Abstract
A rapid and simple method based on the coupling of supercritical fluid extraction and ultra-high-performance liquid chromatography combined with Q-Exactive Orbitrap tandem mass spectrometry (SFE-UHPLC-Q-Exactive Orbitrap-MS) detection for the identification of compounds from Citrus reticulata semen (CRS) was developed for the first time in this study. Through the optimization of the SFE parameters including extractive pressure, extractive temperature, and time, most of the compounds were successfully extracted at 50 °C, 33 MPa, and 2 h without an entraining agent, among which 32 compounds were successfully identified. Moreover, the operating conditions of UHPLC-Q-Exactive Orbitrap-MS were also optimized for the analysis of the SFE extracts, and the extracts in the CRS showed good separation performance in 20 min. A total of 28 compounds from the SFE extract were identified by comparing the standard sample together with full scan and related literature data, among which esters and flavonoids were the major compounds identified in the CRS extracts. In addition, 2 phenols, 2 aldehydes, 2 triterpenes, and 5 other compounds were identified. The SFE-UHPLC-Q-Exactive Orbitrap-MS method was successfully validated and applied for the identification of compounds from the CRS.
1. Introduction
The Citrus reticulata semen (CRS) is the dry, mature seed of C. reticulata Blanco and its cultivated varieties and has been consumed as a traditional Chinese medicine for centuries with its properties including qi regulation, knot dispersion, pain relief, and so on.1,2 However, compared with Citri Reticulatae Pericarpium, the chemical composition of CRS has not been fully studied; most Citrus seeds are discarded rather than being exploited, resulting in resource waste and environmental pollution. Therefore, the study of its chemical components is of great significance for its further utilization in medicine and raw materials. Therefore, the purpose of this study is to establish a fast and efficient method for the comprehensive analysis and identification of phytochemical components in supercritical fluid extraction (SFE) extracts of CRS.
As a kind of low-temperature extraction technology, supercritical fluid CO2 extraction (SFE) has the advantages of low toxicity, noncombustibility, high diffusion, gas-like viscosity, liquid-like density, sample processing automation, and high extraction efficiency.3 In addition, liquid chromatography (LC)–MS is often used for the analysis of phytochemical components, and in recent years, ultra-high-performance LC combined with Q-Exactive Orbitrap tandem mass spectrometry (UHPLC-Q-Exactive Orbitrap-MS) has become a faster, more efficient, and more sensitive tool than HPLC–MS for the rapid analysis of plant extracts and bioactive compounds in some traditional Chinese medicines. As far as we know, only a few analytical methods are used for the determination of CRS at present. Previously reported analytical methods were only used to analyze the oil contained in it and one or several specific chemical components such as limonin and obacunone by GC–MS analysis or HPLC method, lacking systematic analysis. Thus, in order to evaluate the quality of CRS more comprehensively, an efficient method based on the coupling of SFE-UHPLC-Q-Exactive Orbitrap-MS detection was developed in this paper, and the chemical components in the SFE extracts of CRS were systematically analyzed. This study is of great significance and provides a valuable basis for further scientific research and application of CRS in food, medicine, health care products, and other fields.
2. Results and Discussion
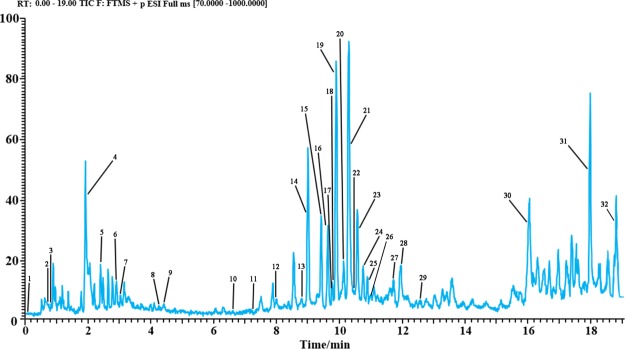
The SFE extracts of CRS were analyzed by UHPLC-Q-Exactive Orbitrap-MS analysis. By optimizing a series of parameters such as flow rate and elution gradient of mobile phase, better analytical conditions are obtained. Under the optimal conditions, the components of SFE extracts in CRS could be separated in the positive mode within 19 min. The method is fast and efficient. The TIC diagram is shown in Figure 1. A total of 32 compounds were identified and separated and numbered 1–32 according to the sequence of peak time (Table 1). Totally, 29 SFE extracts constituents were divided into the following groups: 10 esters, 7 flavones, 2 phenols, 2 aldehydes, 2 triterpenes, and 6 other compounds. The identification of all these compounds was mainly based on MS/MS fragments and the analysis of literature. For a compound with a standard, the retention times and fragments of the compound should be compared with the standard under the same test conditions. Among them, 8 compounds were identified through comparison with standard products, and 20 compounds were identified based on MS/MS-positive ion fragments and relevant data from previous studies. Their chemical structures were illustrated in Figure 2.
Figure 1.
Total ion chromatogram in positive mode of CRS.
