Abstract
Risk-based treatment of onsite wastewaters for decentralized reuse requires information on the occurrence and density of pathogens in source waters, which differ from municipal wastewater due to scaling and dilution effects in addition to variable source contributions. In this first quantitative report of viral enteric pathogens in onsite-collected graywater and wastewater, untreated graywater (n = 50 samples) and combined wastewater (i.e., including blackwater; n = 28) from three decentralized collection systems were analyzed for two norovirus genogroups (GI/GII) and human adenoviruses using droplet digital polymerase chain reaction (ddPCR). Compared to traditional quantitative PCR (qPCR), which had insufficient sensitivity to quantify viruses in graywater, ddPCR allowed quantification of norovirus GII and adenovirus in 4% and 14% of graywater samples, respectively (none quantifiable for norovirus GI). Norovirus GII was routinely quantifiable in combined wastewater by either PCR method (96% of samples), with well-correlated results between the analyses (R2 = 0.96) indicating a density range of 5.2–7.9 log10 genome copies/L. These concentrations are greater than typically reported in centralized municipal wastewater, yet agree well with an epidemiology-based model previously used to develop pathogen log-reduction targets (LRTs) for decentralized non-potable water systems. Results emphasize the unique quality of onsite wastewaters, supporting the previous LRTs and further quantitative microbial risk assessment (QMRA) of decentralized water reuse.
Keywords: norovirus, graywater, wastewater, ddPCR, QMRA, water reuse
INTRODUCTION
Decentralized water reuse can provide economic and environmental benefits; however, expansion of its application has been hindered by uncertain treatment and management requirements (National Academies of Sciences, Engineering, and Medicine, 2016). A fundamental barrier to the development of treatment guidance for decentralized reuse systems has been lack of pathogen characterization data for onsite or locally-collected wastewaters, including graywater (water from bathroom sinks, bathtubs/showers, and clothes washing machines), blackwater (water from toilets and kitchen sinks), and combined wastewater (mixed graywater and blackwater). These water sources experience greater variability in pathogen densities than municipal wastewater due to both sporadic pathogen infections among small populations and lack of wastewater dilution by non-domestic sources such as stormwater and industrial discharges (O’Toole et al., 2014; Schoen and Garland, 2015). Intermittent occurrences present a practical challenge for the interpretation of non-detections during pathogen monitoring, particularly in the case of graywater where pathogen densities may approach measurement sensitivity limits. Indeed, previous efforts to quantify pathogens in graywater have been largely unsuccessful (Christova-Boal et al., 1996; Winward et al., 2008; Benami et al., 2015).
Epidemiology-based pathogen modeling has been proposed as an alternative method for generating decentralized wastewater characterizations (Fane et al., 2002; Ottoson and Stenström, 2003; Barker et al., 2013; Schoen et al., 2014; Jahne et al., 2017). In these models, reported illness incidence rates for enteric pathogens are used to simulate the occurrence of pathogen infections among a population of given size. For each such infection, the pathogen shedding duration and daily concentration in feces is then modeled based on reported shedding characteristics during clinical infections. Separately, levels of fecal contamination in combined wastewater or graywater are modeled based on their reported fecal indicator concentrations relative to raw feces. Results of the pathogen shedding and fecal contamination models are then combined to simulate pathogen occurrences and densities in source waters as a function of the population size contributing to a given wastewater or graywater collection. This method has been used to develop influent pathogen characterizations for a quantitative microbial risk assessment (QMRA) model of pathogen log-reduction targets (LRTs) for decentralized non-potable water systems (Jahne et al., 2017; Schoen et al., 2017; Sharvelle et al., 2017). However, given the lack of pathogen measurement data for onsite-collected wastewaters, the pathogen simulation results underlying these LRTs have not been validated in the context of actual pathogen observations.
To improve upon previous detection methods for low-level pathogen densities in wastewaters, this study presents droplet digital polymerase chain reaction (ddPCR) quantification of viral enteric pathogens (norovirus genogroups GI and GII and human adenoviruses) in untreated graywater and combined wastewater. Measurement results from three different building-scale decentralized collection systems are compared to the epidemiology-based pathogen simulation used to model LRTs for non-potable onsite wastewater reuse, and to traditional quantitative PCR (qPCR) analysis methods. To our knowledge, this is the first quantitative report of norovirus concentrations in graywater or in onsite-collected combined wastewater. Results will support future QMRAs evaluating the safety of decentralized water systems and the continued development of risk-based treatment guidance for decentralized water reuse.
MATERIALS AND METHODS
Sample Collection and Processing
Untreated water samples were collected from two office facilities and one residential facility in the United States (CA, CO, and OH). One office facility supporting approximately 900–1000 persons collects combined wastewater from all sources in the building (WW1) and one office facility supporting approximately 700–800 persons collects graywater primarily from bathroom sinks, with additional contributions from janitorial sinks, water fountains, and several showers in the building (GW1). The residential facility collects graywater from bathroom sinks and showers in a university residence hall supporting approximately 500 persons (GW2). Twice-weekly sampling targeted the winter season (December through April), i.e. the peak season for norovirus (Eftim et al., 2017). To assess potential seasonal effects, additional samples were collected from WW1 during the summer season (June and July). A total of 28, 33, and 17 samples were collected from WW1, GW1, and GW2, respectively.
