Abstract
To improve and focus preclinical testing, we combine tumor models based on a decellularized tissue matrix with bioinformatics to stratify tumors according to stage-specific mutations that are linked to central cancer pathways. We generated tissue models with BRAF-mutant colorectal cancer (CRC) cells (HROC24 and HROC87) and compared treatment responses to two-dimensional (2D) cultures and xenografts. As the BRAF inhibitor vemurafenib is—in contrast to melanoma—not effective in CRC, we combined it with the EGFR inhibitor gefitinib. In general, our 3D models showed higher chemoresistance and in contrast to 2D a more active HGFR after gefitinib and combination-therapy. In xenograft models murine HGF could not activate the human HGFR, stressing the importance of the human microenvironment. In order to stratify patient groups for targeted treatment options in CRC, an in silico topology with different stages including mutations and changes in common signaling pathways was developed. We applied the established topology for in silico simulations to predict new therapeutic options for BRAF-mutated CRC patients in advanced stages. Our in silico tool connects genome information with a deeper understanding of tumor engines in clinically relevant signaling networks which goes beyond the consideration of single drivers to improve CRC patient stratification.
Keywords: in silico simulation, 3D tissue models, colorectal cancer, BRAF mutation, targeted therapy, stratification
1. Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide in men, second in women, and 55% of the cases occur in developed countries [1]. About 15% of CRCs develop as a consequence of microsatellite instability (MSI) and therefore represent a specific subgroup of tumors. Recent analysis of sequence data from over 9000 tumor samples of “The Cancer Genome Atlas” demonstrated that genetic alterations concentrate in 10 common signaling pathways composing over 30 tumor types. CRC with MSI showed high alteration rates in nearly all of these pathways with most changes occurring in the mitogen-activated protein kinase (MAPK) and Wnt/β-catenin signaling pathways [2].
The activating BRAF missense mutation V600E is part of the MAPK pathway and occurs in 59% of melanoma patients and in at least 10% of CRC [3] being a powerful prognostic factor [4,5]. In CRC, the frequency of BRAF V600E mutation is 78% for MSI tumors as compared to only 8% in tumors with chromosomal instability pathway (CIP) [6]. The selective FDA-approved BRAF inhibitor vemurafenib (PLX4032), achieves an impressive clinical response in melanoma [7,8], but fails in most CRCs [9,10]. This may be due to a feedback activation of the epidermal growth factor receptor (EGFR) as its additional inhibition by the small molecule gefitinib (Iressa®) improved suppression of the MAPK signaling pathway in vitro [11,12]. The combination-therapy of BRAF and EGFR inhibition showed first but not yet fully conclusive success [13,14,15]. This stresses the urgent need for a deeper understanding of dependencies in common cancer pathways. To achieve this in a mathematical way, we pursue Boolean models that represent in silico the “yes or no logic” of tumor associated pathways [2] and the corresponding mutational regulation [3,4,5]. Moreover, the “switch on and off” of central connected driving forces of the tumor—the engines— [2] and the complex balances dominating cellular decision processes regarding proliferation, apoptosis, inflammation or growth arrest can be integrated within this approach. Boolean models have proven to reflect biological systems [16,17]. We use the topology of relevant signaling pathways to set up a Boolean interaction network as calculation base for refined new dynamic models allowing more quantitative predictions. For example, these models reveal the extent of proliferation or apoptosis without detailed knowledge of the involved kinetics of individual kinases and other molecular rate constants [18,19].
Commercially available cell lines grown under conventional two-dimensional (2D) in vitro cell culture conditions often do not reflect important characteristics of the tumor [20,21]. However, the in vivo situation relies on a 3D microenvironment that alters cell morphology and organization, proliferation, signaling, differentiation, gene expression and cell surface receptors, generally leading to a higher drug resistance compared to 2D conditions [22,23,24,25,26]. We used MSI CRC cell lines directly derived from patient-derived xenografts (PDX) [27]. These cell lines, HROC24 and HROC87 (Hansestadt ROstock—Colon), harbor a BRAF mutation with further co-mutations: HROC24 with APC mutation and HROC87 with p53 mutation [27,28,29]. For our CRC models, we use an avascular part of a decellularized porcine gut [30,31,32,33]—called small intestine submucosa with preserved mucosa (SISmuc) [34], that is also applied for lung cancer and breast cancer models [18,19,35,36,37]. The preserved basal membrane structure enables physiological anchorage of epithelial cells and the analysis of invasive processes. In lung cancer, we observed better predictivity compared to 2D as we could, for example, mirror the clinical attrition of an anti-HSP90 therapy in patients with a KRAS mutation that was suggested by other preclinical models [19,35]. Moreover, these 3D models were combined with an in silico tool for the development of signaling- and mutational-based signatures for therapy predictions in stratified patient groups [18,35].
In this study, we created CRC tissue models and an in silico topology integrating database information and scientific literature to simulate the transformation of healthy colon epithelial cells to a metastasizing carcinoma including critical mutations, interconnected subnetworks [38], processes such as the epithelial to mesenchymal transition (EMT), and different cellular responses. The in silico topology was adapted to experimentally measured differences in proliferation, apoptosis, and cell signaling upon drug treatment. Subsequently, we used this topology to predict individualized anticancer drug response. This therapeutic strategy provides insights into general mechanisms that can be exploited for better patient stratification in CRC, extending mutational profiles to functional tumor stages.
2. Results
2.1. Characterization of Low Passage Cell Lines and Comparison of 3D Tissue Morphology with Cancerous Specimens
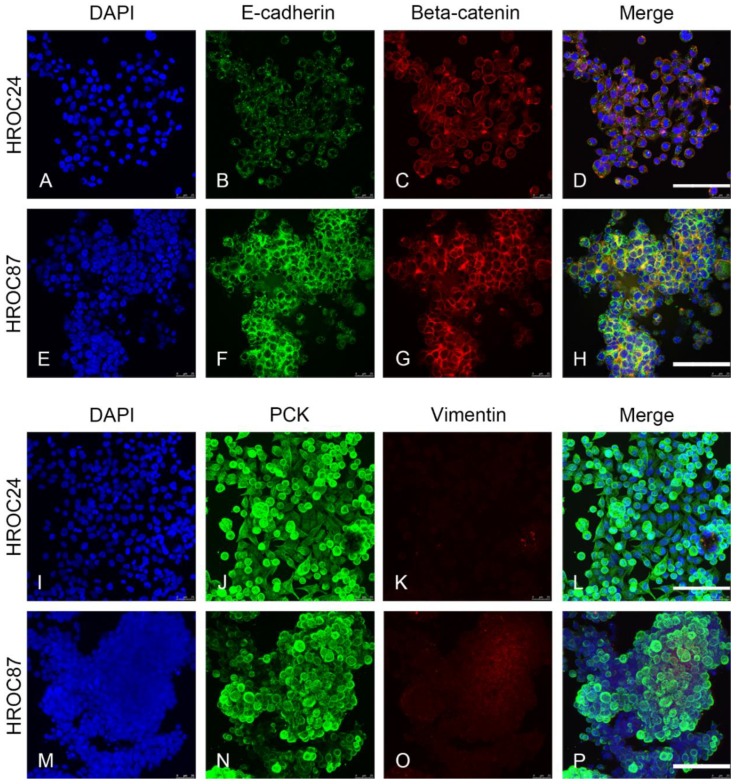
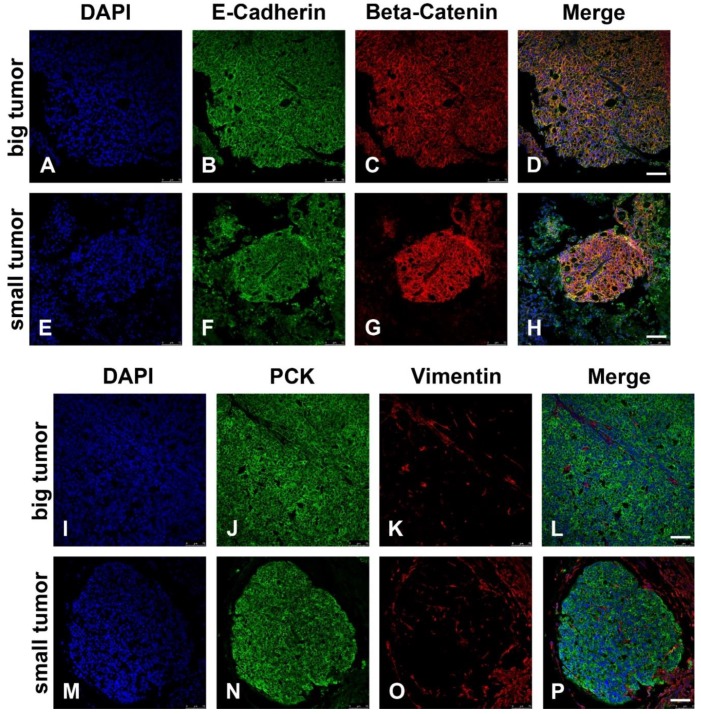
For 3D models, HROC24 and HROC87 cell lines were seeded onto the SISmuc scaffold derived from porcine gut and cultured for 14 days. To assess malignancy, EMT status of the cells was evaluated by immunohistochemical staining of E-cadherin, β -catenin, Pan-cytokeratin (PCK) and vimentin in 2D (Figure 1) and 3D (Figure 2).
Figure 1.
Characterization of 2D-cultured HROC24 and HROC87 cells. Both cell lines express E-cadherin ((B,F), green in (D,H)) and β-catenin ((C,G), red in (D,H)) as well as PCK ((J,N), green in (L,P)), but no vimentin ((K,O), red in (L,P)). E-cadherin and β-catenin clearly co-localize at the cell-to-cell borders in HROC87 tumor cells (yellow in (H)). Cell nuclei were counterstained with DAPI (A,E,I,M). Scale bars in (D,H,L,P): 75 µm for (A–P).
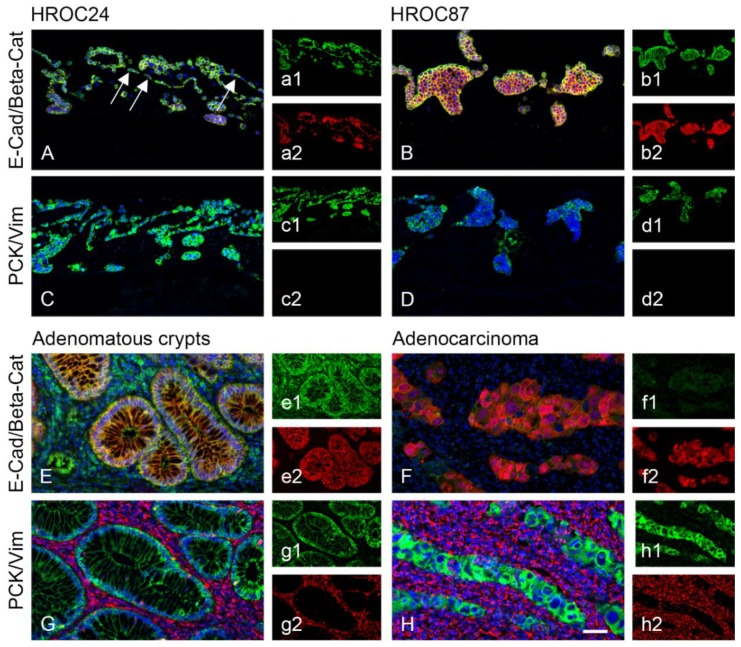
Figure 2.
