Abstract
Adaptation in spatially heterogeneous environments results from the balance between local selection, mutation, and migration. We study the interplay among these different evolutionary forces and demography in a classical two-habitat scenario with asexual reproduction. We develop a new theoretical approach that goes beyond the Adaptive Dynamics framework, and allows us to explore the effect of high mutation rates on the stationary phenotypic distribution. We show that this approach improves the classical Gaussian approximation, and captures accurately the shape of this equilibrium phenotypic distribution in one- and two-population scenarios. We examine the evolutionary equilibrium under general conditions where demography and selection may be nonsymmetric between the two habitats. In particular, we show how migration may increase differentiation in a source–sink scenario. We discuss the implications of these analytic results for the adaptation of organisms with large mutation rates, such as RNA viruses.
Keywords: local adaptation, migration-selection balance, gene flow, Adaptive Dynamics, Quantitative Genetics, skew
SPATIALLY heterogeneous selection is ubiquitous, and constitutes a potent evolutionary force that promotes the emergence and the maintenance of biodiversity. Spatial variation in selection can yield adaptation to local environmental conditions; however, other evolutionary forces, like migration and mutation, tend to homogenize the spatial patterns of differentiation, and thus impede the build up of local adaptation. Understanding the balance between these contrasting evolutionary forces is a major objective of evolutionary biology theory (Slatkin 1978; Savolainen et al. 2013; Whitlock 2015) and could lead to a better understanding of the speciation process and the evolutionary response to global change (Doebeli and Dieckmann 2003; Leimar et al. 2008. In this article, we consider a two-habitat model with explicit demographic dynamics as in Meszéna et al. (1997), Day (2000), Ronce and Kirkpatrick (2001), and Débarre et al. (2013). We assume that adaptation is governed by a single quantitative trait where individuals reproduce asexually. Maladapted populations have a reduced growth rate, and, consequently, lower population size. In other words, selection is assumed to be “hard” (Christiansen 1975; Débarre and Gandon 2010) as the population size in each habitat is affected by selection, mutation, and migration. These effects are complex because, for instance, nonsymmetric population sizes affect gene flow and adaptation feeds back on demography and population sizes (Nagylaki 1978; Meszéna et al. 1997; Day 2000; Ronce and Kirkpatrick 2001; Lenormand 2002; Débarre et al. 2013). To capture the complexity of these feed backs, it is essential to keep track of both the local densities and the distributions of phenotypes in each habitat. Note that this complexity often led to the analysis of the simplest ecological scenarios, where the strength of selection, migration, and demographic constraints are assumed to be the same in the two habitats (we will refer to such situations as symmetric scenarios). See, however, Holt and Gaines (1992), García-Ramos and Kirkpatrick (1997), Gomulkiewicz et al. (1999), and Holt et al. (2003) for the analysis of the effect of asymmetric migration from a source habitat on the dynamics of adaptation in a peripheral (i.e., sink) habitat. Three different approaches have been used to analyze these two-population models. Each of these approaches rely on a set of restrictive assumptions regarding the relative influence of the different evolutionary forces acting on the evolution of the population.
First, under the assumption that the rate of mutation is weak relative to selection, it is possible to use the Adaptive Dynamics framework [see Meszéna et al. (1997); Day (2000); Szilágyi and Meszéna (2009); Débarre et al. (2013), but also Results for a presentation of this framework]. This analysis captures the effect of migration and selection on the long-term evolutionary equilibrium. In particular, this approach shows that weak migration relative to selection promotes the coexistence of two specialist strategies (locally adapted to each habitat). In contrast, when migration is strong relative to selection, a single generalist strategy is favored. The main limitation of this approach is that it relies on the assumption that the mutation rate is vanishingly small, which results in a very limited amount of genetic variability. At most, two genotypes can coexist in this two-habitat model.
Second, Quantitative Genetics formalism, based on computation of the moments of phenotypic distribution, has been used to track evolutionary dynamics in heterogeneous habitats when there is a substantial level of phenotypic diversity in each population (Ronce and Kirkpatrick 2001). This model considers sexual reproduction with a quantitative trait [considering multiple loci with small effects (Fisher 1919)]. However, similar types of equations, describing the dynamics of the moments of phenotypic distribution, can also be derived in the case of asexual reproduction considering large mutation rates [see Débarre et al. (2013) and below]. This formalism allows the recovery of classical migration thresholds below which specialization is feasible. But the analysis of Ronce and Kirkpatrick (2001) also reveals the existence of evolutionary bistability, where transient perturbations of the demography can have long-term evolutionary consequences for specialization. Yet, the assumption of the shape of the phenotypic distribution (assumed to be Gaussian in each habitat) is a major limitation of this formalism.
Third, attempts to account for other shapes of phenotypic distributions in heterogeneous environments have been developed recently (Yeaman and Guillaume 2009; Débarre et al. 2013, 2015). These models highlight that calculations based on the Gaussian approximation, which neglects the skewness of the equilibrium phenotypic distribution, underestimate the level of phenotypic divergence and local adaptation. Yet, there is currently no model able to accurately describe the build up of non-Gaussian distributions. The only attempts to model this distribution describe the phenotypic distributions in each habitat as the sum of two Gaussian distributions Yeaman and Guillaume 2009; Débarre et al. 2013. These models, however, yield only approximate predictions on long-term evolutionary equilibria.
Here, we develop an alternative formalism that yields population size and phenotypic distribution in each habitat at the equilibrium between selection, mutation, and migration. In Materials and Methods, we present our two-population model. For heuristic reasons, we next provide analysis of the equilibrium between selection and mutation in a single population. This provides an illustration of our approach in a simple scenario, and shows how this analysis can go beyond the classical Gaussian approximation. Next, we extend this approach to a two-population scenario, where migration can also influence phenotypic distribution, and we derive approximations for the level of adaptation under a migration–selection–mutation balance. We also explore the effects of nonsymmetric constraints on selection, migration, or demography between the two habitats. We evaluate the accuracy of these approximations by comparing them to numerical solutions of our deterministic model, and we show that our approach improves previous attempts to study the interplay between adaptation and demography in heterogeneous environments. We contend that our results are particularly relevant for organisms with high mutation rates and may help to understand the within-host dynamics of chronic infections by RNA viruses (Drake and Holland 1999; Sanjuán et al. 2010).
The present work has been prepared in parallel to the mathematical article Mirrahimi 2017 where we provide the mathematical basis and proofs for the method used here. See also Gandon and Mirrahimi (2017), where those mathematical results were announced. The aim of the present paper is to show how this approach can help to understand the balance between different evolutionary forces. We present several new biological scenarios, and derive new results that help grasp the interplay between different evolutionary forces and demography.
