Abstract
XY C57BL/6J (B6) mice harboring a Mus musculus domesticus-type Y chromosome (YPOS), known as B6.YPOS mice, commonly undergo gonadal sex reversal and develop as phenotypic females. In a minority of cases, B6.YPOS males are identified and a proportion of these are fertile. This phenotypic variability on a congenic B6 background has puzzled geneticists for decades. Recently, a B6.YPOS colony was shown to carry a non-B6-derived region of chromosome 11 that protected against B6.YPOS sex reversal. Here. we show that a B6.YPOS colony bred and archived at the MRC Harwell Institute lacks the chromosome 11 modifier but instead harbors an ∼37 Mb region containing non-B6-derived segments on chromosome 13. This region, which we call Mod13, protects against B6.YPOS sex reversal in a proportion of heterozygous animals through its positive and negative effects on gene expression during primary sex determination. We discuss Mod13’s influence on the testis determination process and its possible origin in light of sequence similarities to that region in other mouse genomes. Our data reveal that the B6.YPOS sex reversal phenomenon is genetically complex and the explanation of observed phenotypic variability is likely dependent on the breeding history of any local colony.
Keywords: Sex determination, testis, ovary, Y chromosome, ovotestis, Genetics of Sex
IN mammals, the Y chromosome is a dominant male determinant due to the presence of the SRY gene. SRY’s occupancy of key enhancers causes the upregulation of SOX9, which itself drives the differentiation of Sertoli cells and the formation of testis cords (Sekido and Lovell-Badge 2008; Gonen et al. 2018). In XX gonads, in the absence of SRY, other pathways, including canonical WNT signaling and the transcription factor FOXL2, induce ovary development (Pannetier et al. 2016). Many years of study in the mouse model system indicate that the timing of Sry expression is crucial in testis development: any significant reduction or delay in Sry expression during a critical “window of opportunity” for testis determination may result in gonadal sex reversal (i.e., XY ovary or ovotestis development), due to a failure to inhibit the activity of pro-ovarian genes (Hiramatsu et al. 2008). Mutations that disrupt molecules required for normal spatiotemporal expression of Sry can cause XY gonadal sex reversal due to such a delay [reviewed in Carré and Greenfield (2014); Larney et al. (2014)]. However, the exact mechanisms by which this diverse class of molecules regulate Sry expression remain unclear.
One model system for examining the role of Sry in sex determination, and its interaction with genes on the autosomes, is B6.YDOM sex reversal. The C57BL6/J (B6) genetic background is sensitized to disruptions to testis determination due to precocious pro-ovarian gene expression profiles during the sex-determining period of gonadogenesis (Munger et al. 2009; Correa et al. 2012). Introgression of the Y chromosome from certain Mus musculus domesticus strains into the B6 genetic background (B6.YDOM) disrupts testis determination to varying degrees. One classic example involves the wild-derived M. d. poschiavinus Y chromosome: when on the B6 genetic background (B6.YPOS), where all autosomes and the X chromosome are predicted to be B6-derived, XY female development is the norm and examination of B6.YPOS fetal gonad development reveals XY ovary or ovotestis development as the commonest developmental outcomes (Eicher et al. 1982, 1996). While differences exist in the SRY protein isoforms encoded by distinct YDOM alleles, these cannot alone account for the different phenotypic outcomes of YDOM chromosome introgression (Albrecht and Eicher 1997; Albrecht et al. 2003). Rather, it has been reported that altered expression of the relevant SryDOM allele also plays a role (Nagamine et al. 1999). The peak of SryPOS expression has been reported to be delayed in B6.YPOS gonads, when compared to peak expression of SryB6 (Bullejos and Koopman 2005).
With respect to B6.YPOS, a complex model of sex reversal has emerged from a number of studies, invoking misregulation of Sry expression as a consequence of a mismatch between B6 autosomal and/or X-linked gene products required for its normal spatiotemporal expression and regulatory elements of the Sry locus; and altered stability and/or functionality of the SRYDOM protein isoforms in the context of the B6 genome. The exact molecular basis of the proposed functional mismatches remains unclear, as do the molecular events underlying Sry’s unique spatiotemporal profile of expression. Understanding has been hampered by the use, over many years, of different techniques to examine and quantify gene expression. Indeed, recent studies have called into question whether the B6.YPOS genome can be considered a unitary genetic background in the traditional sense.
The degree of gonadal sex reversal is typically variable between B6.YPOS individuals. Around 70–75% of XY individuals develop as phenotypic females and 25–30% as males. B6.YPOS males (once classified as hermaphrodites) have varying amounts of testicular tissue in either gonad, but enough to masculinize external genitalia. Given that any B6.YPOS colony is maintained by crossing fertile males to B6 females at each generation, the basis of this phenotypic variability has been unclear. Recently, a dominantly acting, autosomal modifier of B6.YPOS sex reversal, which protects against sex reversal, was reported in a United States colony of this strain (Arboleda et al. 2014). The 4.5 Mb region of chromosome 11 was identified by virtue of it not being B6-derived: instead, it is thought to derive from the M. d. poschiavinus (POS A) genome that was originally used to establish the B6.YPOS strain. The region in question encompasses the regulatory sequences upstream of the Sox9 gene and is thought to promote Sox9 expression, even in the presence of the “weaker” SryPOS allele, thus protecting against B6.YPOS sex reversal. This, in turn, is thought to account for its persistence in the B6.YPOS colony in question: at each generation, fertile males selected for further breeding are highly likely to be heterozygous for this region because in its absence XY female, or infertile hermaphrodite development, will most likely ensue.
Here, we report an analysis of the genetics of sex reversal in a B6.YPOS colony rederived from the frozen embryo archive at Harwell, which was originally investigated in the 1980–90s before archiving. This colony exhibits familiar ratios of XY female and male development, but lacks the chromosome 11 modifier locus. Instead, whole-genome sequence analysis of a B6.YPOS stud male reveals heterozygosity across an ∼37 Mb region of chromosome 13 for segments not derived from B6. This region, termed Mod13, is of unknown origin and confers protection against gonadal sex reversal in the fetal period by positively affecting Sry expression and negatively affecting pro-ovarian genes; but it contains no known primary sex-determining genes in its non-B6-derived segments. Our data reveal that B6.YPOS colonies in different parts of the world, if maintained in the absence of an Sry transgene or similar, may contain distinct modifiers that protect against sex reversal in a significant proportion of XY animals, depending on breeding history. We discuss the significance of these data for our understanding of B6.YPOS sex reversal.
