Abstract
Kaempferol is a ubiquitous flavonoid, found in various plants having a wide range of known pharmacological activities, including antioxidant, antiinflammatory, anticancer, antiallergic, antidiabetic, neuroprotective, cardioprotective and antimicrobial activities. Nonetheless various evidence suggest that kaempferol is also able to interact with many unknown therapeutic targets modulating signalling pathways, thus providing an opportunity to explore the potential target space of kaempferol. In this study, we have employed various ligand-based approaches to identify the potential targets of kaempferol, followed by validations using modelling and docking studies. Molecular dynamics, free energy calculations, volume and residue contact map analyses were made to delineate the cause of drug-resistance among mutants. We have discovered dihydropteroate synthase (DHPS) as a novel potential therapeutic target for kaempferol. Further studies employing molecular dynamics simulations and binding free energies indicate that kaempferol has potential to inhibit even the sulfone-resistant DHPS mutants, which makes it a very attractive antibiotic agent. The identification of natural-product based kaempferol opens up the door for the design of antibiotics in a quick and high throughput fashion for identifying antibiotic leads.
Keywords: Pharmaceutical science, Pharmaceutical chemistry, Bioinformatics, Computational biology, Biochemistry, Kaempferol, Shape-based screening, Dihydropteroate synthase, DHPS, Docking, Molecular dynamics simulations, Residue contact map, Binding free energy, Antibiotic
Pharmaceutical science; Pharmaceutical chemistry; Bioinformatics, Computational biology; Biochemistry; Kaempferol; Shape-based screening; Dihydropteroate synthase; DHPS; Docking; Molecular dynamics simulations; Residue contact map; Binding free energy; Antibiotic.
1. Introduction
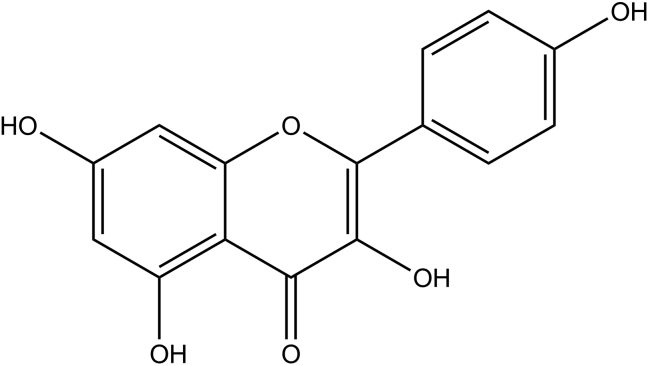
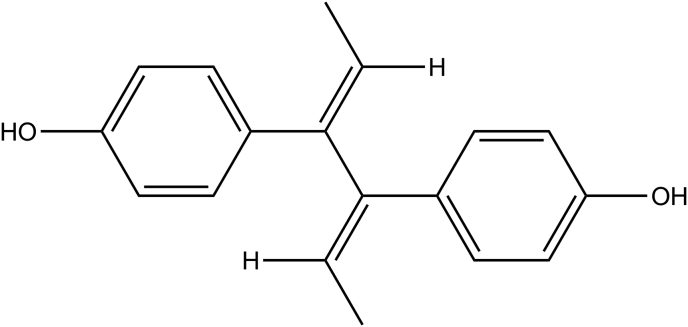
Flavonoids are plant secondary metabolites, which are distributed widely in the plant kingdom. They are commonly found in fruits, vegetables and various medicinal plants. Flavonoids are recognized generally for their antioxidant properties, anticarcinogenic, antiinflammatory and neuroprotective effects (Arredondo et al., 2010; Du et al., 2018; Gui et al., 2018; Spencer, 2009; Vidya Priyadarsini et al., 2010). Several lines of evidence also suggest that certain flavonoids found in plants, which are used for traditional medicines to treat various diseases, are the common bioactive constituents in plants (Barbosa et al., 2007; Calzada et al., 2017; Cooper and Ma, 2017; Kumar and Pandey, 2013; Osama et al., 2017; Panche et al., 2016). Among them, kaempferol (Figure 1) - a member of the flavonols abundantly found in fruits vegetables and medicinal plants - is known to modulate apoptosis, angiogenesis, inflammation and metastasis by targeting specific receptors or enzymes (Chen and Chen, 2013). However, a vast target space for flavonoids especially kaempferol remains to be discovered.
Figure 1.
2D diagram of Kaempferol.
Sulfonamide or Sulfone group containing drugs are widely used in treating various bacterial diseases caused by both gram-positive and gram-negative bacteria (Jayachandran et al., 2010; Smith et al., 2017). Sulfone class of drugs target the essential folate pathway by inhibiting the dihydropteroate synthase enzyme (DHPS), which catalyses the condensation of 6-hydroxymethyl-7,8–dihydropterin-pyrophosphate (DHPP) with p-aminobenzoic acid (pABA) to produce folate intermediate, 7,8-dihydropteroate (Achari et al., 1997; Roland et al., 1979; Then and Angehrn, 1973). However, extensive use of antibacterial agents has led to the emergence of drug resistance strains (Alanis, 2005; Appelbaum, 2012). Since DHPS activity is essential for bacteria and the synthetic pathways are mostly absent in human, it has been a potential target of interest for antibacterial agents.
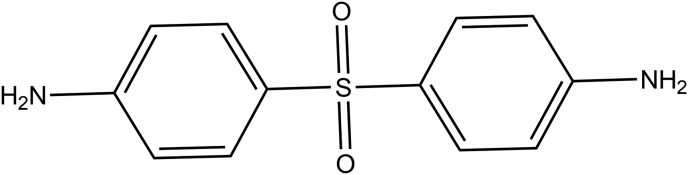
Mycobacterium leprae is one of the bacteria extensively targeted by sulfone drugs, which causes leprosy, a chronic, progressive bacterial infection affecting about a quarter million people worldwide (WHO Weekly Epidemiological Record, 2017). One of the sulfone drugs, dapsone has been primarily used for the treatment of leprosy (Zhu and Stiller, 2001), which block the condensation of pABA and DHPP, but with the emergence of dapsone-resistant strains of M. leprae, it has become less effective against leprosy (Friedmann, 1973). This has led to the use of Multi-Drug Therapy (MDT) with the addition of Rifampin (also referred to as Rifampicin) and Clofazimine as the second line of antibacterial agents (Bullock, 1983; Smith et al., 2017).
To assess the effectiveness of new compounds, it is crucial to gain insight into the DHPS catalytic mechanism and the basis of sulfone drug resistance. Bacterial DHPS exists as a homodimer and each of the subunits has a TIM barrel α/β structure with the α-helices surrounding a central barrel made of 8 parallel β-stands (Baca et al., 2000). There are three DHPS catalytic active sites, pterin, pABA and anion-binding pocket, of which sulfa drugs mostly target pABA active site pocket (Achari et al., 1997; Babaoglu et al., 2004; Roland et al., 1979; Woods, 1940; Zhu and Stiller, 2001). The point mutations (at position 53 and 55) leading to drug resistance in M. Leprae are located near the pABA active site particularly at the flexible and conserved loops, which might influence the binding environment of the active site (Maladan et al., 2019; Matsuoka et al., 2007; Nakata et al., 2011). Moreover, the lack of availability of the crystal structure of M.leprae DHPS hampered the efforts to understand the structural basis of sulfa drug resistance and to discover novel inhibitors.
In our work by using in silico methods, we predicted dihydropteroate synthase enzyme (DHPS) as one of the novel potential target enzymes for kaempferol. To the best of our knowledge, this is the first attempt to study the effect of kaempferol on DHPS, and hence it serves as a potential alternative to targeting sulfone-resistant strains of Mycobacterium leprae. The study mainly focuses on the effectiveness of kaempferol on the sulfone-resistant strain of Mycobacterium leprae.
In this study, different in silico approaches were implemented. First, shape-based screening was used to explore possible therapeutic targets for kaempferol. A new target of kaempferol i.e., DHPS was selected for further study. Homology modelling and molecular docking studies were performed to validate the shape-based screening result. To understand the dynamics of mutations leading to structural deviations in the DHPS active site and ultimately to the drug resistance, molecular dynamics studies, pocket volume and residue network analyses were performed. Further, the effects of kaempferol and dapsone on different mutants of DHPS were also investigated using implicit-solvation based binding free energy calculation.
2. Material and methods
2.1. Ligand-target database generation
Currently, various target prediction methods are available, such as ligand-based, structure-based, machine learning and network-based (Huang et al., 2018). For our study, we have focused on the first case, where the prediction of the plausible target protein was made by comparing a query compound against a library of compounds known to bind to the proteins, which are retrieved by association (Klabunde, 2007). For our study, the target-molecule library was prepared from DrugBank (Wishart et al., 2006), which consist of FDA approved and experimental (discovery-phase) drug molecules. Molecule preparation such as the addition of hydrogen atoms, 2D to 3D conversion etc. was carried out using Openbabel (O'Boyle et al., 2011). Energy minimization was carried by using MMFF94 force field implementing Steepest Descent followed by Conjugate Gradient method for 2600 steps with default convergence criteria. A maximum of 100 lowest-energy conformers were generated for each of the query molecules. Moreover, a relational table was created from the local copy of DrugBank, which consists of information corresponding to the drug molecules such as target, Gene name, Species, UniProt ID, GenBank Gene ID and GenBank Protein ID.
2.2. Ligand-based search
3D ligand-based approaches like shape-based methods, are reported to play a very important role in drug discovery (Ballester et al., 2010; Oyarzabal et al., 2010, 2009; Rush et al., 2005). Shape-based techniques work on the “Similar Property Principle” (Klopmand, 1992), which states that molecules with similar structure should show similar bioactivities. To perform shape-based screening, the geometrical profiles such as pharmacophore features, molecular shapes and molecular fields of the query molecules are calculated and compared with the profiles of the known drug molecules. In this study, SHAFTS (SHApe- FeaTure Similarity) (Liu et al., 2011) was implemented for the shape-based screening of Kaempferol against the conformers generated from the ligand-target library. It implements a hybrid similarity metrics including both molecular shape and pharmacophoric features such as hydrophobic center, positive or negative charge center, hydrogen bond acceptor and donor, and aromatic rings.
A feature triplet hashing method is used for the fast rigid alignment of molecular structure. It finally returns a sorted list of molecule identifiers associated with a structural similarity score against the query and the corresponding structural alignment. In the present study, the cutoff for the hybrid score was set to 1 and only 5 best hits were retained. The predicted hits were cross-verified by the literature survey for any experimental report showing biological effects of kaempferol on the predicted targets.
2.3. Homology modelling
The amino acid sequence of M. leprae DHPS (ID: P0C0X1) was retrieved from the Universal Protein resource (UniProt) database. The monomer unit of DHPS comprises of 286 amino acids. Template selection was made using NCBI-BLAST by performing protein search against Protein Data Bank (PDB), considering (a) higher query coverage (c) sequence identity (d) and structure resolution. MODELLER program (Sali and Blundell, 1993) was used to build the DHPS enzyme model, altogether 10 models were generated for the wild type (WT). Mutations (T53A, T53V, T53I, P55L, P55R) was introduced to generate the models of DHPS from mutated (MT) strains using Swiss-PdbViewer (Guex and Peitsch, 1997). Energy minimization for each of models was performed to remove steric clashes using Schrödinger PRIME (Schrödinger, 2019-1). Truncated Newton and Conjugant gradient algorithms with 100 cycles each were applied in the minimization. The backbone conformations of the generated models were validated using PROCHECK (Laskowski et al., 1993) and the quality of the models was evaluated by qualitative model energy analysis (QMEAN) (Benkert et al., 2009). The QMEAN score represents the global score of the model. The score ranges from 0 to 1 with the higher score for a more reliable model. The scores are calculated based on a linear combination of six structural descriptors. Further, QMEAN Z-score gives the quality of the model by comparing with the high-resolution X-ray crystallographic structures. QMEAN Z-score provides an estimation of the ‘degrees of nativeness’ of the structural features observed in a model and indicates model quality comparable to experimental structures. The model, which had the best validation results was selected for further studies.
2.4. Molecular docking
For molecular docking investigation, Glide (Friesner et al., 2004) from Schrödinger Suite was employed. Protein Preparation Wizard was used for pre-processing DHPS enzymes before docking study. Hydrogen atoms were added at pH 7.0 to maintain the appropriate ionization states for the acidic and basic amino acid residues. Further, the most probable positions for –OH and –SH hydrogen atoms were selected and proper assignment of the protonation states of charged residues and tautomers of His as well as Chi ‘flip’ for Asn, Gln and His amino acid residues were also performed. To avoid steric clashes among the residues due to the addition of hydrogen atoms, energy minimization was carried out using an OPLS-2005 force field with a convergence criterion of root mean square deviation (RMSD) value of 0.30 Å for the heavy-atom displacement.
