved in the regulation of HSCs by TGF-β. In particular, miR-29 is downregulated by TGF-β in HSCs with the consequent upregulation of ECM proteins.64 In addition, TGF-β-dependent downregulation of miR-30c and miR-193 in HSCs has an effect on ECM remodeling.65 Interestingly, it has been shown that the TGF-β pathway is inhibited by BAMBI and Smad7 which interact with and negatively regulate the TGF-β type I receptor.66 Consequently, it was demonstrated that inhibiting TGF-β synthesis or overexpressing BMP-7, which is an opposing factor of TGF-β, resulted in reduction of liver fibrosis.67,68
Platelet-derived growth factor (PDGF) is produced by platelets, macrophages, myofibroblasts, and HSCs. PDGF is the most potent mitogen for HSCs and its levels are elevated in fibrotic livers where, synergistically with TGF-β, it acts as a fibrogenic factor.69,70 PDGF is present in four isoforms (PDGF-A, B, C, and D) which interact with the PDGFR-α and -β, a receptor with intrinsic tyrosine kinase activity. In a rat model of biliary injury by BDL, the expression of PDGF-B, PDGF-D, and PDGFR-β was elevated compared to the other isotypes.71 Another study showed that overexpression of PDGF-B in transgenic mice increased liver fibrosis and HSCs activation.72 These results were confirmed by a study which reported that the PDGFR-β-mediated PDGF-B and PDGF-D signaling pathway are critical in the development of liver fibrosis.73 Therefore, strategies aimed at inhibiting both TGF-β and PDGF signaling can potentially result in promising antifibrotic therapies.
Oxidative stress in liver fibrosis
In the injured liver, apoptotic or necrotic hepatocytes, activated Kupffer cells, activated HSCs, and neutrophils produce reactive oxygen species (ROS) which facilitate HSC activation and migration.74 ROS are produced by nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) by transferring electron from nicotinamide adenine dinucleotide phosphate to molecular oxygen.75 Mammalian cells have seven NOX isoforms: NOX1, NOX2, NOX3, NOX4, NOX5, DUOX1, and DUOX2. HSCs express NOX1, NOX2, and NOX4.76,77 Mice lacking a regulatory component of NOX are not able to produce ROS in response to angiotensin II, PDGF, leptin, or apoptotic bodies. Consequently, those mice show less liver fibrosis after BDL or CCl4 treatment.78,79 In agreement with this result, deficiency of NOX1 and NOX4 protected mice from developing liver inflammation and fibrosis by inhibiting hepatic stellate cells activation.80 Recently, it was also reported that NOX1 expression in macrophages promotes hepatic tumorigenesis by inducing the production of inflammatory cytokines.81 Promising in vivo studies using the dual NOX1/NOX4 inhibitor GTK137831 have demonstrated that the compound can suppress ROS production, inhibit HSCs activation, and improve liver fibrosis in CCl4, BDL, and fast-food diet mouse model of liver injury.77,82–84 Interestingly, the cross talk between ROS and TGF-β is now very well recognized. On one hand, TGF-β inhibits the expression of several antioxidant enzymes, and on the other hand, it has been reported that ROS increases the expression of TGF-β in different organs.85,86
Role of mitochondria and metabolic reprogramming of HSC in fibrosis
Mitochondria play critical roles in energy and ROS production and homeostasis. About 90% of cellular ROS is produced in the mitochondria87 and as mentioned above, has important roles in hepatic fibrogenesis. Although many studies have delineated the role of mitochondria in hepatocytes during disease progression,88 very little is known about mitochondrial status in HSCs and how it plays a role during fibrogenesis. ECM production and secretion by activated HSCs demand abundant supply of intracellular ATP. Mild mitochondrial uncouplers that reduced ATP and ROS levels were able to prevent activation and proliferation of both mouse and human HSCs.89
Moreover, a recent study indicated an increase in mitochondrial respiration and membrane potential in fibrogenic HSCs.90 The elevated mitochondrial membrane potential in activated HSCs (compared to quiescent HSCs) sensitized them to mitochondria delivery of doxorubicin (Dox) via triphenylphosphonium (TPP) conjugation (TPP-Dox). While activated HSCs were killed by TPP-Dox, non-fibrogenic HSCs were spared.90
The activation of HSCs is accompanied by metabolic reprogramming similar to the Warburg effect in tumor cells, in which the cells tend to favor energy production via glycolysis rather than the much more efficient oxidative phosphorylation pathway. The metabolic switch to glycolysis during HSCs activation happens rapidly within the first 48 h of activation. Inhibition of glycolysis by 2-Deoxy-D-glucose (2DG) converts the activated HSCs to a more quiescent state.91 Pyruvate, the end product of glycolysis, is converted to lactate by the enzyme lactate dehydrogenase. Lactate accumulated in activated HSCs, and pharmacologic inhibition of lactate dehydrogenase suppressed HSCs activation.91
The amino acid glutamine is the most abundant circulating amino acid in blood and has important metabolic functions. Glutamine acts as a building block for several molecules such as for the synthesis of non-essential amino acids, synthesis of metabolites that are important in maintaining mitochondrial metabolism, and generation of antioxidants that counteract ROS.92 Similar to the reliance of cancer cells on glutamine metabolism (glutaminolysis),92 HSCs activation also depends on it.93,94 Expression of genes that regulate glutaminolysis increased during activation of HSCs that result in increased glutamine consumption. Consequently, either depletion of glutamine or inhibiting enzymes involved in glutaminolysis disrupted transdifferentiation of HSCs.93,94 Strategies that are targeted at lactate and glutamine metabolism may represent novel therapeutic approaches for the treatment of liver fibrosis. Hedgehog and YAP signaling were identified as important regulators of HSC metabolic switch during activation91,93,94 and also provide potential therapeutic avenues to explore.
Adenosine 5ʹ monophosphate-activated protein kinase (AMPK) is a master regulator of metabolism that maintains cellular energy homeostasis.95 Absence of adiponectin (an AMPK activator) in mice augmented CCl4-induced fibrosis.96 Moreover, activation of AMPK in human HSCs by adiponectin or AICAR inhibited HSCs activation and migration in response to PDGF.97 Similarly, AMPK silencing increased PDGF-induced HSCs proliferation, migration, and activation.97 It was indicated that AMPK activation can inhibit HSCs activation by dampening the activation of either NFkB or mTOR signaling.97
Angiogenesis and liver fibrosis
During chronic liver injury, two distinct pathways are responsible for the functional changes in liver vascular architecture. First, inflammation and fibrosis cause hypoxia which promotes angiogenesis.98,99 Second, the wound-healing process that is typical of chronic liver disease promotes the release of proangiogenic cytokines and growth factors.98–101 HSCs function as the pericytes of the liver because of their anatomical location and their ability to regulate sinusoid contraction.102 Once activated, HSCs promote angiogenesis by producing vascular endothelial growth factor (VEGF) and angiopoietin-1103,104 and increasing the expression of their receptors VEGFR-2 and Tie-2. Interestingly, it has been shown that VEGF directly stimulates HSCs proliferation, migration, and chemotaxis.105 In support to these findings, blocking VEGF or angiopoietin-1 reduces liver fibrosis.104,106 In addition to VEGF, PDGF can also contribute to the angiogenic phenotype of HSCs.107 Moreover, HSCs can secrete several CXC chemokines such as CXCL8, CXCL9, CXCL10, and CXCL12 which have been shown to stimulate angiogenesis during liver fibrosis.108,109
Role of iron overload in liver fibrosis
Iron is one of the most abundant element on Earth and is essential for all living organisms as it plays a critical role in various processes, such as oxygen transport, various enzymatic reactions including DNA synthesis and electron transport chain.110 The human body lacks proper physiological mechanisms to excrete iron110 and therefore needs to be closely regulated. On one hand where iron deficiency can cause anemia, excess iron can lead to severe oxidative stress and cellular damage by fueling the “Fenton reaction” that generates ROS and hydroxyl free radicals.111 In the human body, iron is primarily found bound to hemoglobin or stored in the liver by hepatocytes or Kupffer cells, which are especially prone to the toxic effects of iron overload. The clearest example is the toxic effect of iron on hepatocytes in hereditary hemochromatosis, which eventually leads to cirrhosis and HCC.111–113 Iron overload is a risk factor and might play a causal role in the development and progression of metabolic syndrome, type-2 diabetes, non-alcoholic fatty liver disease (NAFLD), ALD, NASH, and fibrosis.111,114
Hepcidin, a 25 amino acid polypeptide hormone produced by hepatocytes, is a central regulator of iron homeostasis.115 Hepcidins also have anti-microbial properties.115 Binding of hepcidin to the transmembrane iron exporter ferroportin on the iron-storing cells such as hepatocytes and Kupffer cells leads to ferroportin internalization and degradation and thereby prevents release of stored iron into the circulation.115,116 Similarly, ferroportin degradation by hepcidin in intestinal epithelial cells results in reduction of iron transfer into circulation.115 Although ferroportin expression was reported in both murine117 and human118 HSCs, very little is known about the role of ferroportin and hepcidin on HSC activation and fibrosis. Emerging evidence indicate that hepcidin has anti-fibrotic effects and its expression is inversely related to the extent of fibrosis in patients.118 Adenoviral-mediated hepcidin overexpression inhibited fibrosis in CCl4 and bile duct ligation models of mice.118 Ferroportin expression increases in activated HSC and the anti-fibrotic action of hepcidin in HSC is primarily mediated by degradation of ferroportin.118 Mechanistically, hepcidin inhibits TGF-β-mediated Smad3 activation which is linked to ferroportin-mediated regulation of AKT signaling. Ferroportin is a negative regulator of AKT activation. Therefore, hepcidin-mediated ferroportin downregulation increases P-AKT levels. Activated AKT sequesters unphosphorylated Smad3 in the cytosol and prevents heterodimerization with Smad4 and subsequent nuclear translocation thereby blunting the TGF-β-mediated signaling.118,119 Consequently, treatment of mice with either hepcidin or deletion of ferroportin inhibited experimental fibrosis in mice.118 Interestingly, bone morphogenic protein 6 (BMP6), a known inducer of hepcidin, which belongs to the TGF-β superfamily of growth factors, also inhibits hepatic fibrosis.120 Compared to WT mice, Bmp6−/− mice are prone to hepatic inflammation and fibrosis when fed MCD diet and recombinant BMP6 prevented HSC activation.120 Taken together, excessive iron can induce HSC activation and fibrosis by inducing oxidative stress in hepatocytes and Kupffer cells, thereby inducing inflammation and fibrosis.