Materials and Methods
We model an environment containing two habitats that we label 1 and 2 (Figure 1). The population is structured by a quantitative trait z. In each habitat i there is stabilizing selection on the trait z for an optimal value (for habitat ). The growth rate in habitat i is denoted by which has its maximum when . In the following, we will focus mainly on the following quadratic stabilizing selection function (Bürger 2000, pp. 117–121 and chapter VI):
| (1) |
We denote by the selection pressure in habitat i. Without loss of generality, we assume that . But our approach could be used with other stabilizing selection functions (see Equation 12 below, where another selection function is studied in the case of one population).
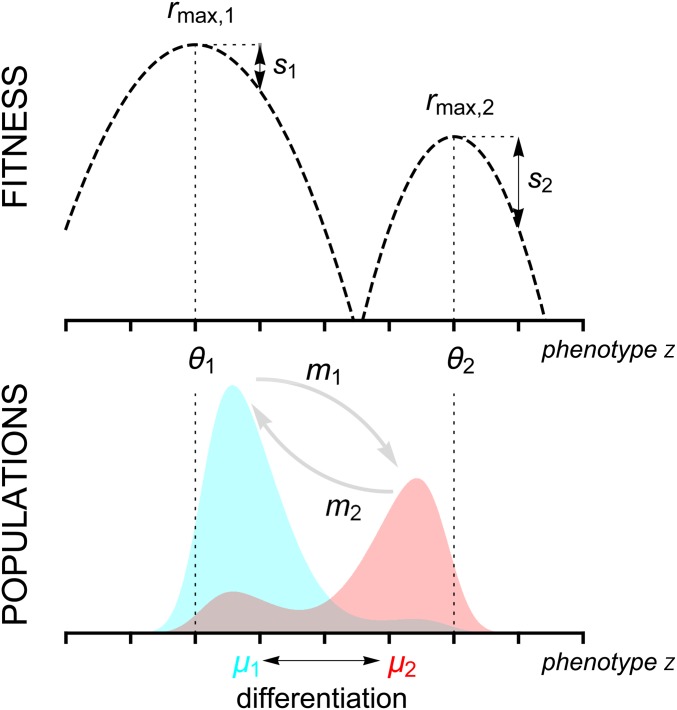
Figure 1.
Schematic representation of the two habitat model. The top figure shows the growth rate (fitness) in each habitat as a function of the phenotypic trait z. In habitat i, the growth rate is assumed to be maximized at and the strength of selection is governed by (see Equation 1). Here, we illustrate a scenario with nonsymmetric fitness functions. The bottom figure shows the phenotypic density in each habitat (light blue and light red in habitats 1 and 2, respectively). Migration from population i is governed by the parameter and tends to reduce the differentiation (i.e., the difference between the mean phenotypes) between populations.
Reproduction is assumed to be asexual. Offspring inherit the phenotype of their parent (i.e., no environmental variance), and we consider a continuum of alleles model (Kimura 1965). Mutations occur with a constant rate U (i.e., mutations are not associated with reproduction), and add an increment y to the parents’ phenotype; we assume that the distribution of these mutational effects is given by , with mean 0 and variance equal to . We also assume that individuals disperse out of habitat i with rate . Rates of migration are assumed to be independent of individuals’ phenotypes.
Let be the phenotypic density in habitat i at time t. The dynamics of this density in each habitat is given by (for and ):
| (2) |
The first term on the right-hand side of the above equation corresponds to the effect of mutations. The second term corresponds to the logistic growth that results from the balance between reproduction given by Equation 1 and density dependence, where measures the intensity of competition within each habitat. The last term corresponds to the dispersal of individuals between habitats.
If we assume that the variance of the mutation distribution is small relative to the mutation rate U, we can consider an approximate model where we replace the mutation term in Equation 2 by a diffusion [see Kimura (1965); Lande (1975) and the more recent article by Champagnat et al. 2008, where the diffusion term has been derived directly from a stochastic individual based model]. See also Bürger (2000, pp. 239–241) for a discussion on the domain of the validity of such model. Our model then becomes:
| (3) |
The total population sizes in each habitat is given by:
| (4) |
In other words, refers to the density of individuals with phenotype z in habitat i, while refers to the total density of the polymorphic population in habitat i.
Data availability statement
No biological data are provided in this article. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11365982.
Results
One population: the selection–mutation equilibrium
In this section, we start by a simple scenario with no migration. This one-population example provides a good introduction to our method. The dynamics of the phenotypic density in a single habitat is given by:
| (5) |
where is the total population size:
For this scenario, we consider a more general form of growth rate than in Equation 1. We suppose only that is maximized for an optimal trait . In the following, we present our two-step approach. First, we analyze the evolutionary equilibria of the problem when the rate of mutation is small, and we identify the evolutionary stable strategy (ESS). Second, we use this ESS to derive an approximation for the stationary solution of Equation 5 when mutation is more frequent and maintains a standing variance at equilibrium.
Adaptive dynamics and ESSs
In this section, we assume that mutations are very rare, such that a mutation is fixed or goes extinct before a new mutation arises in the population. The phenotypic distribution results from a collection of spikes. Such spikes are gradually replaced by others with the arrival of new mutations and through a competitive procedure. The theory of Adaptive Dynamics (Geritz et al. 1998) is based on the study of the stable equilibrium distribution and the localization of the spikes of such equilibrium, known as ESSs. Note that, in this first step, we do not make any assumption regarding the effects of these mutations on the phenotype. We are interested in the identification of the global ESSs, i.e., when the resident population cannot be invaded by any mutation, no matter what its effect.
In absence of migration, the phenotype constitutes a globally stable evolutionary strategy. Indeed, when such monomorphic population reaches its demographic equilibrium, the total population size is given by . The fate of a mutant with phenotype introduced into such a resident population is determined by its fitness (i.e., per capita growth rate minus density dependence) given by:
| (6) |
No mutant trait can indeed invade the population since takes its maximum at .
Equilibrium distribution with mutation
The ESS corresponds to the long-term evolutionary outcome in a scenario where all phenotypic strategies are present initially, but where mutation is absent. In the following, we study the impact of mutation on the ultimate evolutionary equilibrium of the population.
We introduce a new parameter, . Hence, we replace by and we approximate the phenotypic density , the equilibrium of Equation 5, in terms of ε (where the subscript ε in indicates the dependence of the phenotypic density on the parameter ε). Our objective is to provide an approximation of the phenotypic density when the effect of mutation (measured by ε) is small, while the mutation rate can be large.
To study we will use a method based on Hamilton-Jacobi equations (see Equation A.5), which was developed by the mathematical community during the last decade to study selection–mutation models, when the effect of mutations is vanishingly small. This method was first suggested by Diekmann et al. (2005), and was developed for the case of homogeneous environments in Perthame and Barles (2008) and Barles et al. (2009). However, those works, which are addressed to the mathematical community, were focused mainly on the limited case where the effect of mutations, ε, is vanishingly small. Here, we go further than previous studies, and characterize the phenotypic distribution when the mutations have non-negligible effects.
The method is based on the following transformation:
| (7) |
The introduction of the function is a mathematical trick. It is indeed easier to first provide an approximation of rather than directly studying .