The inhibitors were built using the builder panel in Maestro and 3D structures were prepared using the Ligprep module in Schrödinger. OPLS-2005 force field was used for computing partial atomic charges. The correct bond lengths and angles, as well as chirality, stereochemistry, tautomers, ring conformations and ionization states at pH 7.0, were obtained using Ligprep. To generate energy-minimized 3D structures, OPLS-2005 force field was used with default settings (Macro Model, Schrödinger, LLC, USA) until it reached an RMSD cutoff of 0.01 Å.
Grid box was generated at the pABA binding site with a grid box size of 20 × 20 × 20 Å. The van der Waals radius scaling to soften the potential for non-polar parts of the receptor was retained as default and the partial charges were assigned from the OPLS 2005 force field. All the designed inhibitors were docked into the active site defined by the Grid generation protocol implementing extra precision (XP) Glide scoring function to rank the docking poses and to estimate the protein–ligand binding affinities. The Glide algorithm carries out a systematic search of positions, orientations and conformations for the ligand in the active site of the enzyme using a funnel-type approach (Friesner et al., 2004). The output files were analyzed using Maestro's Pose viewer & XP-visualizer utilities.
2.5. Molecular dynamics simulations
The molecular dynamics simulations of the Apoenzyme, mutated and DHPS-inhibitor complexes were performed using Desmond (Bowers et al., 2006) with OPLS-AA 2005 force field (Jorgensen et al., 1996; Kaminski et al., 2001). The apoenzyme and complexes were solvated with TIP3P water model. An orthorhombic periodic boundary box was selected to build the required systems for subsequent MD simulations. To maintain charge neutrality of the system, an appropriate number of counter ions were added. Further, the distance between the box and wall was set greater than 10Å to avoid direct interaction with its own periodic image of the protein complex. The potential energies of the systems were minimized by the steepest descent method using a maximum of 5000 steps with a gradient threshold of 25 kcal/mol/Å. This was followed by L-BFGS (Low-memory Broyden-Fletcher-Goldfarb-Shanno quasi-Newtonian) minimizer until convergence criteria of 1 kcal mol−1 Å−1 was achieved.
The pressure and temperature equilibrations were done with the default parameters provided in Desmond. The MD simulations were carried out on the equilibrated systems for 50 ns at a constant temperature of 300⁰ K and constant pressure of 1 atm with a time step of 2 fs. The particle-mesh Ewald method (PME) (Essmann et al., 1995) was applied to compute long-range electrostatic interactions with a grid spacing of 0.8 Å. Van der Waals and short-range electrostatic interactions were smoothly truncated at 9.0 Å.
2.6. Pocket volume calculation
The volume of the binding site cavities were calculated by using POVME (Wagner et al., 2017). In order to include the entire pocket, ‘inclusion sphere’ were defined by identifying the center atoms of the binding site cavities. In order to remove cavities, which are exterior to the inhibitor binding pocket, ‘exclusion spheres’ were added. A grid spacing of 1.0 was provided to generate field of equidistant points for volume calculation.
2.7. Residue contact map
To analyse the effect of the mutations on the surrounding residues, the protein is depicted as a residue interaction network (RIN), where nodes are represented by Cβ atoms of each residue (Cα in case of glycine) and edges between nodes are considered to exist if it is within a user-defined cut-off distance (6.5–7.5 Å) of each other. MD-TASK (Brown et al., 2017) was used to construct the RIN from the MD trajectories, where the constructed dynamic RIN are representative of simulation time point (frames) in the MD trajectories.
2.8. Binding free energy
Prime MM-GBSA (Molecular Mechanics with Generalized Born and Surface Area Solvation) technique implemented in Schrödinger (Schrödinger, LLC, USA) was used to calculate the free energy of the simulation trajectories of the complexes. The binding energy was calculated for a total of 100 frames of the MD trajectory starting from the last 25 ns stable simulation until the end of the trajectory. The binding free energy was calculated according to the following equation:
ΔEMM is the energy difference between the protein-ligand complexes and sum of the energies of the ligand and protein. ΔGSolv is the difference in GBSA solvation energies of the complex and sum of the solvation energies for the unbound ligand and protein. ΔGSA is the difference in surface area energy for the complex and sum of the surface area energies for ligand and protein.
3. Results and discussion
3.1. FDA approved ligand-target database and shape screening analysis
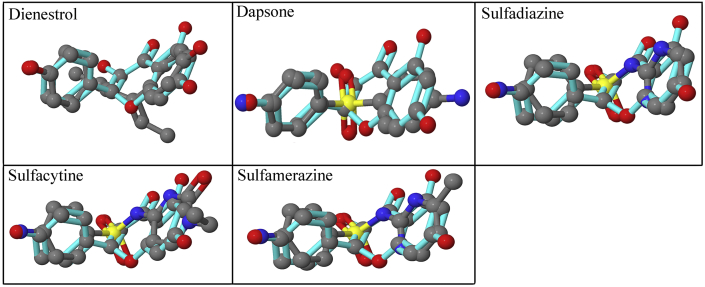
The reference database built from Drugbank consists of unique 2,618 approved and 6,338 experimental (discovery-phase) compounds. The drug-target library was stored according to the format required by SHAFTS. A relational table was created from local copy of DrugBank, which consists of information corresponding to the drug molecules such as target and Gene name, Species, UniProt ID, GenBank Gene ID and GenBank Protein ID. The final hits were processed according to the required format, and the results were extracted as performed in our previous study (Potshangbam et al., 2019). Five potential therapeutic targets were obtained from the shape screening of kaempferol, the corresponding drug molecules with predicted scores are shown in Table 1. The superimposed query and corresponding drug molecules are displayed in Figure 2. The highest scoring drug is dienestrol, it is used in the treatment of vulvar atropy, atrophic vaginitis and atrophic urethritis. It is a synthetic, non-steroidal estrogen and exerts its drug effect by acting as estrogen receptor agonist. It is reported in the literature that kaempferol can also bind and act as an estrogen receptor agonist (Harris et al., 2005; Kuiper et al., 1998). The remaining four hits, dapsone, sulfadiazine, sulfacytine and sulfamerazine are all sulfone drugs and target the same enzyme dihydropteroate synthase for its antibacterial activity. However, no literature was available that can establish the effect of kaempferol on the predicted therapeutic target i.e. dihydropteroate synthase. Therefore we selected dihydropteroate synthase for further study.
Table 1.
Potential therapeutic targets for Kaempferol identified by using shape-based screening method.
| Rank | Drug Name | Structure | Hybrid Score | Shape Score | Feature Score | Target | Validation |
|---|---|---|---|---|---|---|---|
| 1 | Dienestrol | Figure 23 | 1.381 | 0.8505 | 0.5307 | Estrogen receptor | Experimental |
| 2 | Dapsone | Figure 24 | 1.344 | 0.8834 | 0.460 | Dihydropteroate synthase | In silico |
| 3 | Sulfadiazine | Figure 25 | 1.296 | 0.8761 | 0.42 | Dihydropteroate synthase | In silico |
| 4 | Sulfacytine | Figure 26 | 1.289 | 0.8566 | 0.432 | Dihydropteroate synthase | In silico |
| 5 | Sulfamerazine | Figure 27 | 1.271 | 0.8679 | 0.403 | Dihydropteroate synthase | In silico |
Figure 2.
Overlay of query and target molecules shown in cyan and grey carbons respectively from shape-based screening of kaempferol. The drug name of the corresponding hits are displayed in each cell.
Moreover, to perform a comparative study, we selected the second highest-scoring hit dapsone. The sulfone drug is widely used as an antibiotic for a varietiy of bacterial strains. It is mainly used against M. leprae and targets the folic acid synthesis pathway by inhibiting DHPS enzyme.
3.2. Homology model of M. leprae DHPS
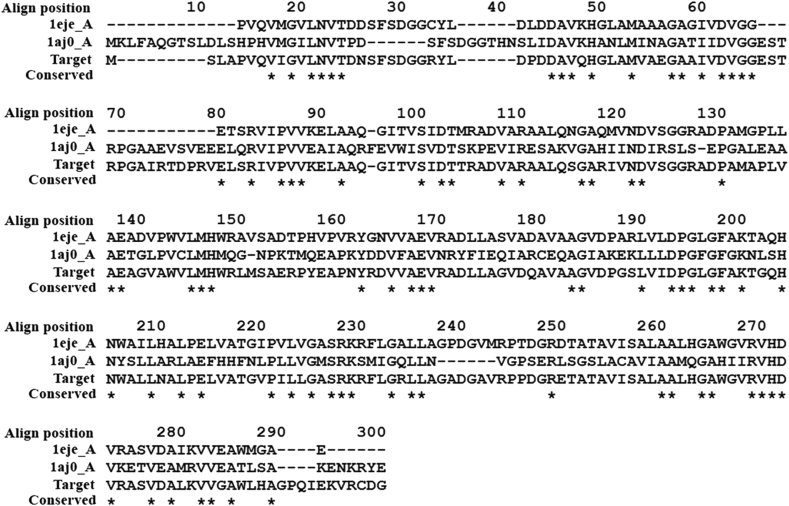
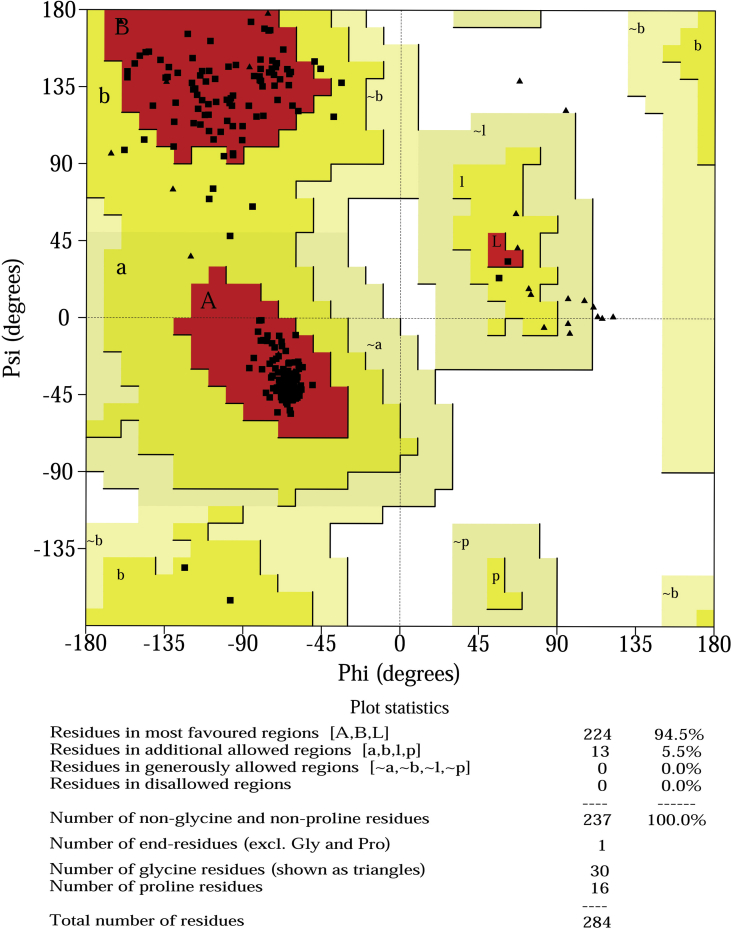
The 3D structure of M. leprae DHPS was modelled by implementing the multiple template selection approach using MODELLER to improve the quality of the model. The selected templates, PDB ID: 1EYE and 1AJ0 showed 77% and 39% sequence identity with 100 % and 96 % sequence coverage, respectively. The templates were aligned by implementing the structure-dependent gap penalty function of MODELLER, which incorporates structural information during the process. The final alignment is shown in Figure 3. We generated 10 models. PROCHEK was used to verify the stereochemical properties of the model and QMEAN server was used to assess the model quality. The predicted model for further study was chosen based on the highest percentage residues in favourable/allowed regions of Ramachandran map and QMEAN score (shown in Table 2). Ramachandran plot generated using PROCHECK for the best model 4 showed 94.5% of the residues in the most favoured region; only 5.5% were in the additionally allowed region and none in the generously allowed and disallowed regions as shown in Figure 4. The QMEAN global score of 0.760 and QMEAN Z-score of -0.378 suggest that the predicted model has good correlation with the experimental structures.