Fibrosis regression
For long time, scientists believed that liver fibrosis is an irreversible process. However, studies performed in patients and rodent models have indicated that fibrosis can regress when the fibrotic insult (e.g. HBV, HCV, alcohol, chemicals, biliary obstruction, and obesity) is removed.121–124 The time needed to obtain a significant regression varies according to the cause and severity of the liver disease. However, whether cirrhosis, characterized by the most advanced stage of fibrosis, vascularized septae, and nodular parenchymal regeneration, can be reversed is still a matter of debate.125 Maturation of ECM scar makes it less accessible and resistant to proteases.122 The presence of non-reducible cross-linked collagen and changes in hepatic vasculature suggested that the cirrhotic process is irreversible.125,126 However, it is now clear that fibrosis and even cirrhosis may regress. When risk factors for chronic liver diseases are no longer present or the underlying conditions are successfully treated, hepatic reparative mechanisms can slowly improve the hepatic architecture.127–130 During the regression process, fibrous septa of cirrhotic livers may become thinned and perforated and the characteristic parenchymal nodularity can disappear with time.127
Increased collagen degradation is the main mechanism of fibrosis resolution.121 During fibrosis resolution, the loss of TGF-β signaling is critical. Then the reduction in the expression of tissue inhibitor of metalloproteinases 1 (TIMP-1) enables matrix metalloproteinases (MMPs) secreted by Kupffer cells/macrophage to degrade fibrillar collagen types I and III.131 Partial degradation of collagen in turn triggers HSCs apoptosis by several mechanisms, such as activation of death receptor-mediated pathways (Fas or TNFR-1 receptors), caspase 8 and 3, upregulation of pro-apoptotic proteins (e.g. p53, Bax, caspase 9), and downregulation of anti-apoptotic proteins (e.g. Bcl-2).132 Overexpression of peroxisome proliferator-activated receptor γ (PPARγ) or treatment with a PPARγ agonist reverted activated HSCs to a quiescent phenotype.133 Additionally, during liver fibrosis resolution, about 50% of activated HSCs reverted to a more quiescent state.5,134 These inactivated HSCs are more sensitive to further fibrogenic stimuli than quiescent HSCs. However, it is still not very well understood whether inactivation is a common feature of all activated myofibroblasts.
Liver fibrosis and HCC
HCC is the most common type of liver cancer and the second leading cause of cancer deaths worldwide with very limited treatment options.135 Furthermore, HCC is the fastest growing cancer in the United States.136 The main risk factors for HCC are chronic hepatitis B virus (HBV) or hepatitis C virus (HCV), excessive alcohol consumption, diabetes, and NASH.137 Advanced liver fibrosis and cirrhosis are major risk factors for HCC, with up to 90% of cases occurring on the background of a cirrhotic liver.138,139 Rarely, HCC can also develop in the absence of cirrhosis and advanced fibrosis such as those induced by inherited metabolic disorders, e.g. hemochromatosis, porphyria, and type-1 glycogen storage diseases.139,140 Compared to HCV-induced HCC that relies more on the cirrhotic microenvironment and the necroinflammatory response, HBV-induced HCC can progress more frequently in the absence of cirrhosis, most likely due to the differential ability of HCV and HBV to integrate into the host genome and directly modulate cellular oncogenesis.140,141 Nevertheless, HBV patients with cirrhosis more frequently progresses to HCC compared to non-cirrhotic HBV patients.142
Although the present main risk factors for HCC are HBV and HCV infections, their relative contribution, especially in developed countries, is rapidly declining due to the effectiveness of HBV vaccines and anti-HCV drugs.143–145 Non-alcoholic fatty liver disease (NAFLD) is now the most common cause of chronic liver disease in the United States.146 About 30% of the US adult population has NAFLD that increases to 90% in morbidly obese individuals.147 NAFLD is characterized by the histological presence of >5% macrovesicular steatosis of hepatocytes in an individual without significant alcohol use or other known causes of chronic liver disease. About 10–20% of NAFLD patients exhibit NASH, which is a severe and chronic liver inflammation that includes ballooning degeneration of hepatocytes, fibrosis, and liver damage.146,148 Quite a few of the patients suffering from NASH progress to end-stage liver disease (cirrhosis), liver failure, and HCC. Although NAFLD is currently the major cause of HCC without advanced fibrosis or cirrhosis in the US population,149 the risk of NAFLD patients for severe liver disease and mortality increased with the stage of fibrosis.150
Fibrosis and cancer-associated fibroblasts (CAF) can influence liver cancer development by modulating the tumor microenvironment.151,152 Fibrosis is a dynamic process. Deposition of ECM in fibrotic liver not only alters the mechanical properties of the liver but also has the ability to modulate and co-ordinate multiple signaling networks in epithelial, endothelial, stromal, immune, and tumor cells by either directly binding specific receptors such as integrins and growth factor receptors or by forming complexes with ligands that augment their activity and promote binding to their receptors.153 For example, fibronectin and vitronectin derived from ECM can either bind their cognate integrin receptors154 or can also bind hepatocyte growth factor (HGF) and modulate signaling by complexing c-Met (HGF receptor) and integrin receptors thereby stimulating the growth and survival of transformed cells.155
HSCs play both direct and indirect roles in HCC development. Besides being a key cell type responsible for fibrosis development, activated HSCs can indirectly support hepatic tumorigenesis by secreting angiogenic factors such as VEGF and angiopoietin-1, and CXC chemokines that can influence tumor vascularization.103,104,108,109 Additionally, HSCs can influence the immune landscape of the liver, tilting the scale towards malignant transformation. Besides being able to secrete pro-inflammatory cytokines, and stimulate Kupffer cells, activated HSCs express PDL-1 (B7-H1)156 and B7-H4157 that can induce either exhaustion or anergy of activated T-cells, respectively, in the liver thereby protecting malignant cells from the adaptive immune system and creating an environment conducive for survival and proliferation of pre-malignant cells. Additionally, HSCs may induce a tolerogenic environment in the liver by expanding immunosuppressive Treg cells through IL-2158 or retinoic acid-dependent manner.159
Not only activation of HSCs but also HSCs senescence may strongly influence the tumor development. Senescence of activated HSCs has been shown to improve liver fibrosis,160 that in effect would be against tumor development. Senescent cells develop a secretory profile composed mainly of inflammatory chemokines, cytokines, and proteases, which is known as senescence-associated secretory phenotype (SASP).161 Senescent HSCs have an inflammatory phenotype,162 and SASP from HSCs promotes HCC in obese mice treated with carcinogen.161 HSCs SASP can skew macrophage polarization towards a tumor-inhibiting M1-state, and inhibiting the HSCs SASP by p53 deletion results in M2 macrophage polarization and HCC promotion.163
