Abstract
Dilated cardiomyopathy (DCM) is one of the most common causes of heart failure. Several studies have used RNA-sequencing (RNA-seq) to profile differentially expressed genes (DEGs) associated with DCM. In this study, we aimed to profile gene expression signatures and identify novel genes associated with DCM through a quantitative meta-analysis of three publicly available RNA-seq studies using human left ventricle tissues from 41 DCM cases and 21 control samples. Our meta-analysis identified 789 DEGs including 581 downregulated and 208 upregulated genes. Several DCM-related genes previously reported, including MYH6, CKM, NKX2–5 and ATP2A2, were among the top 50 DEGs. Our meta-analysis also identified 39 new DEGs that were not detected using those individual RNA-seq datasets. Some of those genes, including PTH1R, ADAM15 and S100A4, confirmed previous reports of associations with cardiovascular functions. Using DEGs from this meta-analysis, the Ingenuity Pathway Analysis (IPA) identified five activated toxicity pathways, including failure of heart as the most significant pathway. Among the upstream regulators, SMARCA4 was downregulated and prioritized by IPA as the top affected upstream regulator for several DCM-related genes. To our knowledge, this study is the first to perform a transcriptomic meta-analysis for clinical DCM using RNA-seq datasets. Overall, our meta-analysis successfully identified a core set of genes associated with DCM.
Keywords: dilated cardiomyopathy, heat failure, meta-analysis, RNA-seq, human
1. Introduction
Cardiovascular diseases are the leading cause of death in the United States and accounted for 840,000 deaths in 2016 [1]. Heart failure is among the cardiovascular diseases with not only a high risk of death but also with other adverse symptoms along with poor quality of life. It limits daily physical and social activities and leads to various physical and emotional distress [2]. Dilated cardiomyopathy (DCM) is one of the most common causes of heart failure, with an estimated prevalence of 1 in 2500 individuals [3]. DCM is an irreversible form of heart muscle disease and is characterized as left ventricle enlargement and systolic dysfunction. Decline in left ventricle contractile function can cause inefficient blood pumping and heart weakness, eventually leading to sudden or heart failure-related death [3].
Genetic mutations in cytoskeletal/sarcolemmal, nuclear envelope, sarcomere and transcriptional coactivator genes can cause DCM [3]. More than 20 genomic loci and genes were reported to contribute to familial DCM accounting for 20% to 35% of DCM cases [3]. Therefore, the identification of genes and their genetic variants for DCM is important and urgently needed. Advances in next-generation sequencing have led to easier and faster discovery of biomarkers for DCM. Several genes responsible for DCM, including MYH7, TNNT2, LMNA, DES, TTN, PLN, ACTC1, SCN5A, NKX2-5 and TBX5, have been identified for clinical diagnosis and evaluation of genetic susceptibility [4,5,6]. RNA-seq, as a revolutionary tool in transcriptomics, enables biomarker identification by profiling differentially expressed genes (DEGs) associated with a specific disease. However, different RNA-seq studies of the same disease show inconsistent gene expression patterns and discordant results due to different sample collection and sequencing protocols and inconsistent data analysis strategies [7]. Systematic and quantitative combined analysis of multiple RNA-seq studies using a meta-analysis approach could largely eliminate the inconsistency in individual studies by increasing the sample size and statistical power for enlisting more robust disease-associated genes [8].
In this study, we performed a quantitative meta-analysis of three independent RNA-seq studies using human left ventricle tissues to identify robust candidate biomarkers for DCM. A total number of 41 DCM and 21 non-failing (NF) samples were analyzed. To our knowledge, this study is the first to perform an RNA-seq meta-analysis in the field of clinical DCM.
2. Materials and Methods
NCBI GEO [9] database (https://www.ncbi.nlm.nih.gov/geo/) was queried for RNA-seq studies of dilated cardiomyopathy as summarized in Table 1. The clinical information of DCM patients and their controls has been reported in Study_1 [10], Study_2 [11] and Study_3 [12]. Only studies using tissue samples obtained from human left ventricle were included in our meta-analysis.
Table 1.
Summary of GEO datasets for the meta-analysis.
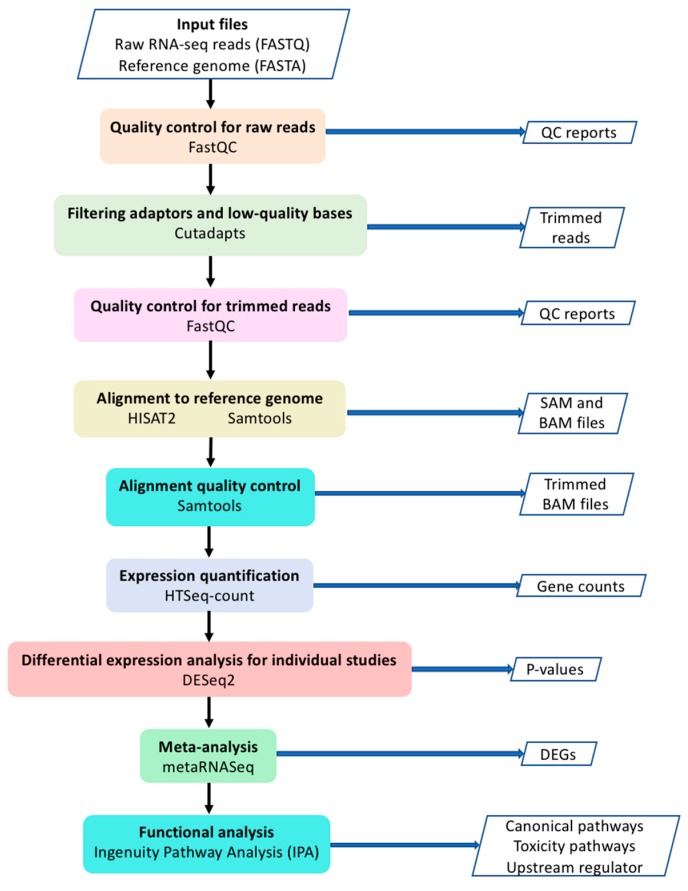
The workflow of our current study is shown in Figure 1. Raw data (FASTQ files) were downloaded from European Nucleotide Archive website (https://www.ebi.ac.uk/ena). Quality control (QC) for raw reads was performed using FastQC [16]. Adaptors and low-quality bases (Phred quality score < 10) were filtered by Cutadapt [17]. Trimmed reads were aligned to human reference genome (GRCh38) using HISAT2 software [18]. Samtools was used to sort the sequences and convert the SAM file to BAM file [19]. Only uniquely mapped reads were used for expression quantification with HTSeq-count [20]. Differential expression analysis for individual studies was performed using DESeq2 package [21]. Genes with low read counts were filtered out based on the mean of normalized counts as the filter statistic using independent filtering function in DESeq2 with default parameters. MetaRNASeq [7] package was used to perform Fisher’s combined probability test for meta-analysis. In this method, p-values for each gene from individual studies are combined through the following formula [7,22].
