Abstract
Nearly all motile cilia and flagella (terms here used interchangeably) have a ‘9+2’ axoneme containing nine outer doublet microtubules and two central microtubules. The central pair of microtubules plus associated projections, termed the central apparatus (CA), is involved in the control of flagellar motility and is essential for the normal movement of ‘9+2’ cilia. Research using the green alga Chlamydomonas reinhardtii, an important model system for studying cilia, has provided most of our knowledge of the protein composition of the CA, and recent work using this organism has expanded the number of known and candidate CA proteins nearly threefold. Here we take advantage of this enhanced proteome to examine the genomes of a wide range of eukaryotic organisms, representing all of the major phylogenetic groups, to identify predicted orthologues of the C. reinhardtii CA proteins and explore how widely the proteins are conserved and whether there are patterns to this conservation. We also discuss in detail two contrasting groups of CA proteins—the ASH-domain proteins, which are broadly conserved, and the PAS proteins, which are restricted primarily to the volvocalean algae.
This article is part of the Theo Murphy meeting issue ‘Unity and diversity of cilia in locomotion and transport’.
Keywords: Chlamydomonas, flagella, central microtubules, axoneme evolution, ASH-domain proteins, PAS-domain proteins
1. Background
Most motile cilia and flagella (terms here used interchangeably) contain a ‘9+2’ axoneme consisting of nine outer doublet microtubules plus a pair of central singlet microtubules. Attached to the outer doublets are substructures that repeat periodically along the axoneme, including outer dynein arms, inner dynein arms, nexin–dynein regulatory complexes (N-DRCs) and radial spokes. Extending from the central singlet microtubules (termed C1 and C2) are at least 11 architecturally distinct projections that are attached periodically with a 32 nm unit repeat along the microtubules ([1]; and see figure 1) and interact with the heads of the radial spokes [4]. The central pair of microtubules and its projections, hereafter referred to as the central apparatus (CA), work together with the outer doublet substructures to generate and control ciliary motility.
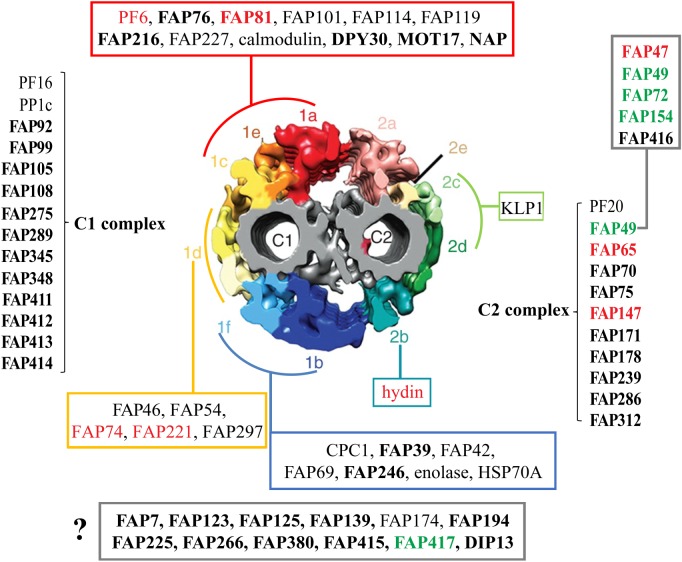
Figure 1.
Summary of CA proteins and their predicted locations in the C1 and C2 microtubules. Diagram of cross-section of the Chlamydomonas reinhardtii CA (modified from [1]) showing predicted locations of CA proteins including novel candidate or confirmed CA proteins identified by Zhao et al. [2] (bold font); FAP76 and FAP216 were localized by Fu et al. [3]. ‘1a’–‘1f’ and ‘2a’–‘2e’ indicate projections C1a to C1f and C2a to C2e, respectively. ASH-domain proteins are in red font; PAS-domain proteins are in green font. Some proteins are predicted to be associated with either the C1 or C2 microtubule, but their specific locations are not yet determined; others (red, green, turquoise, dark blue and yellow boxes) are predicted to be associated with specific projections, pairs of projections or a supercomplex consisting of the C1a, C1e and C1c projections. The FAP47 complex (box, upper right) is likely to be associated with C2 based on solubility properties of FAP49. The question mark indicates proteins whose locations in the CA are not yet known. Modified from Zhao et al. [2]. (Online version in colour.)
The ultrastructure of the ‘9+2’ axoneme has been highly conserved throughout evolution, indicating that the basic machinery for force generation has been conserved since the last eukaryotic common ancestor (LECA) [5]. However, there is a major bifurcation in the way that the CA interacts with the radial spokes, with the CAs of members of the SAR (stramenopiles, alveolates and Rhizaria) and Plantae clades having a CA that rotates within the axoneme during ciliary beating, whereas unikonts have a CA that maintains a fixed orientation relative to the nine outer doublet microtubules (reviewed in [5,6]). Also, although the CA is essential for the normal motility of those flagella that naturally have one, there are many instances where motility has been retained despite loss of the CA and other parts of the ‘9+2’ machinery during evolution. For example, the nodal cilia of mammals lack the CA but are motile [7]. Similarly, sperm of the Asian horseshoe crabs Tachypleus tridentatus and Tachypleus gigas have ‘9+0’ axonemes lacking the CA but retaining outer and inner arms and, surprisingly, radial spokes, whereas sperm of the American horseshoe crab Limulus polyphemus have a normal ‘9+2’ axoneme [8]. Even more reduced are axonemes of sperm of the eel Anguilla anguilla, which lack the CA, radial spokes and outer arms [9,10], and of the centric diatom Thalassiosira pseudonana, which, based on comparative genomic analysis, are predicted to lack the CA, radial spokes and inner arms [11]. Other examples of diversification and reduction of structure of the CA of motile cilia are discussed by Satir et al. [12]. Clearly, there has been ample opportunity for modification of the CA during eukaryotic evolution.
As a complement to structural studies, additional insight into the conservation and diversification of the axonemal motile machinery can be derived from a detailed knowledge of the proteins making up its various substructures. Because the axonemes of Chlamydomonas reinhardtii can be readily isolated and fractionated for biochemical studies, and because many mutants are available lacking specific axonemal substructures, C. reinhardtii has been particularly useful for identifying the proteins of the axonemal machinery and elucidating their functions. The proteomes of the outer arms, the inner arms, the N-DRCs and the radial spokes of C. reinhardtii have been well studied [13–17], and knowledge of the sequences of the proteins of these structures coupled with gene searches has made possible identification of orthologues in other organisms. This in turn has enabled reconstructions of the likely steps in the evolution of the dynein arms [18,19]. However, until recently, the composition of the CA was less well known. It has not yet been possible to isolate and purify the CA from the cilia of any organism, and for decades this precluded a global identification of CA proteins. Early studies using one- and two-dimensional polyacrylamide gel electrophoresis (PAGE) to compare C. reinhardtii wild-type axonemes with those of mutants lacking the CA estimated that the CA contained at least 18 proteins [20], but the methods available did not permit identification of the specific proteins. In the nearly four decades between then and 2019, numerous intrepid investigators solubilized all or part of the C. reinhardtii CA and then used affinity chromatography, co-sedimentation, or co-immunoprecipitation followed by mass spectrometry (MS) to identify individual proteins or small groups of proteins, resulting in the characterization of 22 CA proteins, of which 18 are specific to the CA (figure 1, and see [21] for review). Although this number was in good agreement with the earlier estimates from PAGE, results from a proteomics analysis of the C. reinhardtii wild-type flagellum [22] suggested that the number of CA proteins might be twice as great as previously estimated. More recently, a structural study of the C. reinhardtii CA by cryo-electron tomography (cryo-ET) estimated the mass of each individual projection on the C1 and C2 microtubule; the sum of these masses was greater than 14 MDa [1]. However, the combined masses of the known CA proteins localized to those projections was just over 3 MDa [2]. The 11 MDa of unaccounted mass also suggested that there were many more CA proteins still to be found.