During each sampling event, 5 L (for 25/28 combined wastewater samples) or 10 L (for the remaining combined wastewater and all graywater samples) of water was collected into a sterile polypropylene container and filtered through an Asahi Kasei Rexeed®−15S ultrafilter with 30 kDa molecular weight cutoff (Dial Medical Supply, Chester Springs, PA) upon receipt at the laboratory (<24 h at 4 °C). Ultrafiltration has been demonstrated to provide efficient recovery compared to alternative methods (Cashdollar and Wymer, 2013) and in this study achieved a mean recovery of 60% based on wastewater samples evaluated both with and without the concentration step. An inline vacuum gauge was used to ensure that the transmembrane pressure of the ultrafilter did not rise above the manufacture’s stated limit of 66 kPa. Ultrafilters were stored at 4 °C and eluted within 72 h using a filter-sterilized elution solution consisting of 0.01% sodium polyphosphate (Sigma-Aldrich, St. Louis, MO), 0.01% Tween-80 (Sigma-Aldrich), and 0.001% Y-30 antifoam (Sigma-Aldrich). Elution solution (200 mL) was circulated clockwise through each ultrafilter for 2 min, with a 1 min counter-clockwise cycle between the two clockwise cycles. The entire sample was then centrifuged at 1500×g for 15 min at 4 °C. A solvent extraction was implemented with GW1 samples, where a 15 mL aliquot of supernatant was added to an equal volume of Vertrel XF (The Chemours Company, Wilmington, DE) in a sterile 50 mL polypropylene conical tube and vortexed vigorously for 1 min. The sample was then centrifuged at 5000×g for 15 min at 4 °C. At least 10 mL of the aqueous layer was stored at −70 °C for nucleic acid extraction. The solvent extraction was omitted from processing samples collected at WW1 and GW2 as it did not meaningfully impact virus recovery (2–32% difference in process control trials); instead, the supernatant produced from these samples was used directly in nucleic acid extraction.
Nucleic acids were extracted from 10 mL of samples with the QIAamp DNA Blood Maxi Extraction Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, except Buffer AVL (Qiagen) was substituted for Buffer AL as the lysing agent and the protease digestion was omitted. Nucleic acids were eluted from spin columns twice with the same 1 mL volume of Buffer AE. Negative extraction controls using 10 mL of nuclease-free water were performed with each extraction set. Extracted nucleic acids were stored at −70 °C.
qPCR/Reverse Transcription (RT)-qPCR
Human adenoviruses were quantified in samples using qPCR and the TaqMan assay described previously (Jothikumar et al., 2005). Reactions consisted of GeneAmp 10X PCR Buffer II (diluted to 1X; ThermoFisher Scientific, Waltham, MA), 5 mM MgCl2 (ThermoFisher Scientific), nuclease-free water (Promega Corporation, Madison, WI), 0.4 mM dNTPs (Promega Corporation), 50X ROX reference dye (diluted to 1X; ThermoFisher Scientific), 1.25 U AmpliTaq Gold DNA Polymerase (ThermoFisher Scientific), 400 nM primers, 150 nM probe, and 5 μL nucleic acid extracted sample in 25 μl reactions. PCR occurred on a StepOnePlus Real-Time PCR System (ThermoFisher Scientific) by heating to 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min.
Noroviruses were quantified in samples using a two-step RT-qPCR and the norovirus GIB and GII primer-probe assays described previously (Brinkman et al., 2013; Shay et al., 2014). Reverse transcription reactions were prepared in 15 μL, consisting of GeneAmp 10X PCR Buffer II (diluted to 1X; ThermoFisher Scientific), 1.5 mM MgCl2 (ThermoFisher Scientific), 0.66 mM dNTPs (Promega Corporation), nuclease-free water, 0.833 nM reverse primer, 25 U MuLV Reverse Transcriptase (ThermoFisher Scientific), 15 U RNasin Plus Ribonuclease Inhibitor (Promega Corporation, Madison, WI), and 5 μL nucleic acid extracted sample. Reverse transcription was conducted at 43 °C for 60 min followed by 94 °C for 5 min. The entire volume of the RT reaction was used in qPCR, with the addition of components at final concentrations of 1X PCR Buffer II, 5 mM MgCl2, 1X ROX reference dye, 500 nM primers, 100 nM probe and 1.25 U AmpliTaq Gold DNA Polymerase in a 25 μL reaction. Cycling was run on the StepOne® Plus Real-Time PCR System using the PCR conditions stated above.
A master standard curve was generated for each virus target by analyzing dilutions of wild-type human adenovirus, serotype 5 (Ad5; O.D. 260, Inc., Boise, ID) or a custom Armored RNA (EPA-1615; Asuragen, Austin, TX; Shay et al., 2014) after nucleic acid extraction using the QIAamp DNA Blood Mini Extraction Kit (Qiagen) according to the manufacturer’s instructions, except for the substitution of Buffer AVL (Qiagen) for Buffer AL and omission of the protease step. The concentration of each dilution used to construct the standard curve was determined using ddPCR (described below) and quantification cycle (Cq) values of triplicate reactions were pooled after three independent qPCR or RT-qPCR assessments. The dynamic range of the assays was 0.95–4.79 log10 genome copies/reaction for norovirus GI and GII and 0.63–4.73 log10 genome copies/reaction for adenovirus.
Every qPCR and RT-qPCR plate contained positive controls (one of the dilutions used to make the standard curve) and negative controls (10 mM Tris-HCl, pH 8.5; Qiagen). Wastewater and graywater samples were diluted in a 5-fold series and each dilution was analyzed with 5 replicate reactions. Concentration agreement among multiple dilutions of positive samples (<30% coefficient of variation) indicated lack of PCR inhibition by the sample matrix.
ddPCR/RT-ddPCR
Adenoviruses were quantified in samples using a duplexed ddPCR reaction with the adenovirus assay used in qPCR (labeled with FAM) and the assay for an internal amplification control (IAC; labeled with VIC). The ddPCR IAC consisted of a linearized custom gene (Integrated DNA Technologies, Coralville, IA) containing the amplicon of the Hepatitis G primer-probe assay described previously (Shay et al., 2014). The 25 μL reactions contained ddPCR Supermix for Probes (no dUTP; Bio-Rad Laboratories, Inc., Hercules, CA), 900 nM primers, 250 nM probes, nuclease-free water, approximately 2.8 × 104 copies of the IAC, and 5 μl of exacted sample. Droplets were made using the QX200 AutoDG (Bio-Rad Laboratories, Inc.) according to the manufacturer instructions. PCR was performed in a C1000 Touch Thermal Cycler with 96-Deep Well Reaction Module (ThermoFisher Scientific) by heating to 95 °C for 5 min, followed by 50 cycles of 95 °C for 30 s and 55 °C for 1 min, then a final incubation at 98 °C for 10 min. Amplification in each droplet was assessed with the QX200 Droplet Reader (Bio-Rad Laboratories, Inc.).