Figure 1.
The workflow of the meta-analysis.
When p-values are independent, Fg has a Chi-squared distribution with 2S degrees of freedom. Smaller p-values result in larger Fg and null hypothesis rejection. Where Pgs is the raw p-value obtained for gene g in a differential analysis for study S. p-values were adjusted for Benjamini–Hochberg false discovery rate (FDR). Adjusted p-value smaller than 0.05 was considered as statistically significant. Genes with inconsistent expression directions (upregulated or downregulated) among individual studies were removed from the DEGs list.
Using DEGs from meta-analysis, the Ingenuity Pathway Analysis (IPA, Qiagen, Redwood City, California, USA) was performed to investigate enriched canonical pathways, toxicity functions (IPA-Tox) and upstream regulators [23]. IPA-Tox uses the current knowledge to link DEGs to clinical pathological endpoints. IPA upstream regulator tool also uses current knowledge to identify the upstream transcriptional regulators which explain the observed gene expression changes in a dataset. A Venn diagram of the number of DEGs from individual studies and the meta-analysis was generated using the VennDiagram package [24] in R. The Ohio Supercomputer Center (OSC) was used to perform the compute-intensive tasks [25].
3. Results
3.1. Datasets Analyzed in This Study
Based on our search criteria (RNA-seq, heart failure, dilated cardiomyopathy and human), three RNA-seq datasets (GEO accession numbers GSE116250, GSE57344, and GSE71613) were included in our study. Total numbers of DCM and NF samples were 41 and 21, respectively. The summary of the datasets is shown in Table 1.
3.2. Differentially Expressed Genes
The human gene set, Homo_sapiens.GRCh38.96.gtf, including a total of 58,884 coding and non-coding genes, was used for expression quantification. Our meta-analysis identified 789 DEGs including 581 downregulated and 208 upregulated genes at a false discovery rate (FDR) < 0.05 (Supplementary Table S1). The top 50 DEGs sorted by p-value are shown in Table 2.
Table 2.
Top 50 differential expressed genes identified in meta-analysis of DCM vs. NF.
| Ensembl_ID | Gene_Symbol | Adj_P 1 | Average_Log2FCm 2 | Effectm 3 |
|---|---|---|---|---|
| ENSG00000076351 | SLC46A1 | 0.00E + 00 | 0.37 | Up |
| ENSG00000113389 | NPR3 | 0.00E + 00 | 2.14 | Up |
| ENSG00000126106 | TMEM53 | 0.00E + 00 | −0.60 | Down |
| ENSG00000176293 | ZNF135 | 0.00E + 00 | 0.37 | Up |
| ENSG00000177575 | CD163 | 0.00E + 00 | −1.57 | Down |
| ENSG00000189060 | H1F0 | 0.00E + 00 | −1.01 | Down |
| ENSG00000227495 | AC004771.1 | 0.00E + 00 | −0.40 | Down |
| ENSG00000263986 | AC087393.2 | 0.00E + 00 | 0.49 | Up |
| ENSG00000179526 | SHARPIN | 1.68E−13 | −0.54 | Down |
| ENSG00000197616 | MYH6 | 1.68E−13 | −1.85 | Down |
| ENSG00000157388 | CACNA1D | 3.03E−13 | −1.10 | Down |
| ENSG00000229867 | STEAP3-AS1 | 5.87E−13 | −0.71 | Down |
| ENSG00000167646 | DNAAF3 | 1.28E−12 | 1.30 | Up |
| ENSG00000170448 | NFXL1 | 7.58E−12 | −2.33 | Down |
| ENSG00000186187 | ZNRF1 | 2.44E−11 | −0.56 | Down |
| ENSG00000259661 | AC068831.4 | 2.57E−11 | −0.59 | Down |
| ENSG00000142156 | COL6A1 | 3.39E−11 | −0.78 | Down |
| ENSG00000235910 | APOA1-AS | 4.28E−11 | 0.90 | Up |
| ENSG00000109099 | PMP22 | 4.81E−11 | −0.86 | Down |
| ENSG00000163220 | S100A9 | 6.09E−11 | −2.13 | Down |
| ENSG00000174437 | ATP2A2 | 8.49E−11 | −0.83 | Down |
| ENSG00000175221 | MED16 | 1.06E−10 | −0.83 | Down |
| ENSG00000198624 | CCDC69 | 1.60E−10 | −1.04 | Down |
| ENSG00000133800 | LYVE1 | 1.72E−10 | −2.06 | Down |
| ENSG00000267092 | AC027307.1 | 1.85E−10 | 0.63 | Up |
| ENSG00000129250 | KIF1C | 2.27E−10 | −0.61 | Down |
| ENSG00000256469 | AP002383.2 | 2.84E−10 | 0.82 | Up |
| ENSG00000233098 | CCDC144NL-AS1 | 3.19E−10 | 0.37 | Up |
| ENSG00000105679 | GAPDHS | 3.53E−10 | 0.65 | Up |
| ENSG00000156463 | SH3RF2 | 3.66E−10 | −1.05 | Down |