Materials and Methods
Mouse strains
All mouse experimentation was approved by the Animal Welfare and Ethical Review Body at MRC Harwell. Mice used were bred with licensed approval from the UK Home Office (PPL 70/8898). Mice were housed in individually ventilated cages in a specific pathogen-free environment. Further details of micro- and macroenvironmental conditions are available on request. All lines were maintained on C57BL/6J (B6). The use of the name M. d. poschiavinus is contested (Silver 1995); phylogenetic studies place this strain firmly within the M. m. domesticus fold. However, for the sake of consistency with the existing literature on the Y chromosome from this strain and its effect on testis determination, we continue to use M. d. poschiavinus (and B6.YPOS) here. Genotyping of Mod13 single nucleotide polymorphisms (SNPs) was performed by analysis of SNPs listed in Supplemental Material, Table S2.
Generation of embryos
Noon on the day of the copulatory plug was counted as 0.5 days post coitum (dpc). Adult mice were humanely killed by dislocation of the neck, confirmed by palpation, and embryos were decapitated in ice-cold, phosphate-buffered saline solution. Embryos collected at 11.5 dpc were staged accurately based on the number of tail somites (ts).
Whole-mount in situ hybridization
Whole-mount in situ hybridization (WMISH) analysis of embryonic tissues and probes for Sry, Sox9, Insl3, and Stra8 have been previously described (Bogani et al. 2009; Warr et al. 2009). At least three independent biological samples from a given group were analyzed with a particular marker.
Quantitative RT-PCR
Total RNA was extracted using RNeasy plus micro kit (QIAGEN, Valencia, CA) from gonads separated from the mesonephros. Reverse transcription was carried out with 250 ng of total RNA using the High Capacity cDNA RT Kit (Applied Biosystems, Foster City, CA). Quantitative RT-PCR (qRT-PCR) was performed with Fast SYBR Green Master Mix (Life Technologies) on a 7500 Fast Real-Time PCR system (Applied Biosystems). RNA expression levels were normalized to those of Hrpt1 (endogenous control) using the ΔΔCt method (Livak and Schmittgen 2001). At least three samples for each genotype were analyzed. Primer sequences are available on request.
Whole-genome sequencing and bioinformatic analyses
For sequencing of a B6.YPOS stud male, DNA was extracted from mouse tail tissue and sequencing (Illumina HiSeq2000) performed at the Wellcome Trust Centre for Human Genetics High Throughput Genomics Facility (Oxford, UK). This generated a mean read-depth of 13× from a single lane of 100 nucleotide paired-end reads. Reads were aligned to the reference genome (NCBIM38/mm10) using BWA (Li and Durbin 2009). Detection of SNPs in the alignment was made using a customized version of the Genome Analysis Toolkit (DePristo et al. 2011) with default parameters. We adopted a filtering strategy to identify high-confidence SNPs and reduce the incidence of false positives. SNP sites that failed the Genome Analysis Toolkit internal status check and had a mapping quality <200 were removed.
Data availability
All reagents are available upon request. B6.YPOS sequence data analyzed here have been submitted to the NCBI (BioSample accession: SAMN12896768). Supplemental material available at figshare: https://doi.org/10.25386/genetics.11323655.
Results
Identification of Mod13 and its influence on B6.XYPOS maleness
A B6.YPOS colony was bred at Harwell in the mid-1980s and subsequently archived in the 1990s. We have been unable to determine its precise origins, but B6.YPOS was originally generated in the United States (Eicher et al. 1982). Before this study, the colony had been maintained on B6 for ∼6 years, in the absence of any protective transgene, by selecting rare fertile stud males at each generation to cross with B6 females. Around 65–70% of XY mice were routinely scored as female at weaning.
The report of Arboleda et al. (2014), describing a dominantly acting modifier on chromosome 11 that confers protection against B6.YPOS sex reversal, prompted us to determine whether the Harwell B6.YPOS colony carried the same non-B6-derived region, possibly accounting for the presence of fertile males at each generation. It is the contention of Arboleda et al. that, in the absence of this region, the large majority of XY B6.YPOS mice would develop as females. Characterization of the appropriate SNPs indicated that the Harwell colony lacked this region (Table S1). We then performed whole-genome next-generation sequencing of a B6.YPOS stud male from the Harwell colony to determine whether any significant segments of its genome were not derived from B6. Bioinformatic analyses (see Materials and Methods) revealed that, in addition to the Y chromosome, an ∼37 Mb segment of chromosome 13 from ∼38.2 to 75.7 Mb contained a high density of non-B6 SNPs in the heterozygous state (Figure 1). Other, much smaller, non-B6 sequence clusters were detected, scattered throughout the genome, such as that detected on chromosome 4 (Figure 1). These could not be independently validated (data not shown); we interpret these as likely sequencing errors due to sequence context, such as structural or copy number variations indicated by abnormally high base coverage, or as variants that have arisen in the Harwell B6 colony and are not found in the B6 reference genome used to map the sequence reads.
Figure 1.
Identification of the Mod13 variant region on chromosome 13. The distribution of SNPs in the B6.YPOS genome of a Harwell stud male is shown through comparison with the C57BL/6J reference genome, with chromosome identifiers on the left. Each blue point represents a detected SNP at a given chromosome position. The y-axis of each chromosome plot shows the base-quality scores (BQS) for each SNP, ranging from 200 to 1000. The largest cluster of heterozygous SNPs was found on chromosome 13 between 38 and 76 Mb. Much smaller regions exhibiting a cluster of non-B6 SNPs could not be independently validated and were attributed to sequence context, sequencing errors or genetic drift. Note also the large number of SNPs on the Y chromosome, revealing differences between the M. m. domesticus YPOS chromosome and the C57BL/6J Y chromosome.
We then sought to validate the existence of the non-B6 region on chromosome 13 by performing Sanger sequencing of PCR products encompassing nine putative SNPs identified across the region (Table S2), using all XY mice in the Harwell colony. This analysis confirmed the existence of these non-B6 SNPs, in the heterozygous form, in a proportion of mice in the colony (Figure 2). Moreover, while XY male mice were identified that lacked the region, and XY female mice were identified that carried the region, the non-B6 sequences were present at a significantly higher frequency in B6.YPOS male mice than in B6.YPOS female mice (Figure 2A). These data suggest that this region of chromosome 13 affords some degree of protection against B6.YPOS sex reversal. We called the region Mod13.
