Abstract
Gastrulation is fundamental to the development of multicellular animals. Along with neurulation, gastrulation is one of the major processes of morphogenesis in which cells or whole tissues move from the surface of an embryo to its interior. Cell internalization mechanisms that have been discovered to date in Caenorhabditis elegans gastrulation bear some similarity to internalization mechanisms of other systems including Drosophila, Xenopus, and mouse, suggesting that ancient and conserved mechanisms internalize cells in diverse organisms. C. elegans gastrulation occurs at an early stage, beginning when the embryo is composed of just 26 cells, suggesting some promise for connecting the rich array of developmental mechanisms that establish polarity and pattern in embryos to the force-producing mechanisms that change cell shapes and move cells interiorly. Here, we review our current understanding of C. elegans gastrulation mechanisms. We address how cells determine which direction is the interior and polarize with respect to that direction, how cells change shape by apical constriction and internalize, and how the embryo specifies which cells will internalize and when. We summarize future prospects for using this system to discover some of the general principles by which animal cells change shape and internalize during development.
Keywords: apical constriction, C. elegans, cell polarity, cell shape, gastrulation, morphogenesis, WormBook
Gastrulation is studied in Caenorhabditis elegans because the topic involves major unsolved and partially solved problems that are of wide interest in cell and developmental biology; problems including how cells polarize with respect to external cues, how cell fates direct cell behaviors, how motors are locally controlled in specific cells and specific parts of those cells, how cytoskeletal networks dynamically connect to cell junctions, and how cells integrate all of this information to change shape in a complex, in vivo context. Each of these questions is of strong interest in cell and developmental biology.
The ability to study this diversity of interesting biological questions in C. elegans, a model organism with a well-stocked toolbox of experimental approaches, including straightforward gene manipulation and excellent microscopy in vivo, makes C. elegans gastrulation an attractive model. Because the various questions of interest intersect with each other, the system promises a richly integrated understanding of complex biology.
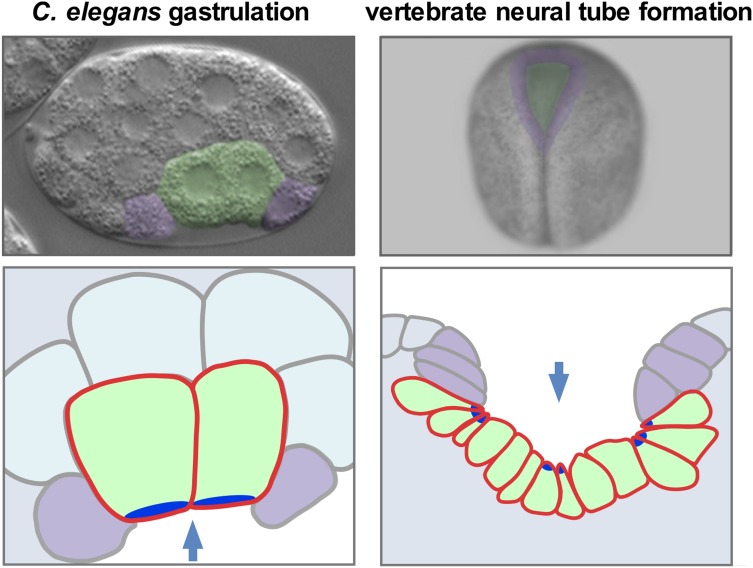
Gastrulation is an example of morphogenesis, which involves the integration of spatial and temporal patterning mechanisms of development to deploy cytoskeletal, force-producing mechanisms with precision: development biologically tells the cell what to do, where, and when. The exploration of specific links between the spatial and temporal patterning mechanisms and force-producing mechanisms is of interest, in part, because it can shed light on how animals are shaped. In addition, the use of a genetic model system to study how cells polarize and then internalize from an embryo’s surface has the potential to shed light on mechanisms that drive analogous morphogenetic events, such as mammalian neural tube formation, an especially error-prone process that has critical implications for human health [see Nikolopoulou et al. (2017) for review]. Neural tube formation in vertebrates depends on apical constriction (as does C. elegans gastrulation; Figure 1), which contributes to the internalization of the future brain and spinal cord from an embryo’s surface. Defects in neural tube closure (spina bifida and anencephaly, for example) constitute the second-most common class of human birth defects, resulting in significant suffering and monetary costs: ∼$1 billion in total in hospitalization costs for all US patients each year (Christianson et al. 2006; Arth et al. 2016). In addition, some treatments to reduce neural tube defects have been demonstrated in mice to be gene-specific. For example, folic acid is beneficial in some genetic backgrounds, but it is strongly detrimental in others (Greene and Copp 2005; Marean et al. 2011). Therefore the use of genetic model systems to identify key genes, and to dissect basic mechanisms by which cells change shape and internalize from an embryo’s surface, may have long-term benefits regarding the diagnosis, treatment, and tailoring of treatments to specific genetic conditions.
Figure 1.
C. elegans gastrulation and vertebrate (Xenopus) neural tube formation. Photos (A and B) and cross-section diagrams (C and D)) of C. elegans and Xenopus embryos, with internalizing cells (just the endodermal precursor cells in C. elegans and neural plate cells in Xenopus) colored green. Apical neighbors that are represented in the cross-section diagrams are colored purple. Arrows mark the direction of internalization in the views shown. Bright blue represents the apical parts of cells that undergo apical constriction. Modified from Sullivan-Brown et al. (2016).
In this review, we describe the cell dynamics associated with gastrulation and address some of the questions that research has begun to answer: how gastrulating cells determine which way is in, how cells change shape, and how it is determined which cells will gastrulate and when. Results from C. elegans have identified mechanisms that may be widely used beyond this model system. As with neural tube formation in vertebrates, many genes have been identified as contributing to gastrulation in C. elegans, only a small proportion of which have been used to date to identify mechanisms. We identify some promising future areas of research based on the questions of interest above, the large number of involved genes identified to date, and the amenability of the system to diverse experimental tools.
Summary of Gastrulation Events in C. elegans
C. elegans gastrulation has been traced at the level of individual cells. These cells internalize in a stereotypical pattern, i.e., in a pattern that is essentially invariant between individual embryos. Nearly all cells that will form the interior of the worm are derived from precursor cells that originate at the surface of the embryo and internalize during gastrulation. These cells include precursors of the entire endoderm, germline, muscles, and pharynx, and many of the neurons (Sulston et al. 1983; Nance and Priess 2002; Harrell and Goldstein 2011). Note that we define gastrulating cells as cells that internalize from the embryo’s surface before embryonic cell divisions are complete, distinguishing gastrulation from the later internalization of certain postmitotic cells, for example during ventral enclosure (Chisholm and Hardin 2005; Harrell and Goldstein 2011).
Given the essentially invariant cell lineage of C. elegans, each cell that gastrulates can be identified in any given embryo before it begins to move, and then the cell can be followed throughout its internalization. Sixty-six cells have been identified to internalize during C. elegans gastrulation, starting with the two precursors of the entire endoderm at the 26–28-cell stage, continuing with germline and mesodermal precursors, and finally with neuronal precursors around the 300-cell stage (Sulston et al. 1983; Nance and Priess 2002; Harrell and Goldstein 2011). Most of these cells internalize from positions along the embryo’s ventral surface (Figure 2) and together form a continuous ventral stripe, although the stripe does not internalize all at once as occurs in Drosophila and some other animals (Stern 2004). Instead, groups of cells—often just pairs—internalize at many distinct, stereotypical times (Figure 2). The space that the cells move into is in general populated by other cells, rather than by a fluid-filled blastocoel as in some animal embryos including certain nematodes (Stern 2004; Schulze and Schierenberg 2011). Only small blastocoel spaces form in C. elegans, together adding up to just 2% of the embryo volume (Nance and Priess 2002). For this reason, in still images, gastrulating cells are only evident when cell identities are labeled to make apparent where the precursors of ultimately internalized tissues lie (Figure 3).