HSC activation can also directly influence HCC development by secreting critical cytokines and chemokines such as HGF, TGF-β, PDGF, IL-6, and Wnt ligands that can act directly on the tumor cells.164–166 Cross-talk between HSCs and HCC cells has been shown in both in vitro and in vivo experiments.165,167 Co-culture with HSCs as well as conditioned media from HSCs culture promotes proliferation, migration, and inflammatory phenotypes of HCC cell lines,165,167 and the cross-talk between HSCs and HCC cells is bidirectional.167 Additional evidence of a tumor promoting role of HSCs is that HCC xenograft tumors grow better in the presence of HSCs.165
PNPLA3, fibrosis, and HCC
The patatin-like phospholipase domain-containing 3 (PNPLA3), also known as adiponutrin (ADPN), is a nutritionally regulated protein whose expression is controlled by the sterol regulatory element-binding protein 1c (SREBP-1c) and possess triglyceride (TG) hydrolase activity.168,169 In a cell, PNPLA3 is found localized in the endoplasmic reticulum and lipid droplets.170 The human PNPLA3 polymorphism (I148M), rs738409, has been strongly implicated in the development and progression of NAFLD, NASH, and fibrosis.170,171 Genome-wide association studies indicate that homozygous PNPLA3 (I148M) mutation increases the risk of NAFLD-associated HCC by 12-fold.172,173 However, whether PNPLA3 (I148) SNP promotes HCC development by triggering a specific oncogenic pathway or by creating a conducive microenvironment by promoting steatosis, inflammation, and fibrosis needs further investigation.174
The I148M SNP leads to a functional loss of the enzymatic activity and leads to TG accumulation in hepatocytes. However, the loss of PNPLA3 enzymatic activity is not sufficient to describe the functional consequence of PNPLA3 (I148M) mutation since Pnpla3−/− mice do not develop NAFLD.175,176 PNPLA3 (I148M) mutant protein is resistant to proteasome-mediated degradation and accumulates on the surface of hepatic lipid droplets which might interfere with the activity of other triglyceride metabolizing enzymes.170 Expression of an enzymatically active but ubiquitylation-resistant isoform of PNPLA3 accumulated on the lipid droplets and increased hepatic triglyceride levels when expressed in livers of mice.177 Therefore, not the loss of enzymatic activity but rather the property of PNPLA3 (I148M) mutant protein to accumulate on the lipid droplets might explain the observed phenotype associated with it. Consequently, Pnpla3 silencing reduces hepatic steatosis, NASH, and fibrosis.177,178
In the liver, PNPLA3 is expressed in both hepatocytes and HSCs and its expression is much higher in HSCs compared to hepatocytes.179 While PNPLA3 modulates TG metabolism in hepatocytes, PNPLA3’s primary role in HSCs is the hydrolysis and release of retinyl esters.174,179 Therefore, loss of PNPLA3 function would lead to intracellular retention of retinol in HSCs. Consequently, PNPLA3 148 M carriers with fatty liver or obesity have lower fasting circulating retinol concentrations180 and retinyl-palmitate was found elevated in the livers of homozygous PNPLA3 I148M carriers.181 HSCs are the primary storage depot of retinol (Vitamin A) in the body which are stored in HSCs lipid droplets.182 Retinol (Vitamin A) is known to influence cell growth, apoptosis, and differentiation183 and has also been associated with NAFLD.184,185 Upon activation, HSCs lose their retinol content and activate into myofibroblasts182; however, it is not clear whether loss of retinol plays any role in HSCs activation. PNPLA3 was shown to increase during HSCs activation in a TGFβ-dependent manner186 and may play a role during the activation process since PNPLA3 ablation may suppress HSCs activation.187 Moreover, PNPLA3 (I148M) mutant HSCs have a more inflammatory and fibrogenic phenotype and therefore can directly contribute to fibrosis development.187 Increased activation of PNPLA3 (I148M) HSCs is attributed to the suppression of PPARγ activity by JNK-mediated phosphorylation of the inhibitory Serine 84 residue and increase in pro-fibrogenic AP-1 activity.187
Besides classical TG and retinol metabolism, PNPLA3 might also affect other metabolic pathways. Overexpression of PNPLA3 I148M in HCC cell line (Huh7) led to two-fold increase in lactic acid suggesting a shift to anaerobic metabolism and mitochondrial dysfunction188 which, as discussed above, favors HSC activation. Additional studies are needed to confirm and better understand mechanisms of PNPLA3 (I143M) in HSC activation and whether the critical function of PNPLA3 (I148M) is in the hepatocyte, the HSCs, or both.
Conclusions and future prospects
Liver fibrosis is a major health problem that still lacks effective therapeutic strategies. Therefore, understanding the mechanisms underlying this process is crucial for translating basic research into new clinical therapies. In particular, translational research should focus on studying fibrosis in different types of human liver diseases such as ASH, NASH, and HCC in order to design targeted therapeutic interventions.
Since several liver cell types are involved in liver fibrosis, identifying the origin of myofibroblasts and a better understanding of how HSCs get activated and inactivated is of paramount importance. Moreover, uncovering the detailed crosstalk between hepatocytes, HSCs, and Kupffer cells during liver fibrosis progression and regression will definitely help to provide new therapeutic strategies.
How CAFs/HSCs, fibrosis, and the tumor microenvironment influence hepatocarcinogenesis also need to be fully elucidated. Although several studies have demonstrated that HSCs and fibrosis promote development of HCC, an alternative hypothesis is that CAFs may limit cancer progression. Therefore, future studies should aim at investigating whether HSCs and CAFs can also exert tumor-suppressive functions in the context of liver cancer. Answering this question may open new therapeutic paths for the treatment of HCC.
Metabolic reprogramming of HSCs during activation (Figure 3) is another area that needs further investigation. Metabolic addiction of HSCs to glycolytic and glutaminolytic pathways could be exploited to develop therapeutic strategies. Additionally, although the importance of PNPLA3 (I143M) in hepatocytes in promoting NAFLD and ALD is well established, its potential role in HSC activation remains enigmatic.
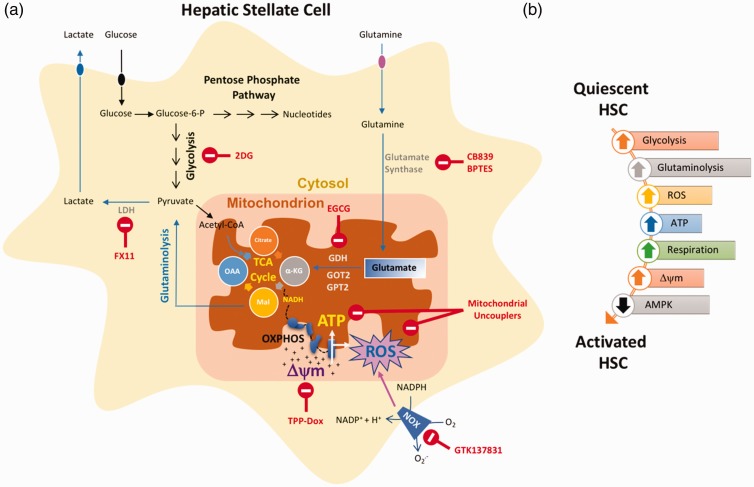
Figure 3.
Metabolic reprogramming of HSC. (a) Multiple aspects of glucose and glutamine metabolism are altered during HSC activation that can provide potential therapeutic avenues (indicated by red font). (b) A simplified depiction of metabolic pathways altered during HSC activation. Up arrows indicate upregulated pathway and down arrows mean downregulated. Δψm: Mitochondrial membrane potential; LDH: lactate dehydrogenase; GDH: glutamate dehydrogenase; GOT2: glutamic-oxaloacetic transaminase 2; GPT2: glutamate pyruvate transaminase 2.
Authors’ contributions
All authors contributed to the planning and writing of the manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the NIH/NIAAA P50AA011999 and NIH/NIEHS Superfund Training Core P42ES010337 grants to D.A.B. and ALF Liver Scholar award and NIH/UCSD 1KL2TR001444 to D.D.