Characterization of the cell lines HROC24 as well as HROC87 cultured on the scaffold SISmuc. While HROC24 cells grow in monolayers and fill the former crypts of the gut (A,C), HROC87 cells grow in dense clusters (B,D). E-cadherin is highly expressed at the cell-to-cell contacts of HROC24 cells (green in (A,a1)) as well as HROC87 cells (green in (B,b1)). β-catenin is more strongly expressed in HROC87 cells (red in (B,b2)) than in HROC24 cells (red in (A,a2)). Both cell lines are positive for PCK (green in (C,D), (c1,d1)) and do not express vimentin (c2,d2). For comparison, adenomatous crypts as well as adenocarcinoma tissue were stained with E-cadherin (green in (E,F), (e1,f1)), β-catenin (red in (E,F), (e2,f2)), PCK (green in (G,H), (g1,h1)) and vimentin (red in (G,H), (g2,h2)). Scale bar in (H): 50 µm for (A–H).
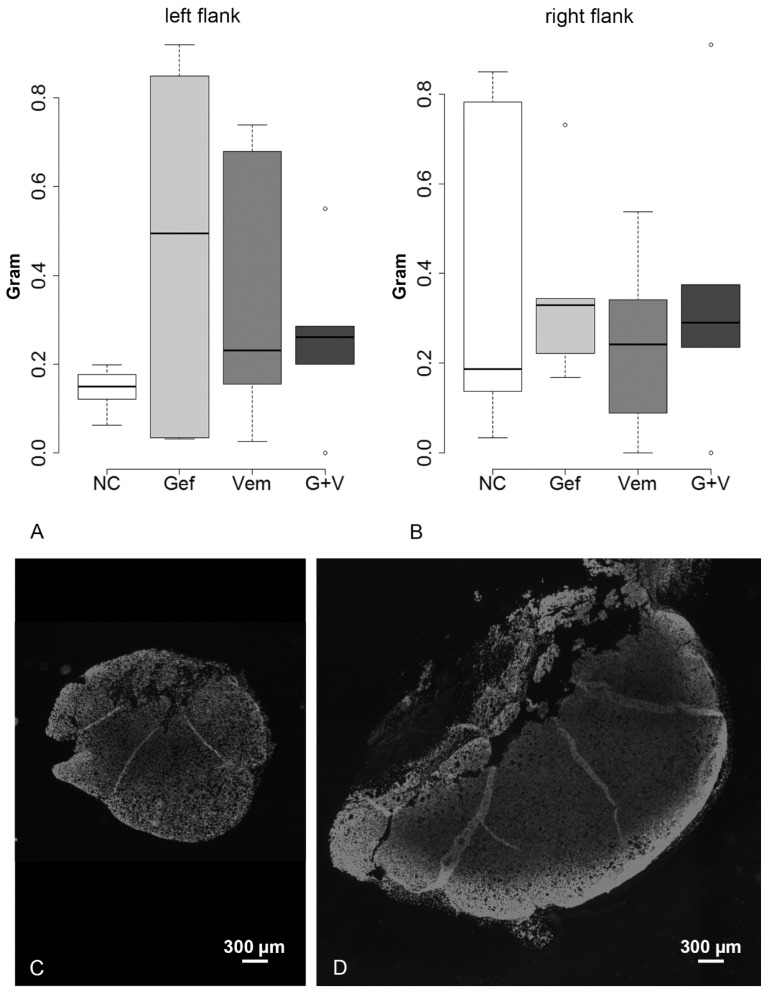
In 2D and 3D, all cells were clearly epithelial (positive for E-cadherin, β-catenin and PCK, negative for vimentin). In 3D, HROC24 cells built a monolayer on the surface (arrows in Figure 2A) of the SISmuc and filled single crypts with cells. In contrast, HROC87 cells formed no monolayer but tight cell aggregates on the matrix surface and grew only in some of the former crypts of the intestine (Figure 2B). Both cell lines thereby nicely reflect the differentiation state of their tumors of origin. β -catenin staining was stronger in HROC87 than in HROC24 cells but remained localized at the cell borders. PDX of HROC87 showed the same expression of all markers (Figure A1) and were of different size (Figure A2) and weight (Figure A3), independent of treatment conditions.
In comparison to patient specimens, our 3D models from HROC24 cells morphologically resembled adenomatous crypts, HROC87 cells the more advanced stage of an adenocarcinoma. Adenomatous crypts expressed E-cadherin at the cell-to-cell contacts of the crypts colocalized with β-catenin (Figure 2E). In an adenocarcinoma, E-cadherin expression was reduced and β -catenin was localized in the cytoplasm and also in the nuclei of the specimen (Figure 2F). As HROC87 cells derive from an adenocarcinoma without β-Catenin translocation [27], our model reflects the original condition in the patient. Both tumors showed no vimentin expression similar to the in vitro results.
Next, we were interested in treatment responses of the different models. Therefore, we compared the effects of the BRAF inhibitor vemurafenib when applied to 2D- and 3D-cultured cells and to PDX as monotherapy and in combination with the EGFR inhibitor gefitinib. To link these results to common pathways, we measured not only cell survival rates, but also proliferation and apoptosis as these responses emerge from different signaling cascades.
2.2. Proliferation and Apoptosis Responses upon Treatment
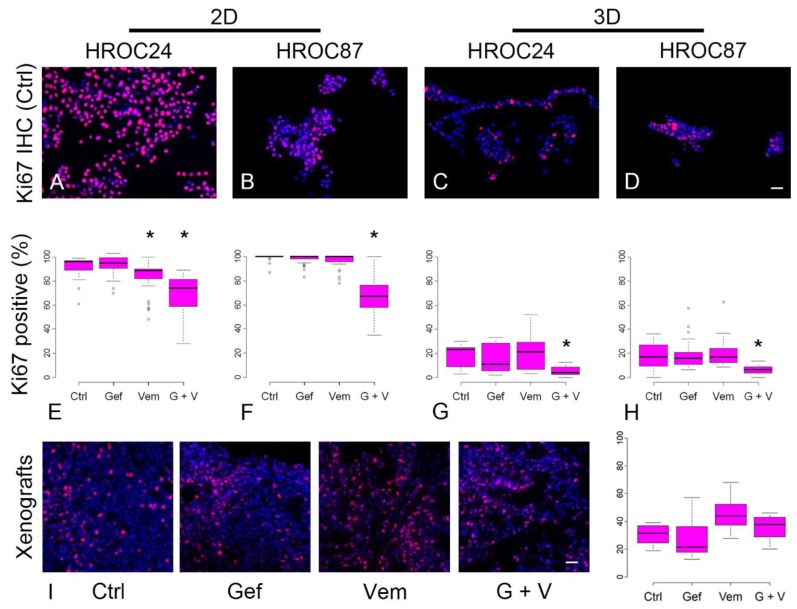
We assessed proliferation in 2D and 3D cell cultures by counting Ki67-stained cells and observed in general a lower proliferation in 3D than in 2D (Figure 3A–H).
Figure 3.
Proliferation of HROC24 and HROC87 cells is influenced differentially by vemurafenib, gefitinib or the combination-therapy. (A–D): Ki67 (red) and DAPI (blue) staining of cells in 2D (A,B) and 3D (C,D) cell culture. In 3D, proliferation is reduced compared to 2D. While under 2D culture conditions, HROC24 cells respond to vemurafenib and to the combination-therapy by reduced proliferation (E), HROC87 cells respond only to the combination-therapy (F). Cultured in 3D, proliferation is in both cell lines only reduced by the combination-therapy (G,H). PDX of HROC87 were stained with Ki67/DAPI and proliferation was evaluated (I). No changes were detected after different treatments in these PDX. *: p < 0.05. Ctrl: untreated control, Gef = gefitinib, Vem = vemurafenib, G + V = combination-therapy with gefitinib and vemurafenib. Scale bars in D and I: 50 µm for A to D and I. ° denotes outliers.
Proliferation was significantly decreased by vemurafenib and the combination-therapy in 2D-cultured HROC24 cells (Figure 3E), whereas proliferation of HROC87 cells was only significantly decreased by combining vemurafenib and gefitinib (Figure 3F). In 3D, HROC24 cells and HROC87 cells both showed significantly reduced proliferation rates only after combination-therapy (Figure 3G, H). Proliferation rates of HROC87 PDX were not influenced by different treatments (Figure 3I).
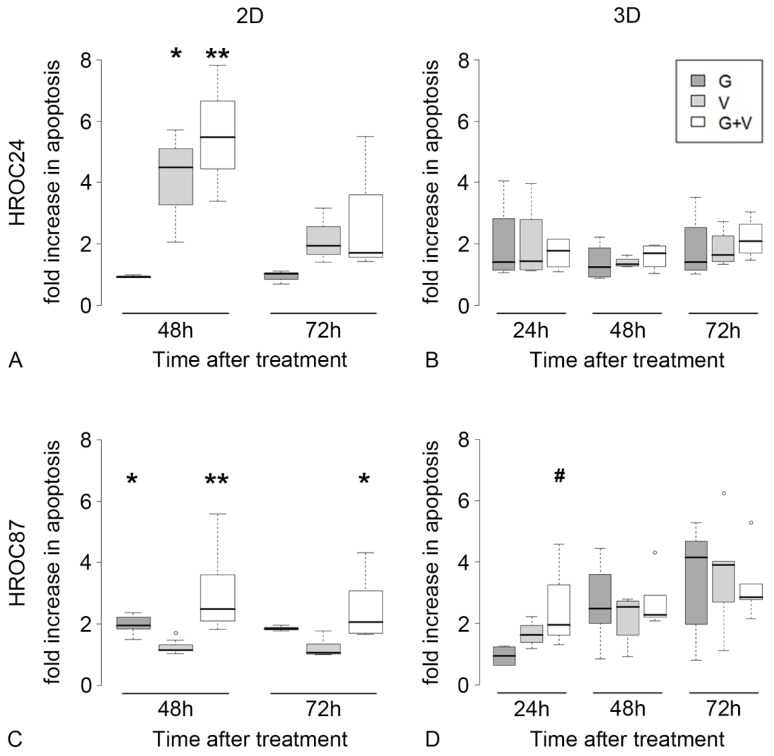
In 2D, the monotherapy with vemurafenib and the combination-therapy significantly increased the apoptosis rate in HROC24 cells after 48h (Figure 4A).
Figure 4.
Apoptosis rates in HROC24 versus HROC27 cells in 2D and 3D. We compared apoptosis rates (fold increase to untreated controls) of gefitinib treatment (G), vemurafenib (V) or their combination (G + V). HROC24 cells respond to monotherapy with vemurafenib in 2D as well as to the combination-therapy (A). In contrast, HROC87 cells respond to gefitinib monotherapy in 2D as well as to the combination-therapy (C). In 3D, both drugs are not able to significantly induce apoptosis, neither in monotherapy nor in combination-therapy in HROC24 (B) and in HROC87 cells (D). #: 0.05 < p < 0.1, *: 0.01 < p < 0.05, **: p < 0.01. ° denotes outliers.
HROC87 cells responded significantly to gefitinib and the combination-therapy by increased apoptosis (Figure 4C). In contrast, in 3D we observed only a trend but no significant apoptosis induction in HROC24 (Figure 4B) and HROC87 cells (Figure 4D).
2.3. Comparison of Signaling Changes upon Treatment between Different Models
Next, we analyzed if in signaling cascades centrally involved regulators of apoptosis and proliferation differ between the applied in vitro models (Figure 5; original blots in Figure A4, Figure A5 and Figure A6, densitometry analysis in Table A1, Figure A7).