In an effort to identify more CA proteins, we recently used quantitative MS to compare the proteomes of isolated axonemes of wild-type C. reinhardtii and a mutant lacking the CA [2]. We identified 44 novel candidate CA proteins (figure 1) that were present in the former and not the latter. For many of the proteins, we were able to predict whether they are associated specifically with the C1 or C2 microtubule, and in some cases we could even predict the specific projection with which they are associated. We confirmed a CA localization for five out of five of the proteins that were investigated in more detail [2]; more recently, three more of the proteins were localized by cryo-ET and all three were confirmed to be CA proteins [3]. Thus, it is likely that most of the other candidates also are CA subunits. In this report, we will refer to both confirmed CA proteins and candidate CA proteins as simply ‘CA proteins’ unless there is reason to be more specific.
Here, we examine the genomes of a wide range of eukaryotic organisms, representing all of the major phylogenetic groups, to identify predicted orthologues of the C. reinhardtii CA proteins and explore how widely the proteins are conserved and whether there are patterns to this conservation. We also discuss in detail two groups of CA proteins—the ASH (ASPM, SPD-2, Hydin)-domain proteins and the PAS (Per-ARNT-Sim) proteins—that are particularly intriguing in the context of evolution and/or function.
2. Methods
(a). Orthologue identification
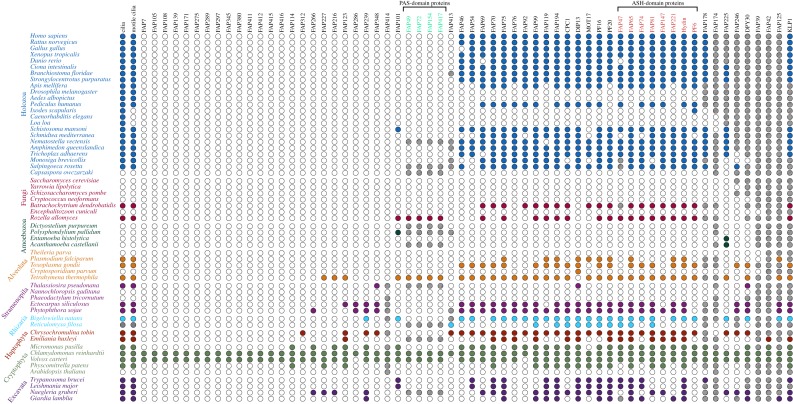
To infer orthologues of C. reinhardtii CA-specific proteins, we used the predicted proteomes for 57 species spanning nine groups (Holozoa, Fungi, Amoebozoa, Alveolata, Stramenopila, Rhizaria, Haptophyta, Cryptophyta, Excavata) downloaded from the websites in electronic supplementary material, table S1. OrthoFinder v2.3.3 was then used with the default parameters to identify orthologous gene groups [23,24]; the DIAMOND sequence aligner was used in the initial ‘any-versus-any’ BLAST step [25]. Orthologues inferred in this way are identified in table S2 in the supplementary material. Orthogroup trees were examined manually to identify most likely orthologues based on grouping in the same clade as the C. reinhardtii query sequence. For example, C. reinhardtii KLP1 is a member of the kinesin-9 family in the kinesin superfamily of proteins [26]. OrthoFinder returned over 2000 KLP1-related sequences which included kinesins from many other kinesin families. Therefore, only proteins in the same clade as KLP1 were considered likely orthologues. Dot (figure 2) and Coulson (electronic supplementary material, figure S1) plots indicating presence of orthologues in each species were constructed using the Coulson plot generator [27].
Figure 2.
Dot plot showing phylogenetic distribution of orthologues of C. reinhardtii CA-specific proteins. Species belonging to the same major taxonomic group are indicated by text with the same colour. Coloured dots indicate either the presence of cilia or motile cilia (first and second columns, respectively) or the presence of at least one likely orthologue in the indicated species. Empty dots indicate the absence of cilia, motile cilia or orthologues. Grey dots for Reticulomyxa filosa indicate uncertainty with regard to the presence of cilia (see text). Hatched dots indicate that sequences from these species were present in the OrthoFinder gene tree but eliminated as likely orthologues because they were in a different clade from the C. reinhardtii query protein (see Methods). Proteins most affected in this way were members of large families or contained common conserved motifs and included KLP1 and FAP125 (kinesin superfamily), FAP42 (transferase family), FAP39 (ATPase), DPY30 (DPY-30 domain), FAP246 (LRR repeats and EF-hands), FAP225 (EF-hands), FAP174 (MYC-binding) and FAP178 (calponin domain). PAS- and ASH-domain proteins (green and red font, respectively) are indicated. (Online version in colour.)
(b). ASH-domain identification
We used two different approaches to identify flagellar proteins containing ASH domains (NCBI Conserved Protein Domain Database accession number: cl13764) (ASH domains are members of the PapD-like superfamily; [28]; http://pfam.xfam.org/clan/PapD-like). In the first approach, BLAST was used to search the 12 ASH/PapD-like domains annotated for C. reinhardtii hydin (http://www.genome.jp/dbget-bin/www_bget?cre:CHLREDRAFT_116240) against the C. reinhardtii predicted protein database v5.5 in Phytozome 12 [target type, proteome; BLASTP; Expect (E) threshold, 100]; similarly, the 12 ASH/PapD-like domains of hydin were searched by pHMMER against the C. reinhardtii reference proteome (https://www.ebi.ac.uk/Tools/hmmer/search/phmmer/; sequence E-values 0.01, bit score 25). In each case, the top 10 proteins from each of the 12 searches were combined and checked against the C. reinhardtii flagellar proteome (http://chlamyfp.org/index.php); only flagellar proteins were considered further. Each of the putative ASH domains in each candidate ASH-domain-containing flagellar protein was then verified by a BLAST search of the Phytozome C. reinhardtii predicted protein database and a pHMMER search of the HMMER C. reinhardtii reference proteome using the above parameters. If only hydin was returned, the protein was rejected as unlikely to be a true ASH-domain-containing protein. If other ASH-domain proteins were returned, it was concluded that the protein was likely to be a true ASH protein. The ASH-domain-containing proteins or ASH-domain sequences thus identified were aligned using MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/), and then used for iterative searches against the C. reinhardtii reference proteome in the hidden Markov models-based jackHMMER with default parameters (https://www.ebi.ac.uk/Tools/hmmer/search/jackhmmer; sequence E-values 0.01, bit score 25). The search was repeated until convergence, and the proteins identified were combined and checked against the C. reinhardtii flagellar proteome (http://chlamyfp.org/index.php); only flagellar proteins were considered further.
In the second approach, the sequence alignments corresponding to the Pfam ASH domain (PF15780; https://pfam.xfam.org/family/ASH#tabview=tab3) generated using hidden Markov models were iteratively searched against the C. reinhardtii reference proteome with jackHMMER using default parameters (sequence E-values 0.01, bit score 25). The search was repeated until convergence, and all the proteins identified were combined and checked against the C. reinhardtii flagellar proteome (http://chlamyfp.org/index.php); only flagellar proteins were considered further. The two approaches identified the same eight flagellar ASH-domain proteins (figure 3).
Figure 3.