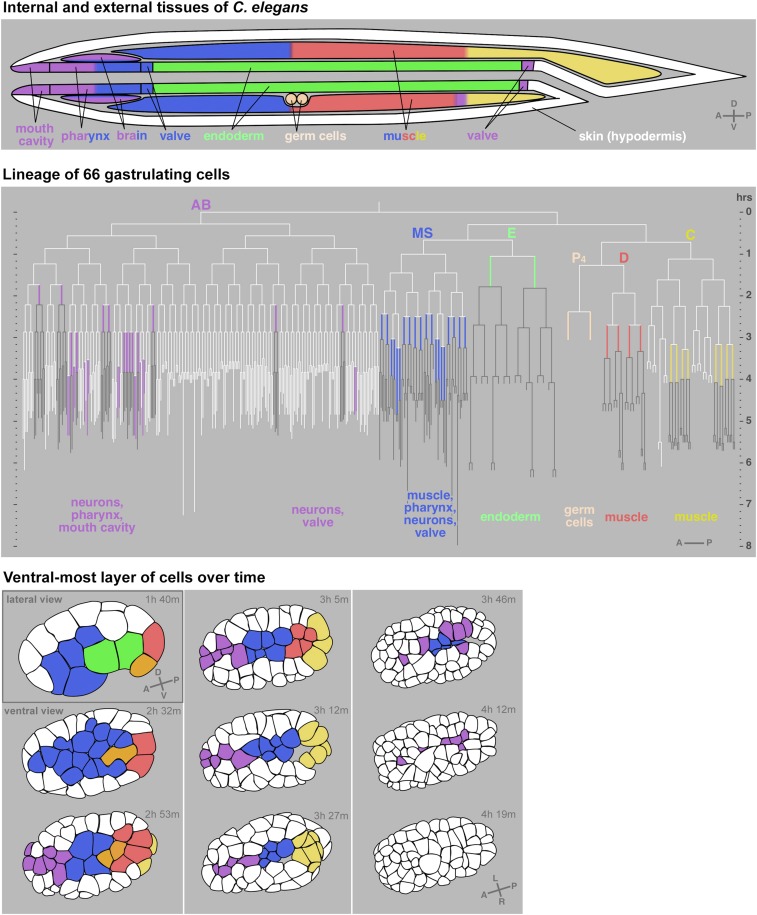
Figure 2.
Movement of C. elegans cells from the surface to the interior during gastrulation. Diagram of internal and external tissues of C. elegans at the end of embryogenesis (top). Lineage with all 66 cells identified to gastrulate marked in colors and their cell fates indicated (middle). Horizontal lines are cell divisions and vertical lines are cells. White lines are exterior cells; colored lines are gastrulating cells; and the progeny of gastrulating cells, most or all of which are interior, are in gray. First nine rounds of embryonic cell divisions are drawn to a total of 409 cells, based on cell division timing data from WormBase release WS170. Tracings of one plane of an embryo over time depicting the ventral surface cells at all but the first timepoint (bottom). Times are marked in hours and minutes after the one-cell-stage division. Some anterior AB lineage-derived cells are not shown because they internalize from the side not shown in the embryos at the bottom. Progenitors of cells that will internalize are also colored, except in lineages that produce some external cells (AB and C lineages), which for clarity are left white until the last or second-to-last cell cycle before internalization in the AB lineage. Gray letters in lower right of some illustrations indicate axes: A (anterior) and P (posterior), D (dorsal) and V (ventral), and L (left) and R (right). White represents cells on the exterior and cells that internalize are marked in color. For movie, see https://youtu.be/BaV63cLO1Tg. Modified from Harrell and Goldstein (2011), which can be referred to for more detailed information.
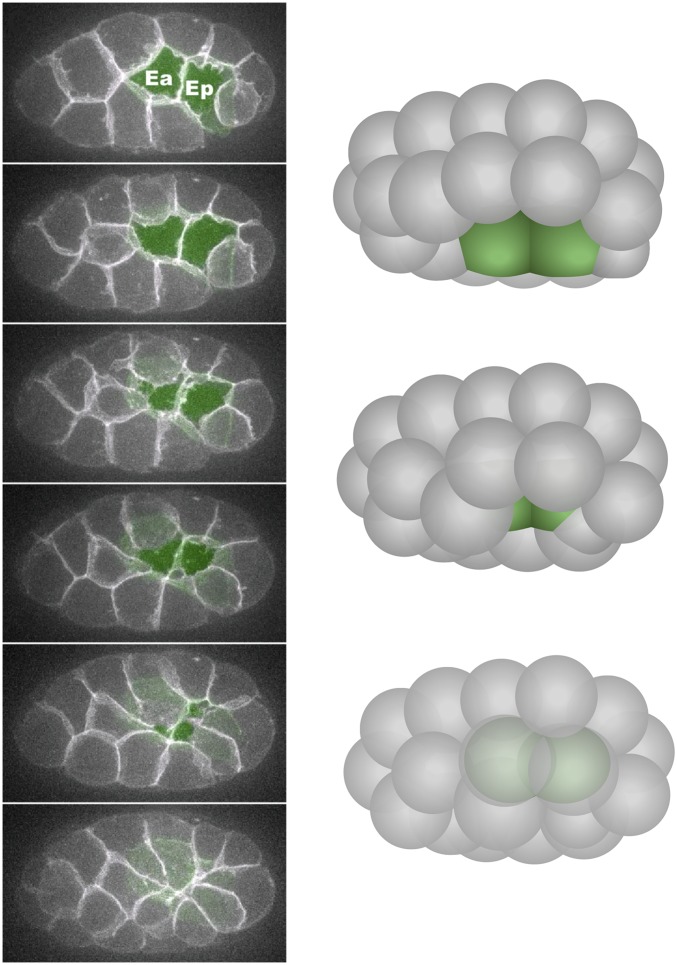
Figure 3.
Endoderm internalization. Left: six frames from a film showing narrowing of the apical surfaces of the endodermal precursor cells (Ea and Ep) from a ventral view (projection of 10 ventral 1-µm z-planes at each timepoint, with endodermal precursor cells false-colored green in each plane). Modified from Roh-Johnson et al. (2012). Right: three-dimensional illustrations of three stages from a ventro–lateral view.
Among the 66 internalizing cells, the two endodermal precursors have been studied most thoroughly because they internalize at an early stage, when cells are relatively large, allowing the dynamics of subcellular components to be resolved by imaging in vivo. During a 15–20-min period starting at the 26-cell stage, the two endodermal precursor cells move from the embryo’s surface to its interior (Figure 3), and a ring of six neighboring cells seals the gap left behind at the embryo’s surface. The mechanisms driving endoderm precursor cell internalization are addressed in a separate section below.
No one stage n C. elegans is generally referred to as the gastrula stage because gastrulation occurs over a long period, during which other events like cell fate specification and cell migrations are occurring. This feature, of multiple cells with different fates internalizing at different times, has resulted in C. elegans gastrulation being used as a model for multiple biological questions. For example, the study of endoderm internalization has unveiled unexpected subcellular dynamics as cells prepare to change shape by apical constriction (Roh-Johnson et al. 2012), and the germline internalizes in a different manner, taking advantage of a conserved physical association with the endoderm (Chihara and Nance 2012). Mechanisms used by these cells and other cells are discussed further below.
Gastrulation appears to play a large part in moving cells toward their final positions in the worm. An eventual outer layer of cells, deriving mostly from the AB lineage in the anterior part of the embryo, originates largely in the anterior side of the embryo and covers an ever-larger portion of the embryo as gastrulation occurs (white cells in Figure 2). After cells internalize, the relative positions of internalizing cell lineages are, for the most part, fixed [with some notable exceptions of long-range migrations described by Sulston et al. (1983)] through the rest of the development of the animal (Figure 2).
The thorough descriptions of gastrulation movements, at the level of individual, identifiable cells that internalize at distinct times, form a platform for dissecting the mechanistic bases of these movements in a model system.
How Gastrulating Cells Determine Which Way Is in: Cell Polarization with Respect to the Outer/Inner Embryo Axis
Gastrulation movements are directional in that cells move from the embryo’s surface toward its interior. The direction of these cell movements can be traced to an earlier polarization of the cytoskeletal machinery. Prior to gastrulation, each cell in the embryo has a contact-free surface that faces the eggshell (the apical side of each cell) and contacted surfaces, touching adjacent cells (the basolateral sides). As we describe in more detail below, nonmuscle myosin II (referred to as just “myosin” hereafter) accumulates at the apical surface of endodermal precursors and generates forces that drive these cells inward. How is this cytoskeletal asymmetry achieved? An early clue came from the observation that several PAR polarity proteins develop apical–basal asymmetries in cells prior to gastrulation. PAR proteins were originally identified for their roles in polarizing the one-cell embryo along its anterior–posterior axis [reviewed in Goldstein and Macara (2007) and Lang and Munro (2017)]. In response to sperm-derived cues, PAR-3, PAR-6, and PKC-3 accumulate in the anterior cortex, whereas PAR-1 and PAR-2 accumulate at the posterior cortex. Both groups of proteins signal to a variety of downstream effectors to generate asymmetries within the cell. During the four-cell stage, the axis of PAR protein asymmetry changes when PAR-6 becomes enriched at apical (external) surfaces (Figure 4) together with PAR-3 and PKC-3, and PAR-1 and PAR-2 become enriched at cell contacts (Etemad-Moghadam et al. 1995; Guo and Kemphues 1995; Boyd et al. 1996; Hung and Kemphues 1999; Nance and Priess 2002). Removing eggshells and placing pairs of embryos in contact revealed that the cue for apical–basal PAR asymmetries is cell–cell contact, rather than an external source such as the eggshell (Nance and Priess 2002). Depleting PAR-3 or PAR-6 from early embryonic cells after polarization of the one-cell embryo, but prior to gastrulation, resulted in slowed endodermal cell internalization, suggesting that these proteins contribute partially redundantly to gastrulation (Nance et al. 2003). Myosin failed to enrich apically in PAR-3-depleted endodermal cells, suggesting that PAR proteins function at least in part by translating cell contact cues into an apical–basal polarization of myosin that is used for internalization. Cells were also less tightly packed together following PAR-3 or PAR-6 depletion, suggesting a role for PAR proteins in regulating cell adhesion.