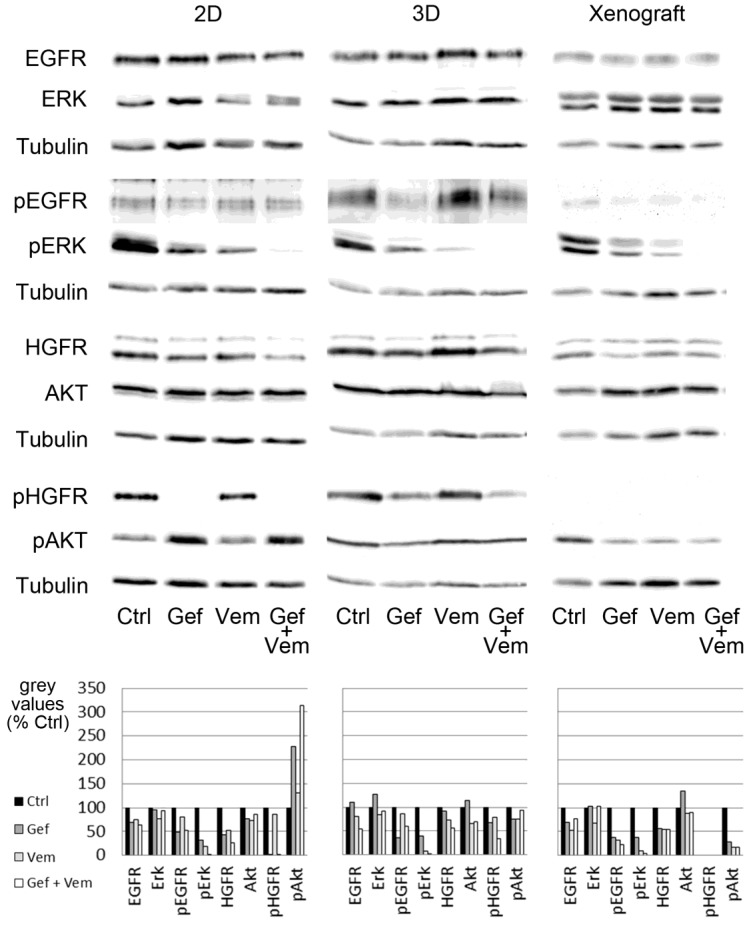
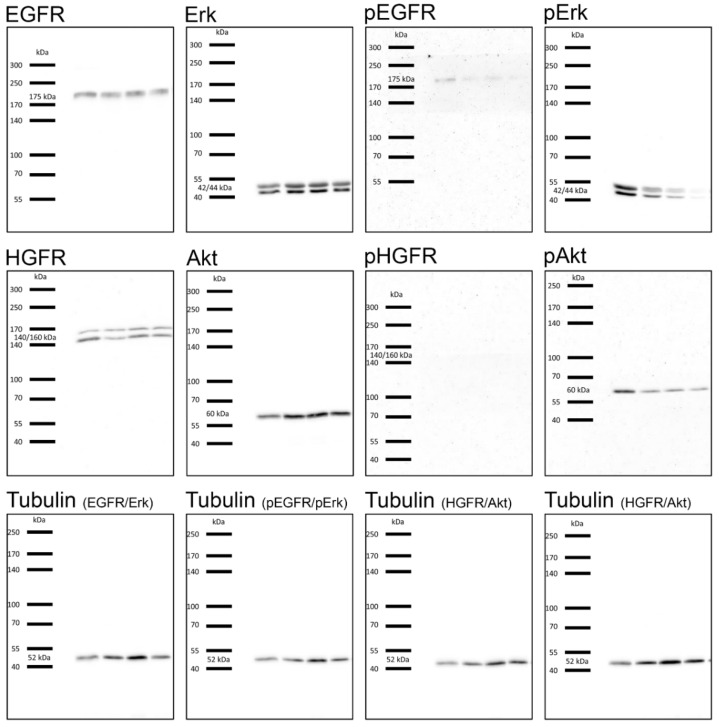
Figure 5.
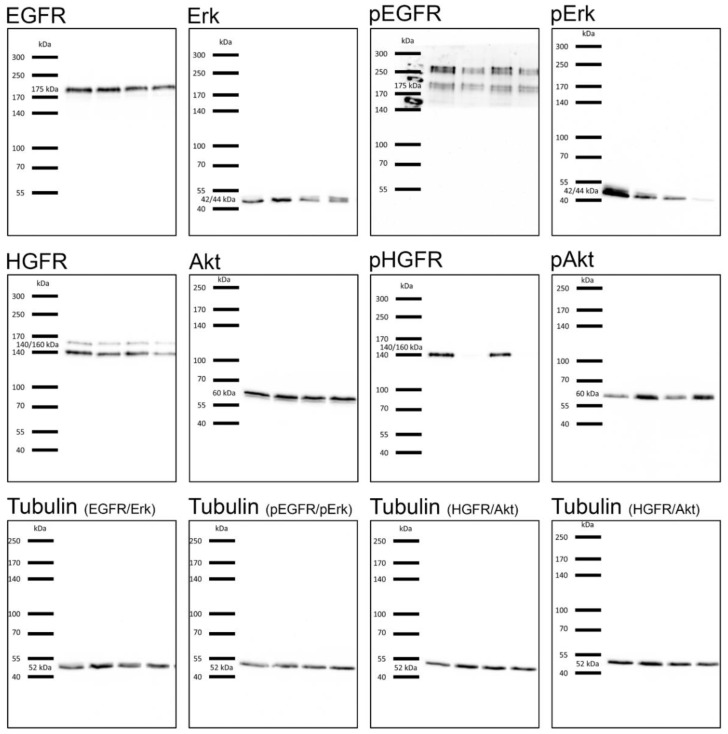
Signaling in HROC87 cells upon treatment with vemurafenib and/or gefitinib. Shown is the Western blot analysis of the phosphorylation status of molecules involved in signal transduction for proliferation (ERK) and apoptosis (AKT) and important receptors in HROC87 cells cultured under 2D and 3D conditions as well as in matching xenografts. In 2D, 3D and PDX, ERK is partly inhibited by gefitinib and vemurafenib in monotherapy, but nearly completely inhibited by the combination-therapy. In contrast to 2D, AKT phosphorylation show no clear upregulation under 3D culture conditions. HGFR could not be activated in the PDX at all. Only in the 2D model, gefitinib and the combination-therapy results in a strong inactivation of the HGFR. Ctrl: untreated control, Gef: gefitinib, Vem: vemurafenib, Gef + Vem: combination-therapy of gefitinib and vemurafenib. Grey values of densitometric analysis of the representative blots are shown in the diagrams (% of Ctrl). Blots with weight markers are shown in Figure A4, Figure A5 and Figure A6. Densitometric analyses of grey values from all 4 to 6 experiments (Figure A7) as well as from the representative blot depicted here (Table A1) are shown in the appendix.
We focused on HROC87 cells because these cells derive from a more advanced tumor stage. Beside the two receptors EGFR and HGFR we investigated ERK, a protein downstream of BRAF at the end of the MAPK cascade which is a main regulator of proliferation, and AKT (Ser473), one of the most important components regulating cell survival and apoptosis [39]. A simplified overview of these central pathways is given in Figure 6.
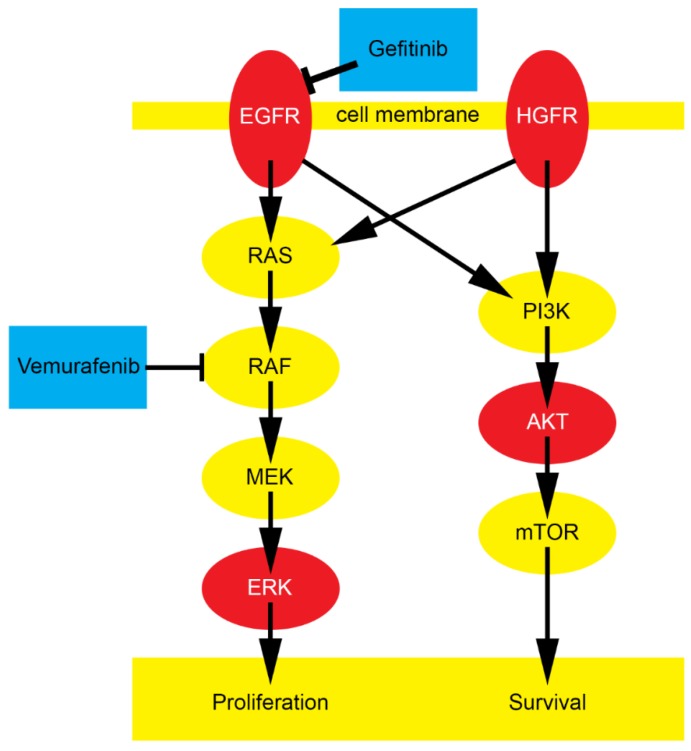
Figure 6.
Simplified overview of central pathways involved in proliferation and survival in CRC. EGFR is inhibited by gefitinib, but there is an interference with HGFR. While vemurafenib only blocks the left pathway in case of BRAF mutation, this interplay between the two pathways could be a reason for the low efficacy of vemurafenib in CRC even in case of an existing BRAF mutation.
All three model types (2D, 3D and PDX) showed decreased ERK phosphorylation after addition of gefitinib or vemurafenib that further diminished after the co-treatment (Figure 5, Table A1 and Figure A7).
We observed AKT activation in 2D after all treatments being strongest after combination-therapy (Figure 5, Table A1 and Figure A7). Contradictory, we measured in 2D conditions significant apoptosis induction upon gefitinib therapy and the combination-therapy. Under 3D conditions, AKT was active after all treatments, but the densitometric evaluation of all experiments revealed a slight reduction in activity upon gefitinib treatment and a tendency to be activated in response to the combination-therapy (Figure A7). Consistently, we observed no significant induction of apoptosis in 3D. In the PDX no strong differences were found in AKT phosphorylation in comparison of the different treatment conditions.
We also assessed the activation level of the EGFR as a target of gefitinib and the activation level of HGFR as an additional important tyrosine kinase receptor. In 2D, EGFR phosphorylation (pEGFR) decreased after gefitinib treatment. This effect was slightly stronger in 3D than in 2D and could only be seen to some extent in the PDX. Regarding vemurafenib, we observed no clear increase of pEGFR after treatment in 2D and 3D (Table A1 and Figure A7). Notably, we could not detect any activation of the HGFR in the animal model. In comparison to 2D culture, the HGFR was more active in 3D after gefitinib treatment and the combination-therapy, thus underlining the higher chemoresistance of the 3D approach.
2.4. In silico System Responses Reflecting 3D In Vitro Data by Integrating HROC87 Specific Mutations and Signaling Cascades
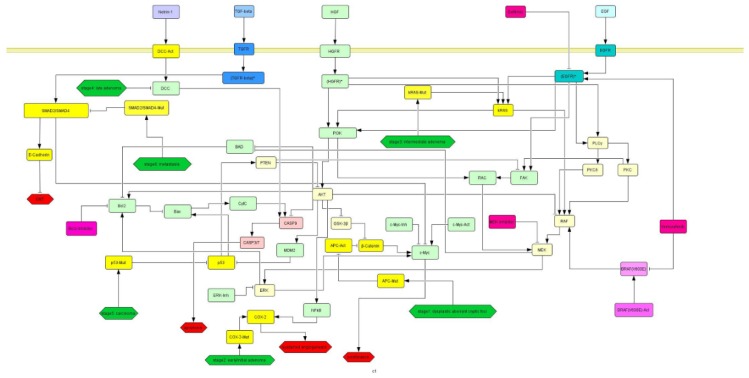
To gain a deeper understanding of common cancer pathways and interdependencies during tumorigenesis, we created an in silico topology based on information from databases and scientific literature to encapsulate relevant pathways in silico [38,39]. The resulting topology integrates centrally involved proteins of the common signaling pathways and generally accepted cancer drivers in CRC—such as APC, p53m, KRAS, DCC, TGFβR, PTEN and SMAD4 allowing a simplified view on the progression from normal cells to tumor cells (Table A2, Figure A8) [2,40,41,42].
A central view of signaling cascades and nodes of the network connecting MAPK with BRAF and vemurafenib is given in Figure 7A, the full network is in the Appendix (Figure A8). For in silico modeling, we considered the HROC87 mutations p53 and BRAF as well as the experimentally measured changes in activation of EGFR, HGFR, ERK and AKT upon different therapies for data-driven modeling using the Standardized QUAlitative Dynamical Modeling (SQUAD) software (Table A3).