Table 1. Summary of Chemical Constituents About Supercritical CO2 Fluid Extraction from CRS by Ultra-HPLC Combined with Q-Exactive Orbitrap Tandem Mass Spectrometry.
| no. | tR (min) | [M + H]+ (m/z) | fragment ions (m/z) | compound formula | identification |
|---|---|---|---|---|---|
| 1 | 0.03 | 195.0875 | 177.0912, 167.0847, 153.0544, 146.9612, 138.0662, 135.0442, 123.0432, 114.6795, 110.0716, 107.0856, 89.0602, 79.0550, 69.0455, 58.0659 | C8H10N4O2 | caffeine |
| 2 | 0.70 | 124.0394 | 119.0366, 106.0655, 101.0255, 96.0448, 90.0198, 80.0501, 78.0343, 68.0503, 62.9905, 53.0393 | C6H5NO2 | nicotinic acid |
| 3 | 0.82 | 123.0553 | 118.0349, 108.9585, 105.0374, 100.0247, 96.0448, 90.5265, 88.0236, 86.0262, 80.0500, 76.5157, 67.5105, 62.9906, 56.9656, 53.0392 | C6H6N2O | nicotinamide |
| 4 | 1.92 | 156.1019 | 138.0913, 122.8464, 113.0840, 110.0967, 108.0810, 96.0812, 88.0762, 86.0606, 84.0451, 82.0657, 71.0498, 69.0342, 67.0548, 53.0394 | unknown | unknown |
| 5 | 2.40 | 137.1073 | 122.0837, 96.0811, 88.2350, 80.0502, 69.0706, 60.5088, 55.0550 | C8H12N2 | tetramethylpyrazine |
| 6 | 2.91 | 153.0546 | 144.6615, 134.0600, 125.0597, 122.4801, 113.0483, 111.0442, 107.0494, 97.0652, 93.0339, 88.9528, 79.0548, 70.9424, 65.0393, 63.0239 | C8H8O3 | vanillin |
| 7 | 3.01 | 183.0652 | 173.9848, 165.0906, 159.9690, 155.0702, 145.9897, 140.0467, 131.9742, 123.0442, 113.9639, 105.0450, 98.9845, 95.0495, 90.9481, 86.9537, 81.0341, 72.9378, 67.0550, 65.0393, 56.9430 | C9H10O4 | 3,5-dimethoxy-4-hydroxybenzaldehyde |
| 8 | 4.21 | 207.0652 | 201.9320, 192.0416, 179.0704, 175.0391, 163.0390, 151.0754, 148.0517, 121.0650, 107.0495, 91.0547, 79.0548, 67.0548 | C11H10O4 | scoparone |
| 9 | 4.41 | 147.0440 | 137.9799, 129.0700, 123.9644, 119.0857, 105.9538, 103.0545, 95.0496, 91.0547, 84.0813, 73.0845, 64.9280, 53.0393 | C9H6O2 | coumarin |
| 10 | 6.61 | 133.0648 | 119.9576, 117.9598, 115.0544, 109.5154, 105.0701, 103.0546, 98.0076, 96.0099, 91.0547, 89.0023, 83.8491, 79.0548, 77.0390, 72.0814, 65.0394, 58.0336 | C9H8O | cinnamaldehyde |
| 11 | 7.24 | 303.0859 | 285.0751, 262.1411, 243.0657, 219.0645, 201.0540, 185.0447, 177.0546, 163.0389, 153.0182, 135.0441, 117.0337, 111.0080, 89.0390, 67.0185 | C16H14O6 | hesperitin |
| 12 | 8.13 | 223.0964 | 214.0120, 205.0857, 199.9964, 186.0551, 177.0546, 163.0389, 149.0597, 145.0284, 135.0441, 121.0650, 117.0337, 107.0859, 93.0703, 89.0391, 79.0548, 67.0550 | C12H14O4 | ethyl ferulate |
| 13 | 8.99 | 471.2012 | 453.1918, 425.1955, 409.2001, 383.2280, 367.1904, 339.1947, 321.1846, 279.1379, 225.0913, 213.0910, 187.0753, 161.0597, 133.0648, 105.0702, 95.0132, 79.0548 | C26H30O8 | limonina |
| 14 | 9.07 | 151.1117 | 146.0297, 139.0752, 128.0193, 123.0806, 121.0650, 113.9638, 109.0650, 105.0700, 95.0859, 87.0045, 81.0704, 79.0547, 67.0550, 65.0393, 55.0549 | C10H14O | 2-adamantanone |
| 15 | 9.41 | 245.1170 | 227.1066, 217.1214, 203.1063, 199.1116, 189.0544, 171.1166, 156.0934, 145.0647, 131.0492, 119.0857, 105.0702, 95.0495, 81.0341, 76.9062, 67.0550, 55.0186 | C15H16O3 | linderalactone |
| 16 | 9.63 | 403.1385 | 388.1148, 373.0914, 358.0676, 327.0854, 301.0701, 284.0666, 258.0519, 229.0339, 211.0235, 193.0125, 183.0287, 165.0546, 1227.0389, 99.0443, 69.0341 | C21H22O8 | nobiletina |