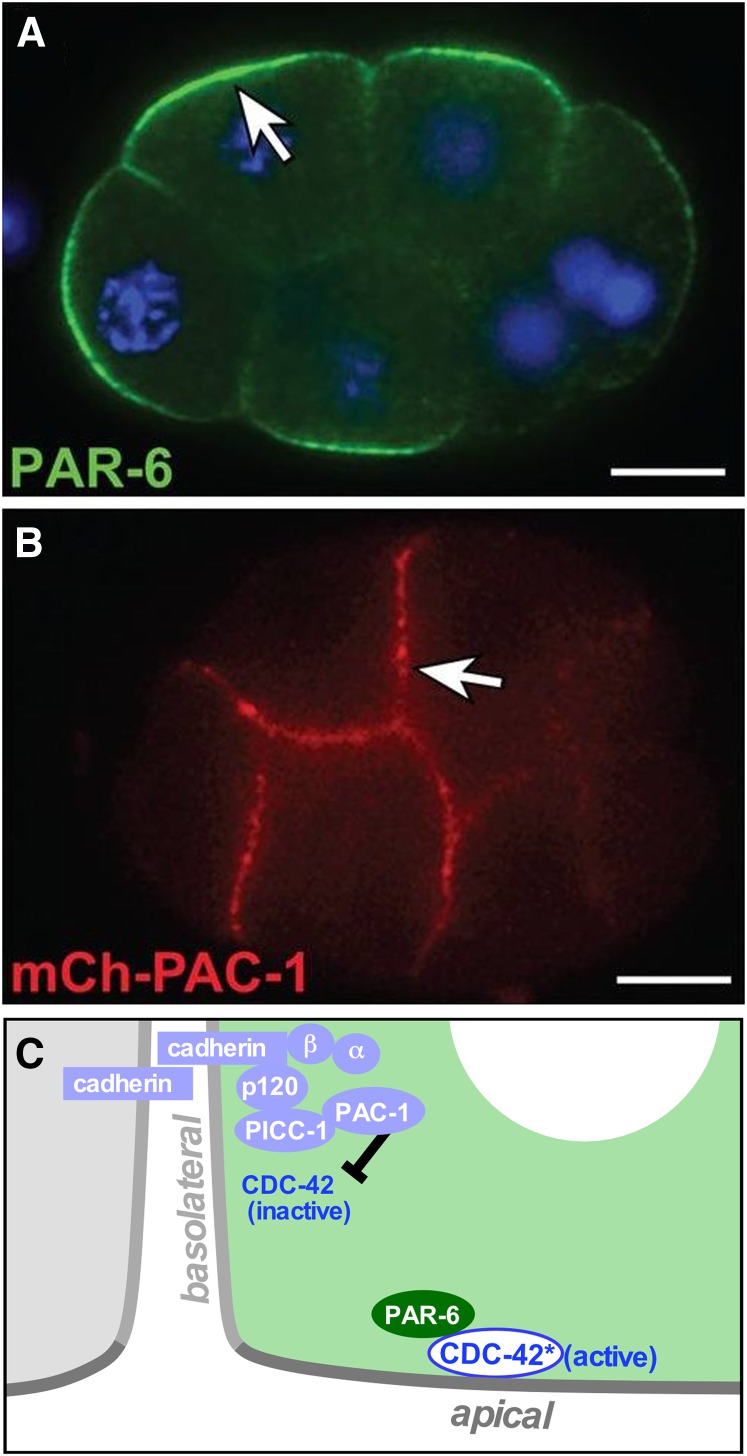
Figure 4.
Apical–basal cell polarization. (A) Anti-PAR-6 localization to apical domains in an eight-cell embryo. (B) mCherry-PAC-1 localization to basolateral domains in an eight-cell embryo. (C) Model discussed in text. Bar, 10µm. Micrographs are from Klompstra et al. (2015).
How do cell contacts lead to PAR protein asymmetry? In diverse types of polarized cells, PAR protein localization is regulated by the Rho family GTPase Cdc42, a signaling protein that cycles between active GTP-bound and inactive GDP-bound states [reviewed in Pichaud et al. (2019)]. C. elegans CDC-42 localizes to both apical and basolateral surfaces of early embryonic cells, and two RhoGEF (Rho-guanine nucleotide exchange factor) proteins that activate CDC-42 (ECT-2 and CGEF-1) show a similar, unpolarized localization (Chan and Nance 2013). However, a Rho-GTPase-activating protein family protein that can negatively regulate CDC-42, called PAC-1/ARHGAP21, is recruited by cell contacts to basolateral surfaces (Anderson et al. 2008) (Figure 4). PAR polarity depends on CDC-42 and its negative regulator PAC-1; apical–basal PAR polarity fails to develop in embryos expressing constitutively active CDC-42, in pac-1 mutant embryos, and in embryos in which PAC-1 is forced to localize to all cell surfaces (Anderson et al. 2008; Klompstra et al. 2015). These findings suggest that PAC-1 breaks CDC-42’s symmetry within cells by concentrating at cell contacts where it locally inactivates CDC-42, leaving CDC-42 active at apical surfaces where it is able to recruit PAR proteins. PAR-6 can bind directly to active CDC-42 through its semi-CRIB (Cdc42/Rac-interactive binding) domain, which is required for PAR-6 apical enrichment (Anderson et al. 2008). As we describe below, CDC-42 can also recruit another CRIB domain protein that is critical for apical myosin activation, providing a direct link between active CDC-42 at the apical surface and local cytoskeletal functions that drive directed cell movements.
A remaining question that has been partially answered is how cell contact results in the local recruitment of PAC-1. Adhesion proteins provide a conceptually appealing link between cell contacts and PAC-1 recruitment; such proteins could enrich at cell contacts through stable homophilic or heterophilic interactions with partners on contacting cells, in turn recruiting PAC-1 to cell contact sites through direct or indirect interactions with their cytoplasmic tail. Among two adhesion proteins that have been implicated in gastrulation (Grana et al. 2010), one protein, HMR-1/E-cadherin, plays just such a role (Klompstra et al. 2015) (Figure 4). HMR-1 enriches at contacts through homophilic interactions and recruits PAC-1 through the p120 catenin JAC-1, which binds directly to the HMR-1 cytoplasmic tail. In turn, JAC-1 recruits the linker protein PICC-1/CCDC85A-C, which binds the N-terminal domain of PAC-1. JAC-1’s role in PAC-1 localization is partially redundant; the α-catenin HMP-1 also plays a role in PAC-1 localization. Moreover, all of these HMR-1-dependent mechanisms appear redundant with other mechanism(s): whereas HMR-1 that is forced to ectopically localize at apical surfaces is sufficient to recruit PAC-1, HMR-1 is not necessary for PAC-1 localization to cell contacts, indicating the involvement of additional HMR-1-independent PAC-1 recruitment mechanisms that remain to be identified. Together, these results implicate an E-cadherin and as-yet-unidentified player(s) in positioning a negative regulator of CDC-42 at cell–cell contacts, providing apical–basal positional information to cells in the form of apically enriched CDC-42 activity (Figure 4). The results also highlight multiple redundant mechanisms, which could feasibly enhance the robustness with which gastrulating cells become polarized.
How Cells Change Shape: Local Deployment of Forces that Drive Movement
The internalization of endodermal precursor cells has been seen to occur after the eggshell is removed from C. elegans embryos, opening up the study of gastrulation mechanisms in a genetic model system to experiments involving the direct manipulation of cells (Lee and Goldstein 2003), as has been successful historically in other systems, like amphibian embryos (Keller et al. 2003). This ability to use such techniques of experimental embryology is unusual among genetic model organisms. After isolating cells of C. elegans embryos in culture, even a single file of cultured cells bends at the time when gastrulation would normally occur, mimicking the cell rearrangements of gastrulation but in a minimal system (Lee and Goldstein 2003). This result suggests that a major cell movement of gastrulation does not require large numbers of cells working in concert in complex three-dimensional topology, pointing us instead toward mechanisms like apical constriction or cell crawling, which depend only on small numbers of cells contacting each other, driving gastrulation (Lee and Goldstein 2003). Cell manipulation experiments also argued against roles for hypothesized chemotactic cues released by neighbors of the endodermal precursor cells to guide their movement toward each other (Lee and Goldstein 2003).