ORCID iD
Debanjan Dhar https://orcid.org/0000-0001-8739-3684
References
- 1. Friedman SL. Liver fibrosis – from bench to bedside. J Hepatol 2003; 38(Suppl 1):S38–53 [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol 2011; 6:425–56 [DOI] [PubMed] [Google Scholar]
- 3.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest 2005; 115:209–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabele E, Brenner DA, Rippe RA. Liver fibrosis: signals leading to the amplification of the fibrogenic hepatic stellate cell. Front Biosci 2003; 8:d69–77 [DOI] [PubMed] [Google Scholar]
- 5.Kisseleva T, Cong M, Paik Y, Scholten D, Jiang C, Benner C, Iwaisako K, Moore-Morris T, Scott B, Tsukamoto H, Evans SM, Dillmann W, Glass CK, Brenner DA. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci U S A 2012; 109:9448–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hauff P, Gottwald U, Ocker M. Early to phase II drugs currently under investigation for the treatment of liver fibrosis. Expert Opin Investig Drugs 2015; 24:309–27 [DOI] [PubMed] [Google Scholar]
- 7.Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun 2013; 4:2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson NC, Arnold TD, Katamura Y, Giacomini MM, Rodriguez JD, McCarty JH, Pellicoro A, Raschperger E, Betsholtz C, Ruminski PG, Griggs DW, Prinsen MJ, Maher JJ, Iredale JP, Lacy-Hulbert A, Adams RH, Sheppard D. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med 2013; 19:1617–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puche JE, Lee YA, Jiao J, Aloman C, Fiel MI, Muñoz U, Kraus T, Lee T, Yee HF, Friedman SL. A novel murine model to deplete hepatic stellate cells uncovers their role in amplifying liver damage in mice. Hepatology 2013; 57:339–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi SS, Diehl AM. Epithelial-to-mesenchymal transitions in the liver. Hepatology 2009; 50:2007–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest 2009; 119:1417–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taura K, Miura K, Iwaisako K, Osterreicher CH, Kodama Y, Penz-Osterreicher M, Brenner DA. Hepatocytes do not undergo epithelial-mesenchymal transition in liver fibrosis in mice. Hepatology 2010; 51:1027–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholten D, Weiskirchen R. Questioning the challenging role of epithelial-to-mesenchymal transition in liver injury. Hepatology 2011; 53:1048–51 [DOI] [PubMed] [Google Scholar]
- 14.Chu AS, Diaz R, Hui JJ, Yanger K, Zong Y, Alpini G, Stanger BZ, Wells RG. Lineage tracing demonstrates no evidence of cholangiocyte epithelial-to-mesenchymal transition in murine models of hepatic fibrosis. Hepatology 2011; 53:1685–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munker S, Wu YL, Ding HG, Liebe R, Weng HL. Can a fibrotic liver afford epithelial-mesenchymal transition? World J Gastroenterol 2017; 23:4661–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prockop DJ. Inflammation, fibrosis, and modulation of the process by mesenchymal stem/stromal cells. Matrix Biol 2016; 51:7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bucala R, Spiegel LA, Chesney J, Hogan M, Cerami A. Circulating fibrocytes define a new leukocyte subpopulation that mediates tissue repair. Mol Med 1994; 1:71–81 [PMC free article] [PubMed] [Google Scholar]
- 18.Abe R, Donnelly SC, Peng T, Bucala R, Metz CN. Peripheral blood fibrocytes: differentiation pathway and migration to wound sites. J Immunol 2001; 166:7556–62 [DOI] [PubMed] [Google Scholar]
- 19.Quan TE, Cowper S, Wu SP, Bockenstedt LK, Bucala R. Circulating fibrocytes: collagen-secreting cells of the peripheral blood. Int J Biochem Cell Biol 2004; 36:598–606 [DOI] [PubMed] [Google Scholar]
- 20.Kisseleva T, Uchinami H, Feirt N, Quintana-Bustamante O, Segovia JC, Schwabe RF, Brenner DA. Bone marrow-derived fibrocytes participate in pathogenesis of liver fibrosis. J Hepatol 2006; 45:429–38 [DOI] [PubMed] [Google Scholar]
- 21.Scholten D, Reichart D, Paik YH, Lindert J, Bhattacharya J, Glass CK, Brenner DA, Kisseleva T. Migration of fibrocytes in fibrogenic liver injury. Am J Pathol 2011; 179:189–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lua I, Li Y, Pappoe LS, Asahina K. Myofibroblastic conversion and regeneration of mesothelial cells in peritoneal and liver fibrosis. Am J Pathol 2015; 185:3258–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutsaers SE. Mesothelial cells: their structure, function and role in serosal repair. Respirology 2002; 7:171–91 [DOI] [PubMed] [Google Scholar]
- 24.Asahina K, Zhou B, Pu WT, Tsukamoto H. Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology 2011; 53:983–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinkevich Y, Mori T, Sahoo D, Xu PX, Bermingham JR, Weissman IL. Identification and prospective isolation of a mesothelial precursor lineage giving rise to smooth muscle cells and fibroblasts for mammalian internal organs, and their vasculature. Nat Cell Biol 2012; 14:1251–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Wang J, Asahina K. Mesothelial cells give rise to hepatic stellate cells and myofibroblasts via mesothelial-mesenchymal transition in liver injury. Proc Natl Acad Sci U S A 2013; 110:2324–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lua I, Li Y, Zagory JA, Wang KS, French SW, Sévigny J, Asahina K. Characterization of hepatic stellate cells, portal fibroblasts, and mesothelial cells in normal and fibrotic livers. J Hepatol 2016; 64:1137–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang L, Tanaka Y, Marumo F, Sato C. Phenotypic change in portal fibroblasts in biliary fibrosis. Liver 1994; 14:76–82 [DOI] [PubMed] [Google Scholar]
- 29.Herbst H, Frey A, Heinrichs O, Milani S, Bechstein WO, Neuhaus P, Schuppan D. Heterogeneity of liver cells expressing procollagen types I and IV in vivo. Histochem Cell Biol 1997; 107:399–409 [DOI] [PubMed] [Google Scholar]
- 30.Cassiman D, Libbrecht L, Desmet V, Denef C, Roskams T. Hepatic stellate cell/myofibroblast subpopulations in fibrotic human and rat livers. J Hepatol 2002; 36:200–9 [DOI] [PubMed] [Google Scholar]
- 31.Desmoulière A, Darby I, Costa AM, Raccurt M, Tuchweber B, Sommer P, Gabbiani G. Extracellular matrix deposition, lysyl oxidase expression, and myofibroblastic differentiation during the initial stages of cholestatic fibrosis in the rat. Lab Invest 1997; 76:765–78 [PubMed] [Google Scholar]
- 32.Yata Y, Scanga A, Gillan A, Yang L, Reif S, Breindl M, Brenner DA, Rippe RA. DNase I-hypersensitive sites enhance alpha1(I) collagen gene expression in hepatic stellate cells. Hepatology 2003; 37:267–76 [DOI] [PubMed] [Google Scholar]
- 33.Dranoff JA, Wells RG. Portal fibroblasts: underappreciated mediators of biliary fibrosis. Hepatology 2010; 51:1438–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fausther M, Goree JR, Lavoie É, Graham AL, Sévigny J, Dranoff JA. Establishment and characterization of rat portal myofibroblast cell lines. PLoS One 2015; 10:e0121161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwaisako K, Jiang C, Zhang M, Cong M, Moore-Morris TJ, Park TJ, Liu X, Xu J, Wang P, Paik YH, Meng F, Asagiri M, Murray LA, Hofmann AF, Iida T, Glass CK, Brenner DA, Kisseleva T. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A 2014; 111:E3297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyama Y, Wang P, Liang S, Iwaisako K, Liu X, Xu J, Zhang M, Sun M, Cong M, Karin D, Taura K, Benner C, Heinz S, Bera T, Brenner DA, Kisseleva T. Mesothelin/mucin 16 signaling in activated portal fibroblasts regulates cholestatic liver fibrosis. J Clin Invest 2017; 127:1254–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marra F. Chemokines in liver inflammation and fibrosis. Front Biosci 2002; 7:d1899–914 [DOI] [PubMed] [Google Scholar]
- 38.Shi Z, Wakil AE, Rockey DC. Strain-specific differences in mouse hepatic wound healing are mediated by divergent T helper cytokine responses. Proc Natl Acad Sci U S A 1997; 94:10663–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwabe RF, Bataller R, Brenner DA. Human hepatic stellate cells express CCR5 and RANTES to induce proliferation and migration. Am J Physiol Gastrointest Liver Physiol 2003; 285:G949–58 [DOI] [PubMed] [Google Scholar]