Note that a first approximation of the population’s phenotypic density, which is commonly used in the theory of Quantitative Genetics, is a Gaussian approximation of the following form around :
| (8) |
The Gaussian approximation is as if we had imposed to be a quadratic function of z, that is . Our objective, however, is to obtain more accurate results than Equation 8, and to approximate without making an a priori Gaussian assumption. To this end, we postulate an expansion for in terms of ε:
| (9) |
and we try to compute the coefficients and . These terms can indeed be computed explicitly, and they lead to an approximation of the total population size and the phenotypic density which we will henceforth call our first approximation (see the supplementary information A.1.1 for these derivations).
In order to provide more explicit formula for the moments of order of the population’s distribution in terms of the parameters of the model, we also provide a second approximation. This second approximation, instead of using the values of and in the whole domain, is based on the Taylor expansions of and around the ESS points (see the supplementary information, Section A.1.2). Our second approximation is, by definition, less accurate than the first. We illustrate below the quality of these different approximations under two different scenarios.
Quadratic growth rate:
We first consider a quadratic growth rate as in Equation 1:
| (10) |
In this case, our first approximation yields the Gaussian distribution (Equation 8) with:
| (11) |
Note that this Gaussian distribution is actually an exact equilibrium of Equation 3, and the above signs can indeed be replaced by equalities [see Kimura (1965) and Bürger (2000, Chapter IV)]. For the derivation of this result, see the supplementary information, Section A.1.1.
An asymmetric growth rate:
We next consider a growth rate that is not symmetric:
| (12) |
In this case, the phenotypic distribution does not have a Gaussian profile, and our approximation yields:
where the values of and can be computed explicitly (see the supplementary information, Section A.1.1). In Figure 2 we plot this first approximation, and compare it with the exact distribution that we derived numerically.
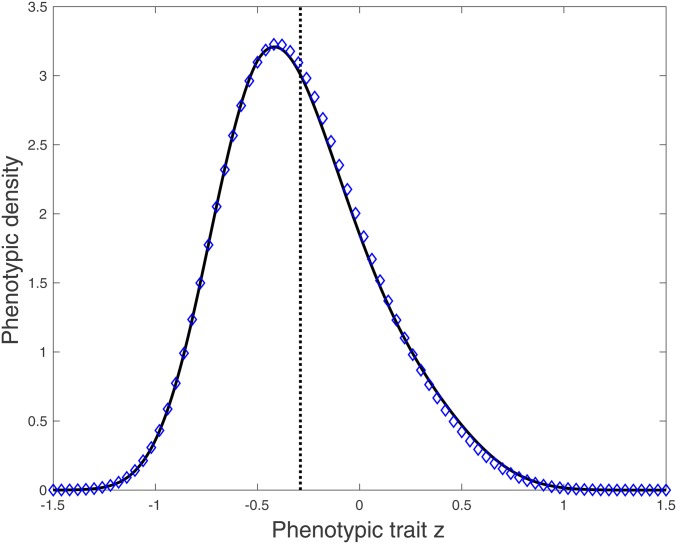
Figure 2.
The selection–mutation equilibrium of the phenotypic density in a single population. We plot the exact phenotypic density at equilibrium obtained from numerical computations of the equilibrium of Equation 5 (blue dots) together with our first approximation (full black line) with the growth rate given in Equation 12. The vertical dotted line indicates the mean of the phenotypic distribution. Note the skewness of the equilibrium distribution that is accurately captured with our approximation (see also Table 1). In this figure, to compute the equilibrium numerically, we solved the dynamic problem numerically (Equation 5) and kept the solution obtained for a long time after the equilibrium has been reached. Parameter values: , ; , , , , , .
We can also use our second approximation to obtain analytic expressions for the mean phenotypic trait (see the supplementary information, Section A.1.2 for the derivation):
the mean variance:
and the third central moment:
In Table 1, we show that our two approximations capture accurately the first three moments of the equilibrium distribution using the parameters that we used in Figure 2. As expected, the first approximation is more accurate, but the analytic expressions of the second approximation given above allow us to capture the influence of the parameters of the model.
Table 1. First three moments of the phenotypic distribution at mutation–selection equilibrium in a single population.
| Exact value | First approximation | Second approximation | |
|---|---|---|---|
| Mean: | −0.29 | −0.29 | −0.35 |
| Variance: | 0.13 | 0.14 | 0.09 |
| Third central moment: | 0.02 | 0.02 | 0.01 |
We compare the values from the exact numerical resolution of Equation 5 and our two approximations using the growth rate given in Equation 12 (see also Figure 2). Parameter values: , ; , , , , , and
Two populations: the selection–mutation–migration equilibrium
Next, we return to analysis of the stationary solution of Equation 3, which results from the equilibrium between selection, mutation, and migration in each habitat. Using Equation 1 and Equation 3, one can derive dynamical equations for the size of the population and the mean phenotype :
where and are the variance and the third central moment of the phenotypic distribution, respectively. These two quantities are also dynamical variables, and their dynamics are governed by higher moments of the phenotypic distribution. But this dynamical system is not closed, and these higher moments are also dynamical variables that depend on additional moments. Various approximations, however, have been used to capture its behavior. Typically, many results are based on the Gaussian approximation, which focuses on the dynamics of the mean and the variance and discards all higher cumulants of the distribution (Bürger 2000; Rice 2004). Yet several authors have pointed out that neglecting the skewness of the distribution can underestimate the amount of differentiation and local adaptation Yeaman and Guillaume 2009; Débarre et al. 2013, 2015. Indeed, in the case of symmetrical habitats, that is when , one can readily obtain the size and the mean trait of the population at equilibrium (the equilibrium is indicated by a superscript *). Using the fact that , , and , we obtain:
The differentiation between the two habitats is thus (Figure 1):
| (13) |
There is, however, no analytic prediction of the magnitude of the different moments of the phenotypic distribution, except in the limiting case when the mutation rate is extremely low (Débarre et al. 2013).
Next, we follow the two-step approach we used to obtain the stationary phenotypic distribution in a single population. First, we analyze the evolutionary equilibria of the system when mutations are rare using the Adaptive Dynamics framework. We identify monomorphic or dimorphic globally ESSs. Second, we use these ESSs to derive approximations of the stationary solution of Equation 3 when mutation is more frequent and maintains a standing variance at equilibrium.
Adaptive dynamics and evolutionary stable strategies
We consider a resident population at a demographic equilibrium set by the phenotypic densities of the resident in both habitats (see the supplementary information, Section A.2.1). We want to determine the fate of a mutant with phenotype introduced into this resident population. The ability of the mutant to invade is determined by its fitness, given by:
| (14) |
To take into account migration between habitats, we introduce an effective fitness, which corresponds to the growth rate of a trait in the whole environment (see Caswell 1989; Metz et al. 1992; Meszéna et al. 1997). The effective fitness which corresponds to the effective growth rate associated with trait in the resident population is the largest eigenvalue of the following matrix:
| (15) |
After some time, the dynamical system will reach a globally stable demographic equilibrium. Because there are two habitats, we expect that, at most, two distinct traits can coexist. With an analysis of the effective fitness W, we characterize such equilibrium corresponding to the evolutionary stable strategy (see the supplementary information, Section A.2.1.1). This equilibrium is indeed either monomorphic with phenotype and total population size or dimorphic with phenotypes and and total population size , where the subscripts and indicate that the phenotype is best adapted to habitat 1 and 2, respectively.