One of the earliest-observed structures suggesting how cells might move in C. elegans gastrulation was a set of cellular extensions found on a specific set of cells. Light and electron microscopy of gastrulating embryos revealed that three cells that neighbor the endodermal precursors make extensions across the outer, apical surface of the endodermal precursors (Nance and Priess 2002). These extensions are rich in filamentous actin and their formation depends on Arp2/3 (Roh-Johnson and Goldstein 2009). In these respects, the extensions on these cells resemble lamellipodia, critical force-producing structures that drive the crawling of cells in diverse biological systems (Insall and Machesky 2009; Fritz-Laylin et al. 2018). The extensions may be induced by the internalizing cells in their neighbors, because in experiments in which ectopic cells are made to internalize, ectopic neighboring cells also make at least superficially similar extensions (Pohl et al. 2012). However, it seems unlikely that the extensions on these cells are indicators of cell crawling because the extensions are short (∼1–2 µm across a roughly 13-µm opening) and highly transient (lasting for only seconds, not minutes at a time) through much of gastrulation (Roh-Johnson and Goldstein 2009). Perhaps most critically, the extensions do not overtake fiducial marks in the cortex of the underlying endodermal precursors (Roh-Johnson et al. 2012) as the leading edge of a crawling cell would do. It is possible that, instead, the actin-rich extensions are specializations that might prime rapid sealing of the ring of cells, much as filopodia have been proposed to do at a later stage in C. elegans development (Raich et al. 1999), although from just one side of the closing ring of cells (the side where these three cells lie). On the opposite side of the ring, the germline precursor cell has been observed to produce blebs in the direction in which it moves, and it has been proposed that blebbing-based motility could contribute to the movement of the germline precursor across its neighboring endodermal precursor (Pohl et al. 2012). To date, it has not been possible to eliminate the extensions nor germline precursor cell blebs without also disrupting the embryo more generally (Severson et al. 2002; Roh-Johnson and Goldstein 2009), so whether either form of cellular protrusions has any critical functions in gastrulation is not yet clear.
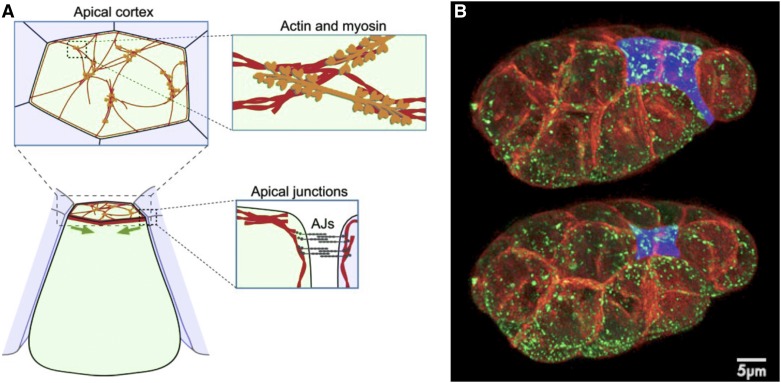
Myosin localization has provided a clue that, in retrospect, has been more telling than the extensions described above. Actin filaments are required for gastrulation (Lee and Goldstein 2003) and myosin is concentrated in the endodermal cells’ apical cortex (Nance and Priess 2002). This pattern raises the possibility that endodermal cells might internalize by actomyosin-driven shrinking of apical surfaces, i.e., by apical constriction (Figure 5). Apical constriction drives morphogenesis in a diversity of other animals, and in principle, constriction of the apical surfaces of cells could be sufficient to internalize endodermal precursor cells by pulling other cells into contact over the surface of apically constricting cells [for review, see Sawyer et al. (2010)].
Figure 5.
Actomyosin, apical junctions, and apical constriction. (A) Components involved in apical constriction include F-actin (red) and nonmuscle myosin II (orange), which form contractile networks. Network is shown at sparse density for purpose of illustration. Shrinkage of the apical cortex (green arrows) is driven by contraction of apical actin–myosin networks linked to apical adherens junctions (AJs, gray), resulting in tissue shape changes. Modified from Martin and Goldstein (2014). (B) Bessel beam structured plane illumination images of an embryo at two timepoints (5 min, 40-sec apart) during endodermal precursor cell internalization, with membranes in red, myosin in green, and apical surfaces of endodermal precursor cells false-colored blue. From Roh-Johnson et al. (2012).
Despite evidence that apical constriction drives such critical events of morphogenesis as gastrulation or neurulation in diverse organisms (Sawyer et al. 2010), and the fact that apical constriction was proposed to drive cell shape change as early as 1902 (Rhumbler 1902), over the next century, to our knowledge the apical sides of cells were not demonstrated directly to constrict in any system. Rather, it remained possible that the reduction in apical area observed in cells of diverse organisms was caused not by the constriction of apical cell surfaces but by the simple bending of the apical membrane and cortex into basolateral regions, perhaps associated with sliding of junctional complexes. The constriction of the apical surface was first demonstrated directly by observing the movements of fluorescent beads or fluorescent Quantum Dots placed onto the apical surfaces of C. elegans endodermal precursor cells. These fluorescent markers were seen to converge near the center of the apical surface of each cell and/or near the apical sites of contact between cells, demonstrating that the reduction in apical surface is indeed accompanied by a constriction of that surface (Lee and Goldstein 2003; Roh-Johnson et al. 2012). Together with observations that plasma membrane marks move in concert with myosin (Roh-Johnson et al. 2012), these results suggest that apical constriction involves concerted centripetal contraction of the actomyosin cortex along with associated plasma membrane and any surface glycocalyx.
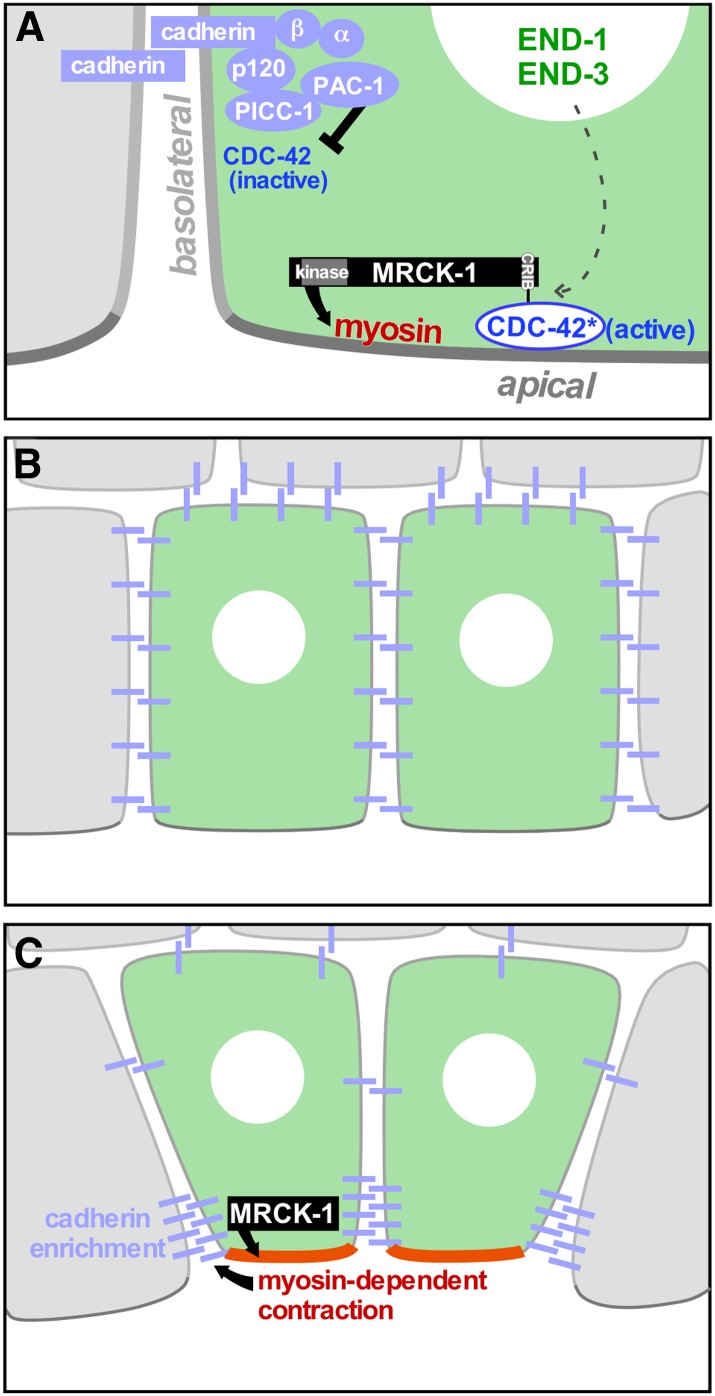
Myosin function has been implicated in apical constriction in C. elegans embryos by experiments disrupting myosin activity and by visualization of activated myosin enriched in the apical cortex of apically constricting cells (Lee and Goldstein 2003; Lee et al. 2006; Harrell and Goldstein 2011). Myosin enrichment in the apical cortex (Nance and Priess 2002) is accompanied by phosphorylation of a conserved epitope in the myosin regulatory light chain (Lee et al. 2006). Phosphorylation of this epitope on myosins in diverse systems suggests that apical myosin is active, and likely to be forming bipolar filaments that can bind to and walk on actin filaments (Bresnick 1999). The apical accumulation and activation of myosin are dependent on signaling cues, the study of which has suggested the outlines of a model by which embryonic patterning results in precise local upregulation of contractile forces, i.e., in the apical cortex of specific cells and at specific stages (Lee et al. 2006; Grana et al. 2010; Marston et al. 2016). In this model, a kinase required for phosphorylation of the regulatory light chain on the apical side of endodermal precursor cells, the CRIB domain-containing kinase MRCK-1, appears to serve as a hub that integrates spatial and temporal patterning information (Marston et al. 2016). At the 26-cell stage, MRCK-1 is recruited apically by active CDC-42, specifically in certain cells (discussed below). MRCK-1 is required for contractile actomyosin dynamics. Laser microsurgery experiments have demonstrated that, consistent with a role in activating myosin, MRCK-1 is also required to locally increase tension in the apical cortex of these cells (Marston et al. 2016). Hence the apical–basal polarity that derives from cell contacts is translated into myosin becoming activated specifically at the contact-free, apical cell surfaces; these surfaces are recognized via the homotypic adhesion protein cadherin localizing to cell–cell contacts and contributing to the local recruitment of PAC-1. PAC-1 locally inactivates CDC-42 at cell–cell contact sites (i.e., basolaterally), and then the active, apical CDC-42 recruits MRCK-1, which in turn activates myosin (Figure 6).