Figure 2.
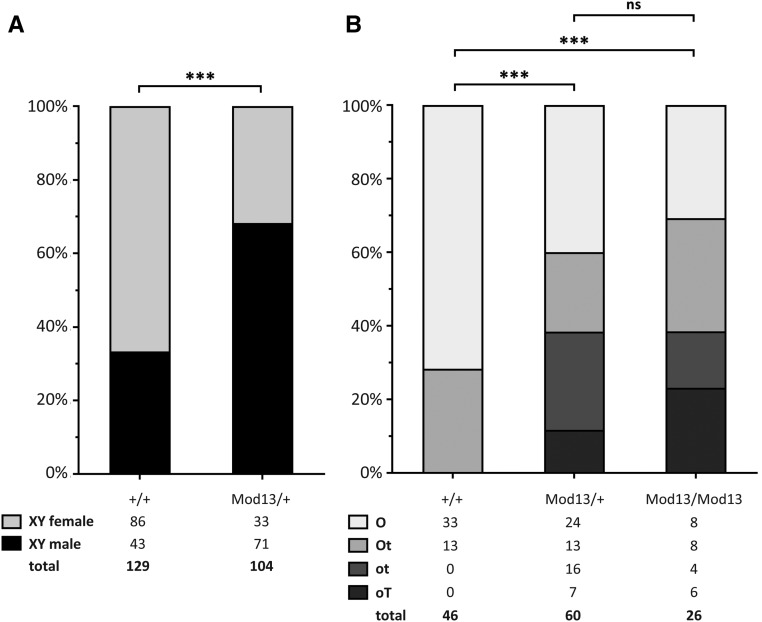
Mod13 promotes male phenotypic sex and testis determination. (A) The proportions of mice with male or female phenotypic sex in cohorts of the Harwell B6.YPOS colony, grouped by genotype (+/+ and Mod13/+) following scoring at weaning; the numbers of mice in each group are shown beneath each bar; a significant difference was found in the male:female ratio between the two genotypic groups (***P < 0.0001, Fisher’s exact test). (B) Scoring of gonadal phenotype in B6.YPOS wild-type (+/+), heterozygous (Mod13/+), and homozygous (Mod13/Mod13) fetuses at 14.5 dpc. Gonads were scored as ovaries (O), OVOtestes (Ot), ovotestes (ot), or ovoTESTES (oT), depending on the proportion and quality of testicular tissue observed and the overall gonad shape and morphology; numbers of gonads in each group are indicated. B6.YPOS+/+ fetal gonads are significantly different to those in the Mod13/+ and Mod13/Mod13 groups (***P < 0.0001, Fisher’s exact test). No normal testes (T) were observed in any B6.YPOS fetuses examined here.
Mod13 influences primary sex determination in B6.YPOS mice
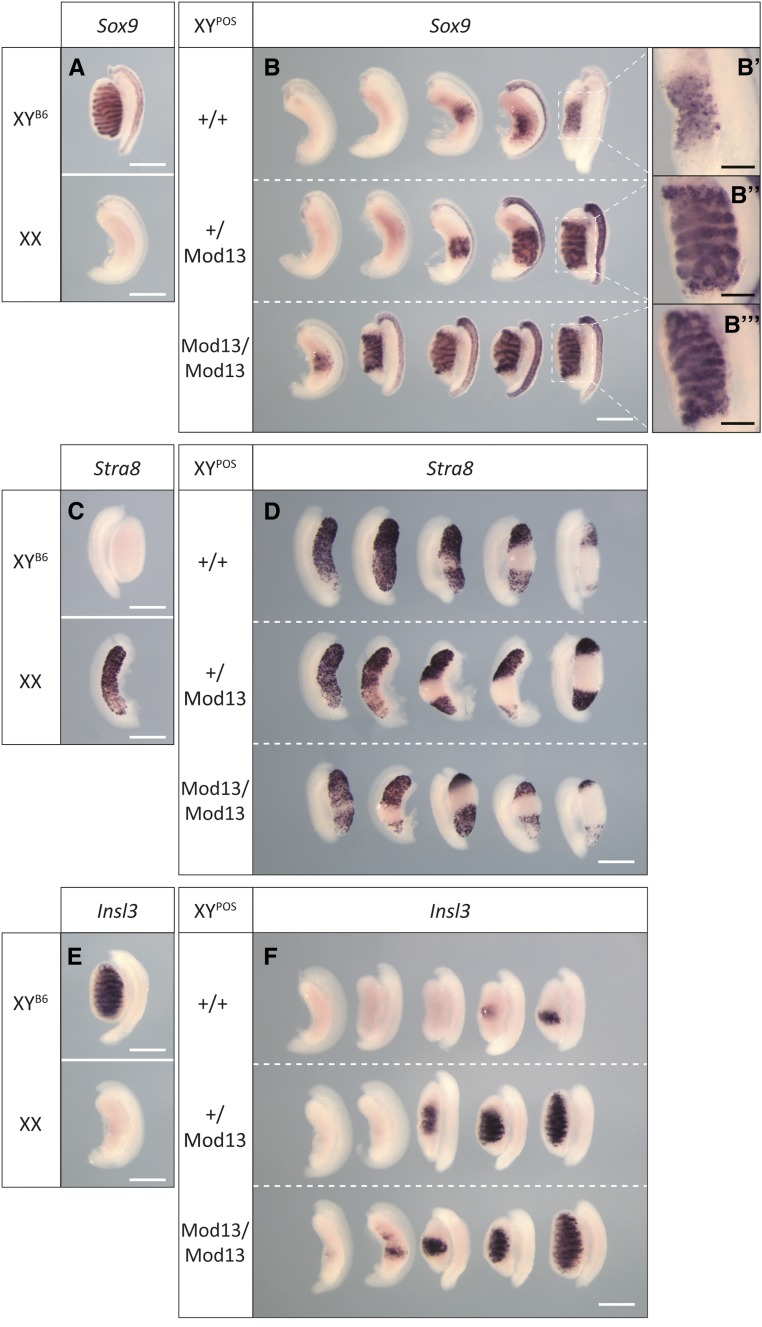
We then tested whether the presence of Mod13 affects primary sex determination. We identified XY males and XX females carrying Mod13 (Mod13/+), and performed timed-mates between these to generate B6.YPOS fetuses that were +/+, Mod13/+, and Mod13/Mod13. Remarkably, there was a clear effect of Mod13 on the morphology of the XY fetal gonads at 14.5 dpc (Figure 2B and Figure 3). B6.YPOS fetuses lacking the region (+/+) formed ovaries or ovotestes with minimal Sox9 expression (Figure 3, A and B), high levels of the meiotic germ cell marker Stra8 (Figure 3, C and D), and little evidence of significant testis cord formation or Leydig cell differentiation, the latter judged on Insl3 expression (Figure 3, E and F). Mod13/+ fetuses also developed ovaries, but there were larger numbers of ovotestes with significant testis cord formation (Figure 3, A–D). Finally, Mod13/Mod13 homozygotes formed ovotestes with the greatest amount of testicular tissue (Figure 2B and Figure 3, A–D), including some gonads with clear evidence of high-quality cord formation throughout the majority of the tissue (Figure 3, B’–B’’’). These data indicate that the presence of Mod13 influences the amount and quality of testicular tissue formed. We conclude from these data that Mod13 affords protection against B6.YPOS sex reversal by enhancing the amount of testicular tissue formed in the fetal period. It behaves, therefore, like a classical quantitative trait locus: its presence is no guarantee of testis development and its absence is not a guarantee of complete gonadal sex reversal. Rather, it positively influences the probability of formation of testicular tissue and its morphological quality.