- 40.Hammerich L, Heymann F, Tacke F. Role of IL-17 and Th17 cells in liver diseases. Clin Dev Immunol 2011; 2011:345803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabre T, Molina MF, Soucy G, Goulet JP, Willems B, Villeneuve JP, Bilodeau M, Shoukry NH. Type 3 cytokines IL-17A and IL-22 drive TGF-beta-dependent liver fibrosis. Sci Immunol 2018; 3:pii: eaar7754 [DOI] [PubMed] [Google Scholar]
- 42.Meng F, Wang K, Aoyama T, Grivennikov SI, Paik Y, Scholten D, Cong M, Iwaisako K, Liu X, Zhang M, Osterreicher CH, Stickel F, Ley K, Brenner DA, Kisseleva T. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology 2012; 143:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kronenberger B, Rudloff I, Bachmann M, Brunner F, Kapper L, Filmann N, Waidmann O, Herrmann E, Pfeilschifter J, Zeuzem S, Piiper A, Mühl H. Interleukin-22 predicts severity and death in advanced liver cirrhosis: a prospective cohort study. BMC Med 2012; 10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin S, Ma S, Huang X, Lu D, Zhou Y, Jiang H. Th22 cells are associated with hepatocellular carcinoma development and progression. Chin J Cancer Res 2014; 26:135–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang R, Tan Z, Deng L, Chen Y, Xia Y, Gao Y, Wang X, Sun B. Interleukin-22 promotes human hepatocellular carcinoma by activation of STAT3. Hepatology 2011; 54:900–9 [DOI] [PubMed] [Google Scholar]
- 46.Radaeva S, Sun R, Pan HN, Hong F, Gao B. Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology 2004; 39:1332–42 [DOI] [PubMed] [Google Scholar]
- 47.Ki SH, Park O, Zheng M, Morales-Ibanez O, Kolls JK, Bataller R, Gao B. Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: role of signal transducer and activator of transcription 3. Hepatology 2010; 52:1291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng D, Wang Y, Wang H, Weng H, Kong X, Martin-Murphy BV, Li Y, Park O, Dooley S, Ju C, Gao B. Acute and chronic effects of IL-22 on acetaminophen-induced liver injury. J Immunol 2014; 193:2512–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong X, Feng D, Wang H, Hong F, Bertola A, Wang FS, Gao B. Interleukin-22 induces hepatic stellate cell senescence and restricts liver fibrosis in mice. Hepatology 2012; 56:1150–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J, Zhang Z, Luan Y, Zou Z, Sun Y, Li Y, Jin L, Zhou C, Fu J, Gao B, Fu Y, Wang FS. Pathological functions of interleukin-22 in chronic liver inflammation and fibrosis with hepatitis B virus infection by promoting T helper 17 cell recruitment. Hepatology 2014; 59:1331–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Cobleigh MA, Lian JQ, Huang CX, Booth CJ, Bai XF, Robek MD. A proinflammatory role for interleukin-22 in the immune response to hepatitis B virus. Gastroenterology 2011; 141:1897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng D, Kong X, Weng H, Park O, Wang H, Dooley S, Gershwin ME, Gao B. Interleukin-22 promotes proliferation of liver stem/progenitor cells in mice and patients with chronic hepatitis B virus infection. Gastroenterology 2012; 143:188–98.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McHedlidze T, Waldner M, Zopf S, Walker J, Rankin AL, Schuchmann M, Voehringer D, McKenzie AN, Neurath MF, Pflanz S, Wirtz S. Interleukin-33-dependent innate lymphoid cells mediate hepatic fibrosis. Immunity 2013; 39:357–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan Z, Liu Q, Jiang R, Lv L, Shoto SS, Maillet I, Quesniaux V, Tang J, Zhang W, Sun B, Ryffel B. Interleukin-33 drives hepatic fibrosis through activation of hepatic stellate cells. Cell Mol Immunol 2018; 15:388–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marvie P, Lisbonne M, L'Helgoualc'h A, Rauch M, Turlin B, Preisser L, Bourd-Boittin K, Theret N, Gascan H, Piquet-Pellorce C, Samson M. Interleukin-33 overexpression is associated with liver fibrosis in mice and humans. J Cell Mol Med 2010; 14:1726–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oo YH, Shetty S, Adams DH. The role of chemokines in the recruitment of lymphocytes to the liver. Dig Dis 2010; 28:31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marra F, DeFranco R, Grappone C, Milani S, Pastacaldi S, Pinzani M, Romanelli RG, Laffi G, Gentilini P. Increased expression of monocyte chemotactic protein-1 during active hepatic fibrogenesis: correlation with monocyte infiltration. Am J Pathol 1998; 152:423–30 [PMC free article] [PubMed] [Google Scholar]
- 58.Karlmark KR, Weiskirchen R, Zimmermann HW, Gassler N, Ginhoux F, Weber C, Merad M, Luedde T, Trautwein C, Tacke F. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 2009; 50:261–74 [DOI] [PubMed] [Google Scholar]
- 59.Baeck C, Wehr A, Karlmark KR, Heymann F, Vucur M, Gassler N, Huss S, Klussmann S, Eulberg D, Luedde T, Trautwein C, Tacke F. Pharmacological inhibition of the chemokine CCL2 (MCP-1) diminishes liver macrophage infiltration and steatohepatitis in chronic hepatic injury. Gut 2012; 61:416–26 [DOI] [PubMed] [Google Scholar]
- 60.Tacke F. Targeting hepatic macrophages to treat liver diseases. J Hepatol 2017; 66:1300–12 [DOI] [PubMed] [Google Scholar]
- 61.Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 2017; 17:306–21 [DOI] [PubMed] [Google Scholar]
- 62.Dooley S, ten Dijke P. TGF-β in progression of liver disease. Cell Tissue Res 2012; 347:245–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li L, Wang JY, Yang CQ, Jiang W. Effect of RhoA on transforming growth factor β1-induced rat hepatic stellate cell migration. Liver Int 2012; 32:1093–102 [DOI] [PubMed] [Google Scholar]
- 64.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M, Tacke F, Trautwein C, Luedde T. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011; 53:209–18 [DOI] [PubMed] [Google Scholar]
- 65.Roy S, Benz F, Vargas Cardenas D, Vucur M, Gautheron J, Schneider A, Hellerbrand C, Pottier N, Alder J, Tacke F, Trautwein C, Roderburg C, Luedde T. miR-30c and miR-193 are a part of the TGF-β-dependent regulatory network controlling extracellular matrix genes in liver fibrosis. J Dig Dis 2015; 16:513–24 [DOI] [PubMed] [Google Scholar]
- 66.Yan X, Lin Z, Chen F, Zhao X, Chen H, Ning Y, Chen YG. Human BAMBI cooperates with Smad7 to inhibit transforming growth factor-beta signaling. J Biol Chem 2009; 284:30097–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arias M, Sauer-Lehnen S, Treptau J, Janoschek N, Theuerkauf I, Buettner R, Gressner AM, Weiskirchen R. Adenoviral expression of a transforming growth factor-beta1 antisense mRNA is effective in preventing liver fibrosis in bile-duct ligated rats. BMC Gastroenterol 2003; 3:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kinoshita K, Iimuro Y, Otogawa K, Saika S, Inagaki Y, Nakajima Y, Kawada N, Fujimoto J, Friedman SL, Ikeda K. Adenovirus-mediated expression of BMP-7 suppresses the development of liver fibrosis in rats. Gut 2007; 56:706–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pinzani M, Gesualdo L, Sabbah GM, Abboud HE. Effects of platelet-derived growth factor and other polypeptide mitogens on DNA synthesis and growth of cultured rat liver fat-storing cells. J Clin Invest 1989; 84:1786–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Breitkopf K, Godoy P, Ciuclan L, Singer MV, Dooley S. TGF-beta/Smad signaling in the injured liver. Z Gastroenterol 2006; 44:57–66 [DOI] [PubMed] [Google Scholar]
- 71.Kinnman N, Hultcrantz R, Barbu V, Rey C, Wendum D, Poupon R, Housset C. PDGF-mediated chemoattraction of hepatic stellate cells by bile duct segments in cholestatic liver injury. Lab Invest 2000; 80:697–707 [DOI] [PubMed] [Google Scholar]
- 72.Czochra P, Klopcic B, Meyer E, Herkel J, Garcia-Lazaro JF, Thieringer F, Schirmacher P, Biesterfeld S, Galle PR, Lohse AW, Kanzler S. Liver fibrosis induced by hepatic overexpression of PDGF-B in transgenic mice. J Hepatol 2006; 45:419–28 [DOI] [PubMed] [Google Scholar]
- 73.Borkham-Kamphorst E, Meurer SK, Van de Leur E, Haas U, Tihaa L, Weiskirchen R. PDGF-D signaling in portal myofibroblasts and hepatic stellate cells proves identical to PDGF-B via both PDGF receptor type α and β. Cell Signal 2015; 27:1305–14 [DOI] [PubMed] [Google Scholar]
- 74.Luangmonkong T, Suriguga S, Mutsaers HAM, Groothuis GMM, Olinga P, Boersema M. Targeting oxidative stress for the treatment of liver fibrosis. Rev Physiol Biochem Pharmacol 2018; 175:71–102 [DOI] [PubMed] [Google Scholar]
