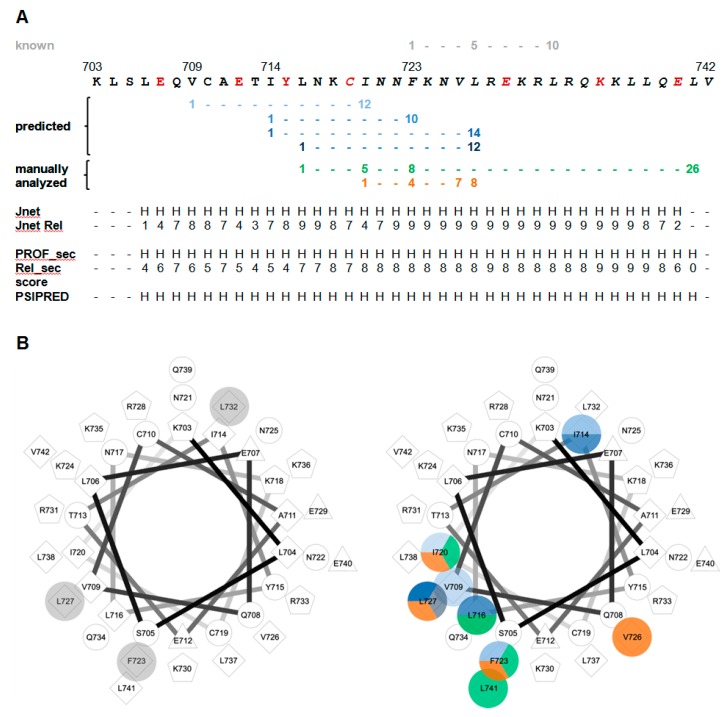
Figure 8.
Analysis of potential binding motifs in bMunc13-2 segment-C. Only CaM binding motifs starting N-terminally to position 1 of the proposed 1–5–10 binding motif were analyzed. The known 1–5–10 CaM binding motif is colored in gray, CaM binding motifs identified by database comparison are shown in different shades of blue, manually identified CaM binding motifs are colored green and orange. (A) Amino acid sequence, predicted CaM binding motifs and secondary structure prediction of segment-C. Amino acids identified in cross-linking studies are highlighted in red. Three different prediction servers were used: Jpred (www.compbio.dundee.ac.uk/jpred), PredictProtein (www.predictprotein.org), and PSIPRED (bioinf.cs.ucl.ac.uk/psipred). The algorithms Jnet and PROF_sec provide a score for α-helical propensity between 0 (lowest propensity) and 9 (highest propensity). H, predicted to be α-helical. (B) Helical wheel projection of segment-C. The known CaM binding motif is displayed on the left; novel CaM binding motifs are highlighted on the right.