Noroviruses were quantified in samples using a duplexed RT-ddPCR reaction with either of the primer-probe assays used in qPCR (labeled with FAM) and the assay for a second IAC (labeled with VIC). The RT-ddPCR IAC consisted of Luciferase Control RNA (Promega Corporation) and was detected with the primer-probe assay described previously (Johnson et al., 2005). The one-step RT-ddPCR reactions contained One-Step RT-ddPCR Advanced Kit for Probes (BioRad Laboratories), 20 U Reverse transcriptase (BioRad Laboratories), 15 mM DTT (BioRad Laboratories), 900 nM primers, 250 nM probes, nuclease-free water, approximately 2.8 × 104 copies of the RT-ddPCR IAC, and 5 μl extracted sample. The QX200 AutoDG was used to generate droplets and RT-ddPCR was performed in the C1000 Touch Thermal Cycler with 96-Deep Well Reaction Module by incubating at 50 °C for 60 min, then 95 °C for 10 min, followed by 50 cycles of 95 °C for 30 s and 55 °C for 1 min, then final incubation at 98 °C for 10 min. Amplification was determined for each droplet with the QX200 Droplet Reader (Bio-Rad Laboratories).
Quality control samples were run on every plate. Positive PCR controls consisted of Ad5 or the custom Armored RNA and negative controls consisted of 10 mM Tris-HCl, pH 8.5; all quality controls were run in triplicate. Samples with <10,000 droplets generated were excluded from analysis and re-run. PCR inhibition was monitored using the respective IACs, with <65% recovery vs. control wells indicating severe inhibition and rejection of the sample. Droplet fluorescence amplitude data (.csv) were exported from the manufacturer software as described by Trypsteen et al. (2015) for analysis in R (Version 3.2.3; R Core Team, 2015).
Data Analysis
For each qPCR plate, the Cq threshold was determined using Equation 1, where ΔRn is the average ROX-normalized change in fluorescence data for cycles 3–15 of amplification and SD is the standard deviation:
| [1] |
Template concentrations (genome copies/reaction) were determined by interpolation of Cq values to the fitted standard curves (Supplementary Material Fig. S1). The lower limit of quantification (LOQ) was defined as the lowest concentration of standard for which ≥ 95% of replicates were positive (i.e., the limit of detection following Bustin et al., 2009), adjusted by factors used in determining sample concentrations (described below). For each sample dilution, typically ranging from undiluted to 1:25 for graywater or 1:625 for combined wastewater, concentrations in each replicate reaction (n = 5) were averaged, adjusted for dilution of nucleic acids, and multiplied by a factor of 4000 to account for the splitting of sample volumes during processing steps (½0th of the eluted sample was used in nucleic acid extraction and ½00th of the extract was analyzed in each PCR reaction). Final sample concentrations (genome copies/L wastewater or graywater) were determined by averaging adjusted concentrations from each dilution for which ≥ 3 of 5 qPCR reactions were within the range of quantification and dividing by volume of the original water sample. For both qPCR and ddPCR assays, LOQs are reported assuming 10 L samples; 5 L sample LOQs are 0.3 log10 genome copies/L higher.
Exported ddPCR amplitude data were analyzed using the ddpcRquant R package, which models the fluorescence threshold for droplet classification based on extreme value theory and the distribution of fluorescence observed in no-template control (NTC) droplets; see Trypsteen et al. (2015) for theoretical and technical details. During data analysis, negative extraction controls (n = 21) were treated as NTCs to include potential impacts of sample processing and cross-talk among channels of the multiplexed reaction on background fluorescence of negative droplets (Jacobs et al., 2017). To further minimize possible false-positive droplets, plates for each PCR target were analyzed simultaneously using all defined NTC wells (n = 63). Threshold and block size parameters were initially set to default values of 0.9995 and 150, respectively; where visual inspection of NTC threshold plots suggested suboptimal thresholding (e.g., thresholds drawn within the main cluster of NTC droplets), threshold percentile was increased iteratively by factors of 10 until only outlier droplets remained positive by visual inspection. In the event that threshold percentile adjustment did not yield satisfactory thresholding at the maximum recommended setting (0.9999995), the block size was also adjusted to its minimum value (100); if thresholding remained unsatisfactory, a manual threshold was applied. Selected parameter settings and associated threshold plots are provided in Supplementary Material Fig. S2. Following extreme-value thresholding based 10,000 algorithm iterations, classified droplet counts for each well were pooled across technical ddPCR replicates (n = 3) of each sample.
ddPCR sample densities were estimated using a Bayesian model implemented in JAGS 4.2.0 (Plummer, 2003) through the R package R2jags (Su and Yajima, 2015). This method allowed for estimation of both sample concentrations and associated credible intervals (CrI). For each sample, concentration λ (mean genome copies/droplet) was modeled to follow a Poisson distribution (Hindson et al., 2011):
| [2] |
where p is the probability of success (a positive reaction) in a binomial distribution with k positive and n total droplets:
| [3] |
A noninformative Jefferys prior for p was assumed (Jeffreys, 1946):
| [4] |
with initial values drawn from a unit uniform distribution. Markov Chain Monte Carlo (MCMC) was performed using 1,000,000 iterations, thinned by a factor of 10, of 3 independent chains following a burn-in of 1,000,000 iterations (each). Convergence was checked based on visual inspection of trace plots for stability and Gelman-Rubin statistics ≤1.001 (Gelman and Rubin, 1992). Sample concentrations were estimated using the median posterior value of λ, and 95% CrI were based on its 2.5th and 97.5th posterior percentiles. To avoid potential false-positive sample determinations given the rare occurrence of positive droplets in NTC wells (Supplementary Material Fig. S2 and Table S1), positive samples were defined as those for which 95% CrI did not overlap with 95% CrI of the merged NTCs. Final concentrations of positive samples (genome copies/L wastewater or graywater) were determined based on the volume of each droplet (0.85 nL; Corbisier et al., 2015); the sample-processing factor of 4000; and the volume of the original water sample, as described for qPCR above. The lower LOQ was defined as the 97.5th percentile NTC concentration following adjustment by these factors. All simulations were performed twice to ensure model run consistency within an acceptable tolerance of 1% difference.