- 75.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev 2007; 87:245–313 [DOI] [PubMed] [Google Scholar]
- 76.Paik YH, Iwaisako K, Seki E, Inokuchi S, Schnabl B, Osterreicher CH, Kisseleva T, Brenner DA. The nicotinamide adenine dinucleotide phosphate oxidase (NOX) homologues NOX1 and NOX2/gp91(phox) mediate hepatic fibrosis in mice. Hepatology 2011; 53:1730–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paik YH, Kim J, Aoyama T, De Minicis S, Bataller R, Brenner DA. Role of NADPH oxidases in liver fibrosis. Antioxid Redox Signal 2014; 20:2854–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Minicis S, Brenner DA. NOX in liver fibrosis. Arch Biochem Biophys 2007; 462:266–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.De Minicis S, Seki E, Paik YH, Osterreicher CH, Kodama Y, Kluwe J, Torozzi L, Miyai K, Benedetti A, Schwabe RF, Brenner DA. Role and cellular source of nicotinamide adenine dinucleotide phosphate oxidase in hepatic fibrosis. Hepatology 2010; 52:1420–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lan T, Kisseleva T, Brenner DA. Deficiency of NOX1 or NOX4 prevents liver inflammation and fibrosis in mice through inhibition of hepatic stellate cell activation. PLoS One 2015; 10:e0129743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang S, Ma HY, Zhong Z, Dhar D, Liu X, Xu J, Koyama Y, Nishio T, KD, Karin G, Mccubbin R, Zhang C, Hu R, Yang G, Chen L, Ganguly S, Lan T, Karin M, Kisseleva T, Brenner DA. NADPH oxidase 1 in liver macrophages promotes inflammation and tumor development in mice. Gastroenterology 2019; 156:1156–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aoyama T, Paik YH, Watanabe S, Laleu B, Gaggini F, Fioraso-Cartier L, Molango S, Heitz F, Merlot C, Szyndralewiez C, Page P, Brenner DA. Nicotinamide adenine dinucleotide phosphate oxidase in experimental liver fibrosis: GKT137831 as a novel potential therapeutic agent. Hepatology 2012; 56:2316–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang JX, Chen X, Serizawa N, Szyndralewiez C, Page P, Schröder K, Brandes RP, Devaraj S, Török NJ. Liver fibrosis and hepatocyte apoptosis are attenuated by GKT137831, a novel NOX4/NOX1 inhibitor in vivo. Free Radic Biol Med 2012; 53:289–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bettaieb A, Jiang JX, Sasaki Y, Chao TI, Kiss Z, Chen X, Tian J, Katsuyama M, Yabe-Nishimura C, Xi Y, Szyndralewiez C, Schröder K, Shah A, Brandes RP, Haj FG, Török NJ. Hepatocyte nicotinamide adenine dinucleotide phosphate reduced oxidase 4 regulates stress signaling, fibrosis, and insulin sensitivity during development of steatohepatitis in mice. Gastroenterology 2015; 149:468–80.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.De Bleser PJ, Xu G, Rombouts K, Rogiers V, Geerts A. Glutathione levels discriminate between oxidative stress and transforming growth factor-beta signaling in activated rat hepatic stellate cells. J Biol Chem 1999; 274:33881–7 [DOI] [PubMed] [Google Scholar]
- 86.Barnes JL, Gorin Y. Myofibroblast differentiation during fibrosis: role of NAD(P)H oxidases. Kidney Int 2011; 79:944–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell 2005; 120:483–95 [DOI] [PubMed] [Google Scholar]
- 88.Grattagliano I, Russmann S, Diogo C, Bonfrate L, Oliveira PJ, Wang DQ, Portincasa P. Mitochondria in chronic liver disease. Curr Drug Targets 2011; 12:879–93 [DOI] [PubMed] [Google Scholar]
- 89.Guimaraes EL, Best J, Dolle L, Najimi M, Sokal E, van Grunsven LA. Mitochondrial uncouplers inhibit hepatic stellate cell activation. BMC Gastroenterol 2012; 12:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gajendiran P, Vega LI, Itoh K, Sesaki H, Vakili MR, Lavasanifar A, Hong K, Mezey E, Ganapathy-Kanniappan S. Elevated mitochondrial activity distinguishes fibrogenic hepatic stellate cells and sensitizes for selective inhibition by mitotropic doxorubicin. J Cell Mol Med 2018; 22:2210–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Y, Choi SS, Michelotti GA, Chan IS, Swiderska-Syn M, Karaca GF, Xie G, Moylan CA, Garibaldi F, Premont R, Suliman HB, Piantadosi CA, Diehl AM. Hedgehog controls hepatic stellate cell fate by regulating metabolism. Gastroenterology 2012; 143:1319–29.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang L, Venneti S, Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng 2017; 19:163–94 [DOI] [PubMed] [Google Scholar]
- 93.Li J, Ghazwani M, Liu K, Huang Y, Chang N, Fan J, He F, Li L, Bu S, Xie W, Ma X, Li S. Regulation of hepatic stellate cell proliferation and activation by glutamine metabolism. PLoS One 2017; 12:e0182679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Du K, Hyun J, Premont RT, Choi SS, Michelotti GA, Swiderska-Syn M, Dalton GD, Thelen E, Rizi BS, Jung Y, Diehl AM. Hedgehog-YAP signaling pathway regulates glutaminolysis to control activation of hepatic stellate cells. Gastroenterology 2018; 154:1465–79.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Garcia D, Shaw RJ. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell 2017; 66:789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kamada Y, Tamura S, Kiso S, Matsumoto H, Saji Y, Yoshida Y, Fukui K, Maeda N, Nishizawa H, Nagaretani H, Okamoto Y, Kihara S, Miyagawa J, Shinomura Y, Funahashi T, Matsuzawa Y. Enhanced carbon tetrachloride-induced liver fibrosis in mice lacking adiponectin. Gastroenterology 2003; 125:1796–807 [DOI] [PubMed] [Google Scholar]
- 97.Caligiuri A, Bertolani C, Guerra CT, Aleffi S, Galastri S, Trappoliere M, Vizzutti F, Gelmini S, Laffi G, Pinzani M, Marra F. Adenosine monophosphate-activated protein kinase modulates the activated phenotype of hepatic stellate cells. Hepatology 2008; 47:668–76 [DOI] [PubMed] [Google Scholar]
- 98.Valfrè di Bonzo L, Novo E, Cannito S, Busletta C, Paternostro C, Povero D, Parola M. Angiogenesis and liver fibrogenesis. Histol Histopathol 2009; 24:323–41 [DOI] [PubMed] [Google Scholar]
- 99.Fernández M, Semela D, Bruix J, Colle I, Pinzani M, Bosch J. Angiogenesis in liver disease. J Hepatol 2009; 50:604–20 [DOI] [PubMed] [Google Scholar]
- 100.Sanz-Cameno P, Trapero-Marugán M, Chaparro M, Jones EA, Moreno-Otero R. Angiogenesis: from chronic liver inflammation to hepatocellular carcinoma. J Oncol 2010; 2010:272170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marra F, Tacke F. Roles for chemokines in liver disease. Gastroenterology 2014; 147:577–94.e1 [DOI] [PubMed] [Google Scholar]
- 102.Rockey DC. Hepatic blood flow regulation by stellate cells in normal and injured liver. Semin Liver Dis 2001; 21:337–49 [DOI] [PubMed] [Google Scholar]
- 103.Aleffi S, Petrai I, Bertolani C, Parola M, Colombatto S, Novo E, Vizzutti F, Anania FA, Milani S, Rombouts K, Laffi G, Pinzani M, Marra F. Upregulation of proinflammatory and proangiogenic cytokines by leptin in human hepatic stellate cells. Hepatology 2005; 42:1339–48 [DOI] [PubMed] [Google Scholar]
- 104.Taura K, De Minicis S, Seki E, Hatano E, Iwaisako K, Osterreicher CH, Kodama Y, Miura K, Ikai I, Uemoto S, Brenner DA. Hepatic stellate cells secrete angiopoietin 1 that induces angiogenesis in liver fibrosis. Gastroenterology 2008; 135:1729–38 [DOI] [PubMed] [Google Scholar]
- 105.Kukla M. Angiogenesis: a phenomenon which aggravates chronic liver disease progression. Hepatol Int 2013; 7:4–12 [DOI] [PubMed] [Google Scholar]
- 106.Yoshiji H, Kuriyama S, Yoshii J, Ikenaka Y, Noguchi R, Hicklin DJ, Wu Y, Yanase K, Namisaki T, Yamazaki M, Tsujinoue H, Imazu H, Masaki T, Fukui H. Vascular endothelial growth factor and receptor interaction is a prerequisite for murine hepatic fibrogenesis. Gut 2003; 52:1347–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang YQ, Luk JM, Ikeda K, Man K, Chu AC, Kaneda K, Fan ST. Regulatory role of vHL/HIF-1alpha in hypoxia-induced VEGF production in hepatic stellate cells. Biochem Biophys Res Commun 2004; 317:358–62 [DOI] [PubMed] [Google Scholar]
- 108.Strieter RM, Burdick MD, Gomperts BN, Belperio JA, Keane MP. CXC chemokines in angiogenesis. Cytokine Growth Factor Rev 2005; 16:593–609 [DOI] [PubMed] [Google Scholar]
- 109.Sahin H, Wasmuth HE. Chemokines in tissue fibrosis. Biochim Biophys Acta 2013; 1832:1041–8 [DOI] [PubMed] [Google Scholar]