| 17 | 9.76 | 287.0910 | 269.0804, 245.0814, 203.0699, 179.0332, 171.0286, 161.0596, 153.0181, 133.0647, 118.0414, 103.0547, 67.0185 | C16H14O5 | isosakuranetin |
| 18 | 9.78 | 343.1173 | 327.0856, 313.0701, 299.0909, 282.0881, 267.0650, 239.0699, 199.0233, 171.0285, 153.0181, 133.0647, 122.0363, 86.0967, 69.0339 | C19H18O6 | 6-demethoxytangeretin |
| 19 | 9.89 | 515.2271 | 469.2221, 455.2037, 419.1846, 411.2158, 393.2051, 349.1804, 321.1849, 279.1374, 243.0992, 215.1061, 187.0752, 161.0596, 133.0647, 95.0131, 81.0340 | unknown | unknown |
| 20 | 10.16 | 433.1488 | 418.1254, 403.1019, 385.0911, 345.0599, 317.0650, 289.0700, 255.0640, 211.0230, 183.0290, 165.0544, 149.0235, 127.0387, 94.0416, 70.9067 | C22H24O9 | 3,5,6,7,8,3′,4′-heptamethoxyflavonea |
| 21 | 10.31 | 137.0596 | 122.0734, 119.0493, 116.5949, 109.0650, 107.0493, 105.0451, 95.0495, 93.0703, 91.0547, 81.0705, 79.0549, 67.0549, 54.4142, 53.0394 | C8H8O2 | p-anisaldehyde |
| 22 | 10.53 | 231.1378 | 213.1274, 189.0910, 185.1324, 175.0754, 163.0752, 143.0855, 135.0442, 119.0857, 105.0702, 91.0547, 69.0706, 55.0550 | C15H18O2 | atractylenolide-I |
| 23 | 10.57 | 373.1280 | 358.1041, 343.0809, 328.0570, 297.0753, 283.0602, 271.0599, 254.0569, 229.0319, 211.0235, 193.0132, 183.0287, 168.0051, 135.0440, 99.0444, 68.9978 | C20H20O7 | tangeretina |
| 24 | 10.76 | 455.2060 | 437.1934, 419.1848, 409.2013, 391.1895, 359.1282, 349.1430, 315.1386, 303.1373, 263.068, 235.1124, 197.0964, 175.0754, 161.0596, 133.0648, 105.0702, 95.0132, 79.0548, 67.0550 | C26H30O7 | obacunone |
| 25 | 10.82 | 183.0804 | 173.9850, 159.9689, 141.9586, 131.9742, 127.9794, 118.9675, 113.9638, 105.0338, 100.9570, 95.0495, 81.0705, 72.9378, 67.0549, 59.9310, 56.9430 | C13H10O | atractylodin |
| 26 | 11.10 | 389.1227 | 374.0992, 359.0757, 341.0652, 331.0807, 316.0574, 285.0761, 260.0672, 227.0544, 215.0184, 197.0080, 187.0235, 169.0130, 163.0752, 148.0518, 113.0237, 85.0291, 55.0185 | C20H20O8 | 5-hydroxy-6,7,8,3′,4′-pentamethoxyflavonea |
| 27 | 11.72 | 191.1064 | 173.0959, 163.1115, 149.0596, 145.1010, 135.0438, 130.0778, 121.0651, 117.0700, 105.0701, 93.0702, 91.0546, 81.0706, 79.0548, 69.0704, 67.0549, 55.0550 | C12H14O2 | ligustilides |
| 28 | 11.91 | 189.0907 | 171.0802, 166.0290, 156.0927, 153.0697, 143.0854, 133.1015, 128.0620, 125.0960, 117.0698, 105.0701, 97.1014, 91.0547, 83.0860, 67.0550, 55.0550 | C12H12O2 | N-butylidenephthalide |
| 29 | 12.52 | 233.1533 | 215.1428, 205.1582, 197.1323, 187.1479, 177.0908, 173.0961, 159.0803, 151.0752, 145.1010, 131.0854, 119.0857, 105.0701, 95.0859, 81.0704, 67.0549 | C15H20O2 | atractylenolide-II |
| 30 | 16.07 | 295.2263 | 277.2158, 259.2050, 241.1943, 223.1689, 205.1582, 187.1482, 173.1321, 161.1323, 147.1165, 135.1167, 121.1012, 107.0858, 95.0859, 81.0704, 67.0549 | Unknown | unknown |
| 31 | 17.98 | 324.2893 | 307.2624, 263.2367, 245.2264, 161.1329, 133.1013, 109.1013, 95.0858, 67.0549, 62.0608 | Unknown | unknown |
| 32 | 18.79 | 307.2627 | 261.2208, 243.2102, 233.2252, 219.2103, 187.1481, 173.1322, 145.1009, 137.1322, 123.1168, 109.1013, 95.0858, 81.0704, 67.0549 | C20H34O2 | linolenic acid ethyl ester |
Further confirmation in comparison with standard compound.