Equilibrium distribution with mutation
In the following, we allow the mutation rate to increase and we study the impact of mutations on the ultimate evolutionary equilibrium of the phenotypic densities, i.e., the stationary solution of Equation 3. We present below the general principle of the approach before examining specific case studies.
As above, we introduce the parameter and we approximate the phenotypic density , the equilibrium of Equation 3 with , in terms of ε. Our objective is to provide an approximation of the phenotypic density in each habitat when the effect of mutation (measured by ε) is small, while the mutation rate can be large.
We use analogous transformation to (7):
| (16) |
Our objective is then to estimate . We proceed as in the one-population scenario, and we postulate an expansion for in terms of ε:
| (17) |
and we try to compute the coefficients and . First, we can show that, when there is migration in both directions (i.e., for ), the zero-order terms are the same in both habitats: (see the supplementary information, Section A.2.1.2). We can indeed compute explicitly which is given by (A.23) in the monomorphic case and by (A.24) in the dimorphic case. As we observe in the equations (A.23) and (A.24), attains its maximum (which is equal to 0) at the ESS points identified in the previous subsection. This means that the peaks of population distribution are around the ESS points ( in the case of the monomorphic ESS and for the dimorphic ESS). Note that the fact that means that the peaks of the population distribution are placed approximately at the same points (ESS points) in both habitats. However, the size of the peaks may be different since is not necessarily equal to .
We are also able to compute the first-order term (see the supplementary information, Section A.2.1.2). This allows us to obtain a first approximation of the phenotypic density . This approximation of the stationary distribution is very accurate (see, for instance, Figure 4).
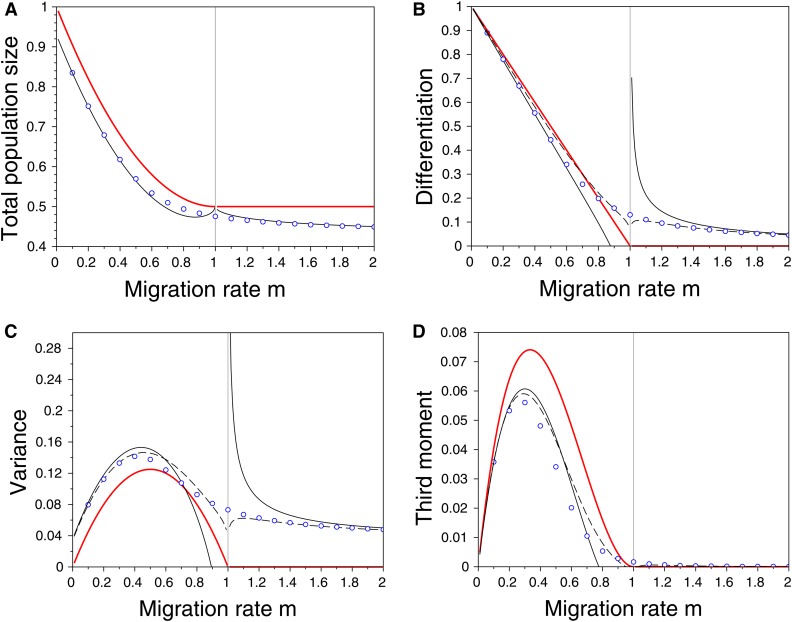
Figure 4.
Effects of migration in a symmetric scenario on (A) the total population size () in habitat 1, (B) the differentiation between habitats (), (C) the variance (), and (D) the third central moment of the phenotypic distribution () in habitat 1 (see Equation 19 and Section B.3 for the definition of these quantities and the analytic formula obtained from our second approximation). The dots refer to the numerical resolutions of the problem with , the red line indicates the case where , while the lines in black refer to our two approximations when (the dashed line for the first approximation and the full line for the second approximation). The vertical gray line indicates the critical migration rate below which dimorphism can evolve in the Adaptive Dynamics scenario. Note that both approximations predict the same total population size. Other parameter values: , , , , and .
As in the one population scenario, we derive more explicit formula for the moments of order of the stationary phenotypic distribution. This second approximation, instead of using the values of and in the whole domain, is based on the computation of the Taylor expansions of and around the ESS points (see the supplementary information, Section A.2.1.3).
Case studies
Symmetric fitness landscapes:
We focus first on a symmetric scenario, where, apart from the position of the optimum, the two habitats are identical: . In this special case, it is possible to fully characterize the evolutionary equilibrium.
When migration rate is higher than the critical migration threshold:
| (18) |
migration prevents the differentiation of the trait between the two habitats (see the supplementary information, Subsection A.2.1.1). The only evolutionary equilibrium, when the mutation rate is vanishingly small, is monomorphic, and satisfies and , where is the dirac delta function and .
Monomorphic case:
Let us suppose that . Then, is the only ESS and . Then, we can provide our first approximation of the phenotypic density following the method introduced above (Figure 3). Moreover, defining , we can use the second approximation to obtain an analytic formula for the moments of the stationary state:
| (19) |
These results are consistent with Equation 13. Note that the equilibrium variance in each habitat, is larger than the equilibrium variance maintained in the absence of heterogeneity between the habitats (cf. Equation 11). This increase in the equilibrium variance comes from φ, which depends on dispersion and the heterogeneity between the two habitats. The variance of the distribution increases as φ decreases. When the approximation for the variance becomes infinitely large. Indeed, this corresponds to the threshold value of migration, below which the above approximation collapses because the distribution becomes bimodal. In this case, we have to switch to analysis of the dimorphic case. Note that the differentiation between habitats also depends on φ. Some differentiation emerges even when the migration rate is above the critical migration rate, (Figures 3 and 4). In Figure 4 we provide a comparison of the results from the first and the second approximations. Our second approximation yields convincing results when the parameters are such that we are far from the transition zone from monomorphic to dimorphic distribution. This approximation is indeed based on an integral approximation, which is relevant only when the population distribution is relatively sharp around the ESS points. This is not the case in the transition zone unless the effect of the mutations, i.e., ε, is very small.