Figure 3.
Cell marker analysis of the effect of Mod13 on fetal XY gonad development using WMISH. (A, C, and E) Marker gene expression in control gonads, XYB6 and XX. (B) Sox9 expression in B6.YPOS wild-type (+/+), heterozygous (Mod13/+) and homozygous (Mod13/Mod13) fetuses at 14.5 dpc. (B’–B’’’) Magnified images of relevant gonads showing details of testis cord morphology. (D) Stra8 expression and (F) Insl3 expression in the same genotypes as shown in B. White scale bar, 500 μm; black scale bar, 200 μm.
Mod13 positively affects Sry and Sox9 expression and negatively affects Rspo1 and Foxl2 at the sex-determining stage of gonadogenesis
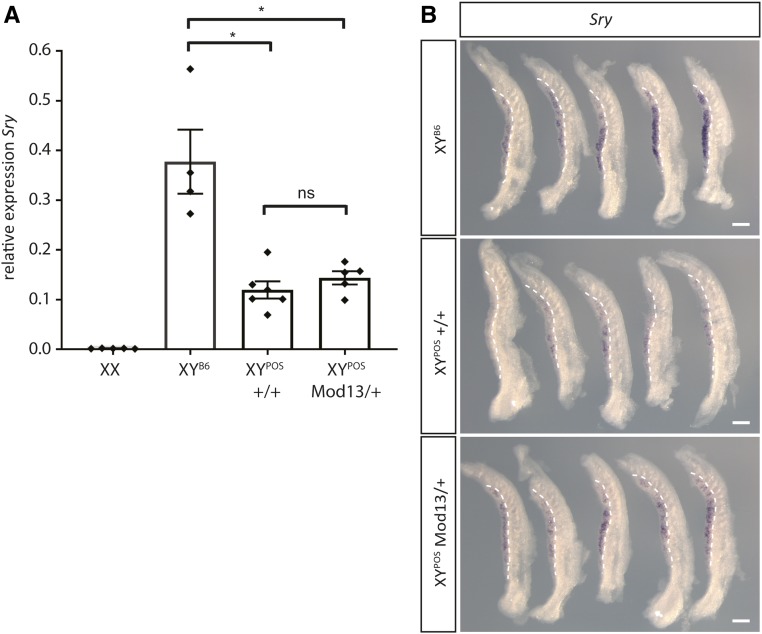
We first examined Sry and Sox9 expression, two key genes in initiating testis development, in developing B6.YPOS gonads. Sry expression in B6.YPOS embryonic gonads has been reported to be delayed in comparison to B6.YB6 gonads when analyzed by WMISH (Bullejos and Koopman 2005). We focused our analysis of Sry expression in B6.YPOS+/+ and Mod13/+ gonads on the 16 ts stage (∼11.25 dpc), at which point expression is reported to be close to its peak in B6.YB6, but less advanced in B6.YPOS (Bullejos and Koopman 2005). qRT-PCR did not reveal any significant difference in Sry levels between the two genotypes (Figure 4A). However, Sry expression is especially spatiotemporally dynamic, and in our experience WMISH can offer a better indication of the progression of its expression. Because the variable phenotypes observed at 14.5 dpc, we analyzed five B6.YPOS gonads that were +/+ and five that were Mod13/+ by WMISH, to allow semiquantitative assessment of variation in expression levels and spatial patterns between distinct gonads. B6.YB6 gonads were also included for comparison; these showed Sry expression throughout, although levels appeared to vary (Figure 4B). WMISH indicated that while a spectrum of Sry expression was observed within each B6.YPOS genotypic class, there was a clear tendency for expression to reach higher levels in Mod13/+ gonads, and for that expression to occupy a larger proportion of the gonad (Figure 4B).
Figure 4.
The influence of Mod13 on Sry expression in B6.YPOS gonads. (A) qRT-PCR analysis of Sry expression in control (XX, n = 5, XYB6, n = 4) and B6.YPOS wild-type (+/+, n = 6) and heterozygous (Mod13/+, n = 5) fetuses at 16/17 tail-somite (ts) stage (11.25–11.5 dpc). Black diamonds denote individual gonad sample measurements. (B) WMISH analysis of Sry expression at the same stage and in the same XY genotypes as in A. *P < 0.05; two-tailed Welch’s t-test. ns, not significant. Bar, 200 μm.