Figure 3.
Alignment of the target and templates (1eje and 1aj0) sequences. Fully-conserved residues are indicated with asterisks (*) and gaps are represented by dash (-).
Table 2.
Score calculated from PROCHECK and Qmean for model validation. Percentage values indicate the fraction of total residues in specific region of the Ramachandran map. Bold font represent selected model.
| Modela | PROCHECKb |
QMEAN SCOREc | |||
|---|---|---|---|---|---|
| Favorable | Additional allowed | Generously allowed | Disallowed | ||
| 1 | 92.80% | 6.8% | 0.4% | 0.0% | 0.753 |
| 2 | 94.1% | 5.9% | 0.0% | 0.0% | 0.750 |
| 3 | 94.5% | 5.5% | 0.0% | 0.0% | 0.753 |
| 4 | 94.5% | 5.5% | 0.0% | 0.0% | 0.760 |
| 5 | 94.5% | 5.1% | 0.4% | 0.0% | 0.73 |
| 6 | 92.8% | 6.8% | 0.0% | 0.4% | 0.751 |
| 7 | 93.2% | 6.8% | 0.0% | 0.0% | 0.746 |
| 8 | 94.5% | 5.5% | 0.0% | 0.0% | 0.751 |
| 9 | 94.1% | 5.9% | 0.0% | 0.0% | 0.756 |
| 10 | 94.5% | 5.1% | 0.4% | 0.0% | 0.729 |
Homology models of DHPS enzyme.
Values calculated using PROCHECK corresponding to favorable, additional allowed, generously allowed and disallowed regions of Ramachandran plot.
QMEAN global score.
Figure 4.
Ramachandran plot of DHPS model showing different areas of the plot, represented by different colors: beige, yellow, brown and red indicating disallowed, generously allowed, additional allowed and the most favoured regions, respectively.
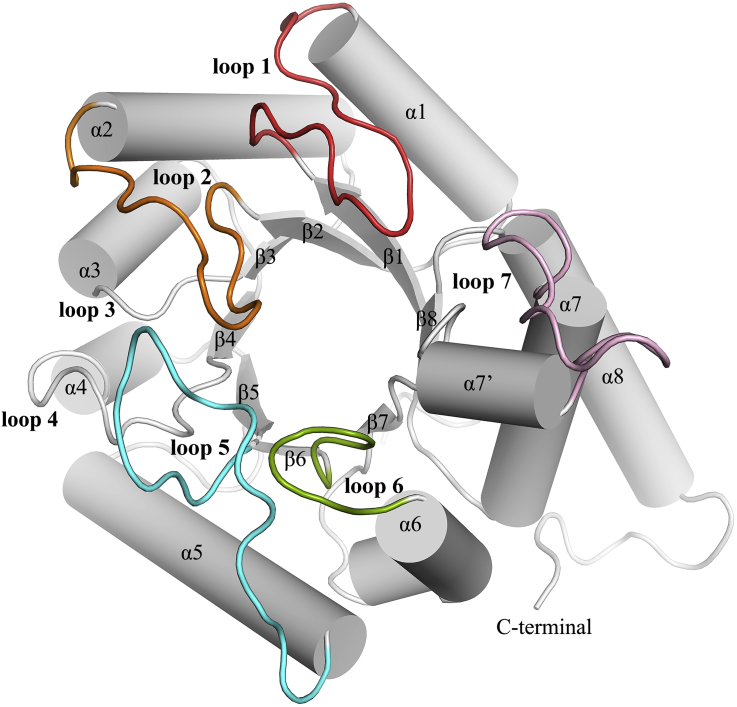
The predicted model is a monomer unit and adopts the typical DHPS structural organisation, which consists of “TIM-barrel” fold having eight α-helices, surrounding a central barrel comprising of eight parallel β-strands as shown in Figure 5. Towards the N-terminal side, the loops that form the α-β connections are short and are not involved in active site formation. However, loops at the C-terminal side are reported to play an active role in sulfa drug activity (Baca et al., 2000). Out of the 8 loops, loops 3, 4 and 8 are away from the pocket, however, loops 1, 2, 5, 6 and 7 are reported to play significant role in DHPS activity (Baca et al., 2000).
Figure 5.
Three-dimensional (3D) structure of DHPS (M. leprae) generated using homology modeling. Helices and beta strands are represented by cylinders and sheets (with arrows), respectively. Loops contributing to the active site pocket are shown in red (loop1), orange (loop2), cyan (loop5), green (loop6) and magenta (loop7).
The pterin-binding pocket is formed by the residues Asn13, Asp21, Asp86, Asn105, Val107, Met130, Gly181, Phe182, Lys213, Arg253 and His255 (Baca et al., 2000). While the pABA binding site is formed mainly by the residues of loops 1, 2, 5 and 7 (Baca et al., 2000). The entire pABA active pocket is enclosed by the distal end of loop1 formed by residues Asp16 to Gly23 (Yun et al., 2012). The highly conserved residues Phe19 from loop1, Pro55 from loop 2, Lys213 from loop7 and Phe182 from loop6 together form the pABA binding pocket (Yun et al., 2012).
3.3. Docking and binding free energy analysis of WT and MT DHPS
Although shape-based screening of kaempferol predicted DHPS as its novel therapeutic target, nevertheless, it is important to carry out further validation as shape-based screening does not account for interactions or binding at the target active site. Moreover, a comparative study with the drug molecule dapsone, considering its effect on wild type and mutants of DHPS enzyme may provide clues on the future direction of using kaempferol as an alternative drug to sulfone resistant strains. Consequently, detail study was carried out by implementing more rigorous in silico methods such as docking and molecular dynamics simulations.
3.4. WT DHPS
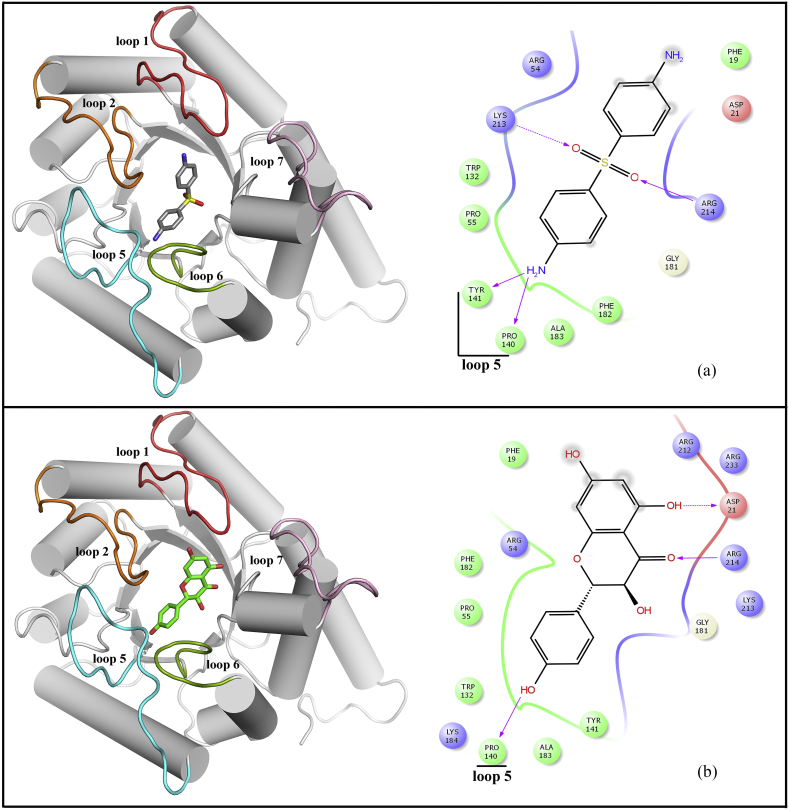
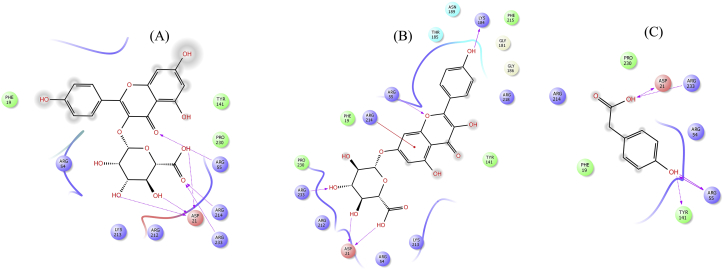
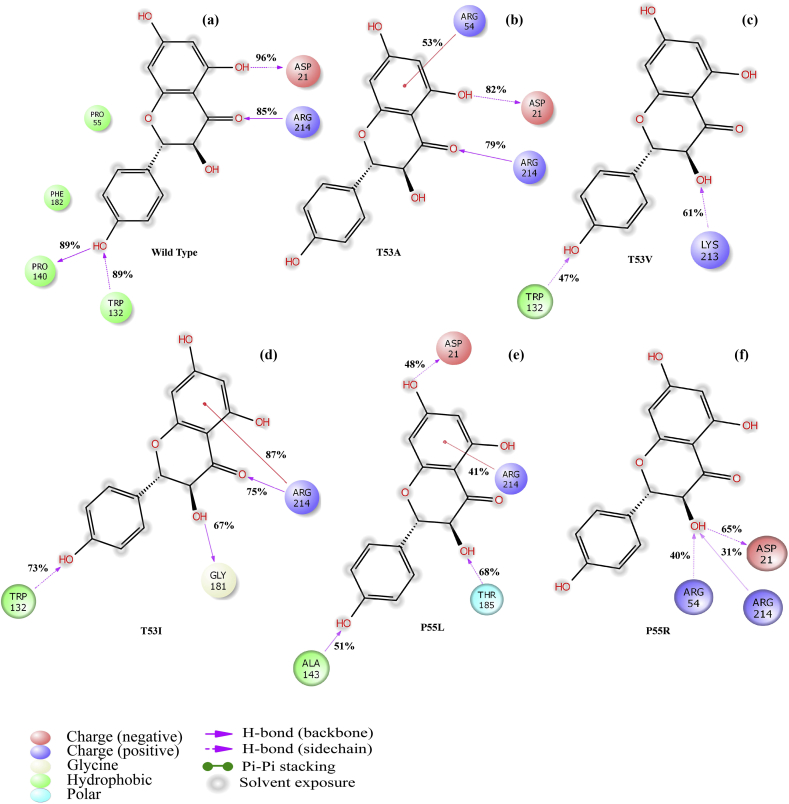
Molecular docking studies of dapsone and kampferol at the pABA pocket of WT and MT DHPS active site revealed kaempferol to be a better predicted inhibitor of DHPS. At the pABA pocket of WT DHPS enzyme, four hydrogen bond interactions were observed with dapsone (shown in Figure 6a). Pro140 and Tyr141 backbone (part of loop 5) make hydrogen bond interactions with H9 and H10 (amine group) with distances of 1.99Å and 2.59Å, respectively. The backbone of highly conserved residue Arg214 of α-helix 7 and side chain of Lys213 formed hydrogen bond interactions with O1 and O2 of sufonyl group (distances 2.4 Å & 2.16Å), respectively. In the case of kaempferol also, three hydrogen bond interactions stabilize the complex as shown in Figure 6b. At loop 5 region, Pro140 backbone interacts with H9 (hydroxyl group) of the phenol ring through hydrogen bond (distance 1.73 Å). Further, O5 of carbonyl oxygen and H11 (hydroxyl group) of the naphthalene ring of kaempferol makes hydrogen bond contacts with conserved residues Arg214 backbone and Asp21 side chain (part of the pterin-binding pocket) with distances of 2.12 Å and 2.66 Å, respectively.
Figure 6.
Docked conformation of the dapsone (a) and kaempferol (b) into the DHPS pABA pocket. The left panel displays 3D conformation of the inhibitors (dapsone in grey carbon and kaempferol in green carbon) and the right panel shows 2D representation of enzyme-inhibitor interactions. The dotted pink arrows indicate hydrogen bond interactions through amino acid side chains and the solid pink arrows represent hydrogen bond interactions through backbone atoms.