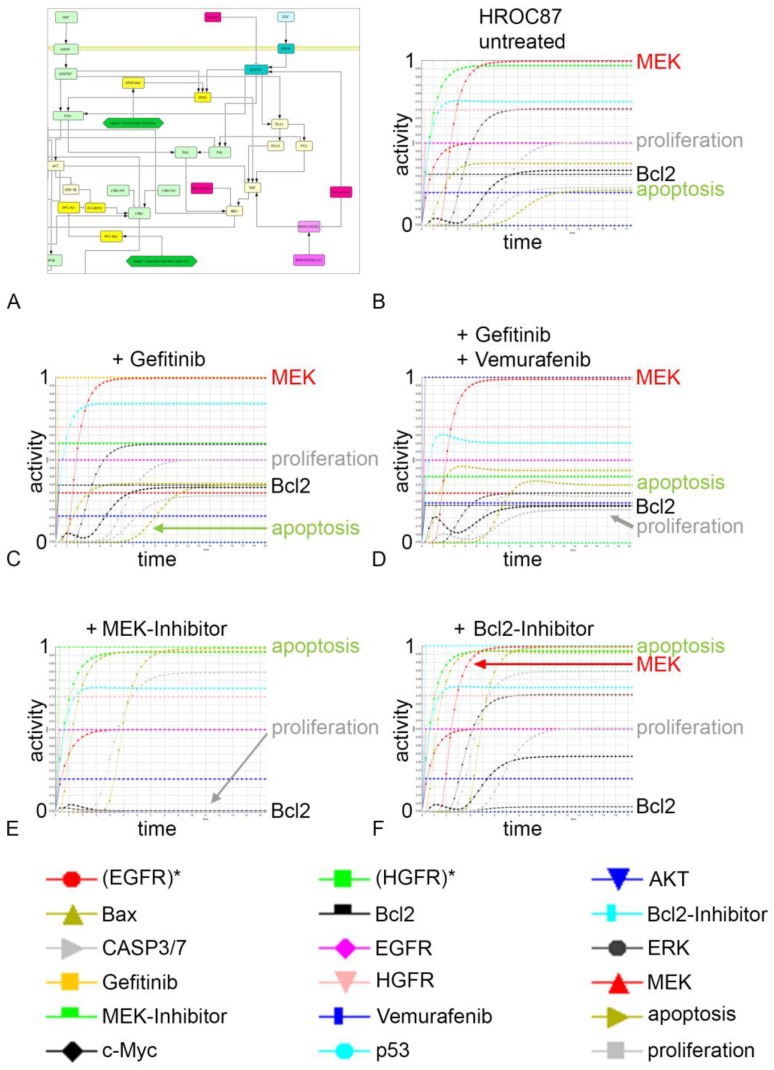
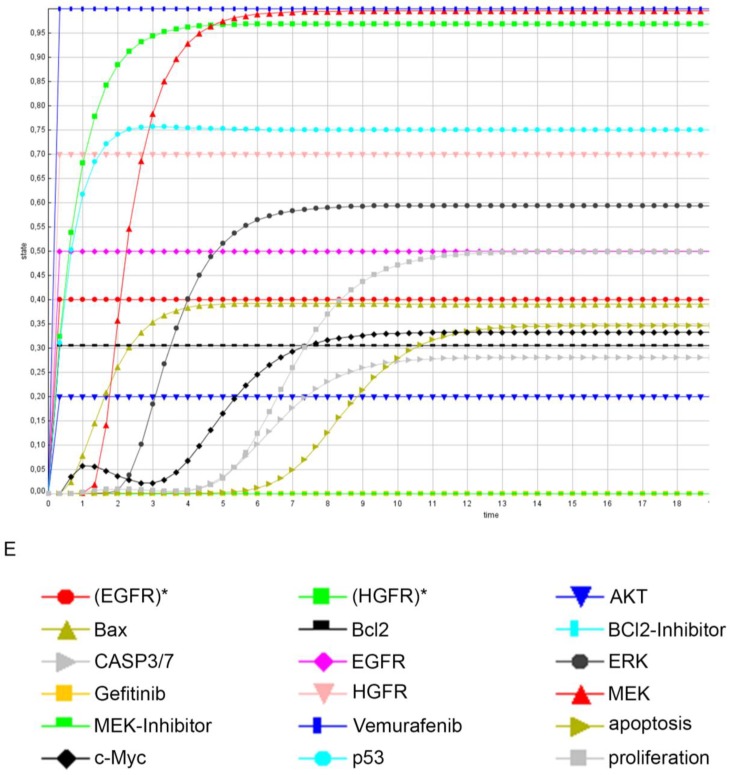
Figure 7.
In silico modeling of drug-responses of in 3D cultured HROC87 cells. (A) Part of the topology (the complete network is given in Figure A8 of the appendix). Our in silico topology accommodates well-documented major growth and apoptotic cascades in CRC and mimics as close as possible the experimental treatment responses as well as the behavior regarding untreated growth. Directed arrows in the network topology represent activation while blunted arrows represent inhibition. Rectangles: genes/proteins. (B) Untreated condition. HROC87 cells show a malignant phenotype with a weak activation of apoptosis (olive-green triangle curve) and high proliferation (grey square curve) signal. Visible in the different curves is also the EGFR (EGFR*), the HGFR (HGFR*), ERK, MEK and additional relevant curves such as Casp3/7. (C) Gefitinib treatment. HROC87 cells show only a weak induction of apoptosis (olive-green triangle curve), but no changes in proliferation (grey square curve) upon gefitinib (yellow square curve, activation: 1). (D) Combination treatment (gefitinib plus vemurafenib). HROC87 cells show only a weak induction of apoptosis (olive-green triangle curve) and reduction of proliferation (grey square curve). Treatments: gefitinib (yellow square curve, activation: 1, not seen because hidden by the curve for vemurafenib) and vemurafenib (blue rectangle curve, activation: 1). (E) MEK inhibition. This shows an induction of apoptosis (olive-green triangle curve) and reduction of proliferation (grey square curve) in HROC87 cells. Notably, there is an inactivation of ERK and Bcl2. (F) Bcl2 inhibition. As expected from blocking an anti-apoptotic protein (Bcl2), the simulation suggests an induction of apoptosis (olive-green triangle curve, positive control) but proliferation stays unchanged as there is no connection with MEK-ERK. * = activated form.
This required manually iterative refinement of the in silico model, until the activation level of key signaling nodes as well as proliferation and apoptosis of in vitro models are correctly mirrored. The pre-simulation values of our in silico model are given in Table 1.
Table 1.
Pre-stimulation activities of the in silico model as starting point for the therapy simulation.
| Name of Network Node | untr. | +gef | +vem | +combi | +MEK-Inh | +BCL2-Inh |
|---|---|---|---|---|---|---|
| BRAF(V600)-Act | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 |
| EGFR | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| HGFR | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 |
| ERK-Inh | 0.26 | 0.3 | 0.3 | 0.41 | 0.26 | 0.26 |
| p53-Mut | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 |
| Bcl2 | 0.31 | 0.3467 | 0.305 | 0.2252 | ||
| c-Myc-Inh | 0.17 | 0.097 | 0.097 | 0.17 | 0.17 | |
| AKT | 0.2 | 0.16 | 0.2 | 0.24 | 0.2 | 0.2 |
| (EGFR) * | 0.3 | 0.4 | 0.3 | |||
| (HGFR) * | 0.6 | 0.4 | ||||
| c-Myc-Act | 0.09 | |||||
| Gefitinib | 1.0 | 1.0 | ||||
| Vemurafenib | 1.0 | 1.0 | ||||
| MEK-Inhibitor | 1.0 | |||||
| Bcl2-Inhibitor | 1.0 |
* denotes that the protein for which this node stands for is the activated form of the receptor.
These pre-simulation values represent the activation levels of the mutations, experimental nodes and associated signaling pathway nodes to start the simulations, subsequently leading to the different treatment outcomes in silico (Table 2), reflecting results of the in vitro model.
Table 2.
Simulation outcome of the pre-stimulation settings 1 to mimic the cell culture response behavior of HROC87 cells.
| Name of Network Node | untr. | +gef | +vem | +combi | +MEK-Inh | +BCL2-Inh |
|---|---|---|---|---|---|---|
| apoptosis | 0.21 | 0.35 | 0.35 | 0.35 | 0.99 | 0.99 |
| proliferation | 0.5 | 0.5 | 0.5 | 0.2 | 0.0 | 0.5 |
| (HGFR)* | 0.97 | 0.6 | 0.97 | 0.4 | 0.97 | 0.97 |
| ERK | 0.71 | 0.59 | 0.59 | 0.3 | 0.0 | 0.71 |
| (EGFR)* | 0.5 | 0.3 | 0.4 | 0.3 | 0.5 | 0.5 |
| AKT | 0.2 | 0.16 | 0.2 | 0.24 | 0.2 | 0.2 |
1 perturbation function of SQUAD, pre-stimulations in Table 1. * denotes that the protein for which this node stands for is the activated form of the receptor.
This is done in a simplified way by considering specific activation levels according to the experimental data as we only model a subnetwork of the complete cancer cell. The software SQUAD [43] models semi-quantitatively the cellular network dynamics of receptor activation, signal processing and cellular output including selective inhibition/activation of individual proteins by drugs to predict tumor-specific responses. The results of dynamical therapy simulations including changes of every node of interest in the network was visualized graphically (Figure 7B–D). In our example, we simulated the untreated situation (Figure 7B), the inhibition of EGFR by gefitinib (Figure 7C), the inhibition of BRAF by vemurafenib (Figure A9) and their combination (Figure 7D). However, even the combination-therapy was not completely effective in our simulations reflecting thereby the results of 3D models and also clinical results. As possible effective targets our simulation suggests MEK and Bcl2, because these stay upregulated over time (red and black lines in Figure 7B–D).
2.5. Breaking Resistance: In Silico Mode-of-Action Analysis in p53 and BRAF Mutation Background
We established our combined in vitro/in silico model with the vision to translate individual genomic mutational profiles into predictions of targeted therapy efficacy in order to prevent therapy failure and to help the patients in case of therapy resistances.
As induction of apoptosis is often hindered in therapy-resistant tumor cells, but is important for an effective therapy, we sought how to connect relevant nodes of the in silico topology to the known apoptosis pathways. We experimentally observed major differences between the 2D and 3D models concerning HGFR and AKT activation upon different therapies together with significant apoptosis induction exclusively in 2D but not in 3D.
Anti-apoptotic Bcl2 is activated by ERK and AKT. Next to RAS and RAF, HGFR activates also AKT, which explains why Bcl2 could still be activated by AKT when ERK is strongly downregulated as shown in the Western Blot (Figure 5) resulting in a weak apoptotic effect in the 3D system and illustrated by the simulations in Figure 7B–D. However, P53 activates apoptosis via BAX (inhibited by Bcl2) and PTEN (inhibits AKT). In the patient group with a p53 mutation, this tumor suppressor does not activate the phosphatase PTEN anymore so that AKT stays active as also experimentally observed and apoptosis is inhibited. Furthermore, MEK and AKT inhibit pro-apoptotic BAD, in which the latter one inhibits Bcl2. This could further explain that we can see no strong apoptosis induction in the 3D system after treatment due to the missing inhibition of Bcl2 via BAD.
The cell line HROC87 harbors also a BRAF mutation. Beside apoptosis, the RAS-RAF-MEK-ERK cascade regulates proliferation. In these cells, a downregulation of ERK correlates with a significant lower proliferation after the combination-therapy that was reflected in silico.