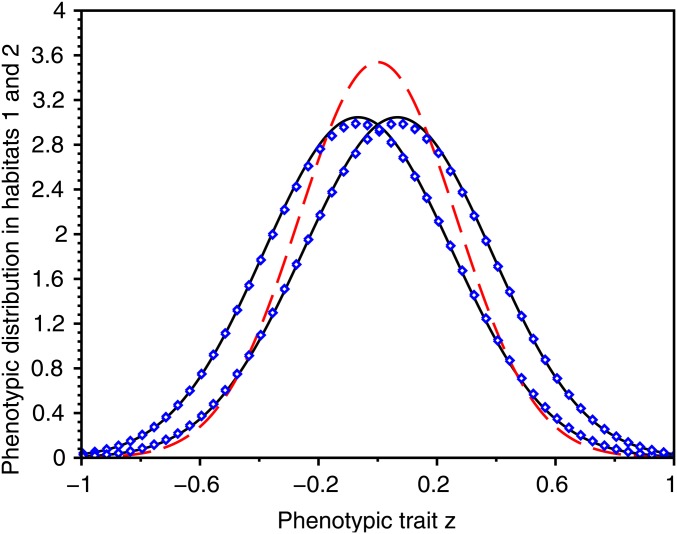
Figure 3.
Selection–mutation–migration equilibrium of the phenotypic densities in the two habitats in a symmetric scenario. We plot the exact phenotypic densities at equilibrium obtained from numerical computations of the equilibrium of Equation 3 (blue dots) together with our first approximation (full black line) in a case where the distribution is unimodal in each habitat. We also plot the approximation given in Débarre et al. (2013) (red dashed line). Note that our approximation captures the emergence of some differentiation, even though we are above the critical migration rate leading to the evolution of a dimorphic population. In the presence of large mutation rates, the population distribution is indeed shifted to the left (respectively right) in the first (respectively second) habitat, while Débarre et al. (2013) provided the same approximation for both habitats. Our calculation yields also better approximations for the variance of the distribution in each habitat (Débarre et al. 2013 underestimates this variance). In this and the following figures, to compute the equilibrium numerically, we have solved the dynamic problem numerically (Equation 3) and kept the solution obtained for a long time after equilibrium has been reached. Parameter values: , , ; , , , .
Dimorphic case:
When , the only globally stable evolutionary equilibrium is dimorphic, which yields the following ESS: with and . When this yields the following phenotypic densities at equilibrium: (analytic expressions for and are given in the supplementary information, Section B.1). When we can use our first and second approximations to obtain convincing approximations of the phenotypic distribution and its moments (see Figure 4). Our first approximation improves the Adaptive Dynamics predictions in a broad range of the parameter space, and, as pointed out above, our second approximation is pertinent when the parameters are such that we are far from the transition zone from dimorphic to monomorphic distribution. The analytic expressions for the local moments of the stationary distribution in each habitat, obtained from our second approximation, are given in the supplementary information, Section B.3.
Nonsymmetric scenarios:
A general nonsymmetric scenario:
In a nonsymmetric scenario, there is also a unique globally stable evolutionary strategy that is either monomorphic or dimorphic. There is still a threshold value of migration, above which the maintenance of a dimorphic polymorphism is impossible: . Note that this condition generalizes the condition in the symmetric case (i.e., when and ). However, for the ESS to be dimorphic, the condition is not enough and two other conditions should also be satisfied. These conditions (i.e., and with the constants , and depending on the parameters , , , , , and θ, see the supplementary information, Section B.2) guarantee that the qualities of the habitats are not very different. Indeed, if one habitat has a higher quality it is likely to overwhelm the dynamics of adaptation in the other habitat. This will yield a monomorphic equilibrium biased toward the high-quality habitat. Figure 5 illustrates that a polymorphism is maintained only in a range of parameter values where the two habitats are relatively similar. Interestingly, in spite of the asymmetry of the two habitats, the locations of the two peaks of the phenotypic distribution are always symmetric, and, consequently, where: . The symmetric locations of the peaks is indeed a consequence of the choice of the quadratic stabilizing selection (Equation 1). See the supplementary information, Section B.1 for the expressions of the densities in each habitat, and Section A.2.1.1 for the conditions leading to this stable equilibrium.
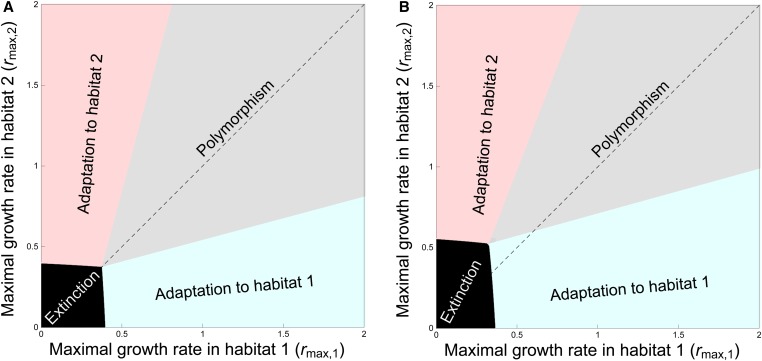
Figure 5.
Maintenance of polymorphism and nonsymmetric adaptation as a function of the maximal growth rates and in the two habitats. In (A) we examine a symmetric situation where all the parameters are identical in the two habitats: , , and . In (B) we show a nonsymmetric case with the same parameters as in (A) except and . The black area indicates the parameter space, where the population is driven to extinction because the maximal growth rates are too low. In the gray area, some polymorphism can be maintained in the two-habitat population as long as the difference in the maximal growth rates are not too high. When this difference reaches a threshold, polymorphism cannot be maintained and the single type that is maintained is more adapted to the good-quality habitat (the habitat with the highest maximal growth rate).
A source–sink scenario:
An extreme case of asymmetry occurs when one population (the source) does not receive any migrant from the second population (the sink). For instance, when and there is no immigration in habitat 1. Note that this is a degenerate case. In contrast with the above analysis, where both and are positive, we have to provide an analysis of ESS for each habitat separately. Moreover, computation of the equilibrium in presence of mutations is slightly different because of this degeneracy (see the supplementary information Section A.2.2).
The evolutionary outcome in the first habitat is obvious because it depends only on selection acting in habitat 1: the ESS is and
| (20) |
Moreover, the population’s phenotypic density, can be computed explicitly: , where and is the probability density of a normal distribution .