| ENSG00000210127 | MT-TA | 3.66E−10 | −1.00 | Down |
| ENSG00000183072 | NKX2-5 | 9.88E−10 | −0.96 | Down |
| ENSG00000125733 | TRIP10 | 1.08E−09 | −0.88 | Down |
| ENSG00000141905 | NFIC | 1.11E−09 | −0.61 | Down |
| ENSG00000135447 | PPP1R1A | 1.14E−09 | −1.00 | Down |
| ENSG00000107796 | ACTA2 | 1.35E−09 | −1.38 | Down |
| ENSG00000140675 | SLC5A2 | 1.75E−09 | 0.41 | Up |
| ENSG00000101187 | SLCO4A1 | 2.57E−09 | −1.91 | Down |
| ENSG00000103710 | RASL12 | 3.61E−09 | −1.34 | Down |
| ENSG00000155659 | VSIG4 | 3.85E−09 | −2.04 | Down |
| ENSG00000253549 | CA3-AS1 | 4.04E−09 | 2.41 | Up |
| ENSG00000105698 | USF2 | 5.02E−09 | −0.50 | Down |
| ENSG00000196642 | RABL6 | 5.44E−09 | −0.52 | Down |
| ENSG00000122034 | GTF3A | 8.93E−09 | −0.62 | Down |
| ENSG00000235790 | AC114488.2 | 9.04E−09 | 0.83 | Up |
| ENSG00000104879 | CKM | 1.05E−08 | −0.95 | Down |
| ENSG00000114867 | EIF4G1 | 1.21E−08 | −0.80 | Down |
| ENSG00000260469 | INSYN1-AS1 | 1.33E−08 | −0.96 | Down |
| ENSG00000260755 | AC010542.2 | 1.49E−08 | 0.38 | Up |
| ENSG00000257453 | AC011611.3 | 1.60E−08 | 1.39 | Up |
1: FDR-adjusted p-value, “E” represents “times ten raised to the power of”; 2: Average of log2FC from individual studies; 3: ‘Up’ or ‘Down’ indicates whether the gene was upregulated or downregulated.
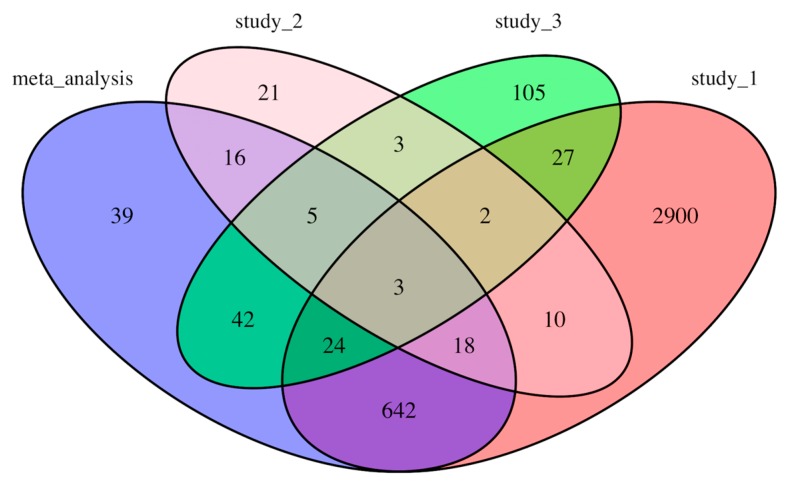
The counts of the common genes shared among individual studies (results from our analyses) and meta-analysis are shown in a Venn diagram (Figure 2). There are 39 DEGs that were only identified through the meta-analysis but not in individual analyses (Table 3). Interestingly, a significant number of DEGs from each individual study did not appear in other studies or meta-analysis. For example, 2900 genes out of total 3626 DEGs in Study_1 were not identified to be differentially expressed in other studies or in our meta-analysis (Figure 2). Only three genes, TUBA3D, LCN10, and NPR3, were common among all three individual analyses and the meta-analysis (Figure 2).
Figure 2.
Venn diagram showing the number of differential expressed genes from individual studies and meta-analysis.
Table 3.
Differential expressed genes newly identified in the meta-analysis of DCM vs. NF.
| Ensembl_ID | Gene_Symbol | Adj_p 1 | Average_Log2FC 2 | Effect 3 |
|---|---|---|---|---|
| ENSG00000158859 | ADAMTS4 | 1.44E−03 | −1.15 | Down |
| ENSG00000185201 | IFITM2 | 6.57E−03 | −1.03 | Down |
| ENSG00000162551 | ALPL | 8.13E−03 | −1.28 | Down |
| ENSG00000151365 | THRSP | 8.81E−03 | 1.71 | Up |
| ENSG00000142733 | MAP3K6 | 8.95E−03 | −0.91 | Down |
| ENSG00000158246 | TENT5B | 1.03E−02 | −1.35 | Down |
| ENSG00000087495 | PHACTR3 | 1.38E−02 | −1.57 | Down |
| ENSG00000132205 | EMILIN2 | 1.53E−02 | −0.95 | Down |
| ENSG00000103316 | CRYM | 1.65E−02 | 0.88 | Up |
| ENSG00000224934 | AL391684.1 | 1.65E−02 | 0.73 | Up |
| ENSG00000198542 | ITGBL1 | 1.68E−02 | 1.02 | Up |
| ENSG00000174429 | ABRA | 1.72E−02 | −1.02 | Down |
| ENSG00000172935 | MRGPRF | 1.76E−02 | −0.98 | Down |
| ENSG00000196154 | S100A4 | 2.03E−02 | −1.09 | Down |
| ENSG00000131386 | GALNT15 | 2.08E−02 | −1.16 | Down |
| ENSG00000255750 | AC022509.1 | 2.12E−02 | 1.14 | Up |
| ENSG00000198517 | MAFK | 2.48E−02 | 0.68 | Up |
| ENSG00000143537 | ADAM15 | 2.51E−02 | −0.77 | Down |
| ENSG00000134470 | IL15RA | 2.52E−02 | −0.92 | Down |
| ENSG00000170458 | CD14 | 2.75E−02 | −0.88 | Down |
| ENSG00000242396 | AC096536.2 | 3.26E−02 | −1.14 | Down |
| ENSG00000203883 | SOX18 | 3.39E−02 | −0.74 | Down |
| ENSG00000132481 | AC087289.1 | 3.44E−02 | −0.74 | Down |
| ENSG00000162458 | FBLIM1 | 3.54E−02 | −0.78 | Down |
| ENSG00000106809 | OGN | 3.61E−02 | 1.18 | Up |
| ENSG00000196923 | PDLIM7 | 3.76E−02 | −0.98 | Down |
| ENSG00000167797 | CDK2AP2 | 3.94E−02 | −0.81 | Down |
| ENSG00000160801 | PTH1R | 4.02E−02 | −0.87 | Down |