The in silico support a central role of MEK, AKT and Bcl2 in the apoptotic regulation in the HROC87 cells. We subsequently investigated the systemic effect of MEK or Bcl2 inhibition. The simulation of MEK inhibition led to full induction of apoptosis due to inhibition of Bcl2 even though AKT and HGFR stay active according to our experimental results (Figure 7E). Moreover, the MEK inhibition also leads to a reduction of proliferation due to the full ERK inhibition (Figure 7E). As a control of network specificity, in silico Bcl2 inhibition led to an induction of apoptosis but had no effect on proliferation (Figure 7F). Moreover, this in silico response behavior is at least supported by current clinical trials [44,45,46].
3. Discussion
In melanoma and non-small cell lung cancer (NSCLC) biomarker-based targeted therapies have entered the clinical routine. In this study, we try to translate these growing insights into common cancer pathways and explore the relevance of individual mutations to CRC. We introduce a tissue engineered human CRC 3D model and an in silico tool for the systemic understanding of mutational steps in cancer progression and their impact on drug efficacy that can support patient stratification.
Cell responses depend on their environment [19,34,47] as also demonstrated by other 3D methods like matrigel coating assays, spheroid cultures or models on synthetic or natural matrices [48,49]. Recently established organoid cultures maintain different cell types and enable personalized drug testing [50]. Our models aim at the detection of general interdependencies between different signaling pathways by selecting early passage cell lines with defined mutations. We have established a 3D test system for BRAF-mutant CRC using cells with different co-mutations [27,28] and show a good molecular and phenotypic reproduction of the original tumors. Advantages of our model are (I) lower proliferation similar to that in patients, (II) higher chemoresistance compared to 2D models, and (III) a comprehensive signaling profile for the tested therapies. In the last step (IV), we reflect results from 3D models by bioinformatical in silico models that connect the two most frequently impaired common signaling pathways in this CRC subgroup to allow focused target predictions.
In general, the high proliferation rates in 2D lead to an overestimation of cytostatic compounds’ efficacy [19,34]. Culturing tumor cells on a tissue matrix adjusted proliferation to in vivo levels. Furthermore, we noted in 2D models to some extent already efficacy of monotherapies, which does not reflect the clinic in CRC [9,10,51]. As we aimed at understanding network effects in a certain mutational background and at models reflecting them, we analyzed differences in central signaling cascades important for proliferation and apoptosis. Whereas in all of the three tested experimental models ERK, as a crucial protein downstream of BRAF at the end of the MAPK pathway, was inhibited in the same way by the different applied drugs, we observed differences in the central apoptosis regulator AKT. Previous studies showed that in BRAF-mutant cells RAF inhibition can lead to increased or unchanged AKT phosphorylation hindering thereby apoptosis induction [52]. In our 2D signaling analysis, we observed indeed an increase of AKT activation after vemurafenib mono- and combination-therapy with gefitinib, but together with apoptosis induction—contradicting former observations. Our 3D CRC models and the PDX showed only weak regulations of pAKT, going in line with no strong apoptosis induction by all therapies in 3D. Additionally, we investigated activation changes in the EGFR as important starting point of the cascades and HGFR as a receptor that is often coregulated with EGFR [53,54,55]. The EGFR showed over all models similar patterns of activation upon the different therapies. Other studies with various BRAF-mutant CRC cell lines could also show the failure of vemurafenib as monotherapy and suggest additional EGFR inhibition as a promising therapeutic approach to avoid a feedback activation of EGFR after BRAF inhibition with vemurafenib [11,56,57]. In our experiments, we did not observe a strong activation of the EGFR upon vemurafenib treatment. Interestingly, in 2D the HGFR was strongly deactivated by vemurafenib and combination-therapy, which was less prominent in 3D conditions. This could be one reason for the observed higher chemoresistance in 3D than in 2D as the activation of the HGF signaling pathway protects tumor cells from apoptosis [58], which is also reported in gefitinib-resistant cell lines [59,60]. PDX could not mirror the human in vivo situation properly because the murine HGF deriving from the tumor microenvironment did not activate the human HGFR as reported also by others [61,62]. These differences in phosphorylation patterns could explain why preclinical xenograft studies often fail in drug efficacy testing and stress the urgent need to develop fully human models.
To model CRC signaling and drug responses in specific genetic backgrounds, our in silico model integrates central cascades of commonly impaired cancer pathways [2]. We connected two common cancer pathways, i.e., MAPK and Wnt/β-catenin-pathway within our in silico topology as in the selected MSI CRC subgroup these pathways are most frequently mutated. For further improvement of patient stratification of MSI tumors with BRAF mutation as part of the MAPK pathway, we focused on alterations driving tumor progression to an invasive state across different defined intermediate states in particular on p53 in case of HROC87. Our analysis revealed in this mutational background Bcl2-Bax-BAD regulation as an important axis or even engine in the network that decides whether apoptosis is induced independently of HGFR, ERK and AKT activation. We demonstrate that in silico modeling allows to simulate targeted therapies, i.e., vemurafenib for BRAF and gefitinib for EGFR inhibition. More importantly, we can provide new suggestions in case of therapy failure, here illustrated by targeting the network-mediated activity of MEK as well as the anti-apoptotic activity of Bcl2 [63,64]. However, the Boolean logic of our in silico simulation allows the integration of several signaling cascades into the topology. This prepares the ground for possible target predictions in patient groups with frequently mutated genes in pathways such as Wnt/β-catenin, even though this was not part of the present study.
There are clinically tested combination-therapies such as ae triple combination-therapy using an EGFR inhibitor, a BRAF inhibitor as well as a MEK inhibitor. This combination is the subject of an open phase I/II study combining trametinib, dabrafenib and panitumumab [44,45] as well as an open-label, phase 3 trial with encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated CRC [46]. On the other hand, venetoclax is already used as Bcl-2-specific inhibitor for chronic lymphocytic leukemia treatment [65]. This supports our therapeutic suggestion from the here established in silico model that targeting the MEK-Bcl2 axis is beneficial.
In our work, we follow the idea of tumor engines first propagated by Gerard Evans. These engines are more fundamental for tumor maintenance than drivers as they are the motor to control whole functional settings. In silico these are incorporated as network pace-makers: they are central, well-connected signaling proteins such as EGFR, KRAS, BRAF or c-Myc. Central signaling pathways induce tumor transformation by communicating with surrounding cells to generate a highly malignant tumor stroma and an immune-evasive phenotype [66]. Targeting these machineries promises longer lasting therapeutic success. The involved network checks and balances can only be appropriately modeled in sufficient complex models to understand and treat cancer-specific differences; here exemplified by the failure of therapeutic success of vemurafenib in case of BRAF mutation in CRC. We are aware that our study delivers only a proof of principle by connecting signaling, apoptosis and proliferation of one early passage cell line representing one individual patient in the in silico model. For the purpose of effective translation to the clinic further in vitro models with more cell lines harboring specific mutation signatures should be generated as well as models with primary cells available from organoid cultures. However, we tried to meet the actual demand on mode-of-action studies with this new combined in silico/in vitro tool.
4. Materials and Methods
4.1. SISmuc Preparation
SISmuc was prepared from porcine gut as previously described [31,67] followed by removal of the mesentery and the vascular tree from the completely decellularized explant. The matrix is registered under the trade mark BioVaSc-TERM® (TERM, Würzburg, Germany) and after seeding with tumor cells under OncoVaSc-TERM®. All explantations were in compliance with the German Animal Protection Laws (§4 Abs. 3) and all animals received humane care in compliance with the guidelines by the FELASA, WHO and FDA (WHO-TRS978 Annex3 und FDA-OCTGT Preclinical Guidance) after approval from our institutional animal protection board (registration reference number #2532-2-12, Ethics Committee of the District of Unterfranken, Würzburg, Germany).
4.2. 2D Cell Culture and Cell Lines
We used HROC24 and HROC87 cells [29] cultured in DMEM/F12 medium supplemented with 10% FCS under standard culture conditions (37 °C, 5% CO2).
4.3. 3D Cell Culture of In Vitro Models
For static 3D cell culture, cells were seeded onto SISmuc and fixed between two metal rings as described previously [34]. All 3D cultures were performed under standard conditions (37 °C, 5% CO2) within DMEM/F12 medium supplemented with 10% FCS as described before [18,19]. Remarkably, we also recognized a trend in AKT activation upon combination-therapy in another FCS batch (N° BS210601.5), but also no induction of apoptosis in 3D conditions. We integrated only data from the FCS batch N° 8SB016. For monocultures, 100,000 cells in 500 µL medium were seeded onto the former luminal side of the SISmuc. After adhesion time (2 h), cell crowns with reseeded scaffolds were completely filled with medium and cultured for 14 days. On day 11 and day 13, cells were treated with 1 µM gefitinib (Iressa, Astra Zeneca, Germany) and 1 µM vemurafenib (PLX4032, S1267, Selleck Chemicals, Houston, TX, USA) as previously described [19]. Then, the tissue was washed with PBS and fixed in 4% PFA for 2h followed by paraffin embedding for immunohistochemical analysis. For WB analysis, tissue was washed twice with ice-cold PBS and lysed (30 min, 4 °C) with modified RIPA buffer. After centrifugation (14,000 rpm, 10 min, 4 °C) the supernatant of the lysate was stored (−80 °C) until use.
4.4. Animal Models
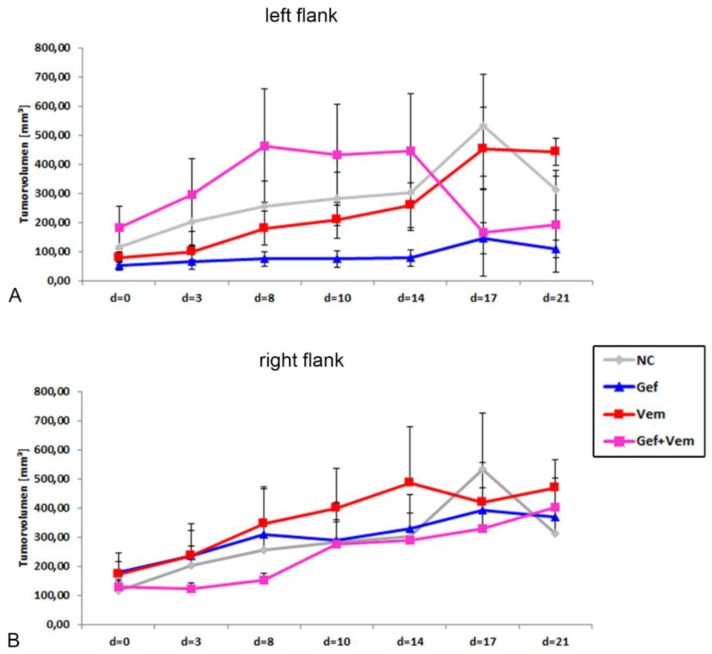
Male adult NMRI Foxn1nu mice with HROC87 T2 PDX in both flanks (mean volume of tumors at the beginning of therapy: 13,062 mm3) were treated with gefitinib (100 mg/kg body weight, i.p., 3×/week), vemurafenib (PLX4720 (kindly provided by Plexxikon, Berkeley, CA, USA) and oral, ad libitum, 417 mg/kg body weight or both, D10030403i Rodent Diet (obtained from a local distributor, Brogaarden, Denmark). A control group received 28 µL DMSO (i.p., 3×/week). Tumor volume was measured on day 0, 3, 8, 10, 14, 17 and 21 after the beginning of therapy. On day 21, mice were sacrificed; tumors were resected and fixed in 4% PFA.