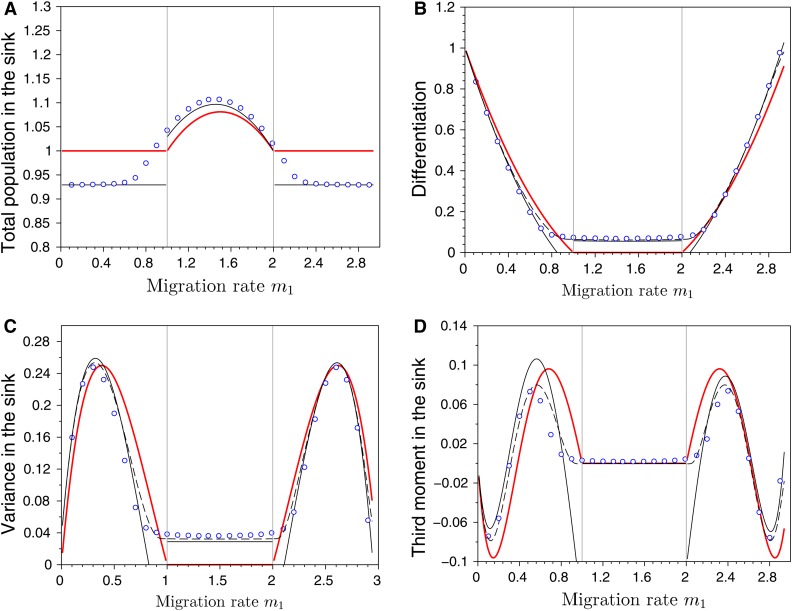
In habitat 2, the evolutionary outcome results from the balance between migration from habitat 1 and local selection. Interestingly, migration has a nonmonotonic effect on adaptation in the sink (See Figure 6). Indeed, Figure 6A shows that the population size in the sink is maximized for intermediate values of migration. More migration from the source has a beneficial effect on the demography of the sink, but it prevents local adaptation. Yet, when migration from the source becomes very large it limits the size of the population in the source (see Equation 20). This limits the influence of the source on the sink, and can even promote adaptation to the sink. In fact, it is worth noting that differentiation between the two habitats can actually increase with migration (Figure 6B). The level of migration from the source that prevents local adaptation in the sink is given by the condition:
| (21) |
Indeed, when condition Equation 21 is verified, the migration from the source overwhelms local selection, and the evolutionary stable strategy in the sink is . In contrast, when condition Equation 21 does not hold, the effective growth rate of the optimal trait θ in the sink habitat is high enough to compete with the trait coming from the source, allowing coexistence between the two strategies. Note again that our two approximations (see supplementary information sections A.2.2.2 for the derivation of the first approximation and B.4 for the analytic formula for the moments of the phenotypic distribution derived from our second approximation) provide very good predictions for the moments of the phenotypic distribution in the sink (Figure 6).
Figure 6.
Effects of migration in a source–sink scenario on (A) the total population size in the sink habitat, (B) the differentiation between habitats, (C) the variance, and (D) the third central moment of the phenotypic distribution in sink. The dots refer to exact numerical computations when , the red line indicates the case where while the lines in black refer to our two approximations when (dashed line for the first approximation and the full line for the second approximation). The vertical gray lines, at and , indicate the critical migration rates where transition occurs between monomorphism and dimorphism in the Adaptive Dynamics framework (see condition Equation 21). Note that both approximations predict the same total population size. Other parameter values: , , , , , , and .
Discussion
The balance between selection, migration, and mutation drives the dynamics of local adaptation in heterogeneous environments. Here, we present a new theoretical approach to obtain accurate approximations for the equilibrium phenotypic densities in a two-habitat environment. This analysis goes beyond the Adaptive Dynamics framework because it allows us to account for the effect of large mutation rates. This analysis does not rely on the Gaussian approximation that underlies many Quantitative Genetics models. Our analysis yields analytic approximations that help provide a good understanding of the balance between the different evolutionary forces in both symmetric and nonsymmetric scenarios.
In the symmetric scenario, we recover the classical results from Quantitative Genetics in a single population (Lande 1975; Bürger 2000; Rice 2004) but expand this to spatially heterogeneous scenarios. In particular, we capture the emergence of differentiation between habitats when the migration rate decreases. When migration is strong relative to selection, the stationary phenotypic density is unimodal in each habitat but heterogeneous selection increases phenotypic variance and differentiation (see Equation 19 and Figure 4). When migration is close to the critical migration rate (see condition Equation 18), we observe a shift between the phenotypic distributions of the two habitats. This pattern was not detected in previous studies, but our method captures this shift and improves the approximation of the variance of the phenotypic distributions [see Débarre et al. (2013) and Figure 3]. When the migration rate is much smaller than and selection is sufficiently strong between habitats, the equilibrium distribution in each habitat can be well approximated as the sum of two distributions. But, unlike previous approximations (Yeaman and Guillaume 2009; Débarre et al. 2013) these two distributions are non-Gaussian. We derive approximations for the moments of these distributions. In other words, this work generalizes previous attempts to derive the distribution of a phenotypic trait at the mutation–selection–migration equilibrium. Our results confirm the importance of the skewness in the phenotypic distribution, and improve predictions of measures of local adaptation in a heterogeneous environment.
In the nonsymmetric scenario, we show that the condition for the maintenance of two specialized strategies is more restrictive (Figure 5). Indeed, asymmetries promote a single strategy that is more locally adapted to the habitat with larger population size and/or lower immigration rate. The impact of biased migration rates from a source population into the adaptation of peripheral populations has been discussed before (Holt and Gaines 1992; García-Ramos and Kirkpatrick 1997; Gomulkiewicz et al. 1999; Holt et al. 2003; Akerman and Bürger 2014). Our approach, however, yields a quantitative description of the shape of the phenotypic distributions in both the source and the sink habitats. These accurate predictions are key to understanding the effect of different evolutionary forces on the level of adaptation in the two habitats. For instance, the analysis of an extreme case with source–sink dynamics reveals the complex interplay between migration, demography, and local selection. The maintenance of a polymorphic equilibrium is possible when migration from the source is either very weak or very strong. This result challenges the classical prediction where migration is always a homogenizing force reducing the differentiation among populations (Figure 6).
Our work illustrates the potential of a new mathematical tool in the field of evolutionary biology. In this work, we use an approach based on Hamilton-Jacobi equations [see (A.22)], which has been developed, mostly by the mathematical community, during the last decade to describe the asymptotic solutions of selection–mutation models, as the effect of mutation vanishes. We refer to Diekmann et al. (2005), Perthame and Barles (2008), and Mirrahimi (2011) for the establishment of the basis of this approach. Note, however, that previous studies were focused mainly on the limit case where the effects of mutations are vanishingly small. In particular, they do not provide approximations of phenotypic density and its moments when the effect of mutations, ε, is nonzero. In the present work, we go further than the previous studies and characterize the phenotypic distributions when the influx of mutations can alter significantly the shape of the stationary distribution. Understanding the build up of this distribution is particularly important when studying the effect of mutation on adaptation. Although mutation is the ultimate source of adaptive variation, the accumulation of deleterious mutations generates a load on the average fitness of populations. This is particularly relevant in organisms like RNA viruses that are characterized by very large mutation rates (Drake and Holland 1999; Sanjuán et al. 2010). In fact, the mutation loads of RNA virus is so high that it may even lead some populations to extinction (Bull et al. 2007; Martin and Gandon 2010). Our model can be used to accurately capture the effect of increasing mutation rates on the mutation load of a population living in a heterogeneous environment (Figure 7). This heterogeneity may be particularly relevant in chronic infections by pathogenic viruses that can adapt to different organs (Kemal et al. 2003; Ducoulombier et al. 2004; Sanjuán et al. 2004; Jridi et al. 2006). A better understanding of the phenotypic distribution at equilibrium in heterogeneous environments may thus provide more accurate prediction on the critical mutation rates that can ultimately lead within-host dynamics to pathogen extinction.