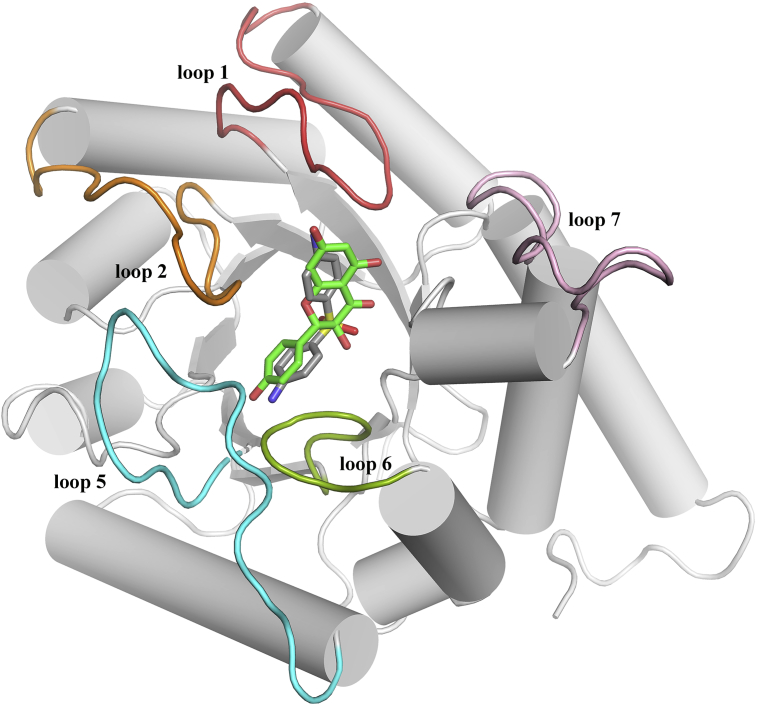
The superimposed structure of dapsone and kaempferol at the pABA binding site is shown in Figure 7. It is clear that both compounds have similar binding mode and conformation with similar stabilizing interactions. However, the predicted dockings scores of dapsone (XP GScore = -4.13 kcal/mol, Glide energy = -35.06 kcal/mol, Glide evdw = -22.34 kcal/mol and Glide ecoul = -12.71 kcal/mol) as compared to kaempferol (XP GScore = -6.29 kcal/mol, Glide energy = -41.28 kcal/mol, Glide evdw = -29.88 kcal/mol and Glide ecoul = -11.40 kcal/mol) suggest that kaempferol could be a better inhibitor. Moreover, the calculated binding free energy of kaempferol (-47.77 kcal/mol) in comparison to dapsone (-38.33 kcal/mol) reaffirms the docking result.
Figure 7.
Superimposed 3D structure of dapsone and kaempferol at the pABA binding site. Dapsone is shown in grey carbon and kaempferol in green carbon.
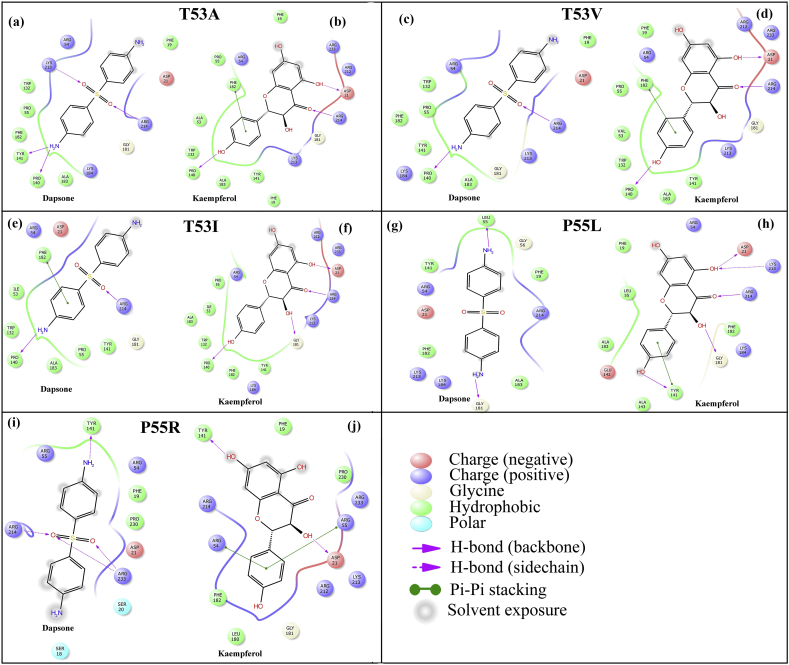
3.5. Mutant T53A
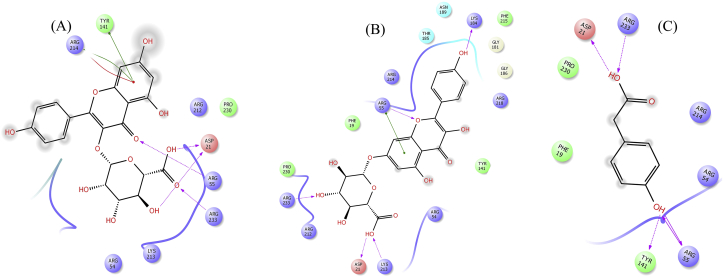
Mutation was introduced at position 53 and threonine was mutated to alanine. Dapsone makes similar hydrogen bond interactions as compared to the wild type (shown in Figure 8a). Backbone atoms of Pro140 and Tyr141 of loop 5 make hydrogen bond interactions with H9 and H10 (amine group) of dapsone with distances of 2.43Å and 2.03Å, respectively. Lys213 and Arg214 established hydrogen bond interactions with O1 and O2 atoms of sufonyl group (distances 2.16 Å & 2.48Å), respectively. Likewise, in the case of kaempferol at the T53A pABA pocket, three hydrogen bond interactions were observed similar to the wild type (Figure 8b). The loop 5 residue, Pro140 (backbone) makes hydrogen bonding contact with H9 (hydroxyl group) of the phenol ring (distance 1.96Å). Additionally, O5 carbonyl oxygen and H11 (hydroxyl group) of the naphthalene ring of kaempferol makes hydrogen bond interactions through Arg214 backbone and Asp21 side chain with distances of 2.18Å and 1.63Å, respectively. Additionally π- π interaction was predicted between Phe182 ring and phenol ring of kaempferol (distance 5.46 Å).
Figure 8.
2D interaction diagram of DHPS mutants T53A, T53V, T53I, P55L and P55R resulting from docking dapsone (a, c, e, g and i) and kaempferol (b, d, f, h and j) into the pABA active site, respectively.
Further, from Table 3 it was observed that there are no significant changes in predicted docking score and binding free energy calculation due to T53A mutation for both dapsone and kaempferol. Nevertheless, as observed earlier, kaempferol showed better predicted XP score (-6.27 kcal/mol, -4.11 kcal/mol) as well as binding free energy (-49.49 kcal/mol, -38.06 kcal/mol) compared to dapsone, respectively.
Table 3.
Docking score and MM-GBSA binding free energy results of dapsone and kaempferol for wild type and mutant DHPS. Dapsone & Kaempferol are indicated by D & K, respectively.
| Parameters | Wild Type |
T53A |
T53V | T53I | P55L | P55R | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutants (DHPS) | ||||||||||||
| D | K | D | K | D | K | D | K | D | K | D | K | |
| XP GScorea | -4.13 | -6.29 | -4.11 | -6.27 | -3.37 | -6.28 | -3.36 | -7.57 | -2.62 | -5.77 | -2.14 | -4.87 |
| Glide energyb | -35.06 | -41.28 | -35.07 | -40.92 | -42.50 | -56.43 | -35.64 | -39.39 | -25.7 | -42.43 | -28.75 | -36.42 |
| Glide evdwc | -22.34 | -29.88 | -22.26 | -29.24 | -25.00 | -29.31 | -29.1 | -25.89 | -22.6 | -25.63 | -19.0 | -27.29 |
| Glide ecould | -12.71 | -11.40 | -12.81 | -11.68 | -10.61 | -11.70 | -6.52 | -13.5 | -3.13 | -16.80 | -9.66 | -9.13 |
| MM-GBSA ΔG Binde | -38.33 | -47.77 | -38.06 | -49.49 | -37.97 | -49.65 | -36.65 | -49.80 | -27.35 | -45.7 | -26.8 | -41.76 |
Total GlideScore.
Modified Coulomb-van der Waals interaction energy.
Van der Waals energy.
Electrostatic energy.
Total binding energy calculated by MM-GBSA method.
3.6. T53V
In this mutation, threonine is replaced by valine at position 53. Due to this mutation, substantial changes were observed for dapsone at the pABA binding pocket. Only two hydrogen bonds were formed by Pro140 and Arg214 backbone atoms with H10 (amine group) and O2 atoms (sufonyl group) of dapsone with distances of 2.31Å and 2.13Å, respectively (Figure 8c). However, in the case of kaempferol, no significant changes were observed due to T53V mutation. Similar to the wild type, three hydrogen bond interactions were predicted between: Pro140 (backbone) and H9 (hydroxyl group of phenol ring), Arg214 backbone and O5 of carbonyl oxygen, Asp21 side chain and H11 (hydroxyl group) of the naphthalene ring with distances of 1.96Å, 2.18Å, and 1.63Å, respectively (Figure 8d). The π-π interaction was also predicted between Phe182 ring and phenol ring of kaempferol (distance 5.46 Å).
The XP docking score and binding free energy values (shown in Table 3) indicated low binding affinity of dapsone at the pABA binding site due to T53V mutation. In the case of kaempferol no significant change was observed due to T53V mutation. Therefore, kaempferol (XP score = -6.28 kcal/mol, MM-GBSA ΔGBind = -49.65 kcal/mol) was predicted to have better binding affinity than dapsone (XP score = -3.37 kcal/mol, MM-GBSA ΔGBind = -37.97 kcal/mol).
3.7. T53I
Threonine is replaced by Isoleucine in this mutation at position 53. At the pABA binding pocket of T53I, dapsone was able to maintain two hydrogen bond interactions, established through Pro140 and Arg214 backbone atoms with H10 (amine group) and O2 atoms (sufonyl group) with distances of 2.30Å and 2.30Å, respectively (Figure 8e). A weak π- π interaction was also observed between Phe182 and aniline ring of dapsone with distance of 5.15Å from the centroid of the rings. On the other hand, kaempferol still maintained the conformation and interactions at the T53I pABA binding pocket similar to the wild type. Pro140 (backbone) forms hydrogen bond contact with H9 (hydroxyl group) of the phenol ring (distance 1.84Å) as shown in Figure 8f. Arg214 backbone and Asp21 side chain make hydrogen bond interactions with O5 of carbonyl oxygen and H11 (hydroxyl group) of the naphthalene ring with distances of 2.09Å and 1.87Å, respectively. Additionally, a hydrogen bond contact was established between Gly181 backbone and H12 (hydroxyl group of naphthalene ring) with distance of 2.53 Å.
As shown in Table 3, dapsone showed a low XP GScore (-3.36 kcal/mol) and binding free energy (-36.65 kcal/mol) as compared to kaempferol (XP GScore = -7.57 kcal/mol, MM-GBSA ΔGBind = -49.80 kcal/mol) suggested kaempferol to be a better predicted inhibitor.
In all the three mutations T53A, T53V and T53I, binding of dapsone to the pABA pocket indicated substantial differences in interaction pattern as compared to the wild type, which presumably led to more variations in the predicted scores. However, binding of kaempferol to pABA pockets of mutants was consistent with the wild type and this resulted in similar predicted scores.
3.8. P55L
Mutation at position 55 from proline to leucine has a profound effect on the binding of dapsone and kaempferol to DHPS active site. At the pABA binding pocket (P55L strain), Leu55 and Gly181 backbone atoms formed hydrogen bond contacts through H12 and H10 (amine group) of dapsone with distances of 2.26Å and 2.04Å, respectively (Figure 8g). Moreover, the interacting amino acid residues are completely different as compared to the wild type and hence showed different conformation. On the other hand, kaempferol retained five hydrogen bond interactions at the pABA binding pocket of P55L mutant (Figure 8h). Tyr141 from loop 5 has one hydrogen bond contact through the main chain atom with H9 (hydroxyl group) of the phenol ring (distance 1.99Å), another π- π interaction was observed between phenol rings of Tyr141 and kaempferol (distance 5.43Å). The backbone atoms of Gly181 and Arg214 makes hydrogen interactions with H12 hydroxyl group and O5 carbonyl group (naphthalene ring) of kaempferol with distances of 1.85 Å and 2.67 Å, respectively. Further, Lys213 and Asp21 makes hydrogen bond contacts through side chain N…..Hζ3 and Oδ2 with O6 and H12 (hydroxyl group) of naphthalene ring having bond lengths of 2.43 Å and 1.86 Å, respectively.