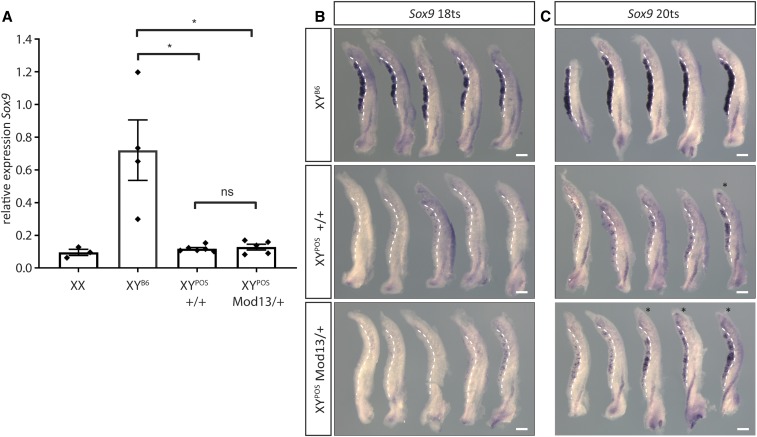
Sox9 expression was then analyzed in B6.YPOS+/+ and Mod13/+ gonads using qRT-PCR and WMISH. At the 18 ts stage, expression was relatively weak in both genotypes, in contrast to strong expression in B6.YB6 controls (Figure 5, A and B), possibly reflecting delayed SryPOS expression and the SRYPOS protein having reduced stability and/or functionality on B6 Sox9 gonadal enhancers. However, at the 20 ts stage, WMISH indicated that Sox9 expression was strong in three out of five Mod13/+ gonads; expression remained low in all but one of five +/+ samples (Figure 5C). Sox9 expression was very strong throughout B6.YB6 gonads at this stage (Figure 5C). These data are consistent with the positive effects of Mod13 heterozygosity on testicular morphology observed at 14.5 dpc.
Figure 5.
The effect of Mod13 on Sox9 expression at the sex-determining stage of gonad development. (A) qRT-PCR analysis of Sox9 expression in gonads from control (XX, n = 3 and XYB6, n = 4) and B6.YPOS wild-type (+/+, n = 6) and heterozygous (Mod13/+, n = 5) fetuses at 16–17 tail-somite (ts) stage. Black diamonds denote individual gonad sample measurements. (B) WMISH analysis of Sox9 expression in same XY genotypes as above, but at 18 ts stage. (C) WMISH analysis of Sox9 expression in same XY genotypes as above, but at 20 ts stage. B6.YPOS gonads showing strong Sox9 expression throughout most of gonad are marked by asterisks. Dotted lines indicate the gonad-mesonephros boundary, with gonad on the left. *P < 0.05; two-tailed Welch’s t-test. ns, not significant. Bar, 200 μm.
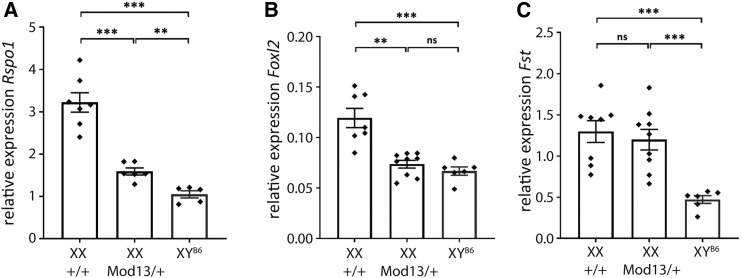
Given its positive effect on testis determination, we then examined whether the presence of Mod13 affected ovary development in XX fetuses. First, analysis of XX+/+, Mod13/+, and Mod13/Mod13 B6 gonads at 14.5 dpc with Sox9 and Stra8 using WMISH revealed no overt abnormalities at this stage (Figure S1A); nor was there any positive effect on Sox9 expression in XX gonads at the sex-determining stage, 11.5 dpc, due to the presence of Mod13 (Figure S1, B and C). To determine whether the presence of Mod13 negatively affected the pro-ovarian pathway gene expression at the sex-determining stage, we carefully quantified expression of Rspo1 and Foxl2, both markers of granulosa cell differentiation, in XX gonads that were either Mod13/+ or +/+. qRT-PCR revealed that levels of pro-ovarian Rspo1 and Foxl2 were significantly reduced in Mod13/+ gonads at 11.5 dpc, by ∼2-fold, to levels closer to those found in control XY gonads at the same stage (Figure 6, A and B); levels of follistatin (Fst) were not similarly affected by Mod13 (Figure 6C). These reductions in Rspo1 and Foxl2 gene expression at 11.5 dpc, apparently not mediated by elevated Sox9 in this context, are consistent with the overtly normal XX gonads observed at 14.5 dpc in Mod13/+ fetuses (see Figure S1). Mouse knockouts for both of these genes, and for Wnt4, only result in gonadal cell lineage reprogramming (sex reversal) in the perinatal/postnatal period (Pannetier et al. 2016). Nevertheless, the capacity of Mod13 to reduce, perhaps transiently, expression of these genes at the critical sex-determining stage could, when allied to its positive effects on Sry and Sox9 in the context of XY gonads, further enhance testis determination.
Figure 6.
Mod13 negatively affects pro-ovarian gene expression in XX gonads at 11.5 dpc. qRT-PCR analysis reveals expression levels of (A) Rspo1, (B) Foxl2, and (C) Fst in XX control (+/+), heterozygous B6.YPOS (Mod13/+) and control XY (XYB6) gonads at 11.5 dpc (17 tail-somite stage). **P < 0.005; ***P < 0.0005; two-tailed Welch’s t-test.