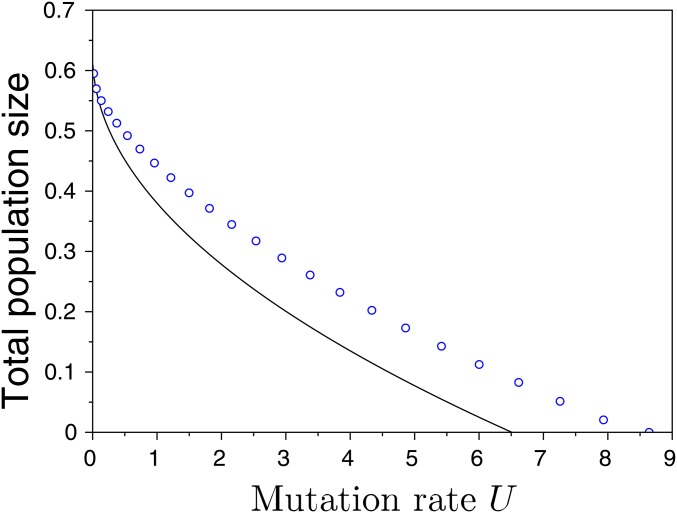
Figure 7.
Effect of increasing the mutation rate U on the total population size in the symmetric scenario used in Figure 3 with . The full line indicates the approximation, and the dots are the results of exact numerical computations. This figure illustrates that our approximation given by the first line of Equation 19 captures reasonably well the effect of large mutation rates on the mutation load in a two-habitat scenario where there is differentiation and some local adaptation.
Our analysis of the equilibrium between selection, migration, and mutation could be extended in several new directions. More than two habitats could be considered, or different growth rates and/or mutation kernels could be used [see Mirrahimi (2013) and the supplementary information, Section (A.3)]. The approach could also be used to analyze situations away from the equilibrium. For instance, it would be possible to track the dynamics of the distribution as the population adapts to a new environment, or to a time-varying environment (Lande and Shannon 1996). Hamilton-Jacobi equations have indeed also been used to study time-varying (but space homogeneous) environments (see, for instance, Mirrahimi et al. 2015; Figueroa Iglesias and Mirrahimi 2018). Finally, it is interesting to note that the generalization of the present ecological scenario to model the adaptation of sexual species in heterogeneous environments remains to be carried out. Our method could be extended to allow for sexual reproduction within the framework of the infinitesimal model (see Fisher 1919; Calvez et al. 2019). But this analysis falls beyond the scope of the present paper.
Acknowledgments
The first author is grateful for partial funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 639638), held by Vincent Calvez, and from the French Agence Nationale de la Recherche (ANR) projects KIBORD ANR-13-BS01-0004 and MODEVOL ANR-13-JS01-0009.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11365982.
Communicating editor: J. Hermisson
Literature Cited
- Akerman A., and Bürger R., 2014. The consequences of gene flow for local adaptation and differentiation: a two-locus two-deme model. J. Math. Biol. 68: 1135–1198. 10.1007/s00285-013-0660-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barles G., Mirrahimi S., and Perthame B., 2009. Concentration in Lotka-Volterra parabolic or integral equations: a general convergence result. Methods Appl. Anal. 16: 321–340. [Google Scholar]
- Bull J. J., Sanjuan R., and Wilke C. O., 2007. Theory of lethal mutagenesis for viruses. J. Virol. 81: 2930–2939. 10.1128/JVI.01624-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger R., 2000. The Mathematical theory of selection, recombination and mutation. Wiley, New-York. [Google Scholar]
- Calvez V., Garnier J., and Patout F., 2019. A quantitative genetics model with sexual mode of reproduction in the regime of small variance. arXiv: 1811.01779v3.
- Caswell H., 1989. Matrix Population Models, Sinauer Associates, Sunderland, MA. [Google Scholar]
- Champagnat N., Ferrière R., and Méléard S., 2008. Individual-based probabilistic models of adaptive evolution and various scaling approximations. Progress in Probability 59: 75–113. 10.1007/978-3-7643-8458-6_6 [DOI] [Google Scholar]
- Christiansen F. B., 1975. Hard and soft selection in a subdivided population. Am. Nat. 109: 11–16. 10.1086/282970 [DOI] [Google Scholar]
- Day T., 2000. Competition and the effect of spatial resource heterogeneity on evolutionary diversification. Am. Nat. 155: 790–803. 10.1086/303356 [DOI] [PubMed] [Google Scholar]
- Débarre F., and Gandon S., 2010. Evolution of specialization in a spatially continuous environment. J. Evol. Biol. 23: 1090–1099. 10.1111/j.1420-9101.2010.01966.x [DOI] [PubMed] [Google Scholar]
- Débarre F., Ronce O., and Gandon S., 2013. Quantifying the effects of migration and mutation on adaptation and demography in spatially heterogeneous environments. J. Evol. Biol. 26: 1185–1202. 10.1111/jeb.12132 [DOI] [PubMed] [Google Scholar]
- Débarre F., Yeaman S., and Guillaume F., 2015. Evolution of quantitative traits under a migration-selection balance: when does skew matter? Am. Nat. 186: S37–S47. 10.1086/681717 [DOI] [PubMed] [Google Scholar]
- Diekmann O., Jabin P.-E., Mischler S., and Perthame B., 2005. The dynamics of adaptation: an illuminating example and a Hamilton-Jacobi approach. Th. Pop. Biol. 67: 257–271. 10.1016/j.tpb.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Doebeli M., and Dieckmann U., 2003. Speciation along environmental gradients. Nature 421: 259–264. 10.1038/nature01274 [DOI] [PubMed] [Google Scholar]
- Drake J. W., and Holland J., 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96: 13910–13913. 10.1073/pnas.96.24.13910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducoulombier D., Roque-Afonso A.-M., Di Liberto G., Penin F., Kara R. et al. , 2004. Frequent compartmentalization of hepatitis c virus variants in circulating B cells and monocytes. Hepatology 39: 817–825. 10.1002/hep.20087 [DOI] [PubMed] [Google Scholar]
- Figueroa Iglesias S., and Mirrahimi S., 2018. Long time evolutionary dynamics of phenotypically structured populations in fluctuating environments. SIAM J. Math. Anal. 50: 5537–5568. 10.1137/18M1175185 [DOI] [Google Scholar]
- Fisher R. A., 1919. Xv.-the correlation between relatives on the supposition of Mendelian inheritance. Earth and Environmental Science Transactions of the Royal Society of Edinburgh 52: 399–433. 10.1017/S0080456800012163 [DOI] [Google Scholar]
- Gandon S., and Mirrahimi S., 2017. A Hamilton-Jacobi method to describe the evolutionary equilibria in heterogeneous environments and with non-vanishing effects of mutations. C. R. Math. 355: 155–160. 10.1016/j.crma.2016.12.001 [DOI] [Google Scholar]
- García-Ramos G., and Kirkpatrick M., 1997. Genetic models of adaptation and gene flow in peripheral populations. Evolution 51: 21–28. 10.1111/j.1558-5646.1997.tb02384.x [DOI] [PubMed] [Google Scholar]