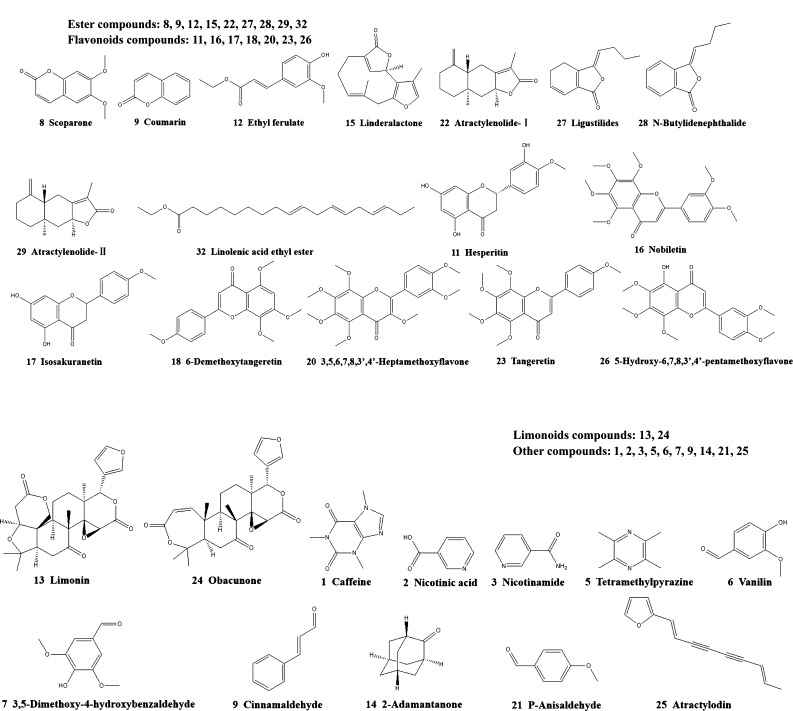
Figure 2.
Chemical structures of compounds identified in CRS.
2.1. Identification of Esters
Esters are major constituents extracted from the SFE extracts of CRS. Ten esters were identified in SFE extracts including N-butylidenephthalide, ligustilides, atractylenolide-I, atractylenolide-II, scoparone, ethyl ferulate, linolenic acid ethyl ester, and linderalactone. In positive mode, characteristic fragment ions such as CO, H2O, and alkyl side chain were prone to lose in these kinds of compounds in high-energy collisions.4
In this paper, for example, identification and characterization of structures were discussed about ligustilides, atractylenolide-I, and atractylenolide-II. Peak 27 produced a relatively strong molecular ion peak at m/z 191.1064 for [M + H]+ and relatively low intensity peaks at m/z 173.0959 for [M + H – H2O]+, 163.1115 for [M + H – CO]+, 145.1010 for [M + H – H2O – CO]+, and at m/z 117.0700 for [M + H – H2O – CO – C2H4]+. Based on the characteristic fragmentation profiles, peak 27 were identified as ligustilides by comparing with the characteristic ions from previous study.5
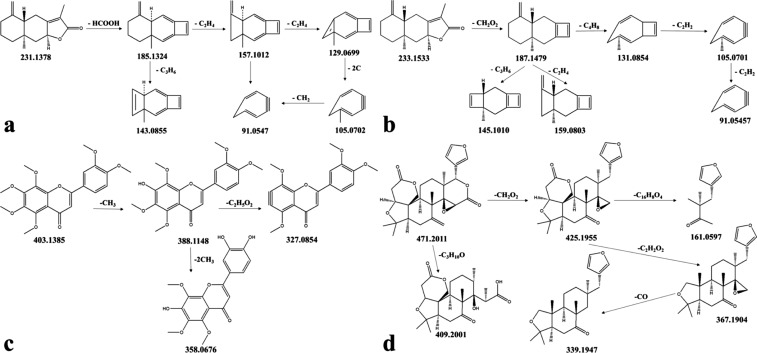
Peak 22 was assigned to be atractylenolide-I by comparison with literature,6 which gave a molecular ion peak [M + H]+ at m/z 231.1378 and fragment ion peak at m/z 213.1274 for [M + H – H2O]+ and m/z 189.0910, 185.1324, 175.0754, 163.0752, 143.0855, 105.0702, and 91.0547. The detailed fragmentation process is shown in Figure 3a.
Figure 3.
Proposed fragmentation pathways of main fragment ions for atractylenolide-I (a), atractylenolide-II (b), nobiletin (c), and limonin (d) in positive ion mode.
Peak 29 provided precursor ion peak of [M + H]+ at m/z 233.1533 and fragment ion peaks at m/z 215.1428 for [M + H – H2O]+, m/z 187.1479 for [M + H – H2O – CO]+, m/z 159.0803 for [M + H – H2O – CO – C2H4]+, m/z 145.1010 for [M + H – H2O – CO – C3H6]+, m/z 131.0854 for [M + H – H2O – CO – C4H8]+, and m/z 105.0701 for [M + H – H2O – CO – C6H10]+. Furthermore, combined with literature7 and reference formula provided by MS, it was therefore confirmed as atractylenolide-II. The detailed fragmentation pattern of peak 29 is shown in Figure 3b.