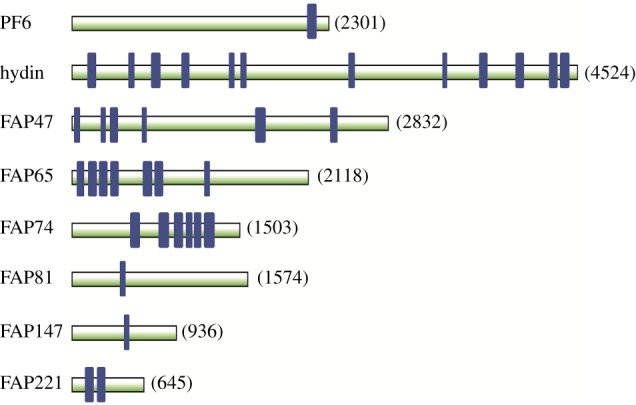
Predicted ASH domains of C. reinhardtii CA proteins. ASH domains are indicated by blue rectangles. Proteins are drawn to scale; the number to the right of each protein indicates the number of amino acids in the protein. ASH domains were identified as described in the Methods. (Online version in colour.)
To confirm identification of the ASH domains, each of these proteins was then examined using the NCBI Conserved Domains tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi; threshold 100). All ASH domains were indeed annotated as ASH or PapD-like except for that of FAP147. The FAP147 ASH domain (amino acids 471–505) identified in our analysis was part of a larger region annotated as a MYCBPAP domain. The human homologue of FAP147 is Myc-binding protein-associated protein (MYCBPAP), which Ponting [29] reported had a single ASH domain at amino acids 432–538. Examination of MYCBPAP using the NCBI Conserved Domains tool showed that amino acids 432–538 are similarly contained in a longer region annotated as a MYCBPAP domain. This confirmed our identification of the ASH domain in FAP147.
Domain predictions based on NCBI sequences for the flagellar ASH-domain proteins can be found at:
PF6 (http://www.genome.jp/dbget-bin/www_bget?cre:CHLREDRAFT_181848),
hydin (http://www.genome.jp/dbget-bin/www_bget?cre:CHLREDRAFT_116240),
FAP47 (https://www.genome.jp/dbget-bin/www_bget?cre:CHLREDRAFT_189076),
FAP65 (https://www.genome.jp/dbget-bin/www_bget?cre:CHLREDRAFT_153955),
FAP74 (http://www.genome.jp/dbget-bin/www_bget?cre:CHLREDRAFT_167096),
FAP81 (http://www.genome.jp/dbget-bin/www_bget?cre:CHLREDRAFT_188960),
FAP147 (http://www.genome.jp/dbget-bin/www_bget?cre:CHLREDRAFT_196787),
FAP221 (http://www.genome.jp/dbget-bin/www_bget?cre:CHLREDRAFT_176588).
(c). PAS-domain identification
To identify all flagellar proteins containing PAS domains (NCBI Conserved Protein Domain Database accession number: cd00130), the following seven known PAS-domain-containing flagellar protein sequences were downloaded from the NCBI (https://www.ncbi.nlm.nih.gov/) or Phytozome websites (version 12, https://phytozome.jgi.doe.gov/pz/portal.html) (NCBI accession number, Phytozome gene number): FAP49 (XP_001696011.1, Cre08.g362050.t1.2); FAP72 (XP_001696096.1, Cre08.g362000.t1.2); FAP153 (Cre08.g361950.t1.1); FAP154 (XP_001696012.1, Cre08.g362100.t1.1); FAP260 (XP_001691551.1, Cre17.g708000.t1.1); FAP3611 (Cre08.g362150.t1.1); PHOT (XP_001693387.1, Cre03.g199000.t1.2). Each protein was then searched for domain predictions at the SMART website (http://smart.embl-heidelberg.de/), and sequences annotated as PAS domains were each searched against the C. reinhardtii predicted protein database v5.5 in Phytozome 12 using the BLASTP function, or searched against the HMMER C. reinhardtii reference proteome using the HMMER website service (https://www.ebi.ac.uk/Tools/hmmer/search/phmmer) with the default parameters (sequence E-value 0.01, bit score 25.0). The PAS-domain-containing proteins or PAS-domain sequences thus identified were aligned using MUSCLE, and then used for iterative searches in the hidden Markov models-based jackHMMER with default parameters (sequence E-value 0.01, bit score 25.0). The search was repeated until convergence, and the proteins identified were combined and checked against the C. reinhardtii flagellar proteome (http://chlamyfp.org/index.php); only flagellar proteins were considered further.
Independently, the sequence alignments corresponding to the Pfam PAS domain (PF00989; https://pfam.xfam.org/family/PAS#tabview=tab4) were searched iteratively against the C. reinhardtii reference proteome with jackHMMER (sequence E-value 0.01, bit score 25.0). The search was repeated until convergence, and all the proteins were checked against the C. reinhardtii flagellar proteome (http://chlamyfp.org/index.php); only flagellar proteins were considered further. The two approaches identified the same 11 flagellar PAS-domain proteins (figure 4), including four not previously recognized as PAS proteins (FAP417 (Cre10.g429750.t1.1), Cre01.g003950.t1.1, Cre01.g004124.t2.1 and Cre01.g004157.t1.2). FAP417 had been identified as a candidate CA protein by Zhao et al. [2]; the other three proteins are not CA proteins but were found by MS in axonemes of wild type and pf18 by Zhao et al. [2] and in a flagellar fraction by Jordan et al. [30] (http://chlamyfp.org/readcsvfile_js.php).
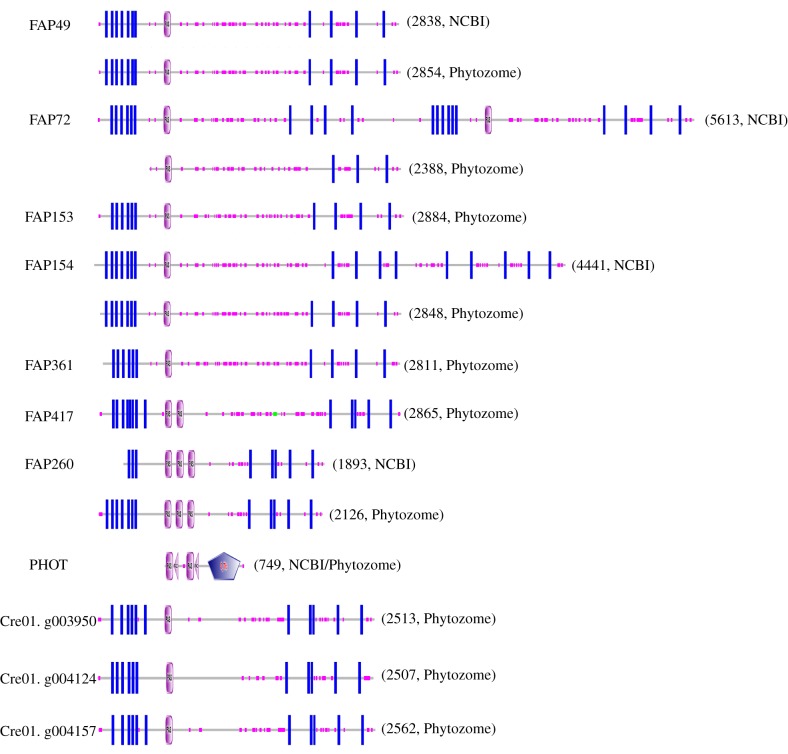
Figure 4.