- Geritz S. A. H., Kisdi E., Mészena G., and Metz J. A. J., 1998. Evolutionarily singular strategies and the adaptive growth and branching of the evolutionary tree. Evol. Ecol. 12: 35–57. 10.1023/A:1006554906681 [DOI] [Google Scholar]
- Gomulkiewicz R., Holt R. D., and Barfield M., 1999. The effects of density dependence and immigration on local adaptation and niche evolution in a black-hole sink environment. Th. Pop. Biol. 55: 283–296. 10.1006/tpbi.1998.1405 [DOI] [PubMed] [Google Scholar]
- Holt R. D., and Gaines M. S., 1992. Analysis of adaptation in heterogeneous landscapes: implications for the evolution of fundamental niches. Evol. Ecol. 6: 433–447. 10.1007/BF02270702 [DOI] [Google Scholar]
- Holt R. D., Gomulkiewicz R., and Barfield M., 2003. The phenomenology of niche evolution via quantitative traits in a ‘black-hole’ sink. Proc. R. Soc. Lond. B Biol. Sci. 270: 215–224. 10.1098/rspb.2002.2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jridi C., Martin J.-F., Marie-Jeanne V., Labonne G., and Blanc S., 2006. Distinct viral populations differentiate and evolve independently in a single perennial host plant. J. Virol. 80: 2349–2357. 10.1128/JVI.80.5.2349-2357.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal K. S., Foley B., Burger H., Anastos K., Minkoff H. et al. , 2003. Hiv-1 in genital tract and plasma of women: compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc. Nat. Aca. Sci. USA. 100: 12972–12977. 10.1073/pnas.2134064100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1965. A stochastic model concerning the maintenance of genetic variability in quantitative characters. Proc. Natl. Acad. Sci. USA 54: 731–736. 10.1073/pnas.54.3.731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1975. The maintenance of genetic variability by mutation in a polygenic character with linked loci. Genet. Res. 26: 221–235. 10.1017/S0016672300016037 [DOI] [PubMed] [Google Scholar]
- Lande R., and Shannon S., 1996. The role of genetic variation in adaptation and population persistence in a changing environment. Evolution 50: 434–437. 10.1111/j.1558-5646.1996.tb04504.x [DOI] [PubMed] [Google Scholar]
- Leimar O., Doebeli M., and Dieckmann U., 2008. Evolution of phenotypic clusters through competition and local adaptation along an environmental gradient. Evolution 62: 807–822. 10.1111/j.1558-5646.2008.00334.x [DOI] [PubMed] [Google Scholar]
- Lenormand T., 2002. Gene flow and the limits to natural selection. Trends Ecol. Evol. 17: 183–189. 10.1016/S0169-5347(02)02497-7 [DOI] [Google Scholar]
- Martin G., and Gandon S., 2010. Lethal mutagenesis and evolutionary epidemiology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1953–1963. 10.1098/rstb.2010.0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszéna G., Czibula I., and Geritz S., 1997. Adaptive dynamics in a 2-patch environment: a toy model for allopatric and parapatric speciation. J. Biol. Syst. 5: 265–284. 10.1142/S0218339097000175 [DOI] [Google Scholar]
- Metz J. A. J., Nisbet R. M., and Geritz S. A. H., 1992. How should we define ‘fitness’ for general ecological scenarios? Trends Ecol. Evol. 7: 198–202. 10.1016/0169-5347(92)90073-K [DOI] [PubMed] [Google Scholar]
- Mirrahimi, S., 2011 Concentration phenomena in PDEs from biology. PhD thesis, University of Pierre et Marie Curie, Paris, France. [Google Scholar]
- Mirrahimi S., 2013. Migration and adaptation of a population between patches. Discrete Continuous Dyn. Syst. Ser. B 18: 753–768. 10.3934/dcdsb.2013.18.753 [DOI] [Google Scholar]
- Mirrahimi S., 2017. A Hamilton-Jacobi approach to characterize the evolutionary equilibria in heterogeneous environments. Math. Models Methods Appl. Sci. 27: 2425–2460. 10.1142/S0218202517500488 [DOI] [Google Scholar]
- Mirrahimi S., Perthame B. and Souganidis P. E., 2015. Time fluctuations in a population model of adaptive dynamics. Annales de l’I.H.P. Analyse non linéaire 32: 41–58. 10.1016/j.anihpc.2013.10.001 [DOI] [Google Scholar]
- Nagylaki T., 1978. A diffusion model for geographically structured populations. J. Math. Biol. 6: 375–382. 10.1007/BF02463002 [DOI] [PubMed] [Google Scholar]
- Perthame B., and Barles G., 2008. Dirac concentrations in Lotka-Volterra parabolic PDEs. Indiana Univ. Math. J. 57: 3275–3302. 10.1512/iumj.2008.57.3398 [DOI] [Google Scholar]
- Rice S. H., 2004. Evolutionary theory: mathematical and conceptual foundations, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- Ronce O., and Kirkpatrick M., 2001. When sources become sinks: migration meltdown in heterogeneous habitats. Evolution 55: 1520–1531. 10.1111/j.0014-3820.2001.tb00672.x [DOI] [PubMed] [Google Scholar]
- Sanjuán R., Codoñer F. M., Moya A., and Elena S. F., 2004. Natural selection and the organ-specific differentiation of hiv-1 v3 hypervariable region. Evolution 58: 1185–1194. 10.1111/j.0014-3820.2004.tb01699.x [DOI] [PubMed] [Google Scholar]
- Sanjuán R., Nebot M., Chirico N., Mansky L., and Belshaw R., 2010. Viral mutation rates. J. Virol. 84: 9733–9748. 10.1128/JVI.00694-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen O., Lascoux M., and Merilä J., 2013. Ecological genomics of local adaptation. Nat. Rev. Genet. 14: 807–820. 10.1038/nrg3522 [DOI] [PubMed] [Google Scholar]
- Slatkin M., 1978. Spatial patterns in the distributions of polygenic characters. J. Theor. Biol. 70: 213–228. 10.1016/0022-5193(78)90348-X [DOI] [PubMed] [Google Scholar]
- Szilágyi A., and Meszéna G., 2009. Two-patch model of spatial niche segregation. Evol. Ecol. 23: 187–205. 10.1007/s10682-007-9212-6 [DOI] [Google Scholar]
- Whitlock M. C., 2015. Modern approaches to local adaptation. Am. Nat. 186: S1–S4. 10.1086/682933 [DOI] [PubMed] [Google Scholar]
- Yeaman S., and Guillaume F., 2009. Predicting adaptation under migration load: the role of genetic skew. Evolution 63: 2926–2938. 10.1111/j.1558-5646.2009.00773.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No biological data are provided in this article. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11365982.