Figure 6.
Model for how the force-producing mechanisms that drive apical constriction are spatially regulated. (A) Active CDC-42, restricted to apical membranes by basolateral inhibition (black inhibitory arrow), recruits MRCK-1 apically via MRCK-1’s Cdc42/Rac-interactive binding domain. MRCK-1 activates myosin and increases tension in the apical cortex. MRCK-1 apical enrichment occurs specifically in the two endodermal precursor cells, dependent on the END-1/END-3 transcription factors (dotted arrow) by unknown mechanisms. (B and C) MRCK-1-dependent myosin activity contributes through as-yet-unexplored mechanisms to junctional cadherin enrichment. Modified from Marston et al. (2016).
Shortly before apical constriction begins, HMR-1/cadherin localization becomes refined from a basolateral pattern to a classical, junctionally enriched pattern typical of epithelial cells (Marston et al. 2016). This relocalization of cadherin depends on MRCK-1 and on myosin activity, suggesting a role for contraction of the apical actomyosin network in cadherin relocalization in this system, as in other systems (Dawes-Hoang et al. 2005; Marston et al. 2016; Weng and Wieschaus 2016) (Figure 6).
Once myosin is localized and activated under the contact-free, apical surface of specific cells, it might seem obvious that the resulting contraction of the apical actomyosin cortex would pull on cell–cell junctions and hence shrink a cell’s apical surface. Indeed, the plasma membrane overlying the apical cortex has been demonstrated to move in concert with the cortex (Roh-Johnson et al. 2012). However, live-embryo microscopy of cytoskeletal and membrane dynamics revealed that the apical actomyosin cortex contractions occur at first in a conveyer belt-like fashion, with actomyosin flowing centripetally from junctions to near the center of the apical surface, but failing to shrink the apical surface for several minutes (Roh-Johnson et al. 2012). During this period, the apical actomyosin network contracts and turns over at the same time, with new myosin particles appearing near the edge of the apical network (at apical junctions) and streaming centripetally. This observation has suggested that the trigger for cell shape change in apical constriction cannot be the activation of cortical contraction, and instead is likely to be the engagement of an as-yet-unidentified molecular clutch that connects this continually contracting actomyosin network to cadherin-containing junctions (adherens junctions). It is possible that these unexpected dynamics are a general feature of apical constriction in diverse organisms, because similar dynamics have been seen at a stage when weak actomyosin contractions occur before cells begin to apically constrict in Drosophila (Roh-Johnson et al. 2012). These findings have focused the search for triggers of apical constriction to mechanisms that can connect cytoskeletal networks to cell–cell junctions (Razzell and Martin 2012; Chanet and Martin 2014; Takeichi 2014; Blanchard et al. 2018).
Why actomyosin contractions would occur without any apparent productive pulling in of junctions at first is not clear. The conveyer belt-like contraction behavior of the actomyosin cortex occurs for long enough to remodel the entire apical cortex multiple times over (Roh-Johnson et al. 2012), much more than might be expected to be needed to take up any possible slack in an actomyosin network. It is possible that the early contractions serve to concentrate cadherin and other junctional components, although the observed early actomyosin contractions occur largely after junctional molecules have been observed in apically enriched patterns (Marston et al. 2016). Alternatively, it is possible that the early contractions serve another function, contributing to cell polarization before being used to constrict the apical cell surface; apical actomyosin flows have been implicated in moving PAR proteins to near the center of cells’ apical surfaces (Munro et al. 2004).
Which Cells Will Gastrulate and When: Cell Fate Specification and Its Links to Gastrulation
Mechanistic links must exist from the genes that specify fate to the force-producing mechanisms that can move cells, since force-producing mechanisms are deployed in specific cells at specific times. For example, although many cells show some centripetal movement of apical actomyosin before and during the 26–28-cell stage (Munro et al. 2004; Roh-Johnson et al. 2012), only in the endodermal precursor cells is MRCK-1 recruited apically, and myosin is recruited and activated apically to a much greater degree in these cells, leading to more pronounced centripetal actomyosin movement (Roh-Johnson et al. 2012; Marston et al. 2016). Similarly, in cells that internalize later, myosin is activated and recruited apically only in specific cells at any one time (Nance and Priess 2002; Harrell and Goldstein 2011). Indeed in diverse organisms, cell fate-specification genes, which are expressed in specific cell lineages and determine these lineages’ fates, are required for the timely internalization of cells (Wieschaus 1996; Sawyer et al. 2010). In C. elegans, multiple cells were recognized to internalize at times better predicted by their cell fates than by their lineal origin, as with other aspects of C. elegans development that are better predicted by cell fate than by lineage (Labouesse and Mango 1999), providing an early clue that cell fates might control cell internalization in this organism (Nance and Priess 2002). Roles for cell fate specification in gastrulation were later demonstrated in multiple experiments in which cells were transformed from one cell fate to another and effects on gastrulation were examined, as detailed below. In many cases, these cells were transformed from one gastrulating cell fate to another gastrulating fate, but cells of each fate normally gastrulate at distinct times. As expected, these fate transformations disrupted the specific, stereotypical temporal patterns of cell internalization. Specific examples are outlined below.
Cells of endoderm, mesoderm, and germline fate have each been transformed in experiments in which effects on gastrulation patterns were examined. The endodermal precursors, which are the first cells to gastrulate, are specified by the endoderm-specific GATA transcription factors END-3 and END-1 [see Maduro (2009) and McGhee (2013) for reviews]. Loss of these transcription factors results in a gastrulation defect in which endodermal precursors fail to internalize at the 26–28-cell stage (Zhu et al. 1997; Nance and Priess 2002; Maduro et al. 2005; Lee et al. 2006). In a mutant in which ectopic cells take on the endodermal fate in an end-1- and end-3-dependent manner (Mello et al. 1992; Maduro et al. 2005), these additional cells enrich MRCK-1 apically and then gastrulate soon after the normal endodermal precursors do (Lee et al. 2006; Marston et al. 2016). One aspect of endodermal precursor cells’ fate is the introduction of a gap phase in cells that otherwise rapidly cycle between mitosis and DNA synthesis (Edgar and McGhee 1988); mutant embryos that lack this gap phase have cells that divide when they should be internalizing and exhibit internalization failure (Knight and Wood 1998; Lee et al. 2006; Sullivan-Brown et al. 2016). This finding suggests that intrinsic to endodermal cell fate is the introduction of a pause in the cell cycle before internalization. This cell cycle pause and its role in permitting cell internalization is a feature of other internalizing cells, for example in Drosophila gastrulation, as well as in diverse types of cells that leave epithelia and invade through a basement membrane (Grosshans and Wieschaus 2000; Kohrman and Matus 2017).