As evident from the docking score and binding free energy calculation (Table 3), P55L mutation significantly affect the binding of dapsone at the pABA pocket. Compared to the wild type, predicted XP GScore (-2.62 kcal/mol) and MM-GBSA binding free energy (-27.35 kcal/mol) of dapsone was drastically reduced. Similarly, binding affinity of kaempferol at the pABA pocket was also decreased but marginally having the predicted XP GScore (-5.77 kcal/mol) and MM-GBSA binding free energy (-45.7 kcal/mol) due to P55L mutation. It is apparent that the kaempferol could sustain the resistance effect due to the mutation as compared to dapsone.
3.9. P55R
Mutation of proline to arginine at position 55 resulted in significant changes in the predicted binding of both dapsone and kaempferol. As shown in Figure 8i due to the P55R mutation, binding conformation of dapsone to pABA is considerably different from the wild type. Instead of binding to the pABA pocket cavity, it favoured to binding at the surface, dangling on to four hydrogen bond interactions. Tyr141 and Arg214 makes hydrogen bond contacts through sidechain atoms Oη and Hη12 with H10 (amine group) and O2 atoms (sufonyl group) of dapsone with distances of 2.20Å and 2.13Å, respectively. Two more hydrogen bonds were observed between the Arg233 sidechain atoms Hη12 and Hη22 with O2 and O1 sufonyl group of dapsone with bond lengths of 2.09 Å and 2.59 Å, respectively. Similar changes at the pABA binding site were also observed in kaempferol binding due to P55R mutation, however, it remained in the pABA pocket cavity, resulting in more stable interaction than dapsone (Figure 8j). Tyr141 and Asp21 make hydrogen bond interactions through sidechain atom of Oη and Oδ2 with H10 and H11 (hydroxyl groups of naphthalene ring of kaempferol) with distances of 1.94 Å and 1.74 Å, respectively. Two π-π interactions were observed between the Arg54 and Arg55 sidechain (methanetriamine) through phenol ring of kaempferol with distances of 5.21 Å and 3.41 Å, respectively.
The P55R mutation prominently affects binding of both dapsone and kaempferol to the pABA pocket with large conformation deviation from the wild type than other mutations (T53A, T53V, T53I and P55L). The docking score and binding free energy calculation clearly showed drastically different results from the wild type. In the P55R mutation, the XP GScore (-2.14 kcal/mol) and MM-GBSA binding free energy (-26.80 kcal/mol) are the lowest among all the other mutations for dapsone. Likewise, the P55R mutation effects kaempferol binding to pABA pocket, however, the XP GScore (-4.87 kcal/mol) and MM-GBSA binding free energy (-41.76 kcal/mol) are comparable to dapsone binding to wild type, suggesting that kaempferol could overcome antibiotic resistant due to the mutations.
Many metabolites of kaempferol have been identified and characterised such as kaempferol-3-glucuronide (K3), kaempferol 7-O-glucuronide (K7) and 4-hydroxyphenylacetic acid (4H). It would be worthwhile to examine how these metabolites interact with wild type enzyme and its mutants. Therefore, these three metabolites, K3, K7 and 4H were also docked into wild type and 5-mutants, T53A, T53V, T53I, P55L and P55R. The results are given in Table 4, and interactions shown in Figure 9, Figure 10, 11, 12, 13, and 14. For the wild type DHPS, the XP GScore of -9.28 kcal/mol, -9.08 kcal/mol, -4.39 kcal/mol, and MM-GBSA binding free energy values of -53.41 kcal/mol, -50.94 kcal/mol, -38.97 kcal/mol, respectively, for K3, K7 and 4H matches with kaempferol with 4H having poor score and K3 and K7 comparatively better scores with respect to kaempferol. Furthermore, these metabolites demonstrate trend, similar to kaempferol, with respect to wild type and mutants.
Table 4.
Docking score and MM-GBSA binding free energy results of Kaempferol-3-glucuronide (K3), kaempferol 7-O-glucuronide (K7) and 4-Hydroxyphenylacetic acid (4H) for wild type and mutant DHPS.
| Wild Type |
T53A |
T53V | T53I | P55L | P55R | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutant (DHPS) | ||||||||||||||||||
| Parameters | K3 | K7 | 4H | K3 | K7 | 4H | K3 | K7 | 4H | K3 | K7 | 4H | K3 | K7 | 4H | K3 | K7 | 4H |
| XP GScorea | -9.28 | -9.08 | -4.39 | -8.76 | -8.29 | -4.29 | -8.73 | -8.33 | -4.41 | -9.10 | -8.71 | -4.4 | -7.66 | -7.44 | -3.49 | -5.70 | -5.36 | -3.97 |
| Glide energyb | -59.67 | -58.35 | -27.45 | -56.98 | -61.92 | -26.82 | -57.4 | -59.18 | -26.9 | -55.5 | -57.6 | -27.0 | -63.03 | -56.18 | -23.88 | -47.4 | -52.00 | -27.34 |
| Glide evdwc | -32.87 | -35.90 | -18.67 | -32.7 | -41.1 | -19.12 | -37.3 | -37.5 | -19.1 | -34.4 | -37.5 | -18.7 | -38.3 | -32.84 | -9.78 | -25.60 | -29.68 | -10.90 |
| Glide ecould | -26.80 | -22.44 | -8.77 | -24.2 | -20.81 | -7.70 | -20.07 | -21.6 | -7.79 | -21.08 | -20.05 | -8.34 | -24.63 | -23.33 | -14.09 | -21.84 | -22.32 | -16.43 |
| MM-GBSA ΔG Binde | -53.41 | -50.94 | -38.97 | -58.38 | -60.50 | -38.06 | -57.41 | -59.18 | -26.98 | -60.15 | -51.68 | -39.06 | -60.33 | -59.30 | -28.20 | -41.08 | -48.15 | -38.54 |
Total GlideScore.
Modified Coulomb-van der Waals interaction energy.
Van der Waals energy.
Electrostatic energy.
Total binding energy calculated by MM-GBSA method.
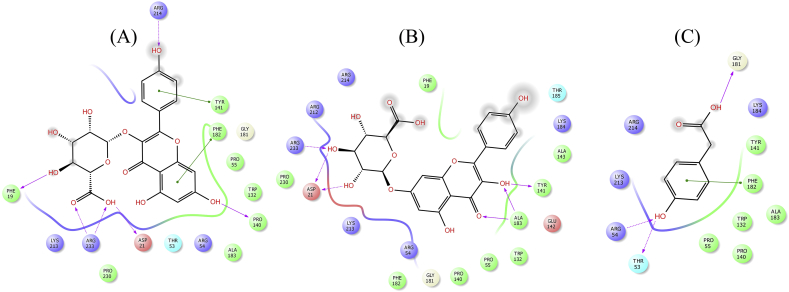
Figure 9.
2D interaction diagram showing docking results of kaempferol-3-glucuronide (A), kaempferol 7-O-glucuronide (B) and 4-Hydroxyphenylacetic acid (C) for wild type DHPS. Pink dotted lines represent hydrogen bonds and green lines denote π-π interactions.
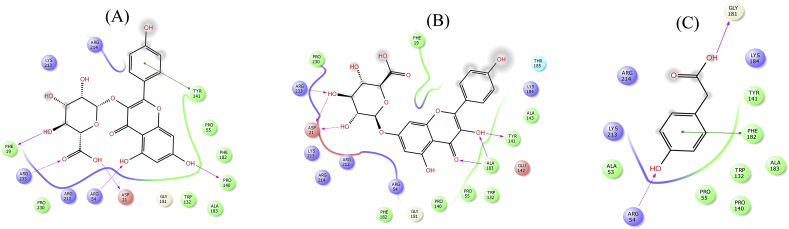
Figure 10.
2D interaction diagram showing docking results of kaempferol-3-glucuronide (A), kaempferol 7-O-glucuronide (B) and 4-Hydroxyphenylacetic acid (C) for DHPS mutant T53A. Pink dotted lines represents hydrogen bonds and green lines denotes π-π interactions.
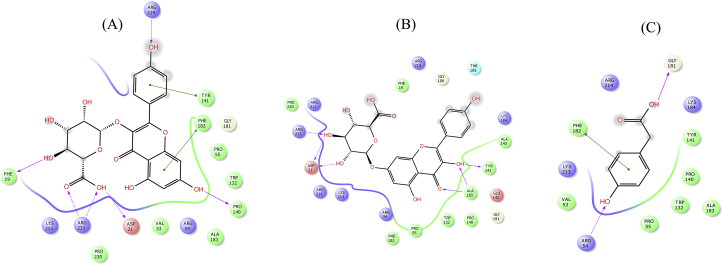
Figure 11.
2D interaction diagram showing docking results of kaempferol3-glucuronide (A), kaempferol 7-O-glucuronide (B) and 4-Hydroxyphenylacetic acid (C) for DHPS mutant T53V. Pink dotted lines represent hydrogen bonds and green lines denote π-π interactions.
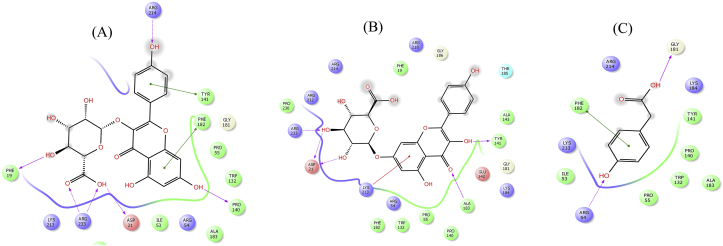
Figure 12.
2D interaction diagram showing docking results of kaempferol3-glucuronide (A), kaempferol 7-O-glucuronide (B) and 4-Hydroxyphenylacetic acid (C) for DHPS mutant T53I. Pink dotted lines represent hydrogen bonds, green lines denote π-π interactions and red line displays cation- π interactions.
Figure 13.
2D interaction diagram showing docking results of kaempferol-3-glucuronide (A), kaempferol 7-O-glucuronide (B) and 4-Hydroxyphenylacetic acid (C) for DHPS mutant P55L. Pink dotted lines represent hydrogen bonds, green lines denote π-π interactions and red line displays cation- π interactions.
Figure 14.
2D interaction diagram showing docking results of kaempferol-3-glucuronide (A), kaempferol 7-O-glucuronide (B) and 4-Hydroxyphenylacetic acid (C) for DHPS mutant P55R. Pink dotted lines represent hydrogen bonds, green lines denote π-π interactions and red line displays cation- π interactions.
The docking studies do not account for the dynamic changes of the enzyme-inhibitor complex. Hence, detail molecular dynamics study was performed to understand the changes induced by the mutation in the pABA pocket, changes in the volume size, overall changes regarding the loop regions and analysis of binding free energy, to see if kaempferol could overcome the mutations' effect.
3.10. Molecular dynamics study
Molecular dynamics simulations revealed insights into the dynamics of the enzyme as well as interactions with inhibitors in contrast to the docking studies of dapsone and kaempferol, which provided the static view only. The modelled 6-structures of wild type, and mutants:- T53A, T53V, T53I, P55L and P55R, and their respective 12-complexes with dapsone and kaempferol were studied with explicit solvent molecular dynamics simulations. Thus, in total 18 molecular dynamics simulations were performed for this study.
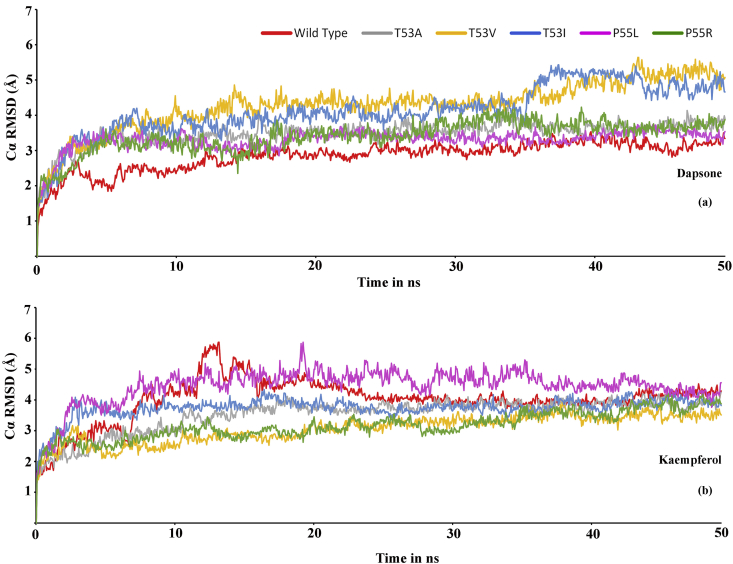
To evaluate the stability of the simulation systems, root mean square deviation (RMSD) of the Cα atoms of the enzyme and complexes were calculated. As shown in Figure 15a, the dapsone bound enzyme complexes showed rapid increase in RMSD values and fluctuate around 3–3.5 Å and achieved stable fluctuation after 30 ns at around 3Å, except for T53V and T53I mutants. They showed higher fluctuations but in constant range (3.5–4 Å) and attained stability after 35 ns at around 5 Å. In the case of kaempferol bound DHPS enzymes (shown in Figure 15b), the RMSD initially fluctuates at around 2.5–4 Å, however, as it closes to 35ns, the RMSD of all the complexes converges to around 3Å and gained stability thereafter. By comparing dapsone and kaempferol bound complexes, it is clear that there are significant differences in RMSD values, suggesting substantial conformational changes, leading to less stability in the enzyme-inhibitor complex in case of dapsone.