| ENSG00000147155 | EBP | 4.05E−02 | −1.18 | Down |
| ENSG00000177679 | SRRM3 | 4.18E−02 | −1.07 | Down |
| ENSG00000106823 | ECM2 | 4.22E−02 | 1.00 | Up |
| ENSG00000173272 | MZT2A | 4.33E−02 | −0.66 | Down |
| ENSG00000106819 | ASPN | 4.40E−02 | 1.27 | Up |
| ENSG00000180999 | C1orf105 | 4.76E−02 | −1.41 | Down |
| ENSG00000175287 | PHYHD1 | 4.85E−02 | −0.87 | Down |
| ENSG00000279296 | PRAL | 4.88E−02 | 0.50 | Up |
| ENSG00000187955 | COL14A1 | 4.90E-02 | 0.93 | Up |
| ENSG00000134684 | YARS | 4.91E-02 | -0.68 | Down |
| ENSG00000228804 | AC072022.1 | 4.96E-02 | -0.71 | Down |
1: FDR-adjusted p-value, “E” represents “times ten raised to the power of”; 2: Average of log2FC from individual studies; 3: ‘Up’ or ‘Down’ indicates whether the gene was upregulated or downregulated.
3.3. Toxicity Pathway Analysis
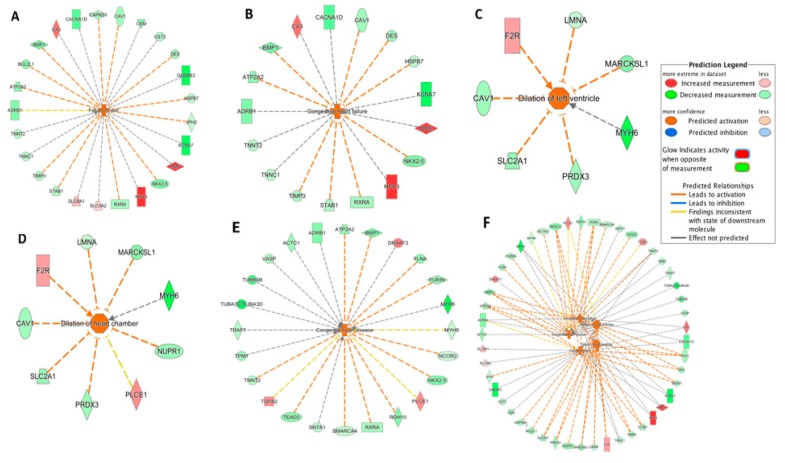
The IPA-Tox tool was used to identify enriched toxicity pathways. Out of 184 significant toxicity pathways (p < 0.05), only five pathways were identified as activated (z-score > 2.0). These were failure of heart, congestive heart failure, dilation of left ventricle, dilation of heart chamber and congenital heart disease. Responsible DEGs for each toxicity pathway and their direction of change are shown in the Figure 3A–E. Failure of heart and congestive heart failure had the highest activation z-score (3.06 and 2.96, respectively). The five pathways and their corresponding DEGs were further integrated (Figure 3F). Among the 39 DEGs from the integrated network (Figure 3F), 8 DEGs were shared in three or more pathways. Interestingly, these eight DEGs were all downregulated in the DCM group (Table 4). ATP2A2, MYH6, and NKX2-5 were among the top 50 DEGs (Table 2).
Figure 3.
Toxicity pathways significantly activated in DCM through IPA-Tox: (A) failure of heart; (B) congestive heart failure; (C) dilation of left ventricle; (D) dilation of heart chamber; (E) congestive heart disease; (F) integrated toxicity pathways. As an example, in Figure 3A, the DES gene was downregulated as indicated by the green color and the downregulation of DES further promoted (indicated by the orange dash line) the activation of failure of heart, as indicated by the orange color. Other indicators are explained in the Prediction Legend section.
Table 4.
DEGs common among three or more toxicity pathways through IPA-Tox analysis.
| DEG | Log2FC 1 | Tox Pathway |
|---|---|---|
| ADRB1 | −0.90 | Failure of heart |
| Congenital heart disease | ||
| Congestive heart failure | ||
| ATP2A2 | −0.83 | Failure of heart |
| Congenital heart disease | ||
| Congestive heart failure | ||
| BMP1 | −1.04 | Failure of heart |
| Congenital heart disease | ||
| Congestive heart failure | ||
| CAV1 | −0.72 | Failure of heart |
| Congestive heart failure | ||
| Dilation of heart chamber | ||
| Dilation of left ventricle | ||
| MYH6 | −1.85 | Congenital heart disease |
| Dilation of heart chamber | ||
| Dilation of left ventricle | ||
| NKX2-5 | −0.96 | Failure of heart |
| Congenital heart disease | ||
| Congestive heart failure | ||
| RXRA | −0.68 | Failure of heart |
| Congenital heart disease | ||
| Congestive heart failure | ||
| TNNT2 | −0.62 | Failure of heart |
| Congenital heart disease | ||
| Congestive heart failure |
1: Average of log2FC from individual studies.
3.4. Upstream Regulator Analysis
Upstream regulator analysis was performed through IPA. Differential expression data from the meta-analysis were used for constructing upstream regulator-targeted genes networks. These regulators could be transcription factors, non-coding RNA, enzymes or other molecules. A total of 876 potential upstream regulators were identified as significant (p-value of overlap < 0.05). Further, 38 of those regulators were differentially expressed in our dataset (29: downregulated; nine: upregulated in DCM) (Supplementary Table S2).