Epidemiology-based modeling
Measurement results (detection rates, concentration ranges, and concentration medians) were compared to previous results of the epidemiology-based pathogen simulation reported by (Jahne et al., 2017); no new simulations were performed. In the simulation, fecal loading to combined wastewater and graywater from household sources (wet g feces/L) was modeled based on the relative concentration of Escherichia coli reported in fresh samples of these waters (i.e., not stored) compared to that reported in human feces. Variable E. coli concentration inputs were modeled using a meta-analysis of available peer-reviewed literature. Separately, occurrences of pathogen shedding among cohort groups representative of potential decentralized system sizes (5-, 100-, or 1000-person) were simulated as a modified compound binomial process based on reported distributions of population incident rates (illnesses/person/year) and infection durations (days/infection). This model predicted the number of pathogen shedders on each day of a 10,000-year simulation. Pathogen concentrations in wastewater and graywater were then modeled by coupling the daily numbers of pathogen shedders with random draws from fecal contamination models and reported distributions of pathogen shedding densities during an infection (#/wet g feces). Refer to (Jahne et al., 2017) for a complete discussion of the model and its inputs.
RESULTS AND DISCSSION
Virus quantification
To our knowledge, this study provides the first quantitative report of viral enteric pathogens in graywater and locally-collected combined wastewater, which differs from municipal wastewater due to scaling and dilution effects (see O’Toole et al. (2014) and (Jahne et al., 2017) for a complete discussion). Pathogens were quantifiable by ddPCR in each of the three decentralized wastewater and graywater collection systems studied (Fig. 1). Norovirus GII was the most frequently quantifiable virus in combined wastewater (27/28 samples, LOQ = 2.0 log10 genome copies/L; 3/50 quantifiable in graywater), whereas adenovirus was the most frequently quantifiable in graywater (7/50 samples, LOQ = 1.8 log10 genome copies/L; 4/28 quantifiable in combined wastewater). Norovirus GI was quantifiable in 1½8 combined wastewater samples (LOQ = 2.0 log10 genome copies/L) but was not quantifiable in graywater (0/50 samples). Among quantifiable samples, combined wastewater concentrations of adenovirus, norovirus GI, and norovirus GII were 2.2–3.2, 2.1–4.0, and 5.2–7.9 log10 genome copies/L, respectively; graywater concentrations of adenovirus and norovirus GII were 2.0–3.8 and 2.1–2.5 log10 genome copies/L, respectively. Complete PCR results are provided in Supplementary Material Table S2.
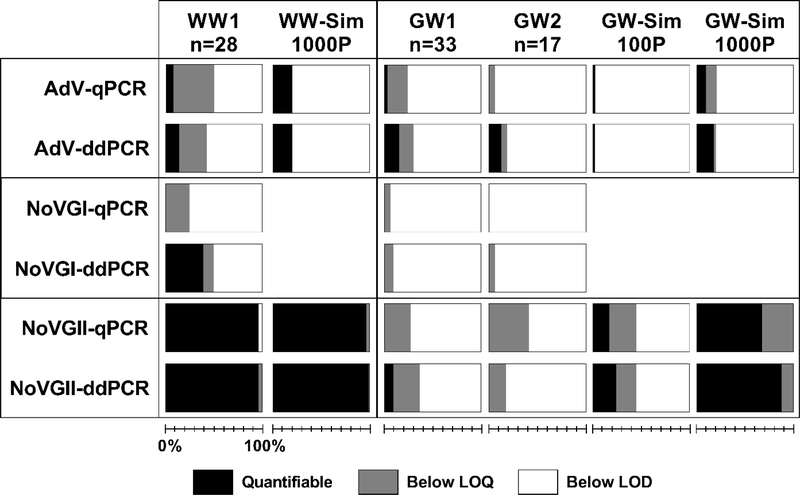
Figure 1.
Percentage of samples quantifiable for adenovirus (AdV), norovirus genogroup I (NoVGI), and NoVGII in three decentralized combined wastewater (WW1) and graywater (GW1 and GW2) collection systems by quantitative polymerase chain reaction (qPCR) and droplet digital PCR (ddPCR). Also shown are putative detections below the lower limit of quantification (LOQ) and non-detections below the limit of detection (LOD). WW-Sim and GW-Sim columns present expected results based on previously published epidemiology-based simulations of these pathogens in comparably sized collection systems,11 i.e. 1000-person for combined wastewater (WW-Sim 1000P) and 100- or 1000-person for graywater (GW-Sim 100P and GW-Sim 1000P), where bars represent the percentage of daily simulations (n=365×10,000) that would fall within each category given respective LOQs for each PCR assay. Since the simulation did not distinguish between norovirus genogroups, expected results for NoVGII represent NoVGI and NoVGII combined.
As previously reported (Hindson et al., 2011; Cavé et al., 2016), ddPCR methods were more sensitive than qPCR; qPCR LOQs (3.2 log10 genome copies/L for both norovirus genogroups and 3.4 log10 genome copies/L for adenovirus) were 1.2–1.6 log10 genome copies/L higher than LOQs for the ddPCR method. Of note, ddPCR data analysis methods applied here were based on the observed distribution of fluorescence in NTC droplets (Trypsteen et al., 2015), rather than automated or manual thresholds (e.g., using Bio-Rad Laboratories on-board QuantaSoft software) or an inaccurate normal assumption (e.g., Strain et al., 2013; Dreo et al., 2014; Jones et al., 2014). While it is acknowledged that the iterative procedure used to determine appropriate settings for extreme-value threshold models introduced subjectivity to the analysis, the method allows reproducibility of results using reported settings (Supplementary Material Fig. S2) and is based on appropriate mathematical assumptions. Importantly, it also allowed for the objective handling of rain (i.e., droplets that fall between negative and positive populations), since thresholding was based only on NTC wells and did not require analyst judgement about the appropriate level of rain to include (or exclude rain entirely, e.g., Strain et al., 2013; Jones et al., 2014). See Trypsteen et al. (2015) for a complete discussion of the extreme value thresholding approach. Furthermore, use of the Bayesian model for determining sample concentrations facilitated estimation of CrI about concentration estimates; in turn, this enabled determination of LOQs based on NTC CrI. The method therefore statistically differentiated likely true positive samples (95% credibility) from sporadic false-positive droplets, which have been observed by others to limit sensitivity (Strain et al., 2013; Kiselinova et al., 2014). This is a critical consideration given low anticipated pathogen concentrations highly sensitivity to method LOQs.