4.5. Fluorescence Immunohistochemistry
Fixation was done with 4% PFA (glass slides: 10 min, SISmuc samples: 2 h). Glass slides were either directly stained or stored in PBS (4 °C, up to 1 week) prior to staining. SISmuc samples were stained as paraffinized sections (3–5 µm) and primary antibodies (E-cadherin (610181, BD Bioscience, San Jose, CA, USA), β -catenin (ab32572, Abcam, Cambridge, UK), vimentin (ab92547, Abcam), PCK (C2562, Sigma Aldrich, St. Louis, MO, USA), Ki67 (ab16667, Abcam) were diluted 1:100 in 5% BSA in PBS (w/v) and incubated overnight (4 °C). Secondary antibodies were diluted 1:400 and incubated for 1h at room temperature (RT). Double staining were performed using primary antibodies of two different species and secondary antibodies marked with fluorescent dyes (Alexa-647, Alexa-555, Life Technologies, Carlsbad, CA, USA). Nuclei were counterstained (DAPI in Mowiol embedding solution). Pictures were taken using a confocal laser scanning microscope (SP-8, Leica, Wetzlar, Germany) or a digital microscope (BZ-9000, Keyence, Neu-Isenburg, Germany).
4.6. Total Cell Number and Proliferation Rate
To determine the total cell number per image (20× magnification), all DAPI-positive cells were evaluated. Ki67-positive cells were counted and presented as percentage of the total cell number per image.
4.7. Protein Lysate Preparation and WB Analysis
Cells on SISmuc or in Petri dishes were washed twice with ice-cold PBS and lysed (30 min, 4 °C) on a rocking platform with modified RIPA buffer containing 137 mM NaCl, 50 mM NaF, 20 mM Tris (pH 8), 2 mM EDTA, 10% (v/v) glycerol, 1.0% (v/v) NP40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, and freshly added 1 mM Na3VO4 and protease inhibitor cocktail tablets (cOmplete™ mini EDTA-free, Roche, Basel, Switzerland). The lysate was centrifuged (14,000 rpm, 10 min, 4 °C) and the supernatant transferred into clean tubes for quantitation of the protein concentration using DC Protein Assay (500-0116, BioRad, Hercules, CA, USA) or stored (−80 °C) until further use. Before SDS gel electrophoresis, a protein precipitation with methanol/chloroform according to Wessel and Flügge [68], was performed. Antibodies used for WB: EGFR (4267, CST), p-EGFR (Tyr1068, Y68) (32430, Abcam), Met (8198, CST), p-Met (Tyr1234/1235) (3077, CST, Danvers, MA, USA), Erk1/2 (05-1152, Millipore, Burlington, MA, USA), p-Erk1/2 (Thr202/Tyr204) (4370, CST), Akt (pan) (4691, CST), p-Akt (Ser473) (4060, CST), α-Tubulin (3873, CST). We used the Protein Ladder_ProSieve QuadColor Protein Marker (Lonza, Basel, Switzerland).
4.8. M30 CytoDeathTM ELISA
M30 CytoDeath™ ELISA (10900, Tecomedical, Bünde, Germany) is used for the quantitative determination of the apoptosis-associated K18Asp396 (M30) neo-epitope in cultured human, monkey or bovine cells. The M30 antibody recognizes caspase-cleaved cytokeratin 18 (ccK18). Cell culture medium supernatants were collected 24h before treatment as well as 24 h, 48 h and 72 h after treatment and stored (−80 °C) until measurement. M30 CytoDeath™ ELISA was performed according to the manufacturer’s instructions.
4.9. Statistical Analysis
All statistical analysis was performed with the open-source software R [69]. Differences with a value of p < 0.05 were considered statistically significant. All experiments were performed at least in triplicates and the results presented as boxplots; statistical significance was calculated by Student’s t-test.
4.10. In Silico Simulations
For generation of the CRC in silico topology, we used CellDesigner (Version 3.5). For semi-quantitative analyses, we used SQUAD [43]. The in silico topology was created on the basis of literature and databank information followed by simulations of 3D in vitro data. The appendix provides details on network setup of the colon cell-specific model and in silico simulation of different therapies for HROC87 cells.
5. Conclusions
We consider a systemic view on targeted treatment approaches as well as so-called tumor engines of CRC instead of isolated drivers and non-relevant or bystander alterations. Furthermore, signaling in cancer cells depends on the cells’ environment and cannot be reflected accurately by oversimplified 2D culture or animal experiments due to species differences in signaling cascades such as HGFR. We meet these challenges by the identification of cellular network changes in the functional context of refined 3D tissue models mirroring better clinical responses and by a deeper understanding of common signaling alterations in CRC and its specific subtypes in silico that can be translated to the clinic by improving patient stratification.
Acknowledgments
We would like to thank Heide Häfner for technical assistance in the experiments and Jutta Habedanck-Nietzer and Ulrike Rapp-Galmiche for native-speaker corrections.
Appendix A
For all experiments we used PDX generated from a patient tumor (of patient HROC87) in passage 3 in immunodeficient mice.
Figure A1.
HROC87 PDX characterization. E-cadherin is highly expressed at the cell-to-cell contacts of HROC87 cells (green in (B,D,F,H)) in the xenografts. β -catenin is highly expressed in HROC87 cells (red in (C,D,G,H)). Cells are positive for PCK (green in (J,L,N,P)) and the tumor cells do not express vimentin while the murine stroma is stained (red in (K,L,O,P)). Scale bar in (D,H,L,P): 50 µm for (A–P).
Figure A2.
HROC87 PDX volumes. Volumes of HROC87 xenografts do not differ between the various treatment conditions. Moreover, there is considerable variation and hence in these animal experiments treatment differences do not really become apparent. This was the case in the left (A) as well as the right (B) flank. NC: untreated control, Gef: gefitinib, Vem: vemurafenib, Gef + Vem: combination-therapy with gefitinib and vemurafenib.
Figure A3.
Tumor weight difference of different tumor groups. Tumor weights were very variable within the different treatment groups in the left (A) as well as in the right (B) flanks. Tumor size as well as weight could be different in the two tumors of the same mouse (C,D). Again, with this variation in animal experiments treatment differences do not really become apparent.
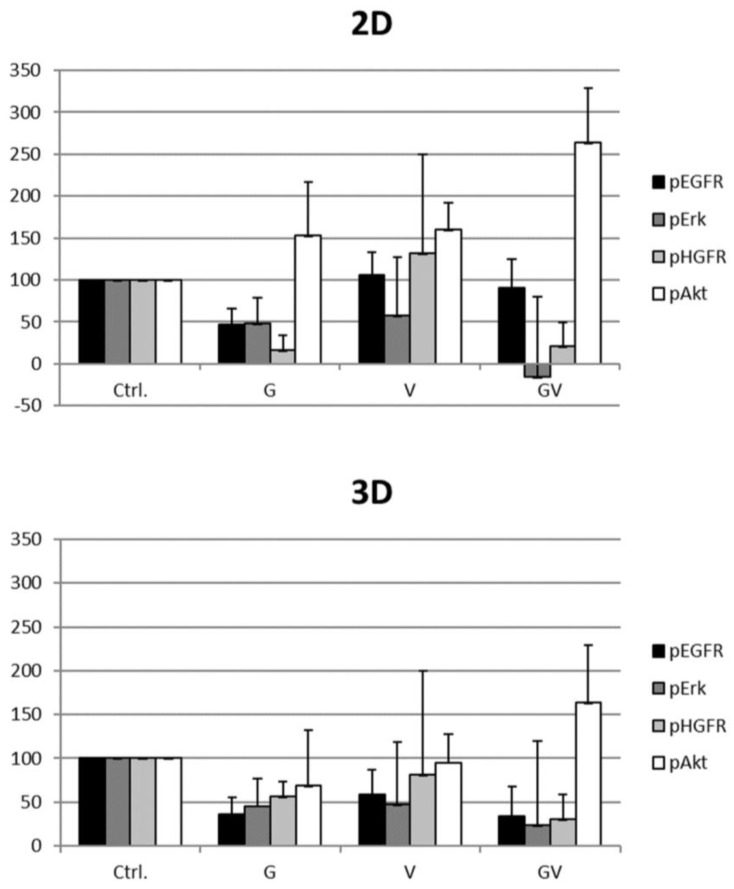
Table A1.
Grey values (in % of control) of the representative blot in Figure 5.
| 2D | ||||||||
| Figure 5 | EGFR | Erk | pEGFR | pErk | HGFR | Akt | pHGFR | pAkt |
| Ctrl. | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| G | 70 | 96 | 49 | 32 | 44 | 76 | 1 | 227 |
| V | 75 | 77 | 80 | 18 | 52 | 73 | 86 | 130 |
| GV | 63 | 94 | 53 | 2 | 26 | 86 | 2 | 314 |
| 3D | ||||||||
| Figure 5 | EGFR | Erk | pEGFR | pErk | HGFR | Akt | pHGFR | pAkt |
| Ctrl. | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| G | 111 | 128 | 36 | 40 | 91 | 114 | 67 | 74 |
| V | 81 | 83 | 86 | 7 | 72 | 65 | 78 | 75 |
| GV | 55 | 91 | 61 | 2 | 55 | 70 | 34 | 93 |
| PDX | ||||||||
| Figure 5 | EGFR | Erk | pEGFR | pErk | HGFR | Akt | pHGFR | pAkt |
| Ctrl. | 100 | 100 | 100 | 100 | 100 | 100 | - | 100 |
| G | 70 | 102 | 37 | 38 | 55 | 134 | - | 27 |
| V | 53 | 67 | 32 | 10 | 55 | 88 | - | 17 |
| GV | 77 | 104 | 22 | 4 | 55 | 89 | - | 18 |
Figure A4.
Blots with molecular weight marker of the 2D experiments shown in Figure 5.
Figure A5.
Blots with molecular weight marker of the 3D experiments shown in Figure 5.
Figure A6.
Blots with molecular weight marker of the PDX experiments shown in Figure 5.
Figure A7.
Grey values (in % of control) of 4–6 western blots. For 2D, the values of 4–5 western blots and for 3D, the values of 5–6 western blots were analyzed. Bars: arithmetic mean. Error bars: standard deviation. Ctrl = untreated control, G = gefitinib, V = vemurafenib, GV = combination-therapy of gefitinib and vemurafenib. pEGFR = phospho-EGFR, pErk = phospho-Erk, pHGFR = phospho-HGFR, pAkt = phospho-Akt.
Appendix A.1. Ideal-Typic Transformation from a Healthy Epithelium to a Colon Carcinoma
Transformation to malignant stages usually happens step-wise over time involving different activation processes (Table A2). Here, we give a simplified example and route of the suppressor pathway. In a first step of cancer progression, the β-catenin signaling pathway is changed by a mutation. In 85% of all early stages, APC is changed by mutation which leads to increased cell proliferation [69,70]. Therefore, the normal epithelium (stage 0) transforms by dysplasia to APC (stage 1). An aberration of COX-2 then leads to the early adenoma stage (stage 2) [71]. Afterwards, mutations of KRAS lead to the intermediate adenoma (stage 3), detectable in 55 % of all cases [72]. The late adenoma (stage 4) is based on loss of 18q / DCC [38]. Mutations in the p53 gene lead to the carcinoma (stage 5) [38]. Metastasis (stage 6) is induced possibly via E-cadherin, the activation of molecules involved in the TGFβ signaling pathway (Smad2, Smad4, TGFβR). Further tumor progression selects for more resistant cells and may include activation of antiapoptotic pathways e.g., by the protein BAX [73,74].
Table A2.
Stepwise transformation of a normal colon epithelial cell to colon carcinoma.
| Stage 0 | Stage 1 | Stage 2 | Stage 3 | Stage 4 | Stage 5 | Stage 6 |
|---|---|---|---|---|---|---|
| normal epithelium | dysplastic aberrant cryptic foci | early/initial adenoma | intermediate adenoma | late adenoma | carcinoma | metastasis |
| APC (β-catenin) | COX-2 | KRAS | DCC/loss of 18q | p53 | E-cadherin/BAX |
Appendix A.2. Bioinformatics Mod$eling and Systems Biology Data
Bioinformatics modeling of the in vitro data-uses data-driven modeling based on Boolean semi-quantitative models. These start from (i) a Boolean model (yes and no decisions) of the network and then (ii) approximates by exponential functions the behavior of the individual kinases, pathways and cellular reactions (e.g., apoptosis, growth, differentiation) over time.