Domain architecture of predicted C. reinhardtii flagellar proteins containing PAS domains. Pink ovals marked ‘PAS’ indicate predicted PAS domains. Blue rectangles indicate predicted transmembrane domains. Small pink boxes along each protein indicate regions of low sequence complexity. In PHOT, pink triangles marked ‘PAC’ indicate ‘motif C-terminal to PAS motifs (likely to contribute to PAS structural domain)’ and the blue pentagon marked ‘S_TKc’ indicates a ‘serine/threonine protein kinases, catalytic domain’. The number to the right of each architecture indicates the number of amino acids in the protein sequence. PAS domains were identified as described in the Methods. When there is a discrepancy between sequences from NCBI and Phytozome, predictions for both sequences are shown. The locations of exclusive unique peptides suggested that some Phytozome models were more accurate than the corresponding NCBI models; e.g. for FAP72, unique peptides were predicted to originate from throughout the N-terminal half of the NCBI model (most of which is identical to the Phytozome model) but not the C-terminal half. (Online version in colour.)
The NCBI and Phytozome databases differ in their sequences for many of these proteins (figure 4). There is no sequence for FAP361 in the NCBI database, and a search for ‘FAP153’ returns the sequence for FAP49 with the annotation that FAP153 is likely a part of FAP49. However, when the Phytozome sequences for FAP153 and FAP361 were added to the database of C. reinhardtii proteins downloaded from NCBInr and the database then re-searched using the MS spectra from our proteomic analyses of C. reinhardtii flagellar axonemes, we found more than 20 unique peptides for FAP49 and one unique peptide for FAP153 [2], indicating that they are distinct proteins that are expressed. To our knowledge, no peptides unique to FAP361 have been identified in any proteomic study of C. reinhardtii flagellar fractions. The Phytozome model for FAP72 lacks sequence at its N-terminus that is present in the NCBI model, but this sequence is encoded in the Phytozome C. reinhardtii genome immediately upstream of the FAP72 sequence and almost certainly is part of FAP72. Based on the locations of unique peptides in the NCBI and more recent Phytozome protein models, the latter appear to be more accurate (see legend to figure 4 for details).
(d). Maximum-likelihood tree of PAS proteins
The Orthofinder output of homology relationships included one orthogroup containing all of the flagella-associated PAS-domain proteins except PHOT. This orthogroup had a total of 133 sequences, some of which were duplicates. Examination of the predicted domain architecture of each of these sequences using SMART revealed some that seemed unlikely to be true orthologues because they lacked predicted transmembrane domains or PAS domains. Therefore, candidates for construction of a maximum-likelihood tree of C. reinhardtii flagellar PAS proteins and related sequences were selected from the orthogroup as follows. First, each protein in the orthogroup was subjected to a reverse-BLASTP search (cut-off E = 1 × 10−5) against the C. reinhardtii flagellar PAS proteins (excluding PHOT). Proteins passing this test were then examined for predicted domain architecture similar to that of the C. reinhardtii flagellar PAS proteins—i.e. the presence of at least one PAS domain flanked N-terminally by a group of closely spaced transmembrane domains and C-terminally by multiple widely spaced transmembrane domains, and the absence any other functional domain. Of the 133 orthogroup proteins, 74 passed the reverse-BLASTP search and the manual inspection; 58 proteins were included in the tree after duplicate sequences were removed.
The tree was constructed using IQ-TREE [31]. Sequence evolution model (LG+F+I+G4) was selected using the IQ-TREE onboard model finder (http://www.iqtree.org/doc/Substitution-Models#protein-models; [32]) and bootstrapping support was performed with Ultrafast Bootstrap [33]. The tree was rooted with Rozella allomycis as the outgroup and was displayed with iTOL [34]. Multiple sequence alignment for tree calculation was performed with Clustal Omega [35].
3. Results and discussion
(a). Conservation of CA proteins
Because of the cilium's importance for human health, most previous studies of C. reinhardtii CA proteins focused only on identifying their mammalian orthologues (e.g. [2,21]). To obtain insight into the conservation of CA proteins over a wider range of phylogenetic groups, we used the orthogroup inference algorithm OrthoFinder to infer homology relationships between each of the 62 C. reinhardtii CA-specific proteins and all predicted proteins of 57 ciliated and non-ciliated species representing all of the major eukaryotic lineages (supplementary material, table S2). The results are presented as a dot plot (figure 2) and, for a subset of the proteins that are subunits of specific projections or share ASH domains, as a Coulson plot (electronic supplementary material, figure S1). The first one or two columns of the figures indicate whether an organism has cilia or motile cilia. Arthropods lack motile cilia except for the sperm of some members of the group, including insects, where there is considerable variation in the structure of the axoneme [36,37]. Mosquitos, including those of the genus Aedes, have a motile spermatozoan with a ‘9+1’ axoneme in which the two central microtubules are replaced by a single, apparently solid, fibre with no obvious projections [38,39]. Pediculus humanus is reported to have sperm flagella with a ‘9+9+2’ axoneme [36,40], but we have been unable to find published electron micrographs that would provide detail on its CA ultrastucture. The arachnid Ixodes scapularis, which has non-flagellated sperm [41], lacks motile cilia. Nematodes such as Caenorhabditis elegans and Loa loa have only non-motile ‘9+0’ cilia [42,43]. The marine centric diatom T. pseudonana has a motile flagellated spermatozoan [44] that previously was predicted to lack a CA based on comparative genomic analyses [11]. Flagella have not been observed directly in Reticulomyxa filosa, but R. filosa may have flagellated gametes based on the presence in its genome of sequences predicted to encode flagellar proteins ([45], and this study).
Several observations can be drawn from these analyses:
— First, many CA proteins are conserved in all ciliated lineages, consistent with the inference from cilia ultrastructure that the LECA had a ‘9+2’ axoneme. Importantly, all C. reinhardtii CA projections to which CA-specific proteins have been localized (figure 1) contain proteins that have predicted orthologues across the diversity of eukaryotes. These include C1a/e/c (PF6, FAP76, FAP81, FAP119, MOT17), C1b/f (CPC1, FAP69), C1d (FAP54, FAP74, FAP221), C2b (hydin) and C2c/d (KLP1). Although one cannot conclude that these orthologues are localized to the exact same CA projections in all eukaryotes in which they occur, the results suggest that they have fundamental conserved roles in CA architecture and function. This conclusion is consistent with the observations that loss of hydin causes loss of the C2b projection in both C. reinhardtii and mice [46,47], and that the overall architecture of the CA as determined by cryo-ET is very similar between C. reinhardtii and Strongylocentrotus purpuratus [1].
— Conversely, some proteins that have been localized to specific CA projections in C. reinhardtii are not highly conserved, including FAP114 and FAP227 in C1a/e and FAP297 in C1d. This is consistent with the observations from cryo-ET that the CAs of C. reinhardtii and S. purpuratus differ in some structural details, such as the number of longitudinal connections between projections [1]. Such proteins presumably represent adaptation to the specific needs of the CA in C. reinhardtii.
— Orthologues of nearly all C. reinhardtii CA proteins are consistently absent in organisms lacking cilia or having only ‘9+0’ cilia. For those candidate CA proteins whose orthologues otherwise have a wide phylogenetic distribution (specifically FAP65, FAP70, FAP75, FAP81, FAP147, FAP194, MOT17), this provides supporting evidence that they are indeed components of the CA and are likely to function only in the CA.
— Orthologues of C. reinhardtii CA proteins appear to be completely absent from Aedes albopictus, suggesting that the central fibre in the A. albopictus ‘9+1’ axoneme has no compositional similarity to the CA of C. reinhardtii.