2.2. Identification of Flavonoids
Flavonoids are important components of natural flavonoids in Citrus. According to the classification of flavonoids, they were divided into normal flavones, flavanones, and polymethoxylated flavones.8,9
With the high-resolution UHPLC-Q-Exactive Orbitrap-MS system, a total of seven flavonoids were identified in the SFE extracts of CRS including peaks 11, 16, 17, 18, 20, 23, and 26. Among them, peaks 26, 20, 16, and 23 were unambiguously identified as 5-hydroxy-6,7,8,3′,4′-pentamethoxyflavone, 3,5,6,7,8,3′,4′-heptamethoxyflavone, nobiletin, and tangeretin by comparing with the literature and the reference standard with the same fragmentation pattern. Nobiletin was selected as a typical example of flavonoids to illustrate the fragmentation pathways. Nobiletin is a polymethoxylated flavone with six methoxy substituents in its structure, which produces strong molecular ion peaks at m/z 403.1385 and formed relatively lower intensities of MS2 fragments at m/z 388.1148 for [M + H – CH3]+, m/z 373.0914 for [M + H – 2CH3]+, m/z 358.0676 for [M + H – 3CH3]+, and m/z 327.0854 for [M + H – 3CH3 – CH3O]+, among others, with a series of characteristic losses of CH3 and CH3O. Figure 3c depicts the proposed fragmentation pattern.
2.3. Identification of Limonins
Limonoids are secondary metabolites of tetracyclic triterpenes in Rutaceae and Meliaceae widely distributed in the seeds and membranes of Citrus fruits, which are a major group of chemical constituents from CRS. Two limonoids were identified in SFE extracts, including limonoid and obacunone, after comparison with fragments of the standard reference and literature.10,11
Taking limonin, for example, according to the literature, the fragmentation model of the standard substance and the fragment ions provided by MS can be known, and the positive ion of limonin was detected at m/z 471.2012. The generation of m/z 425.1955 fragment ions was due to neutral loss of the ester group in lactonic ring D. The fragment ion peak at m/z 409.2001 indicates that the epoxy group can transform into dihydroxyl after deleting the carboxyl group. The loss of the epoxy group and the lactone ring A produced fragments of ions at m/z 367.1904 and m/z 339.1947, respectively. In addition, the fragment ions at m/z 161.0597 were generated because of the cleavage of ring C. The detailed fragmentation schemes are shown in Figure 3d.
2.4. Identification of Others
In addition, except for four unknown compounds, nine other compounds including peak 25, peak 1, peak 3, peak 2, peak 5, peak 14, peak 6, peak 7, peak 21, and peak 10 were identified as atractylodin, caffeine, nicotinamide, nicotinic acid, tetramethylpyrazine, 2-adamantanone, two phenols (vanillin, 3,5-dimethoxy-4-hydroxybenzaldehyde), and two aldehydes (p-anisaldehyde, cinnamaldehyde), respectively, combined with literature12−21 and reference formula provided by MS. Using peak 25 as example, peak 25 showed an ion signal at m/z 183.0804 [M + H]+ in the MS spectrum. In the MS2 spectrum, the major fragment ion peaks were at m/z 159.9689, m/z 131.9742, m/z 113.9638, and m/z 105.0338 [M + H – C6H6]+. Peak 25 was identified as atractylodin by comparing the reference formula and previous research.
3. Conclusions
In the present study, all samples from CRS were extracted by supercritical carbon dioxide under optimized conditions, and a total of 32 compounds released from all of these varieties of SFE extracts were separated and identified by UHPLC-Q-Exactive Orbitrap-MS including 10 esters, seven flavonoids, two phenols, two aldehydes, two triterpenes, and nine other compounds, among which nine esters were first identified in CRS. The analysis results showed that the chemical composition of SFE extracts and methanol extracts was significantly different. According to our previous studies, methanol extracts of CRS mainly consist of hesperidin, neohesperidin, and other compounds with greater polarity, as well as medium-polarity compounds such as nobiletin and tangeretin, hardly containing ester and other low-polarity compounds. The SFE extracts used in this study were mainly low-polarity compounds such as esters and medium-polarity compounds such as flavonoids.
According to the literature studies, the triterpenoids and ester compounds in CRS have good pharmacological activities. For example, the triterpenoid limonin has antitumor,22,23 analgesic, and anti-inflammatory effects,24 while obacunone has anti-inflammatory,25 antioxidation,26 and antitumour effects.27,28 Esters, moreover, as ligustilides, were effective for limiting the oxidative stress29 and anti-inflammatory effect.30
Since ancient times, China has a history of using CRS as a traditional Chinese medicine. However, because of the lack of systematic research on the chemical components contained in CRS and the lack of research on the biological activity of its chemical components, the application of CRS is limited and the quality is varied. Therefore, the composition of compounds in the middle- and low-polar compounds in CRS was investigated through the SFE-UHPLC-Q-Exactive Orbitrap-MS method for the first time in this study, which laid a foundation for further scientific research and application of the CRS.