(i) We started our modeling by setting-up a simplified network model of the colon cancer cell using literature (see also Table A2) and databases. The model includes three parts: (a) central tumor signaling cascades such as MAPK and its connection to apoptosis and proliferation, (b) tumor-specific mutations (HROC87: BRAF, p53) and experimental measured nodes (experimental data summarized in Table A3) and (c) the treatments.
The network was visualized using CellDesigner Software (Figure A8). (ii) Next, the network model was step-wise iteratively refined looking at the experimental data of cell culture treatment and cellular reaction as well as the specific modifications to closely correspond to the behavior of the HROC87 tumor cells in cell culture regarding apoptosis, proliferation and signaling (in particular its specific mutations as well as different reactivity to different drugs). This was modeled by basic pre-stimulation (activation, inhibition) of the pathways and specific nodes as we only focus on the central part of the cellular network involving not all modulatory nodes available in the cell (Table 1). The pre-stimulation mimics hence stronger or lower responsiveness of the tumor pathways. The stimulation settings were step-wise optimized to follow as close as possible the experimental data and taking note of the Boolean network topology logic of the signaling pathways as shown in Figure A8.
Table A3.
Pre-stimulation activities of the in silico model as starting point for the therapy simulation.
| Western Blot Data (in Comparison to Untreated Control) | Proliferation Value in 3D | Apoptosis in 3D Compared to Ctrl | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Ctrl | Gef | Vem | Gef+Vem | HROC87 | HROC87 | ||||
| pEGFR | medium | ↓↓ | ↓ | ↓↓ | Ctrl | 0.5 | Ctrl | weak | |
| pErk | strong | ↓ | ↓ | ↓↓ | Gef | 0.5 | Gef | ↑ | |
| pHGFR | strong | ↓ | 0 | ↓↓ | Vem | 0.5 | Vem | ↑ | |
| pAkt | weak | ↓ | 0 | ↑ | Gef + Vem | 0.2 | Gef+Vem | ↑ | |
The pre-stimulation was done using the perturbation function in SQUAD. The simulations are shown in Figure 7 and in Figure A9 for vemurafenib treatment. The activation outcomes after the pre-stimulation settings are listed in Table 2, reflecting the cell culture response behavior of the HROC87 cells from Table A3.
Figure A8.
General idealized topology for CRC. The topology includes several relevant signaling cascades shown in different colors. Mutations that lead to cancer progression are marked in yellow, corresponding stages as green hexagons. Possible drugs applied in this study for in silico modeling are added in pink. Cellular responses such as apoptosis and proliferation are indicated in red. Activation of nodes (proteins or systemic responses) is illustrated by arrows, inhibition by blunted arrows.
Appendix A.3. Simulation of Different Treatment Scenarios
Several treatment in silico simulations are already given in the results section, we show here additionally the simulation of vemurafenib treatment.
Figure A9.
Simulation of vemurafenib treatment of HROC87 cells. The cells show in 3D culture only a weak induction of apoptosis (olive-green triangle curve) but no changes on proliferation (grey square curve) upon vemurafenib (blue rectangle curve, activation: 1). * = activated form.
Author Contributions
Conceptualization: G.D. and T.D.; methodology: F.B., S.L.N., M.K., F.S., M.L. and H.W.; software: F.S., M.K. and T.D., formal analysis: F.B., S.L.N. and M.K.; investigation: F.B., S.L.N., F.S., M.K., J.J. and S.M.; resources: M.L.; data curation: F.B. and M.K.; writing—original draft preparation: S.L.N., M.K., T.D. and G.D.; writing—review and editing: M.K., M.L. and H.W.; visualization: F.B., M.K. and S.L.N.; supervision: S.L.N., T.D. and G.D.; project administration: S.L.N., T.D. and G.D.; funding acquisition: T.D. and H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Bayern-Fit program until 2019 (F.B., S.L.N.) and the Federal Ministry of Education and Research Era-Net grant 01KT1801 (M.K., T.D.). Furthermore, this publication was funded by the German Research Foundation (DFG) and the University of Würzburg in the funding program Open Access Publishing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.World Health Organization Globocan 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012 v1.0. [(accessed on 23 December 2019)]; Available online: https://publications.iarc.fr/Databases/Iarc-Cancerbases/GLOBOCAN-2012-Estimated-Cancer-Incidence-Mortality-And-Prevalence-Worldwide-In-2012-V1.0-2012ISBN-13(Database)978-92-832-2447-1.
- 2.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Pietrantonio F., Petrelli F., Coinu A., Di Bartolomeo M., Borgonovo K., Maggi C., Cabiddu M., Iacovelli R., Bossi I., Lonati V., et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: A meta-analysis. Eur. J. Cancer. 2015;51:587–594. doi: 10.1016/j.ejca.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 5.Yokota T., Ura T., Shibata N., Takahari D., Shitara K., Nomura M., Kondo C., Mizota A., Utsunomiya S., Muro K., et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br. J. Cancer. 2011;104:856–862. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiel A., Heinonen M., Kantonen J., Gylling A., Lahtinen L., Korhonen M., Kytölä S., Mecklin J.P., Orpana A., Peltomäki P., et al. BRAF mutation in sporadic colorectal cancer and Lynch syndrome. Virchows Arch. 2013;463:613–621. doi: 10.1007/s00428-013-1470-9. [DOI] [PubMed] [Google Scholar]
- 7.Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O’Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagaraja A.K., Bass A.J. Hitting the target in BRAF-mutant colorectal cancer. J. Clin. Oncol. 2015;33:3990–3992. doi: 10.1200/JCO.2015.63.7793. [DOI] [PubMed] [Google Scholar]
- 10.Kopetz S., Desai J., Chan E., Hecht J.R., O’Dwyer P.J., Lee R.J., Morris V., Janku F., Dasari A., Chung W., et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. J. Clin. Oncol. 2010;28:3534. doi: 10.1200/jco.2010.28.15_suppl.3534. [DOI] [Google Scholar]
- 11.Prahallad A., Sun C., Huang S., Di Nicolantonio F., Salazar R., Zecchin D., Beijersbergen R.L., Bardelli A., Bernards R. Unresponsiveness of colon cancer to BRAF (V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 12.Corcoran R.B., Ebi H., Turke A.B., Coffee E.M., Nishino M., Cogdill A.P., Brown R.D., Della Pelle P., Dias-Santagata D., Hung K.E., et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly K., Brungs D., Szeto E., Epstein R.J. Anticancer activity of combination targeted therapy using cetuximab plus vemurafenib for refractory BRAF (V600E)-mutant metastatic colorectal carcinoma. Curr. Oncol. 2014;21:e151–e154. doi: 10.3747/co.21.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capalbo C., Marchetti P., Coppa A., Calogero A., Anastasi E., Buffone A., Belardinilli F., Gulino M., Frati P., Catalano C., et al. Vemurafenib and panitumumab combination tailored therapy in BRAF-mutated metastatic colorectal cancer: A case report. Cancer Biol. Ther. 2014;15:826–831. doi: 10.4161/cbt.28878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaeger R.D., Cercek A., O’Reilly E.M., Reidy D.L., Kemeny N.E., Wolinsky T., Gollub M.J., Lacouture M.E., Rosen N., Vakiani E., et al. Pilot study of vemurafenib and panitumumab combination therapy in patients with BRAF V600E mutated metastatic colorectal cancer. J. Clin. Oncol. 2015;33:611. doi: 10.1200/jco.2015.33.3_suppl.611. [DOI] [Google Scholar]
- 16.Schlatter R., Philippi N., Wangorsch G., Pick R., Sawodny O., Borner C., Timmer J., Ederer M., Dandekar T. Integration of Boolean models exemplified on hepatocyte signal transduction. Brief Bioinform. 2012;13:365–376. doi: 10.1093/bib/bbr065. [DOI] [PubMed] [Google Scholar]
- 17.Buiga P., Elson A., Tabernero L., Schwartz J.M. Regulation of dual specificity phosphatases in breast cancer during initial treatment with Herceptin: A Boolean model analysis. BMC Syst. Biol. 2018;12:11. doi: 10.1186/s12918-018-0534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlich C., Muller L.C., Kunz M., Schmitt F., Walles H., Walles T., Dandekar T., Dandekar G., Nietzer S.L. A combined 3D tissue engineered in vitro/in silico lung tumor model for predicting drug effectiveness in specific mutational backgrounds. J. Vis. Exp. 2016;6:e53885. doi: 10.3791/53885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stratmann A.T., Fecher D., Wangorsch G., Gottlich C., Walles T., Walles H., Dandekar T., Dandekar G., Nietzer S.L. Establishment of a human 3D lung cancer model based on a biological tissue matrix combined with a Boolean in silico model. Mol. Oncol. 2014;8:351–365. doi: 10.1016/j.molonc.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danes B.S., Deangelis P., Traganos F., Melamed M.R., Alm T. Demonstration of altered cellular DNA content distribution in long-term colon epithelial cell lines with colon cancer genotypes. Scand. J. Gastroenterol. 1988;23:840–846. doi: 10.3109/00365528809090770. [DOI] [PubMed] [Google Scholar]
- 21.Bocsi J., Zalatnai A. Establishment and long-term xenografting of human pancreatic carcinomas in immunosuppressed mice: Changes and stability in morphology, DNA ploidy and proliferation activity. J. Cancer Res. Clin. Oncol. 1999;125:9–19. doi: 10.1007/s004320050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J.M., Mhawech-Fauceglia P., Lee N., Parsanian L.C., Lin Y.G., Gayther S.A., Lawrenson K. A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab. Invest. 2013;93:528–542. doi: 10.1038/labinvest.2013.41. [DOI] [PubMed] [Google Scholar]
- 23.Zschenker O., Streichert T., Hehlgans S., Cordes N. Genome-wide gene expression analysis in cancer cells reveals 3D growth to affect ECM and processes associated with cell adhesion but not DNA repair. PLoS ONE. 2012;7:e34279. doi: 10.1371/journal.pone.0034279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meli L., Jordan E.T., Clark D.S., Linhardt R.J., Dordick J.S. Influence of a three-dimensional, microarray environment on human cell culture in drug screening systems. Biomaterials. 2012;33:9087–9096. doi: 10.1016/j.biomaterials.2012.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komoto C., Nakamura T., Ohmoto N., Kobayashi H., Yagami T., Nishiguchi K., Iwaki K., Kuwahara A., Yamamori M., Okamura N., et al. Three-dimensional, but not two-dimensional, culture results in tumor growth enhancement after exposure to anticancer drugs. Kobe J. Med. Sci. 2007;53:335–343. [PubMed] [Google Scholar]
- 26.Kalluri R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 27.Maletzki C., Stier S., Gruenert U., Gock M., Ostwald C., Prall F., Linnebacher M. Establishment, characterization and chemosensitivity of three mismatch repair deficient cell lines from sporadic and inherited colorectal carcinomas. PLoS ONE. 2012;7:e52485. doi: 10.1371/journal.pone.0052485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maletzki C., Klier U., Marinkovic S., Klar E., Andra J., Linnebacher M. Host defense peptides for treatment of colorectal carcinoma—A comparative in vitro and in vivo analysis. Oncotarget. 2014;5:4467–4479. doi: 10.18632/oncotarget.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullins C.S., Micheel B., Matschos S., Leuchter M., Burtin F., Krohn M., Hühns M., Klar E., Prall F., Linnebacher M. Integrated biobanking and tumor model establishment of human colorectal carcinoma provides excellent tools for preclinical research. Cancers. 2019;11:1520. doi: 10.3390/cancers11101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groeber F., Engelhardt L., Lange J., Kurdyn S., Schmid F.F., Rucker C., Mielke S., Walles H., Hansmann J. A first vascularized skin equivalent for as an alternative to animal experimentation. ALTEX. 2016;33:415–422. doi: 10.14573/altex.1604041. [DOI] [PubMed] [Google Scholar]