— Similarly, no orthologues were found in Drosophila melanogaster, which was unexpected given that the spermatozoan of D. melanogaster has a ‘9+2’ axoneme. However, published transmission electron micrographs of fixed and plastic-embedded D. melanogaster sperm tails (e.g. [48–51]) do not provide convincing evidence for CA projections structurally similar to those of C. reinhardtii or mammals. Moreover, Laurençon et al. [52] compiled a list of D. melanogaster homologues of proteins in datasets of cilia and basal body proteins of other organisms; their list contains homologues of proteins of the outer and inner dynein arms, the N-DRCs and radial spokes, but, consistent with our finding, no homologues of CA-specific proteins. The results suggest that details of the D. melanogaster CA—specifically the projections of the central microtubules—are not conserved. It may be that a highly adapted CA has evolved to support the unusual motility of the extremely long sperm tail during migration through the D. melanogaster female reproductive tract [53].
— R. filosa has orthologues of several C. reinhardtii CA proteins, but fewer than any other ciliated organism except D. melanogaster, A. albopictus and Plasmodium falciparum. The results are in good agreement with those of Glöckner et al. [45], who analysed the genome of R. filosa and found that, compared with Naegleria gruberi, it had a reduced set of genes encoding flagellar proteins. They suggested three possible explanations for this finding: ‘the basic requirements to produce a flagellar apparatus are smaller than estimated…, the detected orthologs are only remnants of a life stage with a functional flagella, or other proteins fulfil analogous functions in R. filosa’ [45, p. 14]. Our analysis suggests that R. filosa does indeed have a flagellated gamete, but with a greatly simplified or highly adapted CA.
— Orthologues of the C. reinhardtii CA proteins containing ASH domains are present in all eukaryotic lineages, suggesting that they were present in the LECA and have important, conserved roles in CA assembly or function. These proteins are discussed in more detail in the next section.
— As noted in Background (§1), the CA of cilia of the SAR and Plantae rotates during ciliary beating, whereas the CA of unikont cilia maintains a fixed orientation relative to the outer doublet microtubules [5]. Orthologues of C. reinhardtii FAP123, FAP125, FAP239, FAP348 and DPY30 are present in the former but not the latter, and thus are candidates for having a role related to CA rotation. Orthologues of all but FAP125 likewise are present in the Haptophyta; it would be interesting to know if members of this group also have a rotating CA. Orthologues of FAP125, FAP239 and DPY30 are present in Excavata, in which the orientation of the CA has been reported as invariant (trypanosomatids; [54]), variable but not random (Euglena gracilis; [55]), or appears—based on serial thin sections—to rotate relative to the outer doublets (Ophirina amphinema; [56]).
— Six C. reinhardtii CA proteins have orthologues in organisms lacking cilia or a CA. Orthologues of DPY30, DIP13 and FAP348 are present in T. pseudonana, which is predicted to lack a CA. DIP13 (for deflagellation-inducible protein of 13 kDa) was identified first in C. reinhardtii by Pfannenschmid et al. [57], who localized it to C. reinhardtii flagella, basal bodies and cytoplasmic microtubules by immunofluorescence microscopy; partial knockdown of DIP13 expression in C. reinhardtii resulted in a cell-division phenotype in a small proportion of cells. They concluded that this small protein likely had a general function in stabilizing microtubules or linking microtubules to motile systems. If, in addition to its function in the CA, C. reinhardtii DIP13 has a role in the cytoplasm as proposed by Pfannenschmid and colleagues, it could explain the presence of its orthologues in T. pseudonana as well as in the non-ciliated Cryptosporidium parvum. Similarly, the orthologues of DPY30, a confirmed CA protein in C. reinhardtii, and FAP348 may have roles in the cytoplasm of T. pseudonana. Orthologues of C. reinhardtii FAP225 are present in the non-ciliated Entamoeba histolytica and Acanthamoeba castellanii, raising the possibility that these proteins have functions in the cytoplasm of some members of the Amoebozoa. An apparent orthologue of C. reinhardtii PF6 (a confirmed CA protein) was identified in I. scapularis, and an apparent orthologue of FAP101 was identified in Polysphondylium pallidum; despite their similarities to the C. reinhardtii proteins, these may not be true orthologues.
— Seventeen of the C. reinhardtii CA proteins had orthologues only in Volvox carteri. These could be CA proteins that recently evolved in this lineage and are specifically adapted to flagellar motility in the Volvocales. Alternatively, given that most C. reinhardtii CA proteins are widely conserved, it is possible that some of these nonconserved proteins were false positives in our analysis of the CA proteome [2]. Nine of these proteins (FAP39, FAP108, FAP139, FAP275, FAP289, FAP380, FAP412, FAP415, FAP416) were among a total of only 12 that were identified in just one replicate in our proteomic analysis; that combined with their limited distribution in eukaryotic lineages raises the possibility that they are volvocalean-specific proteins that contaminated our wild-type preparation and were incorrectly identified as CA proteins. Further study of the proteins will be necessary to resolve this.
(b). Flagellar ASH-domain proteins are concentrated in the CA
Ponting [29] identified over a dozen ASH-domain-containing proteins in humans and mice. Most were associated with centrioles or cilia. Based on this, Ponting proposed that the ASH domains may have a microtubule-binding function. More recently, Schou et al. [28] showed that four subunits of the human transport particle or TRAPP complex have ASH domains combined with TPR domains; these newly identified proteins may represent a distinct sub-family of ASH-domain proteins. Schou and colleagues showed that the ASH domains target fusion proteins to centrosomes and bundle microtubules, supporting a role for these domains in microtubule binding.
The C. reinhardtii flagellum contains a total of eight of these proteins (figure 3). The mammalian homologues of five of these were identified by Ponting [29], although he did not know that they were all ciliary proteins. Remarkably, all eight are confirmed or candidate CA proteins (see table 1 of [21], and table 2 of [2]). The ASH-domain proteins (indicated in red font in figure 1) occur in multiple locations within the C. reinhardtii CA. Of those whose locations are known, PF6 and FAP81 are in the C1a/e/c supercomplex [3,58,59], FAP74 and FAP221 are in the C1d projection [60] and hydin is in the C2b projection [46]. FAP47 is likely to be in yet another CA structure [2].
The conservation of the ASH-domain proteins throughout eukaryotic evolution, their concentration in the CA and their distribution in several projections there raise intriguing questions about their roles. If the ASH domains have a microtubule-binding function, as proposed by Ponting [29] and supported by Schou et al. [28], then they likely function in attaching the CA projections to the C1 and C2 microtubules. Such a function would be consistent with their locations in multiple projections, and also consistent with more than one ASH-domain protein being in any given projection, since several projections, including C1d and the C1a/e/c supercomplex, have multiple connections to the microtubule with which they are associated [1,3]. A role in binding to the central microtubules is supported by the findings that knockdown of C. reinhardtii FAP74 and hydin causes loss of the C1d and C2b projections, respectively [46,60]. It also would explain why the CA ASH-domain proteins are so widely conserved, since a means to anchor the projections to the central microtubules would be universally required for microtubule-based CAs with projections. Indeed, the physical connections between the CA projections and their underlying microtubules appear to be very similar in C. reinhardtii and sea urchin [1]. There are likely to be additional proteins attached to or embedded in the central microtubules that specify precisely where each projection will attach to the microtubules, and these could interact with the ASH-domain proteins to confer specificity on this assembly process.
Other possibilities for the functions of the ASH-domain proteins include interaction with the intraflagellar transport machinery to move pre-assembled projections into the flagella, or interactions with the radial spoke heads. These possibilities are not mutually exclusive with each other or with a role in binding the projections to the CA. Further study will be necessary to definitively clarify the functions of these widely conserved proteins. In any case, the ASH-domain proteins are very likely to have similar important functions in the CAs of diverse organisms.