Figure 15.
Root mean square deviation (RMSD) of wild type DHPS and mutants in the presence of bound inhibitors dapsone (a) and kaempferol (b) during the 50 ns dynamics simulation.
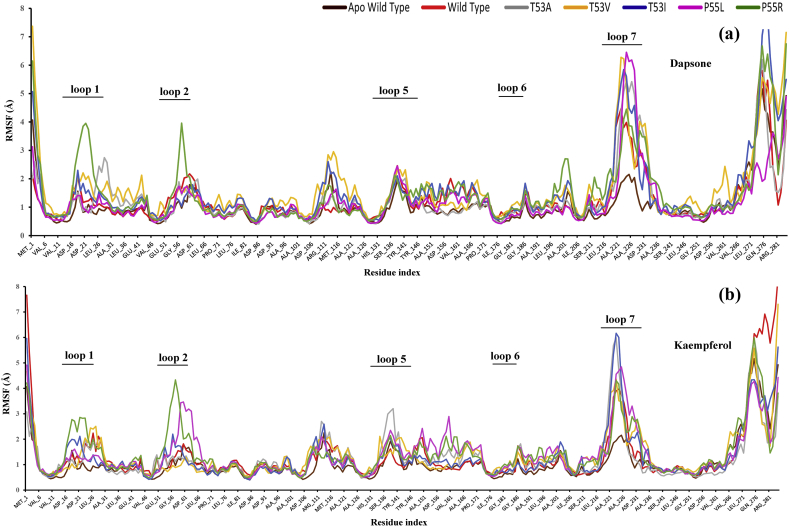
Root-mean-square fluctuations (RMSF) of Cα atoms were evaluated to reveal the fluctuations in local regions of the enzyme complexes, which may provide insight into the stability of the wild type and mutated systems and also between dapsone and kaempferol complexes. As shown in Figure 16, significant fluctuations of the mutant complexes were found around loop 1, 2 (mutation position) and 7, which mostly contribute to the pABA binding pocket. In contrast, very small fluctuations were observed in the case of wild type complexes.
Figure 16.
Root mean square fluctuation (RMSF) of wild type DHPS and mutants in the presence of bound inhibitors dapsone (a) and kaempferol (b) during the dynamics simulation.
In dapsone bound complexes, particularly P55R, high fluctuations in the loop 1 and loop 2 regions, with a maximum peak of around 4Å (in both the regions) were observed as compared to wild type (1.5 and 1.6 Å), respectively (Figure 16a). Similarly, kaempferol bound complexes also showed high fluctuation around loop 1 and loop 2 regions, but the highest was found in P55R, with maximum peaks around 2.8 Å and 4.4 Å as compared to wild type (1.1 Å and 1.2 Å), respectively (Figure 16b). The fluctuations of the residues were more pronounced in the pABA binding loop regions, mainly loop 1 and loop 2, suggesting of highly flexible and less stable mutant complexes, particularly P55R as compared to the wild type.
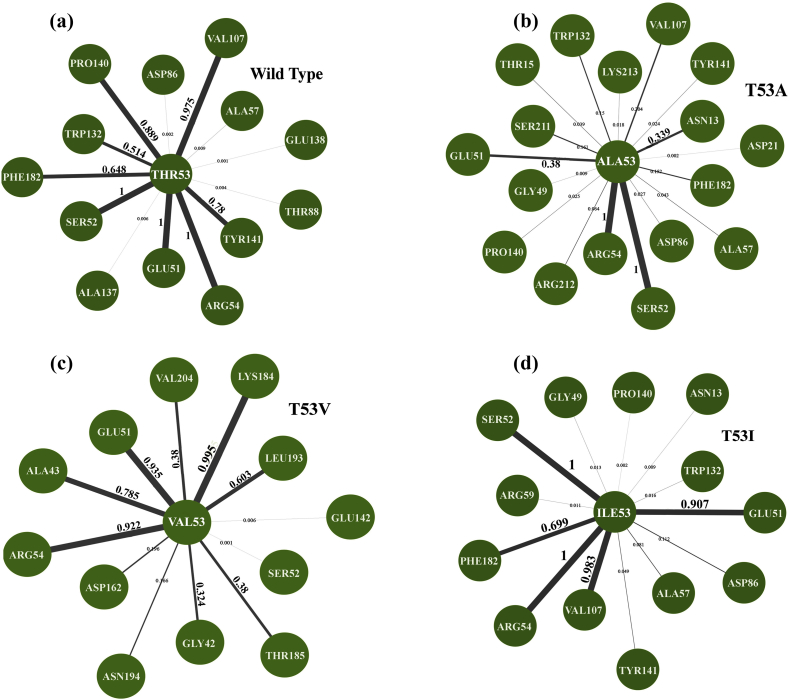
3.11. Residue contact map analysis
The contact map analysis allows studying how frequently a residue interacts with its surrounding residues. At position 53, the contact map was calculated for three mutated residues Ala53, Val53 and Ile53. Similarly, at position 55, contact maps for two mutated residues Leu55 and Arg55 were calculated. As shown in Figure 17a, in the residue network of the Wild type (Thr53), strong contacts were made by Glu51, Ser52, and Arg54 constantly throughout the simulation for 100% of the time. Val107 and Pro140 also made contact with the wild type for 97.5% and 88 % of simulation time, respectively. In mutant T53A, consistent contacts were maintained by Ser52 and Arg54 for 100%, however, Val107 and Pro140 contacts were reduced to 20.4 % and 2.5 % of the time, respectively (Figure 17b). Mutants T53V maintained contacts with Glu51 and Arg54 for 93.5 % and 100 % of simulation time. It also makes new contacts with Ala43 and Lys184 for 78% and 95.5% during the entire simulation period (Figure 17c). Interestingly, T53I mutant was able to maintain most of the prominent contacts observed in the wild type, namely; Glu51, Ser52, Arg54 and Val107 for 90.7%, 100%, 100% and 98.3% of the simulation time, respectively (Figure 17d). At position 55 (shown in Figure 18a), the wild type Pro55 make prominent contact with Gly56, Arg54, Phe19, Asn17 and Tyr141 consistently for 100%, 100%, 86.2%, 77.2% and 71.8% throughout the simulation time, respectively. In the case of P55L (Fig. 18b) and P55R (Figure 18c) mutants, only one prominent contact Gly56 was found which was also observed in the wild type for 100% of the time. New contacts Pro197, Leu199 and Ile206 were found in P55L mutant for 99.8%, 96.6%, 89.9% and 75.5 of the simulation time. In P55R mutant, additional contacts Leu196 and Pro205 were observed for 99.5% and 75.5 during the simulation period.
Figure 17.
Residue contact map of wild type (a) and mutants T53A (b), T53V (c) and T53I (d).
Figure 18.
Residue contact map of wild type (a) and mutants P55L (b) and P55R (c).
It can be seen that most of the prominent contacts at positions, 53 and 55 in wild type were found in the loop regions, which significantly contributed to the pABA binding pocket. Therefore, loss or gain of contacts in mutants would consequently affect the binding of the inhibitors, perhaps by interfering with the pocket size of the active site. Further investigation considering the dynamics of the pABA binding pocket relative to the mutations and its effect on the inhibitor binding are being studied.
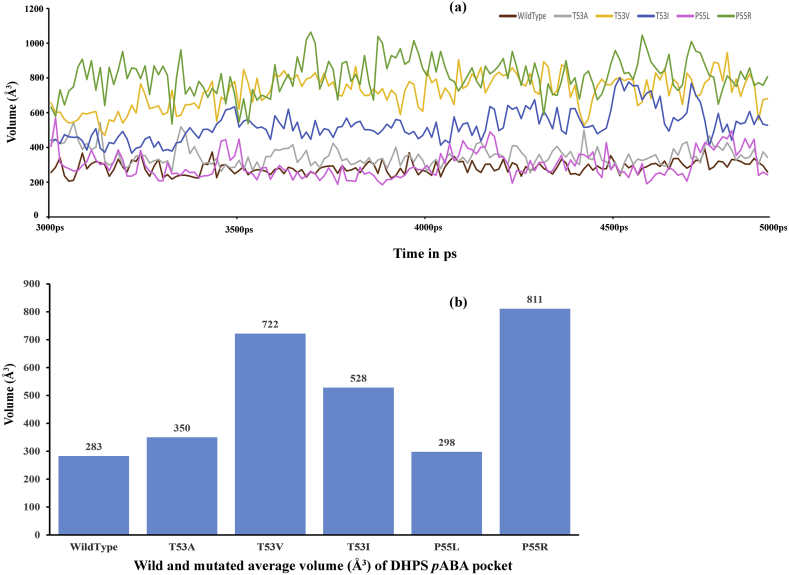
3.12. Volume analysis
Investigation on the pABA binding pocket volume revealed important insights into the binding of dapsone and kaempferol and their inhibitory effect on wild type and mutants. As shown in Figure 19, it is seen that the size of the pocket volume increases in all the mutants as compared to the wild type. Similar results were also reported earlier (Chaitanya et al., 2015; Chowdhury et al., 2014). As discussed earlier, the loss or gain of the prominent contacts (in residue contact map) in the mutants could result in different volume sizes of the binding pocket, which ultimately might affect the binding of the inhibitors relative to the wild type. The volume calculations were carried only for the equilibrated trajectories (30 ns–50 ns). The calculated volume along the simulation period is shown in Figure 19a and the average volumes of wild type and the mutants are shown in Figure 19b. The wild type pABA binding pocket volume is the smallest having an average volume of 283 Å3. The mutant enzyme P55L showed relatively smaller pocket size (average volume of 298 Å3) as compared to other mutants T53A and T53I (average volumes of 350 Å3 and 528 Å3, respectively). The second-largest pocket volume is formed by the mutant T53V and the largest by the P55R (average pocket volumes of 722 Å3 and 811 Å3, respectively).
Figure 19.
Plot of DHPS pABA pocket volume for wild type and mutants during the simulation period (a). Histogram of average pABA pocket volume size for wild type and DHPS mutants (b).
Interestingly, the variations in pocket volume size consistently correlate with the docking and MM-GBSA (single docked pose) results of dapsone and kaempferol binding to pABA pocket. As the pocket size increases among the mutants, the docking scores and MM-GBSA values decrease, except for kaempferol bound T53I mutant, which showed increased scores regardless of the increased volume of the pocket as compared to the wild type. Nevertheless, it will be more reasonable to compare data derived from dynamics studies as opposed to the static results from docking studies. Therefore, molecular interactions of ligand-enzyme complexes, as well as MM-GBSA scores were calculated along the trajectories to find out relative stability of the complexes, based on the interactions and binding free energies during the simulation period for dapsone and kaempferol bound complexes.
3.13. Protein-ligand interactions analysis
Protein-ligand binding and stable complex formations are mainly characterized by the strong hydrogen bond and/or hydrophobic interactions. The interactions were monitored throughout the simulation period and the outputs were given in percentage interaction during the simulation time.