4. Materials and Methods
4.1. Materials and Reagents
About 10 kg of fresh Xinhui Citrus was collected from Nantan Island, Xinhui District, Jiangmen City, Guangdong Province, in October 2017 and identified as the fruit of C. reticulata “Chachi” by Professor Zheng Guodong from the school of Pharmacy, Guangzhou Medical University. After removing the peel and pulp of the fresh Citrus, the seeds were collected, washed, dried, and stored in the Pharmacognosy Laboratory, School of Pharmacy, Guangzhou Medical University. Eight compounds were used as reference standards with purity greater than 98%. Among them, 3,5,6,7,8,3′,4′-heptamethoxyflavone and 5-hydroxy-6,7,8,3′,4′-pentamethoxyflavone were purchased from Sichuan Weikeqi Biotechnology Co., Ltd and Nanjing Chunqiu Biological Engineering Co. Ltd, respectively. All the others were purchased from Chengdu Mansite Biotechnology Co. Ltd. In addition, acetonitrile and MS-grade formic acid were bought from Thermo Fisher Scientific (China) Co. and Merck KGaA Company of Germany, Ltd., respectively. Other reagents were all of analytical grade.
4.2. Sample Preparation
The 50 g powder samples of CRS were directly loaded into the SFE kettle having a specification of 50 g without the use of any kind of support material and entrainer for the SFE extraction. The SFE conditions were as follows: extraction temperature: 50 °C, extractive pressure: 33 MPa, extractive time: 2 h, and CO2 flow rate: 0.05 L/min. The SFE sample was collected, placed into a vial, and stored at 4 °C. Prior to analysis, the reserved solution was further diluted with dichloromethane to an appropriate concentration and filtered through a needle-type microporous membrane with an aperture of 0.22 μm.
4.3. Analytical System
The ultra-high-performance LC was carried out on an Dionex UltiMate 3000 UHPLC system (Thermo Scientific, USA) equipped with an online degasser, a quaternary pump, an autosampler, and a column temperature compartment, which was connected to a Q-Exactive Orbitrap tandem mass spectrometer (Thermo Scientific, USA) via an electrospray ionization interface and high-resolution MS detection was performed in the positive ion mode, and the data were acquired in full scan mode. A ZORBAX Eclipse Plus C18 (2.1 mm × 50 mm, 1.8 μm) was utilized at 40 °C and eluted with a gradient solvent from A/B (90:10) to A/B (10:90) at a flow rate of 0.50 mL/min, where A is 0.1% (v/v) formic acid solution and B is 0.1% (v/v) formic acid solution of acetonitrile. The linear gradient elution was as follows: 0–2 min, 10–25% B; 2–4 min, 25–25% B; 4–10 min, 25–50% B; 10–14 min, 50–50% B; 14–18 min, 50–85% B; and 18–19 min, 85–90% B.
Acknowledgments
The authors greatly appreciate the financial support from the National Key R&D Program of China (nos. 2017YFC1701103 and 2017YFC1701105), National Modern Agricultural Industrial Park of China (no. njf [2017] 110), National Natural Science Foundation of China (no. 31401613), and Cultivation Plan for High-level University Academic Backbone of Guangzhou Medical University in 2017 (no. gydf [2017] 210).
Author Contributions
⊥ G.Z. and X.Y. contributed equally to the manuscript.
The authors declare no competing financial interest.
References
- Shimizu S.; Miyamoto S.; Fujii G.; Nakanishi R.; Onuma W.; Ozaki Y.; Fujimoto K.; Yano T.; Mutoh M. Suppression of intestinal carcinogenesis in Apc-mutant mice by limonin. J. Clin. Biochem. Nutr. 2015, 57, 39–43. 10.3164/jcbn.15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-C.; Yang Y.; Liu J.; Jiang A.-D.; Chu Z.-X.; Chen S.-Y.; Gong G.-Q.; He G.-W.; Xu Y.-G.; Zhu Q.-H. Discovery of novel limonin derivatives as potent anti-inflammatory and analgesic agents. Chin. J. Nat. Med. 2018, 16, 231–240. 10.1016/s1875-5364(18)30052-9. [DOI] [PubMed] [Google Scholar]
- Belo Y. N.; Al-Hamimi S.; Chimuka L.; Turner C. Ultrahigh-pressure supercritical fluid extraction and chromatography of Moringa oleifera and Moringa peregrina seed lipids. Anal. Bioanal. Chem. 2019, 411, 3685–3693. 10.1007/s00216-019-01850-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Xu H.-Y.; Ma Y.; Wang X.-G.; Shi Y.; Huang B.; Tang S.-H.; Zhang Y.; Li D.-F.; Liang R.-X.; Yang H.-J. Characterization and rapid identification of chemical constituents of NaoXinTong capsules by UHPLC-linear ion trap/Orbitrap mass spectrometry. J. Pharm. Biomed. Anal. 2015, 111, 104–118. 10.1016/j.jpba.2015.01.020. [DOI] [PubMed] [Google Scholar]