Sequence comparisons indicate that Mod13 has greater similarity to other inbred strains than C57BL/6J
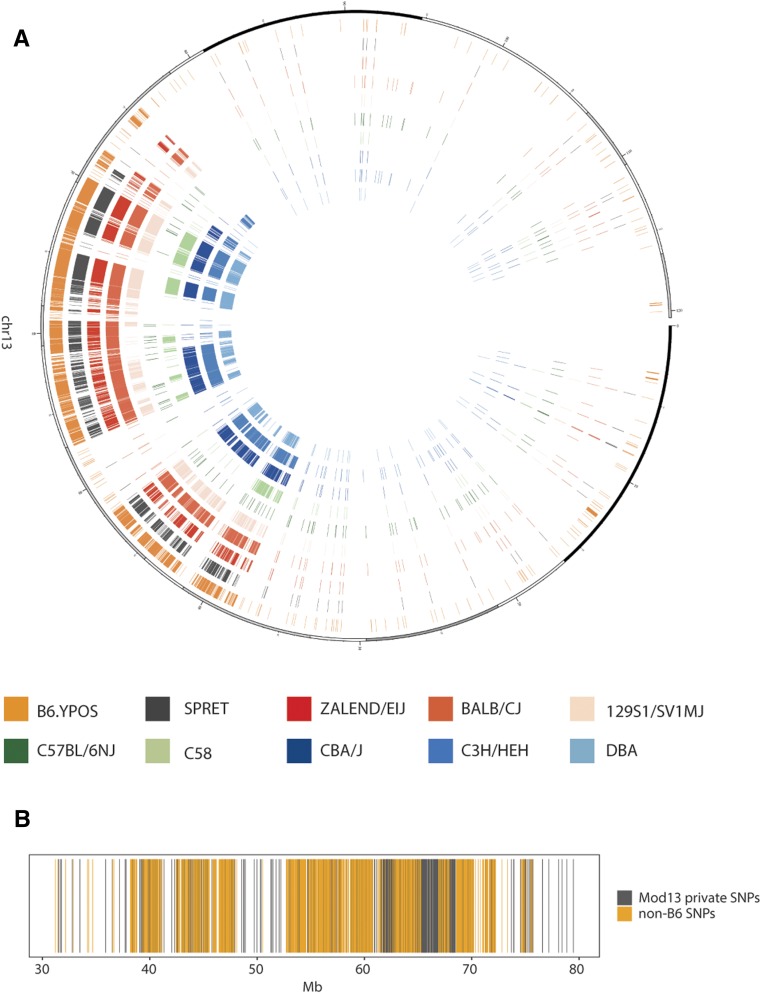
We performed a comparative analysis of chromosome 13 sequences, including the Mod13 genomic region identified in the Harwell B6.YPOS colony, in 36 laboratory mouse strains, comprising classical inbred strains and wild-derived mouse strains: a comparison between Mod13 and nine of these is shown in Figure 7A. The B6.YPOS Mod13 region is complex, containing segments of B6 homozygosity in addition to heterozygous regions (Figure 7B), an organization that has presumably arisen by successive rounds of recombination over multiple generations of backcrossing to the B6 background. Remarkably, the non-B6 Mod13 sequences showed significant similarity with most other inbred strains analyzed, with the greatest similarity to BALB/cJ; the exception was C57BL/6N (B6N), suggesting that B6J and B6N are both outliers in respect of sequence composition in this region of mouse chromosome 13 (Figure 7B). Moreover, the distribution of heterozygous SNPs across the region is not smooth: there are clusters of non-B6 SNPs as well as SNPs private to Mod13, where private is defined as SNPs found uniquely in Mod13 (Figure 7B). These Mod13 private SNPs cluster primarily between 60 and 70 Mb of chromosome 13 (Figure 7B).
Figure 7.
Heterozygous SNP density in Mod13. (A) Circos plot showing sequence comparisons between Harwell B6.YPOS chromosome 13 and cognate sequences in nine other mouse strains. Chromosome 13 is represented as a circle, with Harwell B6.YPOS sequences shown at the boundary and other genomes in concentric circles. Each line represents an individual SNP. Note the similarity between Mod13 sequences (left) of Harwell B6.YPOS and all of the other nine strains analyzed, with the exception of C57BL/6N. (B) Summary plot of SNP density grouped in 1 Mb windows over a 50 Mb interval containing the Mod13 region of chromosome 13. Colors denote SNPs that are either private to Mod13 when compared with 36 other mouse strains (gray) or are present in at least one other mouse strain (orange).
The non-B6-derived segments of Mod13 contain no known primary sex-determining genes but contain genes expressed in supporting cells at 11.5 dpc
There are 268 protein-coding genes within the Mod13 region, harboring 48 predicted mis-sense SNPs, from position 38.2 to 75.7 Mb of chromosome 13 (assembly GRCm38), which are not B6-derived (see Table S3). Only two of these genes have been previously implicated in sexual development: Ptch1, which is required for Leydig cell specification, but not testis determination (Clark et al. 2000; Yao et al. 2002); and Adamts16, which is linked to testis development in a number of contexts but is also not required for testis determination (Livermore et al. 2019). The known testis-determining gene Gadd45g (Warr et al. 2012) is located at position 51.8 Mb. However, the gene harbors no predicted deleterious SNPs and resides in a 4.5 Mb region of Mod13 devoid of any significant clusters of non-B6 SNPs. We analyzed data from recent single-cell RNA-sequencing analyses of XY and XX gonads (Stévant et al. 2018, 2019) to determine whether any of these genes are expressed in the supporting cell lineage of the developing gonad (XY, XX, or both) at 11.5 dpc, a minimal requirement for a putative role in sex determination. No outstanding candidates were identified based on a combination of expression profile and biological link to the known sex-determining pathways, but a number of genes exhibit expression in supporting cell precursors at 11.5 dpc (Figure S2 and Table S3). Interestingly, the Mod13 region does contain a number of key genes functioning in androgen biosynthesis, spermatogenesis, and male fertility (Table S3), such as Srd5a1 (Mahendroo et al. 2001), Hsd17b3 (Baker et al. 1997), and Spata31 (Wu et al. 2015), which may promote fertility in B6.YPOS Mod13/+ males independently of Mod13’s effect on primary sex determination.
Discussion
Through the identification and phenotypic analysis of the variant Mod13 region, we have established that the B6.YPOS sex reversal phenomenon is likely to have distinct genetic foundations depending on the breeding history of colonies held in different parts of the world. Interestingly, this possibility was predicted by Eicher and Washburn (2001), who suggested that an autosomal gene in a non-B6 state, which enhanced testicular development, would be kept in a forced heterozygous state by virtue of the breeding scheme used in B6.YPOS colonies (Eicher and Washburn 2001). This phenomenon is also consistent with the proposals of Arboleda et al. (2014), who predict that B6.YPOS colonies maintained by the selection of a transgene that protects against sex reversal, such as Sry, would, over time, lose modifiers that also enhanced the chances of male development, since they would be redundant in the presence of the transgene (Arboleda et al. 2014). However, they did not go on to predict the existence of distinct modifiers in geographically separated B6.YPOS colonies. We have no definitive explanation for the origin of the Harwell Mod13 variant of chromosome 13. The non-B6 sequence regions of Mod13 do not precisely match any particular inbred strain, but sequence similarity to BALB/cJ is especially high. Moreover, all other inbred strains analyzed show similarity to Mod13 sequences. Thus, sequence variants in this region are frequently unique to B6J and B6N, suggesting that these strains are outliers when it comes to sequence composition in this part of mouse chromosome 13.