- 110.Abbaspour N, Hurrell R, Kelishadi R. Review on iron and its importance for human health. J Res Med Sci 2014; 19:164–74 [PMC free article] [PubMed] [Google Scholar]
- 111.Philippe MA, Ruddell RG, Ramm GA. Role of iron in hepatic fibrosis: one piece in the puzzle. World J Gastroenterol 2007; 13:4746–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bonkovsky HL. Iron and the liver. Am J Med Sci 1991; 301:32–43 [DOI] [PubMed] [Google Scholar]
- 113.Mehta KJ, Farnaud SJ, Sharp PA. Iron and liver fibrosis: mechanistic and clinical aspects. World J Gastroenterol 2019; 25:521–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dongiovanni P, Fracanzani AL, Fargion S, Valenti L. Iron in fatty liver and in the metabolic syndrome: a promising therapeutic target. J Hepatol 2011; 55:920–32 [DOI] [PubMed] [Google Scholar]
- 115.Lee PL, Beutler E. Regulation of hepcidin and iron-overload disease. Annu Rev Pathol 2009; 4:489–515 [DOI] [PubMed] [Google Scholar]
- 116.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004; 306:2090–3 [DOI] [PubMed] [Google Scholar]
- 117.Zhang AS, Xiong S, Tsukamoto H, Enns CA. Localization of iron metabolism-related mRNAs in rat liver indicate that HFE is expressed predominantly in hepatocytes. Blood 2004; 103:1509–14 [DOI] [PubMed] [Google Scholar]
- 118.Han CY, Koo JH, Kim SH, Gardenghi S, Rivella S, Strnad P, Hwang SJ, Kim SG. Hepcidin inhibits Smad3 phosphorylation in hepatic stellate cells by impeding ferroportin-mediated regulation of Akt. Nat Commun 2016; 7:13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang L, Zhou F, ten Dijke P. Signaling interplay between transforming growth factor-beta receptor and PI3K/AKT pathways in cancer. Trends Biochem Sci 2013; 38:612–20 [DOI] [PubMed] [Google Scholar]
- 120.Arndt S, Wacker E, Dorn C, Koch A, Saugspier M, Thasler WE, Hartmann A, Bosserhoff AK, Hellerbrand C. Enhanced expression of BMP6 inhibits hepatic fibrosis in non-alcoholic fatty liver disease. Gut 2015; 64:973–81 [DOI] [PubMed] [Google Scholar]
- 121.Arthur MJ. Reversibility of liver fibrosis and cirrhosis following treatment for hepatitis C. Gastroenterology 2002; 122:1525–8 [DOI] [PubMed] [Google Scholar]
- 122.Issa R, Zhou X, Constandinou CM, Fallowfield J, Millward-Sadler H, Gaca MD, Sands E, Suliman I, Trim N, Knorr A, Arthur MJ, Benyon RC, Iredale JP. Spontaneous recovery from micronodular cirrhosis: evidence for incomplete resolution associated with matrix cross-linking. Gastroenterology 2004; 126:1795–808 [DOI] [PubMed] [Google Scholar]
- 123.Parés A, Caballería J, Bruguera M, Torres M, Rodés J. Histological course of alcoholic hepatitis. Influence of abstinence, sex and extent of hepatic damage. J Hepatol 1986; 2:33–42 [DOI] [PubMed] [Google Scholar]
- 124.Dixon JB, Bhathal PS, Hughes NR, O'Brien PE. Nonalcoholic fatty liver disease: improvement in liver histological analysis with weight loss. Hepatology 2004; 39:1647–54 [DOI] [PubMed] [Google Scholar]
- 125.Desmet VJ, Roskams T. Cirrhosis reversal: a duel between dogma and myth. J Hepatol 2004; 40:860–7 [DOI] [PubMed] [Google Scholar]
- 126.Iredale JP. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest 2007; 117:539–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wanless IR, Nakashima E, Sherman M. Regression of human cirrhosis. Morphologic features and the genesis of incomplete septal cirrhosis. Arch Pathol Lab Med 2000; 124:1599–607 [DOI] [PubMed] [Google Scholar]
- 128.Marcellin P, Gane E, Buti M, Afdhal N, Sievert W, Jacobson IM, Washington MK, Germanidis G, Flaherty JF, Aguilar Schall R, Bornstein JD, Kitrinos KM, Subramanian GM, McHutchison JG, Heathcote EJ. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a 5-year open-label follow-up study. Lancet 2013; 381:468–75 [DOI] [PubMed] [Google Scholar]
- 129.Chang TT, Liaw YF, Wu SS, Schiff E, Han KH, Lai CL, Safadi R, Lee SS, Halota W, Goodman Z, Chi YC, Zhang H, Hindes R, Iloeje U, Beebe S, Kreter B. Long-term entecavir therapy results in the reversal of fibrosis/cirrhosis and continued histological improvement in patients with chronic hepatitis B. Hepatology 2010; 52:886–93 [DOI] [PubMed] [Google Scholar]
- 130.D'Ambrosio R, Aghemo A, Rumi MG, Ronchi G, Donato MF, Paradis V, Colombo M, Bedossa P. A morphometric and immunohistochemical study to assess the benefit of a sustained virological response in hepatitis C virus patients with cirrhosis. Hepatology 2012; 56:532–43 [DOI] [PubMed] [Google Scholar]
- 131.Duarte S, Baber J, Fujii T, Coito AJ. Matrix metalloproteinases in liver injury, repair and fibrosis. Matrix Biol 2015; 44-46:147–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Iredale JP. Hepatic stellate cell behavior during resolution of liver injury. Semin Liver Dis 2001; 21:427–36 [DOI] [PubMed] [Google Scholar]
- 133.She H, Xiong S, Hazra S, Tsukamoto H. Adipogenic transcriptional regulation of hepatic stellate cells. J Biol Chem 2005; 280:4959–67 [DOI] [PubMed] [Google Scholar]
- 134.Troeger JS, Mederacke I, Gwak GY, Dapito DH, Mu X, Hsu CC, Pradere JP, Friedman RA, Schwabe RF. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology 2012; 143:1073–83.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Laursen L. A preventable cancer. Nature 2014; 516:S2–3 [DOI] [PubMed] [Google Scholar]
- 136.Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 2015; 12:408–24 [DOI] [PubMed] [Google Scholar]
- 137.Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin Gastroenterol Hepatol 2015; 13:2140–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology 2004; 127:S35–50 [DOI] [PubMed] [Google Scholar]
- 139.O'Rourke JM, Sagar VM, Shah T, Shetty S. Carcinogenesis on the background of liver fibrosis: implications for the management of hepatocellular cancer. World J Gastroenterol 2018; 24:4436–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Trevisani F, Frigerio M, Santi V, Grignaschi A, Bernardi M. Hepatocellular carcinoma in non-cirrhotic liver: a reappraisal. Dig Liver Dis 2010; 42:341–7 [DOI] [PubMed] [Google Scholar]
- 141.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006; 6:674–87 [DOI] [PubMed] [Google Scholar]
- 142.Ieluzzi D, Covolo L, Donato F, Fattovich G. Progression to cirrhosis, hepatocellular carcinoma and liver-related mortality in chronic hepatitis B patients in Italy. Dig Liver Dis 2014; 46:427–32 [DOI] [PubMed] [Google Scholar]
- 143.Chang MH, You SL, Chen CJ, Liu CJ, Lee CM, Lin SM, Chu HC, Wu TC, Yang SS, Kuo HS, Chen DS. Taiwan hepatoma study G. Decreased incidence of hepatocellular carcinoma in hepatitis B vaccines: a 20-year follow-up study. J Natl Cancer Inst 2009; 101:1348–55 [DOI] [PubMed] [Google Scholar]
- 144.Kimer N, Dahl EK, Gluud LL, Krag A. Antiviral therapy for prevention of hepatocellular carcinoma in chronic hepatitis C: systematic review and meta-analysis of randomised controlled trials. BMJ Open 2012; 2:e001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology 2019; 156:477–91.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cohen JC, Horton JD, Hobbs HH. Human Fatty liver disease: old questions and new insights. Science 2011; 332:1519–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Torres DM, Harrison SA. Diagnosis and therapy of nonalcoholic steatohepatitis. Gastroenterology 2008; 134:1682–98 [DOI] [PubMed] [Google Scholar]
- 148.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology 2010; 52:1836–46 [DOI] [PubMed] [Google Scholar]
- 149.Mittal S, El-Serag HB, Sada YH, Kanwal F, Duan Z, Temple S, May SB, Kramer JR, Richardson PA, Davila JA. Hepatocellular carcinoma in the absence of cirrhosis in United States veterans is associated with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2016; 14:124–31.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hagstrom H, Nasr P, Ekstedt M, Hammar U, Stal P, Hultcrantz R, Kechagias S. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J Hepatol 2017; 67:1265–73 [DOI] [PubMed] [Google Scholar]