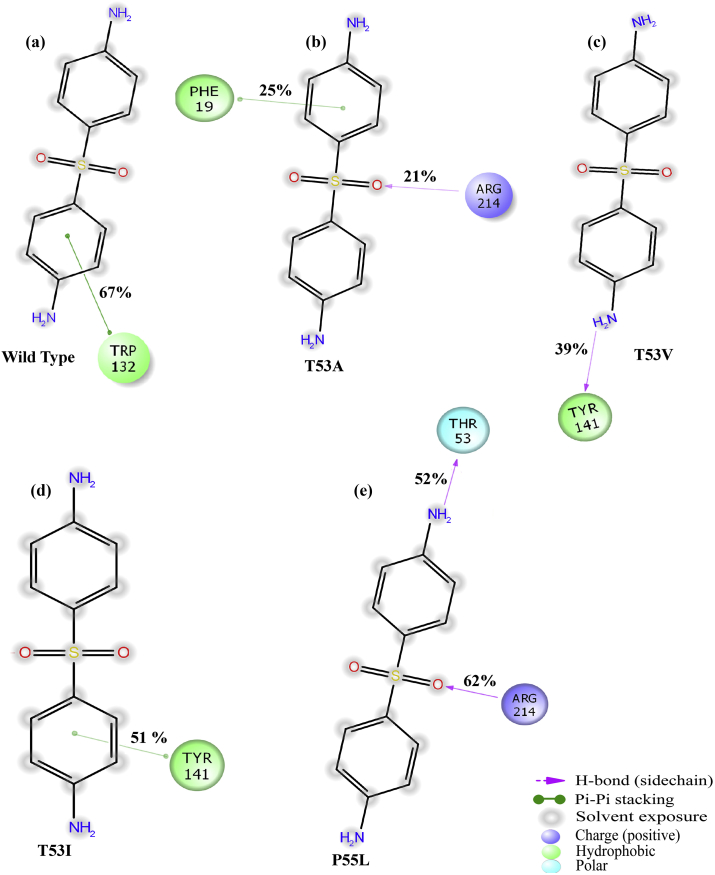
The four hydrogen bond interactions observed in the docked conformation of dapsone bound to the wild type DHPS were lost during the simulation (shown in Figure 20a). However, a new π- π interaction was observed for 67% of the simulation time between the indole ring of Trp132 and aniline ring of dapsone. In mutant T53A, only for the 21% of the simulation time, original interaction was observed between Arg214 backbone and O2 atoms (sufonyl group) of dapsone (displayed in Figure 20b). A new weak π- π interaction was also detected for only 25% of the simulation time. As shown in Fig. 20c and d, in dapsone bound mutants, T53V and T53I, all the interactions observed in docked conformations were lost during the course of simulation, however new hydrogen bonding and π- π interaction were formed by backbone atom of Tyr141 and phenol ring of Tyr141 with H10 (amine group) and aniline ring of dapsone for 39% and 51% of simulation time, respectively. Further, in mutant P55L, all the interactions seen in docked conformation were absent along the simulation period (Figure 20e). However, new hydrogen bonding interactions were made by Arg214 backbone atom with O2 of sufonyl group, and also between Thr53 backbone atom and H12 amine group of dapsone for 62% and 52% of simulation time, respectively. In the case of mutant P55R, binding of dapsone does not produce any significant interactions during the simulation period.
Figure 20.
2D plot of dapsone interacting at the pABA pocket with wild type DHPS (a) and mutants, T53A (b), T53V (c), T53I (d), and P55L (e). The strength of the interactions during the 50 ns period are given in percentage.
In contrast, binding of kaempferol to the pABA pocket of wild type and DHPS mutants were relatively more or less strong due to the fact that higher percentage of interactions were observed throughout the simulation time (shown in Figure 21). In the kaempferol bound to wild type, all the hydrogen bond interactions observed in the docking results were retained for 89% (Pro140 with H9), 85% (Arg214 with O5 of carbonyl oxygen) and 96% (Asp21 with H11) as shown in Figure 21a. An additional stable hydrogen bond interaction was found between side chain atom of Trp132 and O2 of phenol group of kaempferol for 89% of simulation time. In mutant T53A, only two hydrogen bond interactions remained during the simulation period between Arg214 and carbonyl group (O5), and Asp21 and hydroxyl group (H11) of kaempferol for 79% and 82% simulation time, respectively (Figure 21b). New cation- π interaction was formed by Arg54 side chain and naphthalene ring of kamepferol for 53% of simulation time. In the case of T53V mutant, none of the interactions observed in docked conformation endured during the simulation period. However, new hydrogen bond interactions were established between Trp32 side chain and hydroxyl (H9), and Lys213 side chain and hydroxyl (O6) of naphthalene ring for 47% and 61% of simulation time, respectively (Figure 21c). Further, two hydrogen bond interactions (as observed in docking) remained intact throughout the simulation run for 67% (Gly181 backbone and H12) and 75% (Arg214 backbone atoms with H10) of time for mutant T53I (Figure 21d). Strong and new hydrogen bond (between Trp132 side chain and O2 of phenol group) and cation- π interaction (Arg214 side chain with naphthalene ring) were also observed for 73% and 87% of simulation time, respectively. On the other hand, in the P55L mutant, no interaction was observed similar to those found in docked conformation. However, additional interactions were found, which compensated the interactions lost in the initial stage. Three observed hydrogen bonds are between: backbone atom of Ala143 with H9 (hydroxyl atom of phenol), Thr185 side chain with H12 (hydroxyl atom of naphthalene ring), Asp21 side chain with H10 (hydroxyl atom of naphthalene ring) for 51%, 68% and 48% of simulation time, respectively. Moreover, Arg214 side chain formed a π- π interaction with naphthalene ring for 41% of simulation time (Figure 21e). Lastly, in the case of P55R mutant, only one hydrogen bond interaction (Asp21 side chain with H12) was retained up to 65% of the simulation time. Two additional hydrogen bonds were found between Arg54 side chain with O6 (hydroxyl of naphthalene ring), and Arg214 backbone with O6, for 40% and 31% of the simulation periods, respectively (Figure 21f).
Figure 21.
2D plot of kaempferol interacting at the pABA pocket with wild type DHPS (a) and mutants, T53A (b), T53V (c), T53I (d), P55L (e) and P55R (f). The strength of the interactions during the 50 ns period is shown in percentage.
It is evident in the present analysis that in dapsone and kaempferol bound wild type and mutant DHPS enzymes, several original interactions were replaced by the new interactions during the simulation, presumably to compensate the lost contacts and to contribute in stabilizing the complexes. In the case of dapsone, the interactions were consistently correlated with the pocket volume of the pABA binding site. As the volume of the pocket increases, the percentage interaction also decreases. The most affected mutant interactions due to pocket-size are T53V and P55R. However, in the case of kaempferol, even though original interactions were lost during the simulation period, new and stable interactions were also formed, which relatively stabilizes the complex better in the mutants. The higher interaction percentage of kaempferol also seems to circumvent the binding pocket volume effect. However, similar to the dapsone, mutants T53V and P55R were observed to have relatively lower percentages of interaction throughout the simulation, which interestingly have the largest pocket volume sizes.
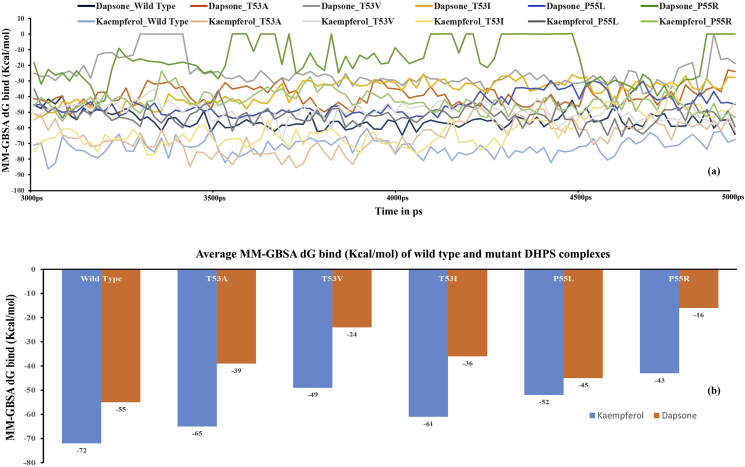
3.14. MM-GBSA binding free energy analysis
The binding free energy of dapsone and kaempferol, bound to the pABA pocket of wild type and mutated DHPS, were calculated to quantitatively examine the differences in energetics, which may bring further insight into the drug-resistance mechanism of the mutants. MM-GBSA analyses were carried out only on the equilibrated trajectories (30 ns–50 ns). The calculated binding free energies along the simulation period is shown in Figure 22a and the average binding free energies of wild type and the mutants, relative to the in dapsone and kaempferol are shown in Figure 22b. It is evident from the MM-GBSA results that both dapsone and kaempferol showed a similar trend of binding free energies for wild type and the mutants. The highest average binding free energy for daspone (-55 kcal/mol) and kaempferol (-72 kcal/mol) were observed when bound to the wild type. Similarly, the lowest binding free energy for both dapsone (-16 kcal/mol) and kaempferol (-43 kcal/mol) were predicted, when bound to the P55R mutant. Further, similar pattern of predicted binding free energies were seen for both daspone (-39 kcal/mol, -24 kcal/mol, -36 kcal/mol and -45 kcal/mol) and kaempferol (-65 kcal/mol, -49 kcal/mol, -61 kcal/mol and -52 kcal/mol) when bound to mutants T53A, T53V, T53I and P55L, respectively.
Figure 23.
2D structure of Dienestrol.
Figure 24.
2D structure of Dapsone.
Figure 25.
2D structure of Sulfadiazine.
Figure 26.
2D structure of Sulfacytine.
Figure 27.
2D structure of Sulfamerazine.
Figure 22.
Plot of binding free energy of DHPS wild type and mutants complexed with dapsone and kaempferol during the simulation period (a) and the average binding free energies of wild type and the mutants (b).
The difference in binding free energy due to the binding of dapsone and kaempferol in wild type and mutants can be explained in relation to the pABA pocket volume sizes. As shown in Table 5, the predicted binding free energies correspond to the sizes of the pABA pocket volume. In short, the predicted binding free energy is inversely proportional to pABA pocket volume. However, kaempferol bound T53I mutant showed higher predicted binding free energy than P55L, even though the pocket volume size is larger in the former case. Since the stability of the protein-ligand complex depends primarily on the hydrogen bonds and hydrophobic interactions, kaempferol bound to mutant T53I showed more stable hydrogen bonds and hydrophobic interactions as compared to mutant P55L (Figure 21d & e, respectively). Similarly, kaempferol bound wild type DHPS and mutants showed higher binding free energies as compared to corresponding complexes with dapsone, because of the more stable hydrogen bonds and hydrophobic interactions observed in DHPS-kaempferol complexes (Figure 20, Figure 21).
Table 5.
Calculated volume for wild type and mutant DHPS pABA pocket and predicted binding free energy of dapsone and kaempferol to pABA pocket.
| DHPSa | Volumeb | MM-GBSA ΔG bindc (Dapsone) | MM-GBSA ΔG bindd (Kaempferol) |
|---|---|---|---|
| Wild type | 283 Å3 | -55 kcal/mol | -72 kcal/mol |
| T53A | 350 Å3 | -39 kcal/mol | -65 kcal/mol |
| T53V | 722 Å3 | -24 kcal/mol | -49 kcal/mol |
| T53I | 528 Å3 | -36 kcal/mol | -61 kcal/mol |
| P55L | 298 Å3 | -45 kcal/mol | -52 kcal/mol |
| P55R | 811 Å3 | -16 kcal/mol | -43 kcal/mol |
Wild type DHPS and mutants.
Volume of DHPS pABA binding pocket in Å3.
Total binding free energy calculated by MM-GBSA method for dapsone.
Total binding free energy calculated by MM-GBSA method for kaempferol.
4. Conclusion
The present investigation for the first time elucidates the role of kaempferol as a potent antibiotic agent with dihydropteroate synthase (DHPS) as its target. Also, using docking as well as molecular dynamics simulation studies, we demonstrated that the mutants in DHPS (M. leprae) leading to drug resistance were probably due to the increased pABA volume size. Comparison of calculated interactions as well as binding free energies suggests that kaempferol could bind more favourably than dapsone and has a potential to overcome the drug resistance due to mutations.
The drug discovery efforts require huge investment and over ten-years period. However, rapid emergence of drug-resistant strains has thwarted any such efforts in case of discovery of new antibiotics, which is not economically viable for the industry. The identification of natural-product based kaempferol opens the door for the design of antibiotics in a quick and high throughput fashion for identifying antibiotic leads, as well as for other diseases. The accuracy of results of such structural studies would definitely improve with the availability of enzyme crystal structure from M. leprae as well as multiple validations of the results including co-crystallization of kaempferol with DHPS enzyme. Attempts to in this direction are underway.