The mesoderm in C. elegans derives from multiple cell lineages, including the MS lineage. The MS lineage (Figure 2) develops its unique pattern of cell fates dependent on the T-box transcription factor TBX-35 and the NK-2 homeodomain transcription factor CEH-51 (Broitman-Maduro et al. 2009). Deletion of the genes encoding these two proteins results in the one cell lineage behaving like another (the MS lineage behaving like a C lineage; Broitman-Maduro et al. 2009), and the normally invariant pattern of gastrulation is disrupted in a manner consistent with this fate transformation (Harrell and Goldstein 2011). Similarly, transforming the germline precursors to a somatic cell fate disrupts the normal pattern of gastrulation (Harrell and Goldstein 2011).
Gastrulation is controlled by different cell fate-specification mechanisms in different cells, and even different cell polarity mechanisms, since PAR proteins that specify the polarity of endodermal precursors do not have an important function in later-internalizing lineages (Nance et al. 2003; Harrell and Goldstein 2011). Despite the varied cell fate and cell polarity inputs, common cytoskeletal mechanisms appear to be used by diverse cells: myosin becomes enriched and activated in the apical cortex of later-internalizing cells, and the disruption of myosin activity using a conditional mutant prevents most cells from internalizing (Nance and Priess 2002; Harrell and Goldstein 2011). To date, only the germline precursors are known to gastrulate by a distinct mechanism from that found in other cells. The germline precursors at the embryo’s surface adhere to already-internalized endodermal precursors, and when the endodermal precursors partially envelop the germline precursors and move dorsally as a layer of more dorsal cells thins through divisions, the germline precursors appear to be internalized by “hitchhiking” on these endodermal precursor cell movements (Chihara and Nance 2012). The thinning and spreading of the surface layer of cells, via cell divisions in the plane of the embryo’s surface, might contribute more generally throughout gastrulation by contributing compressive forces in the surface layer, much as has been described during epiboly in other organisms (Trinkaus 1969; Pohl et al. 2012). The distinct gastrulation strategy employed by germline precursors may reflect the fact that they are transcriptionally quiescent (Seydoux and Fire 1994; Seydoux et al. 1996), and therefore cannot induce gastrulation movements using the transcriptional triggers that are important in endodermal and mesodermal lineages. Indeed, germline precursor cells require HMR-1/E-cadherin to adhere to and internalize with endodermal precursor cells, and upregulate levels of HMR-1 just prior to gastrulation using a post-transcriptional mechanism (Chihara and Nance 2012).
As yet, no targets of any of the fate-specifying transcription factors described above have been implicated in gastrulation. Endoderm fate appears both necessary and sufficient for the timely recruitment of the myosin activator MRCK-1 to a specific subcellular site (Marston et al. 2016), but the mechanisms by which endodermal transcription factors affect the myosin activator’s recruitment are, for the most part, undiscovered to date, and little is known about links from fate specification to internalization in other lineages. Such links from fate-specifying transcription factors to morphogenesis mechanisms are currently better understood in Drosophila gastrulation [for review, see Manning and Rogers (2014)]. The degree to which lessons learned from Drosophila apply widely is incompletely known. In Drosophila, gastrulation is orchestrated by two mesoderm-specifying transcription factors, and these transcription factors drive mesoderm-specific expression of both an apical RhoGEF-binding protein and an apically secreted signal/receptor pair that regulates a second apical RhoGEF-binding protein. The apically recruited RhoGEF locally activates Rho, leading to apically active Rho-kinase, which in turn activates myosin apically, contributing to the contraction of the apical sides of these cells. Drosophila gastrulation involves unusual, radial sarcomere-like arrangements of myosin and Rho-kinase (Coravos and Martin 2016), but otherwise similar themes to apical constriction in C. elegans and other systems (Martin and Goldstein 2014). Vertebrates similarly use Rho-kinase to locally activate myosin, except that it is Rho-kinase itself (rather than its upstream activator RhoGEF, as in Drosophila) that is recruited apically in specific cells, through local recruitment by a Rho-kinase-binding protein Shroom3 [for review, see Martin and Goldstein (2014)]. Further work on how fate specification controls local tissue shapes in Drosophila, C. elegans, vertebrates, and other systems is of interest toward understanding the core, conserved mechanisms, the diversity of mechanisms used in animals, and perhaps the evolutionary innovations that have shaped different animals.
Unsolved Problems
C. elegans lends itself to powerful genetic approaches for the identification of key molecules. Genetic approaches have succeeded in identifying some of the gene products that are critical for morphogenesis in diverse model organisms, but they have not done so as efficiently as they have for other processes such as cell fate specification (Wieschaus 1996). For this reason, it has been proposed that morphogenesis often depends on (1) redundant cellular mechanisms, “belt-plus-suspenders” situations in which disruption of any one cellular mechanism produces only a subtle phenotype, and (2) cellular mechanisms that are important for other processes as well, such as cell division, the disruption of which can result in embryos that fail to reach certain stages of morphogenesis (Wieschaus 1996). Therefore, the ability to detect subtle phenotypes and dissect redundancy is likely to be important for understanding the mechanisms of morphogenesis. The ability to view subtle phenotypes directly in C. elegans by microscopy of optically clear embryos as they gastrulate makes it possible to recognize subtle defects such as delayed internalization (Sawyer et al. 2011; Sullivan-Brown et al. 2016). One risk of such close observation is the identification of overly subtle phenotypes that hardly contribute to major mechanisms, but to date, the delayed internalization of C. elegans endodermal precursors has often been associated with specific cellular defects, such as decreased apical myosin activation (Lee et al. 2006) or a failure of adherens junctions to move in concert with apical actomyosin contractions (Roh-Johnson et al. 2012). Moreover, the targeting of multiple genes together has identified specific combinations that lead to complete failures of cells to internalize, with the endoderm remaining on the exterior as a terminal phenotype (Sawyer et al. 2011). This makes the identification of new gastrulation genes possible by screening for specific enhancers of sensitized backgrounds (Sawyer et al. 2011; Sullivan-Brown et al. 2016).
As with mammalian neural tube formation (Harris and Juriloff 2010), many genes have been identified as contributing to C. elegans gastrulation, i.e., live imaging of embryos after disruption of these genes has shown that normal internalization fails in some embryos. Yet many such genes—indeed in both systems, the vast majority of the contributing genes—have not yet been studied to understand specific mechanisms of interest (Sawyer et al. 2011; Sullivan-Brown et al. 2016; Nikolopoulou et al. 2017). Identifying genes is no longer a rate-limiting step in discovery, particularly in C. elegans given the ease with which RNA interference screens can be done. For both mammalian neural tube formation and C. elegans gastrulation, we view these troves of contributing genes as having strong potential for the future identification and dissection of additional mechanisms contributing to cell shape change and cell internalization in vivo.
Some genes contributing to C. elegans gastrulation have well-defined or strongly predicted biochemical roles, but are not yet tied to any specific functions in or near the cells that internalize. Examples include a Rho-associated kinase without as-yet-identified gastrulation-relevant targets (Marston et al. 2016), and predicted or validated ubiquitin ligase complex members (Sawyer et al. 2011), which may function in signaling or in the degradation of as-yet-unknown target proteins. Other proteins that contribute to C. elegans gastrulation have orthologs that contribute to vertebrate neural tube formation, including a transcription factor that affects lineage-specific expression of genes in multiple C. elegans embryonic cells and a member of the WAVE complex, which functions in actin regulation in diverse systems and exhibits tissue-enriched expression during neurulation in the neural plate of Xenopus, where apical constriction occurs (Sullivan-Brown et al. 2016). A Wnt–Frizzled signaling pathway contributes to C. elegans gastrulation at least in part through MRCK-1 localization and myosin activation, although by an as-yet-undefined cell–cell signaling route, i.e., it is unknown which cells are signaling to which, as well as whether this signaling conveys critical spatial or temporal information to gastrulating cells (Lee et al. 2006; Marston et al. 2016).