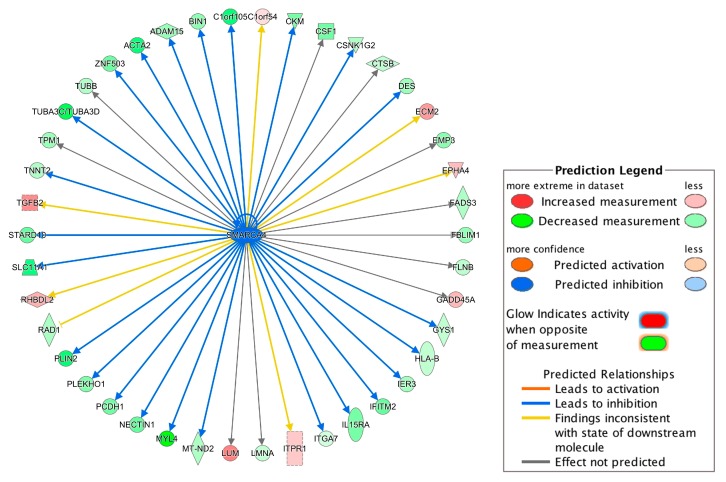
Among the 38 regulators, SMARCA4 (also known as Brg1) had the highest absolute prediction score (z-score = −3.00) and it was downregulated in the DCM group (Supplementary Table S2). Figure 4 summarized SMARCA4-regulated genes. Interestingly, the downregulation of SMARCA4 resulted in DES and TNNT2 downregulation (Figure 4), which could further promote failure of heat, congestive heart failure and congestive heart disease based on the IPA-Tox analysis (Figure 3). Therefore, SMARCA4 is a promising upstream regulator candidate regulating multiple downstream genes contributing to heart failure-related phenotypes.
Figure 4.
Network of the upstream regulator SMARCA4 and its targeted genes. The network was constructed using the differentially expressed genes identified in our meta-analysis. For example, the downregulation of SMARCA4 led to the inhibition (indicated by the blue arrow line) of the DES expression and the downregulation of DES is indicated by the green color. Other indicators are explained in the Prediction Legend section.
3.5. Canonical Pathway Analysis
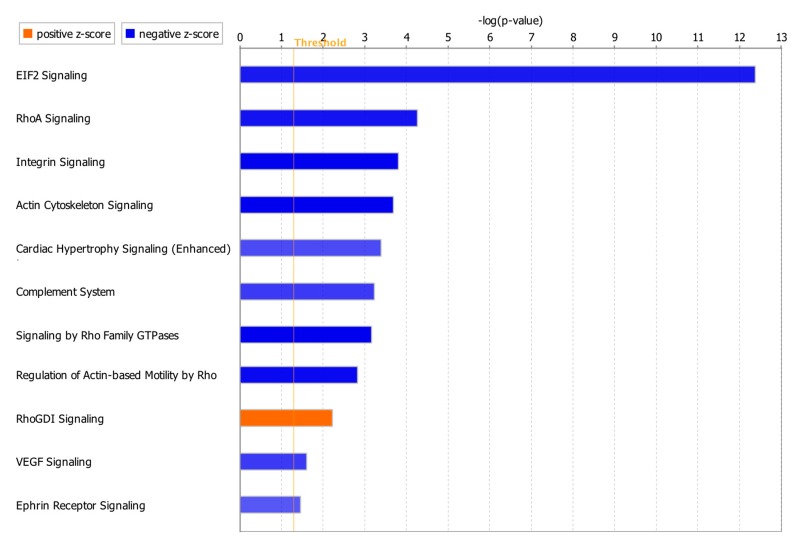
Canonical pathway analysis through IPA identified 96 significant pathways (p-value < 0.05). Considering z-score with more than 2.0 for significant activation or less than −2.0 for significant inhibition status in the pathway analysis, only 11 of the total 96 pathways had absolute z-score more than 2.0 with 10 pathways as significantly inhibited and one pathway as significantly activated (Figure 5). DEGs involved in each pathway are summarized in Supplementary Table S3. NKX2-5, CAV1 and ATP2A2 in the integrin signaling and cardiac hypertrophy signaling (enhanced) pathways (Supplementary Table S3) were also involved in the toxicity pathways such as failure of heart, congenital heart disease, congestive heart failure, dilation of heart chamber and dilation of left ventricle (Figure 3, Table 4). IL15RA, MAP3K6 and CD14 were among the genes which were identified only in our meta-analysis, but not in the individual studies (Table 3).
Figure 5.
Significant canonical pathways with absolute z-score > 2.0. The z-score represents the activation or inhibition state of a canonical pathway. A z-score greater than 2.0 was considered as significantly activated. A z-score smaller than −2.0 was considered as significantly inhibited.
4. Discussion
In this study, we performed a quantitative meta-analysis of three independent RNA-seq studies using human left ventricle tissues to profile gene expression signatures and identify novel genes associated with DCM. To better integrate the RNA-seq results from different studies, we applied a consistent bioinformatics pipeline (Figure 1) to analyze the raw RNA-seq data (FASTQ files) from the three independent studies in which three different pipelines were used [10,11,12]. Among a total of 58,884 genes used for expression quantification, 789 genes were identified as differentially expressed in meta-analysis including 581 downregulated and 208 upregulated genes (Supplementary Table S1). Only three genes, LCN10, TUBA3D and NPR3, were common between all three individual analyses and the meta-analysis (Figure 2). Interestingly, these three genes were among the genes with the high fold change in our analysis (log2FC = −2.64, −1.75 and 2.14, respectively) (Supplementary Table S1). LCN10 is a member of the lipocalin family that generally binds to small hydrophobic ligands and transport them to specific cells [26]. LCN10 has been shown to be significantly downregulated in patients with right ventricular heart failure [27]. TUBA3D, also known as TUBA2, encodes a member of the α tubulin family which is a major component of microtubules. TUBA3D along with other genes encoding microtubule subunits has been reported previously to be downregulated in DCM patients [28]. NPR3 (also known as NPR-C) encodes one of three receptors of natriuretic peptide which are small peptides responsible for regulating blood volume and pressure [29]. Several variations of NPR3 have been previously shown to be related to essential hypertension and coronary artery disease [30,31]. Follow-up functional studies are needed to investigate the mechanisms by which these genes contribute to the development of cardiomyopathy.