It is our contention that one of following occurred: either M. d. poschiavinus-derived Mod13 sequences were present in the earliest crosses performed when the YPOS chromosome was introgressed onto B6 and have been maintained by selection ever since, which cannot be tested given the absence of M. d. poschiavinus sequences in the mouse genome database; or, more likely, at some point after the establishment of the B6.YPOS strain at Harwell, an outcross was performed, to a strain thought likely to be protective against sex reversal, such as BALB/cJ, in an attempt to rescue a, perhaps small, colony in which very few B6.YPOS fertile males were being produced—this might be the fate of any colony lacking Mod13 or another modifier/transgene. F1 males from such a cross would then have been crossed to B6 for subsequent generations, gradually resulting in the B6 background being restored, but with segments of the introgressed Mod13 region being maintained. However, we can find no evidence of such an outcross in the breeding records at Harwell. In this context, it is interesting to note that several of the inbred strains showing sequence similarity to Mod13 variants, such as BALB/cJ, C3H, and DBA, are known to protect against the disruptive effects of YPOS, i.e., introgression of YPOS into these backgrounds does not overtly disrupt testis determination (Eicher et al. 1995). We hypothesize that the B6-unique region of chromosome 13 contributes to the particular sensitivity shown by this strain to testis determination defects.
We show that Mod13 positively affects Sry, at least as evidenced by semiquantitative WMISH analysis, and Sox9 expression at 11.25–11.5 dpc in XY gonads. Sry and Sox9 levels appear enhanced, but variable, at this stage in XY Mod13/+ gonads, as predicted from the effects of Mod13 on testis determination observed at 14.5 dpc. But the effect of Mod13 is not restricted to this: we also report a negative effect on pro-ovarian expression at the same stage in XX gonads, not apparently mediated by changes to Sox9 expression (nor, of course, Sry). Taken together, these data suggest a possibly complex, oligogenic effect of Mod13 on testis determination during B6.YPOS gonadogenesis, simultaneously altering protestis and pro-ovary gene networks in direct and indirect fashion. We cannot, however, exclude the possibility that the positive effect on Sry expression is mediated by Mod13’s negative effect on pro-ovarian gene expression, by an unknown mechanism. Our data also indicate that the SRYPOS protein can upregulate Sox9 through the B6 version of the Sox9 regulatory region, although some degree of functional mismatch might account for the apparent delay in Sox9 reaching peak levels, even in the Mod13/+ B6.YPOS fetal gonad.
The large size of Mod13 (∼37 Mb) suggests that there may be more than one causative gene responsible for its protective effect against XY gonadal sex reversal, with loci possibly distributed throughout the region, some with sequence variants directly promoting protestis pathways, and some opposing pro-ovarian pathways. Our data reveal an effect of Mod13 on the amount and quality of testicular tissue formed in the fetal period, and it seems reasonable to conclude that this is a major determinant of the likelihood of adult males being fertile. Gadd45g would be an outstanding candidate for a gene exerting such effects, but no non-B6 variant clusters were detected in the region containing this testis-determining gene. We cannot rule out the possibility that the region also contains gene variants that promote male fertility, independently of effects on testis determination. In this context, it is interesting to note that the region contains genes such as Srd5a1 and Hsd17b3 that are implicated in adult male reproductive function.
Establishing causality for particular gene variants in the region is likely to be a daunting challenge. Our bioinformatic analysis indicates potential candidate genes on the basis of their expression in the supporting cell lineage at 11.5 dpc, the time at which, and cell lineage in which, sex determination is initiated in the mouse. None of these are previously implicated in sex determination, and each would need to be assessed for a role independently by inactivation and phenotypic assessment of the consequences. In addition to the possibility that multiple genes may play small, but additive, roles in testis determination in B6.YPOS mice, it would be very difficult to identify particular Mod13 sequence variants that are responsible for its protective effects. Some variants may not result in a loss-of-function effect, and so loss-of-function modelling may not reveal a role. Mod13 is perhaps best viewed as a potential source of a novel sex-determining gene(s). In the meantime, the precise molecular basis of the B6.YPOS sex reversal phenomenon remains unclear. We are now, however, able to examine B6.YPOS sex reversal, and the effect of specific transgenes on this phenotype, in the absence of the confounding effects of Mod13.
Acknowledgments
We thank the husbandry team in Ward 5 of the Mary Lyon Centre (MLC) at Harwell, particularly Jackie Harrison, Gemma Atkins, and Michelle Sandell. We also thank Martin Fray and the MLC (Medical Research Council) FESA (Frozen Embryo & Sperm Archive) team. We thank Valerie Arboleda and Eric Vilain for kindly providing tissues and genotyping details for the chromosome 11 modifier. We dedicate this paper to Bruce Cattanach, whose pioneering research on mouse sex determination at Harwell remains an inspiration.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11323655.