Since concentrations quantifiable by ddPCR often fell close to ddPCR LOQs and below the qPCR LOQs, the numbers of samples with quantifiable concentrations of each target were greater for ddPCR than qPCR (Fig. 1). Indeed, with the exception of one sample quantifiable for adenovirus, the viruses were unquantifiable by qPCR in all 50 samples of graywater. In combined wastewater, norovirus GII densities were high enough for quantification by either method (Fig. 2), with well correlated results (R2 = 0.96) supporting accuracy of the newer ddPCR approach in wastewater matrices. This agreement also indicates that PCR inhibition did not meaningfully affect performance of either method, since the volumes of extract analyzed per reaction were vastly different. For the two samples, one combined wastewater and one graywater, where adenovirus was quantifiable by both methods, concentrations determined by qPCR were 1.6 and 0.3 log10 genome copies/L greater than concentrations determined by ddPCR using the same DNA extracts and primer/probe set. In the case of the more extreme difference, both qPCR and ddPCR measurements were 0.6 log10 genome copies/L above their respective LOQs, highlighting remaining challenges to sample quantification near method sensitivity limits.
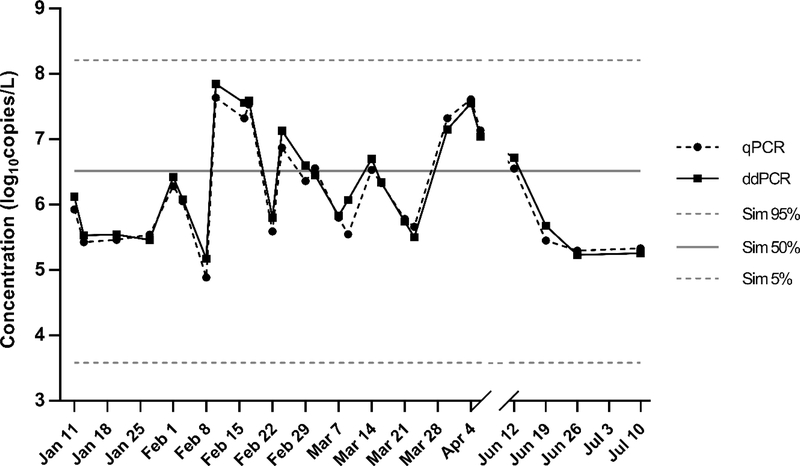
Figure 2.
Norovirus genogroup II concentrations in decentralized combined wastewater collections as determined by quantitative polymerase chain reaction (qPCR) and droplet digital PCR (ddPCR). Results are compared to a previously published epidemiology-based simulation of norovirus genogroups in a comparably sized 1000-person wastewater collection;11 Sim 50% indicates the median of daily simulations (n=365×10,000) and Sim 5% and 95% are respective percentiles.
Additionally, both qPCR and ddPCR analysis methods resulted in putative detections below their respective LOQs (Fig. 1). For ddPCR, these represented samples with 95% CrI overlapping with NTC CrI, typically equating to a single positive droplet. For qPCR, these represented samples with Cq values < 40 yet beyond the standard curve range of quantification, typically equating to <1 copy per reaction. In either case, putative detections could not be confirmed as positive samples following the relatively strict analysis criteria described in Methods, e.g. qPCR LOQs based on 95% positive standards; combined analysis of ddPCR plates to include maximum NTC variability; and comparison of respective 95% CrI for ddPCR samples and NTCs when making positive sample determinations. These criteria were specifically designed to minimize false-positive occurrences in the dataset given the public health implications of reported pathogen detection. However, noting that an unknown subset of the putative samples is indeed likely positive, it is worthwhile to consider potential gains that could be made with improved measurement sensitivity. Although current methods could not characterize these samples, they should not be discounted as entirely negative; this is supported by positive detections near the respective LOQs.
Reuse implications
Previous QMRA of decentralized water reuse has been limited by unavailability of direct pathogen monitoring data, in part due to insufficient method sensitivity for low-level pathogen detection in onsite-collected waters such as graywater and rainwater (roof runoff) (Schoen et al., 2017). This study provides new quantitative enteric virus data to support future such efforts, with complete measurement results provided in Supplementary Material Table S2 for subsequent analysis and use. Data generated from the two graywater sites may be combined to reduce site-specificity and pool limited detection results, although it is noted that appropriate statistical methods for left-censored data must be applied. Densities of norovirus GII in the combined wastewater can be utilized directly yet remain site-specific. As discussed below, general agreement of measurement results with the epidemiology-based model presented by (Jahne et al., 2017) also supports further use of these simulated values in QMRA, particularly given the continued method sensitivity limitations observed here.
Since quantitative measurements of enteric viruses in decentralized wastewater and graywater were previously unavailable, results are compared to the epidemiology-based simulations previously used to develop pathogen LRTs for these source waters (Jahne et al., 2017; Schoen et al., 2017; Sharvelle et al., 2017). In order to generate broadly-applicable treatment targets, the previous model was based on a meta-analysis of fecal contamination data (i.e., fecal indicator bacteria) in onsite-collected waters from the United States, Australia, Europe, and Israel and national or state-level illness incidence rates reported by the Centers for Disease Control and Prevention. Therefore, while population and source characteristics may differer from site-to-site, simulation results are intended to capture this inherent variability and be generally representative of onsite systems such as those monitored in the current study. It should be noted, however, that use of reported illness rates in the model neglects asymptomatic infections that may contribute to measured pathogen loads (Jahne et al., 2017).