- 151.Affo S, Yu LX, Schwabe RF. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Annu Rev Pathol 2017; 12:153–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Baglieri J, Brenner DA, Kisseleva T. The role of fibrosis and liver-associated fibroblasts in the pathogenesis of hepatocellular carcinoma. Int J Mol Sci 2019; 20:pii: E1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hynes RO. The extracellular matrix: not just pretty fibrils. Science 2009; 326:1216–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Charo IF, Nannizzi L, Smith JW, Cheresh DA. The vitronectin receptor alpha v beta 3 binds fibronectin and acts in concert with alpha 5 beta 1 in promoting cellular attachment and spreading on fibronectin. J Cell Biol 1990; 111:2795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rahman S, Patel Y, Murray J, Patel KV, Sumathipala R, Sobel M, Wijelath ES. Novel hepatocyte growth factor (HGF) binding domains on fibronectin and vitronectin coordinate a distinct and amplified met-integrin induced signalling pathway in endothelial cells. BMC Cell Biol 2005; 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yu MC, Chen CH, Liang X, Wang L, Gandhi CR, Fung JJ, Lu L, Qian S. Inhibition of T-cell responses by hepatic stellate cells via B7-H1-mediated T-cell apoptosis in mice. Hepatology 2004; 40:1312–21 [DOI] [PubMed] [Google Scholar]
- 157.Chinnadurai R, Grakoui A. B7-H4 mediates inhibition of T cell responses by activated murine hepatic stellate cells. Hepatology 2010; 52:2177–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jiang G, Yang HR, Wang L, Wildey GM, Fung J, Qian S, Lu L. Hepatic stellate cells preferentially expand allogeneic CD4+ CD25+ FoxP3+ regulatory T cells in an IL-2-dependent manner. Transplantation 2008; 86:1492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dunham RM, Thapa M, Velazquez VM, Elrod EJ, Denning TL, Pulendran B, Grakoui A. Hepatic stellate cells preferentially induce Foxp3+ regulatory T cells by production of retinoic acid. J Immunol 2013; 190:2009–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW. Senescence of activated stellate cells limits liver fibrosis. Cell 2008; 134:657–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013; 499:97–101 [DOI] [PubMed] [Google Scholar]
- 162.Schnabl B, Purbeck CA, Choi YH, Hagedorn CH, Brenner D. Replicative senescence of activated human hepatic stellate cells is accompanied by a pronounced inflammatory but less fibrogenic phenotype. Hepatology 2003; 37:653–64 [DOI] [PubMed] [Google Scholar]
- 163.Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, Zhao Z, Thapar V, Joyce JA, Krizhanovsky V, Lowe SW. Non-cell-autonomous tumor suppression by p53. Cell 2013; 153:449–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van Zijl F, Mair M, Csiszar A, Schneller D, Zulehner G, Huber H, Eferl R, Beug H, Dolznig H, Mikulits W. Hepatic tumor-stroma crosstalk guides epithelial to mesenchymal transition at the tumor edge. Oncogene 2009; 28:4022–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Amann T, Bataille F, Spruss T, Muhlbauer M, Gabele E, Scholmerich J, Kiefer P, Bosserhoff AK, Hellerbrand C. Activated hepatic stellate cells promote tumorigenicity of hepatocellular carcinoma. Cancer Sci 2009; 100:646–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 2008; 88:125–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Coulouarn C, Corlu A, Glaise D, Guenon I, Thorgeirsson SS, Clement B. Hepatocyte-stellate cell cross-talk in the liver engenders a permissive inflammatory microenvironment that drives progression in hepatocellular carcinoma. Cancer Res 2012; 72:2533–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bruschi FV, Tardelli M, Claudel T, Trauner M. PNPLA3 expression and its impact on the liver: current perspectives. Hepat Med 2017; 9:55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Huang Y, He S, Li JZ, Seo YK, Osborne TF, Cohen JC, Hobbs HH. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci U S A 2010; 107:7892–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pingitore P, Romeo S. The role of PNPLA3 in health and disease. Biochim Biophys Acta Mol Cell Biol Lipids 2019; 1864:900–6 [DOI] [PubMed] [Google Scholar]
- 171.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008; 40:1461–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu YL, Patman GL, Leathart JB, Piguet AC, Burt AD, Dufour JF, Day CP, Daly AK, Reeves HL, Anstee QM. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol 2014; 61:75–81 [DOI] [PubMed] [Google Scholar]
- 173.Burza MA, Pirazzi C, Maglio C, Sjoholm K, Mancina RM, Svensson PA, Jacobson P, Adiels M, Baroni MG, Boren J, Ginanni Corradini S, Montalcini T, Sjostrom L, Carlsson LM, Romeo S. PNPLA3 I148M (rs738409) genetic variant is associated with hepatocellular carcinoma in obese individuals. Dig Liver Dis 2012; 44:1037–41 [DOI] [PubMed] [Google Scholar]
- 174.Trepo E, Romeo S, Zucman-Rossi J, Nahon P. PNPLA3 gene in liver diseases. J Hepatol 2016; 65:399–412 [DOI] [PubMed] [Google Scholar]
- 175.Basantani MK, Sitnick MT, Cai L, Brenner DS, Gardner NP, Li JZ, Schoiswohl G, Yang K, Kumari M, Gross RW, Zechner R, Kershaw EE. Pnpla3/adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res 2011; 52:318–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chen W, Chang B, Li L, Chan L. Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology 2010; 52:1134–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.BasuRay S, Wang Y, Smagris E, Cohen JC, Hobbs HH. Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc Natl Acad Sci U S A 2019; 116:9521–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Linden D, Ahnmark A, Pingitore P, Ciociola E, Ahlstedt I, Andreasson AC, Sasidharan K, Madeyski-Bengtson K, Zurek M, Mancina RM, Lindblom A, Bjursell M, Bottcher G, Stahlman M, Bohlooly YM, Haynes WG, Carlsson B, Graham M, Lee R, Murray S, Valenti L, Bhanot S, Akerblad P, Romeo S. Pnpla3 silencing with antisense oligonucleotides ameliorates nonalcoholic steatohepatitis and fibrosis in Pnpla3 I148M knock-in mice. Mol Metab 2019; 22:49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pirazzi C, Valenti L, Motta BM, Pingitore P, Hedfalk K, Mancina RM, Burza MA, Indiveri C, Ferro Y, Montalcini T, Maglio C, Dongiovanni P, Fargion S, Rametta R, Pujia A, Andersson L, Ghosal S, Levin M, Wiklund O, Iacovino M, Boren J, Romeo S. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet 2014; 23:4077–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mondul A, Mancina RM, Merlo A, Dongiovanni P, Rametta R, Montalcini T, Valenti L, Albanes D, Romeo S. PNPLA3 I148M variant influences circulating retinol in adults with nonalcoholic fatty liver disease or obesity. J Nutr 2015; 145:1687–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kovarova M, Konigsrainer I, Konigsrainer A, Machicao F, Haring HU, Schleicher E, Peter A. The genetic variant I148M in PNPLA3 is associated with increased hepatic Retinyl-Palmitate storage in humans. J Clin Endocrinol Metab 2015; 100:E1568–74 [DOI] [PubMed] [Google Scholar]
- 182.Lee YS, Jeong WI. Retinoic acids and hepatic stellate cells in liver disease. J Gastroenterol Hepatol 2012; 27(Suppl 2):75–9 [DOI] [PubMed] [Google Scholar]
- 183.Fields AL, Soprano DR, Soprano KJ. Retinoids in biological control and cancer. J Cell Biochem 2007; 102:886–98 [DOI] [PubMed] [Google Scholar]
- 184.Chaves GV, Pereira SE, Saboya CJ, Spitz D, Rodrigues CS, Ramalho A. Association between liver vitamin a reserves and severity of nonalcoholic fatty liver disease in the class III obese following bariatric surgery. Obes Surg 2014; 24:219–24 [DOI] [PubMed] [Google Scholar]
- 185.Chen G. The link between hepatic vitamin a metabolism and nonalcoholic fatty liver disease. Curr Drug Targets 2015; 16:1281–92 [DOI] [PubMed] [Google Scholar]
- 186.Pingitore P, Dongiovanni P, Motta BM, Meroni M, Lepore SM, Mancina RM, Pelusi S, Russo C, Caddeo A, Rossi G, Montalcini T, Pujia A, Wiklund O, Valenti L, Romeo S. PNPLA3 overexpression results in reduction of proteins predisposing to fibrosis. Hum Mol Genet 2016; 25:5212–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bruschi FV, Claudel T, Tardelli M, Caligiuri A, Stulnig TM, Marra F, Trauner M. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatology 2017; 65:1875–90 [DOI] [PubMed] [Google Scholar]
- 188.Min HK, Sookoian S, Pirola CJ, Cheng J, Mirshahi F, Sanyal AJ. Metabolic profiling reveals that PNPLA3 induces widespread effects on metabolism beyond triacylglycerol remodeling in huh-7 hepatoma cells. Am J Physiol Gastrointest Liver Physiol 2014; 307:G66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]