(c). PAS proteins
Our BLASTP and HMMER searches for C. reinhardtii PAS proteins found 11 that have been reported in C. reinhardtii flagellar fractions (figures 4 and 5); together they represent an intriguing yet poorly understood group of proteins. The first to be identified was PHOT, a blue-light receptor that is present in both the cell body and axoneme and controls multiple steps in the C. reinhardtii life cycle [61,62]. Additional PAS proteins were identified in our analysis of the C. reinhardtii flagellar proteome [22], where peptides derived from them were found primarily in a KCl extract of the axonemal fraction, despite the fact that they are predicted to have transmembrane domains. This was not particularly surprising, because several flagellar membrane proteins fractionate with and apparently are anchored to the axoneme [22,63]. In our analysis of the CA proteome [2], we identified three of these, FAP49, FAP72 and FAP154 (green font in figure 1), as candidate CA proteins because they were present in wild-type axonemes but greatly reduced in pf18 axonemes, and because their abundances in the wild-type axoneme were similar to that of known CA-specific proteins. A fourth protein, FAP417 (also green font in figure 1), was identified as a candidate CA protein by Zhao et al. [2] and as a PAS protein in the current analysis. Three other PAS proteins (Cre01.g003950.t1.1, Cre01.g004157.t1.2 and Cre01.g004124.t2.1) identified by our BLASTP and HMMER searches are flagellar but not CA proteins; they were found by MS in axonemes of wild type and pf18 by Zhao et al. [2] and in the flagellar fractions of Jordan et al. [30], although they were not specifically commented upon in either publication. Here we focus primarily on the candidate CA proteins.
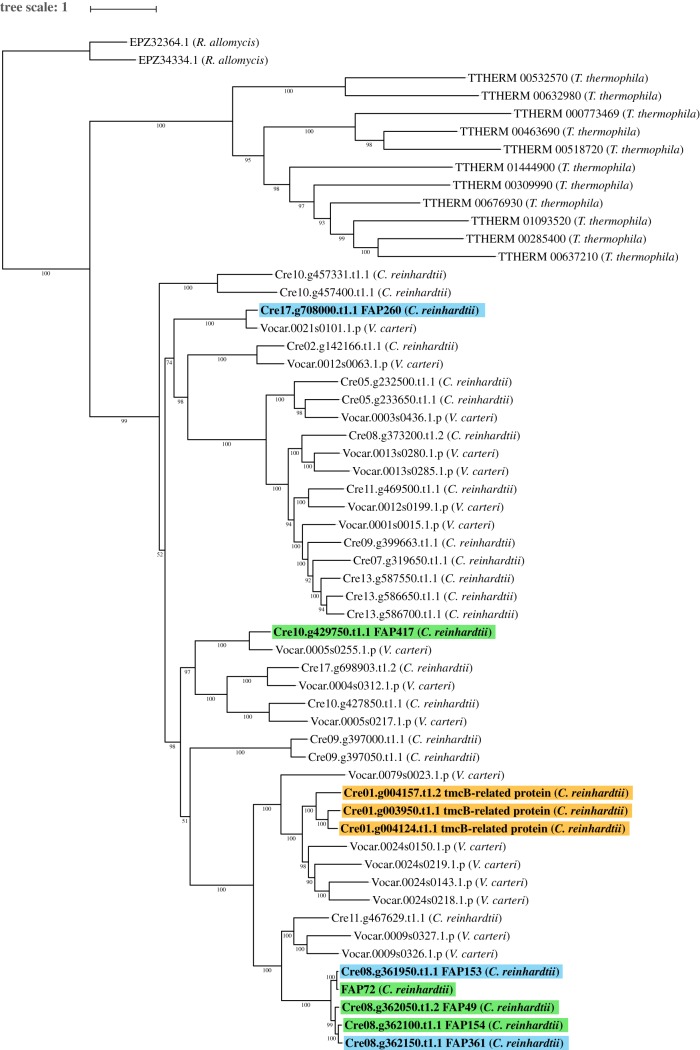
Figure 5.
Maximum-likelihood tree of C. reinhardtii PAS proteins and orthologues. Maximum-likelihood tree of C. reinhardtii flagellar-associated PAS proteins with high-confidence orthologues, rooted with R. allomycis as the outgroup. Green highlighting indicates C. reinhardtii PAS proteins predicted to be associated with the CA; orange highlighting indicates C. reinhardtii flagellar PAS proteins encoded on Chromosome 1; blue highlighting indicates other C. reinhardtii flagellar PAS proteins. For the tree construction only, a model for FAP72 was used that included the N-terminus of NCBI FAP72 (aa 1–471) joined to the N-terminus of Phytozome FAP72 (see Methods). The conjoined model lacks only 15 unmatched amino acids from the C-terminus of Phytozome FAP408. Phytozyme lists two transcripts for the Cre01.g004124 gene: Cre01.g004124.t1.1 (primary) and Cre01.g004124.t2.1; the former includes two amino acids not present in the latter and was used to construct this tree, whereas the latter is the one associated with this gene in the Chlamydomonas Flagellar Proteome Project (http://chlamyfp.org/index.php) and so is referred to in the main text. The tree was constructed with IQ-TREE [31] and was visualized using iTOL [34]. Support values show ultrafast bootstrap data with 1000 iterations [33]. Scale represents average number of substitutions per residue. (Online version in colour.)
FAP49, FAP72 and FAP154, together with FAP153 and FAP361, are predicted to be encoded by successive adjacent genes on Chromosome 8 and have very similar sequences (79–97 or 78–95% identity based on NCBI or Phytozome models, respectively); expression of all of these closely related proteins except FAP361 was confirmed by the identification of exclusive unique peptides in our proteomic analyses. Although structural predictions are tentative because of differences in NCBI and Phytozome gene models (figure 4), all five proteins are likely to have very similar structures consisting of six or seven closely spaced N-terminal predicted transmembrane domains closely followed by an N-terminal PAS domain and then four more widely spaced C-terminal predicted transmembrane domains. The three PAS proteins found in flagellar fractions by Jordan et al. [30] and Zhao et al. [2] are encoded by genes closely spaced on Chromosome 1, have a similar architecture to the PAS proteins encoded on Chromosome 8 and are about 45% similar to the latter in their sequences. FAP417 is encoded on Chromosome 10, is about 30% identical to the PAS proteins encoded on Chromosome 8 and also has a similar architecture except that it is predicted to have two PAS domains. As far as we know, the combination of approximately seven closely spaced N-terminal transmembrane domains closely followed by one or two PAS domains and then approximately four more widely spaced transmembrane domains is an architecture that is distinct from those of other PAS proteins that have been characterized [64]. Another flagellar PAS protein, FAP260, is encoded on Chromosome 17, has a sequence that is about 30% identical to those encoded on Chromosome 8 and is predicted to have three PAS domains flanked by transmembrane domains.
Among the 57 species whose genomes were included in our analysis, closely related orthologues of the flagellar PAS proteins are present only in V. carteri, Tetrahymena thermophila and R. allomycis (figure 2). A maximum-likelihood tree of these proteins reveals a major expansion of this protein family in C. reinhardtii and V. carteri (figure 5). Those PAS proteins that have been found in flagellar fractions are highlighted: the four candidate CA proteins have a green highlight, the three proteins encoded on Chromosome 1 have an orange highlight and other flagellar proteins including FAP361 have a blue highlight. The five adjacent genes on Chromosome 8 appear to have arisen as a result of recent gene duplication events. The tree reveals numerous other closely related PAS proteins that are predicted in C. reinhardtii but have not been detected in flagellar fractions; they may be located in the cytoplasm.