In combined wastewater, the ddPCR quantification rate of norovirus GII (96%) agreed well with the comparable 1000-person simulation (99% for all genogroups combined) (Fig. 1). Densities of detected norovirus GII ranged between 5.2 and 7.9 log10 genome copies/L, within the 90% range of simulation results (3.6–8.2 log10 genome copies/L), and the measurement median (6.1 log10 genome copies/L) reflected the simulation median (6.5 log10 genome copies/L) within 0.4 orders of magnitude (Fig. 2). When norovirus GI was quantifiable in combined wastewater (39% of samples), its concentration was 2.2–4.7 log10 genome copies/L lower than the concentration of norovirus GII and therefore did not contribute meaningfully to overall norovirus concentrations as presented by the model. While it should be noted that the infectivity of noroviruses detected by PCR methods is unknown, it has nonetheless been suggested that the pathogen be specifically considered in the evaluation of water reuse projects (Nappier et al., 2018), as was done for development of the decentralized non-potable system LRTs (Sharvelle et al., 2017). See Nappier et al. (2018) for a complete discussion of norovirus use in risk assessment.
For adenovirus, the ddPCR quantification rate (14%) was slightly lower than previously predicted by the epidemiology-based model (20%) (Fig. 1), but within the 90% range of modeled occurrence rates during individual years of the simulation (11–30%) (Jahne et al., 2017). However, combined wastewater concentrations of adenovirus by ddPCR (2.2–3.2 log10 genome copies/L; median 2.4 log10 genome copies/L) were considerably lower than predicted by the model (90% range 4.0–8.0 log10 genome copies/L; median 6.0 log10 genome copies/L) and comparable to those found in graywater. As noted above, adenovirus concentrations near method LOQs (1.8 log10 genome copies/L for ddPCR) differed among ddPCR and qPCR assays, indicating quantification uncertainty in these low-level detections. Adenovirus was included in this study due to its high fecal shedding during an infection, persistence in aquatic environments, and common detection in municipal wastewater (Allard and Vantarakis, 2017), which were anticipated to result in high frequency of detection and quantification; rather, norovirus appears to be a more consistent viral pathogen in decentralized wastewater at this site. This is particularly relevant because proposed virus LRTs for decentralized non-potable water systems were based largely on norovirus (Sharvelle et al., 2017), for which measurements agreed well with the simulation used to develop them (Fig. 2).
Two recent studies have performed meta-analyses of norovirus concentrations in untreated municipal wastewater, which differs from locally-collected wastewater due to population scaling and wastewater dilution effects (O’Toole et al., 2014; Jahne et al., 2017). Pouillot et al. (2015) estimated mean norovirus GII and GI concentrations of 3.9 and 1.5 log10 genome copies/L, respectively, in the United States and Canada; Eftim et al. (2017) estimated mean 4.7 and 3.3 log10 genome copies/L, respectively, in the same region. As in this work, norovirus GII was greater than GI in both studies. Although these studies differed in inclusion criteria and analysis methods (including handling of non-detects), both sets of mean norovirus GII results were considerably lower than measured in the decentralized system of this study (mean 6.3 log10 genome copies/L). The current measurements therefore support the distinction of pathogen load between centralized and decentralized wastewater collections, a central premise of the epidemiology-based concentration model and associated decentralized system LRTs (Jahne et al., 2017; Schoen et al., 2017; Sharvelle et al., 2017). Nonetheless, both previous studies also report considerable variation within their data and model results, and site-specific differences, as well as spatiotemporal variability in norovirus outbreak dynamics, cannot be discounted (Eftim et al., 2017). It should also be noted that different processing and analytical methods were used among individual studies underlying the meta-analyses, potentially impacting comparability of results. Contrary to the previous reports, norovirus GII showed limited seasonality in wastewater samples; summer sample concentrations were within the range observed during the peak winter season (Fig. 2). However, the sample size in this study was much smaller than the sample sizes of the meta-analyses. Seasonality was not considered in the simulation model.
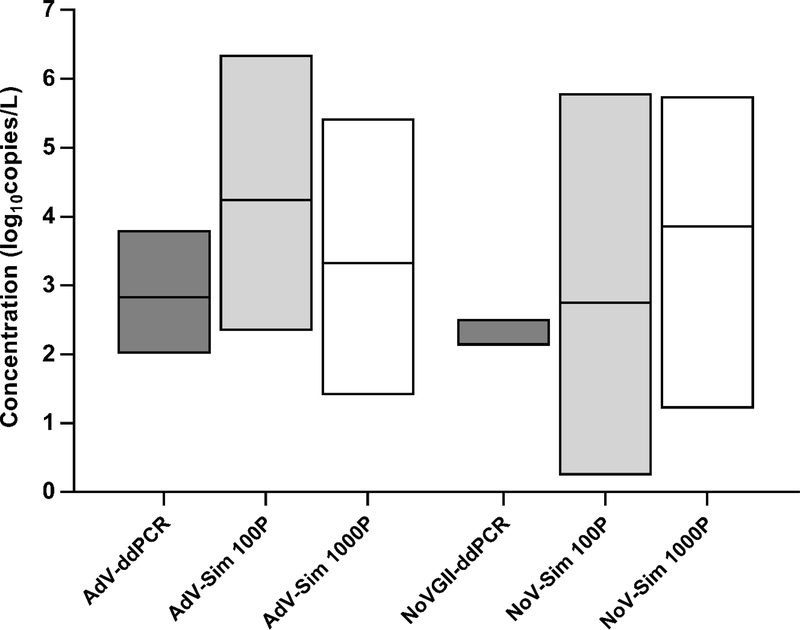
In graywater, ddPCR quantification rates of adenovirus in both systems (12% and 15%) were within the 2–18% range predicted by 100- and 1000-person simulations that bracket their population sizes (Fig. 1). Norovirus GII was quantifiable by ddPCR in fewer graywater samples (9% and 0% for the two systems) than predicted by the simulation (24–88%). However, an additional 27% and 17% of samples from either site had putative detections below ddPCR LOQs (Fig. 1), and the 5th percentile annual norovirus occurrence rate for 100-person collections was half of its median value (22% vs. 45%) (Jahne et al., 2017), indicating that individual years could experience considerable variability in norovirus occurrence among small population sizes. Fecal contamination of graywater is inherently variable with specific characteristics of the water source and its use, including source type and user behavior (Nolde, 2000; Jefferson et al., 2004). Graywater collected from an office building and university dormitory in this study may therefore not completely align with the sources and use patterns of domestic residential systems modeled by (Jahne et al., 2017). The quantification rate of norovirus at the office building site (GW1; 9%), for which bathroom sinks were the primary graywater source, was comparable to that of O’Toole et al. (2012), who detected norovirus GI in 8% of bathroom sink graywater samples using qualitative PCR. Although limited graywater samples were quantifiable for either adenovirus (n = 7/50) or norovirus (n = 3/50), observed concentrations (2.0–3.8 and 2.1–2.5 log10 genome copies/L, respectively) were within the ranges predicted by the 100- and 1000-person simulations (Fig. 3). Further study of additional graywater collection systems with varying source characteristics is warranted.