- Li M.-N.; Dong X.; Gao W.; Liu X.-G.; Wang R.; Li P.; Yang H. Global identification and quantitative analysis of chemical constituents in traditional Chinese medicinal formula Qi-Fu-Yin by ultra-high performance liquid chromatography coupled with mass spectrometry. J. Pharm. Biomed. Anal. 2015, 114, 376–389. 10.1016/j.jpba.2015.05.030. [DOI] [PubMed] [Google Scholar]
- Chen Q.; He H.; Li P.; Zhu J.; Xiong M. Identification and quantification of atractylenolide I and atractylenolide III in Rhizoma Atractylodes Macrocephala by liquid chromatography-ion trap mass spectrometry. Biomed. Chromatogr. 2013, 27, 699–707. 10.1002/bmc.2847. [DOI] [PubMed] [Google Scholar]
- Yan H.; Sun Y.-Y.; Zhang Q.-L.; Yang M.-J.; Wang X.-R.; Wang Y.; Yu Z.-G.; Zhao Y.-L. Simultaneous determination and pharmacokinetic study of Atractylenolide I, II and III in rat plasma after intragastric administration of Baizhufuling extract and Atractylodis extract by UPLC-MS/MS. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2015, 993–994, 86–92. 10.1016/j.jchromb.2015.05.006. [DOI] [PubMed] [Google Scholar]
- Zhao X. J.; Xing T. T.; Li Y. F.; Jiao B. N.; Jiang D. Efficient analysis of phytochemical constituents in the peel of Chinese wild citrus Mangshanju (Citrus reticulata Blanco) by ultra high performance liquid chromatography-quadrupole time-of-flight-mass spectrometry. J. Sep. Sci. 2018, 41, 1947–1959. 10.1002/jssc.201701023. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Cai H.; Cao G.; Duan Y.; Pei K.; Tu S.; Zhou J.; Xie L.; Sun D.; Zhao J.; Liu J.; Wang X.; Shen L. Profiling and analysis of multiple constituents in Baizhu Shaoyao San before and after processing by stir-frying using UHPLC/Q-TOF-MS/MS coupled with multivariate statistical analysis. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2018, 1083, 110–123. 10.1016/j.jchromb.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Liu S.; Chen P.; Zhang N.; Sun L.; Dai G.; Zhu L.; Li C.; Zhao Y.; Zhang L.; Fu H.; Ju W. Comprehensive characterization of the in vitro and in vivo metabolites of limonin in human samples using LC-Q-TOF/MS. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2017, 1068–1069, 226–232. 10.1016/j.jchromb.2017.10.035. [DOI] [PubMed] [Google Scholar]
- Lei X.; Shan G.; Zhang F.; Liu P.; Meng L.; Jia T. Determination and comparison of alkaloids and triterpenes among tissues after oral administration of crude and processed Phellodendri Chinensis Cortex by UPLC-QqQ-MS. Nat. Prod. Res. 2019, 1–4. 10.1080/14786419.2018.1560293. [DOI] [PubMed] [Google Scholar]
- Zhang Y.-H.; Bo C.; Fan Y.-H.; An R.; Chen L.; Zhang Y.-F.; Jia Y.-Q.; Wang X.-H. Qualitative and quantitative determination of Atractylodes rhizome using ultra-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometry with data-dependent processing. Biomed. Chromatogr. 2019, 33, e4443 10.1002/bmc.4443. [DOI] [PubMed] [Google Scholar]
- Viana C.; Zemolin G. M.; Dal Molin T. R.; Gobo L.; Ribeiro S. M.; Leal G. C.; Marcon G. Z.; de Carvalho L. M. Detection and determination of undeclared synthetic caffeine in weight loss formulations using HPLC-DAD and UHPLC-MS/MS. J. Pharm. Anal. 2018, 8, 366–372. 10.1016/j.jpha.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catz P.; Shinn W.; Kapetanovic I. M.; Kim H.; Kim M.; Jacobson E. L.; Jacobson M. K.; Green C. E. Simultaneous determination of myristyl nicotinate, nicotinic acid, and nicotinamide in rabbit plasma by liquid chromatography-tandem mass spectrometry using methyl ethyl ketone as a deproteinization solvent. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2005, 829, 123–135. 10.1016/j.jchromb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Li S.; Lin H.; Qu C.; Tang Y.; Shen J.; Li W.; Yue S.; Kai J.; Shang G.; Zhu Z.; Zhang C.; Liu P.; Yan H.; Zhang L.; Qian L.; Qian D.; Duan J.-A. Urine and plasma metabonomics coupled with UHPLC-QTOF/MS and multivariate data analysis on potential biomarkers in anemia and hematinic effects of herb pair Gui-Hong. J. Ethnopharmacol. 2015, 170, 175–183. 10.1016/j.jep.2015.05.019. [DOI] [PubMed] [Google Scholar]