Specific alleles or change in expression levels of several genes from the top 50 DEG list (Table 2) such as MYH6 [32], CKM [33], NKX2-5 [34,35], and ATP2A2 [36] have been previously reported to be associated with DCM. MYH6 encoding α heavy chain subunit of cardiac myosin was downregulated in the DCM group in our dataset. Mutations in MYH6 have been reported in patients with both dilated and hypertrophic phenotypes of cardiomyopathy [32]. Inhibition of CKM, a cytoplasmic enzyme, which is a serum marker for myocardial infarction and contributes to energy homeostasis, has been also reported to inhibit cardiac contractility and contractility of muscle [29,33]. NKX2-5, which encodes a homeobox-containing transcription factor, was downregulated in our dataset [29]. Mutation in this gene could affect heart formation and development and has been reported in DCM [34,35]. ATP2A2, an ATPase enzyme that regulates the level of calcium ions inside the cells, is found in the sarcoplasmic reticulum involved in muscle contraction and relaxation by releasing and storing calcium ions [29]. Downregulation of ATP2A2 has been reported during human end-stage heart failure and has been suggested to be responsible for change in Ca2+ movements and myocardial relaxation [36].
Our meta-analysis identified 39 unique DEGs that were not identified as DEGs in individual analyses (Table 3), indicating that our meta-analysis further increased the statistical power for identifying new disease-associated genes, which has been reported as an advantage of the meta-analysis approach [7,8]. Some of these genes, including PTH1R, S100A4 and ADAM15, have been reported to be differentially expressed in DCM or other types of cardiomyopathies. PTH1R encodes a protein from G-protein coupled receptor family 2, which is a receptor for parathyroid hormone (PTH) and for parathyroid hormone-like hormone (PTHLH) [29]. This gene was downregulated in our study. Although no report was found for direct association between PTH1R and DCM, PTH has direct effects on heart and calcium homeostasis, which could impact cardiovascular system [37]. Other studies have also observed the downregulation of S100A4 and upregulation of ADAM15 in failing heart in human [38,39]. However, ADAM15 was downregulated in our study, suggesting a complicated pathophysiological role of ADAM15 in heart failure.
For further understanding physiological processes involved in DCM, we performed Ingenuity Pathway Analysis (IPA, Qiagen). Five toxicity pathways, including failure of heart (with the highest activation z-score), were identified as significantly activated in the DCM group. Interestingly, all these activated toxicity pathways were related to heart failure and cardiac dilation, demonstrating that the identified DEGs in our meta-analysis are significant to capture the progression of heart failure.
Among the upstream regulators, SMARCA4 with the highest absolute prediction score (z-score = −3.00) was downregulated in the DCM group and was predicted to target 42 genes (Figure 4). SMARCA4, also known as BRG1, is a member of the SWI/SNF family of proteins with helicase and ATPase activities and is known for regulating the transcription of several genes by altering the chromatin structure [29]. Among the genes involved in progression of toxicity pathways (Figure 3), DES and TNNT2 are targeted by SMARCA4 (Figure 4) and the downregulation of SMARCA4 resulted in the downregulation of DES and TNNT2. The evidence shows that chromatin remodeling is required for DES expression [40] and inhibition or lack of DES could lead to idiopathic DCM [41]. Inhibition of TNNT2 was also previously reported to lead to left ventricular dilation and impaired contractility [42].
Canonical pathway analysis showed that 11 pathways were affected with ten significantly inhibited and one significantly activated (Figure 5). Actin cytoskeleton signaling, Ras homolog family member A (RhoA) signaling, integrin signaling, cardiac hypertrophy signaling (enhanced) and vascular endothelial growth factor (VEGF) signaling pathways have been previously reported to be related to cardiac functions [43,44,45,46,47]. Change in actin cytoskeleton has been considered for the regulation of cardiomyocyte [43]. RhoA in cardiomyocytes can have both detrimental and beneficial effects on the heart as it has been reported to prevent progression of dilation and heart failure, but also promote the cardiac fibrosis [44]. Uncontrolled integrin activation could also result in contractile dysfunction and arrhythmias [45]. Downregulation of VEGF has been observed in dilated cardiomyopathy and heart failure [46]. Interestingly, several genes including NKX2-5, ATP2A2 and CAV1 involved in these pathways were also found in toxicity pathways such as failure of heart, congenital heart disease, congestive heart failure, dilation of heart chamber and dilation of left ventricle. Roles of NKX2-5 and ATP2A2 in developing cardiac diseases were discussed above. Loss of CAV-1 gene expression in mice has been also reported to cause the progression of hypertrophic cardiomyopathy and sudden cardiac death syndrome [48]. Overall, our meta-analysis successfully identified a core set of genes associated with DCM including well-known cardiomyopathy biomarkers as well as newly identified genes. This set of genes from our meta-analysis will be provided as more robust candidates for DCM compared to the genes profiled from individual studies. Our meta-analysis further provided the DCM-associated genes as potential diagnostic biomarkers and therapeutic targets for future clinical application. For most of the new DEGs identified in our study, there is no report of a direct relationship with DCM. Thus, future research on these new DEGs is needed to validate their physiological roles in DCM and further understand their molecular mechanisms in cardiovascular pathophysiology, especially cardiomyopathies.
Acknowledgments
The work was supported by Dean’s Postdoc to Faculty Transition Fellowship from University of Toledo College of Medicine and Life Sciences to Xi Cheng. Patricia Munroe acknowledges support from the National Institute of Health Research Cardiovascular Biomedical Research Centre at Barts and Queen Mary University of London. We appreciate the computational support from Ohio Supercomputer.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/1/60/s1. Table S1: A complete list of differential expressed genes identified in meta-analysis of DCM vs NF. Table S2: Differentially expressed upstream regulators and their target genes. Table S3: Significant canonical pathways with absolute z-score > 2.0 and the involved genes.