Figure 3.
Concentrations of adenovirus (AdV) and norovirus genogroup II (NoVGII) in graywater as measured by droplet digital polymerase chain reaction (ddPCR) and predicted by a previously published epidemiology-based pathogen simulation11 for 100- and 1000-person population sizes (Sim 100P and Sim 1000P, respectively) that bracket observed systems. For ddPCR, AdV n=7 and NoVGII n=3 quantifiable detections. The simulation results include positive pathogen occurrences from n=365×10,000 daily simulations and do not distinguish between norovirus genogroups.
Conclusions
This work demonstrates that previously-reported enteric virus reduction targets for decentralized wastewater collections are appropriate in the context of direct pathogen observations; in general, previously-unavailable measurements agree well with the modeled concentrations underpinning LRT estimates. Moreover, the study provides much-needed empirical data to inform future QMRA efforts. While ddPCR offers improved sensitivity over qPCR, additional methods development remains necessary for routine quantification of enteric viruses, particularly in graywater. Future monitoring efforts should also consider parasitic protozoa (Cryptosporidium spp. and Giardia lamblia) and enteric bacteria (Campylobacter jejuni) to validate proposed LRTs for the respective pathogen classes, as well as other onsite-available alternative water sources including roof runoff and stormwater.
Supplementary Material
Acknowledgements
The authors thank the collaborating facilities and the U.S. EPA Region 9 Laboratory for their assistance with sample collection and analysis, and appreciate the laboratory support provided by Joseph Stevens, Anne Turner, and Varun Rao. The authors also thank Dr. Sharon Nappier (U.S. EPA Office of Water) and Dr. Sandhya Parshionikar (U.S. EPA Office of Research and Development) for their thoughtful reviews of the draft manuscript.
Funding sources
Funding for this research was provided by the U.S. Environmental Protection Agency.
Footnotes
Disclaimers
The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Mention of trade names, products, or services does not convey, and should not be interpreted as conveying, official EPA approval, endorsement, or recommendation. The authors declare no competing financial interest.
Data availability
All original data referenced in this article is available in its Supplementary Material and via the U.S. EPA ScienceHub repository (https://catalog.data.gov/dataset/epa-sciencehub).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
References
- Allard A, Vantarakis A, 2017. Adenoviruses In: Rose JB, Jim_enez-Cisneros B. (Eds.), Global Water Pathogens Project. Michigan State University, E. Lansing, MI, UNESCO, Paris, France. Part 3 Viruses, [Google Scholar]
- Meschke JS; Girones R Barker SF, O’Toole J, Sinclair MI, Leder K, Malawaraarachchi M, Hamilton AJ, 2013. A probabilistic model of norovirus disease burden associated with greywater irrigation of home-produced lettuce in Melbourne, Australia. Water Res. 47 (3), 1421e1432. [DOI] [PubMed] [Google Scholar]
- Benami M, Gillor O, Gross A, 2015. The question of pathogen quantification in disinfected graywater. Sci. Total Environ 506, 496e504. [DOI] [PubMed] [Google Scholar]
- Brinkman NE, Haffler TD, Cashdollar JL, Rhodes ER, 2013. Evaluation of methods using celite to concentrate norovirus, adenovirus and enterovirus from wastewater. J. Virol. Methods 193 (1), 140e146. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 55 (4), 611e622. [DOI] [PubMed] [Google Scholar]
- Cashdollar JL, Wymer L, 2013. Methods for primary concentration of viruses from water samples: a review and meta-analysis of recent studies. J. Appl. Microbiol 115 (1), 1e11. [DOI] [PubMed] [Google Scholar]
- Cav_e L, Brothier E, Abrouk D, Bouda PS, Hien E, Nazaret S, 2016. Efficiency and sensitivity of the digital droplet PCR for the quantification of antibiotic resistance genes in soils and organic residues. Appl. Microbiol. Biotechnol 100 (24), 10597e10608. [DOI] [PubMed] [Google Scholar]
- Christova-Boal D, Eden RE, McFarlane S, 1996. An investigation into greywater reuse for urban residential properties. Desalination 106 (1e3), 391e397. [Google Scholar]
- Corbisier P, Pinheiro L, Mazoua S, Kortekaas A-M, Chung PYJ, Gerganova T, Roebben G, Emons H, Emslie K, 2015. DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Anal. Bioanal. Chem 407 (7), 1831e1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreo T, Pirc M, Ram_sak Z, Pav_si_c J, Milavec M, _Zel J, Gruden K, 2014. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: a case study of fire blight and potato brown rot. Anal. Bioanal. Chem 406 (26), 6513e6528. [DOI] [PubMed] [Google Scholar]
- Eftim SE, Hong T, Soller J, Boehm A, Warren I, Ichida A, Nappier SP, 2017. Occurrence of norovirus in raw sewageeA systematic literature review and meta-analysis. Water Res. 111, 366e374. [DOI] [PubMed] [Google Scholar]
- Fane S, Ashbolt N, White S, 2002. Decentralised urban water reuse: the implications of system scale for cost and pathogen risk.Water Sci. Technol 46 (6e7), 281e288. [PubMed] [Google Scholar]
- Gelman A, Rubin DB, 1992. Inference from iterative simulation using multiple sequences. Stat. Sci 457e472, 1992. [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, 2011. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem 83 (22), 8604e8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs BK, Goetghebeur E, Vandesompele J, De Ganck A, Nijs N, Beckers A, Papazova N, Roosens NH, Clement L, 2017. Model-based classification for digital PCR: your umbrella for rain. Anal. Chem 89 (8), 4461e4467. [DOI] [PubMed] [Google Scholar]