Declarations
Author contribution statement
A.M. Postshangbam: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
R.S. Rathore and P. Nongdam: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work is supported by the DST SYST programme under the Grant No: SP/YO/071/2017(G). Financial support from the DBT-RA Program in Biotechnology and Life Sciences is gratefully acknowledged by Angamba Meetei Potshangbam. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research. The authors also acknowledged computational facilities provided by Bioinformatics Infrastructure Facility, Manipur University.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Achari A., Somers D.O., Champness J.N., Bryant P.K., Rosemond J., Stammers D.K. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- Alanis A.J. Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 2005;36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Appelbaum P.C. 2012 and beyond: potential for the start of a second pre-antibiotic era? J. Antimicrob. Chemother. 2012;67:2062–2068. doi: 10.1093/jac/dks213. [DOI] [PubMed] [Google Scholar]
- Arredondo F., Echeverry C., Abin-Carriquiry J.A., Blasina F., Antúnez K., Jones D.P., Go Y.-M., Liang Y.-L., Dajas F. After cellular internalization, quercetin causes Nrf2 nuclear translocation, increases glutathione levels, and prevents neuronal death against an oxidative insult. Free Radic. Biol. Med. 2010;49:738–747. doi: 10.1016/j.freeradbiomed.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Babaoglu K., Qi J., Lee R.E., White S.W. Crystal structure of 7,8-dihydropteroate synthase from Bacillus anthracis: mechanism and novel inhibitor design. Structure. 2004;12:1705–1717. doi: 10.1016/j.str.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Baca A.M., Sirawaraporn R., Turley S., Sirawaraporn W., Hol W.G. Crystal structure of Mycobacterium tuberculosis 7,8-dihydropteroate synthase in complex with pterin monophosphate: new insight into the enzymatic mechanism and sulfa-drug action. J. Mol. Biol. 2000;302:1193–1212. doi: 10.1006/jmbi.2000.4094. [DOI] [PubMed] [Google Scholar]
- Ballester P.J., Westwood I., Laurieri N., Sim E., Richards W.G. Prospective virtual screening with Ultrafast Shape Recognition: the identification of novel inhibitors of arylamine N-acetyltransferases. J. R. Soc. Interface R. Soc. 2010;7:335–342. doi: 10.1098/rsif.2009.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa E., Calzada F., Campos R. In vivo antigiardial activity of three flavonoids isolated of some medicinal plants used in Mexican traditional medicine for the treatment of diarrhea. J. Ethnopharmacol. 2007;109:552–554. doi: 10.1016/j.jep.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Benkert P., Schwede T., Tosatto S.C. QMEANclust: estimation of protein model quality by combining a composite scoring function with structural density information. BMC Struct. Biol. 2009;9:35. doi: 10.1186/1472-6807-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K.J., Chow E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A., Sacerdoti F.D., others . SC 2006 Conference, Proceedings of the ACM/IEEE. IEEE. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters; p. 43. [Google Scholar]
- Brown D.K., Penkler D.L., Sheik Amamuddy O., Ross C., Atilgan A.R., Atilgan C., Tastan Bishop Ö. MD-TASK: a software suite for analyzing molecular dynamics trajectories. Bioinforma. Oxf. Engl. 2017;33:2768–2771. doi: 10.1093/bioinformatics/btx349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock W.E. Rifampin in the treatment of leprosy. Rev. Infect. Dis. 1983;5(Suppl 3):S606–613. doi: 10.1093/clinids/5.supplement_3.s606. [DOI] [PubMed] [Google Scholar]
- Calzada F., Correa-Basurto J., Barbosa E., Mendez-Luna D., Yepez-Mulia L. Antiprotozoal constituents from annona cherimola miller, a plant used in Mexican traditional medicine for the treatment of diarrhea and Dysentery. Phcog. Mag. 2017;13:148–152. doi: 10.4103/0973-1296.197636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaitanya V.S., Das M., Bhat P., Ebenezer M. Computational modelling of dapsone interaction with dihydropteroate synthase in Mycobacterium leprae; insights into molecular basis of dapsone resistance in leprosy. J. Cell. Biochem. 2015;116:2293–2303. doi: 10.1002/jcb.25180. [DOI] [PubMed] [Google Scholar]
- Chen A.Y., Chen Y.C. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 2013;138:2099–2107. doi: 10.1016/j.foodchem.2012.11.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M.R.H., Bhuiyan M.I., Saha A., Mosleh I.M., Mondol S., Ahmed C.M.S. Identification and analysis of potential targets in Streptococcus sanguinis using computer aided protein data analysis. Adv. Appl. Bioinforma. Chem. AABC. 2014;7:45–54. doi: 10.2147/AABC.S67336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper E.L., Ma M.J. Alzheimer Disease: clues from traditional and complementary medicine. J. Tradit. Complement. Med. 2017;7:380–385. doi: 10.1016/j.jtcme.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G., Zhao Z., Chen Y., Li Z., Tian Y., Liu Z., Liu B., Song J. Quercetin protects rat cortical neurons against traumatic brain injury. Mol. Med. Rep. 2018 doi: 10.3892/mmr.2018.8801. [DOI] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- Friedmann P.S. Dapsone-resistant leprosy. Proc. Roy. Soc. Med. 1973;66:623–624. doi: 10.1177/003591577306600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Gui Y., Zhang J., Chen L., Duan S., Tang J., Xu W., Li A. Icariin, a flavonoid with anti-cancer effects, alleviated paclitaxel-induced neuropathic pain in a SIRT1-dependent manner. Mol. Pain. 2018;14 doi: 10.1177/1744806918768970. 1744806918768970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D.M., Besselink E., Henning S.M., Go V.L.W., Heber D. Phytoestrogens induce differential estrogen receptor alpha- or Beta-mediated responses in transfected breast cancer cells. Exp. Biol. Med. Maywood NJ. 2005;230:558–568. doi: 10.1177/153537020523000807. [DOI] [PubMed] [Google Scholar]
- Huang H., Zhang G., Zhou Y., Lin C., Chen S., Lin Y., Mai S., Huang Z. Reverse screening methods to search for the protein targets of chemopreventive compounds. Front. Chem. 2018;6:138. doi: 10.3389/fchem.2018.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran S., Lleras-Muney A., Smith K.V. Modern medicine and the twentieth century decline in mortality: evidence on the impact of sulfa drugs. Am. Econ. J. Appl. Econ. 2010;2:118–146. [Google Scholar]
- Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- Kaminski G.A., Friesner R.A., Tirado-Rives J., Jorgensen W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. [Google Scholar]
- Klabunde T. Chemogenomic approaches to drug discovery: similar receptors bind similar ligands. Br. J. Pharmacol. 2007;152:5–7. doi: 10.1038/sj.bjp.0707308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopmand G. Concepts and applications of molecular similarity. In: Johnson Mark A., Maggiora Gerald M., editors. J. Comput. Chem. Vol. 13. John Wiley & Sons; New York: 1992. pp. 539–540. 1990, 393. [Google Scholar]
- Kuiper G.G.J.M., Lemmen J.G., Carlsson B., Corton J.C., Safe S.H., van der Saag P.T., van der Burg B., Gustafsson J.-Å. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor β. Endocrinology. 1998;139:4252–4263. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- Kumar S., Pandey A.K. Chemistry and biological activities of flavonoids: an overview [WWW Document] Sci. World J. 2013 doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Liu X., Jiang H., Li H. SHAFTS: a hybrid approach for 3D molecular similarity calculation. 1. Method and assessment of virtual screening. J. Chem. Inf. Model. 2011;51:2372–2385. doi: 10.1021/ci200060s. [DOI] [PubMed] [Google Scholar]
- Maladan Y., Krismawati H., Hutapea H.M.L., Oktavian A., Fatimah R., Widodo A new Mycobacterium leprae dihydropteroate synthase variant (V39I) from Papua, Indonesia. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M., Budiawan T., Aye K.S., Kyaw K., Tan E.V., Cruz E.D., Gelber R., Saunderson P., Balagon V., Pannikar V. The frequency of drug resistance mutations in Mycobacterium leprae isolates in untreated and relapsed leprosy patients from Myanmar, Indonesia and the Philippines. Lepr. Rev. 2007;78:343–352. [PubMed] [Google Scholar]
- Nakata N., Kai M., Makino M. Mutation analysis of the Mycobacterium leprae folP1 gene and dapsone resistance. Antimicrob. Agents Chemother. 2011;55:762–766. doi: 10.1128/AAC.01212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osama A., Awadelkarim S., Ali A. Antioxidant activity, acetylcholinesterase inhibitory potential and phytochemical analysis of Sarcocephalus latifolius Sm. bark used in traditional medicine in Sudan. BMC Compl. Alternative Med. 2017;17:270. doi: 10.1186/s12906-017-1772-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyarzabal J., Howe T., Alcazar J., Andrés J.I., Alvarez R.M., Dautzenberg F., Iturrino L., Martínez S., Van der Linden I. Novel approach for chemotype hopping based on annotated databases of chemically feasible fragments and a prospective case study: new melanin concentrating hormone antagonists. J. Med. Chem. 2009;52:2076–2089. doi: 10.1021/jm8016199. [DOI] [PubMed] [Google Scholar]
- Oyarzabal J., Zarich N., Albarran M.I., Palacios I., Urbano-Cuadrado M., Mateos G., Reymundo I., Rabal O., Salgado A., Corrionero A., Fominaya J., Pastor J., Bischoff J.R. Discovery of mitogen-activated protein kinase-interacting kinase 1 inhibitors by a comprehensive fragment-oriented virtual screening approach. J. Med. Chem. 2010;53:6618–6628. doi: 10.1021/jm1005513. [DOI] [PubMed] [Google Scholar]
- Panche A.N., Diwan A.D., Chandra S.R. Flavonoids: an overview. J. Nutr. Sci. 2016;5 doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potshangbam A.M., Polavarapu R., Rathore R.S., Naresh D., Prabhu N.P., Potshangbam N., Kumar P., Vindal V. MedPServer: a database for identification of therapeutic targets and novel leads pertaining to natural products. Chem. Biol. Drug Des. 2019;93:438–446. doi: 10.1111/cbdd.13430. [DOI] [PubMed] [Google Scholar]
- Roland S., Ferone R., Harvey R.J., Styles V.L., Morrison R.W. The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J. Biol. Chem. 1979;254:10337–10345. [PubMed] [Google Scholar]
- Rush T.S., Grant J.A., Mosyak L., Nicholls A. A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction. J. Med. Chem. 2005;48:1489–1495. doi: 10.1021/jm040163o. [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Smith C.S., Aerts A., Saunderson P., Kawuma J., Kita E., Virmond M. Multidrug therapy for leprosy: a game changer on the path to elimination. Lancet Infect. Dis. 2017;17:e293–e297. doi: 10.1016/S1473-3099(17)30418-8. [DOI] [PubMed] [Google Scholar]
- Spencer J.P.E. Flavonoids and brain health: multiple effects underpinned by common mechanisms. Genes Nutr. 2009;4:243–250. doi: 10.1007/s12263-009-0136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Then R., Angehrn P. Sulphonamide-induced “thymineless death” in Escherichia coli. J. Gen. Microbiol. 1973;76:255–263. doi: 10.1099/00221287-76-2-255. [DOI] [PubMed] [Google Scholar]
- Vidya Priyadarsini R., Senthil Murugan R., Maitreyi S., Ramalingam K., Karunagaran D., Nagini S. The flavonoid quercetin induces cell cycle arrest and mitochondria-mediated apoptosis in human cervical cancer (HeLa) cells through p53 induction and NF-κB inhibition. Eur. J. Pharmacol. 2010;649:84–91. doi: 10.1016/j.ejphar.2010.09.020. [DOI] [PubMed] [Google Scholar]
- Wagner J.R., Sørensen J., Hensley N., Wong C., Zhu C., Perison T., Amaro R.E. POVME 3.0: software for mapping binding pocket flexibility. J. Chem. Theor. Comput. 2017;13:4584–4592. doi: 10.1021/acs.jctc.7b00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Weekly Epidemiological Record . Vol. 95. WHO; 7 February 2020. [WWW Document], n.d. pp. 49–60.http://www.who.int/wer/2020/en/ 35. [Google Scholar]
- Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.D. The relation of p-aminobenzoic acid to the mechanism of the action of sulphanilamide. Br. J. Exp. Pathol. 1940;21:74–90. [Google Scholar]
- Yun M.-K., Wu Y., Li Z., Zhao Y., Waddell M.B., Ferreira A.M., Lee R.E., Bashford D., White S.W. Catalysis and sulfa drug resistance in dihydropteroate synthase. Science. 2012;335:1110–1114. doi: 10.1126/science.1214641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.I., Stiller M.J. Dapsone and sulfones in dermatology: overview and update. J. Am. Acad. Dermatol. 2001;45:420–434. doi: 10.1067/mjd.2001.114733. [DOI] [PubMed] [Google Scholar]