The limited phylogenetic distribution of orthologues of the above PAS proteins suggests that they have a function specific to volvocalean algae and possibly a few other members of the SAR and Plantae. PAS domains in many other proteins are involved in responses to environmental factors such as light, oxygen and redox potential [65,66]. Therefore, it is of interest that the C. reinhardtii mutant fap47-1, the axonemes of which lack or have greatly reduced amounts of the candidate CA PAS proteins FAP49, FAP72 and FAP154, appears to have a defect in phototaxis [2]. It is possible that the PAS proteins function in a sensory or signalling pathway that controls phototactic steering, and that this pathway is unique to the Volvocales.
Still unexplained is the presence of predicted transmembrane domains in the candidate CA PAS proteins. The CA contacts the flagellar membrane at its tip [67,68], but the estimated abundances of the candidate CA PAS proteins is similar to that of confirmed CA proteins [2], whereas proteins located only at the tip of the CA would be expected to be much less abundant. Moreover, the assignment of two of the proteins, FAP72 and FAP417, to the CA was based on their presence in only one of two replicates [2], so there is a possibility that they were contaminants in the wild-type preparation. Further study is needed to confirm that these are indeed CA proteins, and if so, to determine where in the CA they are located and how they function.
4. Summary
Orthologues of many C. reinhardtii CA-specific proteins are present in all eukaryotic lineages, suggesting that the ancestors of these proteins were present in the flagellum of the LECA. These include orthologues of proteins localized to multiple CA projections, further suggesting that these projections were already highly differentiated in the LECA. Among the widely conserved proteins are those containing ASH domains, which within the flagellum are restricted to the CA; these proteins are likely to have important conserved functions, possibly including attachment of the projections to the central microtubules. Further insight into the evolution of the flagellum will benefit from a more detailed understanding of the specific locations and functions of the CA-specific proteins.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Dr G. Pazour (University of Massachusetts Medical School) for help with analysis of the ASH- and PAS-domain-containing proteins, and Dr Pazour and Dr D. Mitchell (SUNY Upstate Medical University, Syracuse) for helpful comments on the manuscript.
Endnote
FAP361 originally was identified as a flagellar protein based on peptides shared with other flagellar PAS proteins [22]; its status currently is unclear because peptides unique to it have not been identified in any Chlamydomonas flagellar fraction (§2c).
Data accessibility
Data not included in figures are available in the electronic supplementary material.
Authors' contributions
Conception and design (L.Z., Y.H. and G.B.W.), and analysis and interpretation of data (L.Z., Y.H., N.A.M. and G.B.W.). Drafting the article and revising it for important intellectual content (L.Z., Y.H., N.A.M. and G.B.W.).
Competing interests
The authors declare no competing financial interests.
Funding
This work was supported by National Institutes of Health grants R37 GM030626 and R35 GM122574 to G.B.W., and by the Robert W. Booth Endowment at the University of Massachusetts Medical School to G.B.W.
References
- 1.Carbajal-Gonzalez BI, Heuser T, Fu X, Lin J, Smith BW, Mitchell DR, Nicastro D. 2013. Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton 70, 101–120. ( 10.1002/cm.21094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao L, Hou Y, Picariello T, Craige B, Witman GB. 2019. Proteome of the central apparatus of a ciliary axoneme. J. Cell Biol. 218, 2051–2070. ( 10.1083/jcb.201902017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu G, et al. 2019. Structural organization of the C1a-e-c supercomplex within the ciliary central apparatus. J. Cell Biol. 218, 4236–4251. ( 10.1083/jcb.201906006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakazawa Y, Ariyoshi T, Noga A, Kamiya R, Hirono M. 2014. Space-dependent formation of central pair microtubules and their interactions with radial spokes. PLoS ONE 9, e110513 ( 10.1371/journal.pone.0110513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell DR. 2017. Evolution of cilia. Cold Spring Harb. Perspect. Biol. 9, a028290 ( 10.1101/cshperspect.a028290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omoto CK, Gibbons IR, Kamiya R, Shingyoji C, Takahashi K, Witman GB. 1999. Rotation of the central pair microtubules in eukaryotic flagella. Mol. Biol. Cell 10, 1–4. ( 10.1091/mbc.10.1.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. 1998. Randomization of left–right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837. ( 10.1016/S0092-8674(00)81705-5) [DOI] [PubMed] [Google Scholar]
- 8.Ishijima S, Sekiguchi K, Hiramoto Y. 1988. Comparative study of the beat patterns of American and Asian horseshoe crab sperm: evidence for a role of the central pair complex in forming planar waveforms in flagella. Cytoskeleton 9, 264–270. ( 10.1002/cm.970090308) [DOI] [Google Scholar]
- 9.Baccetti B, Burrini AG, Dallai R, Pallini V. 1979. The dynein electrophoretic bands in axonemes naturally lacking the inner or the outer arm. J. Cell Biol. 80, 334–340. ( 10.1083/jcb.80.2.334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbons BH, Gibbons IR, Baccetti B. 1983. Structure and motility of the 9+0 flagellum of eel spermatozoa. J. Submicrosc. Cytol. 15, 15–20. [PubMed] [Google Scholar]
- 11.Merchant SS, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. ( 10.1126/science.1143609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satir P, Mitchell DR, Jékely G. 2008. How did the cilium evolve? Curr. Top. Dev. Biol. 85, 63–82. ( 10.1016/S0070-2153(08)00803-X) [DOI] [PubMed] [Google Scholar]
- 13.Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, Porter ME. 2013. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol. Biol. Cell 24, 1134–1152. ( 10.1091/mbc.e12-11-0801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King SM. 2018. Dynein: dynein mechanics, dysfunction, and disease. Oxford, UK: Elsevier. [Google Scholar]
- 15.Lin J, Tritschler D, Song K, Barber CF, Cobb JS, Porter ME, Nicastro D. 2011. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem. 286, 29 175–29 191. ( 10.1074/jbc.M111.241760) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang P, et al. 2006. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119, 1165–1174. ( 10.1242/jcs.02811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witman G. (ed.). 2009. Cell motility and behavior. The Chlamydomonas sourcebook, 2nd edn, vol. 3. Kidlington, UK: Academic Press. [Google Scholar]
- 18.Kollmar M. 2016. Fine-tuning motile cilia and flagella: evolution of the dynein motor proteins from plants to humans at high resolution. Mol. Biol. Evol. 33, 3249–3267. ( 10.1093/molbev/msw213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickstead B. 2018. The evolutionary biology of dyneins. In Dyneins: structure, biology and disease. 1: The biology of dynein motors (ed. King SM.), pp. 101–138. Oxford, UK: Elsevier; ( 10.1016/B978-0-12-809471-6.00003-6) [DOI] [Google Scholar]
- 20.Adams GM, Huang B, Piperno G, Luck DJ. 1981. Central-pair microtubular complex of Chlamydomonas flagella: polypeptide composition as revealed by analysis of mutants. J. Cell Biol. 91, 69–76. ( 10.1083/jcb.91.1.69) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loreng TD, Smith EF. 2017. The central apparatus of cilia and eukaryotic flagella. Cold Spring Harb. Perspect. Biol. 9, a028118 ( 10.1101/cshperspect.a028118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pazour GJ, Agrin N, Leszyk J, Witman GB. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113. ( 10.1083/jcb.200504008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 ( 10.1186/s13059-015-0721-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. bioRXiv, 466201 ( 10.1101/466201) [DOI]