Author Contributions
Conceptualization, X.C.; data curation, A.A.; formal analysis, A.A.; investigation, A.A., P.B.M., B.J. and X.C.; methodology, A.A., P.B.M., B.J. and X.C.; project administration, X.C.; resources, X.C.; supervision, X.C.; validation, A.A. and X.C.; visualization, A.A.; writing—original draft, A.A.; writing—review and editing, A.A., P.B.M., B.J. and X.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Dean’s Postdoc to Faculty Transition Fellowship from University of Toledo College of Medicine and Life Sciences to Dr. Xi Cheng.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Benjamin E.J., Muntner P., Bittencourt M.S. Heart disease and stroke statistics—2019 update: A report from the American Heart Association. Circulation. 2019;139:e56–e528. doi: 10.1161/CIR.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 2.Heo S., Lennie T.A., Okoli C., Moser D.K. Quality of life in patients with heart failure: Ask the patients. Hear. Lung. 2009;38:100–108. doi: 10.1016/j.hrtlng.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maron B.J., Towbin J.A., Thiene G., Antzelevitch C., Corrado D., Arnett D., Moss A.J., Seidman C.E., Young J.B. Contemporary definitions and classification of the cardiomyopathies: An American Heart Association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functio. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 4.Tayal U., Prasad S., Cook S.A. Genetics and genomics of dilated cardiomyopathy and systolic heart failure. Genome Med. 2017;9:20. doi: 10.1186/s13073-017-0410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kayvanpour E., Sedaghat-Hamedani F., Amr A., Lai A., Haas J., Holzer D.B., Frese K.S., Keller A., Jensen K., Katus H.A. Genotype-phenotype associations in dilated cardiomyopathy: Meta-analysis on more than 8000 individuals. Clin. Res. Cardiol. 2017;106:127–139. doi: 10.1007/s00392-016-1033-6. [DOI] [PubMed] [Google Scholar]
- 6.McNally E.M., Mestroni L. Dilated cardiomyopathy: Genetic determinants and mechanisms. Circ. Res. 2017;121:731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rau A., Marot G., Jaffrézic F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinform. 2014;15:91. doi: 10.1186/1471-2105-15-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker E., Hernandez A.V., Kattan M.W. Meta-analysis: Its strengths and limitations. Cleve. Clin. J. Med. 2008;75:431. doi: 10.3949/ccjm.75.6.431. [DOI] [PubMed] [Google Scholar]
- 9.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweet M.E., Cocciolo A., Slavov D., Jones K.L., Sweet J.R., Graw S.L., Reece T.B., Ambardekar A.V., Bristow M.R., Mestroni L. Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genom. 2018;19:812. doi: 10.1186/s12864-018-5213-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y., Morley M., Brandimarto J., Hannenhalli S., Hu Y., Ashley E.A., Tang W.H.W., Moravec C.S., Margulies K.B., Cappola T.P. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105:83–89. doi: 10.1016/j.ygeno.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiano C., Costa V., Aprile M., Grimaldi V., Maiello C., Esposito R., Soricelli A., Colantuoni V., Donatelli F., Ciccodicola A. Heart failure: Pilot transcriptomic analysis of cardiac tissue by RNA-sequencing. Cardiol. J. 2017;24:539–553. doi: 10.5603/CJ.a2017.0052. [DOI] [PubMed] [Google Scholar]
- 13.Sweet M., Ambardekar A., Bristow M., Mestroni L., Taylor M. RNA-seq of heart failure in human left ventricles. GEO Database. 2018 [Google Scholar]
- 14.Liu Y., Morley M., Brandimarto J., Hannenhalli S., Hu Y., Ashley E., Tang W., Moravec C., Margulies K., Cappola T., et al. RNA-Seq Identifies Novel Myocardial Gene Expression Signatures of Heart Failure [RNA-seq] GEO Database. 2014 doi: 10.1016/j.ygeno.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiano C., Costa V., Casamassmi A., Aprile M., Rienzo M., Esposito R., Ciccodicola A., Napoli C. RNA-Sequencing shows novel transcriptomic signatures in failing and non-failing human heart. GEO Database. 2015 [Google Scholar]
- 16.Andrews S. FastQC: A Quality Control Tool for High Throughout Sequence Data. [(accessed on 04 January 2020)];2010 Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 17.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 18.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher R.A. Statistical Methods for Research Workers. Oliver and Boyd; Edinburgh, UK: 1925. [Google Scholar]
- 23.Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2013;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H., Boutros P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohio Supercomputer Center Ohio Supercomputer Center. Columbus OH. [(accessed on 4 January 2020)];1987 Available online: https://www.osc.edu.