Communicating editor: B. Draper
Literature Cited
- Albrecht K. H., and Eicher E. M., 1997. DNA sequence analysis of Sry alleles (subgenus Mus) implicates misregulation as the cause of C57BL/6J-Y(POS) sex reversal and defines the SRY functional unit. Genetics 147: 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht K. H., Young M., Washburn L. L., and Eicher E. M., 2003. Sry expression level and protein isoform differences play a role in abnormal testis development in C57BL/6J mice carrying certain Sry alleles. Genetics 164: 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleda V. A., Fleming A., Barseghyan H., Delot E., Sinsheimer J. S. et al. , 2014. Regulation of sex determination in mice by a non-coding genomic region. Genetics 197: 885–897. 10.1534/genetics.113.160259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P. J., Sha J. H., and O’Shaughnessy P. J., 1997. Localisation and regulation of 17beta-hydroxysteroid dehydrogenase type 3 mRNA during development in the mouse testis. Mol. Cell. Endocrinol. 133: 127–133. 10.1016/S0303-7207(97)00159-7 [DOI] [PubMed] [Google Scholar]
- Bogani D., Siggers P., Brixey R., Warr N., Beddow S. et al. , 2009. Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol. 7: e1000196 10.1371/journal.pbio.1000196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullejos M., and Koopman P., 2005. Delayed Sry and Sox9 expression in developing mouse gonads underlies B6-Y(DOM) sex reversal. Dev. Biol. 278: 473–481. 10.1016/j.ydbio.2004.11.030 [DOI] [PubMed] [Google Scholar]
- Carré G. A., and Greenfield A., 2014. Characterising novel pathways in testis determination using mouse genetics. Sex Dev. 8: 199–207. 10.1159/000358402 [DOI] [PubMed] [Google Scholar]
- Clark A. M., Garland K. K., and Russell L. D., 2000. Desert hedgehog (Dhh) gene is required in the mouse testis for formation of adult-type Leydig cells and normal development of peritubular cells and seminiferous tubules. Biol. Reprod. 63: 1825–1838. 10.1095/biolreprod63.6.1825 [DOI] [PubMed] [Google Scholar]
- Correa S. M., Washburn L. L., Kahlon R. S., Musson M. C., Bouma G. J. et al. , 2012. Sex reversal in C57BL/6J XY mice caused by increased expression of ovarian genes and insufficient activation of the testis determining pathway. PLoS Genet. 8: e1002569 10.1371/journal.pgen.1002569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R. et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher E. M., and Washburn L. L., 2001. Does one gene determine whether a C57BL/6J-Y(POS) mouse will develop as a female or as an hermaphrodite? J. Exp. Zool. 290: 322–326. 10.1002/jez.1072 [DOI] [PubMed] [Google Scholar]
- Eicher E. M., Washburn L. L., Whitney J. B., and Morrow K. E., 1982. Mus poschiavinus Y chromosome in the C57BL/6J murine genome causes sex reversal. Science 217: 535–537. 10.1126/science.7089579 [DOI] [PubMed] [Google Scholar]
- Eicher E. M., Shown E. P., and Washburn L. L., 1995. Sex reversal in C57BL/6J-YPOS mice corrected by a Sry transgene. Philos. Trans. R. Soc. Lond. B Biol. Sci. 350: 263–268; discussion 268–269. 10.1098/rstb.1995.0160 [DOI] [PubMed] [Google Scholar]
- Eicher E. M., Washburn L. L., Schork N. J., Lee B. K., Shown E. P. et al. , 1996. Sex-determining genes on mouse autosomes identified by linkage analysis of C57BL/6-YPos sex reversal. Nat. Genet. 14: 206–209. 10.1038/ng1096-206 [DOI] [PubMed] [Google Scholar]
- Gonen N., Futtner C. R., Wood S., Garcia-Moreno S. A., Salamone I. M. et al. , 2018. Sex reversal following deletion of a single distal enhancer of Sox9. Science 360: 1469–1473. 10.1126/science.aas9408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu R., Matoba S., Kanai-Azuma M., Tsunekawa N., Katoh-Fukui Y. et al. , 2008. A critical time window of Sry action in gonadal sex determination in mice. Development 136: 129–138. 10.1242/dev.029587 [DOI] [PubMed] [Google Scholar]
- Larney C., Bailey T. L., and Koopman P., 2014. Switching on sex: transcriptional regulation of the testis-determining gene Sry. Development 141: 2195–2205. 10.1242/dev.107052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Livermore C., Warr N., Chalon N., Siggers P., Mianne J. et al. , 2019. Male mice lacking ADAMTS-16 are fertile but exhibit testes of reduced weight. Sci. Rep. 9: 17195 10.1038/s41598-019-53900-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendroo M. S., Cala K. M., Hess D. L., and Russell D. W., 2001. Unexpected virilization in male mice lacking steroid 5 alpha-reductase enzymes. Endocrinology 142: 4652–4662. 10.1210/endo.142.11.8510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger S. C., Aylor D. L., Syed H. A., Magwene P. M., Threadgill D. W. et al. , 2009. Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal. Genes Dev. 23: 2521–2536. 10.1101/gad.1835809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine C. M., Morohashi K., Carlisle C., and Chang D. K., 1999. Sex reversal caused by Mus musculus domesticus Y chromosomes linked to variant expression of the testis-determining gene Sry. Dev. Biol. 216: 182–194. 10.1006/dbio.1999.9436 [DOI] [PubMed] [Google Scholar]
- Pannetier M., Chassot A. A., Chaboissier M. C., and Pailhoux E., 2016. Involvement of FOXL2 and RSPO1 in ovarian determination, development, and maintenance in mammals. Sex Dev. 10: 167–184. 10.1159/000448667 [DOI] [PubMed] [Google Scholar]
- Sekido R., and Lovell-Badge R., 2008. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453: 930–934 (erratum: Nature 456: 824). 10.1038/nature06944 [DOI] [PubMed] [Google Scholar]
- Silver L. M., 1995. Mouse Genetics: Concepts and Applications, Oxford University Press, Oxford, UK. [Google Scholar]
- Stévant I., Neirijnck Y., Borel C., Escoffier J., Smith L. B. et al. , 2018. Deciphering cell lineage specification during male sex determination with single-cell RNA sequencing. Cell Rep. 22: 1589–1599. 10.1016/j.celrep.2018.01.043 [DOI] [PubMed] [Google Scholar]
- Stévant I., Kuhne F., Greenfield A., Chaboissier M. C., Dermitzakis E. T. et al. , 2019. Dissecting cell lineage specification and sex fate determination in gonadal somatic cells using single-cell transcriptomics. Cell Rep. 26: 3272–3283.e3. 10.1016/j.celrep.2019.02.069 [DOI] [PubMed] [Google Scholar]
- Warr N., Siggers P., Bogani D., Brixey R., Pastorelli L. et al. , 2009. Sfrp1 and Sfrp2 are required for normal male sexual development in mice. Dev. Biol. 326: 273–284. 10.1016/j.ydbio.2008.11.023 [DOI] [PubMed] [Google Scholar]
- Warr N., Carre G. A., Siggers P., Faleato J. V., Brixey R. et al. , 2012. Gadd45γ and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev. Cell 23: 1020–1031. 10.1016/j.devcel.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. Y., Yang Y., Xu Y. D., and Yu H. L., 2015. Targeted disruption of the spermatid-specific gene Spata31 causes male infertility. Mol. Reprod. Dev. 82: 432–440. 10.1002/mrd.22491 [DOI] [PubMed] [Google Scholar]
- Yao H. H., Whoriskey W., and Capel B., 2002. Desert Hedgehog/Patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 16: 1433–1440. 10.1101/gad.981202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All reagents are available upon request. B6.YPOS sequence data analyzed here have been submitted to the NCBI (BioSample accession: SAMN12896768). Supplemental material available at figshare: https://doi.org/10.25386/genetics.11323655.