The tools for investigating how little-studied genes function in vivo have improved dramatically in recent years. Single-cell transcriptome sequencing of every cell from the one-cell stage through to the birth of endodermal precursor cells (Tintori et al. 2016) and an online resource for mining the resulting massive data set (http://tintori.bio.unc.edu) have helped to identify which cells express which genes. These resources are valuable for identifying candidate regulators of cell-specific behaviors. In addition, modern tools for cell-specific gene disruption and clustered regularly interspaced short palindromic repeats in C. elegans have opened up methods for defining precisely in which cells genes act (Dickinson and Goldstein 2016; Paix et al. 2017). Genes identified as strongly expressed by single-cell RNA sequencing (Tintori et al. 2016) are strong candidate drivers for cell-specific gene expression.
Progress in understanding gastrulation mechanisms and cell internalization mechanisms more generally in various animal models will be enhanced by meeting several current challenges. For genes that affect cell internalization, it can be challenging to distinguish molecular components that drive active changes in cell shape (for example, producing forces that drive apical constriction) from those that affect morphogenesis in other ways, for example affecting viscoelastic properties of the cells that change shape or their neighbors that provide a mechanical context that cells experience (Davidson 2017). Tools to identify the cellular site of action for each protein of interest could help in this regard, and it is increasingly possible to dissect specific roles of molecules that function in cell shape changes, for example in the timely transmission of forces from the cytoskeleton to junctions, or in the polarization of myosin activators. It is also increasingly feasible to visualize the subcellular dynamics of key proteins in vivo by tagging methods that mark complete populations of such proteins and that can be well validated as nondisruptive, for example by full rescue of viability upon tagging essential proteins at their endogenous loci (Dickinson and Goldstein 2016). To understand how cell shape changes are triggered, it will be important in some systems to know which molecules bind to which others in vivo on relevant minute-to-minute timescales. Single-cell biochemistry methods (Dickinson et al. 2017) are likely to help resolve such rapid changes. Tools for automated analysis of in vivo dynamics are improving, and the physics of intracellular, force-producing mechanisms is increasingly well understood [for example, Gross et al. (2017), Ulman et al. (2017), and Moen et al. (2019)]. In general, the recent development of tools for studying complex, in vivo cell dynamics is making it increasingly possible to discover mechanisms of interest.
Study of apical constriction intersects with some of the major themes in cell and developmental biology, including cell polarization, the control of motor activity, and dynamic control of forces, in a complex in vivo context. C. elegans gastrulation has been a fertile ground for research and is well positioned to make long-term contributions to the discovery of basic mechanisms with broad relevance to development in diverse animals, and with a potential for impacts on human health.
Acknowledgments
We thank current and past members of our laboratories who have developed ideas and made discoveries, and Allison Hall, Jeff Hardin, Jon Hibshman, Amy Maddox, Mark Slabodnick, and an anonymous reviewer for comments on the manuscript. Current work on gastrulation in our laboratories is funded by National Institutes of Health grants R01 GM-083071 and R35 GM-134838 to B.G., and R35 GM-118081 to J.N.
Footnotes
Communicating editor: G. Seydoux
Literature Cited
- Anderson D. C., Gill J. S., Cinalli R. M., and Nance J., 2008. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science 320: 1771–1774. 10.1126/science.1156063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arth A., Kancherla V., Pachón H., Zimmerman S., Johnson Q. et al. , 2016. A 2015 global update on folic acid-preventable spina bifida and anencephaly. Birth Defects Res. A Clin. Mol. Teratol. 106: 520–529. 10.1002/bdra.23529 [DOI] [PubMed] [Google Scholar]
- Blanchard G. B., Étienne J., and Gorfinkiel N., 2018. From pulsatile apicomedial contractility to effective epithelial mechanics. Curr. Opin. Genet. Dev. 51: 78–87. 10.1016/j.gde.2018.07.004 [DOI] [PubMed] [Google Scholar]
- Boyd L., Guo S., Levitan D., Stinchcomb D. T., and Kemphues K. J., 1996. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development 122: 3075–3084. [DOI] [PubMed] [Google Scholar]
- Bresnick A. R., 1999. Molecular mechanisms of nonmuscle myosin-II regulation. Curr. Opin. Cell Biol. 11: 26–33. 10.1016/S0955-0674(99)80004-0 [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G., Owraghi M., Hung W. W. K., Kuntz S., Sternberg P. W. et al. , 2009. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development 136: 2735–2746. 10.1242/dev.038307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E., and Nance J., 2013. Mechanisms of CDC-42 activation during contact-induced cell polarization. J. Cell Sci. 126: 1692–1702. 10.1242/jcs.124594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanet S., and Martin A. C., 2014. Mechanical force sensing in tissues. Prog. Mol. Biol. Transl. Sci. 126: 317–352. 10.1016/B978-0-12-394624-9.00013-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara D., and Nance J., 2012. An E-cadherin-mediated hitchhiking mechanism for C. elegans germ cell internalization during gastrulation. Development 139: 2547–2556. 10.1242/dev.079863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A. D., and Hardin J., 2005. Epidermal morphogenesis (December 1, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.35.1, http://www.wormbook.org. 10.1895/wormbook.1.35.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, A., C. P. Howson, and B. Modell, 2006 March of Dimes Global Report on Birth Defects: The hidden toll of dying and disabled children. Available at: https://www.marchofdimes.org/global-report-on-birth-defects-the-hidden-toll-of-dying-and-disabled-children-full-report.pdf. Accessed: [insert date].
- Coravos J. S., and Martin A. C., 2016. Apical sarcomere-like actomyosin contracts nonmuscle Drosophila epithelial cells. Dev. Cell 39: 346–358. 10.1016/j.devcel.2016.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L. A., 2017. Mechanical design in embryos: mechanical signalling, robustness and developmental defects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20150516. 10.1098/rstb.2015.0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes-Hoang R. E., Parmar K. M., Christiansen A. E., Phelps C. B., Brand A. H. et al. , 2005. Folded gastrulation, cell shape change and the control of myosin localization. Development 132: 4165–4178. 10.1242/dev.01938 [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., and Goldstein B., 2016. CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202: 885–901. 10.1534/genetics.115.182162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Schwager F., Pintard L., Gotta M., and Goldstein B., 2017. A single-cell biochemistry approach reveals PAR complex dynamics during cell polarization. Dev. Cell 42: 416–434.e11. 10.1016/j.devcel.2017.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar L. G., and McGhee J. D., 1988. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 53: 589–599. 10.1016/0092-8674(88)90575-2 [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B., Guo S., and Kemphues K. J., 1995. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 83: 743–752. 10.1016/0092-8674(95)90187-6 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin L. K., Lord S. J., Kakley M., and Mullins R. D., 2018. Concise language promotes clear thinking about cell shape and locomotion. Bioessays 40: e1700225 10.1002/bies.201700225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B., and Macara I. G., 2007. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell 13: 609–622. 10.1016/j.devcel.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grana T. M., Cox E. A., Lynch A. M., and Hardin J., 2010. SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Dev. Biol. 344: 731–744. 10.1016/j.ydbio.2010.05.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene N. D. E., and Copp A. J., 2005. Mouse models of neural tube defects: investigating preventive mechanisms. Am. J. Med. Genet. C. Semin. Med. Genet. 135C: 31–41. 10.1002/ajmg.c.30051 [DOI] [PubMed] [Google Scholar]
- Gross P., Kumar K. V., and Grill S. W., 2017. How active mechanics and regulatory biochemistry combine to form patterns in development. Annu. Rev. Biophys. 46: 337–356. 10.1146/annurev-biophys-070816-033602 [DOI] [PubMed] [Google Scholar]
- Grosshans J., and Wieschaus E., 2000. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell 101: 523–531. 10.1016/S0092-8674(00)80862-4 [DOI] [PubMed] [Google Scholar]
- Guo S., and Kemphues K. J., 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620. 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Harrell J. R., and Goldstein B., 2011. Internalization of multiple cells during C. elegans gastrulation depends on common cytoskeletal mechanisms but different cell polarity and cell fate regulators. Dev. Biol. 350: 1–12. 10.1016/j.ydbio.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. J., and Juriloff D. M., 2010. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res. A Clin. Mol. Teratol. 88: 653–669. 10.1002/bdra.20676 [DOI] [PubMed] [Google Scholar]