- 25.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. ( 10.1038/nmeth.3176) [DOI] [PubMed] [Google Scholar]
- 26.Miki H, Okada Y, Hirokawa N. 2005. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 15, 467–476. ( 10.1016/j.tcb.2005.07.006) [DOI] [PubMed] [Google Scholar]
- 27.Field HI, Coulson RM, Field MC. 2013. An automated graphics tool for comparative genomics: the Coulson plot generator. BMC Bioinformatics 14, 141 ( 10.1186/1471-2105-14-141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schou KB, Morthorst SK, Christensen ST, Pedersen LB. 2014. Identification of conserved, centrosome-targeting ASH domains in TRAPPII complex subunits and TRAPPC8. Cilia 3, 6 ( 10.1186/2046-2530-3-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponting CP. 2006. A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics 22, 1031–1035. ( 10.1093/bioinformatics/btl022) [DOI] [PubMed] [Google Scholar]
- 30.Jordan MA, Diener DR, Stepanek L, Pigino G. 2018. The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol. 20, 1250–1255. ( 10.1038/s41556-018-0213-1) [DOI] [PubMed] [Google Scholar]
- 31.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. ( 10.1038/nmeth.4285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. ( 10.1093/molbev/msx281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. ( 10.1093/nar/gkw290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 ( 10.1038/msb.2011.75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips DM. 1970. Insect sperm: their structure and morphogenesis. J. Cell Biol. 44, 243–277. ( 10.1083/jcb.44.2.243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips DM. 1974. Structural variants in invertebrate sperm flagella and their relationship to motility. In Cilia and flagella (ed. Sleigh M.), pp. 379–402. London, UK: Academic Press. [Google Scholar]
- 38.Swan MA. 1981. The generation and propagation of double waves in mosquito (Aedes notoscriptus) sperm-tails. Gamete Res. 4, 241–250. ( 10.1002/mrd.1120040308) [DOI] [Google Scholar]
- 39.Phillips DM. 1969. Exceptions to the prevailing pattern of tubules (9+9+2) in the sperm flagella of certain insect species. J. Cell Biol. 40, 28–43. ( 10.1083/jcb.40.1.28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito S. 1966. Movement and structure of louse spermatozoa. J. Cell Biol. 31, 128A. [Google Scholar]
- 41.Rothschild L. 1961. Structure and movement of tick spermatozoa (Arachnida, Acari). Q. J. Microsc. Sci. 102, 239–247. [Google Scholar]
- 42.Kozek WJ, Orihel TC. 1983. Ultrastructure of Loa loa microfilaria. Int. J. Parasitol. 13, 19–43. ( 10.1016/S0020-7519(83)80063-0) [DOI] [PubMed] [Google Scholar]
- 43.Nechipurenko IV, Sengupta P. 2017. The rise and fall of basal bodies in the nematode Caenorhabditis elegans. Cilia 6, 9 ( 10.1186/s13630-017-0053-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore ER, Bullington BS, Weisberg AJ, Jiang Y, Chang J, Halsey KH. 2017. Morphological and transcriptomic evidence for ammonium induction of sexual reproduction in Thalassiosira pseudonana and other centric diatoms. PLoS ONE 12, e0181098 ( 10.1371/journal.pone.0181098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glöckner G, et al. 2014. The genome of the foraminiferan Reticulomyxa filosa. Curr. Biol. 24, 11–18. ( 10.1016/j.cub.2013.11.027) [DOI] [PubMed] [Google Scholar]
- 46.Lechtreck KF, Witman GB. 2007. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell Biol. 176, 473–482. ( 10.1083/jcb.200611115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lechtreck KF, Delmotte P, Robinson ML, Sanderson MJ, Witman GB. 2008. Mutations in Hydin impair ciliary motility in mice. J. Cell Biol. 180, 633–643. ( 10.1083/jcb.200710162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tokuyasu KT. 1974. Dynamics of spermiogenesis in Drosophila melanogaster. 3. Relation between axoneme and mitochondrial derivatives. Exp. Cell Res. 84, 239–250. ( 10.1016/0014-4827(74)90402-9) [DOI] [PubMed] [Google Scholar]
- 49.Green LL, Wolf N, McDonald KL, Fuller MT. 1990. Two types of genetic interaction implicate the whirligig gene of Drosophila melanogaster in microtubule organization in the flagellar axoneme. Genetics 126, 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen MG, Turner FR, Hutchens JA, Raff EC. 2001. Axoneme-specific β-tubulin specialization: a conserved C-terminal motif specifies the central pair. Curr. Biol. 11, 529–533. ( 10.1016/S0960-9822(01)00150-6) [DOI] [PubMed] [Google Scholar]
- 51.Mottier-Pavie V, Megraw TL. 2009. Drosophila Bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol. Biol. Cell 20, 2605–2614. ( 10.1091/mbc.e08-11-1115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laurençon A, Dubruille R, Efimenko E, Grenier G, Bissett R, Cortier E, Rolland V, Swoboda P, Durand B. 2007. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 8, R195 ( 10.1186/gb-2007-8-9-r195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Lu X. 2011. Drosophila sperm motility in the reproductive tract. Biol. Reprod. 84, 1005–1015. ( 10.1095/biolreprod.110.088773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gadelha C, Wickstead B, McKean PG, Gull K. 2006. Basal body and flagellum mutants reveal a rotational constraint of the central pair microtubules in the axonemes of trypanosomes. J. Cell Sci. 119, 2405–2413. ( 10.1242/jcs.02969) [DOI] [PubMed] [Google Scholar]
- 55.Melkonian M, Robenek H, Rassat J. 1982. Flagellar membrane specializations and their relationship to mastigonemes and microtubules in Euglena gracilis. J. Cell Sci. 55, 115–135. [DOI] [PubMed] [Google Scholar]
- 56.Yabuki A, Gyaltshen Y, Heiss AA, Fujikura K, Kim E. 2018. Ophirina amphinema n. gen., n. sp., a new deeply branching discobid with phylogenetic affinity to jakobids. Scient. Rep. 8, 16219 ( 10.1038/s41598-018-34504-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfannenschmid F, Wimmer VC, Rios RM, Geimer S, Krockel U, Leiherer A, Haller K, Nemcova Y, Mages W. 2003. Chlamydomonas DIP13 and human NA14: a new class of proteins associated with microtubule structures is involved in cell division. J. Cell Sci. 116, 1449–1462. ( 10.1242/jcs.00337) [DOI] [PubMed] [Google Scholar]
- 58.Dutcher SK, Huang B, Luck DJ. 1984. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 98, 229–236. ( 10.1083/jcb.98.1.229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rupp G, O'Toole E, Porter ME. 2001. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell 12, 739–751. ( 10.1091/mbc.12.3.739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DiPetrillo CG, Smith EF. 2010. Pcdp1 is a central apparatus protein that binds Ca2+-calmodulin and regulates ciliary motility. J. Cell Biol. 189, 601–612. ( 10.1083/jcb.200912009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang K, Beck CF. 2003. Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 100, 6269–6274. ( 10.1073/pnas.0931459100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang K, Kunkel T, Beck CF. 2004. Localization of the blue-light receptor phototropin to the flagella of the green alga Chlamydomonas reinhardtii. Mol. Biol. Cell 15, 3605–3614. ( 10.1091/mbc.e04-01-0010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang K, Diener DR, Mitchell A, Pazour GJ, Witman GB, Rosenbaum JL. 2007. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J. Cell Biol. 179, 501–514. ( 10.1083/jcb.200704069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294. ( 10.1016/j.str.2009.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogt JH, Schippers JH. 2015. Setting the PAS, the role of circadian PAS domain proteins during environmental adaptation in plants. Front. Plant Sci. 6, 513 ( 10.3389/fpls.2015.00513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ringo DL. 1967. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J. Cell Biol. 33, 543–571. ( 10.1083/jcb.33.3.543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dentler WL, Rosenbaum JL. 1977. Flagellar elongation and shortening in Chlamydomonas. III. Structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J. Cell Biol. 74, 747–759. ( 10.1083/jcb.74.3.747) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data not included in figures are available in the electronic supplementary material.