- 31.Schanz J., Pusch J., Hansmann J., Walles H. Vascularised human tissue models: A new approach for the refinement of biomedical research. J. Biotechnol. 2010;148:56–63. doi: 10.1016/j.jbiotec.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Scheller K., Dally I., Hartmann N., Munst B., Braspenning J., Walles H. Upcyte® microvascular endothelial cells repopulate decellularized scaffold. Tissue Eng. Part C Methods. 2013;19:57–67. doi: 10.1089/ten.tec.2011.0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moll C., Reboredo J., Schwarz T., Appelt A., Schurlein S., Walles H., Nietzer S. Tissue engineering of a human 3D in vitro tumor test system. J. Vis. Exp. 2013:e50460. doi: 10.3791/50460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nietzer S., Baur F., Sieber S., Hansmann J., Schwarz T., Stoffer C., Häfner H., Gasser M., Waaga-Gasser A.M., Walles H., et al. Mimicking metastases including tumor stroma: A new technique to generate a three-dimensional colorectal cancer model based on a biological decellularized intestinal scaffold. Tissue Eng. Part C Methods. 2016;22:621–635. doi: 10.1089/ten.tec.2015.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gottlich C., Kunz M., Zapp C., Nietzer S.L., Walles H., Dandekar T., Dandekar G. A combined tissue-engineered/in silico signature tool patient stratification in lung cancer. Mol. Oncol. 2018;12 doi: 10.1002/1878-0261.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lubtow M.M., Nelke L.C., Seifert J., Kuhnemundt J., Sahay G., Dandekar G., Nietzer S.L., Luxenhofer R. Drug induced micellization into ultra-high capacity and stable curcumin nanoformulations: Physico-chemical characterization and evaluation in 2D and 3D in vitro models. J. Control. Release. 2019;303:162–180. doi: 10.1016/j.jconrel.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Wallstabe L., Gottlich C., Nelke L.C., Kuhnemundt J., Schwarz T., Nerreter T., Einsele H., Walles H., Dandekar G., Nietzer S.L., et al. ROR1-CAR T cells are effective against lung and breast cancer in advanced microphysiologic 3D tumor models. JCI Insight. 2019;4:e126345. doi: 10.1172/jci.insight.126345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fearon E.R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 39.Song G., Ouyang G., Bao S. The activation of Akt/PKB signaling pathway and cell survival. J. Cell Mol. Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armaghany T., Wilson J.D., Chu Q., Mills G. Genetic alterations in colorectal cancer. Gastrointest. Cancer Res. 2012;5:19–27. [PMC free article] [PubMed] [Google Scholar]
- 41.Chubb D., Broderick P., Dobbins S.E., Frampton M., Kinnersley B., Penegar S., Price A., Ma Y.P., Sherborne A.L., Palles C., et al. Rare disruptive mutations and their contribution to the heritable risk of colorectal cancer. Nat. Commun. 2016;7:11883. doi: 10.1038/ncomms11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang Y.C., Chang J.G., Liu T.C., Lin C.Y., Yang S.F., Ho C.M., Chen W.T., Chang Y.S. Mutation analysis of 13 driver genes of colorectal cancer-related pathways in Taiwanese patients. World J. Gastroenterol. 2016;22:2314–2325. doi: 10.3748/wjg.v22.i7.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Cara A., Garg A., De Micheli G., Xenarios I., Mendoza L. Dynamic simulation of regulatory networks using SQUAD. BMC Bioinform. 2007;8:462. doi: 10.1186/1471-2105-8-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bendell J.C., Atreya C.E., André T., Tabernero J., Gordon M.S., Bernards R., Van Cutsem E., Tejpar S., Sidhu R., Go W.Y., et al. Efficacy and tolerability in an open-label phase I/II study of MEK inhibitor trametinib (T), BRAF inhibitor dabrafenib (D), and anti-EGFR antibody panitumumab (P) in combination in patients (pts) with BRAF V600E mutated colorectal cancer (CRC) J. Clin. Oncol. 2014;32:3515. doi: 10.1200/jco.2014.32.15_suppl.3515. [DOI] [Google Scholar]
- 45.ClinicalTrials.Gov BRAF/MEK/EGFR Inhibitor Combination Study in Colorectal Cancer (CRC) [(accessed on 23 December 2019)]; Available online: https://clinicaltrials.gov/show/NCT01750918.
- 46.Kopetz S., Grothey A., Yaeger R., Van Cutsem E., Desai J., Yoshino T., Wasan H., Ciardiello F., Loupakis F., Hong Y.S., et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N. Engl. J. Med. 2019;381:1632–1643. doi: 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 47.Bissell M.J., Radisky D. Putting tumours in context. Nat. Rev. Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Debnath J., Brugge J.S. Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 49.Fischbach C., Chen R., Matsumoto T., Schmelzle T., Brugge J.S., Polverini P.J., Mooney D.J. Engineering tumors with 3D scaffolds. Nat. Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 50.Van Es J.H., Clevers H. Generation and analysis of mouse intestinal tumors and organoids harboring APC and K-Ras mutations. Methods Mol. Biol. 2015;1267:125–144. doi: 10.1007/978-1-4939-2297-0_6. [DOI] [PubMed] [Google Scholar]
- 51.Corcoran R.B., Atreya C.E., Falchook G.S., Kwak E.L., Ryan D.P., Bendell J.C., Hamid O., Messersmith W.A., Daud A., Kurzrock R., et al. Combined BRAF and MEK Inhibition with dabrafenib and trametinib in BRAF V600-mutant colorectal cancer. J. Clin. Oncol. 2015;33:4023–4031. doi: 10.1200/JCO.2015.63.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fritsche-Guenther R., Witzel F., Kempa S., Brummer T., Sers C., Bluthgen N. Effects of RAF inhibitors on PI3K/AKT signalling depend on mutational status of the RAS/RAF signalling axis. Oncotarget. 2016;7:7960–7969. doi: 10.18632/oncotarget.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puri N., Salgia R. Synergism of EGFR and c-Met pathways, cross-talk and inhibition, in non-small cell lung cancer. J. Carcinog. 2008;7:9. doi: 10.4103/1477-3163.44372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonine-Summers A.R., Aakre M.E., Brown K.A., Arteaga C.L., Pietenpol J.A., Moses H.L., Cheng N. Epidermal growth factor receptor plays a significant role in hepatocyte growth factor mediated biological responses in mammary epithelial cells. Cancer Biol. Ther. 2007;6:561–570. doi: 10.4161/cbt.6.4.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spix J.K., Chay E.Y., Block E.R., Klarlund J.K. Hepatocyte growth factor induces epithelial cell motility through transactivation of the epidermal growth factor receptor. Exp. Cell Res. 2007;313:3319–3325. doi: 10.1016/j.yexcr.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirschi B., Gallmeier E., Ziesch A., Marschall M., Kolligs F.T. Genetic targeting of B-RafV600E affects survival and proliferation and identifies selective agents against BRAF-mutant colorectal cancer cells. Mol. Cancer. 2014;13:122. doi: 10.1186/1476-4598-13-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oddo D., Sennott E.M., Barault L., Valtorta E., Arena S., Cassingena A., Filiciotto G., Marzolla G., Elez E., van Geel R.M., et al. Molecular landscape of acquired resistance to targeted therapy combinations in BRAF-mutant colorectal cancer. Cancer Res. 2016;76:4504–4515. doi: 10.1158/0008-5472.CAN-16-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Provencal M., Berger-Thibault N., Labbe D., Veitch R., Boivin D., Rivard G.E., Gingras D., Béliveau R. Tissue factor mediates the HGF/Met-induced anti-apoptotic pathway in DAOY medulloblastoma cells. J. Neurooncol. 2010;97:365–372. doi: 10.1007/s11060-009-0041-z. [DOI] [PubMed] [Google Scholar]
- 59.Engelman J.A., Zejnullahu K., Gale C.M., Lifshits E., Gonzales A.J., Shimamura T., Zhao F., Vincent P.W., Naumov G.N., Bradner J.E., et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 60.Straussman R., Morikawa T., Shee K., Barzily-Rokni M., Qian Z.R., Du J., Davis A., Mongare M.M., Gould J., Frederick D.T., et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeffers M., Rong S., Vande Woude G.F. Hepatocyte growth factor/scatter factor-Met signaling in tumorigenicity and invasion/metastasis. J. Mol. Med. 1996;74:505–513. doi: 10.1007/BF00204976. [DOI] [PubMed] [Google Scholar]
- 62.Francone T.D., Landmann R.G., Chen C.T., Sun M.Y., Kuntz E.J., Zeng Z., Dematteo R.P., Paty P.B., Weiser M.R. Novel xenograft model expressing human hepatocyte growth factor shows ligand-dependent growth of c-Met-expressing tumors. Mol. Cancer Ther. 2007;6:1460–1466. doi: 10.1158/1535-7163.MCT-06-0466. [DOI] [PubMed] [Google Scholar]
- 63.Hata A.N., Engelman J.A., Faber A.C. The BCL2 family: Key mediators of the apoptotic response to targeted anticancer therapeutics. Cancer Discov. 2015;5:475–487. doi: 10.1158/2159-8290.CD-15-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nehls O., Okech T., Hsieh C.J., Enzinger T., Sarbia M., Borchard F., Gruenagel H.H., Gaco V., Hass H.G., Arkenau H.T., et al. Studies on p53, BAX and Bcl-2 protein expression and microsatellite instability in stage III (UICC) colon cancer treated by adjuvant chemotherapy: Major prognostic impact of proapoptotic BAX. Br. J. Cancer. 2007;96:1409–1418. doi: 10.1038/sj.bjc.6603728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Souers A.J., Leverson J.D., Boghaert E.R., Ackler S.L., Catron N.D., Chen J., Dayton B.D., Ding H., Enschede S.H., Fairbrother W.J., et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 66.Kortlever R.M., Sodir N.M., Wilson C.H., Burkhart D.L., Pellegrinet L., Brown Swigart L., Littlewood T.D., Evan G. Myc cooperates with Ras by programming inflammation and immune suppression. Cell. 2017;171:1301–1315. doi: 10.1016/j.cell.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linke K., Schanz J., Hansmann J., Walles T., Brunner H., Mertsching H. Engineered liver-like tissue on a capillarized matrix for applied research. Tissue Eng. 2007;13:2699–2707. doi: 10.1089/ten.2006.0388. [DOI] [PubMed] [Google Scholar]
- 68.Wessel D., Flugge U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 69.Weinberg R.A. The Biology of Cancer. 1st ed. Garland Science; New York, NY, USA: 2007. [Google Scholar]
- 70.Pinto D., Clevers H. Wnt, stem cells and cancer in the intestine. Biol. Cell. 2005;97:185–196. doi: 10.1042/BC20040094. [DOI] [PubMed] [Google Scholar]
- 71.Wang D., Dubois R.N. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29:781–788. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fodde R., Smits R., Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 73.Sun Q., Hua J., Wang Q., Xu W., Zhang J., Kang J., Li M. Expressions of GRP78 and Bax associate with differentiation, metastasis, and apoptosis in non-small cell lung cancer. Mol. Biol. Rep. 2012;39:6753–6761. doi: 10.1007/s11033-012-1500-8. [DOI] [PubMed] [Google Scholar]
- 74.Philippi N., Walter D., Schlatter R., Ferreira K., Ederer M., Sawodny O., Timmer J., Borner C., Dandekar T. Modeling system states in liver cells: Survival, apoptosis and their modifications in response to viral infection. BMC Syst. Biol. 2009;3:97. doi: 10.1186/1752-0509-3-97. [DOI] [PMC free article] [PubMed] [Google Scholar]