- Jahne MA, Schoen ME, Garland JL, Ashbolt NJ, 2017. Simulation of enteric pathogen concentrations in locally-collected greywater and wastewater for microbial risk assessments. Microb. Risk Anal 5, 44e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson B, Palmer A, Jeffrey P, Stuetz R, Judd S, 2004. Grey water characterization and its impact on the selectilon and operation of technologies for urban reuse. Water Sci. Technol 50 (2), 157e164. [PubMed] [Google Scholar]
- Jeffreys H, 1946. An invariant form for the prior probability in estimation problems. In: Proceedings of the Royal Society of London Series A, Mathematical and Physical Sciences; 1946, pp. 453e461. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Lee PK, Holmes VF, Alvarez-Cohen L, 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol 71 (7), 3866e3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Williams J, G€artner K, Phillips R, Hurst J, Frater J, 2014. Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline,’definetherain’. J. Virol. Methods 202, 46e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothikumar N, Cromeans TL, Hill VR, Lu X, Sobsey MD, Erdman DD, 2005. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol 71 (6), 3131e3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselinova M, Pasternak AO, De Spiegelaere W, Vogelaers D, Berkhout B, Vandekerckhove L, 2014. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One 9 (1), e85999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nappier SP, Soller JA, Eftim S, 2018. Potable water reuse: what are the microbiological risks? Curr. Environ. Health Rep 5 (2), 283e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine, 2016. Using Graywater and Stormwater to Enhance Local Water Supplies: an Assessment of Risks, Costs, and Benefits. National Academies Press, Washington DC. [Google Scholar]
- Nolde E, 2000. Greywater reuse systems for toilet flushing in multi-storey buildingseover ten years experience in Berlin. Urban Water 1 (4), 275e284. [Google Scholar]
- O’Toole J, Sinclair M, Malawaraarachchi M, Hamilton A, Barker SF, Leder K, 2012. Microbial quality assessment of household greywater. Water Res. 46 (13), 4301e4313. [DOI] [PubMed] [Google Scholar]
- O’Toole J, Sinclair M, Barker SF, Leder K, 2014. Advice to risk assessors modeling viral health risk associated with household graywater. Risk Anal. 34 (5), 797e802. [DOI] [PubMed] [Google Scholar]
- Ottoson J, Stenstr€om TA, 2003. Faecal contamination of greywater and associated microbial risks. Water Res. 37 (3), 645e655. [DOI] [PubMed] [Google Scholar]
- Plummer M, 2003. JAGS: a program for analysis of bayesian graphical models using gibbs sampling. In: Hornik K, Leisch F, Zeileis A. (Eds.), Proceedings of the 3rd International Workshop on Distributed Statistical Computing (DSC 2003) March 20e22, 2003, Vienna, Austria. [Google Scholar]
- Pouillot R, Van Doren JM, Woods J, Plante D, Smith M, Goblick G, Roberts C, Locas A, Hajen W, Stobo J, 2015. Meta-analysis of the reduction of norovirus and male-specific coliphage concentrations in wastewater treatment plants. Appl. Environ. Microbiol 81 (14), 4669e4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2015. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Schoen ME, Xue XB, Hawkins TR, Ashbolt NJ, 2014. Comparative human health risk analysis of coastal community water and waste service options. Environ. Sci. Technol 48 (16), 9728e9736. [DOI] [PubMed] [Google Scholar]
- Schoen ME, Garland J, 2015. Review of pathogen treatment reductions for onsite non-potable reuse of alternative source waters. Microb. Risk Anal 5, 25e31. [Google Scholar]
- Schoen ME, Ashbolt NJ, Jahne MA, Garland J, 2017. Risk-based enteric pathogen reduction targets for non-potable and direct potable use of roof runoff, stormwater, and greywater. Microb. Risk Anal 5, 32e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharvelle S, Ashbolt N, Clerico E, Hultquist R, Leverenz H, Olivieri A, 2017. Risk-Based Framework for the Development of Public Health Guidance for Decentralized Non-potable Water Systems. Prepared by the National Water Research Institute for the Water Environment & Reuse Foundation, Alexandria, VA. [Google Scholar]
- Shay F, Brinkman N, Cashdollar J, Griffin S, McMinn B, Rhodes E, Varughese E, Karim M, Grimm A, Spencer S, Borchardt M, 2014. Method 1615 Measurement of Enterovirus and Norovirus Occurrence inWater by Culture and RT-qPCR. U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD, 2013. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8 (4), e55943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y-S, Yajima M, 2015. R2jags: Using R to Run ‘JAGS’. R Package Version 0, pp. 5e7. [Google Scholar]
- Trypsteen W, Vynck M, De Neve J, Bonczkowski P, Kiselinova M, Malatinkova E, Vervisch K, Thas O, Vandekerckhove L, De Spiegelaere W, 2015. ddpcRquant: threshold determination for single channel droplet digital PCR experiments. Anal. Bioanal. Chem 407 (19), 5827e5834. [DOI] [PubMed] [Google Scholar]
- Winward GP, Avery LM, Frazer-Williams R, Pidou M, Jeffrey P, Stephenson T, Jefferson B, 2008. A study of the microbial quality of grey water and an evaluation of treatment technologies for reuse. Ecol. Eng 32 (2), 187e197. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.