- Hung T. J., and Kemphues K. J., 1999. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development 126: 127–135. [DOI] [PubMed] [Google Scholar]
- Insall R. H., and Machesky L. M., 2009. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell 17: 310–322. 10.1016/j.devcel.2009.08.012 [DOI] [PubMed] [Google Scholar]
- Keller R., Davidson L. A., and Shook D. R., 2003. How we are shaped: the biomechanics of gastrulation. Differentiation 71: 171–205. 10.1046/j.1432-0436.2003.710301.x [DOI] [PubMed] [Google Scholar]
- Klompstra D., Anderson D. C., Yeh J. Y., Zilberman Y., and Nance J., 2015. An instructive role for C. elegans E-cadherin in translating cell contact cues into cortical polarity. Nat. Cell Biol. 17: 726–735. 10.1038/ncb3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J. K., and Wood W. B., 1998. Gastrulation initiation in Caenorhabditis elegans requires the function of gad-1, which encodes a protein with WD repeats. Dev. Biol. 198: 253–265. [PubMed] [Google Scholar]
- Kohrman A. Q., and Matus D. Q., 2017. Divide or conquer: cell cycle regulation of invasive behavior. Trends Cell Biol. 27: 12–25. 10.1016/j.tcb.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M., and Mango S. E., 1999. Patterning the C. elegans embryo: moving beyond the cell lineage. Trends Genet. 15: 307–313. 10.1016/S0168-9525(99)01750-3 [DOI] [PubMed] [Google Scholar]
- Lang C. F., and Munro E., 2017. The PAR proteins: from molecular circuits to dynamic self-stabilizing cell polarity. Development 144: 3405–3416. 10.1242/dev.139063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., and Goldstein B., 2003. Mechanisms of cell positioning during C. elegans gastrulation. Development 130: 307–320. 10.1242/dev.00211 [DOI] [PubMed] [Google Scholar]
- Lee J.-Y., Marston D. J., Walston T., Hardin J., Halberstadt A. et al. , 2006. Wnt/Frizzled signaling controls C. elegans gastrulation by activating actomyosin contractility. Curr. Biol. 16: 1986–1997. 10.1016/j.cub.2006.08.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M. F., Hill R. J., Heid P. J., Newman-Smith E. D., Zhu J. et al. , 2005. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev. Biol. 284: 509–522. 10.1016/j.ydbio.2005.05.016 [DOI] [PubMed] [Google Scholar]
- Maduro M., 2009. Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim. Biophys. Acta. 1789: 250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A. J., and Rogers S. L., 2014. The Fog signaling pathway: insights into signaling in morphogenesis. Dev. Biol. 394: 6–14. 10.1016/j.ydbio.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marean A., Graf A., Zhang Y., and Niswander L., 2011. Folic acid supplementation can adversely affect murine neural tube closure and embryonic survival. Hum. Mol. Genet. 20: 3678–3683. 10.1093/hmg/ddr289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston D. J., Higgins C. D., Peters K. A., Cupp T. D., Dickinson D. J. et al. , 2016. MRCK-1 drives apical constriction in C. elegans by linking developmental patterning to force generation. Curr. Biol. 26: 2079–2089. 10.1016/j.cub.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. C., and Goldstein B., 2014. Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development 141: 1987–1998. 10.1242/dev.102228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., 2013. The Caenorhabditis elegans intestine. Wiley Interdiscip. Rev. Dev. Biol. 2: 347–367. 10.1002/wdev.93 [DOI] [PubMed] [Google Scholar]
- Mello C. C., Draper B. W., Krause M., Weintraub H., and Priess J. R., 1992. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell 70: 163–176. 10.1016/0092-8674(92)90542-K [DOI] [PubMed] [Google Scholar]
- Moen E., Bannon D., Kudo T., Graf W., Covert M. et al. , 2019. Deep learning for cellular image analysis. Nat. Methods 16: 1233–1246. 10.1038/s41592-019-0403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E., Nance J., and Priess J. R., 2004. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell 7: 413–424. 10.1016/j.devcel.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Nance J., and Priess J. R., 2002. Cell polarity and gastrulation in C. elegans. Development 129: 387–397. [DOI] [PubMed] [Google Scholar]
- Nance J., Munro E. M., and Priess J. R., 2003. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development 130: 5339–5350. 10.1242/dev.00735 [DOI] [PubMed] [Google Scholar]
- Nikolopoulou E., Galea G. L., Rolo A., Greene N. D. E., and Copp A. J., 2017. Neural tube closure: cellular, molecular and biomechanical mechanisms. Development 144: 552–566. 10.1242/dev.145904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., and Seydoux G., 2017. Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 121–122: 86–93. 10.1016/j.ymeth.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud F., Walther R. F., and Nunes de Almeida F., 2019. Regulation of Cdc42 and its effectors in epithelial morphogenesis. J. Cell Sci. 132: jcs217869. 10.1242/jcs.217869 [DOI] [PubMed] [Google Scholar]
- Pohl C., Tiongson M., Moore J. L., Santella A., and Bao Z., 2012. Actomyosin-based self-organization of cell internalization during C. elegans gastrulation. BMC Biol. 10: 94 10.1186/1741-7007-10-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich W. B., Agbunag C., and Hardin J., 1999. Rapid epithelial-sheet sealing in the Caenorhabditis elegans embryo requires cadherin-dependent filopodial priming. Curr. Biol. 9: 1139–1146. 10.1016/S0960-9822(00)80015-9 [DOI] [PubMed] [Google Scholar]
- Razzell W., and Martin P., 2012. Cell biology. Embryonic clutch control. Science 335: 1181–1182. 10.1126/science.1220388 [DOI] [PubMed] [Google Scholar]
- Rhumbler L., 1902. Zur Mechanik des Gastrulationsvorganges insbesondere der Invagination. Arch. Entwicklungsmech. Org. 14: 401–476. 10.1007/BF02188499 [DOI] [Google Scholar]
- Roh-Johnson M., and Goldstein B., 2009. In vivo roles for Arp2/3 in cortical actin organization during C. elegans gastrulation. J. Cell Sci. 122: 3983–3993. 10.1242/jcs.057562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh-Johnson M., Shemer G., Higgins C. D., McClellan J. H., Werts A. D. et al. , 2012. Triggering a cell shape change by exploiting preexisting actomyosin contractions. Science 335: 1232–1235. 10.1126/science.1217869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J. M., Harrell J. R., Shemer G., Sullivan-Brown J., Roh-Johnson M. et al. , 2010. Apical constriction: a cell shape change that can drive morphogenesis. Dev. Biol. 341: 5–19. 10.1016/j.ydbio.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J. M., Glass S., Li T., Shemer G., White N. D. et al. , 2011. Overcoming redundancy: an RNAi enhancer screen for morphogenesis genes in Caenorhabditis elegans. Genetics 188: 549–564. 10.1534/genetics.111.129486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J., and Schierenberg E., 2011. Evolution of embryonic development in nematodes. Evodevo 2: 18 10.1186/2041-9139-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A. F., Baillie D. L., and Bowerman B., 2002. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr. Biol. 12: 2066–2075. 10.1016/S0960-9822(02)01355-6 [DOI] [PubMed] [Google Scholar]
- Seydoux G., and Fire A., 1994. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development 120: 2823–2834. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Mello C. C., Pettitt J., Wood W. B., Priess J. R. et al. , 1996. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 382: 713–716. 10.1038/382713a0 [DOI] [PubMed] [Google Scholar]
- Stern C. D., 2004. Gastrulation: from cells to embryo, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- Sullivan-Brown J. L., Tandon P., Bird K. E., Dickinson D. J., Tintori S. C. et al. , 2016. Identifying regulators of morphogenesis common to vertebrate neural tube closure and Caenorhabditis elegans gastrulation. Genetics 202: 123–139. 10.1534/genetics.115.183137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., and Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Takeichi M., 2014. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15: 397–410. 10.1038/nrm3802 [DOI] [PubMed] [Google Scholar]
- Tintori S. C., Osborne Nishimura E., Golden P., Lieb J. D., and Goldstein B., 2016. A transcriptional lineage of the early C. elegans embryo. Dev. Cell 38: 430–444. 10.1016/j.devcel.2016.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkaus J. P., 1969. Cells into organs; the forces that shape the embryo, Prentice-Hall, Englewood Cliffs, N.J. [Google Scholar]
- Ulman V., Maška M., Magnusson K. E. G., Ronneberger O., Haubold C. et al. , 2017. An objective comparison of cell-tracking algorithms. Nat. Methods 14: 1141–1152. 10.1038/nmeth.4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M., and Wieschaus E., 2016. Myosin-dependent remodeling of adherens junctions protects junctions from Snail-dependent disassembly. J. Cell Biol. 212: 219–229. 10.1083/jcb.201508056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E., 1996. From molecular patterns to morphogenesis—the lessons from studies on the fruit fly Drosophila (Nobel lecture). Angew. Chem. Int. Ed. Engl. 35: 2188–2194. 10.1002/anie.199621881 [DOI] [Google Scholar]
- Zhu J., Hill R. J., Heid P. J., Fukuyama M., Sugimoto A. et al. , 1997. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11: 2883–2896. 10.1101/gad.11.21.2883 [DOI] [PMC free article] [PubMed] [Google Scholar]