- Dou L.-L.; Duan L.; Guo L.; Liu L.-L.; Zhang Y.-D.; Li P.; Liu E.-H. An UHPLC-MS/MS method for simultaneous determination of quercetin 3-O-rutinoside, kaempferol 3-O-rutinoside, isorhamnetin 3-O-rutinoside, bilobalide and ligustrazine in rat plasma, and its application to pharmacokinetic study of Xingxiong injection. Chin. J. Nat. Med. 2017, 15, 710–720. 10.1016/s1875-5364(17)30101-2. [DOI] [PubMed] [Google Scholar]
- Genta M. T.; Villa C.; Mariani E.; Longobardi M.; Loupy A. Green chemistry procedure for the synthesis of cyclic ketals from 2-adamantanone as potential cosmetic odourants. Int. J. Cosmet. Sci. 2002, 24, 257–262. 10.1046/j.1467-2494.2002.00147.x. [DOI] [PubMed] [Google Scholar]
- Kundu A. Vanillin biosynthetic pathways in plants. Planta 2017, 245, 1069–1078. 10.1007/s00425-017-2684-x. [DOI] [PubMed] [Google Scholar]
- Lv Q.; Chu X.; Yao X.-Y.; Ma K.-L.; Zhang Y.; Deng X.-M. Inhibition of the type III secretion system by syringaldehyde protects mice from Salmonella enterica serovar Typhimurium. J. Cell Mol. Med. 2019, 23, 4679–4688. 10.1111/jcmm.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.; Kurusarttra S.; Ryu J.-Y.; Kanaly R. A.; Hur H.-G. Production of Natural Fragrance Aromatic Acids by Coexpression of trans-Anethole Oxygenase and p-Anisaldehyde Dehydrogenase Genes of Pseudomonas putida JYR-1 in Escherichia coli. J. Agric. Food Chem. 2012, 60, 11972–11979. 10.1021/jf303531u. [DOI] [PubMed] [Google Scholar]
- Ji B.; Zhao Y.; Zhang Q.; Wang P.; Guan J.; Rong R.; Yu Z. Simultaneous determination of cinnamaldehyde, cinnamic acid, and 2-methoxy cinnamic acid in rat whole blood after oral administration of volatile oil of Cinnamoni Ramulus by UHPLC-MS/MS: An application for a pharmacokinetic study. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2015, 1001, 107–113. 10.1016/j.jchromb.2015.07.049. [DOI] [PubMed] [Google Scholar]
- Yao J.; Liu J.; Zhao W. By blocking hexokinase-2 phosphorylation, limonin suppresses tumor glycolysis and induces cell apoptosis in hepatocellular carcinoma. OncoTargets Ther. 2018, 11, 3793–3803. 10.2147/ott.s165220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K. N. C.; Jayaprakasha G.-K.; Patil B.-S. Citrus limonoids and curcumin additively inhibit human colon cancer cells†. Food Funct. 2013, 4, 803–810. 10.1039/c3fo30325j. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Wang X.; Zhu Q.; Gong G.; Luo D.; Jiang A.; Yang L.; Xu Y. Synthesis and pharmacological evaluation of novel limonin derivatives as anti-inflammatory and analgesic agents with high water solubility. Bioorg. Med. Chem. Lett. 2014, 24, 1851–1855. 10.1016/j.bmcl.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Hou R.; Liu F.; Liu H.; Fei Q.; Han Y.; Cai R.; Peng C.; Qi Y. Obacunone causes sustained expression of MKP-1 thus inactivating p38 MAPK to suppress pro-inflammatory mediators through intracellular MIF. J. Cell. Biochem. 2018, 119, 837–849. 10.1002/jcb.26248. [DOI] [PubMed] [Google Scholar]
- Xu S.; Chen W.; Xie Q.; Xu Y. Obacunone activates the Nrf2-dependent antioxidant responses. Protein Cell 2016, 7, 684–688. 10.1007/s13238-016-0297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.; Jayaprakasha G. K.; Patil B. S. Obacunone exhibits anti-proliferative and anti-aromatase activity in vitro by inhibiting the p38 MAPK signaling pathway in MCF-7 human breast adenocarcinoma cells. Biochimie 2014, 105, 36–44. 10.1016/j.biochi.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Murthy K. N. C.; Jayaprakasha G. K.; Patil B. S. Cytotoxicity of obacunone and obacunone glucoside in human prostate cancer cells involves Akt-mediated programmed cell death. Toxicology 2015, 329, 88–97. 10.1016/j.tox.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Bunel V.; Antoine M.-H.; Nortier J.; Duez P.; Stévigny C. Nephroprotective effects of ferulic acid, Z-ligustilide and E-ligustilide isolated from Angelica sinensis against cisplatin toxicity in vitro. Toxicol. Vitro 2015, 29, 458–467. 10.1016/j.tiv.2014.12.017. [DOI] [PubMed] [Google Scholar]
- Huang J.; Lu X.-Q.; Zhang C.; Lu J.; Li G.-Y.; Lin R.-C.; Wang J.-H. Anti-inflammatory ligustilides from Ligusticum chuanxiong Hort. Fitoterapia 2013, 91, 21–27. 10.1016/j.fitote.2013.08.013. [DOI] [PubMed] [Google Scholar]