- 26.Suzuki K., Lareyre J.-J., Sánchez D., Gutierrez G., Araki Y., Matusik R.J., Orgebin-Crist M.-C. Molecular evolution of epididymal lipocalin genes localized on mouse chromosome 2. Gene. 2004;339:49–59. doi: 10.1016/j.gene.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 27.Di Salvo T.G., Yang K.-C., Brittain E., Absi T., Maltais S., Hemnes A. Right ventricular myocardial biomarkers in human heart failure. J. Card. Fail. 2015;21:398–411. doi: 10.1016/j.cardfail.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinig M., Adriaens M.E., Schafer S., van Deutekom H.W.M., Lodder E.M., Ware J.S., Schneider V., Felkin L.E., Creemers E.E., Meder B. Natural genetic variation of the cardiac transcriptome in non-diseased donors and patients with dilated cardiomyopathy. Genome Biol. 2017;18:170. doi: 10.1186/s13059-017-1286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2015;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang P.Y., Liu Z.Y., Qin L., Zhao P. Association of NPRA and NPRC gene variants and hypertension in Mongolian population. Genet. Mol. Res. GMR. 2015;14:18494–18502. doi: 10.4238/2015.December.23.37. [DOI] [PubMed] [Google Scholar]
- 31.Hu Q., Liu Q., Shasha Wang X.Z., Zhang Z., Lv R., Jiang G., Ma Z., He H., Li D., Liu X. NPR-C gene polymorphism is associated with increased susceptibility to coronary artery disease in Chinese Han population: A multicenter study. Oncotarget. 2016;7:33662. doi: 10.18632/oncotarget.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carniel E., Taylor M.R.G., Sinagra G., Di Lenarda A., Ku L., Fain P.R., Boucek M.M., Cavanaugh J., Miocic S., Slavov D. α-Myosin heavy chain: A sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation. 2005;112:54–59. doi: 10.1161/CIRCULATIONAHA.104.507699. [DOI] [PubMed] [Google Scholar]
- 33.Crozatier B., Badoual T., Boehm E., Ennezat P.-V., Guenoun T., Su J., Veksler V., Hittinger L.U.C., Ventura-Clapier R. Role of creatine kinase in cardiac excitation-contraction coupling: Studies in creatine kinase-deficient mice. FASEB J. 2002;16:653–660. doi: 10.1096/fj.01-0652com. [DOI] [PubMed] [Google Scholar]
- 34.Sveinbjornsson G., Olafsdottir E.F., Thorolfsdottir R.B., Davidsson O.B., Helgadottir A., Jonasdottir A., Jonasdottir A., Bjornsson E., Jensson B.O., Arnadottir G.A. Variants in NKX2-5 and FLNC Cause Dilated Cardiomyopathy and Sudden Cardiac Death. Circ. Genom. Precis. Med. 2018;11:e002151. doi: 10.1161/CIRCGEN.117.002151. [DOI] [PubMed] [Google Scholar]
- 35.Hanley A., Walsh K.A., Joyce C., McLellan M.A., Clauss S., Hagen A., Shea M.A., Tucker N.R., Lin H., Fahy G.J. Mutation of a common amino acid in NKX2. 5 results in dilated cardiomyopathy in two large families. BMC Med. Genet. 2016;17:83. doi: 10.1186/s12881-016-0347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercadier J.-J., Lompre A.-M., Duc P., Boheler K.R., Fraysse J.B., Wisnewsky C., Allen P.D., Komajda M., Schwartz K. Altered sarcoplasmic reticulum Ca2 (+)-ATPase gene expression in the human ventricle during end-stage heart failure. J. Clin. Investig. 1990;85:305–309. doi: 10.1172/JCI114429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown S.J., Ruppe M.D., Tabatabai L.S. The parathyroid gland and heart disease. Methodist Debakey Cardiovasc. J. 2017;13:49. doi: 10.14797/mdcj-13-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pleger S.T., Most P., Katus H.A. S100 proteins: A missing piece in the puzzle of heart failure? Cardiovasc. Res. 2007;75:1–2. doi: 10.1016/j.cardiores.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Fedak P., Moravec C., McCarthy P., Altamentova S., Wong A., Skrtic M., Verma S., Weisel R., Li R. Altered expression of disintegrin metalloproteinases and their inhibitor in human dilated cardiomyopathy. Circulation. 2006;17:238–245. doi: 10.1161/CIRCULATIONAHA.105.571414. [DOI] [PubMed] [Google Scholar]
- 40.Ivana L., Roy K., Carlson K.A., Imbalzano A.N. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J. Biol. Chem. 2001;276:41486–41491. doi: 10.1074/jbc.M107281200. [DOI] [PubMed] [Google Scholar]
- 41.Pawlak A., Gil R.J., Grajkowska W., Nasierowska-Guttmejer A.M., Rzezak J., Kulawik T. Significance of low desmin expression in cardiomyocytes in patients with idiopathic dilated cardiomyopathy. Am. J. Cardiol. 2013;111:393–399. doi: 10.1016/j.amjcard.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 42.Ahmad F., Banerjee S.K., Lage M.L., Huang X.N., Smith S.H., Saba S., Rager J., Conner D.A., Janczewski A.M., Tobita K. The role of cardiac troponin T quantity and function in cardiac development and dilated cardiomyopathy. PLoS ONE. 2008;3:e2642. doi: 10.1371/journal.pone.0002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehler E. Actin-associated proteins and cardiomyopathy—The ‘unknown’beyond troponin and tropomyosin. Biophys. Rev. 2018;10:1121–1128. doi: 10.1007/s12551-018-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lauriol J., Keith K., Jaffré F., Couvillon A., Saci A., Goonasekera S.A., McCarthy J.R., Kessinger C.W., Wang J., Ke Q. RhoA signaling in cardiomyocytes protects against stress-induced heart failure but facilitates cardiac fibrosis. Sci. Signal. 2014;7:ra100. doi: 10.1126/scisignal.2005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valencik M.L., Zhang D., Punske B., Hu P., McDonald J.A., Litwin S.E. Integrin activation in the heart: A link between electrical and contractile dysfunction? Circ. Res. 2006;99:1403–1410. doi: 10.1161/01.RES.0000252291.88540.ac. [DOI] [PubMed] [Google Scholar]
- 46.Abraham D., Hofbauer R., Schäfer R., Blumer R., Paulus P., Miksovsky A., Traxler H., Kocher A., Aharinejad S. Selective downregulation of VEGF-A165, VEGF-R1, and decreased capillary density in patients with dilative but not ischemic cardiomyopathy. Circ. Res. 2000;87:644–647. doi: 10.1161/01.RES.87.8.644. [DOI] [PubMed] [Google Scholar]
- 47.Hunter J.J., Chien K.R. Signaling pathways for cardiac hypertrophy and failure. N. Engl. J. Med. 1999;341:1276–1283. doi: 10.1056/NEJM199910213411706. [DOI] [PubMed] [Google Scholar]
- 48.Park D.S., Cohen A.W., Frank P.G., Razani B., Lee H., Williams T.M., Chandra M., Shirani J., De Souza A.P., Tang B. Caveolin-1 null (−/−) mice show dramatic reductions in life span. Biochemistry. 2003;42:15124–15131. doi: 10.1021/bi0356348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.